Background: Aeromonas hydrophila flagellar glycosylation is crucial for flagellar production.
Results: Aeromonas hydrophila strain AH-3 lateral and polar flagellins are modified in O-linkage with a single monosaccharide or a heterogenous glycan (heptasaccharide), respectively. Mutants in pseudaminic biosynthesis abolish flagellar biogenesis and flagellins expression.
Conclusion: Flagellar formation depends on glycosylation.
Significance: O-Glycosylation by two different glycans for polar and lateral flagellins and is crucial for flagellar biogenesis.
Keywords: Bacteria, Genetics, Glycoprotein, Molecular Biology, Mutagenesis, Flagellin, Mass Spectrometry, Pseudaminic Acid
Abstract
Polar and lateral flagellin proteins from Aeromonas hydrophila strain AH-3 (serotype O34) were found to be glycosylated with different carbohydrate moieties. The lateral flagellin was modified at three sites in O-linkage, with a single monosaccharide of 376 Da, which we show to be a pseudaminic acid derivative. The polar flagellin was modified with a heterogeneous glycan, comprised of a heptasaccharide, linked through the same 376-Da sugar to the protein backbone, also in O-linkage. In-frame deletion mutants of pseudaminic acid biosynthetic genes pseB and pseF homologues resulted in abolition of polar and lateral flagellar formation by posttranscriptional regulation of the flagellins, which was restored by complementation with wild type pseB or F homologues or Campylobacter pseB and F.
Introduction
Mesophilic Aeromonas are ubiquitous water-borne bacteria, considered opportunistic pathogens of both aquatic and terrestrial animals, with some species being associated with gastrointestinal and extraintestinal human diseases (1). Flagellar motility represents an important advantage for bacteria in moving toward favorable conditions or in avoiding detrimental environments, and it allows flagellated bacteria to successfully compete with other microorganisms (2). Flagellar morphogenesis is a complex cascade of events that requires coordinated expression of >50 genes encoding structural subunits, regulatory proteins, and chemo-sensor machinery. Mesophilic Aeromonas are aquatic microorganisms that constitutively express a single polar flagellum, although ∼60% of strains most commonly associated with diarrhea (3) also are able to express many lateral flagella when grown in viscous environments or on surfaces (4). Investigations have revealed that both the polar and lateral flagellar systems of the mesophilic Aeromonas are involved in adherence to both biotic and abiotic surfaces, as well as in the biofilm formation (5).
Glycosylation, either N- or O-linked, has been described extensively in eukaryotes but more recently has also been described in bacteria (reviewed in Refs. 6–8). The most commonly reported form of protein glycosylation in bacteria is O-linkage of the sugar moiety to the hydroxyl oxygen of serine (Ser) or threonine (Thr) residues of proteins. This appears to be most commonly found in bacterial flagellins and pili (6) with increasing numbers of bacterial pathogens reported to modify flagellins or pili with diverse carbohydrate moieties. These include bacteria such as, Campylobacter jejuni, Helicobacter pylori, Pseudomonas spp., Neisseria spp., or Clostridum spp. (9–14). Structural studies of the glycan moieties modifying proteins have been carried out in some cases, showing that the glycan structures elaborated vary markedly between species. Such studies have shed light on glycan structural diversity and in some cases aided in understanding the associated biosynthetic pathways. There is some limited understanding of the biological significance of bacterial glycoproteins, but some recent insights as functions affecting surface properties linked to antigenicity have been described (15). The predominant O-glycans attached to the Campylobacter or Helicobacter flagellum are derivatives of nonulosonic acids sugars such as pseudaminic acid (Pse) or legionaminic acid (Leg). These are C9 sugars that are related to sialic acids (16). Reconstitution and biochemical characterization of the Pse biosynthesis showed that H. pylori uses a set of enzymes: PseB, PseC, PseH, PseG, PseI, and PseF (15).
Aeromonas hydrophila AH-3, similar to ∼60% of the clinical isolates of mesophilic Aeromonas produces a single polar flagellum constitutively expressed in both liquid and solid media and also have inducible lateral flagella expressed in high viscosity media (17). We previously described A. hydrophila AH-3 putative homologues of Pse biosynthetic genes, and their mutation impedes flagellar formation (18). We also indicated that both flagella were glycosylated according to a chemical test (18). Also, in Aeromonas caviae Sch3N, the mutation in biosynthetic Pse genes affected either the flagellar biogenesis as well as the O-antigen LPS (19). In this strain, we were able to demonstrate that Pse residues are the component of their specific O-antigen LPS (19).
In this study, for the first time in A. hydrophila strain AH-3, we show that polar and lateral flagellins are modified by different carbohydrate moieties but share one of the sugars (pesudaminic acid derivative). We also studied how the glycosylation type affects the flagellar biogenesis and also the flagellin production at the transcriptional and post-transcriptional level.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Plasmids, and Growth Conditions
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli strains were grown in Luria-Bertani (LB) Miller broth and on LB Miller agar at 37 °C, whereas Aeromonas strains were grown either in tryptic soy broth or on agar at 30 °C. When indicated kanamycin (50 μg/ml), rifampicin (100 μg/ml), spectinomycin (50 μg/ml), tetracycline (20 μg/ml), and chloramphenicol (25 μg/ml) were added to the media.
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid | Relevant characteristics | Ref. or Source |
|---|---|---|
| E. coli strains | ||
| DH5α | F− end A hsdR17 (rK− mK+) supE44 thi-1 recA1 gyr-A96 _80lacZM15 | 28 |
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac (F− proABlacIqZ_M15 Tn10) | Stratagene |
| S17–1 | hsdR pro recA, RP4–2 in chromosome Km::Tn7 (Tc::Mu) | 37 |
| BL21(λD3) | F− ompT hsdSB (rB− mB−) gal dcm (λD3) | Novagen |
| A. hydrophila strains | ||
| AH-3 | O34, wild type | 37 |
| AH-405 | AH-3, spontaneous RifR | 37 |
| AH-3ΔFlrA | AH-3 mutant unable produce polar flagellum, produces lateral flagella | 30 |
| AH-3ΔPseB | AH-3 pseB mutant in frame with pDM4 | This study |
| AH-3ΔPseF | AH-3 pseF mutant in frame with pDM4 | This study |
| Plasmids | ||
| pRK2073 | Helper plasmid, SpcR | 37 |
| pBAD33 | Arabinose-inducible expression vector, CmR | 27 |
| pBAD-PseB | pBAD33 with AH-3 pseB | This study |
| pBAD-PseF | pBAD33 with AH-3 pseF | This study |
| pBAD-PseBCj | pBAD33 with C. jejuni pseB | This study |
| pBAD-PseFCj | pBAD33 with C. jejuni pseF | This study |
| pDM4 | pir dependent with sacAB genes, oriR6K, CmR | 25 |
| pDM-PseB | pDM4 with AH-3 pseB fragment, CmR | This study |
| pDM-PseF | pDM4 with AH-3 pseF fragment, CmR | This study |
DNA Techniques
DNA manipulations were carried out essentially according to standard procedures (20). DNA restriction endonucleases and E. coli DNA polymerase Klenow fragment were obtained from Promega. T4 DNA ligase and alkaline phosphatase were obtained from Invitrogen and GE Healthcare, respectively. PCR was performed using BioTaq DNA polymerase (Ecogen) in a Gene Amplifier PCR System 2400 PerkinElmer thermal cycler. Plasmid DNA for sequencing was isolated by Qiagen plasmid purification kit (Qiagen, Inc. Ltd.) as recommended by the suppliers. Double-strand DNA sequencing was performed by using the Sanger dideoxy-chain termination method (21) with the BigDye Terminator cycle sequencing kit (version 3.1, Applied Biosystem). Custom-designed primers used for DNA sequencing were purchased from Sigma-Aldrich. The DNA sequences were compared with those available in the GenBankTM and EMBL databases at the National Center for Biotechnology Information (NCBI) (22). The Terminator search program in the GCG Wisconsin package was used to search for factor-independent transcriptional terminators. The Neural Network Promoter Prediction, PromScan (23), and PRODORIC (24) were used to search promoter sequences.
Electrospray Liquid Chromatography Mass Spectrometry Analysis of Intact Flagellins
Mass spectrometry studies of intact flagellin proteins were carried out using 50-μl aliquots of protein containing sample as described previously, with some modification (13). The lateral flagellin sample was injected onto a protein microtrap (Michrom Bioresources Inc., Auburn, CA) connected to a gradient HPLC pump (Agilent 1100 HPLC). Solvent A was 0.1% formic acid in HPLC grade water (Fisher), and solvent B was 0.1% formic acid in acetonitrile. An HPLC gradient of 5–60% solvent B (1 ml/min) over 60 min was used to resolve the protein mixture. The more complex polar flagellin sample was injected onto an Agilent mRP Hi recovery column (5 μm; 0.5 mm, inner diameter × 100 mm) also connected to a gradient HPLC pump (Agilent 1100 HPLC). A step gradient was applied to separate the protein mixture as follows: 20% B for 10 min, 35% B for 10 min, 45% B for 10 min, 55% B for 10 min. In both cases, a precolumn splitter was used to direct ∼35 μl/min of the HPLC mobile phase through the trap or column and into the electrospray interface of the QTOF2 (Waters, Milford, MA) to allow real-time monitoring of ion elution profiles. Intact masses of proteins were calculated by spectral deconvolution using MaxEnt (Waters, Beverly, MA) or similar software.
