Background: The late endosomal/lysosomal transmembrane protein LAPTM5 is expressed in hematopoietic cells.
Results: LAPTM5 facilitates activation of NF-κB and MAPK signaling and proinflammatory cytokine release mediated by cytokine and pattern recognition receptors.
Conclusion: LAPTM5 is a positive regulator of proinflammatory responses by macrophages.
Significance: The late endosomal/lysosomal system plays a role in the regulation of inflammatory responses by macrophages.
Keywords: Cytokine, Endosomes, Lysosomes, Macrophages, NF-kappa B (NF-KB), Ubiquitin, A20, Nedd4
Abstract
LAPTM5 (lysosomal-associated protein transmembrane 5) is a protein that is preferentially expressed in immune cells, and it interacts with the Nedd4 family of ubiquitin ligases. Recent studies in T and B cells identified LAPTM5 as a negative regulator of T and B cell receptor levels at the plasma membrane. Here we investigated the function of LAPTM5 in macrophages. We demonstrate that expression of LAPTM5 is required for the secretion of proinflammatory cytokines in response to Toll-like receptor ligands. We also show that RAW264.7 cells knocked down for LAPTM5 or macrophages from LAPTM5−/− mice exhibit reduced activation of NF-κB and MAPK signaling pathways mediated by the TNF receptor, as well as multiple pattern recognition receptors in various cellular compartments. TNF stimulation of LAPTM5-deficient macrophages leads to reduced ubiquitination of RIP1 (receptor-interacting protein 1), suggesting a role for LAPTM5 at the receptor-proximate level. Interestingly, we find that macrophages from LAPTM5−/− mice display up-regulated levels of A20, a ubiquitin-editing enzyme responsible for deubiquitination of RIP1 and subsequent termination of NF-κB activation. Our studies thus indicate that, in contrast to its negative role in T and B cell activation, LAPTM5 acts as a positive modulator of inflammatory signaling pathways and hence cytokine secretion in macrophages. They also highlight a role for the endosomal/lysosomal system in regulating signaling via cytokine and pattern recognition receptors.
Introduction
LAPTM5 (lysosomal-associated protein transmembrane 5) is a 29-kDa protein preferentially expressed in cells of lymphoid and myeloid origin (1, 2). The intracellular domains of LAPTM5 contain three PY ((L/P)PXY) motifs, which bind the WW domains present within the Nedd4 family of ubiquitin ligases, and a ubiquitin-interacting motif (UIM).3 Our earlier studies showed that LAPTM5 trafficking from the Golgi to the late endosome/lysosome requires its association with the ubiquitin ligase Nedd4 and the clathrin adaptor GGA3. Although the Nedd4-LAPTM5 interaction leads to ubiquitination of LAPTM5, this event is not necessary for LAPTM5 sorting. Rather, the Nedd4-LAPTM5 complex recruits ubiquitinated GGA3, which binds the UIM of LAPTM5 and targets it to the lysosome (3). Recent studies have indicated that LAPTM5 negatively regulates T and B cell receptor signaling by directly interacting with these receptors and mediating their down-regulation (4, 5). However, the role of LAPTM5 in other immune cells remains unknown. In this study, we investigated the function of LAPTM5 in macrophages.
Macrophages are at the first line of defense against invading microbial pathogens. In response to infection, activated macrophages produce cytokines to initiate the inflammatory response. This process depends largely on the activation of nuclear factor-κB (NF-κB) family of transcription factors, and ubiquitination has emerged as an important regulatory mechanism for NF-κB signaling (6).
Macrophages express several pattern recognition receptors that detect pathogen-associated molecular patterns present on bacteria, viruses, and parasites. Recognition of pathogen-associated molecular patterns is mediated at the cell surface or intracellularly by a variety of receptors, including transmembrane Toll-like receptors (TLRs) and cytoplasmic nucleotide oligomerization domain (NOD)-like receptors (7). Under resting conditions, the NF-κB heterodimers, composed of p65 and p50 subunits, are retained within the cytoplasm through interaction with the IκBs (inhibitors of κB). The cytosolic NF-κB complexes are activated upon recognition of specific pathogen-associated molecular patterns such as LPS and muramyl dipeptide (MDP), viral nucleic acids, as well as the proinflammatory cytokine TNFα. These ligands trigger distinct signaling cascades that converge at the IκB kinase (IKK) complex composed of IKKα, IKKβ, and the regulatory subunit IKKγ (also known as NEMO). Activated IKK phosphorylates IκBα, triggering Lys48-linked ubiquitination and its subsequent proteasomal degradation, thus allowing the entry of NF-κB into the nucleus to activate transcription of a variety of inflammatory target genes. In the process, the NF-κB subunits are also phosphorylated within their transactivation domain, an event that is necessary for optimal induction of target genes (7–10). In addition to activation of NF-κB, both LPS and TNFα are potent inducers of MAPK pathways (11).
Ubiquitination has been shown to play critical regulatory roles at multiple steps of NF-κB activation. In the TNF receptor 1 (TNFR1) pathway, stimulation with TNFα leads to the recruitment of several signaling proteins, including the adaptor protein TRADD, the TNF receptor-associated factors (TRAF2 and TRAF5), and RIP1 (receptor interacting protein kinase 1). Formation of the receptor complex leads to Lys63-linked polyubiquitination of RIP1, which serves as a docking site for the ubiquitin-binding domain of IKKγ (12). To ensure transient NF-κB activation, the ubiquitin-editing enzyme A20 removes the Lys63-ubiqitin chains, thus eliminating the signal for activation of downstream components (13). In addition to TNFα-mediated NF-κB activation, A20 has emerged as an important component of the negative feedback loop for pattern recognition receptors (14, 15). Because insufficient activation of NF-κB can lead to impaired host defense, whereas persistent NF-κB responses can drive inflammatory disorders and malignancy, proper regulation of this pathway is of critical importance (16).
In this study, we demonstrate that LAPTM5 is a positive regulator of NF-κB and MAPK signaling that allows efficient proinflammatory cytokine production in response to several inducers of macrophage activation. During TNFα stimulation, LAPTM5 is required for proper initiation of NF-κB and MAPK signaling by acting at the receptor-proximate level. Thus, our findings suggest that LAPTM5 is an important component of inflammatory signaling pathways in macrophages and highlight a role for the endosomal/lysosomal system in regulating these cascades.
EXPERIMENTAL PROCEDURES
Antibodies, Reagents, and Constructs
The following antibodies were used: rabbit phospho-NF-κB p65 (93H1), rabbit NF-κB p65 (3034), and rabbit IκBα (44D4) from Cell Signaling; mouse p38 MAPK (Thr(P)180/Tyr(P)182), mouse p38α/SAPK2a, hamster TNFR1 (CD120a/55R-286), and mouse RIP1 (38/RIP) from BD Transduction; mouse β-actin (A2228), mouse anti-FLAG, mouse anti-HA, and rabbit LAMP2 (L0668) from Sigma; mouse TNFR1 (H-5) and mouse A20 (A-12) from Santa Cruz; anti-mouse/rat TNFα (TN3-19), anti-mouse IL12/IL23p40 (C15.6), and anti-mouse IL6 (MP5-20F3) from eBioscience; biotin-conjugated IL12/IL23 p40 (C17.8), IL6 (MP5–32C11), and TNFα from eBioscience; rabbit active MAPK (V8031) and rabbit Erk1/2 (V1141) from Promega; rabbit JNK1 (OPA1-03076) and rabbit phospho-JNK1/2 (Thr(P)183 and Tyr(P)185) from ABR; mouse His (34660) from Qiagen; mouse ubiquitin (MMS-258R) from Covance; rat LAMP1(H4A3) from Developmental Studies Hybridoma Bank, University of Iowa. Alexa 488-labeled transferrin was obtained from Molecular Probes. Polyclonal anti-LAPTM5 antibody was described earlier (3). Secondary antibodies conjugated to fluorophores or horseradish peroxidase were from Molecular Probes. The following reagents and cDNAs were also used: MDP and inactive MDP control (InvivoGen), LPS (L2654; Sigma), CpG DNA (Integrated DNA Technologies), bafilomycin A1 (Sigma), ammonium chloride (Sigma), leupeptin (BioShop), and lactacystin (Sigma). The full-length mouse A20 cDNA (GC-Mm05770) was obtained from GeneCopoeia and cloned into N-terminally tagged mCherry and FLAG expression vectors using Gateway system (Invitrogen). For bacterial expression, the A20 ZnF4–7 (amino acids 550–775) was cloned into pDest17-His6 Gateway vector (Invitrogen), and the C terminus of LAPTM5 (amino acids 206–261) was cloned into pGEX2TK using standard PCR and restriction enzyme cloning. HA-LAPTM5 was previously described (3). For retrovirus production, HA-LAPTM5 was cloned into the LZRS-pBMN (GFP-IRES) retroviral vector using the ClaI and BamHI sites.
