Background: The Tat translocon transports folded proteins across energized prokaryotic cytoplasmic membranes.
Results: The N-terminal transmembrane domain of TatA interacts with the membrane-stabilizing Psp machinery.
Conclusion: Membrane-stabilization takes place where folded proteins are Tat-dependently translocated.
Significance: Membrane stress is directly related to Tat transport.
Keywords: Membrane Function, Membrane Proteins, Membrane Transport, Protein Translocation, Protein-Protein Interactions, Phage Shock Proteins, Twin-arginine Translocation
Abstract
Tat systems transport folded proteins across energized membranes of bacteria, archaea, and plant plastids. In Escherichia coli, TatBC complexes recognize the transported proteins, and TatA complexes are recruited to facilitate transport. We achieved an abstraction of TatA from membranes without use of detergents and observed a co-purification of PspA, a membrane-stress response protein. The N-terminal transmembrane domain of TatA was required for the interaction. Electron microscopy displayed TatA complexes in direct contact with PspA. PspB and PspC were important for the TatA-PspA contact. The activator protein PspF was not involved in the PspA-TatA interaction, demonstrating that basal levels of PspA already interact with TatA. Elevated TatA levels caused membrane stress that induced a strictly PspBC- and PspF-dependent up-regulation of PspA. TatA complexes were found to destabilize membranes under these conditions. At native TatA levels, PspA deficiency clearly affected anaerobic TMAO respiratory growth, suggesting that energetic costs for transport of large Tat substrates such as TMAO reductase can become growth limiting in the absence of PspA. The physiological role of PspA recruitment to TatA may therefore be the control of membrane stress at active translocons.
Introduction
In most prokaryotes as well as in plant plastids, two general protein translocation systems exist that serve to transport proteins across energized biological membranes: The Sec system, which is restricted to unfolded proteins, and the twin-arginine translocation (Tat)2 system, which serves to transport folded proteins (1). The Tat-dependent transport of folded proteins can cope with proteins of very distinct sizes, ranging from small high potential ferredoxins of less than 10 kDa to large oligomeric oxidoreductases of more than 140 kDa. Transport is achieved by the coordinated action of two membrane protein complexes, one consisting of multiple copies of the component TatA, and the other consisting of TatC that often is associated with a third component, TatB (2). Tat(B)C complexes are receptors that specifically recognize the transported proteins and possibly initiate or trigger the transport process (3–8). There is accumulating evidence for a participation of TatA complexes late during the translocation process (8–13). TatA complexes have been shown to vary in size and to be influenced by the association with substrate-bound TatBC complexes (14–16). A high resolution NMR structure of a solubilized TatA protomer of Bacillus subtilis showed that TatA consists of an N-terminal region that includes a short membrane anchor, followed by a hinge region, an amphipathic helix and most likely a largely unstructured C-terminal region that was not resolved (17).
In our efforts to further analyze the TatA complex of Escherichia coli, we found that a significant population of TatA can be stripped off the membrane bilayer during affinity chromatography. This approach resulted in the identification of a TatA-interacting protein, the phage shock protein A (PspA) that is already known to improve Tat transport under substrate saturating conditions (18). We thus analyzed this TatA-PspA interaction in detail, identified the interaction determinants, and found physiological evidence for the functional basis of the TatA-PspA interaction.
EXPERIMENTAL PROCEDURES
Strains and Growth Conditions
The E. coli strains MC4100 (19), its tatAE-deficient derivative JARV16 (20), and strain BW25113 as well as pspA, pspB, pspC, or pspF-deleted derivatives of BW25113 (21) were used for physiological studies, and E. coli XL1-Blue Mrf′ Kan or Tet (Stratagene) were used for cloning. LacZ activity was measured with the transcriptional fusion of the pspA-promoter with lacZ in the E. coli psp-reporter strain MC3 (22). E. coli was grown at 37 °C on LB medium (1% tryptone, 1% NaCl, 0.5% yeast extract) in the presence of the appropriate antibiotics (100 μg/ml ampicillin, 15 or 50 μg/ml kanamycin). TMAO respiration was tested by anaerobic growth on M9 medium supplemented with 0.5% glycerol, trace elements (SL12 (23)), and 1.1% (w/v) TMAO. Cells were also grown anaerobically on LB medium supplemented with 0.5% glycerol and 0.4% nitrate, and TMAO was added to 1.1% for induction of torA expression. For rhamnose-dependent gene expressions, 0.1% rhamnose was added at an OD600 nm of 0.6 and cells were harvested after 3 h of induction.
Plasmids and Genetic Methods
The tatA or tatE genes were expressed from the rhamnose-dependent pBW22-based vectors (24). Strep-tagged TatA was amplified by PCR using the primers Strep-BamHI-F (5′-GTC TCG GAT CCT GGA GCC ACC CGC AGT TCG AA-3′) and AmpR-ScaI-R (5′-TGG TGA GTA CTC AAC CAA GTC ATT CTG-3′), and cloned into the corresponding sites of pBW-tatA-H6 (25). TatA without an N terminus tatA-CT was amplified by PCR using the primers tatA-CT-Nde-F (5′-CTG TAC ATA TGA GTG GCA CCA AAA AGC TCG GCT CC-3′) and tatA-Bam-R (5′-TGA TTG GAT CCC ACC TGC TCT TTA TCG TGG CGC-3′) and cloned into the corresponding sites of pBW-tatA-strep. The vector pBW-tatA was derived from pBW-tatA-strep by insertion of a single stop codon between the coding region of tatA and its Strep-tag II using a PCR with the primer pair tatA-NdeI-F (5′-GAA CAC ATA TGG TGG TAT CAG TAT TTG GC-3′) and tatA-BamHI-1-R (5′-TCA AAG GAT CCT TAC ACC TGC TCT TTA TCG TG-3′).
To verify the importance of the N terminus of TatA various gene fusions of tatA or its domains were either cloned in front of or behind the mature domain-encoding region of hip. pBW-mat-hip-strep served as template (26). For the fusion proteins, the TatA-CT-encoding region was amplified by PCR with the primers tatA-CT-BglII-F (5′-ATA TAA GAT CTA GTG GCA CCA AAA AGC TCG G-3′) and tatA-Bam-R (5′-TGA TTG GAT CCC ACC TGC TCT TTA TCG TGG CGC-3′), restricted with BamHI/BglII and ligated behind the hip-gene in the BamHI restricted vector pBW-mat-hip-strep. A new BamHI restriction site was introduced into the plasmid pBW-tatA-strep using the QuikChange method with the primer pair tatA-BamHI-2-F (5′-GTT GTA CTG CTT TTT GGC GGA TCC AAG CTC GGC TCC ATC GGT TC-3′) and tatA-BamHI-2-R (5′-GAA CCG ATG GAG CCG AGC TTG GAT CCG CCA AAA AGC AGT ACA AC-3′), to combine the N terminus of TatA with the mature domain of HiPIP. For this, the primer pair mat-hip-BglII-F (5′-ATA TAA GAT CTT CCG CTC CCG CCA ATG-3′) and hip-BamHI-R (5′-AAC GGG GAT CCG CCG GCC TTC AGG GTC CAG-3′) was used. The newly designed plasmid pBW-tatA-BamHI-strep was treated with the restriction enzyme BamHI and ligated with the aforementioned PCR-amplified and BglII and BamHI-treated mat-hip gene, resulting in the vector pBW-tatA-NT-mat-hip-strep.
Single copy integration of tatA-strep into the chromosome of the E. coli strain JARV16 was done according to standard protocols (27). Initially, the tatA-strep-gene was amplified by PCR, using the primers tatA-NdeI-F (5′-GAA CAC ATA TGG GTG GTA TCA GTA TTT GGC-3′) and tatA-BssHII-R (5′-ATA TAG CGC GCT TAT TTT TCG AAC TGC GGG-3′), restricted with NdeI/BssHII and ligated with the corresponding sites of pAH120. For wt-level expression of tatA we exchanged PrhaB with PtatA, resulting in the pAH120-Ptat-tatA-strep plasmid, which was then integrated into the chromosome.