For the lateral flagellin, an abundant multiply charged protein ion was selected for MS/MS. The collision energy was increased incrementally, and labile, protein, and glycan-associated fragment ions were observed. For the polar flagellin, front-end collision-induced dissociation (feCID)3 was performed by increasing the cone voltage from 45 to 85 V, and glycan-associated oxonium ions were observed. Tandem MS/MS of a prominent glycan oxonium ion resulting from feCID was also performed by increasing the collision energy incrementally.
Solution Enzymatic Digests and Bottom Up Mass Spectrometry Analysis of Glycopeptides
To identify the type and location of glycosylation sites, flagellin (50 to 200 μg) was digested with trypsin (Promega, Madison, WI) at a ratio of 30:1 (protein/enzyme, w/w) in 50 mm ammonium bicarbonate at 37 °C overnight as described previously (13). Protein digests were analyzed by nano-liquid chromatography MS/MS (nLC-MS/MS) using either a Q-TOF Ultima hybrid quadrupole time-of-flight MS (Waters) as described previously (13) or an LTQ XL orbitrap MS (Thermo Fisher Scientific, Ottawa, Ontario, Canada) coupled to a nanoAcquity ultrahigh-pressure liquid chromatography system (Waters). MS/MS spectra were acquired automatically on doubly, triply, and quadruply charged ions in collision-induced dissociation (CID) mode for initial glycopeptide identification. The lateral flagellin modification site was determined using targeted CID of the m/z 870.42+ glycopeptide ion with alternating high and low collision energies (24 and 20 V). The polar flagellin modification site was determined using targeted electron transfer dissociation (ETD) of the m/z 1060.73+ glycopeptide ion with a 300-s reaction time using an LTQ-XL ion trap MS (Thermo Fisher Scientific). Data were collected between 300 and 3200 m/z (high mass range). Multistage mass spectrometry studies of glycan moieties were performed in high resolution (100,000) mode on the LTQ XL orbitrap MS.
Construction of Defined Mutants
Aeromonas pseB and pseF were formerly named flmA and neuA-like, respectively (16). The chromosomal in-frame pseB and pseF deletion mutants, A. hydrophila AH-3ΔPseB and AH-3ΔPseF, respectively, were constructed by allelic exchange as described by Milton et al. (25). Briefly, upstream (fragment AB) and downstream (fragment CD) of pseB and pseF genes were independently amplified using two sets of asymmetric PCRs for each gene. Primer pairs A-PseB (5′-GAAGATCTGAAAACGGCTGGAACTGG-3′) and B-PseB (5′-TGTTTAAGTTTAGTGGATGGGGACCAGCATCTTGCCAAAC-3′) and C-PseB (5′-CCCATCCACTAAACTTAAACAAGCGACTTCCTGACGGTAG-3′) and D-PseB (5′-GAAGATCTGATGATTTTCACCGGATGG-3′) amplify DNA fragments of 667 (PseB-AB) and 626 (PseB-CD) bp for the pseB in-frame deletion. DNA fragment PseB-AB contains 595 bp upstream pseB and the first 24 codons of pseB. DNA fragment PseB-CD contains from the first base in the 319 codon of pseB to 566 bp downstream pseB. Primer pairs A-PseF (5′-GAAGATCTCAGTTCAAGGGCGAGAAAG-3′) and B-PseF (5′-TGTTTAAGTTTAGTGGATGGGGATATTCTTGCGGGGAATG-3′) and C-PseF (5′-CCCATCCACTAAACTTAAACACACAGGGTGGTCGACATAG-3′) and D-PseF (5′-GAAGATCTTGTTGACGTGCCAGTTG-3′) amplify DNA fragments of 708 (PseF-AB) and 544 (PseF-CD) bp for pseF in-frame deletion. The DNA fragment PseF-AB contains 648 bp upstream pseF and the first 20 codons of pseF. DNA fragment PseF-CD contains from the first base in the 205 codon of pseF to 460 bp downstream pseF. DNA fragments AB and CD of each gene were annealed at the overlapping regions (underlined letters in primers B and C) and amplified as a single fragment using primers A and D. The fusion products were purified, BglII-digested (the BglII site is double-underlined in primers A and D), ligated into BglII-digested and phosphatase-treated pDM4 vector independently (25, 26), electroporated into E. coli MC1061 (λpir), and plated on chloramphenicol plates at 30 °C to obtain pDM-PseB and pDM-PseF plasmids, respectively. Plasmids with mutated genes were transferred into A. hydrophila AH-405 by triparental matings using the E. coli MC1061 (λpir) containing the insertion constructs and the mobilizing strain HB101/pRK2073. Transconjugants were selected on plates containing chloramphenicol and rifampicin. PCR analysis confirmed that the vector had integrated correctly into the chromosomal DNA. After sucrose treatment, transconjugants that were rifampicin-resistant (RifR) and chloramphenicol-sensitive (CmS) were chosen and confirmed by PCR.
Plasmid Constructions
Plasmids pBAD-PseB and pBAD-PseF containing the complete pseB and pseF of A. hydrophila AH-3, respectively, under the arabinose promoter (pBAD) on pBAD33 (27) and plasmids pBAD-PseBCj and pBAD-PseFCj containing the complete pseB and pseF of Campylobacter jejuni NCTC11168, respectively, under the arabinose promoter (pBAD) on pBAD33-Gm (26) were obtained. Oligonucleotides 5′-TCCCCCGGGCATAGTTGGTGCTCTTT-3′ and 5′-GCTCTAGACCTAAGCACCTCGACTACG-3′ generated a band of 1180 bp containing the pseB gene, and oligonucleotides 5′-GATATCATGTTATCGACCACCAAGG-3′ and 5′-GCTCTAGAAGTCGTGGCAGAGGAAG-3′generated a band of 961 bp (SmaI site is underlined, the XbaI site is double-underlined, and EcoRV site is in boldface type) containing the pseF gene. The amplified bands were digested with SmaI and XbaI or EcoRV and XbaI and ligated into SmaI- and XbaI-digested pBAD33 vector to construct the pBAD-PseB and pBAD-PseF recombinant plasmids. Oligonucleotides 5′-TCCCCCGGGCAAGTATCAAGCCCAAAC-3′ and 5′-GCTCTAGAAGA TGAAGGGCTGAAGTG-3′ generate a band of 1285 bp containing the C. jejuni pseB gene and oligonucleotides 5′-TCCCCCGGGTCAATTTCAAAGGTTTGGTT-3′ and 5′-GCTCTAGACAGGATAAGGAATTTCATCG-3′ generate a band of 937 bp containing the C. jejuni pseF gene. (The SmaI site is underlined, and XbaI is double-underlined.) The amplified bands were digested with SmaI and XbaI and ligated into SmaI- and XbaI-digested pBAD33 vector (27) to construct the pBAD-PseBCj and pBAD-PseFCj recombinant plasmids. Plasmids were independently introduced into the E. coli DH5α (28) and sequenced.
Cell Fractions
A. hydrophila cells grown in tryptic soy broth at 25 °C were harvested by centrifugation (5000 × g), washed with 20 mm MgCl2 in 100 mm Tris buffer (pH = 8.0), and resuspended in the same cold buffer, and French press cell lysis was performed. The lysates were centrifuged at 5000 × g to remove unbroken cells. After centrifugation at 4 °C for 1 h at 115,000 × g, the cytoplasmic fraction remained in the supernatant, whereas the whole membrane fraction was retained in the pellet.
Flagellar Purification
A. hydrophila AH-3 was grown in tryptic soy broth for the polar flagellum purification. For the isolation of lateral flagella, the strains were grown on tryptic soy agar plates washed with 100 mm Tris (pH 7.8). Cells were in both cases collected by centrifugation at 5000 × g and suspended in the same Tris buffer. Flagella were removed from the cells by shearing in a vortex with a glass bar for 3–4 min and then passing repetitively (minimum six times) through a syringe. Cells were removed by centrifugation at 8000 × g for 30 min, and the supernatant centrifuged at 18,000 × g for 20 min. From the remaining supernatant, the flagella were pelleted by ultracentrifugation at 100,000 × g for 60 min and resuspended in 100 mm Tris (pH 7.8) plus 2 mm EDTA buffer. This flagella-enriched fraction was purified in a cesium chloride gradient by ultracentifugation at 60,000 × g for 48 h. The band containing the flagella was collected, and cesium chloride was removed by extensive dialysis against the same buffer (100 mm Tris, 2 mm EDTA). Purified flagella were analyzed by SDS-PAGE or by glycosylation chemical studies.
Immunological Methods
Western blot of whole cell, cell fractions, or purified flagella was performed as briefly described. After SDS-PAGE, immunoblotting was carried out by transfer to polyvinylidine fluoride membranes (Millipore Corp., Bedford, MA) at 1.3 A for 1 h in the buffer of Towbin et al. (29). The membranes were then incubated sequentially with 1% bovine serum albumin, specific anti-polar flagellins polyclonal serum (30) (1:2000), alkaline phosphatase-labeled goat anti-rabbit immunoglobulin G, and 5-bromo-4-chloroindolylphosphate disodium-nitroblue tetrazolium. Incubations were carried out for 1 h, and washing steps with 0.05% Tween 20 in phosphate-buffered saline were included after each incubation step.