Cell Culture and Transfections
RAW264.7 (kind gift from Dr. S. Grinstein, The Hospital for Sick Children, Toronto), HEK293T (ATCC), and HeLa (ATCC) cells were grown under standard conditions in DMEM supplemented with 10% FCS, 100 units/ml penicillin, and 100 μg/ml streptomycin. HeLa cells were transfected using Lipofectamine 2000 (Invitrogen), and HEK293T cells were transfected using standard calcium phosphate transfection.
Generation of Stable Knockdown Cell Lines and siRNAs
The hairpin sequence against mouse LAPTM5 (5′-CCCTGTGTCATTGCTTGTGTAT-3′) was generated using the shRNA Retriever program (available online). Template oligonucleotide incorporating miR-30 microRNA sequences was synthesized (Sigma) and inserted into pSM2 vector according to Ref. 17. The shRNA was then cloned from pSM2 into pGIPZ lentiviral vector (Open Biosystems) following the manufacturer's instructions. The mouse pGIPZ nonsilencing control (RHS4346) was purchased from Open Biosystems. Lentivirus production was performed according to the manufacturer's protocol (Open Biosystems). Transduction of RAW264.7 cells was carried out in 6-well plates with the addition of 5 μg/ml polybrene and centrifuged for 2 h at 2500 rpm, 32 °C. The cells were cultured in DMEM as described above and supplemented with 1 μg/ml puromycin to select for pGIPZ-expressing cells. For siRNA-mediated knockdown experiments, negative control siRNA (1027310) was purchased from Qiagen. LAPTM5 siRNA duplexes (5′-CUGUGUCAUUGCUUGUGUA-3′) with 3′-dTdT overhangs were synthesized at Qiagen. RAW264.7 cells were transfected with siRNAs using Lipofectamine 2000 (Invitrogen) following the manufacturer's protocol.
Isolation of Bone Marrow-derived Macrophages (BMDMs) and Retroviral Transduction
BMDMs were prepared by culture of bone marrow isolated from femurs and tibias of age-matched (12–14 weeks old) C57BL/6 wild-type and LAPTM5−/− mice (5). After red blood cell lysis, the cells were cultured in RPMI 1640 medium supplemented with 20% FBS, 100 units/ml penicillin, 100 μg/ml streptomycin, and 40 ng/ml macrophage colony-stimulating factor (R & D Systems). After 7 days of culture, BMDMs were tested for purity by flow cytometry using F4/80 and CD11b antibodies (BD Pharmingen). Macrophage purity was above 80%. LPS stimulations were performed between 7 and 9 days of cell culture. Experimental and animal care were performed in accordance with institutional guidelines. For retroviral transduction, Plat-E packaging cell line (kind gift from Dr. M. Marks, University of Pennsylvania, Philadelphia, PA) was transfected with retroviral vectors. Retrovirus was harvested 48 h after transfection, supplemented with 4 μg/ml polybrene, and added to BMDM cultures. Tissue culture plates were spun in a table top centrifuge at 32 °C, 1200 × g for 2 h. Virus was removed, and fresh medium was added. Expression was assayed 72 h after infection.
Quantitative Real Time PCR
Total RNA was isolated using RNeasy kit (Qiagen), digested on-column with DNase, and 1 μg of total RNA was converted to cDNA using SuperScript VILO (Invirogen) following the manufacturer's protocol. A comparative Ct experiment was performed using TaqMan gene expression assay reagents on a ViiA 7 real time PCR instrument (Applied Biosystems). The following TaqMan probes were used: Laptm5 (Mm04208394_m1), Tnfaip3/A20 (Mm00437121_m1), and Gusb (Mm00446953_m1). Gusb was used as endogenous control.
ELISA and NO Production
RAW264.7 or BMDMs were seeded at density of 5 × 105 cells/ml. The following day cells were left untreated or stimulated with LPS or CpG for 24 h, and the supernatant was subjected to ELISA. The IL6, TNFα, and IL12p40 ELISA were performed according to the manufacturer's instructions (eBioscience). The amount of cytokines was calculated from standard curve derived from recombinant mouse IL6, TNFα, or IL12p40 (R & D Systems). For NO production the supernatants were analyzed for nitrite oxidant using a Griess Reagent kit (Promega) according to the manufacturer's protocol.
Immunofluorescent Confocal Microscopy
RAW264.7 or HeLa cells were fixed in 4% paraformaldehyde, immunostained with the indicated antibodies, and analyzed using LSM510 Zeiss confocal microscope as described previously (3). Co-localization between LAPTM5 and LAMP1 was quantified using Volocity 5.4.1 software and expressed as quantification coefficient (M2), corresponding to degrees of freedom (r = 20) and level of significance (p < 0.05 for unstimulated and p < 0.01 for LPS).
Co-immunoprecipitation, Pulldown, and in Vitro Binding Assays
For analysis of phosphoproteins, RAW264.7 cells or BMDMs were stimulated with LPS, TNFα, or MDP for the indicated amount of time, placed on ice, and washed with ice-cold PBS. The cells were lysed in lysis buffer (150 mm NaCl, 50 mm HEPES, 10% glycerol, 1% Triton X-100, 2 mm EDTA, 10 μg/ml leupeptin, 10 μg/ml aprotinin, 1 μg/ml pepstatin A, 1 mm PMSF, and 1 mm Na3VO4) and cleared by centrifugation at 14,000 rpm for 30 min. Equal amounts of proteins were resolved by SDS-PAGE, transferred to nitrocellulose membrane, and analyzed by immunoblotting with the indicated antibodies, followed by secondary antibodies and ECL detection (GE Healthcare). For co-immunoprecipitation, HEK293T cell lysates expressing transfected FLAG-A20 and/or HA-LAPTM5 (2 mg each) were incubated overnight with 10 μl of anti-FLAG M2 affinity gel (Sigma). Bound proteins were washed once with lysis buffer and three times with HNTG (150 mm NaCl, 20 mm HEPES, pH 7.5, 10% glycerol, and 0.1% Triton X-100), eluted with 1× SDS-PAGE sample buffer. Bound LAPTM5 was detected with anti-HA antibody. To identify RIP1 in the complex with TNFR1, RAW264.7 cells were transfected with control or LAPTM5 siRNAs. At 72 h after transfection, the cells were treated with TNFα for the indicated time intervals, and the cell lysates were prepared as described above. To immunoprecipitate the TNFR1, cell lysates (4 mg each) were incubated overnight at 4 °C with a mixture of mouse and hamster anti-TNFR1 antibodies (7.5 μg of each) and 15 μl of protein G-agarose beads (BioShop). The beads were washed, and the complexes were eluted as described above. For pulldown of endogenous A20, GST fusion proteins were produced in bacteria and purified on glutathione-agarose beads (Sigma). 50 μg of GST or GST C terminus (LAPTM5) were incubated with 2 mg of RAW264.7 cell lysate for 4 h at 4 °C. The beads were washed, and the samples were eluted as described above. For in vitro binding, GST fusion proteins were generated as described above. His-tagged ZnF4–7 (A20) was produced in bacteria and purified on Ni2+-agarose beads (Qiagen). To detect direct binding, 50 μg of His-ZnF4–7 (A20) was incubated with 50 μg of GST or GST-LAPTM5-C-term in PBS with 10% glycerol for 1 h. The complexes were washed four times with HNTG, and His-ZnF4–7 was eluted with 1× elution buffer (0.5 m imidazole, 0.3 m NaCl, and 20 mm Tris, pH 7.9).