Biochemical Methods
SDS-PAGE and protein estimations were carried out by standard methods (28, 29). Immunoblots were developed as described previously (30), using polyclonal antisera against purified hexahistidine-tagged HiPIP, hexahistidine-tagged PspA, or the C-terminal 16 residues of TatA. Blots were developed with Strep-Tactin-HRP or Strep-Tactin-AP conjugate (IBA Göttingen, Germany) according to the manufacturer's instructions.
TatA protomers or fusion proteins with C-terminal Strep-Tag II-fusions were purified from soluble or membrane fractions by Strep-Tactin-based affinity chromatography according to the supplier's protocol (IBA), with the exception that 250 mm NaCl was used in washing and elution buffers. For electron microscopy, elution fractions of Strep-tagged TatA were further purified by sucrose gradient centrifugation and negatively stained with 2% (w/v) uranyl acetate pH 4.5 as described previously (31). Hexahistidine-tagged PspA was purified by affinity chromatography and sucrose gradient centrifugation as described previously (31).
Mass spectrometry and electron microscopy were carried out as described previously (26, 31). TMAO reductase was detected in native gels as described elsewhere (32). Preparation of inverted vesicles (INV) and fluorescence quenching experiments were carried out as described previously (6). The activity of β-galactosidase was determined according to Miller (33).
RESULTS
E. coli TatA Interacts with PspA
TatA is membrane-anchored by a single N-terminal transmembrane domain (TMD) and a subsequent amphipathic helix that associates with the membrane (17, 34). As TatA exists in complexes of very distinct oligomeric states (35, 36), and as TatA has a very unusual and short trans-membrane domain (2, 34, 37), we reasoned that it might be possible to abstract a population of TatA from membrane vesicles without the use of detergents. This would permit an analysis of TatA complexes that are not organized in detergent micelles.
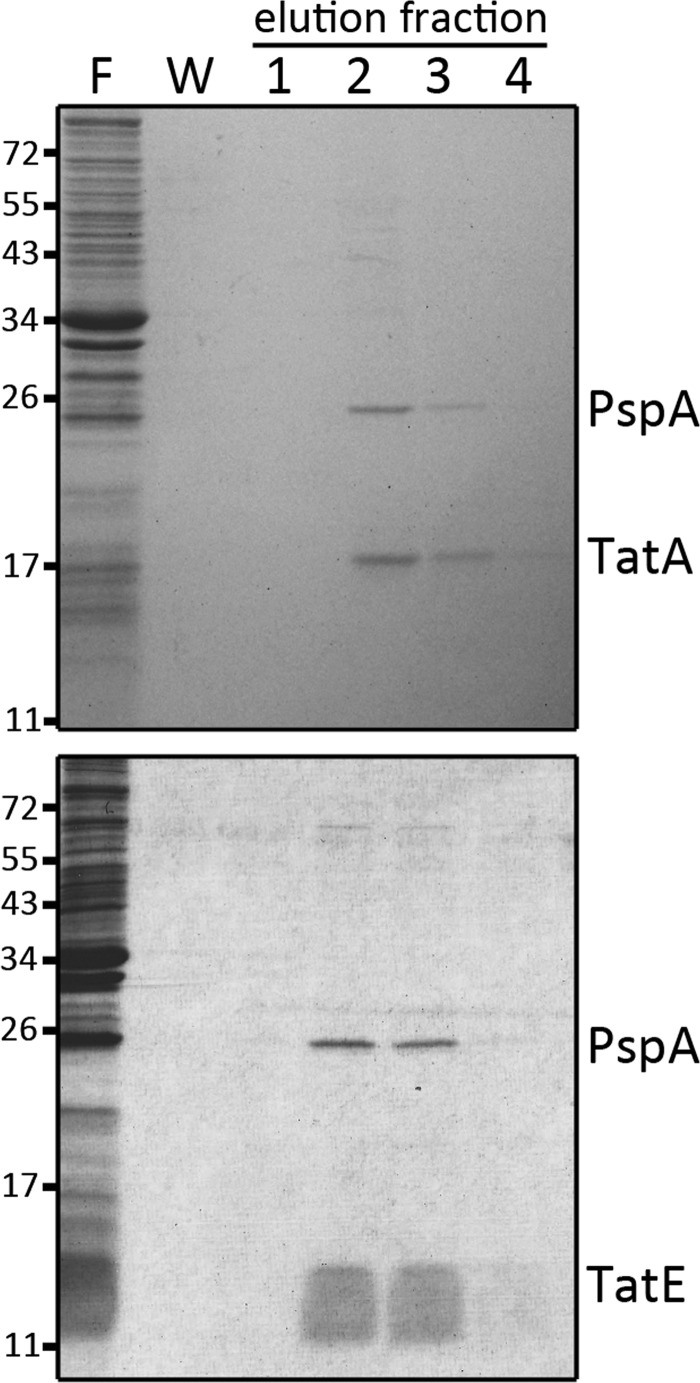
The experiment was carried out using a TatA variant that contained a C-terminal Strep-tag II, resulting in multiple Tags on the homooligomeric functional TatA complexes (38). The complexes could bind to Strep-Tactin affinity matrices with sufficient affinity to be abstracted from membranes. Membrane vesicles were only delayed on the column and could be removed by six successive washing steps. Highly enriched soluble TatA could be obtained by this approach (Fig. 1, upper gel). The membrane-abstracted TatA was remarkably stable in solution and did not show precipitation for over weeks. One protein co-purified with TatA in a Coomassie Blue detectable band. This protein band had an intensity similar to TatA and was identified as PspA by mass spectrometry. PspA is known to have dual function and location: in the cytoplasm it has regulatory functions, and as a membrane-associated protein it stabilizes the membrane integrity upon manifold stress conditions (39, 40). PspA has been reported to physically stabilize the membrane in vitro (41). The PspA interaction was also observed for the second TatA paralog of E. coli, TatE, a protein that can functionally substitute for TatA (Fig. 1, lower gel). The interaction was detectable also at wild type levels after single copy genomic integration of the tatA-strep gene under control of its natural promoter (supplemental Fig. S1). In further control experiments with TatA that was produced without the C-terminal Strep-tag, it was confirmed that the PspA elution indeed depended on the TatA-strep-binding to the affinity column (supplemental Fig. S2). The TatA-PspA interaction immediately raised our interest, as PspA is known to improve Tat transport when the translocation system is saturated (18), and to our knowledge PspA is the first non-Tat interaction partner of the Tat-system.
FIGURE 1.
PspA interacts with TatA and TatE. C-terminally Strep-tagged TatA (upper gel) or TatE (lower gel) were purified from membrane fractions by Strep-tactin affinity chromatography, and fractions were analyzed by SDS-PAGE/Coomassie staining. F, flow-through (unbound material); W, last wash fraction; E1–4, elution fractions. The positions of TatA, TatE, and PspA bands are indicated on the right. Molecular weight markers are indicated on the left.
