Background: Elucidation of the biochemical and biological function of IFN-γ induced mGBP2.
Results: Mutations in the G-domain of mGBP2 inhibit GTPase activity and multimerization resulting in failure in the recruitment to T. gondii.
Conclusion: The GTPase activity is essential for physiological intracellular localization and intersection of T. gondii.
Significance: This study identifies residues relevant for GTP hydrolysis and essential for biological function of mGBP2 in host-pathogen interaction.
Keywords: GTPase, Host-Pathogen Interactions, Immunology, Interferon, Parasite, Interferon-γ-inducible Guanylate-binding Proteins, Parasitophorous Vacuole, Toxoplasma gondii
Abstract
One of the most abundantly IFN-γ-induced protein families in different cell types is the 65-kDa guanylate-binding protein family that is recruited to the parasitophorous vacuole of the intracellular parasite Toxoplasma gondii. Here, we elucidate the relationship between biochemistry and cellular host defense functions of mGBP2 in response to Toxoplasma gondii. The wild type protein exhibits low affinities to guanine nucleotides, self-assembles upon GTP binding, forming tetramers in the activated state, and stimulates the GTPase activity in a cooperative manner. The products of the two consecutive hydrolysis reactions are both GDP and GMP. The biochemical characterization of point mutants in the GTP-binding motifs of mGBP2 revealed amino acid residues that decrease the GTPase activity by orders of magnitude and strongly impair nucleotide binding and multimerization ability. Live cell imaging employing multiparameter fluorescence image spectroscopy (MFIS) using a Homo-FRET assay shows that the inducible multimerization of mGBP2 is dependent on a functional GTPase domain. The consistent results indicate that GTP binding, self-assembly, and stimulated hydrolysis activity are required for physiological localization of the protein in infected and uninfected cells. Ultimately, we show that the GTPase domain regulates efficient recruitment to T. gondii in response to IFN-γ.
Introduction
IFN-γ is an immunomodulatory cytokine with profound and pleiotropic effects on nearly all nucleated cells (1). It confers vertebrate immunity against a diverse spectrum of microbial pathogens due to the initiation of complex transcriptional programs (2).
Two families of GTPases, the 47- and 65-kDa guanylate-binding proteins (GBPs),3 account for a large percentage of the mRNA transcripts induced by IFN-γ, and the GBPs represent classical secondary response genes (3). Recently, their implication in mammalian host defense and potential for pathogen specificity have been demonstrated (4–7). The 47-kDa guanylate-binding proteins, also called immunity-related GTPases (IRGs), have been identified in several mammalian species; nevertheless, the gene family appears to be degenerated in humans (8). In contrast, the GBPs are highly conserved throughout the vertebrate lineage with seven human and 11 murine isoforms in the database (9, 10).
Biochemical and structural data revealed that the GBPs, the IRGs, the IFN-α-induced Mx proteins, and very large inducible GTPases belong to the superfamily of dynamin-related GTP-binding proteins (11). The GBPs share unique biochemical properties such as low affinities for guanine nucleotides, in contrast to the small GTPases such as Ras, Rab, Rho, and α-subunits of heterotrimeric G proteins (12, 13). They are stable without guanine nucleotide, coordinate a chelated Mg2+ ion cofactor, and undergo nucleotide-dependent multimerization and a cooperative mechanism of GTP hydrolysis (14, 15). The high turnover rates are achieved without accessory GTPase-activating proteins, and the products of the hydrolysis reaction are GDP and GMP. This implies a different mechanism of action for the GBPs versus the family of small GTPases (14, 16–21).
The GTP-binding domain (the G-domain) has a universal structure and a universal switch mechanism. The N-terminal 278 residues of human GBP1 (hGBP1) constitute a modified G-domain with a number of extra secondary structural elements compared with the canonical Ras structure (14, 17). It consists of a mixed six-stranded β-sheet and five α-helices that are typical for nucleotide-binding proteins (18). The large G-domain of GBPs contains four conserved sequence elements G1–G4, which are lined up along the nucleotide-binding site. They exhibit the canonical G1 motif or phosphate-binding loop (P-loop) with the GXXXXGK(Ser/Thr) sequence, G2 (Thr), and G3 (DXXG) GTP-binding sites, but they possess the Arg residues in the G4 region instead of the (Asn/Thr)KXD motif found in other GTPases, such as Ras and IRGs (supplemental Fig. 1) (16, 22, 23). The side chain of the aspartic acid in the RD motif is responsible for the guanine nucleotide recognition (16). The conserved residues of the P-loop contribute to binding through the interactions with the α-, β-, and γ-phosphates of the nucleotide and Mg2+ coordination, and the switch regions surrounding the G2 and G3 motifs are involved in conformational rearrangement of the G-domain during hydrolysis (15, 24, 25).
Important roles during pathogen infections were described for the Mx proteins and the IRGs (26, 27). Mx proteins have been shown to possess a potent antiviral activity against a wide range of RNA viruses, including bunya- and orthomyxoviruses, directly interacting with viral particles, thereby inhibiting viral assembly or replication (26). The IFN-γ-induced IRGs are recruited to pathogen-containing phagosomes (28–30) where they confer selective protection against Salmonella typhimurium, Leishmania major, and Mycobacterium. avium. Elimination of several IRGs in mice by gene targeting severely cripples IFN-γ-regulated defense against T. gondii, Listeria monocytogenes, Mycobacterium spp., and other pathogens (27, 29, 31–33).
Nevertheless, little is known about the biological function of GBPs. We have recently demonstrated a strong induction of mGBPs upon IFN-γ stimulation in vitro and in vivo after infection of mice with the Gram-positive bacterium L. monocytogenes or the parasitic protozoan T. gondii and the recruitment of several mGBPs, in particular mGBP2, to the PV of T. gondii (9, 34). Moreover, we generated mGBP2−/− mice that are significantly more susceptible to T. gondii infection than their WT littermates (data not shown). Furthermore, loss-of-function analyses revealed the role for mGBP1, mGBP7, and mGBP10 in cell autonomous immunity to mycobacterial infection in vitro and in vivo (7); therefore, mGBP7 was suggested to be required for IFN-γ-induced oxidant protection against intracellular bacteria by delivering the subunits of the NADPH oxidase to phagosomal membranes and mGBP1 to be involved in the autolysosomal pathway. mGBP5 has been shown to positively regulate Salmonella-induced pyroptosis of RAW 264.7 macrophages through a caspase-1-dependent mechanism as well as inflammasome assembly (35, 36), and hGBP1 was proposed to potentiate the anti-chlamydial effects of IFN-γ (6). Additionally, moderate antiviral activities against vesicular stomatitis virus and encephalomyocarditis virus were detected for mGBP2 and hGBP1 (4, 37). hGBP1 showed antiviral activity against hepatitis C virus replication dependent on its GTPase activity (38). Furthermore, hGBP1 has been reported to contribute to paclitaxel resistance in ovarian cancer cell lines and has been suggested to play a role in the development of multidrug resistance (39). Taken together, the mGBPs play an important role as effector molecules in host defense against intracellular pathogens. Furthermore, GBPs were implicated in the regulation of cell adhesion and proliferation (40, 41).
In this study, we present a systematic characterization of the GTPase activity of mGBP2 to investigate the mechanism of cooperative catalysis and to analyze the cellular localization of the protein. Introduction of point mutations to the key position of the conserved motifs in the GTP-binding domain (15) revealed mGBP2 variants that are deficient in nucleotide binding, multimerization, and/or GTP hydrolysis, respectively. Furthermore, our results indicate that the intracellular localization in vesicle-like structures and the recruitment of mGBP2 to the PV membrane of T. gondii in IFN-γ-stimulated murine embryonic fibroblasts (MEFs) depend on multimerization and the GTPase activity.
EXPERIMENTAL PROCEDURES
Expression Constructs
The WT ORF of mGBP2 (NCBI accession number for mGBP-2, NM_010260.1) was subjected to site-directed mutagenesis (QuikChange II Mutagenesis kit, Stratagene) for the generation of GTPase mutants R48A, K51A, E99A, and D182N in the pEGFP-C2 plasmid (Clontech). The respective genes were then cloned into the pWPXL plasmid (Trono Lab) as N-terminal GFP fusion constructs. The lentiviral envelope vector pLP/VSVG (Invitrogen) and the packaging vector psPAX2 (Trono Lab) were used for the lentiviral genetic transfer. For the recombinant expression in Escherichia coli, the sequences were cloned into the pQE80L plasmid (Qiagen). The proteins were expressed as His6 fusion constructs. All constructs were verified by sequencing (GATC Biotech).
Cell Culture and Transduction
MEFs were cultured in Dulbecco's modified Eagle's medium (DMEM, Invitrogen) supplemented with 10% (v/v) heat-inactivated low endotoxin fetal bovine serum (FBS, Cambrex), 100 units/ml penicillin, 100 μg/ml streptomycin, 2 mm l-glutamine (Biochrom), and 0.05 mm β-mercaptoethanol (Invitrogen). Human foreskin fibroblasts (HS27, ATCC CRL-1634) were held in culture in Iscove's modified Dulbecco's medium (Invitrogen) with the same supplementations. 293FT cells were cultivated in DMEM supplemented with 10% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin. All recombinant lentiviruses were produced by transient transfection of 293FT cells according to standard protocols (42). Briefly, subconfluent 293FT cells were co-transfected with 20 μg of a plasmid vector, 10 μg of psPAX2, and 5 μg of pLP/VSVG by calcium chloride precipitation in FBS-free medium. After 6 h, the medium was changed (10% FBS), and supernatants with recombinant lentivirus vectors were harvested 48 h later. MEFs were seeded in 24-well plates (Corning Inc.) and transduced with 600 μl of lentivirus with 25 μg of Polybrene (Millipore). After 4 h of incubation, the medium was changed. The transduction efficacy was analyzed by flow cytometry. Subsequently, GFP-positive cells were sorted and cultivated.
Tachyzoites from T. gondii strain ME49 were maintained by serial passage in confluent monolayers of HS27 cells. After infection of fibroblasts, parasites were harvested and passaged as described previously (34).
Infection of Murine MEFs with T. gondii
Cells were stimulated with 200 units/ml IFN-γ (R&D Systems) 16 h before infection. For immunofluorescence, MEFs were cultured in 24-well plates (Falcon, BD Biosciences) on coverslips (inner diameter, 13 mm, VWR International) and inoculated with freshly harvested T. gondii at a ratio of 50:1. To remove extracellular parasites, cells were washed with PBS.
Immunofluorescence Analysis
Cells were fixed in 4% paraformaldehyde (Sigma) permeabilized with 0.02% saponin (Calbiochem), blocked in 0.002% saponin with 2% goat serum (DakoCytomation), and stained as described previously (34). For staining of endogenous mGBP2, anti-mGBP2 affinity-purified antiserum (Eurogentec (34)) was used at a concentration of 1:200. The outer membrane of T. gondii was visualized by anti-SAG1 (Abcam) at a concentration of 1:700. As secondary reagents, 1:200 concentrated Cy2-conjugated goat anti-rabbit IgG and Cy3-conjugated goat anti-mouse IgG plus IgM (Jackson ImmunoResearch) were used. Nuclei were counterstained with 1:2500 4′,6-diamidino-2-phenylindole (DAPI, Invitrogen). The coverslips were fixed in fluorescence mounting medium (Fluoromount-G, Southern Biotechnology Associates). Fluorescence was visualized using an LSM510 meta confocal microscope (Zeiss). To avoid cross-talk in the detection of the used fluorophores, a multitracking scanning mode was used. Image analysis and processing were performed with the LSM software (Zeiss).