RESULTS
LPS Stimulation Affects Protein Stability and Localization of LAPTM5 in Macrophages
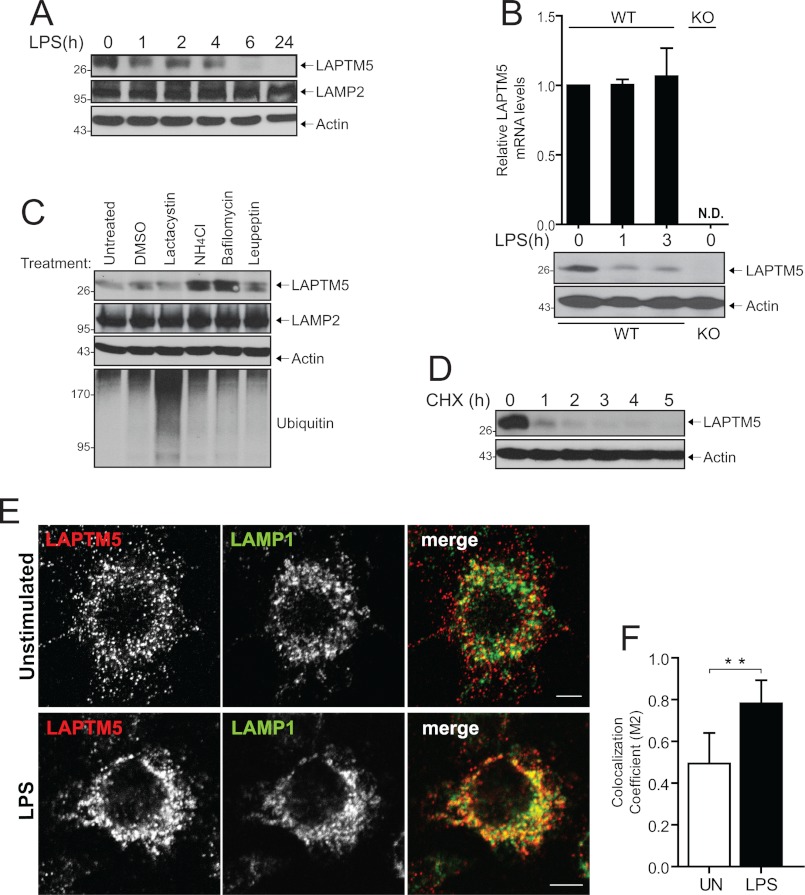
To explore whether LAPTM5 plays a role in the regulation of macrophage activation, we first sought to examine whether inflammatory stimuli affect LAPTM5 expression. For this, we used an anti-LAPTM5 antibody that was previously described by our laboratory (3). To confirm the specificity of the antibody, we silenced the expression of LAPTM5 in RAW264.7 cells, a murine macrophage cell line, by transient transfection of silencing RNA duplexes (siRNA; supplemental Fig. S1A), as well as generated a stable cell line expressing shRNA against LAPTM5 (supplemental Fig. S1B). Interestingly, we found that stimulation of RAW264.7 cells with LPS, the ligand for the cell surface receptor TLR4, resulted in down-regulation of LAPTM5 protein, decreasing slightly within 1 h and disappearing by 6 h of stimulation (Fig. 1A). To confirm the results obtained in RAW264.7 cells, we tested the expression levels of LAPTM5 protein in primary macrophages under acute stimulation conditions. Consistent with the data above, LPS stimulation of BMDMs led to a dramatic decrease of LAPTM5 protein within 1 h of exposure to the ligand (Fig. 1B). In parallel, we performed quantitative real time PCR analysis to establish whether the changes seen at the protein level are caused by changes in mRNA. LAPTM5 mRNA levels remained unchanged over the course of stimulation (Fig. 1B). These data indicate that LPS induces down-regulation of LAPTM5 protein levels, but not mRNA levels. To determine whether the LAPTM5 protein is degraded via the lysosomal or proteasomal pathways, we treated RAW264.7 cells with various inhibitors of degradation. As shown in Fig. 1C, inhibition of lysosomal degradation using ammonium chloride, bafilomycin, or leupeptin led to accumulation of LAPTM5 protein levels, which was not observed upon lactacystin-mediated inhibition of the proteasome. In contrast, the levels of the well characterized lysosomal glycoprotein LAMP2 remained unaffected by these treatments (Fig. 1C). Next, we sought to determine the stability of LAPTM5 in macrophages using cycloheximide to block new protein synthesis. As shown in Fig. 1D, the LAPTM5 pool was almost entirely depleted within 1 h of cycloheximide treatment. Because stimulation of macrophages with LPS affected LAPTM5 expression, we sought to examine whether the cellular distribution of LAPTM5 changes upon macrophage activation. Prior work from our laboratory, as well as others, has demonstrated that LAPTM5 localizes to the late endosomal/lysosomal membranes (1, 3–5). To verify the subcellular localization of endogenous LAPTM5 in macrophages, we performed immunofluorescence analysis in RAW264.7 cells. As seen in Fig. 1 (E, top panels, and F), double immunostaining of resting cells with the late endosomal/lysosomal marker LAMP1 revealed the presence of some of the LAPTM5 molecules within lysosomes. However, a substantial population of LAPTM5-positive vesicles appeared close to but not overlapping with LAMP1-positive vesicles. To address the possibility that LAPTM5 is associated with early endosomes, we performed transferrin uptake. As shown in supplemental Fig. S2, LAPTM5 does not co-localize with internalized transferrin. However, following 15 min of LPS stimulation, we observed a redistribution of LAPTM5-positive vesicles, resulting in a substantial increase in the co-localization of LAPTM5 with LAMP1 (Fig. 1E, bottom panels, and F). Collectively, these results indicate that, in macrophages, LAPTM5 has a rapid turnover and is targeted to the lysosome for degradation rather than being a resident lysosomal transmembrane protein. Moreover, LPS stimulation targets LAPTM5 to the lysosomes for degradation.
FIGURE 1.
