Abstract
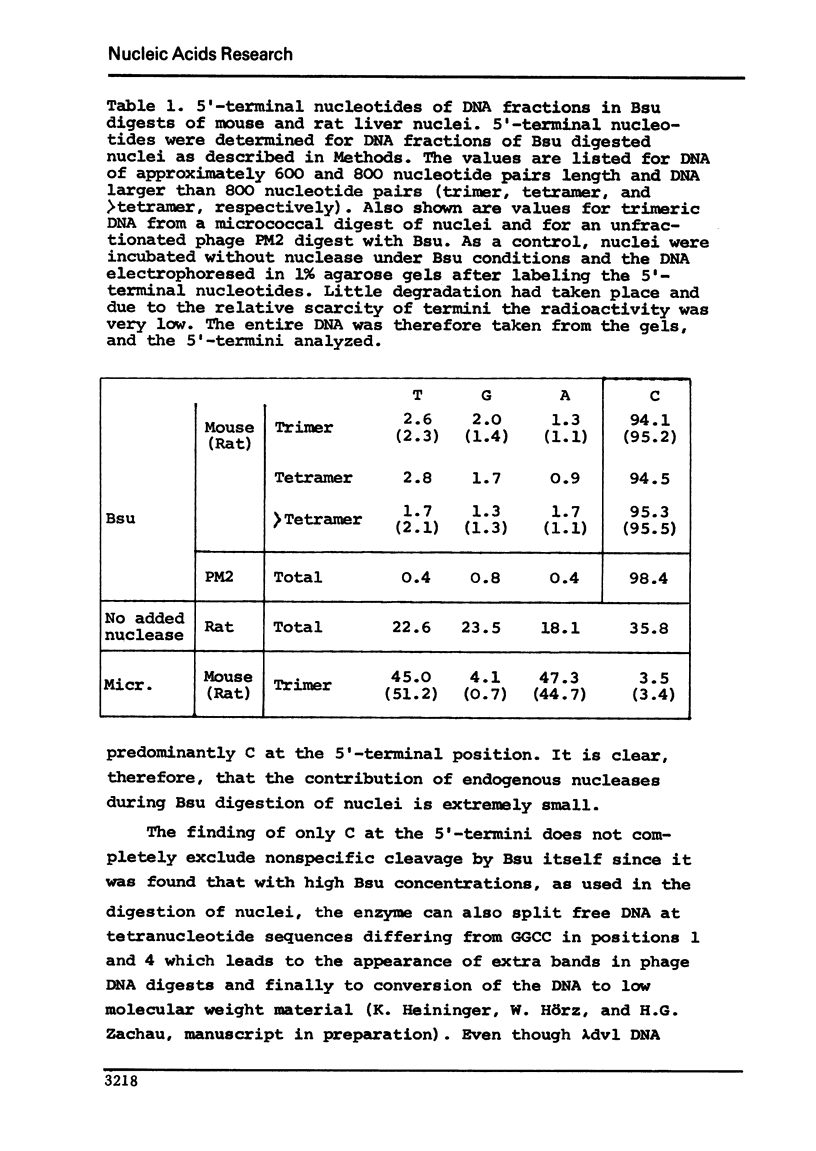
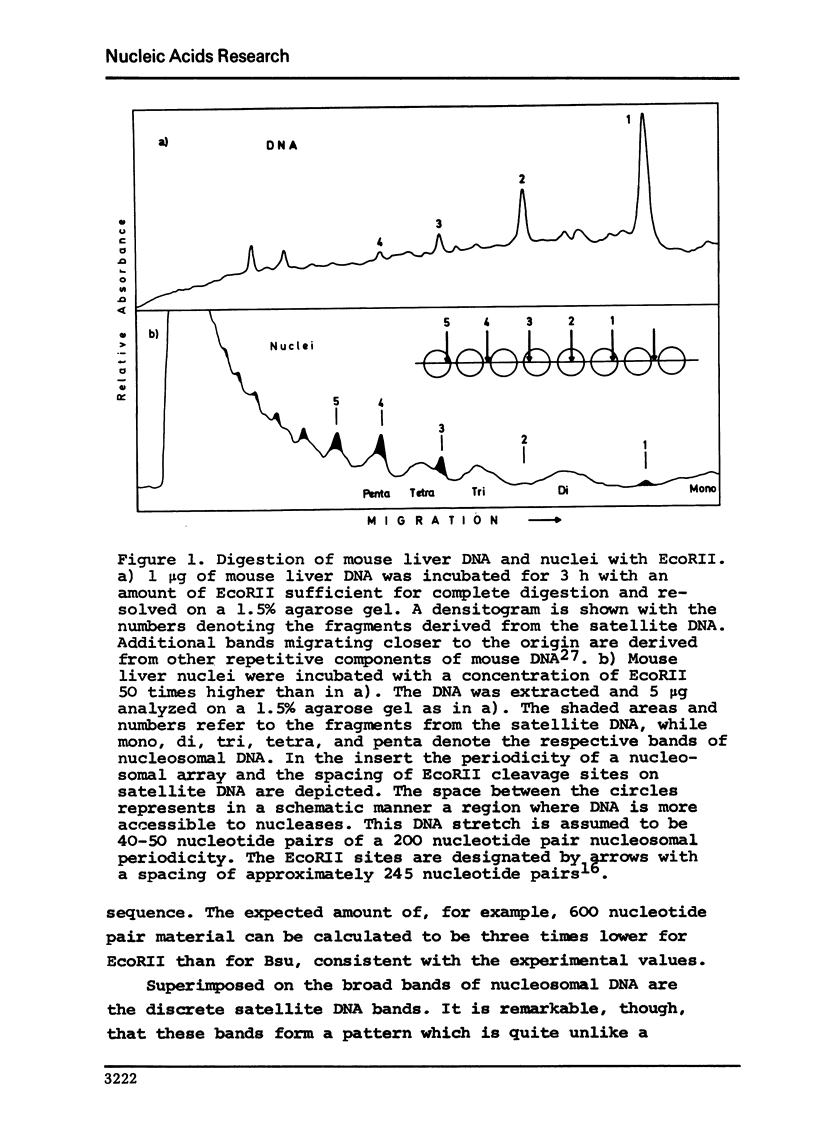
Digestion of mouse and rat liver nuclei with a restriction nuclease from Bacillus subtilis (Bsu) is examined in continuation of previous work from this laboratory (Pfeiffer et al., 1975, Nature 258, 450). The finding of more than 95% C in the 5'-termini of the DNA fragments generated during digestion with Bsu shows that the participation of endogenous nucleases in Bsu digestion is extremely small. The restriction nuclease Hae III, an isoschizomer of Bsu, yields identical degradation patterns. The patterns conform to what one expects from statistical calculations based on a nucleosome structure of chromatin with a region preferentially accessible to the nuclease of 40-50 nucleotide pairs per nucleosome. Integrity of the histones is maintained during digestion with restriction nucleases. Digestion of mouse liver nuclei with EcoRII shows that most if not all of the satellite DNA is organized in a nucleosome structure. Also in rat liver, much of the repetitive DNA appears to be present in nucleosomes.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bokhon'ko A., Reeder R. H. The subunit structure of mouse satellite chromatin. Biochem Biophys Res Commun. 1976 May 3;70(1):146–152. doi: 10.1016/0006-291x(76)91120-7. [DOI] [PubMed] [Google Scholar]
- Bron S., Murray K. Restriction and modification in B. subtilis. Nucleotide sequence recognised by restriction endonuclease R. Bsu R from strain R. Mol Gen Genet. 1975 Dec 30;143(1):25–33. doi: 10.1007/BF00269417. [DOI] [PubMed] [Google Scholar]
- Bron S., Murray K., Trautner T. A. Restriction and modification in B. subtilis. Purification and general properties of a restriction endonuclease from strain R. Mol Gen Genet. 1975 Dec 30;143(1):13–23. doi: 10.1007/BF00269416. [DOI] [PubMed] [Google Scholar]
- Elgin S. C., Weintraub H. Chromosomal proteins and chromatin structure. Annu Rev Biochem. 1975;44:725–774. doi: 10.1146/annurev.bi.44.070175.003453. [DOI] [PubMed] [Google Scholar]
- Fairbanks G., Steck T. L., Wallach D. F. Electrophoretic analysis of the major polypeptides of the human erythrocyte membrane. Biochemistry. 1971 Jun 22;10(13):2606–2617. doi: 10.1021/bi00789a030. [DOI] [PubMed] [Google Scholar]