Protein Expression and Purification
The expression of WT and mutant mGBP2 with an N-terminal His6 tag was induced from pQE80L vector (Qiagen) by adding 100 μm isopropyl β-d-thiogalactoside (MBI Fermentas) in E. coli strain Rosetta 2(DE3)pLysS (Novagen) culture at 37 °C and an A600 of 0.6. 4 h after induction, cells were harvested by centrifugation at 4000 × g, resuspended in the high salt buffer (50 mm Tris-HCl (pH 8.0), 5 mm MgCl2, 300 mm NaCl, 10 mm imidazole, 1 mm β-mercaptoethanol) and 250 μm Pefabloc (Roth), and disrupted in the Constant Cell Disruption System (Constant Cell) at a pressure of 2.7 kbars. After centrifugation for 1 h at 205,000 × g, the supernatant was applied to a Ni-NTA-Superflow column (Qiagen) with a bed volume of 25 ml connected to an ÄKTAprime System (GE Healthcare). After thorough washing with low salt buffer (50 mm Tris-HCl (pH 8.0), 5 mm MgCl2, 150 mm NaCl, 10 mm imidazole, and 1 mm β-mercaptoethanol), mGBP2 and mutant proteins were eluted with a linear gradient of high imidazole buffer (50 mm Tris-HCl (pH 8.0), 5 mm MgCl2, 150 mm NaCl, 300 mm imidazole, and 1 mm β-mercaptoethanol). Fraction purity was verified by SDS-PAGE. Fractions containing mGBP2 were pooled and concentrated using centrifugal ultrafiltration devices with a 50-kDa MWCO (Millipore). 6 ml of concentrated protein was applied to a Superdex200 26/60 column (GE Healthcare) equilibrated in gel filtration buffer (50 mm Tris-HCl (pH 8.0), 5 mm MgCl2, and 2 mm dithioerythritol (DTT, Sigma)). Fractions containing monomeric mGBP2 were pooled, and purity was analyzed by SDS-PAGE and Coomassie staining (R-250, Merck). The protein was concentrated up to 20–40 mg/ml using centrifugal ultrafiltration devices.
Fluorimetry
The 2′/3′-O-(N-methylanthraniloyl (mant-) nucleotides, mant-GMP, mant-GDP, and mant-GTPγS (Jena Bioscience), were used as fluorescent probes. The binding of nucleotide to the protein was monitored as an increase in mant-fluorescence. All measurements were done in gel filtration buffer at 25 °C. In protein titration experiments, the 0.5 μm solution of a mant-nucleotide was excited at 355 nm, and the fluorescence was monitored at 448 nm (Fluorolog, Horiba). The equilibrium dissociation constant Kd was obtained from a fit to the data as described previously (43).
GTP Hydrolysis and Cooperativity
The GTPase activity of WT and mutant mGBP2 was measured at different protein concentrations in gel filtration buffer with 1 mm GTP (Fluka) at 37 °C. Aliquots were taken at intervals, and the progress of GTP hydrolysis was analyzed by reversed-phase chromatography (C-18, Supelco, Sigma; HPLC, Merck/Hitachi) with optical detection. The HPLC running buffer contained 10 mm tetrabutylammonium bromide (Fluka), 0.2 mm sodium azide (Roth), 100 mm potassium phosphate (pH 6.5), and 2% acetonitrile (AppliChem). With this method, the hydrolysis rates as well as the product ratio (GDP/GMP) were quantitatively determined. The rates were derived from a linear fit to the initial rate of the reaction (<30% GTP hydrolyzed) and plotted against the protein concentrations used. If cooperativity occurred, a binding model describing the interaction of two molecules of mGBP2 was fitted to the data, and the dimerization Kd was obtained (15).
Analytical Size-exclusion Chromatography
States of multimerization of WT and mutant mGBP2 proteins were analyzed by size-exclusion chromatography (Superdex200 (3.2/30) GL; ÄKTAmicro, GE Healthcare). 20 μl of mGBP2 at a 30 μm concentration were preincubated for 15 min at 4 °C prior to injection in gel filtration buffer with 5 μm (GTPγS, Gpp(NH)p, and GDP) or 50 μm (GMP) of the respective nucleotide (according to the binding Kd values of the WT protein).
Aluminum fluoride solutions were prepared by the addition of 300 μm aluminum chloride (AlCl3), 10 mm sodium fluoride (NaF), and 50 μm GDP. The formation of AlF3 and AlF4− complexes is a pH-dependent process, which is denoted here as AlFx. Size calibration was carried out using standard proteins with molecular masses between 29 and 200 kDa. The void volume (V0) was measured using blue dextran. Elution was followed by monitoring the absorbance at 280 nm.
Fluorescence Anisotropy Microscopy
Multiparameter fluorescence image spectroscopy (MFIS) was performed using a multiparameter fluorescence detection setup with the corresponding fluorescence anisotropy analyses as described previously (44, 45). Experiments were performed with a confocal laser scanning microscope (Olympus FV1000, IX81 inverted microscope) additionally equipped with a single photon counting device with picosecond time resolution (PicoQuant Hydra Harp 400). GFP was excited at 485 nm with a linearly polarized, pulsed (32 MHz) diode laser (LDH-D-C-485) at 0.9 microwatts at the objective (×60 water immersion, Olympus UPlanSApo NA 1.2, diffraction limited focus). The emitted light was collected in the same objective and separated into its perpendicular and parallel polarization (PBS 101, Thorlabs). Fluorescence was then detected by single-photon avalanche diodes (PD5CTC, Micro Photon Devices, Bolzano, Italy) in a narrow range of the GFP emission spectrum (bandpass filter, HC520/35 (AHF)). Images were taken with 10-μs pixel time and a resolution of 161 nm/pixel (pixel size 318 nm). Series of 40–100 frames were merged to one image and further analyzed (46).
Anisotropy Analyses
The steady state anisotropy is given by Equation 1,
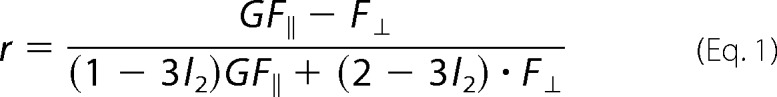 |
The fluorescence signal of GFP with parallel and perpendicular polarizations is denoted as F‖ and F⊥, respectively. The ratio of the detection efficiencies of the perpendicular and parallel polarized light is given as G = g⊥/g‖. The G-factor of the setup was determined to be 1.021 by calibration measurements using the dye rhodamine 110. The factors l1 and l2 above account for polarization mixing due to the objective lens as described in Ref. 47. These factors were determined to be l1 = 0.0308 and l2 = 0.0368, respectively, for the given setup in a separate calibration measurement according to Ref. 48. The fluorescence signal was also corrected for dead time of the detection electronics following Equation 2
 |
and Ref. 49, with sdet for the detected signal, srec for the recorded signal, and td for the dead time of the detector, respectively. The dead time of 80 ns of the setup, including detector dead time, was calculated from linear autocorrelations of detection channels.
RESULTS
Generation of mGBP2 Variants by Mutations in the G-domain
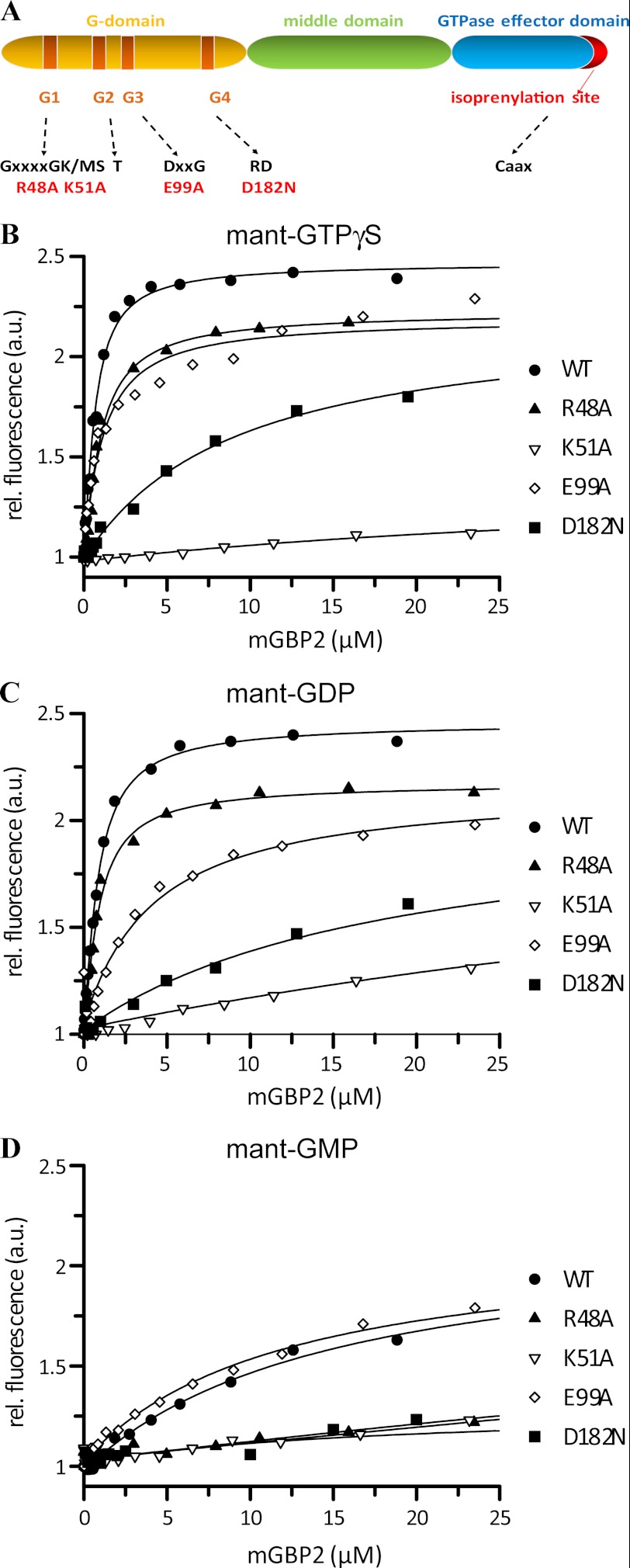
To investigate the biochemical patterns, a number of residues in the G-domain of mGBP2 (Table 1, Fig. 1A, and supplemental Fig. 1) were mutated in analogy to the experiments on hGBP1 (15). Thus, the residues Arg-48 and Lys-51 in the P-loop, Glu-99 in the switch II region, and Asp-182 in the G4 motif were chosen as target residues for mutagenesis with respect to their high level of conservation within the GBP family.
TABLE 1.
Mutated amino acids in conserved motifs of mGBP2
The residues Arg-48 and Lys-51 in the P-loop as well as residue Glu-99 in the switch II region were mutated to alanine, and Asp-182 in the G4 motif was mutated to asparagine.
| P-loop, GXXXXGK(S/T) | Switch II (DXXG) | G4 motif (N/T)KXD | |
|---|---|---|---|
| mGBP2 | GLYR48TGK51S | DTE99GLGDVEKGDNQND | TLRD182 |
FIGURE 1.
Nucleotide binding. A linear map of mGBP2 with a schematic depiction of protein domains, nucleotide-binding motifs G1–G4, and the respective consensus amino acid sequence is shown. The mutated amino acids are indicated (A). A solution containing 0.5 μm mant-GTPγS (B), mant-GDP (C), and mant-GMP (D) was titrated with WT (●), R48A (▴), K51A (▿), E99A (♢) and D182N (■) mGBP2. The fluorescence was excited at 355 nm and measured at 448 nm. The values were normalized to the fluorescence of the nucleotide alone. Dissociation constants are calculated from the fit of the binding curves. The results averaging over two to four experiments each are given in Table 2.
Nucleotide Binding of mGBP2
Differential properties of mGBP2 may be induced by different guanine nucleotides. Therefore, the nucleotide binding affinities of recombinant WT and mutant proteins were determined using fluorescence spectroscopy with guanine nucleotides labeled with the fluorescent mant group.