LAPTM5 stability and localization in macrophages. A, LPS induces down-regulation of LAPTM5. RAW264.7 cells were stimulated with 1 μg/ml LPS for the indicated times. The cells were lysed and immunoblotted with antibodies toward LAPTM5, LAMP2, or actin. B, LAPTM5 protein, but not mRNA, is rapidly down-regulated in BMDMs. BMDMs from WT or LAPTM5 knock-out (KO) mice were stimulated with 100 ng/ml LPS, in duplicate. At the indicated time points, total RNA was isolated, reverse transcribed, and analyzed by quantitative PCR (upper graph), or cells were lysed and total lysates were immunoblotted with anti-LAPTM5 and anti-actin antibodies (lower Western blot panels). N.D. indicates that no LAPTM5 transcripts above base line were detected in KO. The data are presented as the means ± S.D. (n = 4). The results are representative of two independent experiments. C, inhibition of lysosomal degradation leads to accumulation of LAPTM5. RAW264.7 cells were treated for 3 h with 5 μm lactacystine, 20 mm NH4Cl, 500 nm bafilomycin, 500 nm leupeptin, or dimethyl sulfoxide (DMSO) control. Total cell lysates were immunoblotted with antibodies toward LAPTM5, LAMP2, or ubiquitin. Actin is shown as a control for protein loading. D, LAPTM5 is rapidly degraded in macrophages. RAW264.7 cells were treated with 10 μm cycloheximide (CHX). After the indicated time intervals, the cells were lysed, and the levels of LAPTM5 were detected with anti-LAPTM5 antibodies. Actin controls are shown as well. E, LAPTM5 localizes to lysosome-associated compartments under basal conditions, and LPS stimulation targets LAPTM5 to the lysosomes. Confocal fluorescence analysis of unstimulated RAW264.7 cells (top panels) or stimulated with 1 μg/ml LPS for 15 min (bottom panels) immunostained with anti-LAPTM5 and anti-LAMP1 antibodies to label the late endosome/lysosome. Representative confocal images are shown. The merged panels show an overlay of the two channels. Bar, 5 μm. F, quantification of co-localization between LAPTM5 and LAMP1. Quantification of 22 individual cells, randomly selected and unstimulated (UN) or stimulated with 1 μg/ml LPS for 15 min (LPS), as shown in E. The extent of co-localization between LAPTM5 and LAMP1 is indicated as co-localization coefficient (M2). Statistical analysis was performed using Student's t test. The data are expressed as the means ± S.D. **, p < 0.001.
LAPTM5 Is Required for TLR-mediated Inflammatory Cytokine Production in Macrophages
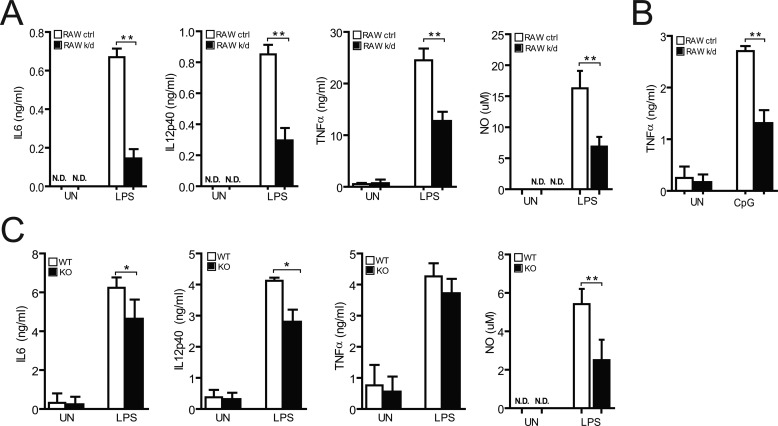
To test whether LAPTM5 is involved in the activation of macrophages, we examined the role of LAPTM5 in the induction of various proinflammatory cytokines and NO in response to LPS. RAW264.7 cells expressing shRNA against LAPTM5 produced 4-fold less IL6 and less than half IL12, TNFα, and NO than did control cells (Fig. 2A). To determine whether LAPTM5 function extends to intracellular TLR family members, we stimulated RAW cells with CpG unmethylated DNA to engage signaling through TLR9, which resides within the endosomal compartments (18). As shown in Fig. 2B, CpG-induced production of TNFα was significantly reduced in LAPTM5-deficient cells. We further explored the effect of LPS on cytokine production in BMDMs from LAPTM5 wild-type (LAPTM5+/+) and knock-out (LAPTM5−/−) mice. Although LAPTM5−/− BMDMs produced only slightly less TNFα after treatment with LPS, the secretion of IL6, IL12, and NO was significantly lower in these cells compared with wild-type macrophages (Fig. 2C). Collectively, these results indicate that LAPTM5 is necessary for efficient production of the proinflammatory cytokines IL6, IL12, TNFα, and NO in response to stimulation of both endosomal and plasma membrane resident Toll-like receptors.
FIGURE 2.
LAPTM5 is required for TLR-mediated cytokine secretion in macrophages. A and B, stable RAW264.7 cells expressing control shRNA (RAW ctrl, white columns) or shRNA against LAPTM5 (RAW k/d, black columns) were unstimulated (UN) or stimulated with 1 μg/ml LPS (to activate TLR4) (A) or 50 μg/ml CpG (to activate TLR9) (B). Culture supernatants were collected after 24 h and the production of IL6 or IL12p40 (A). TNFα was determined by ELISA (A and B) and NO (A) by Griess reaction assay. N.D. indicates that no IL6, IL12p40, or NO above base line was detected (A). C, analysis of proinflammatory mediators from BMDMs. BMDMs form WT (white columns) and LAPTM5 knock-out (KO, black columns) mice were stimulated with 100 ng/ml LPS for 24 h, after which the supernatants were harvested and analyzed by ELISA (IL6, IL12p40, and TNFα), or Griess reaction assay (NO). A–C, statistical analysis was performed using Student's t test. The data show the means ± S.D. of three independent experiments. *, p < 0.01; **, p < 0.001).
LAPTM5 Positively Regulates LPS-induced NF-κB and MAPK Activation in Macrophages
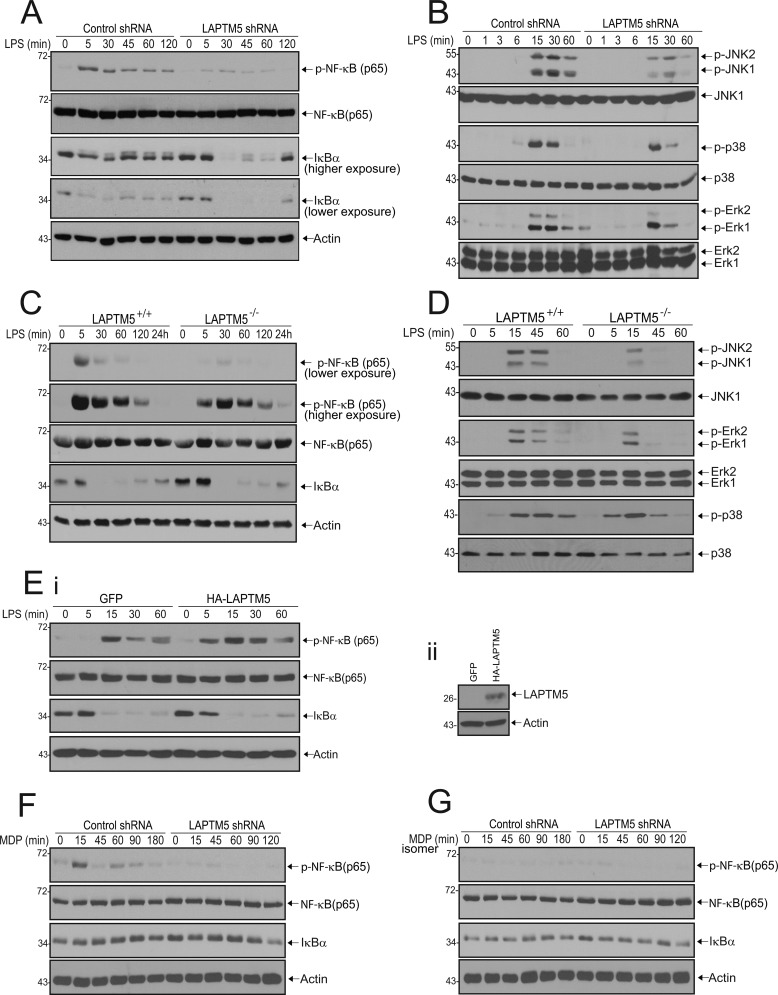
To decipher the mechanisms by which LAPTM5 modulates TLR signaling, we stimulated RAW264.7 cells with LPS, prepared cell lysates at various time points, and examined whether LAPTM5 deficiency affects the phosphorylation status of NF-κB by immunoblotting for the p65 subunit. As seen in Fig. 3A, LPS stimulation of control cells resulted in rapid phosphorylation of p65 (pNF-κB (p65)), peaking at 5 min and gradually decreasing upon prolonged exposure to the ligand. In contrast, in cells expressing shRNA against LAPTM5, p65 phosphorylation was markedly reduced. We next examined the lysates for the presence of IκBα protein. LPS treatment of cells expressing control shRNA triggered moderate degradation of IκBα, followed by resynthesis at 45 min after stimulation, because IκBα is under the transcriptional control of NF-κB (19). However, in LAPTM5-deficient RAW cells, replenishment of IκBα pool was not observed until 120 min after stimulation (Fig. 3A). These results suggest that in RAW264.7 cells, LAPTM5 facilitates activation of NF-κB by TLR4. In addition to NF-κB, LPS is a potent activator of MAPK pathways (11). We thus sought to determine whether LAPTM5 is also required for induction of these signaling cascades. In the presence of control shRNA, LPS triggered transient phosphorylation of JNK, p38, and ERK that persisted for up to 60 min. Knockdown of LAPTM5, however, led to a considerable reduction of JNK, p38, and ERK phosphorylation, already seen at 30 min after stimulation (Fig. 3B). These changes were not caused by protein degradation, because total protein levels were not affected. To further verify the involvement of LAPTM5 in LPS-mediated NF-κB and MAPK activity, we examined its effects in BMDMs from LAPTM5−/− mice. Consistent with our findings in RAW264.7 cells, LPS-induced phosphorylation of p65 in LAPTM5−/− BMDMs was reduced compared with LAPTM5+/+ BMDMs (Fig. 3C). In addition, we observed a delay in NF-κB activation seen by a shift in the peak of p65 phosphorylation from 5 min in LAPTM5+/+ to 30 min in LAPTM5−/− macrophages. Furthermore, LAPTM5−/− BMDMs also exhibited reduced JNK, ERK, and p38 activation (Fig. 3D).