- Frischauf A. M., Eckstein F. Purification of a phosphodiesterase from Bothrops atrox venom by affinity chromatography. Eur J Biochem. 1973 Feb 1;32(3):479–485. doi: 10.1111/j.1432-1033.1973.tb02631.x. [DOI] [PubMed] [Google Scholar]
- Hewish D. R., Burgoyne L. A. Chromatin sub-structure. The digestion of chromatin DNA at regularly spaced sites by a nuclear deoxyribonuclease. Biochem Biophys Res Commun. 1973 May 15;52(2):504–510. doi: 10.1016/0006-291x(73)90740-7. [DOI] [PubMed] [Google Scholar]
- Hörz W., Hess I., Zachau H. G. Highly regular arrangement of a restriction-nuclease-sensitive site in rodent satellite DNAs. Eur J Biochem. 1974 Jun 15;45(2):501–512. doi: 10.1111/j.1432-1033.1974.tb03575.x. [DOI] [PubMed] [Google Scholar]
- Ilyin Y. V., Varshavsky A. Y., Mickelsaar U. N., Georgiev G. P. Studies on deoxyribonucleoprotein structure. Redistribution of proteins in mixtures of deoxyribonucleoproteins, DNA and RNA. Eur J Biochem. 1971 Sep 24;22(2):235–245. doi: 10.1111/j.1432-1033.1971.tb01537.x. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D. Chromatin structure: a repeating unit of histones and DNA. Science. 1974 May 24;184(4139):868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- MELCHERS F., ZACHAU H. G. SPALTUNG VON LOESLICHER RIBONUCLEINSAEURE UND SERIN-SPEZIFISCHEN TRANSFER-RIBONUCLEINSAEURE-FRAKTIONEN MIT PANKREAS-RIBONUCLEASE. Biochim Biophys Acta. 1964 Dec 16;91:559–572. [PubMed] [Google Scholar]
- Murray K. Nucleotide sequence analysis with polynucleotide kinase and nucleotide "mapping" methods. 5'-Terminal sequences of deoxyribonucleic acid from bacteriophages lambda and 424. Biochem J. 1973 Mar;131(3):569–582. doi: 10.1042/bj1310569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noll M. Internal structure of the chromatin subunit. Nucleic Acids Res. 1974 Nov;1(11):1573–1578. doi: 10.1093/nar/1.11.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noll M., Thomas J. O., Kornberg R. D. Preparation of native chromatin and damage caused by shearing. Science. 1975 Mar 28;187(4182):1203–1206. doi: 10.1126/science.187.4182.1203. [DOI] [PubMed] [Google Scholar]