Fig. 1 shows protein titration curves for mant-GTPγS (Fig. 1B), a nonhydrolyzable analog of GTP, mant-GDP (Fig. 1C), and mant-GMP (Fig. 1D). Binding of mant-nucleotides to the WT protein resulted in a dose-dependent increase of fluorescence. A one-site model was applied to the fluorescence data (16, 43). From the calculated curves, the equilibrium dissociation constants were estimated. WT mGBP2 showed comparable affinities to mant-GTPγS and mant-GDP with Kd values of 0.45 and 0.54 μm, respectively, whereas the affinity to mant-GMP was 30-fold weaker (Kd = 14.4 μm) (Table 2).
TABLE 2.
Dissociation constants (Kd) of mant-nucleotides determined by fluorescence titrations at 25 °C and GTPase activity parameters obtained by concentration-dependent hydrolysis at 37 °C for the WT and mutant mGBP2 proteins
n. coop. means not cooperative; ND means not detected.
| Nucleotide binding |
GTP hydrolysis |
|||||
|---|---|---|---|---|---|---|
| mant-GTPγS, Kd | mant-GDP, Kd | mant-GMP, Kd | Maximum hydrolysis | Dimer Kd | GMP | |
| μm | μm | μm | mina | μm | % | |
| WT | 0.45 | 0.54 | 14.4 | 102 | 0.03 | 74 |
| R48A | 0.74 | 0.63 | 19.8 | 0.26 | 0.35 | 35 |
| K51A | 44.1 | 58.9 | 107 | ND | n. coop. | 0 |
| E99A | 0.97 | 3.87 | 13.4 | 3.50 | 0.18 | 7 |
| D182N | 8.77 | 18.0 | > 100 | 3.50 | 3.01 | 26 |
a The % GMP indicates the relative amount of the two products, GDP and GMP.
The P-loop mGBP2 mutant R48A was comparable with WT affinities for mant-GTPγS (Kd = 0.74 μm) and mant-GDP (Kd = 0.63 μm), as well as for mant-GMP (Kd = 19.8 μm). Nevertheless, the increase in mant-fluorescence at saturating concentrations was decreased indicating an ineffective shielding of parts of the nucleotide from the solvent. The other P-loop mutant K51A exhibited strongly decreased binding to all three types of mant-nucleotides. It showed a 97–110-fold weaker affinity for mant-GTPγS (Kd = 44.1 μm) and mant-GDP (Kd = 58.9 μm) compared with the WT. The interaction with mant-GMP was 7-fold decreased, and the mutant showed the weakest affinity to this nucleotide (Kd = 107 μm). The tightest binding was observed for mant-GTPγS. Taken together, the K51A mutation of mGBP2 appeared to cause impairment in nucleotide binding. The switch II region is involved in nucleotide stabilization (18). The mutation E99A in the DXXG motif showed a 7-fold decreased affinity for mant-GDP (Kd = 3.87 μm) and 2-fold for mant-GTPγS (Kd = 0.97 μm) but no significant change in mant-GMP binding (Kd = 13.4 μm), indicating that this residue has a weak effect on nucleotide binding. Finally, the mutation in the G4 motif of mGBP2, D182N, resulted in a protein with a 19-fold weaker interaction with mant-GTPγS (Kd = 5.0 μm) and 33-fold decreased binding to mant-GDP (Kd = 8.2 μm). Interestingly, the affinity to mant-GMP was very weak (Kd > 100 μm). Similar to the K51A, the D182N mutant showed the highest affinity to mant-GTPγS. The observations that the affinities of this mutant to mant-GTPγS, mant-GDP (Table 2), and for another GTP analog mant-Gpp(NH)p (supplemental Fig. 2, supplemental Table 1) were in the same order of magnitude, which suggests a diminished specificity for the guanine base. Additional analysis of binding affinities of the WT protein and remaining GTPase mutants for mant-Gpp(NH)p (supplemental Fig. 2 and supplemental Table 1) yielded overall increased Kd values for this nucleotide but with similar ratios to the WT protein as compared with the affinities for mant-GTPγS, with the R48A mutant representing the only exception (Kd = 0.29 μm).
In summary, binding studies suggest that under cellular conditions with ∼50–150 μm GTP (16, 50), WT mGBP2 as well as the R48A and E99A mutants are expected to be in the GTP-bound form. However, despite decreased nucleotide affinities, the D182N mutant will be almost completely GTP-bound, whereas the K51A mutant will occur predominantly nucleotide-free.
Hydrolysis
The identification of the nucleotide bound and the rate of conversion of GTP to GDP by hydrolysis are crucial factors that determine the functional regulation of GTPases (16, 51, 52). We therefore analyzed the kinetics of GTP hydrolysis for mGBP2 to investigate whether the mutations affect the product ratio and the ability to undergo the nucleotide-dependent multimerization resulting in stimulation of the GTPase activity.
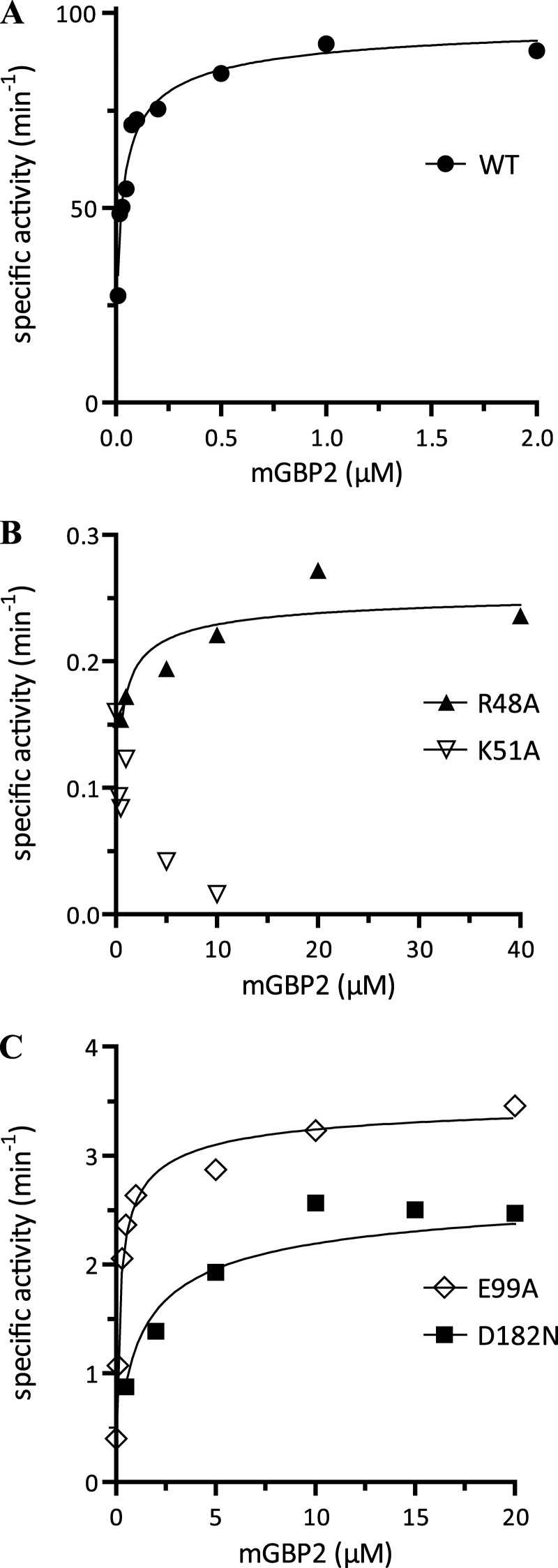
The rate of hydrolysis and product formation was analyzed by reversed-phase HPLC at a fixed GTP concentration and different mGBP2 concentrations (Fig. 2), establishing conditions of pseudo-first order. WT mGBP2 hydrolyzed GTP to GDP and GMP and yielded GMP as the major product (74%, Table 2). Interestingly, offering GDP as a substrate did not result in GMP production (supplemental Fig. 3). The specific GTPase activity of the WT protein increased with increasing protein concentrations (Fig. 2A). Fitting a model describing the two molecule interaction of mGBP2 revealed a cooperative mechanism of GTP hydrolysis (supplemental Fig. 4), yielding a maximum velocity of 102 min−1 and a dimer dissociation constant of 0.03 μm (Table 2), respectively. Thus, the dimer affinity of mGBP2 is 10-fold higher than that for its human counterpart hGBP1 (14, 15).
FIGURE 2.
GTP hydrolysis. Concentration-dependent GTP hydrolysis catalyzed by mGBP2 WT and its GTPase mutants was measured with a fixed concentration of GTP (1 mm) GTP at 37 °C. The initial rates were measured (<30% GTP hydrolyzed) from the linear parts of time course experiments and normalized to the protein concentrations used (specific activity). The specific activities were then plotted against protein concentrations. The data were fitted to a model describing the interaction of two molecules of mGBP2, yielding Kd (μm) and the maximal specific activity Kmax (min−1). The maximum specific GTPase activities, dimer dissociation constants, and the amount of GMP production for WT (●) (A), R48A (▴), K51A (▿) (B), E99A (♢), and D182N (■) (C) are summarized in Table 2.
For the R48A mutant, we detected a 390-fold decreased specific activity (0.26 min−1) compared with the maximum activity of the WT even at much higher protein concentrations (Fig. 2B and Table 2). The GTP hydrolysis rate of the K51A mutant was negligible at all protein concentrations tested. The dimerization constant for the R48A mutant was 11-fold increased, indicating a weak cooperativity of hydrolysis. For K51A, the estimation of the dimer dissociation constant was not possible, and therefore, there is no indication of cooperativity in the reaction (Fig. 2B and Table 2). In contrast to K51A, the R48A mutant retained a GMP production of 35% indicating the cooperative regulation of the second cleavage step (Table 2). The mutant E99A had a 30-fold decreased activity (3.50 min−1) and a very low GMP production (7%) accompanied by a 6-fold increased dimer dissociation constant (Kd = 0.18 μm) that suggests the involvement for the respective glutamic acid in the hydrolysis reaction (Fig. 2C and Table 2). The D182N mutant is distinct from all other mutants with a more than 100-fold lower affinity of the protein dimer (Kd = 3.01 μm), so stimulated GTPase activity occurred at higher protein concentrations. However, the production of GMP by D182N-mGBP2 was detectable but reduced (26%) (Fig. 2C and Table 2). Thus, from these data and the structural model of mGBP2 (supplemental Fig. 5), it can be concluded that the residue Asp-182 will be rather involved in binding of the guanine base than in catalysis.