FIGURE 3.
LAPTM5 is required for TLR4 and NOD2-mediated activation of NF-κB and for TLR4-dependent MAPK signaling. A and B, knockdown of LAPTM5 leads to reduced TLR4-mediated NF-κB and MAPK activation. Stable RAW264.7 cells expressing control or LAPTM5 shRNA were stimulated with 1 μg/ml LPS for the indicated times. The cells were lysed and analyzed by immunoblotting with antibodies toward phosphorylated NF-κB (p-NF-κB (p65)), NF-κB (p65), IκBα, and actin (A) and phosphorylated JNK1/2 (p-JNK1/2), p38 (p-p38), and Erk1/2 (p-Erk1/2) and the respective total levels of these downstream effectors (B). C and D, knock-out of LAPTM5 leads to reduced TLR4-dependent NF-κB and MAPK activation. BMDMs from wild-type (LAPTM5+/+) and LAPTM5 knock-out (LAPTM5−/−) mice were stimulated with 100 ng/ml LPS, lysed at the designated time points, and subjected to immunoblot analysis with antibodies as indicated in A. E, exogenous expression of LAPTM5 in LAPTM5−/− BMDMs partially rescues NF-κB activation. LAPTM5−/− BMDMs were transduced with a retrovirus expressing GFP alone (GFP) or GFP-IRES HA-tagged LAPTM5 (HA-LAPTM5). Panel i, cells were stimulated with 100 ng/ml LPS, lysed at the indicated time points, and subjected to immunoblot analysis with antibodies as indicated in A. Panel ii, expression of HA-tagged LAPTM5. The cell lysates were immunoblotted with anti-HA antibodies to detect exogenous HA-LAPTM5 or actin antibodies as a control for protein loading. F and G, LAPTM5 knockdown results in reduced NOD2-mediated NF-κB activation. Stable RAW264.7 cells expressing control or LAPTM5 shRNA were stimulated with 10 μg/ml MDP (D) or 10 μg/ml inactive MDP isomer (E). After the indicated time periods, the cells were lysed and immunoblotted as in A.
Next, we examined whether exogenous expression of LAPTM5 in BMDMs from LAPTM5 knock-out mice could rescue the defects seen in NF-κB activation. For these studies, HA-tagged LAPTM5 (or GFP alone) was introduced into LAPTM5−/− BMDMs using a retroviral vector. As seen in Fig. 3E, overexpression of LAPTM5 partially rescued NF-κB activation, compared with the GFP-expressing control cells. The observation that only partial rescue was seen could be attributed to the fact that we were unable to restore LAPTM5 expression to the levels seen in wild-type cells. Collectively, these data demonstrate that in macrophages LAPTM5 acts to positively regulate NF-κB and MAPK activation.
LAPTM5 Is Required for NOD2-mediated NF-κB Activation
Activation of NF-κB in macrophages can be triggered by a variety of pattern recognition receptors to allow pathogen recognition at different subcellular compartments. NOD2 is a cytosolic receptor that recognizes bacterial cell wall peptidoglycan MDP (20). Thus, we sought to determine whether LAPTM5 can modulate NOD2-specific NF-κB activation. Because commercial sources of MDP can be contaminated with LPS, we also tested an inactive MDP isomer. As shown in Fig. 3F, in the presence of control shRNA, MDP triggered phosphorylation of p65 that was sustained for 90 min and was specific to NOD2 signaling, because the inactive MDP isomer failed to induce any response (Fig. 3G). By contrast, in cells expressing shRNA against LAPTM5, phosphorylation of p65 was hardly detected after NOD2 stimulation (Fig. 3F). However, in contrast with LPS, we did not observe any changes in IκBα levels, presumably because MDP is a less potent activator of NF-κB in macrophages than LPS (15). Taken together, these results indicate that LAPTM5 regulates NF-κB activation mediated by all of the pattern recognition receptors tested here, independent of their subcellular localization or their specific ligand. These findings also suggest a role for LAPTM5 in a signal transduction event shared by all the three receptors tested.