ELISA was performed by dispensing standardized suspensions of each cell fraction in coating buffer (pH 9.6) into 96-well microtiter plates. The plates were left to stand overnight at 4 °C. The wells were blocked with 1% bovine serum albumin in phosphate-buffered saline for 2 h at 37 °C. Anti-polar flagellins polyclonal serum (1:5000) was added and incubated for 2 h at 37 °C, and detection was performed by using peroxidase-labeled sheep anti-rabbit immunoglobulin G (1:1000) and 2,2′-azino-di-(3-ethylbenzthiazoline sulfonate) as the substrate.
Motility Assays (Swarming and Swimming)
Freshly grown bacterial colonies were transferred with a sterile toothpick into the center of swarm agar (1% tryptone, 0.5% NaCl, 0.5% agar) or swim agar (1% tryptone, 0.5% NaCl, 0.25% agar). The plates were incubated face up for 16–24 h at 25 °C, and motility was assessed by examining the migration of bacteria through the agar from the center toward the periphery of the plate. Swimming motility in liquid media was assessed by phase-contrast microscopy at a magnification of ×400.
Transmission Electron Microscopy
Bacterial suspensions were placed on Formvar-coated grids and negative stained with a 2% solution of uranyl acetate, pH 4.1. Preparations were observed on a Hitachi 600 transmission electron microscope.
RESULTS
nLC-MS/MS Analysis of Wild Type Flagellin Peptide Digests
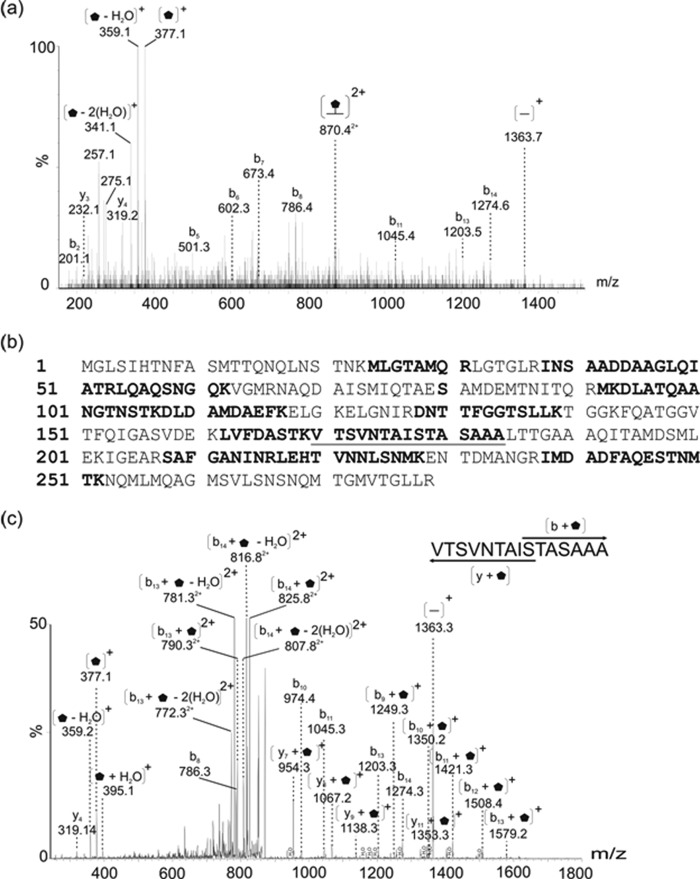
We purified polar and lateral flagellins from the wild type strain as described under “Experimental Procedures” and shown in Fig. 1. In some cases, the isogenic mutant AH-3ΔFlrA (30) was used for isolation of lateral flagellin. This mutant is unable to produce constitutive polar flagellum with unaltered lateral flagella. Analysis of tryptic digests of both polar and lateral flagellin preparations by nLC-MS/MS allowed the identification of many unmodified flagellin peptides. A number of MS/MS spectra were derived from peptides that appeared to possess glycan modifications. For example, Fig. 2a shows the peptide MS/MS spectrum of the lateral flagellin tryptic glycopeptide 170TVTSVNTAISTASAAA184. This peptide is not a typical tryptic cleavage product, but peptide type y and b fragment ions were observed at low intensities in the MS/MS spectrum, confirming the sequence identity. Of note, the low m/z region of the spectrum was dominated by an intense ion at m/z 377, which did not correspond to any predicted peptide type y or b ions. In addition, a neutral loss of water was observed from this ion, giving rise to the dehydrated form of the ion at m/z 359. The total mass of the glycopeptide was 1739 Da, (predicted unmodified peptide mass is 1363 Da), which suggested a total modification of 376 Da. The intense ion observed at m/z 377 therefore likely corresponds to the glycan oxonium ion. The protein sequence coverage for the lateral flagellin is shown in Fig. 2b. Further MS studies were performed, using targeted CID MS/MS of the doubly charged glycopeptide ion at m/z 870.4 with alternating high and low collision energies. This generated a combined MS/MS spectrum containing typical CID peptide fragmentation. In addition, y and b type peptide ions with the glycan attached were also observed (Fig. 2c). Although complex, this spectrum allowed the mapping of the glycan moiety to the serine at position 178 of the flagellin protein sequence.
FIGURE 1.
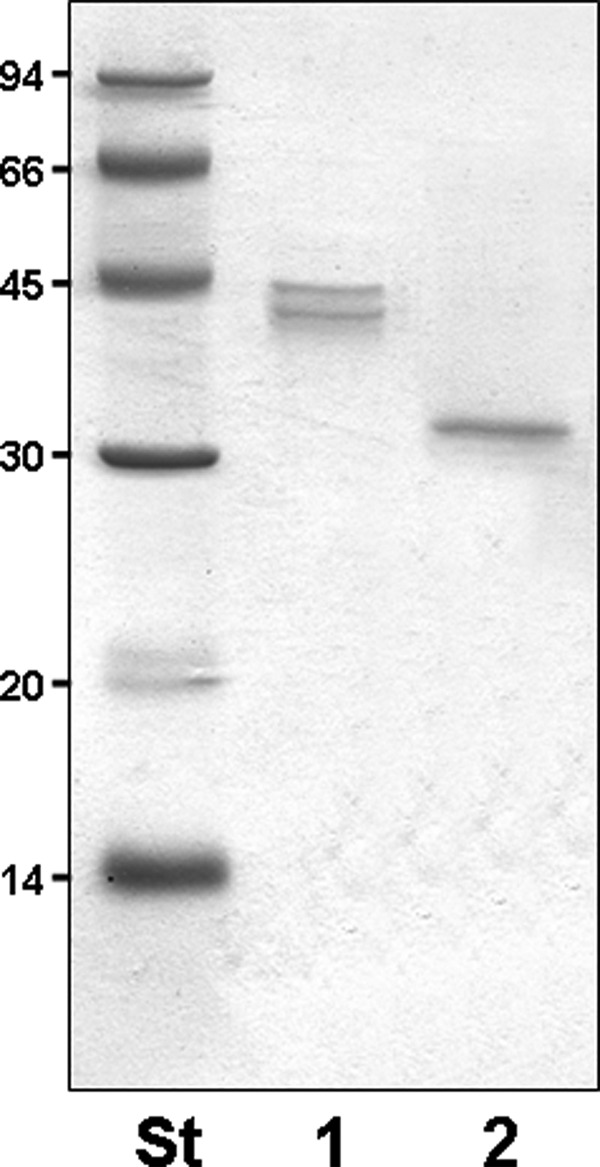
SDS-PAGE of purified flagellins. 1, polar flagellins. 2, lateral flagellin. St, size standard.
FIGURE 2.
MS/MS analysis of modified tryptic peptides from lateral flagellar protein of Aeromonas hydrophila AH-3.
a, nLC-MS/MS spectrum of the doubly protonated glycopeptide ion at m/z 870.4 from lateral flagellin. Visible were peptide fragment ions of low intensity, giving the peptide sequence 170TVTSVNTAISTASAAA184. Also observed was a daughter ion corresponding to the singly charged unmodified peptide ion (m/z 1363.7). The observed mass excess was 376 Da, with a corresponding intense oxonium ion visible at m/z 377, with ion corresponding to likely loss of water at m/z 359. b, protein sequence coverage from mass spectrometry analyses of flagellin protein is shown. Indicated in boldface type are unmodified peptides, boldface type and underlined letters indicate peptides modified with glycan. Of note, de novo sequencing of flagellin peptides showed the amino acid at position 80 to be serine, not glycine as predicted from the gene sequence. c, doubly charged glycopeptide ion at m/z 870.2 was targeted for MS/MS with alternating collision energies of 20 and 24 V in CID mode. The combined spectrum results in a typical y and b ion series as well as y and b ions harboring the intact 376-Da glycan. The singly charged ion at m/z 954.3 (y7 + glycan) and the singly charged ion at m/z 1249.3 (b9 + glycan) suggests that the glycan is attached in O-linkage and that serine 178 is the modified residue.  , 376-Da sugar.
, 376-Da sugar.