Nucleotide-dependent Multimerization
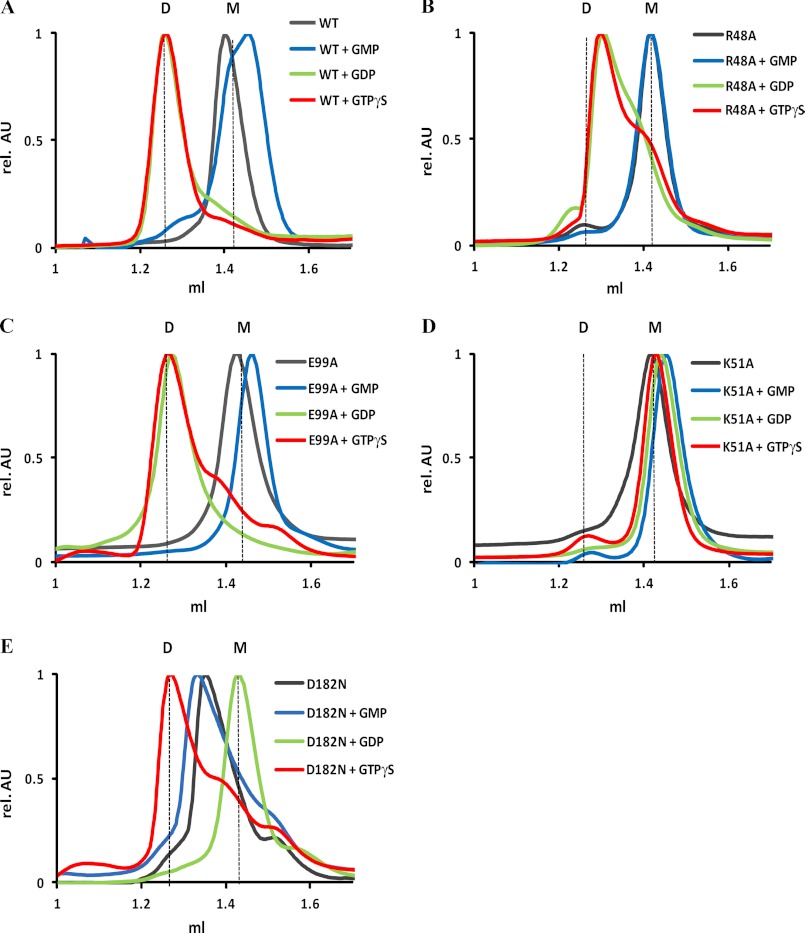
Members of the large GTPase family oligomerize depending on the nucleotide-bound state (11, 21, 53, 54) subsequently leading to concentration-dependent GTP hydrolysis (55). We have shown that the GTPase activity of mGBP2 also increased in a concentration-dependent manner (Fig. 2), so we sought to determine whether mGBP2 forms oligomers as well. Using size-exclusion chromatography, we found that mGBP2 eluted as a monomer at a size of ∼67 kDa in the absence of nucleotide and in complex with GMP but as a dimer in the presence of GTPγS, GDP (Fig. 3A), and Gpp(NH)p (supplemental Fig. 6).
FIGURE 3.
Nucleotide-dependent multimerization. Size-exclusion chromatography of WT (A) and mutant mGBP2 R48A (B), K51A (C), E99A (D), and D182N (E) bound to GTPγS, GDP, GMP, and in the nucleotide-free state at 4 °C. Elution of all proteins was followed using absorbance by 280 nm. The protein size was estimated by appropriate standard proteins, and the absorbance values were normalized to the peaks of the curves. D, dimer, M, monomer.
The elution profiles of GTPase mutants in different nucleotide states are shown in Fig. 3, B–E. Without nucleotide and in the presence of GMP, all mutants eluted as monomers. The R48A mutant showed in the presence of GTPγS, GDP, and Gpp(NH)p mixed populations of monomeric and dimeric proteins (Fig. 3B and supplemental Fig. 6). This is remarkable as the binding affinities to mant-GTPγS and mant-GDP were comparable with the WT (Table 2), and the affinity to mant-Gpp(NH)p was even slightly stronger (supplemental Fig. 2 and supplemental Table 1). In the case of the K51A mutant, no dimer formation was observed independently of the offered nucleotide (Fig. 3C, Fig. 4, and supplemental Fig. 6). The E99A mutant dimerized in the presence of GTPγS, GDP, and Gpp(NH)p, which is in agreement with its binding affinities (Fig. 3D and Table 2). Thus, this mutation in the switch II region does not affect the dimerization behavior of mGBP2. Similar conclusions can be drawn for the Asp-182 mutant. The protein eluted as a monomer in the presence of GDP and tended to a dimeric state in the presence of the respective GTPγS concentration (Fig. 3E). Nevertheless, dimers could be detected at higher nucleotide concentrations (50 μm, data not shown).
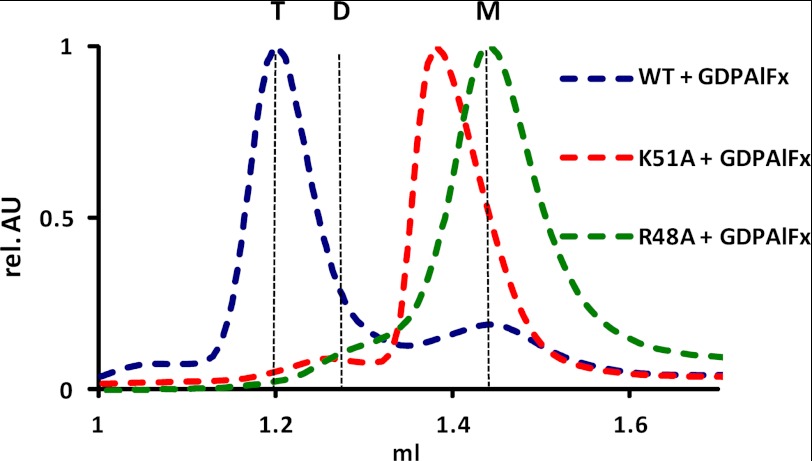
FIGURE 4.
Establishment of the transition state. Size-exclusion chromatography of the WT protein, K51A, and R48A mGBP2 mutants in the GDP·AlFx-bound form. Elution of all proteins was followed using absorbance by 280 nm. The protein size was estimated by appropriate standard proteins, and the absorbance values were normalized to the peaks of the curves. T, tetramer; D, dimer, M, monomer.
Interestingly, WT mGBP2 assembled to approximately twice the molecular weight of the dimer in the presence of GDP and AlFx (AlF3 or AlF4−) (Fig. 4). The GDP·AlFx complex bound to the protein is regarded as an analog of the transition state of the GTPase reaction (14). Additionally, the elution profile of R48A in the presence of GDP·AlFx revealed the protein in the monomeric state (Fig. 4). Thus, we conclude that the mutation of the arginine in the P-loop impairs the nucleotide-dependent multimerization during the hydrolysis reaction.
Taken together, the biochemical analyses showed that the R48A and E99A mutations of mGBP2 affect hydrolysis, whereas the D182N mutant impairs nucleotide binding, and the P-loop lysine mutant K51A leads to a dysfunctional protein.
Subcellular Localization and Homo-FRET Analysis of Multimerization of mGBP2
mGBP2 has been identified as one of the most abundantly expressed genes, on both mRNA and protein levels in murine ANA-1 macrophages after stimulation with IFN-γ (23, 34). It localizes to vesicle-like structures of heterogeneous size within the cytoplasm of IFN-γ-treated bone marrow-derived macrophages, NIH 3T3 fibroblasts, RAW 264.7 macrophages, and MEFs (34, 56).
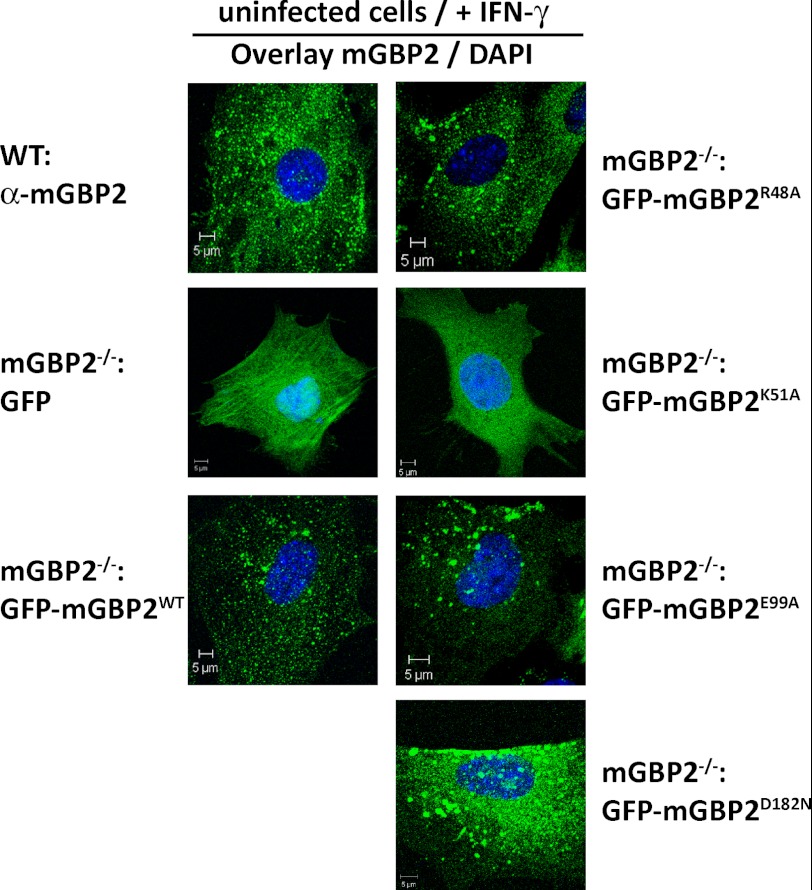
In an effort to investigate the IFN-γ-initiated antimicrobial mechanisms of mGBP2, a knock-out mouse was generated in our laboratory (data not shown). MEFs from these mice were stably transduced with N-terminal GFP fusion constructs of the mGBP2 WT protein or the GTPase mutants introduced above. The localization of the reconstituted WT mGBP2 fusion protein within vesicle-like structures in IFN-γ-stimulated cells was largely comparable with the endogenous protein (Fig. 5), which is in contrast to the IRG proteins, where large fluorescent tags impaired multimerization and localization (57). The effect of mutations in the GTP-binding domain on the subcellular distribution of mGBP2 in MEFs was determined after IFN-γ stimulation of cells. The localization of several GTPase mutant proteins differed from that of the WT (Fig. 5). Comparable with the GFP-WT-mGBP2 protein, the two hydrolytically impaired mutants R48A and E99A accumulated in vesicle-like structures; however, the fluorescence pattern showed a smaller number but a larger size of the vesicle-like structures, and some of the binding mutant D182N appeared to be in the cytosol in addition to a vesicular distribution. The dysfunctional K51A mutant was completely dislocated and occurred evenly distributed throughout the cytoplasm. From these results we conclude that the nucleotide dependent self-assembly of mGBP2 is a prerequisite for the accumulation in vesicle-like structures.
FIGURE 5.
Intracellular localization of WT and mutant mGBP2 proteins. Subcellular localization of mGBP2 and GTPase mutants was analyzed by transduction of GFP fusion constructs in MEFs or by usage of WT MEFs. Cells were stimulated with IFN-γ for 16 h. After fixation, endogenous mGBP2 was stained with a specific α-mGBP2 antibody and the cell nuclei with DAPI. Glass slides were analyzed by confocal microscopy. Bars, 5 μm.