- Petrova M., Philippsen P., Zachau H. G. Partial degradation of transfer RNAs by different preparations of snake venom exonuclease. Biochim Biophys Acta. 1975 Jul 23;395(4):455–467. doi: 10.1016/0005-2787(75)90069-6. [DOI] [PubMed] [Google Scholar]
- Pfeiffer W., Horz W., Igo-Kemenes T., Zachau H. G. Restriction nucleases as probes of chromatin structure. Nature. 1975 Dec 4;258(5534):450–452. doi: 10.1038/258450a0. [DOI] [PubMed] [Google Scholar]
- Philippsen P., Streeck R. E., Zachau H. G. Defined fragments of calf, human, and rat DNA produced by restriction nucleases. Eur J Biochem. 1974 Jun 15;45(2):479–488. doi: 10.1111/j.1432-1033.1974.tb03573.x. [DOI] [PubMed] [Google Scholar]
- RANDERATH K., RANDERATH E. ION-EXCHANGE CHROMATOGRAPHY OF NUCLEOTIDES ON POLY-(ETHYLENEIMINE)-CELLULOSE THIN LAYERS. J Chromatogr. 1964 Oct;16:111–125. doi: 10.1016/s0021-9673(01)82445-6. [DOI] [PubMed] [Google Scholar]
- Roberts R. J., Breitmeyer J. B., Tabachnik N. F., Myers P. A. A second specific endonuclease from Haemophilus aegyptius. J Mol Biol. 1975 Jan 5;91(1):121–123. doi: 10.1016/0022-2836(75)90375-7. [DOI] [PubMed] [Google Scholar]
- Roizés G. Analysis of eucaryotic DNAs with a restriction endonuclease from H. influenzae: isolation of "hidden" satellite DNAs. Nucleic Acids Res. 1974 Sep;1(9):1099–1120. doi: 10.1093/nar/1.9.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw B. R., Herman T. M., Kovacic R. T., Beaudreau G. S., Van Holde K. E. Analysis of subunit organization in chicken erythrocyte chromatin. Proc Natl Acad Sci U S A. 1976 Feb;73(2):505–509. doi: 10.1073/pnas.73.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. T., Whitlock J. P., Jr Chemical evidence that chromatin DNA exists as 160 base pair beads interspersed with 40 base pair bridges. Nucleic Acids Res. 1976 Jan;3(1):117–127. doi: 10.1093/nar/3.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. O., Nathans D. Letter: A suggested nomenclature for bacterial host modification and restriction systems and their enzymes. J Mol Biol. 1973 Dec 15;81(3):419–423. doi: 10.1016/0022-2836(73)90152-6. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Long range periodicities in mouse satellite DNA. J Mol Biol. 1975 May 5;94(1):51–69. doi: 10.1016/0022-2836(75)90404-0. [DOI] [PubMed] [Google Scholar]