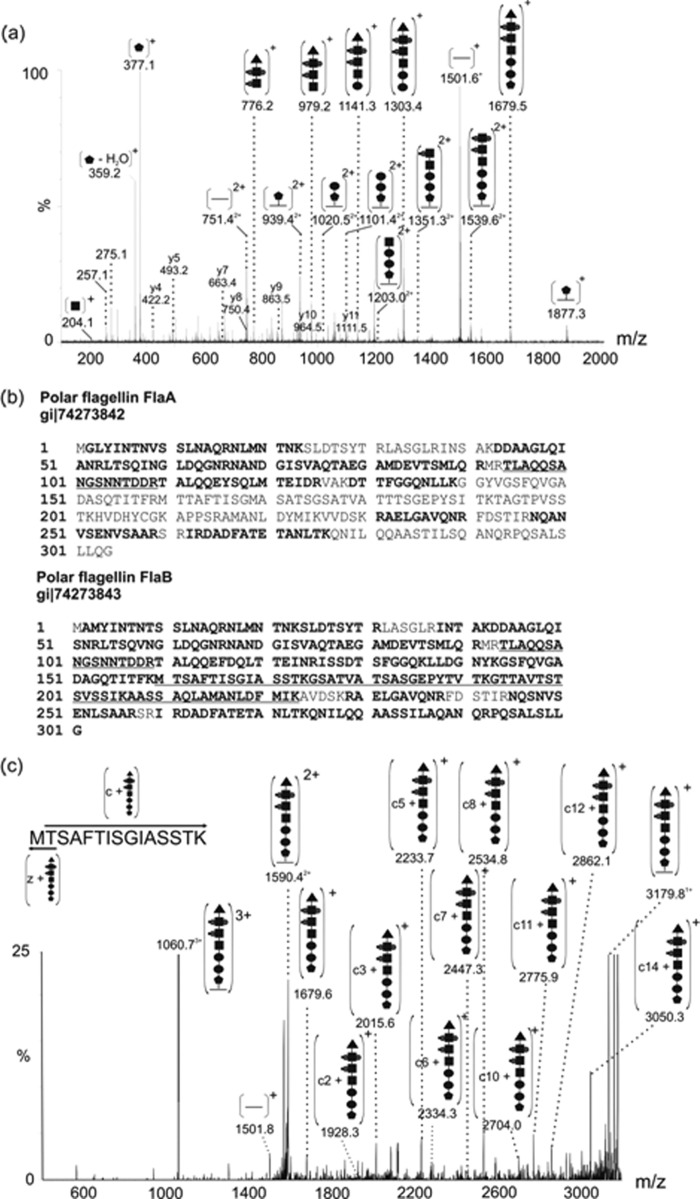
Tandem mass spectrometry studies were also carried out on tryptic digests of purified polar flagellin. The polar flagellin is comprised of two monomers, FlaA and FlaB. In our studies, FlaB appeared to be the more abundant (∼90%) of the two proteins. The overall sequence coverage of both FlaA and FlaB are shown in Fig. 3b. Fig. 3a shows the peptide MS/MS spectrum of a glycopeptide from FlaB (160TMTSAFTISGIASSTK174). Fragmentation of the triply charged precursor ion at m/z 1060.4 gave rise to a series of low intensity peptide type y and b fragment ions. A singly charged ion, corresponding to the unmodified peptide, was observed at m/z 1501, suggesting a mass excess of 1678 Da. The MS/MS spectrum was dominated by a series of high intensity ions that were not peptide related. The complex non-peptide fragmentation pattern was putatively mapped to a series of neutral losses of glycan from either the glycopeptide or a glycan oxonium ion at m/z 1679 and gave a sequence of a heptasaccharide comprised of three N-acetylhexosamine (one of which was additionally modified by two phosphate groups and a methyl group, whereas another was additionally modified by a single phosphate and a methyl group), two hexoses and two unknown glycans of 376 and 102 Da (sequence 376-162-162-203-297-377-102 Da). The heptasaccharide is distinct from the AH-3 O34-antigen, which is comprised of a tetrasaccharide repeat unit of d-mannose, d-GalNAc, and 6-deoxytalose (31). ETD fragmentation of glycopeptides was used to map the sites of glycan modification. Fig. 3c shows the ETD MS/MS spectrum of the same FlaB glycopeptide 160TMTSAFTISGIASSTK174 harboring a 1679-Da glycan. The spectrum was dominated by charged reduced precursor ions. In addition, ions corresponding to both the unmodified peptide (m/z 1501) and glycan oxonium ion (m/z 1680) were observed. Manipulation of the ETD reaction time and collection of data in the high mass region allowed the observation of a series of c type peptide ions with glycan moiety attached. The series of ions suggested the glycan moiety was attached in O-linkage to the threonine at position 161 in the polar flagellin FlaB protein sequence.
FIGURE 3.
MS/MS analysis of modified tryptic peptides from polar flagellar protein of A. hydrophila AH-3.
a, nLC-MS/MS spectrum of the triply protonated glycopeptide ion at m/z 1060.7 from polar flagellin. Visible were peptide fragment ions of low intensity, giving the peptide sequence 160TMTSAFTISGIASSTK174. Also observed was a daughter ion corresponding to the singly charged unmodified peptide ion (m/z 1501.1). The observed mass excess was 1678 Da, and a putative glycan oxonium ion was observed at m/z 1679. Indicated in the MS/MS spectrum are a series of neural losses from the glycan oxonium ion, giving a monosaccharide sequence on 376-162-162-203-297-377-102. It is likely that masses of 203 and 162 Da correspond to N-acetylhexosamine and hexose, respectively. Dominating the low mass region of the peptide MS/MS spectrum were glycan-related ions, at m/z 377 and 359. In addition, a doubly charged ion series was also observed, corresponding to glycopeptide fragments. b, protein sequence coverage from mass spectrometry analyses of flagellin proteins FlaA and FlaB are shown. Indicated in boldface type are unmodified peptides; boldface and underlined type indicates peptide modified with glycan. c, the triply charged glycopeptide ion at m/z 1060.7 was targeted for ETD with a reaction time of 300 s. In the resulting spectrum, the singly charged ion at m/z 1928.2 (c2 + glycan) along with ions corresponding to c3, c5, c6, c7, c8, c10, c11, c12, c14 all with the glycan still attached suggest that the peptide is modified at the threonine in position 161 of the protein sequence in O-linkage.  , 376-Da sugar; ●, hexose; ■, HexNAc; ▴, 102-Da unknown moiety; •, phosphorylation; ■, methylation.
, 376-Da sugar; ●, hexose; ■, HexNAc; ▴, 102-Da unknown moiety; •, phosphorylation; ■, methylation.
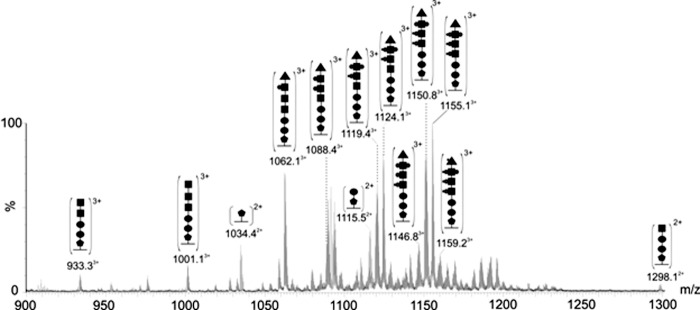
Further investigation of polar flagellin tryptic digests showed clusters of multiply charged ions in the high m/z regions of the MS spectra. These clusters were observed at five distinct retention times. Targeted analyses of ions from each cluster gave MS/MS spectra that showed them to be five different flagellin peptides, each modified with glycans of varying mass. Four of these glycopeptides correspond to the FlaB protein sequence. However, the fifth glycopeptide was observed to have a peptide sequence common to both FlaA and FlaB (94TTLAQQSANGSNNTDDR109). In the case of each glycopeptide, putative glycan-related oxonium ions were observed at m/z 377 and 204, suggesting the presence of the unknown sugar (376 Da) and a HexNAc residue (203 Da). Variable chain lengths were observed, from a single 376 Da moiety to the heptasaccharide mentioned above. It was also observed that at each retention time, the major clusters were separated by 80 Da, strongly suggesting phosphorylation. Within each major cluster, the mass difference between glycopeptide ion peaks was 14 Da, suggesting methylation. De novo sequencing of many of these glycopeptide ion peaks revealed that the each of the three HexNAc residues appears to harbor additional variable modifications of 0–2 phosphates and 0–2 methyl groups, accounting for the extensive glycan heterogeneity and glycopeptides ion clustering. However, it was not determined whether the additional phosphate is linking the HexNAc residues or simply modifying the residue. Further investigation is required to determine whether the methyl group is being added to the phosphate or directly to the HexNAc residue itself. Interestingly, no HexNAc residue was observed to be methylated without phosphorylation. The order in which each of the HexNAc moieties were modified with phosphate or methyl groups was also variable, leading to isobaric species which further complicated the fragmentation spectra. The complexity of these MS/MS spectra made assignment of the sequence of the glycan chain challenging. In each case, however, the sugar linking to the peptide chain was observed to be 376 Da. Although the sequence of the variable glycan chains could not be determined for every glycopeptide identified, the peptide sequence of each glycopeptide listed in Table 2 was determined by de novo sequencing and is shown with the observed glycan mass excess. The glycan chains associated with the 1691-Da peptide (94TTLAQQSANGSNNTDDR109) whose composition was determined by de novo sequencing are indicated in Fig. 4.
TABLE 2.