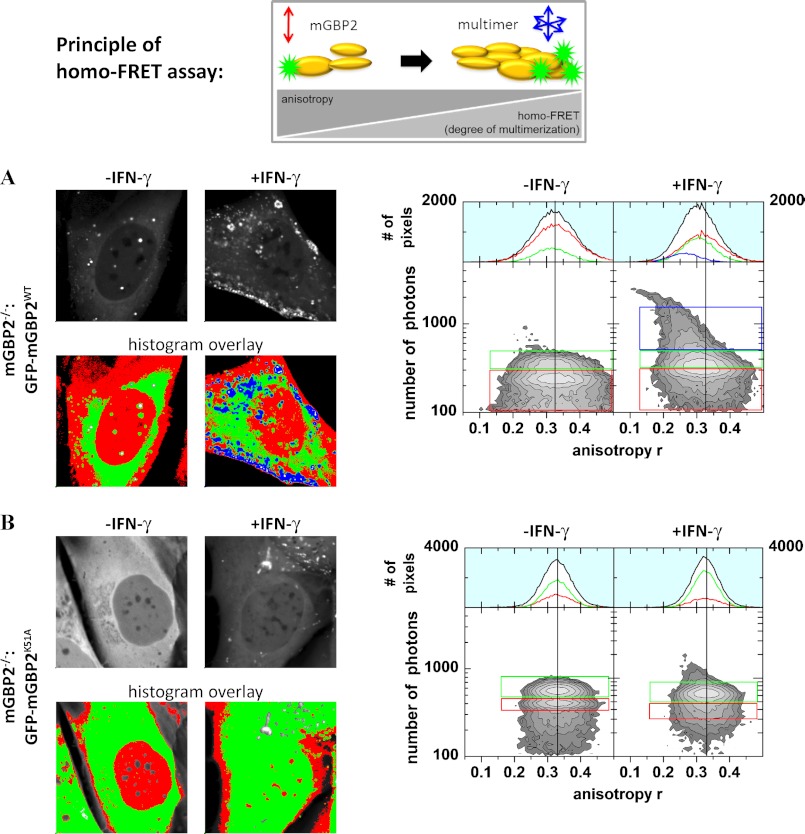
To verify the multimerization data in a physiological context, steady state fluorescence anisotropy-based homo-Förster resonance energy transfer (FRET) detection was employed to study IFN-γ-dependent intracellular multimerization of mGBP2 proteins in cells. homo-FRET can only occur over a very short distance (<10 nm). Thus, the depolarization of the GFP signal due to homo-FRET is an appropriate readout parameter for the multimerization of mGBP2 (58).
mGBP2−/− MEFs expressing the GFP-WT-mGBP2 construct (Fig. 6A) were evaluated in comparison with cells expressing only GFP (supplemental Fig. 7) or the GFP-K51A-mGBP2 mutant (Fig. 6B). Steady state anisotropy MFIS measurements of free GFP delivered almost equal values (Δ = 0.0006, Table 3) in stimulated and unstimulated cells (supplemental Fig. 7); thus, the anisotropy was not influenced by IFN-γ, as clearly shown in the multiparameter two-dimensional histogram, where steady state anisotropies over photon numbers of each analyzed pixel of the MFIS-images are displayed (supplemental Fig. 7). The steady state anisotropy of the WT in unstimulated cells was slightly higher than the anisotropy of free GFP. i.e. according to the Perrin equation (59) this is due to slower rotational diffusion times of GFP-mGBP2 molecules in comparison with smaller GFP molecules (Fig. 6A). After stimulation with IFN-γ, the mean steady state anisotropy of the WT protein over all pixels was significantly reduced (Δ = 0.0196, Table 3) due to energy transfer between GFP fluorophores (homo-FRET). The decrease of anisotropy is a strong indicator for multimerization of the WT protein. The mean anisotropy in vesicles in the stimulated cells was even more decreased (0.2631, Table 3, Fig. 6A population in the blue box), indicating that the degree of multimerization of WT mGBP2 increases in these structures. Thus, the K51A mutant showed slightly higher steady state anisotropy in unstimulated cells than the WT (Table 3 and Fig. 6B). This can be explained by the inability of the K51A mutant to form any multimers. The steady state anisotropy of the K51A mutant represents the anisotropy value of the monomeric mGBP2. The WT protein can already form multimeric complexes in unstimulated cells. The steady state anisotropy of the K51A mutant does not decrease significantly after stimulation with IFN-γ (Table 3). Thus, IFN-γ cannot induce multimerization in this mutant.
FIGURE 6.
Intracellular multimerization of mGBP2. Left panel, confocal (upper panel) and intensity (lower panel) images of living IFN-γ-stimulated (right) and -unstimulated (left) mGBP2−/− MEFs expressing GFP-WT-mGBP2 (A) or GFP-K51A-mGBP2 (B) superimposed with a selection of pixels with high (blue), middle (green), and low (red) fluorescence intensities. Right panel, two-dimensional histograms of anisotropy (r) of GFP on x axis and photon number per pixel on y axis. Color boxes highlight pixels with certain fluorescence intensities.
TABLE 3.
Average anisotropy of GFP-WT-mGBP2, GFP-K51A-mGBP2, and GFP
The intensity values of all significant pixels were summed, and the overall anisotropy per cell was calculated. Additionally, mean anisotropies of several cells before and after stimulation were calculated.
| Average anisotropy |
Δ (IFN-γ) | ||
|---|---|---|---|
| − IFN-γ | + IFN-γ | ||
| GFP- mGBP2WT | |||
| Whole cell | 0.3227 | 0.3031 | 0.0196 |
| Red | 0.3260 | 0.3145 | 0.0115 |
| Green | 0.3165 | 0.3041 | 0.0124 |
| Blue | 0.2631 | ||
| Mean | 0.3152/±0.0038 (n = 9) | 0.2984/±0.0089 (n = 10) | 0.0168 |
| GFP-mGBP2K51A | |||
| Whole cell | 0.3280 | 0.3254 | 0.0026 |
| Red | 0.3289 | 0.3261 | 0.0028 |
| Green | 0.3281 | 0.3267 | 0.0014 |
| Mean | 0.3301/±0.0053 (n = 10) | 0.3272/±0.0036 (n = 10) | 0.0029 |
| GFP | |||
| GFP | 0.3096 | 0.3091 | 0.0006 |
| Mean | 0.3064/±0.0056 (n = 3) | 0.3064/±0.0058 (n = 4) | 0 |
In conclusion, WT mGBP2 proteins form multimers in the cellular context, and IFN-γ enhances this effect significantly. For multimerization, GTPase activity of mGBP2 is essential.
Role of mGBP2 in T. gondii Infection
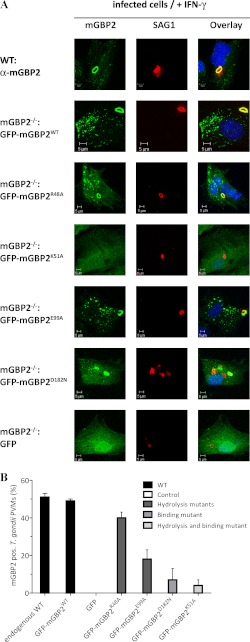
Nevertheless, still little is known about the molecular functions of this protein. Here, we were interested in the impact of GTPase function and multimerization behavior of mGBP2 on the ability to accumulate at the PV membrane of T. gondii. For this purpose, we infected IFN-γ stimulated mGBP2−/− MEFs, which were reconstituted with the WT or GTPase mutants, with the T. gondii strain ME49 and quantified the recruitment efficiency by confocal microscopy (Fig. 7A).
FIGURE 7.
Recruitment of WT and mutant mGBP2 proteins to the PV membrane of T. gondii. Recruitment of mGBP2 and GTPase mutants was analyzed by transduction of GFP fusion constructs in MEFs or by usage of WT MEFs. Cells were stimulated with IFN-γ for 16 h and subsequently infected with the T. gondii strain ME49 for 2 h. After fixation, endogenous mGBP2 was stained with a specific α-mGBP2 antibody, T. gondii with the α-SAG1 antibody and the cell nuclei with DAPI. Glass slides were analyzed by confocal microscopy (A). Recruitment rates were estimated by the ratio of mGBP2-associated T. gondii versus the whole amount of intracellular parasites (B). Bars, 5 μm.
After 30 min of infection, accumulation of mGBP2 was observable around the PV (data not shown) (34). After 2 h, ∼50% of the invaded parasites were completely surrounded by the bulk of mGBP2 protein (Fig. 7, A and B). It is noteworthy that almost identical recruitment rates for the endogenous and the reconstituted WT mGBP2 could be measured. The R48A mutant, which showed a failure in transition state stabilization (see Fig. 4), revealed a slightly decreased ability to relocate to the PV compared with the WT (40%; Fig. 7B), whereas the catalytically incapacitated E99A mutant was significantly affected in the recruitment efficiency (18%). The association rates of the binding mutant D182N were strongly decreased (7%), and the recruitment of the biochemically dysfunctional K51A mutant was almost abolished (4%). Interestingly, without IFN-γ pre-stimulation, no protein co-localization with the PV was detectable for any of the analyzed mGBP2 constructs (data not shown) (34). From these results we conclude that the ability to form multimers and the cooperative GTPase activity regulate the active recruitment of mGBP2 to the PV of T. gondii.
DISCUSSION
The type II interferon (IFN-γ) pathway bridges innate and acquired immunity (2, 34, 60–62). Interestingly, the family of IFN induced 65-kDa GBPs is now beginning to emerge as a major determinant of antimicrobial resistance (2, 3, 7, 34). In this study, we present a systematic characterization of mGBP2, elucidating the relationship of cellular biochemistry and host defense functions.
By sequence homology, mGBP2 is the closest murine ortholog of hGBP1 (68% identity, supplemental Table 2 (10)). Nevertheless, there are some interesting differences. The binding constant of mGBP2 for mant-GMP is reduced by 1 order of magnitude compared with mant-GTPγS and mant-GDP, whereas the affinities of hGBP1 to all three guanylate nucleotides are in the same range (15). This could provide an explanation for the higher turnover rates of mGBP2 in comparison with hGBP1 by reducing the lifetime of GMP-bound states (14).
In the nucleotide-free state, mGBP2 (this study) and hGBP1 are monomers (15), whereas the hGBP5a/b splicing variant constitutes a constitutive dimer that is dependent on the C-terminal domain (63).
mGBP2 forms dimers after binding to GTP analogs as does hGBP1 (14). The dimer assembly of hGBP1 employs an interaction site within the G-domain (24). The most striking difference in the nucleotide-dependent oligomerization behavior between mGBP2 and hGBP1 is that mGBP2 assembles into dimers in the presence of GDP, whereas hGBP1 remains a monomer (15). Nevertheless, GDP is not accepted as a substrate from solution underlining the need for higher oligomerization (tetramerization) for efficient GTP hydrolysis (64, 65). This hypothesis is supported by our anisotropy measurements in living cells. Lower anisotropies in untreated cells expressing GFP-WT-mGBP2 clearly indicate complex formation of several mGBP2 molecules; however, the participation of other proteins within this complex cannot be excluded by this homo-FRET assay.
Binding of the transition state analog GDP·AlFx (66) to mGBP2 leads to the formation of a tetramer, concordant with hGBP1 (15). In contrast, the GBP-related GTPase atlastin 1 remains dimeric in the presence of GDP·AlFx (67), and GDP·AlFxdoes not change the dimeric state of hGBP5a/b (63). Furthermore, the R227E/K228E mutant of hGBP1, which exhibits the loss of contact between the G-domain and the C-terminal part of the protein, forms tetramers after addition of Gpp(NH)p, suggesting that this conformational flexibility allows for exposure of a C-terminal interaction site leading to a constitutive G-domain independent dimer (24).
The cooperative stimulation of the GTPase activity of mGBP2 occurs exclusively as a result of self-assembly without external GTPase-activating protein. mGBP2 has an additional property to hydrolyze GTP to GMP probably in two consecutive cleavage reactions, with GMP as the major product. This resembles hGBP1, and hGBP2 produces GDP rather than GMP, whereas for hGBP5 and atlastin 1, GDP is the only product of GTP hydrolysis (63, 68–70). It is assumed that the flexibility of the nucleotide in the GTP-binding pocket of GBPs allows for conversion from GTP to GMP and that GDP hydrolysis involves the same reaction mechanism as the hydrolysis of GTP (65, 71).
The mutation of Lys-51 in mGBP2 results in a nearly complete loss of function as all steps in the GTPase cycle are impaired. The analogous mutant in hGBP1 leads to a profound loss of GTPase activity (15). The corresponding K44E mutant in dynamin exhibits a decreased affinity for both GTP and GDP (72), whereas the K51N mutant of human MxA is defective in hydrolysis (73).