Detection of TatA-associated PspA Structures by Electron Microscopy
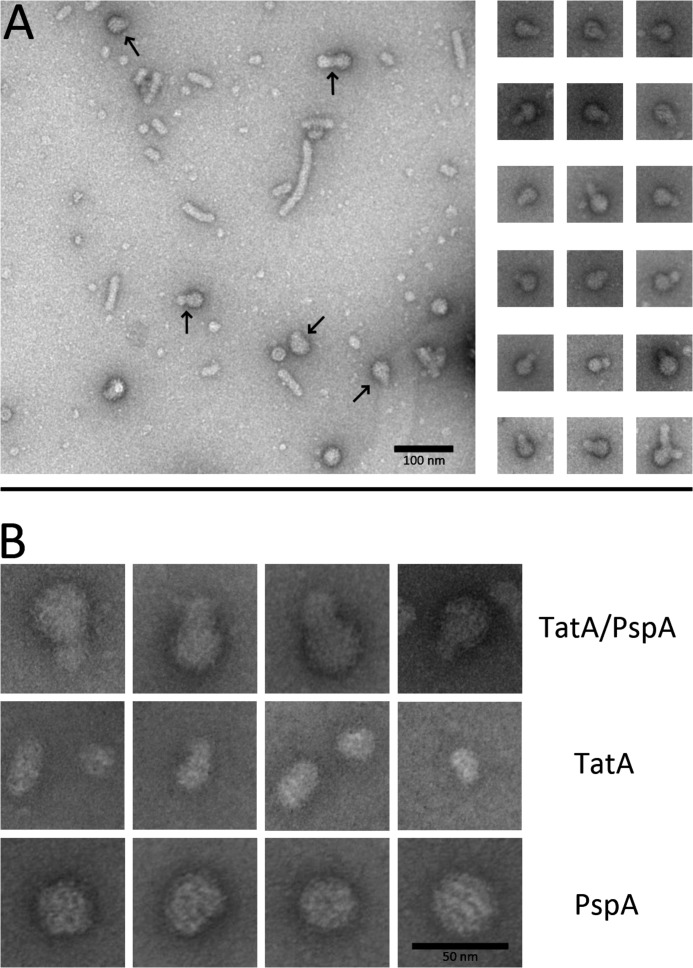
TatA as well as PspA are known to form large complexes that can be detected by electron microscopy (EM) (14, 31, 42). We thus analyzed the co-eluting TatA-PspA fractions by transmission EM. The fractions were compared with TatA-strep, which was prepared from a strain that carried a pspA deletion, and with purified hexahistidine-tagged PspA. PspA alone formed the large approx. 30 nm spheroids that consisted of scaffolds surrounded by intensive stain, as seen before (Ref. 31, Fig. 2). TatA-strep alone formed large thin disks of variable, often about 16 nm in diameter, or elongated structures that most likely represented filamentous stacks of these disks. The TatA structures had a fuzzier surface and showed significantly less contrast than the usually spherical PspA scaffolds. TatA-strep complexes that interacted with PspA were easily recognizable as composite complexes with typical PspA scaffolds in direct contact with TatA (Fig. 2). Most PspA-interacting TatA particles were of the rather small ∼16 nm diameter type, and larger TatA stacks occurred only rarely, suggesting that PspA-associated filamentous stacks are either rare or more fragile.
FIGURE 2.
Detection of the TatA-PspA interaction by electron microscopy. A, overview micrograph of negatively stained affinity-purified Strep-tagged TatA, with co-purified PspA (left) and depicted TatA-PspA complexes (right). In the overview image, arrows point to readily recognizable conjunctions of TatA-PspA complexes. Note that longer filament-like TatA stacks, although frequently observed in the overview, were only rarely seen to interact with PspA. B, higher resolution micrographs of TatA-PspA complexes (upper panel), TatA as purified in the absence of PspA (middle panel), and a TatA-free PspA preparation (lower panel). The smaller ∼16 nm diameter TatA particles are seen as predominant interaction partners of PspA and are therefore depicted in the TatA panel of B.
The Transmembrane Domain of TatA Is Crucial for the PspA Interaction
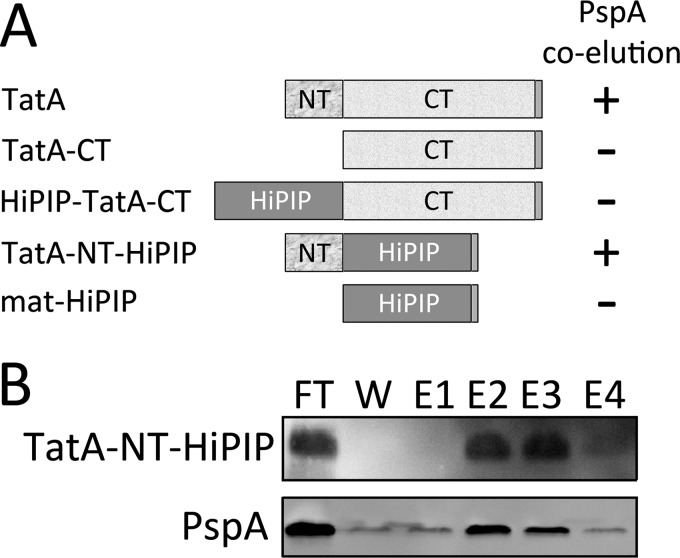
To identify the region(s) of TatA responsible or required for the PspA interaction, we constructed fusions of TatA domains with a soluble and very stable protein, the mature domain of the high potential iron-sulfur protein (HiPIP) (43). The constructs contained either the N-terminal region (TatA-NT; res. 1–21), or the hinge-to-the-C terminus region of TatA (TatA-CT; res. 21–86). These regions were fused either to the N terminus or the C terminus of HiPIP, depending on the natural position of the corresponding domains in TatA, resulting in the constructs TatA-NT-HiPIP and HiPIP-TatA-CT, respectively. In control experiments it was ensured that HiPIP alone, without fused TatA domains, did not interact with PspA (data not shown).
PspA interacted with the TatA-NT-HiPIP construct, indicating that the N-terminal 21 residues of TatA are sufficient for the PspA interaction (Fig. 3). In contrast, the HiPIP-TatA-CT construct, which was soluble due to the lack of the N terminus of TatA, did not show any PspA interaction, although significant PspA can be found in the soluble fraction (supplemental Fig. S3A). Together, these data indicate that the N terminus of TatA mediates the interaction with PspA.
FIGURE 3.
The N terminus of TatA is required and sufficient for the TatA-PspA interaction. A, scheme constructs that resulted in the identification of the N terminus as the region that mediates the interaction with PspA. NT: residues 1–21 of TatA; CT: residues 21–86 of TatA; All constructs were C-terminally Strep-tagged. B, PspA co-elutes with the N terminus of TatA. Purification of TatA-NT-HiPIP, in which the short N-terminal transmembrane domain of TatA is fused to a Strep-tagged mature domain of HiPIP to allow its purification and detection. Membrane fractions of MC4100/pBW-tatA-NT-mat-hip-strep were loaded on Strep-Tactin matrices, and bound protein was eluted as described under “Experimental Procedures.” TatA-NT-HiPIP as well as PspA were detected in fractions of the purification by immunoblot analyses. FT, flow through; E1-E4: elution fractions.
As it was still possible that the N-terminal fusion of HiPIP might have interfered with a PspA interaction, we tested whether a TatA-CT construct without fused HiPIP interacted with PspA. As in the case of the HiPIP fusion, this construct did not show any PspA co-purification (supplemental Fig. S3B), confirming that a membrane association is essential for the PspA interaction.