Assigned oligopeptides
Based on nanoelectrospray liquid chromatography MS/MS analysis of tryptic digests of the AH-3 polar and lateral flagellins.
| Observed Precursor m/z | Monoisotopic precursor m/z | Charge state of glycopeptide | Residues | Sequence of glycopeptide | Unmodified peptide [M+H]+ | Protein (strain) | Mass excess (Da) |
|---|---|---|---|---|---|---|---|
| 870.4 | 870.4 | [M + 2H]2 + | 170–184 | VTSVNTAISTASAAAa | 1363.6 | LafA (AH3) | 376 |
| 1034.4 | 1034.4 | [M + 3H]3 + | 94–109 | TLAQQSANGSNNTDDR | 1691.8 | FlaA/FlaB (AH3) | 376 |
| 1062.1 | 1062.1 | [M + 3H]3 + | 94–109 | TLAQQSANGSNNTDDR | 1691.8 | FlaA/FlaB (AH3) | 1492 |
| 1088.4 | 1088.4 | [M + 3H]3 + | 94–109 | TLAQQSANGSNNTDDR | 1691.8 | FlaA/FlaB (AH3) | 1571 |
| 1093.1 | 1093.1 | [M + 3H]3 + | 94–109 | TLAQQSANGSNNTDDR | 1691.8 | FlaA/FlaB (AH3) | 1585 |
| 1120.4 | 1119.4 | [M + 3H]3 + | 94–109 | TLAQQSANGSNNTDDR | 1691.8 | FlaA/FlaB (AH3) | 1665 |
| 1124.7 | 1124.1 | [M + 3H]3 + | 94–109 | TLAQQSANGSNNTDDR | 1691.8 | FlaA/FlaB (AH3) | 1679 |
| 1151.4 | 1151.0 | [M + 3H]3 + | 94–109 | TLAQQSANGSNNTDDR | 1691.8 | FlaA/FlaB (AH3) | 1759 |
| 1155.5 | 1155.1 | [M + 3H]3 + | 94–109 | TLAQQSANGSNNTDDR | 1691.8 | FlaA/FlaB (AH3) | 1772 |
| 939.4 | 939.4 | [M + 2H]2 + | 160–174 | MTSAFTISGIASSTK | 1501.8 | FlaB (AH3) | 376 |
| 1031.0 | 1029.8 | [M + 3H]3 + | 160–174 | MTSAFTISGIASSTK | 1501.8 | FlaB (AH3) | 1585 |
| 1057.4 | 1056.1 | [M + 3H]3 + | 160–174 | MTSAFTISGIASSTK | 1501.8 | FlaB (AH3) | 1665 |
| 1062.0 | 1060.7 | [M + 3H]3 + | 160–174 | MTSAFTISGIASSTK | 1501.8 | FlaB (AH3) | 1679 |
| 1083.7 | 1082.4 | [M + 3H]3 + | 160–174 | MTSAFTISGIASSTK | 1501.8 | FlaB (AH3) | 1743 |
| 1088.4 | 1087.1 | [M + 3H]3 + | 160–174 | MTSAFTISGIASSTK | 1501.8 | FlaB (AH3) | 1758 |
| 1632.1 | 1630.6 | [M + 2H]2 + | 160–174 | MTSAFTISGIASSTK | 1501.8 | FlaB (AH3) | 1758 |
| 1092.7 | 1091.7 | [M + 3H]3 + | 160–174 | MTSAFTISGIASSTK | 1501.8 | FlaB (AH3) | 1772 |
| 1158.5 | 1157.4 | [M + 3H]3 + | 175–192 | GSATVATSASGEPYTVTK | 1726.9 | FlaB (AH3) | 1743 |
| 1743.9 | 1742.5 | [M + 2H]2 + | 175–192 | GSATVATSASGEPYTVTK | 1726.9 | FlaB (AH3) | 1758 |
| 1167.8 | 1166.8 | [M + 3H]3 + | 175–192 | GSATVATSASGEPYTVTK | 1726.9 | FlaB (AH3) | 1772 |
| 1003.4 | 1001.8 | [M + 3H]3 + | 193–206 | GTTAVTSTSVSSIK | 1338.7 | FlaB (AH3) | 1665 |
| 1020.0 | 1018.7 | [M + 3H]3 + | 193–206 | GTTAVTSTSVSSIK | 1338.7 | FlaB (AH3) | 1715 |
| 1029.1 | 1028.1 | [M + 3H]3 + | 193–206 | GTTAVTSTSVSSIK | 1338.7 | FlaB (AH3) | 1744 |
| 1033.9 | 1032.8 | [M + 3H]3 + | 193–206 | GTTAVTSTSVSSIK | 1338.7 | FlaB (AH3) | 1758 |
| 1038.4 | 1037.4 | [M + 3H]3 + | 193–206 | GTTAVTSTSVSSIK | 1338.7 | FlaB (AH3) | 1772 |
| 1151.1 | 1149.8 | [M + 3H]3 + | 207–223 | AASSAQLAMANLDFMIK | 1781.9 | FlaB (AH3) | 1665 |
| 1181.7 | 1180.5 | [M + 3H]3 + | 207–223 | AASSAQLAMANLDFMIK | 1781.9 | FlaB (AH3) | 1758 |
a Atypical tryptic cleavage product, sequence confirmed with peptide type y and b ion coverage.
FIGURE 4.
Polar flagellin glycopeptide heterogeneity. nLC-MS spectrum of the flagellin tryptic digest at 12.8–13.4 min of elution, showing clusters of multiply charged ions. Further targeted analyses showed these to be flagellin glycopeptide ions as annotated. The glycan chains range from a single 376-Da modification to a 1772-Da heptasaccharide chain, including variably phosphorylated and methylated HexNAc residues.  , 376-Da sugar; ●, hexose; ■, HexNAc; ▴, 102-Da unknown moiety; •, phosphorylation; ■, methylation.
, 376-Da sugar; ●, hexose; ■, HexNAc; ▴, 102-Da unknown moiety; •, phosphorylation; ■, methylation.
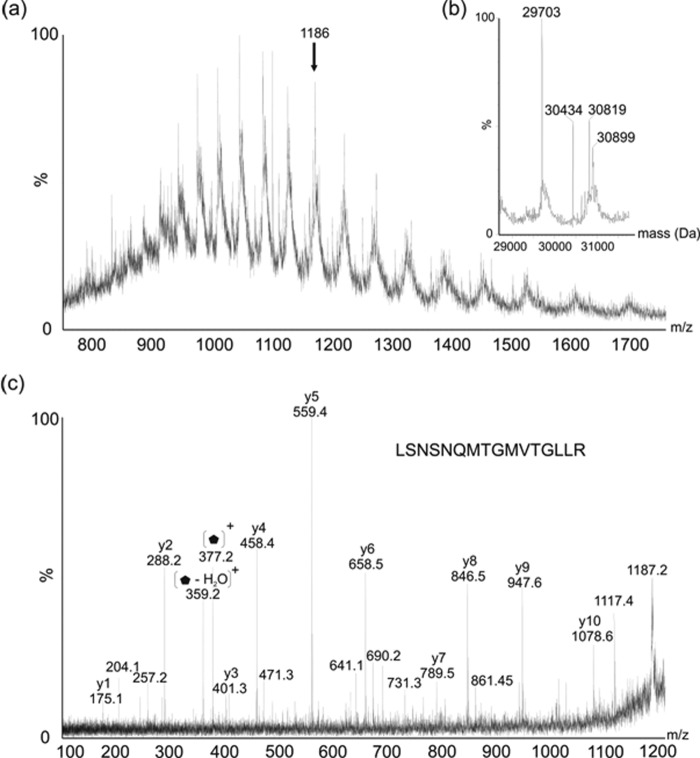
Intact Mass Analysis of Wild Type Lateral and Polar Flagellins
The observed intact masses of flagellin proteins from A. hydrophila AH-3 were obtained from rapid LC-MS of purified proteins into a QTOF2 ESI mass spectrometer. The MS spectrum of purified lateral flagellin showed a multiply charged ion envelope, as shown in Fig. 5a, typical of protein. The spectrum was deconvoluted (Fig. 5b) and showed multiple protein masses of 29.70, 30.43, 30.82, and 30.90 kDa. The mass of the protein predicted from the translated gene sequence is 29.5 kDa, suggesting that the lowest observed mass corresponded to the unmodified form of the protein. The other observed masses likely corresponded to flagellin-harboring modifications of 0.7–1.2 kDa in size. In fact, the observed 30.43 kDa mass likely corresponds to the unmodified protein with two 376-Da glycans attached. Furthermore, 30.82 kDa seems to be the unmodified protein harboring three 376-Da glycans. The largest observed intact mass acquired, 30.90 kDa, is 80 Da greater than the mass of flagellin modified with three 376 Da glycans. The mass difference of 80 Da is strongly suggestive of phosphorylation. A tandem MS experiment was carried out on one multiply charged protein ion (m/z 1186.4), and as shown in Fig. 5c, this MS/MS spectrum was dominated by peptide y-type ions, resulting from nonspecific protein fragmentation. The observed peptide sequence was LSNSNQMTGMVTGLLR, which corresponded to the C-terminal peptide of the lateral flagellin. The weak ion at m/z 1722 likely corresponds to the average mass of the C-terminal peptide ion. In addition, the spectrum showed putative glycan-related ions that could not be attributed to peptide fragment ions, including ions at m/z 377 and 359.