The G4 region in 65-kDa GBPs and atlastins is conserved as a reduced 181RD182 motif and contributes to the specificity for guanine bases (16, 17, 53). In the case of hGBP1, the mutation D184N leads to a change of nucleotide specificity from guanine to xanthine (16). The D182N mutant of mGBP2 decreases the affinity for guanine nucleotides comparable with hGBP1 (15), and it affects the dimer/tetramer formation of mGBP2 providing an explanation for the significant reduction in cooperativity, thus fostering the role of Asp-182 of mGBP2 in guanine nucleotide binding and stabilization. The mutation at the equivalent position in Ras (Asp-119) exhibits a 20-fold decreased affinity for both GTP and GDP and is oncogenic (74).
Arg-48 in the P-loop is highly conserved within GBPs and atlastins (15, 53). The affinity to mant-GMP for the R48A mutant of mGBP2 remains in the range of the WT protein, whereas the affinity of the corresponding mutant in hGBP1 is 15-fold lowered compared with the corresponding WT (15). The weak GTPase activity of the R48A mutant is accompanied by a reversal of the product ratio in favor of GDP, and cooperativity can be estimated only at high protein concentrations. The mutant fails to maintain a tetramer, when bound to the GDP·AlFx complex, which is probably not caused by a lack of binding of aluminum fluoride (15). In hGBP1, the respective arginine stabilizes the negative charge in the transition state and is required for cooperative GTPase activity (14, 21, 65). Upon comparison with the arginine residue positioned by the α-helical domain of Gα proteins (75), we conclude that the residue Arg-48 in the P-loop of mGBP2 is supposed to function as a GTPase-activating “arginine finger.” In the structures of atlastin, the respective arginine participates in dimer formation (67, 70). Thus, we suggest that Arg-48 is involved in the process of multimerization.
Both switch I and switch II regions undergo major structural changes between GTP- and GDP-bound states forming the so-called phosphate cap (15). An invariant glutamic acid is highly conserved in the switch II region of GBPs. The E99A mutant of mGBP2 in the switch II region has moderate effects on nucleotide binding but significant effects on GTP hydrolysis, whereby the most striking difference to the WT is the strongly reduced production of GMP. These results are in agreement with data obtained for the corresponding mutation in hGBP1 (15). We conclude that Glu-99 is directly involved in catalysis and that the mutation abrogates conformational rearrangements required for the second step of hydrolysis. Intriguingly, mutations of residues in the switch I and switch II regions of dynamin also have little impact on GTP binding but decrease the GTPase activity (76).
We suggest that under physiological conditions mGBP2 WT protein and the R48A mutant occur as GTP-bound dimers, whereas the K51A mutant is mostly a monomer, being either nucleotide-free or bound to GTP. The switch II E99A mutant constitutes a dimer and shows a decrease in GTPase activity and GMP production. The D182N mutant is partially GTP-bound, forms dimers, and is able to perform the hydrolysis of GTP; however, it is markedly slower with decreased GMP production.
We investigated the influence of biochemical properties on the importance of localization and function in host defense against T. gondii. Endogenous mGBP2 as well as the reconstituted WT protein reside within vesicle-like structures. Here, we provide evidence that the localization requires GTPase activity. The binding mutants are dislocated to a degree congruent with their impairment in GTP binding and multimerization capacity. In detail, the G4 motif D182N mutant with decreased guanine base specificity partially localizes in punctate structures, whereas the P-loop K51A GTPase activity-deficient mutant shows a mostly homogeneous distribution. The corresponding D208N mutant in dynamin exhibits a similar localization as the WT protein and does not affect endocytosis in overexpressing cells (72). The constitutively active Irga6-K82A mutant, which is locked in the GTP-bound state, aggregates in the cytoplasm and prevents the endogenous protein to localize at the smooth endoplasmic reticulum (77).
Both of the other mutants, which we show to form GTP-dependent dimers (R48A and E99A), accumulate in vesicle-like structures. For human GBPs, membrane association has been shown to be regulated by dimerization (78, 79). Interestingly, some mGBPs lacking an isoprenylation motif are distributed in vesicles as well (34).
According to the detailed MFIS analyses, the increased concentration and multimerization of WT mGBP2 in vesicle-like structures after stimulation provides evidence that additional IFN-γ inducible factors influence the localization of the proteins. This vesicular distribution might be due to multimerization in homo- and hetero-oligomers, where other members of the mGBP family could serve as interaction partners and regulate the ultimate intracellular localization of mGBP2, which is suggested by Britzen-Laurent et al. (78) for hGBPs. Therefore, we hypothesize that the multimerization of mGBP2 may help to assemble complexes acting as a scaffold for binding partners to fulfill their antimicrobial functions.
The in vivo role of mGBPs during infection with the protozoan T. gondii was previously demonstrated in C57BL/6 mice (34), indicating the rapid up-regulation of mGBPs (9, 34). Using the mGBP2−/− mouse, we recently were able to confirm an effector function of mGBP2 in the host defense against T. gondii, as knock-out animals were more susceptible to the infection showing enhanced morbidity and mortality as the WT mice (data not shown).
T. gondii survives in a specialized PV that resists fusion with lysosomes (80). The regulation of mGBP localization by T. gondii under proinflammatory conditions was recently demonstrated by in vivo analyses (34). Several of the IFN-γ-induced mGBPs accumulate at the PV membrane rapidly after infection (34). Interestingly, members of the IRG family, such as Irgm3, Irgm2, Irgb6, Irgd, and Irga6, are reported to relocate from their resting locations to the avirulent T. gondii strain ME49 and to accumulate at the PV in IFN-γ-stimulated astrocytes and gs3T3 cells (5, 77, 81, 82). Here, we investigate the engagement of mGBP2 GTPase mutants in the cell-autonomous defense against T. gondii.
Co-localization analyses revealed that both GTP binding and multimerization as well as the GTPase activity are important for the efficient recruitment to the PV of T. gondii. The recruitment capacities for the binding mutant D182N and the GTPase-defective K51A mutant are almost abolished. For the corresponding Irga6-K82A mutant, strong suppressive effects were shown on T. gondii resistance and PV membrane localization in IFN-γ-induced cells, exhibiting a dominant inhibitory effect over the WT protein (77, 81). The D184N mutant of hGBP1 has a dominant effect on the WT protein showing a rescue of MMP-1 expression and tube formation (83) suggesting the requirement for fast GTP hydrolysis of hGBP1 for its physiological role.
The slightly decreased recruitment efficiency to the PV for the R48A mutant in comparison with the WT may be conceivable by heterodimerization with other endogenous GBP family members, which are induced by IFN-γ (84, 85). This could permit the stabilization of a transition state during hydrolysis reactions, which may serve as the key regulatory event. This hypothesis appears reasonable as the E99A mutant cannot reach the PV recruitment rate of the WT protein. The nonhydrolyzing E99A mutant of hGBP1 fails to constitutively associate with the Golgi apparatus in IFN-γ and AlFx-treated cells (84). This implicates that the conformational changes induced by cleavage reactions might reveal surfaces for possible IFN-γ-induced interacting partners providing biological functions (86). The IRGM subfamily of 47-kDa GTPases (8), including Irgm1, Irgm2, and Irgm3, regulates other IRG proteins through control of their GTPase cycle, permitting GTP-dependent assembly of the GKS motif at the PV membrane of T. gondii (77). These regulatory interactions are associated with direct nucleotide-dependent contact between the GTPases. In dynamin, assembly-incompetent mutants as well as several mutants that are defective in binding and hydrolysis are potent dominant negative inhibitors of receptor-mediated endocytosis (87–89). Mutations in the GTPase domain of human MxA and murine Mx1 that abrogate GTP binding and GTPase activity disrupt the protein's ability to confer viral resistance (26, 90, 91). Nevertheless, the molecular role of GBPs in T. gondii infection remains to be elucidated. However, the accumulation of mGBP2 seems to be relevant for defense against T. gondii because virulent T. gondii can circumvent the accumulation of mGBPs (34).
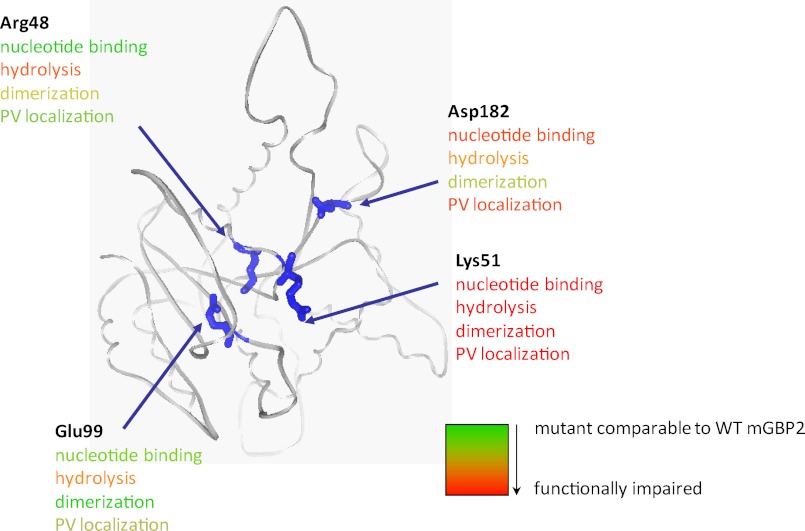
In summary, we generated and characterized a set of mGBP2 mutants that have individual defects in nucleotide binding, hydrolysis, and multimerization that are schematically summed up in Fig. 8. The biochemical and MFIS data demonstrate that IFN-γ is essential for the formation of nucleotide-dependent multimers and that the multimerization is required for localization of mGBP2 in vesicle-like structures, which is a prerequisite for the recruitment to the PV of T. gondii.
FIGURE 8.
A schematic diagram is given summarizing the biochemical and host-pathogen interaction features of mGBP2 mutants with respect to nucleotide binding, dimerization, and PV localization.
Supplementary Material
Acknowledgments
We thank Karin Buchholz, André Abts, Christian Schwarz, Eric Keil, and Ralf Kühnemuth for experimental assistance, technical support, and fruitful discussions.
This work was supported by Deutsche Forschungsgemeinschaft Grants PF259/4-1, FOR 729, GRK 1045, and SFB 590 (to K. P. and C. A. M. S.) and the Jürgen Manchot Foundation.

This article contains supplemental Figs. 1–7 and Tables 1 and 2.
- GBP
- guanylate-binding protein
- MFIS
- multiparameter fluorescence image spectroscopy
- PV
- parasitophorous vacuole
- mant
- 2′/3′-O-(N-methylanthraniloyl)
- GTPγS
- guanosine 5′-3-O-(thio)triphosphate
- Gpp(NH)p
- guanosine 5′-(β,γ-imido)triphosphate
- h
- human
- m
- mouse
- IRG
- immunity-related GTPase
- MEF
- murine embryonic fibroblast.