The TatA-PspA Interaction Is Influenced by PspB and PspC, but Not by PspF
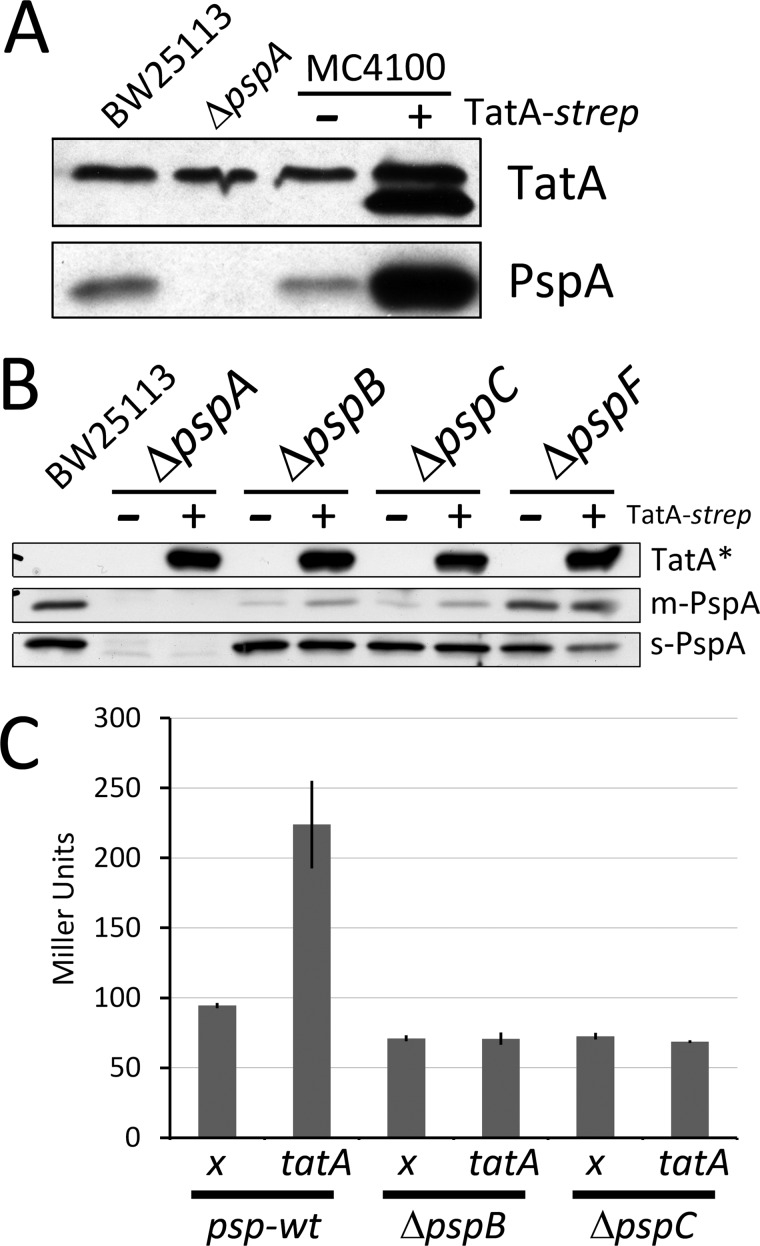
PspA is known to interact with the membrane integral phage shock response proteins PspB and PspC that most likely cooperatively sense membrane stress (44). In addition, PspA is known to interact with the regulatory protein PspF that can activate a σ54-dependent transcription of the psp genes under membrane stress conditions (45). To find out whether these interactions are important for the TatA-PspA contact, we carried out our analyses in E. coli strains that were deleted in the genes encoding PspB, PspC, or PspF (Fig. 4). Without PspB, PspC, or PspF, only basal levels of PspA are present in cells. The deletions of pspB or pspC further resulted in significantly lowered PspA amounts in the membrane fraction, consistent with the view that these proteins contribute to the PspA interaction with the membrane (44, 46–48). In the absence of PspB or PspC no TatA-PspA co-elution was detectable, whereas the deletion of pspF did not have any effect, indicating that selectively the membrane-integral interaction partners of PspA are likely to be important for the stability or formation of the TatA interaction, but not the soluble interaction partner PspF (Fig. 4A). In successful complementation experiments, the co-elution of PspA could be achieved again when the pspB or pspC genes were expressed from a plasmid (Fig. 4B). For these complementations, hexahistidine-tagged PspB or PspC components were used, which allowed for the detection of PspB or PspC in the PspA-TatA association. We clearly could detect PspC that seems to be an integral part of the association. PspB was also detected, but to a much lesser extent (Fig. 4B).
FIGURE 4.
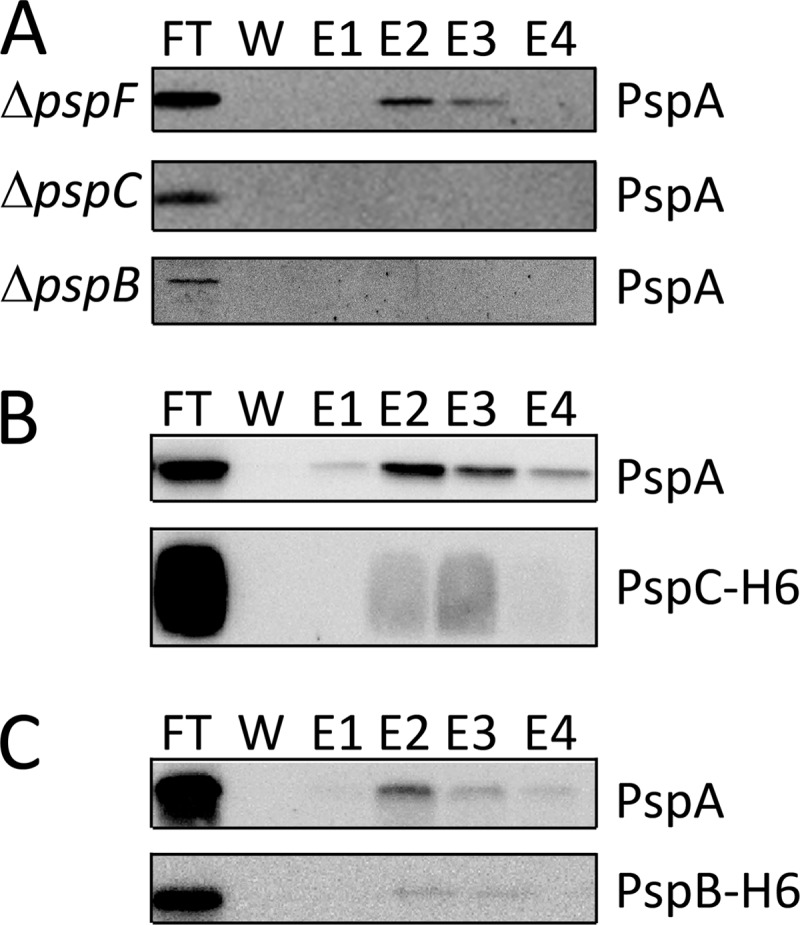
The TatA-PspA interaction depends on PspB and PspC, but not on PspF. A, detection of PspA in purifications of TatA-strep produced in the E. coli psp-deletion strains JWK1296 ( = ΔpspF)/pBW-tatA-strep, JWK1299 ( = ΔpspC)/pBW-tatA-strep and JWK1298 ( = ΔpspB)/pBW-tatA-strep. B, complementation of the PspA-TatA interaction in the pspC deletion strain JWK1299 by expression of pspC in trans. Upper panel, detection of PspA in the TatA-strep purification fractions. Lower panel, detection of PspC in the TatA-strep purification fractions. Note that PspC co-elutes in TatA-strep purifications, as does PspA. C, complementation of the PspA-TatA interaction in the pspB deletion strain JWK1298 by expression of pspB in trans. Upper panel, detection of PspA in the TatA-strep purification fractions. Lower panel, detection of PspB in the TatA-strep purification fractions. All panels show immunoblots with detections of the proteins indicated on the right side.
Recombinant TatA Can Induce the Phage Shock Response
As the PspA interaction suggested that membrane-integral TatA is related to membrane stress, we analyzed whether TatA itself can create such membrane stress. We found that recombinant TatA production resulted in the phage shock response: PspA levels were significantly increased when TatA was produced (Fig. 5A). We noted that the C-terminally linked Strep-tag (GS-WSHPQEFK) compensated in part the unusual slow migration behavior of untagged TatA. The phage shock response induction by recombinant TatA strictly depended on the membrane-stress-sensor PspBC complex as well as on the σ54-activator PspF (Fig. 5B). Also the basal level of soluble PspA was kept remarkably constant in wt and ΔpspB, ΔpspC, or ΔpspF backgrounds, not showing any increase of PspA (in case of the ΔpspF strain even a slight decrease) in response to recombinant TatA. While the absence of PspF did not influence membrane-binding of basal level PspA, only very little PspA was detected in membrane fractions in the absence of PspB or PspC, confirming the important role of PspB and PspC in the membrane interaction of PspA. We also noted that the portion of membrane-associated basal-level PspA was to some extent increased by TatA in the pspB or pspC mutant strains (Fig. 5B). It thus may be that TatA per se can already recruit some PspA to membranes at uninduced levels (there is no induction without PspB or PspC).