FIGURE 5.
Electrospray mass spectrometry of intact lateral flagellin protein from Aeromonas hydrophila AH-3.
a, electrospray mass spectrum of intact lateral flagellin protein, showing an envelope of multiply charged protein ions. b, the reconstructed molecular mass profile of lateral flagellin, showing masses at 29,703, 30,424, 30,819, and 30,899 Da. This corresponds in total to three sites of modification with a 376-Da glycan moiety. c, tandem mass spectrometry analysis of the multiply charged protein ion at m/z 1186. A peptide type y ion fragmentation series was observed, the sequence of which corresponded to the C-terminal peptide of the lateral flagellin LSNSNQMTGMVTGLLR. In addition, several ions were observed that did not correspond to peptide fragment ions, observed at m/z 377, with likely daughter ion at 359 due to loss of water.  , 376-Da sugar.
, 376-Da sugar.
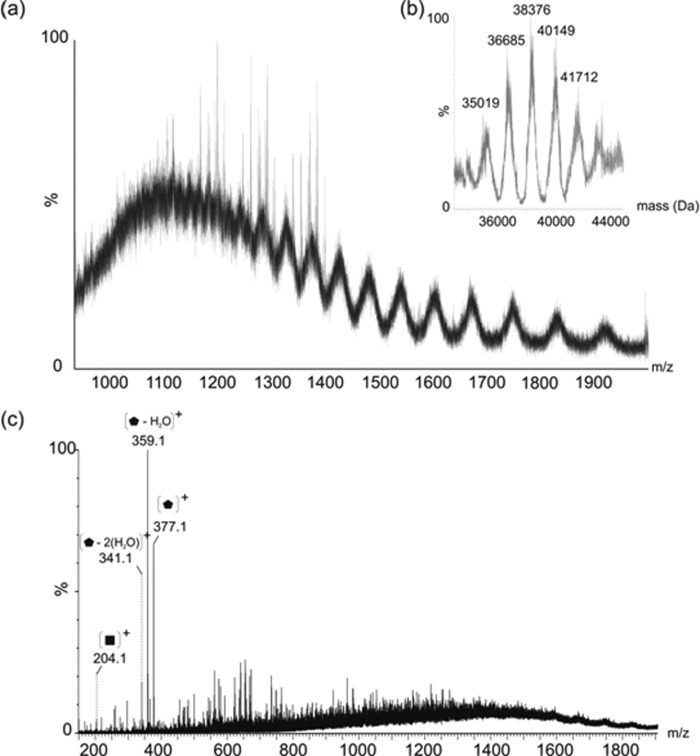
The electrospray mass spectrum resulting from the intact mass analysis of the polar flagellin preparation revealed a substantially more complex ion envelope than that of the lateral flagellin (Fig. 6a). This suggested the presence of multiple protein species, differing slightly in molecular mass, which is also supported by the high m/z range clustering of glycopeptides with variable glycan chain lengths observed in the nLC-MS/MS of tryptic digests. Deconvolution of the complex ion envelope, as shown in Fig. 6b, indicates the presence of five major protein masses: 35.0, 36.7, 38.4, 40.1, and 41.7 kDa. The similarity in the predicted molecular mass of FlaA and FlaB combined with the significant heterogeneity of the observed glycans modifying these proteins makes analysis of the intact mass and the associated mass differences challenging. Based on the predicted mass of the more abundant monomer, FlaB (31.8 kDa), it is plausible that 35.0 kDa represents this protein modified by two of the heptasaccharide chains detailed above. Furthermore, 36.7, 38.4, 40.1, and 41.7 could each plausibly be FlaB harboring three, four, five, and six heptasaccharide glycan chains, respectively. This is supported by the confirmation of five modified flagellin peptides in nLC-MS/MS studies of flagellin tryptic digests, although a sixth modified peptide was not identified. In addition, during feCID, four intense ions were observed in the low mass region of the spectrum at m/z 377, 359, 341, and 204, which likely correspond to nonspecific fragmentation of glycan from the proteins (Fig. 6c).
FIGURE 6.
Electrospray mass spectrometry of intact polar flagellin protein from Aeromonas hydrophila AH-3.
a, electrospray mass spectrum of intact polar flagellin protein showing a complex envelope of multiply charged protein ions. b, the reconstructed molecular mass profile of the polar flagellin is not as well resolved as the lateral flagellin due to the increased complexity, but does clearly show masses at 35,019, 36,685, 38,376, 40,149, and 41,712 Da. This seems to correspond to FlaB, modified with two, three, four, five, and six heptasaccharide glycan chains, ranging in mass from 1563 to 1773 Da. Similar glycan heterogeneity is also observed in the nLC-MS/MS analysis of tryptic digests. c, feCID of polar flagellin, generated by increasing the cone voltage of the QTOF2 mass spectrometer to 85 V. Clear glycan related oxonium ions are present in the low m/z region, resulting from nonspecific fragmentation of the intact flagellin glycoprotein.  , 376-Da sugar; ■ = HexNAc.
, 376-Da sugar; ■ = HexNAc.
Structural Characterization of Unusual O-Linked Glycan Moieties
A major challenge in the elucidation of novel bacterial glycans is the often limiting amounts of protein or glycan that can be isolated. High resolution MS can provide much information regarding glycan structure and composition, although cannot determine the absolute configuration of the sugar moiety. Nuclear magnetic resonance is the gold standard for structural studies; however, the poor sensitivity of the technique often means that sufficient amounts of bacterial glycan cannot be isolated for structural determinations.
Both polar and lateral flagellins were observed to be modified with a novel glycan of mass 376 Da. MS fragmentation of the corresponding glycan oxonium ion, gave rise to an MS/MS characteristic of glycan (Fig. 7). A loss of water (18 amu) gave rise to a daughter ion at m/z 359. Further loss of a mass corresponding to an acetyl group (42 amu) gave rise to an ion at m/z 317. The remainder of the mass spectrum gave a fragment ion pattern similar to that observed for pseudaminic acid (316 Da), with the daughter ions highlighted in Fig. 7 with an asterisk indicating the sugar fragment ions that were also observed in MS/MS fragmentation of pseudaminic acid (Pse5Ac7Ac) (32). These fragment ions included those at m/z 275.1, 257.1, 239.1, 221.1, 180.1, 162.1, and 134.1. These data suggest that the ion at m/z 377 is pseudaminic acid, modified by the addition of an acetyl group and water. An identical sugar fragmentation pattern was recorded for the m/z 377 oxonium ion observed to modify both polar and lateral flagellins. Where possible, accurate mass measurements were performed for the glycan-related fragment ions in glycopeptide MS/MS spectra, and these were used to determine plausible elemental formulae of unknown glycans. The MS/MS spectrum of the triply charged glycopeptide ion at m/z 1057.13+ (MTSAFTISGIASSTK), modified with the heptasaccharide moiety was carried out in high resolution mode. The glycan oxonium ion at m/z 377 gave an m/z of 377.1539 (Table 3), with a top ranked plausible elemental formula of C15H25N2O9. Fragmentation of the ion at m/z 377 in high resolution mode produced the m/z 317 oxonium daughter ion at m/z 317.1332 (Table 3), giving a most plausible elemental formula of C13H21N2O7. These data, along with the fragmentation data detailed above, confirm that the unknown glycan oxonium ion at m/z 377 is a pseudaminic acid derivative (pseudaminic acid is C13H20N2O7).
FIGURE 7.
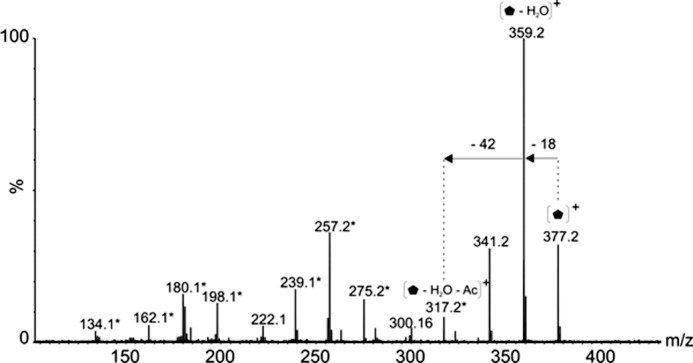
Glycan structural characterization by mass spectrometry. Tandem mass spectrum of singly charged ion at m/z 377 following feCID of intact flagellin protein showed consecutive losses of water and an acetyl group from the parent ion giving rise to an ion at m/z 317. Subsequent losses from this ion gave rise to a fragmentation pattern similar to that observed with nonulosonic acid sugars. Daughters ions marked with an asterisk denote those fragment ions found in MS/MS spectra of pseudaminic acid.  , 376-Da sugar; *, fragment ions common to pseudaminic acid; Ac, acetyl group.
, 376-Da sugar; *, fragment ions common to pseudaminic acid; Ac, acetyl group.
TABLE 3.
Accurate masses of glycan oxonium and glycan-related ion and most plausible empirical formula
MS/MS spectrum of the glycopeptide MTSAFTISGIASSTK was used, and mass spectral calibration was accurate to 1.3 ppm.
| Observed mass | Plausible formula | Predicted mass |
|---|---|---|
| 377.1539a | C15H25N2O9 | 377.15545 |
| 317.1332a | C13H21N2O7 | 317.13433 |
| 203.0789b | C8H13NO5 | 203.07824 |
| 162.0521b | C6H10O5 | 162.05274 |
a Glycan oxonium ion.
b Observed mass difference from neutral loss of sugar.