REFERENCES
- 1. Boehm U., Klamp T., Groot M., Howard J. C. (1997) Cellular responses to interferon-γ. Annu. Rev. Immunol. 15, 749–795 [DOI] [PubMed] [Google Scholar]
- 2. MacMicking J. D. (2004) IFN-inducible GTPases and immunity to intracellular pathogens. Trends Immunol. 25, 601–609 [DOI] [PubMed] [Google Scholar]
- 3. Shenoy A. R., Kim B. H., Choi H. P., Matsuzawa T., Tiwari S., MacMicking J. D. (2007) Emerging themes in IFN-γ-induced macrophage immunity by the p47 and p65 GTPase families. Immunobiology 212, 771–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Anderson S. L., Carton J. M., Lou J., Xing L., Rubin B. Y. (1999) Interferon-induced guanylate binding protein-1 (GBP-1) mediates an antiviral effect against vesicular stomatitis virus and encephalomyocarditis virus. Virology 256, 8–14 [DOI] [PubMed] [Google Scholar]
- 5. Collazo C. M., Yap G. S., Sempowski G. D., Lusby K. C., Tessarollo L., Woude G. F., Sher A., Taylor G. A. (2001) Inactivation of LRG-47 and IRG-47 reveals a family of interferon γ-inducible genes with essential, pathogen-specific roles in resistance to infection. J. Exp. Med. 194, 181–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tietzel I., El-Haibi C., Carabeo R. A. (2009) Human guanylate binding proteins potentiate the anti-chlamydia effects of interferon-γ. PLoS One 4, e6499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kim B. H., Shenoy A. R., Kumar P., Das R., Tiwari S., MacMicking J. D. (2011) A family of IFN-γ-inducible 65-kD GTPases protects against bacterial infection. Science 332, 717–721 [DOI] [PubMed] [Google Scholar]
- 8. Bekpen C., Hunn J. P., Rohde C., Parvanova I., Guethlein L., Dunn D. M., Glowalla E., Leptin M., Howard J. C. (2005) The interferon-inducible p47 (IRG) GTPases in vertebrates. Loss of the cell autonomous resistance mechanism in the human lineage. Genome Biol. 6, R92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kresse A., Konermann C., Degrandi D., Beuter-Gunia C., Wuerthner J., Pfeffer K., Beer S. (2008) Analyses of murine GBP homology clusters based on in silico, in vitro, and in vivo studies. BMC Genomics 9, 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Olszewski M. A., Gray J., Vestal D. J. (2006) In silico genomic analysis of the human and murine guanylate-binding protein (GBP) gene clusters. J. Interferon Cytokine Res. 26, 328–352 [DOI] [PubMed] [Google Scholar]
- 11. Pawlowski N., Khaminets A., Hunn J. P., Papic N., Schmidt A., Uthaiah R. C., Lange R., Vopper G., Martens S., Wolf E., Howard J. C. (2011) The activation mechanism of Irga6, an interferon-inducible GTPase contributing to mouse resistance against Toxoplasma gondii. BMC Biol. 9, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wennerberg K., Rossman K. L., Der C. J. (2005) The Ras superfamily at a glance. J. Cell Sci. 118, 843–846 [DOI] [PubMed] [Google Scholar]
- 13. Coleman D. E., Berghuis A. M., Lee E., Linder M. E., Gilman A. G., Sprang S. R. (1994) Structures of active conformations of Giα1 and the mechanism of GTP hydrolysis. Science 265, 1405–1412 [DOI] [PubMed] [Google Scholar]
- 14. Prakash B., Praefcke G. J., Renault L., Wittinghofer A., Herrmann C. (2000) Structure of human guanylate-binding protein 1 representing a unique class of GTP-binding proteins. Nature 403, 567–571 [DOI] [PubMed] [Google Scholar]
- 15. Praefcke G. J., Kloep S., Benscheid U., Lilie H., Prakash B., Herrmann C. (2004) Identification of residues in the human guanylate-binding protein 1 critical for nucleotide binding and cooperative GTP hydrolysis. J. Mol. Biol. 344, 257–269 [DOI] [PubMed] [Google Scholar]
- 16. Praefcke G. J., Geyer M., Schwemmle M., Robert Kalbitzer H., Herrmann C. (1999) Nucleotide-binding characteristics of human guanylate-binding protein 1 (hGBP1) and identification of the third GTP-binding motif. J. Mol. Biol. 292, 321–332 [DOI] [PubMed] [Google Scholar]
- 17. Prakash B., Renault L., Praefcke G. J., Herrmann C., Wittinghofer A. (2000) Triphosphate structure of guanylate-binding protein 1 and implications for nucleotide binding and GTPase mechanism. EMBO J. 19, 4555–4564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vetter I. R., Wittinghofer A. (2001) The guanine nucleotide-binding switch in three dimensions. Science 294, 1299–1304 [DOI] [PubMed] [Google Scholar]
- 19. Buhrman G., Holzapfel G., Fetics S., Mattos C. (2010) Allosteric modulation of Ras positions Q61 for a direct role in catalysis. Proc. Natl. Acad. Sci. U.S.A. 107, 4931–4936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ahmadian M. R., Stege P., Scheffzek K., Wittinghofer A. (1997) Confirmation of the arginine finger hypothesis for the GAP-stimulated GTP hydrolysis reaction of Ras. Nat. Struct. Biol. 4, 686–689 [DOI] [PubMed] [Google Scholar]
- 21. Praefcke G. J., McMahon H. T. (2004) The dynamin superfamily: universal membrane tubulation and fission molecules? Nat. Rev. Mol. Cell Biol. 5, 133–147 [DOI] [PubMed] [Google Scholar]
- 22. Cheng Y. S., Patterson C. E., Staeheli P. (1991) Interferon-induced guanylate-binding proteins lack an N(T)KXD consensus motif and bind GMP in addition to GDP and GTP. Mol. Cell. Biol. 11, 4717–4725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Boehm U., Guethlein L., Klamp T., Ozbek K., Schaub A., Fütterer A., Pfeffer K., Howard J. C. (1998) Two families of GTPases dominate the complex cellular response to IFN-γ. J. Immunol. 161, 6715–6723 [PubMed] [Google Scholar]
- 24. Vöpel T., Syguda A., Britzen-Laurent N., Kunzelmann S., Lüdemann M. B., Dovengerds C., Stürzl M., Herrmann C. (2010) Mechanism of GTPase activity-induced self-assembly of human guanylate-binding protein 1. J. Mol. Biol. 400, 63–70 [DOI] [PubMed] [Google Scholar]
- 25. Wehner M., Kunzelmann S., Herrmann C. (2012) The guanine cap of human guanylate-binding protein 1 is responsible for dimerization and self-activation of GTP hydrolysis. FEBS J. 279, 203–210 [DOI] [PubMed] [Google Scholar]
- 26. Haller O., Kochs G. (2002) Interferon-induced Mx proteins. Dynamin-like GTPases with antiviral activity. Traffic 3, 710–717 [DOI] [PubMed] [Google Scholar]
- 27. Taylor G. A. (2007) IRG proteins. Key mediators of interferon-regulated host resistance to intracellular pathogens. Cell. Microbiol. 9, 1099–1107 [DOI] [PubMed] [Google Scholar]
- 28. Taylor G. A., Stauber R., Rulong S., Hudson E., Pei V., Pavlakis G. N., Resau J. H., Vande Woude G. F. (1997) The inducibly expressed GTPase localizes to the endoplasmic reticulum, independently of GTP binding. J. Biol. Chem. 272, 10639–10645 [DOI] [PubMed] [Google Scholar]
- 29. MacMicking J. D., Taylor G. A., McKinney J. D. (2003) Immune control of tuberculosis by IFN-γ-inducible LRG-47. Science 302, 654–659 [DOI] [PubMed] [Google Scholar]
- 30. Martens S., Sabel K., Lange R., Uthaiah R., Wolf E., Howard J. C. (2004) Mechanisms regulating the positioning of mouse p47 resistance GTPases LRG-47 and IIGP1 on cellular membranes. Retargeting to plasma membrane induced by phagocytosis. J. Immunol. 173, 2594–2606 [DOI] [PubMed] [Google Scholar]
- 31. MacMicking J. D. (2005) Immune control of phagosomal bacteria by p47 GTPases. Curr. Opin. Microbiol. 8, 74–82 [DOI] [PubMed] [Google Scholar]
- 32. Martens S., Howard J. (2006) The interferon-inducible GTPases. Annu. Rev. Cell Dev. Biol. 22, 559–589 [DOI] [PubMed] [Google Scholar]
- 33. Taylor G. A., Feng C. G., Sher A. (2004) p47 GTPases. Regulators of immunity to intracellular pathogens. Nat. Rev. Immunol. 4, 100–109 [DOI] [PubMed] [Google Scholar]
- 34. Degrandi D., Konermann C., Beuter-Gunia C., Kresse A., Würthner J., Kurig S., Beer S., Pfeffer K. (2007) Extensive characterization of IFN-induced GTPases mGBP1 to mGBP10 involved in host defense. J. Immunol. 179, 7729–7740 [DOI] [PubMed] [Google Scholar]
- 35. Rupper A. C., Cardelli J. A. (2008) Induction of guanylate binding protein 5 by γ interferon increases susceptibility to Salmonella enterica serovar typhimurium-induced pyroptosis in RAW 264.7 cells. Infect. Immun. 76, 2304–2315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shenoy A. R., Wellington D. A., Kumar P., Kassa H., Booth C. J., Cresswell P., MacMicking J. D. (2012) GBP5 promotes NLRP3 inflammasome assembly and immunity in mammals. Science 336, 481–485 [DOI] [PubMed] [Google Scholar]
- 37. Carter C. C., Gorbacheva V. Y., Vestal D. J. (2005) Inhibition of VSV and EMCV replication by the interferon-induced GTPase, mGBP-2. Differential requirement for wild-type GTP binding domain. Arch. Virol. 150, 1213–1220 [DOI] [PubMed] [Google Scholar]
- 38. Itsui Y., Sakamoto N., Kakinuma S., Nakagawa M., Sekine-Osajima Y., Tasaka-Fujita M., Nishimura-Sakurai Y., Suda G., Karakama Y., Mishima K., Yamamoto M., Watanabe T., Ueyama M., Funaoka Y., Azuma S., Watanabe M. (2009) Antiviral effects of the interferon-induced protein guanylate binding protein 1 and its interaction with the hepatitis C virus NS5B protein. Hepatology 50, 1727–1737 [DOI] [PubMed] [Google Scholar]
- 39. Duan Z., Foster R., Brakora K. A., Yusuf R. Z., Seiden M. V. (2006) GBP1 overexpression is associated with a paclitaxel resistance phenotype. Cancer Chemother. Pharmacol. 57, 25–33 [DOI] [PubMed] [Google Scholar]
- 40. Balasubramanian S., Messmer-Blust A. F., Jeyaratnam J. A., Vestal D. J. (2010) Role of GTP binding, isoprenylation, and the C-terminal α-helices in the inhibition of cell spreading by the interferon-induced GTPase, mouse guanylate-binding protein-2. J. Interferon Cytokine Res. 31, 291–298 [DOI] [PubMed] [Google Scholar]
- 41. Vestal D. J., Jeyaratnam J. A. (2011) The guanylate-binding proteins. Emerging insights into the biochemical properties and functions of this family of large interferon-induced guanosine triphosphatase. J. Interferon Cytokine Res. 31, 89–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zufferey R., Nagy D., Mandel R. J., Naldini L., Trono D. (1997) Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat. Biotechnol. 15, 871–875 [DOI] [PubMed] [Google Scholar]
- 43. Herrmann C., Nassar N. (1996) Ras and its effectors. Prog. Biophys. Mol. Biol. 66, 1–41 [DOI] [PubMed] [Google Scholar]
- 44. Kudryavtsev V., Felekyan S., Woźniak A. K., König M., Sandhagen C., Kühnemuth R., Seidel C. A., Oesterhelt F. (2007) Monitoring dynamic systems with multiparameter fluorescence imaging. Anal. Bioanal. Chem. 387, 71–82 [DOI] [PubMed] [Google Scholar]
- 45. Weidtkamp-Peters S., Felekyan S., Bleckmann A., Simon R., Becker W., Kühnemuth R., Seidel C. A. (2009) Multiparameter fluorescence image spectroscopy to study molecular interactions. Photochem. Photobiol. Sci. 8, 470–480 [DOI] [PubMed] [Google Scholar]
- 46. Widengren J., Kudryavtsev V., Antonik M., Berger S., Gerken M., Seidel C. A. (2006) Single-molecule detection and identification of multiple species by multiparameter fluorescence detection. Anal. Chem. 78, 2039–2050 [DOI] [PubMed] [Google Scholar]