The Ubiquitin-editing Enzyme A20 Is Up-regulated in LAPTM5-deficient Macrophages
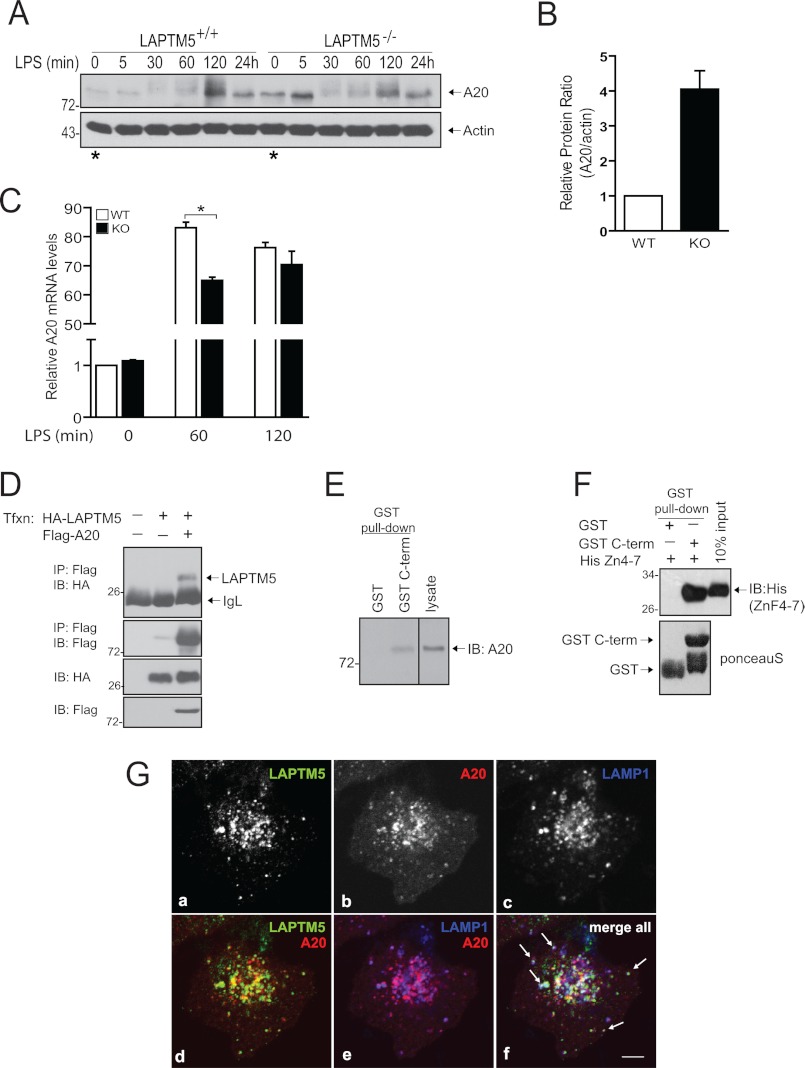
Because LAPTM5 deficiency resulted in a muted inflammatory response, we hypothesized that LAPTM5 may regulate a molecule involved in termination of NF-κB signaling. The ubiquitin-editing enzyme A20 was previously shown to be critical for attenuation of TLR and NOD2-mediated NF-κB responses in macrophages. A20 restricts NF-κB activation by directly removing Lys63-linked polyubiquitin chains from key signal-transducing proteins, leading to destabilization of the central IKK-activating complex (14, 15). Because LAPTM5 was previously reported to interact with A20 in a large scale yeast two-hybrid screen, we first sought to determine whether levels of A20 in macrophages were affected by deletion of LAPTM5 (21). Interestingly, unstimulated LAPTM5−/− BMDMs expressed increased levels of A20 relative to the wild-type BMDMs before stimulation (compare Fig. 4A, time 0 LAPTM5+/+ versus LAPTM5−/− indicated by the asterisks with Fig. 4B). Quantitative real time PCR analysis revealed that the elevated A20 protein level in LAPTM5−/− BMDMs (relative to LAPTM5+/+ cells) was not due to an increase in basal A20 mRNA expression (Fig. 4C). Moreover, in agreement with the defect in NF-κB activation (Fig. 3C), LAPTM5−/− BMDMs displayed (transient) reduced induction of A20 mRNA expression in response to LPS stimulation (Fig. 4C). Similar to results obtained in LAPTM5-deficient BMDMs, we observed increased basal A20 protein levels in RAW264.7 cells following silencing of LAPTM5 with shRNA (supplemental Fig. S3). These results therefore indicate that LAPTM5 is involved in the negative regulation of A20 protein levels in macrophages. To test whether LAPTM5 can bind to A20, we co-expressed full-length HA-LAPTM5 and FLAG-A20 in HEK293T cells and subjected cell lysates to immunoprecipitation with anti-FLAG antibody (A20) and immunoblotting with HA antibodies (LAPTM5). As shown in Fig. 4D, this co-immunoprecipitation experiment revealed the presence of LAPTM5 in complex with A20. To further examine the interaction in macrophages, we performed a GST pulldown assay by incubating purified GST-tagged C terminus of LAPTM5 with RAW264.7 cell lysate. As seen in Fig. 4E, endogenous A20 was able to bind the C terminus of LAPTM5, but not GST alone. Interestingly, this interaction occurred without LPS stimulation, suggesting that LAPTM5 may regulate A20 in a stimulus-independent manner. Next, we aimed to examine whether the binding is direct. Thus, we incubated purified C terminus of LAPTM5 with recombinant His-tagged fragment of A20 that encompasses the last four zinc finger domains (A20 ZnF4–7; the minimal region previously documented to bind LAPTM5 in a yeast two-hybrid screen (21)). As shown in Fig. 4F, A20 ZnF4–7 specifically bound to the C terminus of LAPTM5 but not to control GST beads alone. We further studied the relationship between A20 and LAPTM5 using immunofluorescence. Although A20 is a soluble protein, a recent study has suggested that a fraction of A20 localizes to the late endosomal membranes (22). Therefore, we co-transfected A20 and LAPTM5 in HeLa cells and assessed their localization by co-immunostaining with antibodies toward LAMP1. In addition to cytosolic distribution, A20 clearly localized also to intracellular vesicles (Fig. 4G, panel b) that partially co-localized with LAPTM5 (Fig. 4G, panel d). Moreover, some of these double-positive vesicles also contained LAMP1 (Fig. 4G, panel f). Thus, LAPTM5 could potentially be a direct regulator of A20 through a protein-protein interaction in the endolysosomal system.
FIGURE 4.
LAPTM5 binds A20 and negatively regulates its levels. A, A20 is up-regulated in LAPTM5-deficient macrophages. BMDMs from wild-type (LAPTM5+/+) and LAPTM5 knock-out (LAPTM5−/−) mice were stimulated with 100 ng/ml LPS for the indicated time points. The cells were lysed and subjected to immunoblotting with antibodies toward A20 or actin. Note the increased basal A20 expression in LAPTM5−/− compared with LAPTM5+/+ BMDMs indicated by the asterisks. The data are representative of three independent experiments with similar results. B, quantification of basal A20 protein levels relative to actin from WT (white column) and LAPTM5 knock-out (KO; black column) BMDMs, as shown in A. The graph indicates the increase of A20 protein in LAPTM5 KO relative to WT BMDMs and is presented as the means ± S.D. of three independent experiments. C, quantitative PCR analysis of A20 mRNA levels. WT (white columns) and KO (black columns) BMDMs were stimulated with 100 ng/ml LPS. Total RNA was isolated at the indicated time periods, reverse transcribed, and analyzed by quantitative PCR. Statistical analysis was performed using Student's t test. The data are presented as the means ± S.D. (n = 4; *, p < 0.01 WT versus KO). The results are representative of two independent experiments. D–F, interaction between LAPTM5 and A20. D, co-immunoprecipitation of LAPTM5 with A20. FLAG-tagged A20 was immunoprecipitated (IP) from HEK293T cells co-expressing HA-tagged LAPTM5, and the precipitate was immunoblotted (IB) with anti-HA antibodies to detect LAPTM5. The same membrane was probed with anti-FLAG antibodies to detect A20 (second panel). The bottom panels represent lysate controls for protein expression. IgL, immunoglobulin light chain. E, binding of endogenous A20 to the C terminus (C-term) of LAPTM5. RAW cells lysate was incubated with bacterially purified GST-tagged C terminus of LAPTM5 or GST-beads alone and immunoblotted with anti-A20 antibodies. F, in vitro binding of the last four ZnF motifs of A20 (ZnF4–7) to the C terminus (C-term) of LAPTM5. Upper panel, GST-tagged C terminus of LAPTM5 immobilized on GST beads, or GST beads alone, was incubated with purified His-tagged region of A20 spanning the last four ZnF domains (His (ZnF4–7)) and blotted with anti-His antibodies. Lower panel, Ponceau S staining to demonstrate the amount of GST or GST C-terminal input (50% of the total protein used in the assay). G, A20 co-localizes with LAPTM5 within lysosome-associated compartments. Confocal microscopy analysis of subcellular localization of ectopically expressed HA-LAPTM5 (panel a) and mCherry-tagged A20 (panel b) in HeLa cells. LAMP1 staining was used as a marker for the lysosomes (panel c). Representative confocal image is shown. Panels d and e show co-localization between A20 and, LAPTM5 and LAMP1, respectively. All three channels are merged in panel f, and white arrows indicate vesicles containing all three proteins. Bar, 5 μm.