Also from the MS/MS spectrum of the glycopeptide MTSAFTISGIASSTK, accurate masses of other monosaccharide components of the heptasaccharide were measured. From these data, the neutral losses of 203 and 162 Da gave top ranked plausible elemental formulae corresponding to HexNAc and Hexose respectively (Table 3). An accurate mass for the 102 Da unknown glycan could not be determined, and therefore no plausible elemental formula could be assigned. Further studies will be required before this can be accurately assigned.
Pseudaminic Acid Biosynthetic Mutants
Mutants AH-3ΔPseB and AH-3ΔPseF in frame were unable to produce polar or lateral flagella under induced conditions by transmission electron microscopy, as described previously for insertional mutants in pseudaminic acid biosynthesis homologous genes (18). The introduction of the Aeromonas wild type corresponding genes recovers the production of both kinds of flagella in the mutants. C. jejuni pseB or pseF were fully able to complement A. hydrophila AH-3ΔPseB and AH-3ΔPseF mutants, respectively, as judged by the recovery of either the polar and lateral flagella by transmission electron microscopy as described previously for insertional mutants in pseudaminic acid biosynthesis homologous genes (18). FlaA is the polar flagellin, and FlrA corresponds to the master polar flagellum regulator (30). LafA is the lateral flagellin, and LafK is the master regulator for lateral flagella (33). RT-PCRs confirmed gene transcription for all these genes in the strains tested (wild type strain AH-3 and mutants AH-3ΔPseB and AH-3ΔPseF). Then, the lack of polar and lateral flagellar formation observed in the mutants is not by the lack of flagellin or the master regulator transcription.
These data prompted us to examine the polar flagellin in different bacterial cell locations by immunodetection. Although the polar flagellin in the wild type cytoplasm fraction is clearly observed by Western blot using specific serum against this protein, polar flagellin is observed at greatly reduced levels in the cytoplasmic fraction of the mutant strains AH-3ΔPseB and AH-3ΔPseF using the same technique (Fig. 8). Furthermore, the polar flagellin in the cytoplasm fraction showed a reduced Mr in comparison with the one detected in the wild type strain whole membrane fraction or purified polar flagellum (Fig. 8). No polar flagellin was detected in the whole membrane fraction of the mutant strains AH-3ΔPseB and AH-3ΔPseF (data not shown). Using a quantitative ELISA assay with the cytoplasm fractions of the wild type and the mutants and specific serum against this protein, we could quantify that the AH-3ΔPseB and AH-3ΔPseF mutants produced <8% of polar flagellin than the wild type strain (Table 4). A full recovery of the polar flagellin in the cytoplasm fraction of the AH-3ΔPseB and AH-3ΔPseF was observed when plasmids carrying either these genes from A. hydrophila AH-3 or C. jejuni were introduced in the mutants (data shown for AH-3ΔPseB mutant in Fig. 8). The ELISA results using cytoplasm fractions of several A. hydrophila strains and specific antiserum against polar flagellin shown in Table 4 also confirmed these results.
FIGURE 8.
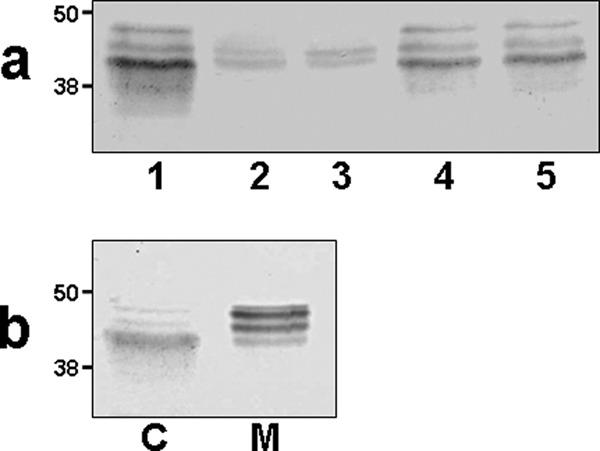
Western blots using specific antiserum against purified polar flagellins. Molecular weights are indicated. a, cytoplasmic fractions of strains: 1, A. hydrophila AH-3 (wild type); 2, AH-3ΔPseB mutant; 3, AH-3ΔPseF mutant; 4, AH-3ΔPseB mutant complemented with pBAD-PseB; and 5, AH-3ΔPseB mutant complemented with pBAD-PseBCj. b, A. hydrophila AH-3 (wild type). C, cytoplasmic fraction; M, whole membrane fraction. The fractions were separated as described under “Experimental Procedures.”
TABLE 4.
Detection of polar flagellin in cytoplasmic fractions
Several A. hydrophila strains were analyzed using an EIA experiment as described under “Experimental Procedures.”
| Strain | A405 (EIA) |
|---|---|
| AH-3 | 1.87 ± 0.09 |
| AH-3ΔPseB | 0.11 ± 0.01 |
| AH-3ΔPseF | 0.10 ± 0.01 |
| AH-3ΔPseB + pBAD-PseB | 1.79 ± 0.11 |
| AH-3ΔPseB + pBAD-PseBCj | 1.82 ± 0.08 |
| AH-3ΔPseB + pBAD33 | 0.09 ± 0.02 |
| AH-3ΔPseF + pBAD-PseF | 1.83 ± 0.10 |
| AH-3ΔPseF + pBAD-PseFCj | 1.80 ± 0.09 |
| AH-3ΔPseF + pBAD33 | 0.11 ± 0.01 |
DISCUSSION
For the first time, we were able to demonstrate that both flagella, the constitutively expressed polar flagellum and the inducible lateral flagella, in A. hydrophila strain AH-3 (serotype O34) showed O-glycosylations with two different glycans. The lateral flagella showed an O-glycosylation with a single monosaccharide of 376 Da linked in at least three different sites to the lateral flagellin, with one site of modification at serine 178. The largest observed intact mass acquired, 30.90 kDa, is 80 Da greater than the mass of flagellin modified with three 376-Da glycans, and the mass difference of 80 Da is strongly suggestive of phosphorylation. From high resolution MS data, we also can conclude that the 376-Da monosaccharide is a derivative of pseudaminic acid. The polar flagellum is also O-glycosylated with a more complex glycan that we were able to determine as an heptasaccharide comprised of three N-acetylhexoasmine (with variable addition of 0–2 phosphate groups and 0–2 methyl groups on each), two hexoses and two unknown monosaccharides of 376 and 102 Da in the sequence 376 Da-Hex-Hex-HexNAc- HexNAc-HexNAc-102 Da. A maximum of six heptasaccharide chains could be observed by the analysis of the intact mass of the polar flagellins, one of them linked to FlaB threonine 161. This heptasaccharide is unrelated to the O34-antigen LPS characterized in this strain consisting of a tetrasaccharide repeat unit of d-mannose, d-GalNAc, and 6-deoxytalose (31). In all of the cases, the 376-Da monosaccharide is the first attached to the polar flagellin.
This 376-Da monosaccharide (shared by both types of flagella) is absolutely necessary for flagellar production because Aeromonas mutants unable to biosynthesize pseudaminic acid (pseB or F mutants, 16), are completely unable to produce both flagella and lack motility. The pseudaminic acid biosynthetic mutants are completely able to transcribe the corresponding flagellin genes as their wild type strain and showed a DNA transcription of the polar or lateral flagellar master regulons similar to the wild type strain. However, we were able to demonstrate that these biosynthetic mutants are unable to produce significant amounts of polar flagellin in their cytoplasm. This fact is not regulated by transcription, and then a post-transcriptional regulation could be the cause for the reduced amount of cytoplasmic polar flagellins. The differences in molecular weight observed between the cytoplasmic or membrane attached polar flagellins in the wild type led us to suggest that cytoplasmic polar flagellins are non glycosylated in the wild type strain. All of these defects are rescued by the reintroduction of the A. hydrophila wild type genes or C. jejuni pseB or pseF, respectively.
Although in the O-glycosylation of Pseudomonas aeruginosa or Nesisseria spp. pilins some monosaccharides are block transfer-linked to oligosaccharide transferases that include a lipid carrier (34, 35), no such activity has been described for flagellar O-glycosylation to our knowledge. It is tempting to speculate that A. hydrophila strain AH-3 showed two different putative genes for linking the glycans to the flagellins (33, 36). The mutation of one of these genes abrogates polar flagellum formation but not lateral flagellar production (Maf-1; 36), and the other one abolishes the lateral but not the polar flagellum (Maf-5; 33). This fact seems to support the fact that O-glycosylation by two different glycans is observed for polar and lateral flagellins. Further studies will be required to understand the complete A. hydrophila strain AH-3 polar glycosylation and to understand the heptasaccharide glycan formation. In the strain AH-3, the cytoplasmic activity of pseudaminic acid like glycosylation is determinant to produce flagellin able to polymerize in the flagellar biogenesis.
As polar and lateral flagellar systems of the mesophilic Aeromonas are involved in adherence to surfaces and biofilm formation (33, 36), glycosylation seems to be crucial for pathogenesis in these motile bacteria. The detailed knowledge of the flagellar glycosylation will also contribute to the study of the pathogenic features from this microorganism.