- 47. Koshioka M., Sasaki K., Masuhara H. (1995) Time-dependent fluorescence depolarization analysis in three-dimensional microspectroscopy. Appl. Spectrosc. 49, 224–228 [Google Scholar]
- 48. Schaffer J., Eggeling C., Subramaniam V., Striker G., Seidel C. A. (1999) Identification of single molecules in aqueous solution by time-resolved fluorescence anisotropy. J. Phys. Chem. A 103, 331–336 [Google Scholar]
- 49. Becker W. (2005) Advanced Time-correlated Single Photon Counting Techniques, 1st Ed., Springer, Berlin, Heidelberg, Germany [Google Scholar]
- 50. Otero A. D. (1990) Transphosphorylation and G protein activation. Biochem. Pharmacol. 39, 1399–1404 [DOI] [PubMed] [Google Scholar]
- 51. Bourne H. R., Sanders D. A., McCormick F. (1991) The GTPase superfamily. Conserved structure and molecular mechanism. Nature 349, 117–127 [DOI] [PubMed] [Google Scholar]
- 52. Uthaiah R. C., Praefcke G. J., Howard J. C., Herrmann C. (2003) IIGP1, an interferon-γ-inducible 47-kDa GTPase of the mouse, showing cooperative enzymatic activity and GTP-dependent multimerization. J. Biol. Chem. 278, 29336–29343 [DOI] [PubMed] [Google Scholar]
- 53. Zhu P. P., Patterson A., Lavoie B., Stadler J., Shoeb M., Patel R., Blackstone C. (2003) Cellular localization, oligomerization, and membrane association of the hereditary spastic paraplegia 3A (SPG3A) protein atlastin. J. Biol. Chem. 278, 49063–49071 [DOI] [PubMed] [Google Scholar]
- 54. Haller O., Gao S., von der Malsburg A., Daumke O., Kochs G. (2010) Dynamin-like MxA GTPase. Structural insights into oligomerization and implications for antiviral activity. J. Biol. Chem. 285, 28419–28424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Warnock D. E., Hinshaw J. E., Schmid S. L. (1996) Dynamin self-assembly stimulates its GTPase activity. J. Biol. Chem. 271, 22310–22314 [DOI] [PubMed] [Google Scholar]
- 56. Vestal D. J., Gorbacheva V. Y., Sen G. C. (2000) Different subcellular localizations for the related interferon-induced GTPases, MuGBP-1 and MuGBP-2. Implications for different functions? J. Interferon Cytokine Res. 20, 991–1000 [DOI] [PubMed] [Google Scholar]
- 57. Zhao Y. O., Könen-Waisman S., Taylor G. A., Martens S., Howard J. C. (2010) Localization and mislocalization of the interferon-inducible immunity-related GTPase, Irgm1 (LRG-47) in mouse cells. PLoS One 5, e8648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lidke D. S., Nagy P., Barisas B. G., Heintzmann R., Post J. N., Lidke K. A., Clayton A. H., Arndt-Jovin D. J., Jovin T. M. (2003) Imaging molecular interactions in cells by dynamic and static fluorescence anisotropy (rFLIM and emFRET). Biochem. Soc. Trans. 31, 1020–1027 [DOI] [PubMed] [Google Scholar]
- 59. Lakowicz J. R. (2006) Springer, [Google Scholar]
- 60. Vestal D. J. (2005) The guanylate-binding proteins (GBPs). Proinflammatory cytokine-induced members of the dynamin superfamily with unique GTPase activity. J. Interferon Cytokine Res. 25, 435–443 [DOI] [PubMed] [Google Scholar]
- 61. Zhao Y. O., Rohde C., Lilue J. T., Könen-Waisman S., Khaminets A., Hunn J. P., Howard J. C. (2009) Toxoplasma gondii and the immunity-related GTPase (IRG) resistance system in mice. A review. Mem. Inst. Oswaldo Cruz 104, 234–240 [DOI] [PubMed] [Google Scholar]
- 62. Hunn J. P., Feng C. G., Sher A., Howard J. C. (2011) The immunity-related GTPases in mammals. A fast-evolving cell-autonomous resistance system against intracellular pathogens. Mamm. Genome 22, 43–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wehner M., Herrmann C. (2010) Biochemical properties of the human guanylate binding protein 5 and a tumor-specific truncated splice variant. FEBS J. 277, 1597–1605 [DOI] [PubMed] [Google Scholar]
- 64. Kunzelmann S., Praefcke G. J., Herrmann C. (2005) Nucleotide binding and self-stimulated GTPase activity of human guanylate-binding protein 1 (hGBP1). Methods Enzymol. 404, 512–527 [DOI] [PubMed] [Google Scholar]
- 65. Ghosh A., Praefcke G. J., Renault L., Wittinghofer A., Herrmann C. (2006) How guanylate-binding proteins achieve assembly stimulated processive cleavage of GTP to GMP. Nature 440, 101–104 [DOI] [PubMed] [Google Scholar]
- 66. Bigay J., Deterre P., Pfister C., Chabre M. (1985) Fluoroaluminates activate transducin-GDP by mimicking the γ-phosphate of GTP in its binding site. FEBS Lett. 191, 181–185 [DOI] [PubMed] [Google Scholar]
- 67. Byrnes L. J., Sondermann H. (2011) Structural basis for the nucleotide-dependent dimerization of the large G protein atlastin-1/SPG3A. Proc. Natl. Acad. Sci. U.S.A. 108, 2216–2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Schwemmle M., Staeheli P. (1994) The interferon-induced 67-kDa guanylate-binding protein (hGBP1) is a GTPase that converts GTP to GMP. J. Biol. Chem. 269, 11299–11305 [PubMed] [Google Scholar]
- 69. Neun R., Richter M. F., Staeheli P., Schwemmle M. (1996) GTPase properties of the interferon-induced human guanylate-binding protein 2. FEBS Lett. 390, 69–72 [DOI] [PubMed] [Google Scholar]
- 70. Bian X., Klemm R. W., Liu T. Y., Zhang M., Sun S., Sui X., Liu X., Rapoport T. A., Hu J. (2011) Structures of the atlastin GTPase provide insight into homotypic fusion of endoplasmic reticulum membranes. Proc. Natl. Acad. Sci. U.S.A. 108, 3976–3981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kunzelmann S., Praefcke G. J., Herrmann C. (2006) Transient kinetic investigation of GTP hydrolysis catalyzed by interferon-γ-induced hGBP1 (human guanylate binding protein 1). J. Biol. Chem. 281, 28627–28635 [DOI] [PubMed] [Google Scholar]
- 72. Herskovits J. S., Burgess C. C., Obar R. A., Vallee R. B. (1993) Effects of mutant rat dynamin on endocytosis. J. Cell Biol. 122, 565–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Horisberger M. A. (1992) Interferon-induced human protein MxA is a GTPase that binds transiently to cellular proteins. J. Virol. 66, 4705–4709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Sigal I. S., Gibbs J. B., D'Alonzo J. S., Temeles G. L., Wolanski B. S., Socher S. H., Scolnick E. M. (1986) Mutant ras-encoded proteins with altered nucleotide binding exert dominant biological effects. Proc. Natl. Acad. Sci. U.S.A. 83, 952–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Majumdar S., Ramachandran S., Cerione R. A. (2006) New insights into the role of conserved, essential residues in the GTP binding/GTP hydrolytic cycle of large G proteins. J. Biol. Chem. 281, 9219–9226 [DOI] [PubMed] [Google Scholar]
- 76. Marks B., Stowell M. H., Vallis Y., Mills I. G., Gibson A., Hopkins C. R., McMahon H. T. (2001) GTPase activity of dynamin and resulting conformation change are essential for endocytosis. Nature 410, 231–235 [DOI] [PubMed] [Google Scholar]
- 77. Hunn J. P., Koenen-Waisman S., Papic N., Schroeder N., Pawlowski N., Lange R., Kaiser F., Zerrahn J., Martens S., Howard J. C. (2008) Regulatory interactions between IRG resistance GTPases in the cellular response to Toxoplasma gondii. EMBO J. 27, 2495–2509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Britzen-Laurent N., Bauer M., Berton V., Fischer N., Syguda A., Reipschläger S., Naschberger E., Herrmann C., Stürzl M. (2010) Intracellular trafficking of guanylate-binding proteins is regulated by heterodimerization in a hierarchical manner. PLoS One 5, e14246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Fres J. M., Müller S., Praefcke G. J. (2010) Purification of the CAAX-modified, dynamin-related large GTPase hGBP1 by coexpression with farnesyltransferase. J. Lipid Res. 51, 2454–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Fux B., Rodrigues C. V., Portela R. W., Silva N. M., Su C., Sibley D., Vitor R. W., Gazzinelli R. T. (2003) Role of cytokines and major histocompatibility complex restriction in mouse resistance to infection with a natural recombinant strain (type I-III) of Toxoplasma gondii. Infect. Immun. 71, 6392–6401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Martens S., Parvanova I., Zerrahn J., Griffiths G., Schell G., Reichmann G., Howard J. C. (2005) Disruption of Toxoplasma gondii parasitophorous vacuoles by the mouse p47-resistance GTPases. PLoS Pathog. 1, e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Butcher B. A., Greene R. I., Henry S. C., Annecharico K. L., Weinberg J. B., Denkers E. Y., Sher A., Taylor G. A. (2005) p47 GTPases regulate Toxoplasma gondii survival in activated macrophages. Infect. Immun. 73, 3278–3286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Guenzi E., Töpolt K., Lubeseder-Martellato C., Jörg A., Naschberger E., Benelli R., Albini A., Stürzl M. (2003) The guanylate binding protein-1 GTPase controls the invasive and angiogenic capability of endothelial cells through inhibition of MMP-1 expression. EMBO J. 22, 3772–3782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Modiano N., Lu Y. E., Cresswell P. (2005) Golgi targeting of human guanylate-binding protein-1 requires nucleotide binding, isoprenylation, and an IFN-γ-inducible cofactor. Proc. Natl. Acad. Sci. U.S.A. 102, 8680–8685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Virreira Winter S., Niedelman W., Jensen K. D., Rosowski E. E., Julien L., Spooner E., Caradonna K., Burleigh B. A., Saeij J. P., Ploegh H. L., Frickel E. M. (2011) Determinants of GBP recruitment to Toxoplasma gondii vacuoles and the parasitic factors that control it. PLoS One 6, e24434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Vöpel T., Kunzelmann S., Herrmann C. (2009) Nucleotide-dependent cysteine reactivity of hGBP1 uncovers a domain movement during GTP hydrolysis. FEBS Lett. 583, 1923–1927 [DOI] [PubMed] [Google Scholar]
- 87. Damke H., Binns D. D., Ueda H., Schmid S. L., Baba T. (2001) Dynamin GTPase domain mutants block endocytic vesicle formation at morphologically distinct stages. Mol. Biol. Cell 12, 2578–2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Song B. D., Leonard M., Schmid S. L. (2004) Dynamin GTPase domain mutants that differentially affect GTP binding, GTP hydrolysis, and clathrin-mediated endocytosis. J. Biol. Chem. 279, 40431–40436 [DOI] [PubMed] [Google Scholar]
- 89. Song B. D., Yarar D., Schmid S. L. (2004) An assembly incompetent mutant establishes a requirement for dynamin self-assembly in clathrin-mediated endocytosis in vivo. Mol. Biol. Cell 15, 2243–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Pitossi F., Blank A., Schröder A., Schwarz A., Hüssi P., Schwemmle M., Pavlovic J., Staeheli P. (1993) A functional GTP-binding motif is necessary for antiviral activity of Mx proteins. J. Virol. 67, 6726–6732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ponten A., Sick C., Weeber M., Haller O., Kochs G. (1997) Dominant-negative mutants of human MxA protein. Domains in the carboxy-terminal moiety are important for oligomerization and antiviral activity. J. Virol. 71, 2591–2599 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.