FIGURE 5.
Recombinant TatA production induces the phage shock response. A, SDS-PAGE/immunoblot detection of TatA (upper panel) and PspA (lower panel) in E. coli strains BW25113, BW25113 ΔpspA, MC4100, and MC4100 producing TatA-strep (migrating below the natural TatA band; anti TatA antibody detects both bands). Note that the PspA level is strongly induced by recombinant TatA-strep. B, induction of the phage shock response by recombinant TatA levels depends on the phage shock proteins PspB, PspC, and PspF. Detection of TatA-strep (upper panel) and PspA in the psp-wt strain BW25113 and its mutants deficient in pspA, pspB, pspC, or pspF, as indicated with or without recombinant TatA production. To gain a more complete view of PspA levels, PspA was detected in membrane fractions (m-PspA; middle panel) and in soluble fractions (s-PspA; lower panel); *, exclusively the Strep-tag and thus recombinant TatA is detected on this blot. C, LacZ activities of the pspA-promoter reporter strain MC3, 1 h after induction of pBW-tatA-strep. Cultures of strains containing the empty vector were used as negative controls (x). Note that the activation of the pspA promoter by TatA-strep production depends on the presence of PspB and PspC.
The regulation that resulted in increased PspA abundance was on a transcriptional level, since a pspA-lacZ promoter reporter gene fusion showed a marked increase of LacZ activity upon expression of the tatA gene (Fig. 5C). As typically found for the phage shock response and as already expected from the protein level experiments, up-regulation strictly depended on the PspB and PspC proteins in the membrane.
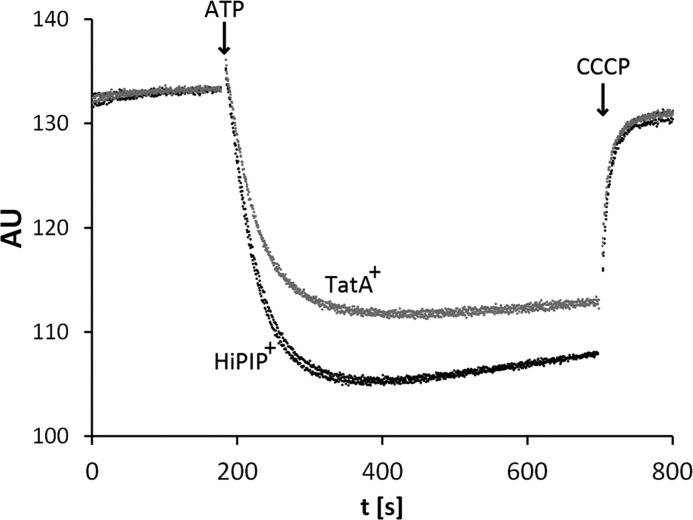
The membrane destabilization by increased TatA levels could be confirmed by direct measurements of the stability of inverted membrane vesicles from strains producing TatA. In control experiments, another protein of similar size (mature HiPIP) was produced. We applied an acridine orange fluorescence quenching approach that monitors the transmembrane proton gradient that can be established with inverted cytoplasmic membrane vesicles (INVs). Acridine orange is trapped inside INVs in the presence of a pH gradient (49) and this accumulation results in a fluorescence quenching that is diminished when the delta pH is affected by membrane destabilization (6). We used whole membrane vesicles in which selectively the INVs can build up a pH gradient by reverse action of ATP synthase. Interestingly, TatA production reduced the fluorescence quenching by about 25% relative to the quenching achieved with the control strain, indicating a partial membrane-destabilization by TatA (Fig. 6).
FIGURE 6.
Membrane stability is compromised by recombinantly overproduced TatA. Fluorescence quenching with vesicles of either E. coli MC4100 producing mature HiPIP from pBW-mat-hip-strep (black traces from two experiments), or E. coli MC4100 overproducing TatA using the plasmid pBW-tatA-strep (gray traces from two experiments). HiPIP has been chosen as negative control to produce a similar size protein with the same rhamnose-inducible vector system. AU: arbitrary fluorescence intensity units; Quenching was started at the indicated time point by addition of 1 mm ATP and the pH gradient was abolished by addition of 25 μm CCCP (uncoupler). Note that the fluorescence quenching is ∼25% weaker when TatA was recombinantly produced.
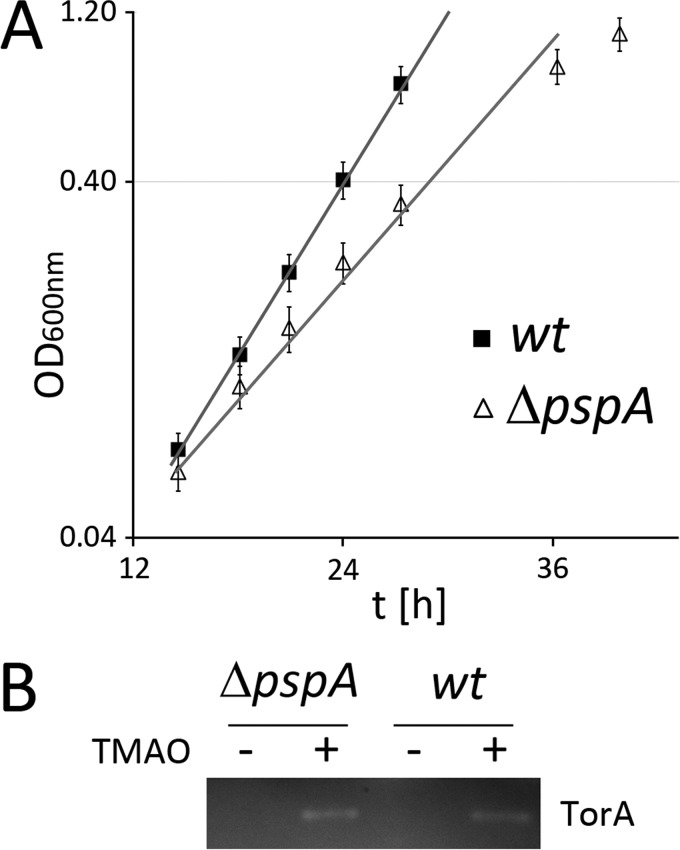
The overall data suggested that TatA is at the site where protein transport affects ionic gradients, which is counterbalanced by PspA. As this effect should be advantageous also in non-recombinant systems, we asked the question whether natural Tat transport results in lower growth rates in the absence of PspA at least under certain growth conditions. We speculated that the transport of the large 92 kDa TMAO reductase TorA during anaerobic TMAO respiration in minimal media could permit the detection of such effects. As shown in Fig. 7A, the growth rate during TMAO respiration was indeed affected. This effect was not observed with aerobic respiratory growth in the same medium (data not shown). Activity stains of periplasmic fractions from cultures that were shifted for 1 h to TMAO respiration showed that the TorA activity was similar in wt or PspA-deficient strains, clearly indicating that PspA is not required for Tat transport per se (Fig. 7B). Therefore, the role of PspA appears to be limited to the minimization of energetic costs, reflected by the growth rate effects, and PspA is not actively involved in Tat transport.
FIGURE 7.
Under TorA transport conditions, optimal E. coli growth requires PspA. A, exponential phase of strict TMAO-respiratory growth of E. coli BW25113 (wt) and its PspA-deficient derivative (ΔpspA). Mean values and standard deviations are from three cultures. B, TorA activity stain in periplasmic fractions of the two strains shown in A. Cells were grown on LB/glycerol/nitrate medium to allow anaerobic growth independently of TorA, and TorA production was induced by TMAO 1 h prior to harvest. Cultures without TMAO addition were analyzed in parallel as negative control.