Acknowledgments
We thank Maite Polo for technical assistance and the Servicios Científico-Técnicos from University of Barcelona. We also thank Luc Tessier for assistance with mass spectrometry instrumentation and Dr. Susan Logan for critical reading of the manuscript.
This work was supported by Plan Nacional de I+D+i (Ministerio de Educación, Ciencia y Deporte and Ministerio de Sanidad, Spain), Generalitat de Catalunya (Centre de Referència en Biotecnologia), and the National Research Council Canada.
- feCID
- front-end collision induced dissociation
- nLC
- nano-liquid chromatography
- ETD
- electron transfer dissociation.
REFERENCES
- 1. Janda J. M., Abbott S. L. (2010) The genus Aeromonas: Taxonomy, pathogenicity, and infection. Clin. Microbiol. Rev. 23, 35–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fenchel T. (2002) Microbial behavior in a heterogeneous world. Science 296, 1068–1071 [DOI] [PubMed] [Google Scholar]
- 3. Kirov S. M., Tassell B. C., Semmler A. B., O'Donovan L. A., Rabaan A. A., Shaw J. G. (2002) Lateral flagella and swarming motility in Aeromonas species. J. Bacteriol. 184, 547–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shimada T., Sakazaki R., Suzuki K. (1985) Peritrichous flagella in mesophilic strains of Aeromonas. Jpn. J. Med. Sci. Biol. 38, 141–145 [DOI] [PubMed] [Google Scholar]
- 5. Kirov S. M., Castrisios M., Shaw J. G. (2004) Aeromonas flagella (polar and lateral) are enterocyte adhesins that contribute to biofilm formation on surfaces. Infect. Immun. 72, 1939–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Logan S. M. (2006) Flagellar glycosylation: A new component of the motility repertoire? Microbiology. 152, 1249–1262 [DOI] [PubMed] [Google Scholar]
- 7. Nothaft H., Szymanski C. M. (2010) Protein glycosylation in bacteria: Sweeter than ever. Nat. Rev. Microbiol. 8, 765–778 [DOI] [PubMed] [Google Scholar]
- 8. Twine S. M., Logan S. M. (2011) in Bacterial Glycomic Current Research Technology and Applications (Reid C. V., Twine S. M., Reid A. N., eds) pp. 69–82, Caister Academic Press, Norfolk, U.K [Google Scholar]
- 9. Guerry P. (2007) Campylobacter flagella: Not just for motility. Trends Microbiol. 15, 456–461 [DOI] [PubMed] [Google Scholar]
- 10. Josenhans C., Vossebein L., Friedrich S., Suerbaum S. (2002) The neuA/flmD gene cluster of Helicobacter pylori is involved in flagellar biosynthesis and flagellin glycosylation. FEMS Microbiol. Lett. 210, 165–172 [DOI] [PubMed] [Google Scholar]
- 11. Arora S. K., Wolfgang M. C., Lory S., Ramphal R. (2004) Sequence polymorphism in the glycosylation island and flagellins of Pseudomonas aeruginosa. J. Bacteriol. 186, 2115–2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Aas F. E., Vik A., Vedde J., Koomey M., Egge-Jacobsen W. (2007) Neisseria gonorrhoeae O-linked pilin glycosylation: Functional analyses define both the biosynthetic pathway and glycan structure. Mol. Microbiol. 65, 607–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Twine S. M., Paul C. J., Vinogradov E., McNally D. J., Brisson J. R., Mullen J. A., McMullin D. R., Jarrell H. C., Austin J. W., Kelly J. F., Logan S. M. (2008) Flagellar glycosylation in Clostridium botulinum. FEBS J. 275, 4428–4444 [DOI] [PubMed] [Google Scholar]
- 14. Twine S. M., Reid C. W., Aubry A., McMullin D. R., Fulton K. M., Austin J., Logan S. M. (2009) Motility and flagellar glycosylation in Clostridium difficile. J. Bacteriol. 191, 7050–7062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chamot-Rooke J., Mikaty G., Malosse C., Soyer M., Dumont A., Gault J., Imhaus A. F., Martin P., Trellet M., Clary G., Chafey P., Camoin L., Nilges M., Nassif X., Duménil G. (2011) Posttranslational modification of pili upon cell contact triggers Neisseria meningitidis dissemination. Science 331, 778–782 [DOI] [PubMed] [Google Scholar]
- 16. Schoenhofen I. C., Vinogradov E., Whitfield D. M., Brisson J. R., Logan S. M. (2009) The CMP-legionaminic acid pathway in Campylobacter: Biosynthesis involving novel GDP-linked precursors. Glycobiology 19, 715–725 [DOI] [PubMed] [Google Scholar]
- 17. Canals R., Vilches S., Wilhelms M., Shaw J. G., Merino S., Tomás J. M. (2007) Non-structural flagellar genes affecting both polar and lateral flagella-mediated motility in Aeromonas hydrophila. Microbiology 153, 1165–1175 [DOI] [PubMed] [Google Scholar]
- 18. Tabei S. M., Hitchen P. G., Day-Williams M. J., Merino S., Vart R., Pang P. C., Horsburgh G. J., Viches S., Wilhelms M., Tomás J. M., Dell A., Shaw J. G. (2009) An Aeromonas caviae genomic island is required for both O-antigen lipopolysaccharide biosynthesis and flagellin glycosylation. J. Bacteriol. 191, 2851–2863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 20. Sanger F., Nicklen S., Coulson A. R. (1977) DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. U.S.A. 74, 5463–5467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Studholme D. J., Dixon R. (2003) Domain architectures of σ54-dependent transcriptional activators. J. Bacteriol. 185, 1757–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Münch R., Hiller K., Grote A., Scheer M., Klein J., Schobert M., Jahn D. (2005) Virtual Footprint and PRODORIC: An integrative framework for regulon prediction in prokaryotes. Bioinformatics 21, 4187–4189 [DOI] [PubMed] [Google Scholar]
- 24. Milton D. L., O'Toole R., Horstedt P., Wolf-Watz H. (1996) Flagellin A is essential for the virulence of Vibrio anguillarum. J. Bacteriol. 178, 1310–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jimenez N., Lacasta A., Vilches S., Reyes M., Vazquez J., Aquillini E., Merino S., Regué M., Tomás J. M. (2009) Genetics and proteomics of Aeromonas salmonicida lipopolysaccharide core biosynthesis. J. Bacteriol. 191, 2228–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Guzman L. M., Belin D., Carson M. J., Beckwith J. (1995) Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177, 4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hanahan D. (1983) Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166, 557–580 [DOI] [PubMed] [Google Scholar]
- 28. Towbin H., Gordon J. (1984) Immunoblotting and dot immunobinding–current status and outlook. J. Immunol. Methods 72, 313–340 [DOI] [PubMed] [Google Scholar]
- 29. Gavıín R., Rabaan A. A., Merino S., Tomás J. M., Gryllos I., Shaw J. G. (2002) Lateral flagella of Aeromonas species are essential for epithelial cell adherence and biofilm formation. Mol. Microbiol. 43, 383–397 [DOI] [PubMed] [Google Scholar]
- 30. Wilhelms M., Molero R., Shaw J. G., Tomás J. M., Merino S. (2011) Transcriptional hierarchy of Aeromonas hydrophila polar-flagellum genes. J. Bacteriol. 193, 5179–5990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Knirel Y. A., Shashkov A. S., Senchenkova S. N., Merino S., Tomás J. M. (2002) Structure of the O-polysaccharide of Aeromonas hydrophila O:34; a case of random O-acetylation of 6-deoxy-l-talose. Carbohydr. Res. 337, 1381–1386 [DOI] [PubMed] [Google Scholar]
- 32. Thibault P., Logan S. M., Kelly J. F., Brisson J. R., Ewing C. P., Trust T. J., Guerry P. (2001) Identification of the carbohydrate moieties and glycosylation motifs in Campylobacter jejuni flagellin. J. Biol. Chem. 276, 34862–34870 [DOI] [PubMed] [Google Scholar]
- 33. Canals R., Altarriba M., Vilches S., Horsburgh G., Shaw J. G., Tomás J. M., Merino S. (2006) Analysis of the lateral flagellar gene system of Aeromonas hydrophila AH-3. J. Bacteriol. 188, 852–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kus J. V., Kelly J., Tessier L., Harvey H., Cvitkovitch D. G., Burrows L. L. (2008) Modification of Pseudomonas aeruginosa Pa5196 type IV Pilins at multiple sites with D-Araf by a novel GT-C family arabinosyltransferase, TfpW. J. Bacteriol. 190, 7464–7478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vik A., Aas F. E., Anonsen J. H., Bilsborough S., Schneider A., Egge-Jacobsen W., Koomey M. (2009) Broad spectrum O-linked protein glycosylation in the human pathogen Neisseria gonorrhoeae. Proc. Natl. Acad. Sci. U.S.A. 106, 4447–4452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Canals R., Ramirez S., Vilches S., Horsburgh G., Shaw J. G., Tomás J. M., Merino S. (2006) Polar flagellum biogenesis in Aeromonas hydrophila. J. Bacteriol. 188, 542–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nogueras M. M., Merino S., Aguilar A., Benedi V. J., Tomás J. M. (2000) Cloning, sequencing, and role in serum susceptibility of porin II from mesophilic Aeromonas hydrophila. Infect. Immun. 68, 1849–1854 [DOI] [PMC free article] [PubMed] [Google Scholar]