LAPTM5 Positively Regulates TNF Receptor Signaling in Macrophages and Acts at the Level of the Receptor Complex
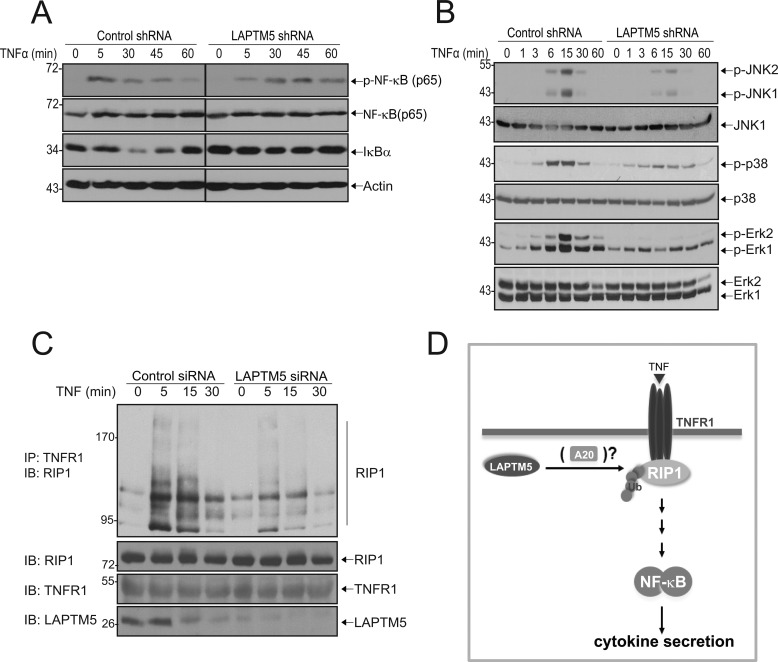
The role of A20 in the termination of NF-κB responses is particularly well described for the TNFR1 signaling pathway. Binding of TNFα to the TNFR1 leads to the recruitment of RIP1 to the receptor complex. This, in turn, promotes Lys63-linked polyubiquitination of RIP1, which serves as a platform for activation of the IKK complex, subsequently leading to the release of NF-κB from IκBα. To attenuate this signaling, the DUB domain of A20 removes the Lys63-linked polyubiquitin chains from RIP1, effectively eliminating the signal for activation of downstream components (13). Thus, we first assessed the role of LAPTM5 in TNFR1-mediated NF-κB activation. As shown in Fig. 5A, TNFα stimulation of control cells resulted in a transient phosphorylation of p65, peaking at 5 min. In contrast, and similar to the defects seen in LPS and MDP-induced signaling (Fig. 3), knockdown of LAPTM5 led to reduced and delayed p65 phosphorylation and impaired down-regulation of IκBα (Fig. 5A). In addition, we observed a substantial defect in the TNFα-mediated phosphorylation of JNK, p38 and ERK (Fig. 5B). Given that A20 deficiency leads to hyperubiquitination of TNFR1-associated RIP1, we hypothesized that knockdown of LAPTM5 would result in reduced ubiquitination of RIP1, because of increased A20 protein levels (13). To test this, we immunoprecipitated the TNFR1 and immunoblotted for RIP1. Numerous previous studies (23–26) have demonstrated a characteristic ladder-like pattern of RIP1, which represents its polyubiquitination. Because direct measurements of RIP1 ubiquitination were not possible in our system (RIP1 ubiquitination by endogenous ubiquitin in RAW cells was below our detection limits, and these cells are not amenable to high efficiency transfection/infection that is necessary for biochemical work, thus preventing us from transfecting/overexpressing tagged ubiquitin), we therefore followed the pattern of RIP1 ladder as surrogate indication of ubiquitination, as routinely done for this protein (25, 26). As shown in Fig. 5C, TNFα stimulation of control cells resulted in polyubiquitination of receptor-associated RIP1, seen by characteristic ladder-like appearance of ubiquitin-modified RIP1 (23, 24). In contrast, we found that TNFα-mediated RIP1 polyubiquitination was reduced in cells when endogenous LAPTM5 was depleted with specific siRNA. These results suggest that LAPTM5 promotes ubiquitination of RIP1 and suggest that LAPTM5 acts at the level of the receptor complex.
FIGURE 5.
LAPTM5 is required for TNFR1-mediated signaling and ubiquitination of RIP1. A and B, knockdown of LAPTM5 leads to reduced TNFR1-mediated NF-κB and MAPK activation. Stable RAW264.7 cells expressing control or LAPTM5 shRNA were stimulated with 50 ng/ml TNFα for the indicated times. The cells were lysed and analyzed by immunoblotting with antibodies toward phosphorylated NF-κB (p-NF-κB (p65)), NF-κB (p65), IκBα, and actin (A) and phosphorylated JNK1/2 (p-JNK1/2), p38 (p-p38), and Erk1/2 (p-Erk1/2) (B), and the respective total levels of these downstream effectors. C, LAPTM5 positively regulates laddering (representing ubiquitination) of RIP1. RAW264.7 cells were transfected with control or LAPTM5 siRNAs. At 72 h after transfection, the cells were treated with TNFα (50 ng/ml) for the indicated times. Immunoprecipitations were performed using anti-TNFR1 antibodies and immunoblotted with anti-RIP1 antibodies. The lower panels represent lysate controls for expression of RIP1, TNFR1, and LAPTM5. D, a model depicting regulation of TNFR1 signaling by LAPTM5. Binding of TNFα to TNFR1 leads to the assembly of receptor signaling complex, resulting in polyubiquitination of RIP1. Activation of downstream components leads to the activation of NF-κB and subsequent release of proinflammatory cytokines. In the absence of LAPTM5, TNFα-induced polyubiquitination of RIP1 is reduced (possibly because of increased A20 protein levels), leading to decreased NF-κB activation and proinflammatory cytokine release. A similar mechanism may apply to other receptors tested in this study.
DISCUSSION
LAPTM5 belongs to a family of late endosomal/lysosomal transmembrane proteins that also includes LAPTM4α and LAPTM4β. Although several studies have implicated the ubiquitously expressed LAPTM4 proteins in mediating drug resistance, as well as function in autophagy, the role of the immune cell specific LAPTM5 has been less clear (27–29). In this study, we present evidence that LAPTM5 acts as a positive regulator of multiple inflammatory signaling cascades in macrophages. The expression of LAPTM5 is required for activation of NF-κB and MAPK signaling and subsequent secretion of proinflammatory cytokines IL6, TNFα, and IL12, as well as NO. Thus, LAPTM5 plays a positive role in macrophage activation.
Loss of LAPTM5 led to reduced activation of NF-κB and MAPK pathways in response to TNF and LPS stimulation, indicating that LAPTM5 acts on a common signaling component. We also show that ubiquitination levels of RIP1 in response to TNF are reduced in LAPTM5 knockdown macrophages, suggesting a role for LAPTM5 at the receptor level or the early assembly phase of the signaling complex containing RIP1 at the plasma membrane (30). The late endosomal localization of LAPTM5 and our failure to detect direct interaction between the TNFR1 and LAPTM5 favor an indirect role of LAPTM5 in controlling TNFR1 signaling. In addition, the stability of the TNFR1 does not seem to be affected by a lack of LAPTM5.