DISCUSSION
Here we report an interaction of the Tat system with a membrane-stress response system, the Psp-System. The detection of this interaction was only possible due to the fact that a portion of TatA, as well as its paralog TatE, can be abstracted from cytoplasmic membranes without the use of detergents. Apparently, the only 3 nm short and not entirely hydrophobic single N-terminal domain of TatA is not the firmest membrane anchor, although it clearly has a transmembrane topology (37). The only TatA interaction partner identified by Coomassie-stained SDS-PAGE and mass spectrometry was PspA, which is a coiled-coil protein that forms multimeric complexes that stabilize cytoplasmic membranes under stress conditions (39).
EM analyses of purified TatA-PspA particles clearly showed typical PspA scaffolds in conjunction with TatA structures (Fig. 2). The transmembrane domain of TatA was responsible and sufficient for the interaction (Fig. 3). The known partners of PspA in the cytoplasmic membrane, PspB and PspC (44, 47), were involved in the interaction: TatA-PspA associations could only be purified in the presence of PspB and PspC, and a co-purification of both components with TatA and PspA was detectable by immunoblot analyses (Fig. 4). These observations indicate that PspB and PspC most likely contribute to the TatA-PspA interaction. This is conceivable considering the fact that PspB and PspC are very important for the recruitment of PspA to the membrane, even under non-stressed conditions, as seen in Fig. 5B. Despite their importance for binding of the majority of PspA to the cytoplasmic membrane, traces of PspA interacted reproducibly with the membrane in the absence of PspB or PspC. Interestingly, we observed that abundant TatA alone, without PspB or PspC, already to some extend enhanced the amount of membrane-associated PspA (Fig. 5B). It is thus possible that TatA alone might interact with PspA already without PspB or PspC present in the membranes, albeit these two components appear to at least strongly enhance this binding. As PspB and PspC clearly stabilize the TatA-PspA interaction to an extent that allows the co-abstraction from membranes (Fig. 4), which depends on the N terminus of TatA, we believe that PspB and PspC could be the membrane integral proteins that mediate this interaction. An additional weaker direct interaction of PspA with TatA might occur via surface-exposed regions. Also a membrane lipid interaction could be involved (41). A PspB- and PspC-independent membrane interaction has also been reported for PspA from Yersinia enterolitica (50).
PspB and PspC are well known to be key players of the phage shock response. In Yersinia enterolitica it is believed that these proteins cooperate to sense membrane stress and to recruit PspA via PspC, and this may well be also the case in E. coli (47, 50). Interestingly, whereas most reported membrane stress situations are externally imposed, the TatA-Psp interaction reported here is not related to external stresses. Tat translocons are sites where membrane stress is continuously generated due to membrane permeabilization that accompanies the transport of fully folded proteins. It has been measured in studies on the thylakoid system that the transport of each folded Tat substrate is accompanied by the transfer of about 80,000 protons (51), which can be regarded as a kind of “natural” membrane stress. As Tat transport is something that takes place under any conditions, it is expected that the Psp system interacts with the Tat system already at uninduced basal levels. This is exactly what we observed: The PspF-mediated signaling pathway for the induction of the psp genes under external membrane stress conditions is not required for the interaction (Fig. 4A). There is a basal psp gene expression that results in a population of PspA at membranes even without external stress (seen for example in Fig. 5, A and B), and a population of these low levels of PspA appears to be interacting with the Tat system. So what is the exact function of PspA at Tat translocons? It is surely not a direct improvement of Tat transport: We did never observe an effect of the lack of PspA on native level Tat substrate transport (e.g. Fig. 7B). In agreement with this, De Lisa et al. did not observe any effect of PspA on non-recombinant Tat transport (18). They instead demonstrated that PspA enhances the transport under overproduction conditions, when the Tat machinery is saturated and when energetic costs for transport are high. A similar positive effect of PspA on recombinant Tat substrate transport was observed with the Streptomyces lividans system (52). Like with E. coli, PspA of S. lividans is important in the membrane stress response (53). Interestingly, PspB or PspC components are missing from S. lividans, which would be in line with the idea that TatA alone may already recruit PspA. The transport-promoting effect of PspA under conditions when energetic costs are high (i.e. when Tat substrates are recombinantly overproduced) already suggests that PspA counterbalances membrane stress at the translocation-site. As we found that PspA had a positive effect on growth when growth depended on the transport of the large Tat substrate TorA, we believe that the energetic benefit of a PspA-TatA interaction is most likely the physiological function.
The exact mechanism by which PspA counterbalances membrane instability and proton leakage remains to be determined. One mechanistic model exists, in which membrane stabilization is proposed to be achieved by multiple interactions of scaffold-like PspA with the membrane (31). This may include protein-protein as well as protein-lipid interactions. As PspA has also been shown to be also important for proper Sec transport (54, 55), it may well be that similar PspA interactions exist for Sec systems, but this and the exact nature of the interaction will have to be analyzed in future studies.
Interestingly, we found that recombinant levels of TatA induce membrane stress, most likely by evoking disorder in the lipid bilayer due to its short and partially flexible transmembrane domain (Fig. 6). In the thylakoid system it has been described that TatA forms higher order oligomers at active translocons that permeabilize the membrane for the substrate passage (16). Similar oligomers might spontaneously form when TatA is highly abundant in the membrane, and it is a feasible speculation that the membrane stress induced by recombinant TatA might mimic the proton leakage at active translocons. The membrane destabilization by TatA complexes may also be the simplest explanation for the observation that TatA can be abstracted from membrane vesicles, which allowed us to identify Psp-system interactions in this study.
A homolog of PspA in plant thylakoids, VIPP1, has been very recently demonstrated to stimulate Tat transport (56). VIPP1 is located on the surface the inner envelope membrane and thylakoids (57, 58). It plays key roles in the biogenesis and structuring of thylakoid membranes (59) and interacts with the protein import machinery of plastids (60). VIPP1 is also found in cyanobacteria, where it is essential for thylakoid formation (61). Interestingly, cyanobacteria contain PspA in addition to VIPP1. In contrast to PspA, all VIPP1 proteins contain a C-terminal ∼30 residues extension that has been suggested to convey thylakoid-specific functions (61). VIPP1 apparently evolved prior to endosymbiosis from a pspA gene duplication in cyanobacteria, followed by a thylakoid-related specialization (61). Recombinant cyanobacterial VIPP1 has been found to improve transport of overproduced Tat substrates similar to PspA, and the C-terminal extension of VIPP1 did not play a role in this (18). Cyanobacterial VIPP1 therefore clearly possesses residual PspA functions, and this might be also expected for VIPP1 from higher plants. In agreement with this, VIPP1 from cyanobacteria is up-regulated upon salt stress (62), and VIPP1 from Chlamydomonas is up-regulated by defects in the photosynthesis apparatus (63), suggesting that functions of PspA might be retained within cyanobacteria and algae.
However, no Tat translocon-binding could be shown in the plant system, and it turned out that VIPP1 promotes Tat transport rather by enhancing productive substrate binding to the thylakoids (56). Lo and Theg provided clear evidence for an improvement of substrate access to productive translocon binding regions early in the Tat pathway, possibly by influencing the exposure of transport-productive membrane surface (56). Such membrane-rearrangements are very much related to the described important role in thylakoid structuring (56, 59). Moreover, a role in membrane stabilization in Tat transport has not been found for plant VIPP1, pointing to clear differences in the Tat-related roles of PspA and VIPP1 (56). In any case, despite of functional specifications, the principle mechanisms of complex assembly and membrane-association may be conserved, as the PspA-domain of VIPP1 is essential for its oligomerization (64).
The herein described recruitment of PspA to TatA sheds light on the membrane stress imposed by Tat transport. It supports the idea that TatA is the membrane-permeabilizing component and attributes an important function to basal PspA levels that minimize energetic costs at membranes under normal growth conditions.
Supplementary Material
Acknowledgment
We thank Vladimir Shevchik for donation of strain MC3.
This work was funded by the Deutsche Forschungsgemeinschaft (Grant BR2285/1–3).

This article contains supplemental Figs. S1–S3.
- Tat
- twin-arginine translocation
- HiPIP
- high potential iron-sulfur protein
- TMAO
- trimethylamine N-oxide
- CCCP
- carbonyl cyanide m-chlorophenylhydrazone
- PspA
- phage shock protein A
- INV
- inverted vesicles.
REFERENCES
- 1. Natale P., Brüser T., Driessen A. J. (2008) Sec- and Tat-mediated protein secretion across the bacterial cytoplasmic membrane–distinct translocases and mechanisms. Biochim. Biophys. Acta 1778, 1735–1756 [DOI] [PubMed] [Google Scholar]
- 2. Hou B., Brüser T. (2011) The Tat-dependent protein translocation pathway. Biomol. Concepts 2, 507–523 [DOI] [PubMed] [Google Scholar]
- 3. Tarry M. J., Schäfer E., Chen S., Buchanan G., Greene N. P., Lea S. M., Palmer T., Saibil H. R., Berks B. C. (2009) Structural analysis of substrate binding by the TatBC component of the twin-arginine protein transport system. Proc. Natl. Acad. Sci. U.S.A. 106, 13284–13289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Holzapfel E., Eisner G., Alami M., Barrett C. M., Buchanan G., Lüke I., Betton J. M., Robinson C., Palmer T., Moser M., Müller M. (2007) The entire N-terminal half of TatC is involved in twin-arginine precursor binding. Biochemistry 46, 2892–2898 [DOI] [PubMed] [Google Scholar]
- 5. Gérard F., Cline K. (2006) Efficient twin arginine translocation (Tat) pathway transport of a precursor protein covalently anchored to its initial cpTatC binding site. J. Biol. Chem. 281, 6130–6135 [DOI] [PubMed] [Google Scholar]
- 6. Richter S., Brüser T. (2005) Targeting of unfolded PhoA to the TAT translocon of Escherichia coli. J. Biol. Chem. 280, 42723–42730 [DOI] [PubMed] [Google Scholar]
- 7. Alami M., Lüke I., Deitermann S., Eisner G., Koch H. G., Brunner J., Müller M. (2003) Differential interactions between a twin-arginine signal peptide and its translocase in Escherichia coli. Mol. Cell 12, 937–946 [DOI] [PubMed] [Google Scholar]
- 8. Mori H., Cline K. (2002) A twin arginine signal peptide and the pH gradient trigger reversible assembly of the thylakoid ΔpH/Tat translocase. J. Cell Biol. 157, 205–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Panahandeh S., Maurer C., Moser M., DeLisa M. P., Müller M. (2008) Following the path of a twin-arginine precursor along the TatABC translocase of Escherichia coli. J. Biol. Chem. 283, 33267–33275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fröbel J., Rose P., Müller M. (2011) Early contacts between substrate proteins and TatA translocase component in twin-arginine translocation. J. Biol. Chem. 286, 43679–43689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cline K., McCaffery M. (2007) Evidence for a dynamic and transient pathway through the TAT protein transport machinery. EMBO J. 26, 3039–3049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dabney-Smith C., Mori H., Cline K. (2006) Oligomers of Tha4 organize at the thylakoid Tat translocase during protein transport. J. Biol. Chem. 281, 5476–5483 [DOI] [PubMed] [Google Scholar]
- 13. Dabney-Smith C., Mori H., Cline K. (2003) Requirement of a Tha4-conserved transmembrane glutamate in thylakoid Tat translocase assembly revealed by biochemical complementation. J. Biol. Chem. 278, 43027–43033 [DOI] [PubMed] [Google Scholar]
- 14. Gohlke U., Pullan L., McDevitt C. A., Porcelli I., de Leeuw E., Palmer T., Saibil H. R., Berks B. C. (2005) The TatA component of the twin-arginine protein transport system forms channel complexes of variable diameter. Proc. Natl. Acad. Sci. U.S.A. 102, 10482–10486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Baglieri J., Beck D., Vasisht N., Smith C. J., Robinson C. (2011) Structure of TatA paralog, TatE, suggests a structurally homogeneous form of Tat protein translocase that transports folded proteins of differing diameter. J. Biol. Chem. 287, 7335–7344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dabney-Smith C., Cline K. (2009) Clustering of C-terminal stromal domains of Tha4 homo-oligomers during translocation by the Tat protein transport system. Mol. Biol. Cell 20, 2060–2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hu Y., Zhao E., Li H., Xia B., Jin C. (2010) Solution NMR structure of the TatA component of the twin-arginine protein transport system from gram-positive bacterium Bacillus subtilis. J. Am. Chem. Soc. 132, 15942–15944 [DOI] [PubMed] [Google Scholar]
- 18. DeLisa M. P., Lee P., Palmer T., Georgiou G. (2004) Phage shock protein PspA of Escherichia coli relieves saturation of protein export via the Tat pathway. J. Bacteriol. 186, 366–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Casadaban M. J. (1976) Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J. Mol. Biol. 104, 541–555 [DOI] [PubMed] [Google Scholar]
- 20. Wexler M., Sargent F., Jack R. L., Stanley N. R., Bogsch E. G., Robinson C., Berks B. C., Palmer T. (2000) TatD is a cytoplasmic protein with DNase activity. No requirement for TatD family proteins in sec-independent protein export. J. Biol. Chem. 275, 16717–16722 [DOI] [PubMed] [Google Scholar]
- 21. Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K. A., Tomita M., Wanner B. L., Mori H. (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2, 2006.0008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bergler H., Abraham D., Aschauer H., Turnowsky F. (1994) Inhibition of lipid biosynthesis induces the expression of the pspA gene. Microbiology 140, 1937–1944 [DOI] [PubMed] [Google Scholar]
- 23. Overmann J., Fischer U., Pfennig N. (1992) A new purple sulfur bacterium from saline littoral sediments, Thiorhodovibrio winogradskyi gen. nov. and sp. nov. Arch. Microbiol. 157, 329–335 [Google Scholar]
- 24. Wilms B., Hauck A., Reuss M., Syldatk C., Mattes R., Siemann M., Altenbuchner J. (2001) High-cell-density fermentation for production of l-N-carbamoylase using an expression system based on the Escherichia coli rhaBAD promoter. Biotechnol. Bioeng 73, 95–103 [DOI] [PubMed] [Google Scholar]
- 25. Berthelmann F., Mehner D., Richter S., Lindenstrauβ U., Lünsdorf H., Hause G., Brüser T. (2008) Recombinant expression of tatABC and tatAC results in the formation of interacting cytoplasmic TatA tubes in Escherichia coli. J. Biol. Chem. 283, 25281–25289 [DOI] [PubMed] [Google Scholar]
- 26. Graubner W., Schierhorn A., Brüser T. (2007) DnaK plays a pivotal role in Tat targeting of CueO and functions beside SlyD as a general Tat signal binding chaperone. J. Biol. Chem. 282, 7116–7124 [DOI] [PubMed] [Google Scholar]
- 27. Datsenko K. A., Wanner B. L. (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 29. Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. (1951) Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193, 265–275 [PubMed] [Google Scholar]
- 30. Brüser T., Yano T., Brune D. C., Daldal F. (2003) Membrane targeting of a folded and cofactor-containing protein. Eur. J. Biochem. 270, 1211–1221 [DOI] [PubMed] [Google Scholar]
- 31. Standar K., Mehner D., Osadnik H., Berthelmann F., Hause G., Lünsdorf H., Brüser T. (2008) PspA can form large scaffolds in Escherichia coli. FEBS Lett. 582, 3585–3589 [DOI] [PubMed] [Google Scholar]
- 32. Silvestro A., Pommier J., Pascal M. C., Giordano G. (1989) The inducible trimethylamine N-oxide reductase of Escherichia coli K12: its localization and inducers. Biochim. Biophys. Acta 999, 208–216 [DOI] [PubMed] [Google Scholar]
- 33. Miller J. H. (1992) A Short Course in Bacterial Genetics, Cold Spring Harbor Laboratory Press, New York [Google Scholar]
- 34. Walther T. H., Grage S. L., Roth N., Ulrich A. S. (2010) Membrane alignment of the pore-forming component TatA(d) of the twin-arginine translocase from Bacillus subtilis resolved by solid-state NMR spectroscopy. J. Am. Chem. Soc. 132, 15945–15956 [DOI] [PubMed] [Google Scholar]
- 35. Leake M. C., Greene N. P., Godun R. M., Granjon T., Buchanan G., Chen S., Berry R. M., Palmer T., Berks B. C. (2008) Variable stoichiometry of the TatA component of the twin-arginine protein transport system observed by in vivo single-molecule imaging. Proc. Natl. Acad. Sci. U.S.A. 105, 15376–15381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Oates J., Barrett C. M., Barnett J. P., Byrne K. G., Bolhuis A., Robinson C. (2005) The Escherichia coli twin-arginine translocation apparatus incorporates a distinct form of TatABC complex, spectrum of modular TatA complexes and minor TatAB complex. J. Mol. Biol. 346, 295–305 [DOI] [PubMed] [Google Scholar]
- 37. Koch S., Fritsch M. J., Buchanan G., Palmer T. (2012) Escherichia coli TatA and TatB proteins have N-out, C-in topology in intact cells. J. Biol. Chem. 287, 14420–14431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Warren G., Oates J., Robinson C., Dixon A. M. (2009) Contributions of the transmembrane domain and a key acidic motif to assembly and function of the TatA complex. J. Mol. Biol. 388, 122–132 [DOI] [PubMed] [Google Scholar]
- 39. Joly N., Engl C., Jovanovic G., Huvet M., Toni T., Sheng X., Stumpf M. P., Buck M. (2010) Managing membrane stress: the phage shock protein (Psp) response, from molecular mechanisms to physiology. FEMS Microbiol. Rev. 34, 797–827 [DOI] [PubMed] [Google Scholar]
- 40. Darwin A. J. (2005) The phage-shock-protein response. Mol. Microbiol. 57, 621–628 [DOI] [PubMed] [Google Scholar]
- 41. Kobayashi R., Suzuki T., Yoshida M. (2007) Escherichia coli phage-shock protein A (PspA) binds to membrane phospholipids and repairs proton leakage of the damaged membranes. Mol. Microbiol. 66, 100–109 [DOI] [PubMed] [Google Scholar]
- 42. Hankamer B. D., Elderkin S. L., Buck M., Nield J. (2004) Organization of the AAA(+) adaptor protein PspA is an oligomeric ring. J. Biol. Chem. 279, 8862–8866 [DOI] [PubMed] [Google Scholar]
- 43. Carter C. W., Jr., Kraut J., Freer S. T., Nguyen-Huu X., Alden R. A., Bartsch R. G. (1974) Two-Angstrom crystal structure of oxidized Chromatium high potential iron protein. J. Biol. Chem. 249, 4212–4225 [PubMed] [Google Scholar]
- 44. Adams H., Teertstra W., Demmers J., Boesten R., Tommassen J. (2003) Interactions between phage-shock proteins in Escherichia coli. J. Bacteriol. 185, 1174–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Elderkin S., Bordes P., Jones S., Rappas M., Buck M. (2005) Molecular determinants for PspA-mediated repression of the AAA transcriptional activator PspF. J. Bacteriol. 187, 3238–3248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jovanovic G., Engl C., Mayhew A. J., Burrows P. C., Buck M. (2010) Properties of the phage-shock-protein (Psp) regulatory complex that govern signal transduction and induction of the Psp response in Escherichia coli. Microbiology 156, 2920–2932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yamaguchi S., Gueguen E., Horstman N. K., Darwin A. J. (2010) Membrane association of PspA depends on activation of the phage-shock-protein response in Yersinia enterocolitica. Mol. Microbiol. 78, 429–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gueguen E., Savitzky D. C., Darwin A. J. (2009) Analysis of the Yersinia enterocolitica PspBC proteins defines functional domains, essential amino acids and new roles within the phage-shock-protein response. Mol. Microbiol. 74, 619–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Warnock D. G., Reenstra W. W., Yee V. J. (1982) Na+/H+ antiporter of brush border vesicles: studies with acridine orange uptake. Am. J. Physiol. 242, F733–F739 [DOI] [PubMed] [Google Scholar]
- 50. Yamaguchi S., Darwin A. J. (2012) Recent findings about the Yersinia enterocolitica phage shock protein response. J. Microbiol. 50, 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Alder N. N., Theg S. M. (2003) Energetics of protein transport across biological membranes. a study of the thylakoid ΔpH-dependent/cpTat pathway. Cell 112, 231–242 [DOI] [PubMed] [Google Scholar]
- 52. Vrancken K., De Keersmaeker S., Geukens N., Lammertyn E., Anné J., Van Mellaert L. (2007) pspA overexpression in Streptomyces lividans improves both Sec- and Tat-dependent protein secretion. Appl. Microbiol. Biotechnol. 73, 1150–1157 [DOI] [PubMed] [Google Scholar]
- 53. Vrancken K., Van Mellaert L., Anné J. (2008) Characterization of the Streptomyces lividans PspA response. J. Bacteriol. 190, 3475–3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kleerebezem M., Crielaard W., Tommassen J. (1996) Involvement of stress protein PspA (phage shock protein A) of Escherichia coli in maintenance of the protonmotive force under stress conditions. EMBO J. 15, 162–171 [PMC free article] [PubMed] [Google Scholar]
- 55. Kleerebezem M., Tommassen J. (1993) Expression of the pspA gene stimulates efficient protein export in Escherichia coli. Mol. Microbiol. 7, 947–956 [DOI] [PubMed] [Google Scholar]
- 56. Lo S. M., Theg S. M. (2012) Role of Vesicle-Inducing Protein in Plastids 1 in cpTat transport at the thylakoid. Plant J., doi: 10.1111/j.1365-313X.2012.05020.x [DOI] [PubMed] [Google Scholar]
- 57. Li H. M., Kaneko Y., Keegstra K. (1994) Molecular cloning of a chloroplastic protein associated with both the envelope and thylakoid membranes. Plant Mol. Biol. 25, 619–632 [DOI] [PubMed] [Google Scholar]
- 58. Liu C., Willmund F., Whitelegge J. P., Hawat S., Knapp B., Lodha M., Schroda M. (2005) J-domain protein CDJ2 and HSP70B are a plastidic chaperone pair that interacts with vesicle-inducing protein in plastids 1. Mol. Biol. Cell 16, 1165–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kroll D., Meierhoff K., Bechtold N., Kinoshita M., Westphal S., Vothknecht U. C., Soll J., Westhoff P. (2001) VIPP1, a nuclear gene of Arabidopsis thaliana essential for thylakoid membrane formation. Proc. Natl. Acad. Sci. U.S.A. 98, 4238–4242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Jouhet J., Gray J. C. (2009) Is chloroplast import of photosynthesis proteins facilitated by an actin-TOC-TIC-VIPP1 complex? Plant Signal Behav. 4, 986–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Westphal S., Heins L., Soll J., Vothknecht U. C. (2001) Vipp1 deletion mutant of Synechocystis: a connection between bacterial phage shock and thylakoid biogenesis? Proc. Natl. Acad. Sci. U.S.A. 98, 4243–4248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Huang F., Fulda S., Hagemann M., Norling B. (2006) Proteomic screening of salt-stress-induced changes in plasma membranes of Synechocystis sp. strain PCC 6803. Proteomics 6, 910–920 [DOI] [PubMed] [Google Scholar]
- 63. Göhre V., Ossenbühl F., Crèvecoeur M., Eichacker L. A., Rochaix J. D. (2006) One of two alb3 proteins is essential for the assembly of the photosystems and for cell survival in Chlamydomonas. Plant Cell 18, 1454–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Aseeva E., Ossenbühl F., Eichacker L. A., Wanner G., Soll J., Vothknecht U. C. (2004) Complex formation of Vipp1 depends on its α-helical PspA-like domain. J. Biol. Chem. 279, 35535–35541 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.