A20 is a powerful negative regulator of NF-κB activation after LPS, CpG, MDP, or TNFα stimulation. In the TNFR1 signaling pathway, A20 deubiquitinates RIP1 and thereby limits signaling intensity. Interestingly, A20 interacts with LAPTM5 by yeast two-hybrid (21). Here, we confirm the interaction of LAPTM5 with A20 in cells and in vitro and find partial co-localization of the two proteins at the late endosomal system when overexpressed in HeLa cells. Moreover, we find that LAPTM5-deficient cells express increased levels of A20 protein, which could possibly explain the decreased ubiquitination of RIP1 after TNFα stimulation. Because A20 mRNA levels are normal in LAPTM5−/− macrophages, LAPTM5 seems to regulate a degradative pathway of A20. Thus, excessive A20 activity could account for the muted NF-κB activation by multiple inflammatory stimuli in LAPTM5-deficient macrophages.
A growing body of evidence suggests that LAPTM5 plays a role in intracellular trafficking; however, the mechanisms remain unknown (4, 5, 21). LAPTM5 contains a functional UIM within its C terminus, which facilitates binding to ubiquitinated proteins. Previous work from our laboratory investigated trafficking of LAPTM5, where we showed that LAPTM5 is sorted from the Golgi to the late endosome/lysosome via PY motif-WW domain interaction with the E3 ubiquitin ligase Nedd4 and ubiquitinated clathrin adaptor GGA3 (3). In the current study we find that in macrophages LAPTM5 has a rapid turnover and is targeted to the lysosome for degradation, rather than being a resident lysosomal transmembrane protein. Moreover, we show that LPS stimulation results in subcellular redistribution of LAPTM5-positive vesicles and targets LAPTM5 to the lysosomes. We speculate that the LPS-induced down-regulation of LAPTM5 protein may ensure efficient termination of signaling to prevent excessive/sustained inflammatory cytokine release. Thus, in addition to transcriptional down-regulation observed upon T and B cell activation (31), LAPTM5 protein levels in macrophages are tightly controlled by a post-transcriptional mechanism.
The behavior of LAPTM5 is reminiscent of that observed for the Nedd4 family interacting proteins (Ndfip1 and Ndfip2), which similarly to LAPTM5 are transmembrane proteins with multiple PY motifs. They localize to the Golgi and late endosomal compartments and show a rapid turnover caused by lysosomal degradation (32, 33). Similar to Ndfip1 and Ndfip2, LAPTM5 may serve as an adaptor for recruitment of Nedd4 family members to their substrates to allow their subsequent ubiquitination. Alternatively, or in addition, LAPTM5, en route from the Golgi to the lysosome, may aid in sorting of ubiquitinated cargo, via its UIM, and facilitate substrate incorporation into the multivesicular body pathway for subsequent degradation. In fact, using immunoelectron microscopy, we previously detected the presence of LAPTM5 within the intraluminal vesicles of multivesicular bodies (3).4 In accord, the UIM of LAPTM5, as well as its PY motifs, were recently reported to be necessary for efficient lysosomal sorting of the TCR (5).
Interestingly, recent studies in mice have identified LAPTM5 as a susceptibility gene for inflammatory bowel disease (34, 35). Deregulation of the NOD2 pathway is a key event in the development of inflammatory bowel disease (36). Therefore, it will be interesting to test the in vivo role of LAPTM5 in activation of innate immune responses, particularly in intestinal homeostasis, via the NOD2 signaling cascade.
In summary, we show here that in macrophages, LAPTM5 is an essential component of several inflammatory pathways and is required for efficient secretion of proinflammatory cytokines. Deficiency in LAPTM5 leads to reduced NF-κB and MAPK activation, strongly suggesting that LAPTM5 acts upstream of multiple signaling pathways. Our findings indicate that LAPTM5 acts as a positive regulator of inflammatory responses by macrophages.
Supplementary Material
Acknowledgments
We thank Drs. Y. Pak for reagents and G. Casallo for technical assistance with quantitative real time PCR.
This work was supported by the Canadian Institute of Health Research Grant MOP-13494 (to D. R.).

This article contains supplemental Figs. S1–S3.
W. K. Glowacka and D. Rotin, unpublished observations.
- UIM
- ubiquitin-interacting motif
- TLR
- Toll-like receptor
- NOD
- nucleotide oligomerization domain
- MDP
- muramyl dipeptide
- IKK
- IκB kinase
- TNFR
- TNF receptor
- BMDM
- bone marrow-derived macrophage.
REFERENCES
- 1. Adra C. N., Zhu S., Ko J. L., Guillemot J. C., Cuervo A. M., Kobayashi H., Horiuchi T., Lelias J. M., Rowley J. D., Lim B. (1996) LAPTM5. A novel lysosomal-associated multispanning membrane protein preferentially expressed in hematopoietic cells. Genomics 35, 328–337 [DOI] [PubMed] [Google Scholar]
- 2. Scott L. M., Mueller L., Collins S. J. (1996) E3, a hematopoietic-specific transcript directly regulated by the retinoic acid receptor α. Blood 88, 2517–2530 [PubMed] [Google Scholar]
- 3. Pak Y., Glowacka W. K., Bruce M. C., Pham N., Rotin D. (2006) Transport of LAPTM5 to lysosomes requires association with the ubiquitin ligase Nedd4, but not LAPTM5 ubiquitination. J. Cell Biol. 175, 631–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ouchida R., Kurosaki T., Wang J. Y. (2010) A role for lysosomal-associated protein transmembrane 5 in the negative regulation of surface B cell receptor levels and B cell activation. J. Immunol. 185, 294–301 [DOI] [PubMed] [Google Scholar]
- 5. Ouchida R., Yamasaki S., Hikida M., Masuda K., Kawamura K., Wada A., Mochizuki S., Tagawa M., Sakamoto A., Hatano M., Tokuhisa T., Koseki H., Saito T., Kurosaki T., Wang J. Y. (2008) A lysosomal protein negatively regulates surface T cell antigen receptor expression by promoting CD3ζ-chain degradation. Immunity 29, 33–43 [DOI] [PubMed] [Google Scholar]
- 6. Mosser D. M., Edwards J. P. (2008) Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 8, 958–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hayden M. S., Ghosh S. (2011) NF-κB in immunobiology. Cell Res. 21, 223–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rothwarf D. M., Karin M. (1999) The NF-κB activation pathway. A paradigm in information transfer from membrane to nucleus. Sci. STKE 1999, RE1. [DOI] [PubMed] [Google Scholar]
- 9. Häcker H., Karin M. (2006) Regulation and function of IKK and IKK-related kinases. Sci. STKE 2006, re13. [DOI] [PubMed] [Google Scholar]
- 10. Viatour P., Merville M. P., Bours V., Chariot A. (2005) Phosphorylation of NF-κB and IκB proteins. Implications in cancer and inflammation. Trends Biochem. Sci. 30, 43–52 [DOI] [PubMed] [Google Scholar]
- 11. Oeckinghaus A., Hayden M. S., Ghosh S. (2011) Crosstalk in NF-κB signaling pathways. Nat. Immunol. 12, 695–708 [DOI] [PubMed] [Google Scholar]
- 12. Skaug B., Jiang X., Chen Z. J. (2009) The role of ubiquitin in NF-κB regulatory pathways. Annu. Rev. Biochem. 78, 769–796 [DOI] [PubMed] [Google Scholar]
- 13. Wertz I. E., O'Rourke K. M., Zhou H., Eby M., Aravind L., Seshagiri S., Wu P., Wiesmann C., Baker R., Boone D. L., Ma A., Koonin E. V., Dixit V. M. (2004) De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-κB signalling. Nature 430, 694–699 [DOI] [PubMed] [Google Scholar]
- 14. Boone D. L., Turer E. E., Lee E. G., Ahmad R. C., Wheeler M. T., Tsui C., Hurley P., Chien M., Chai S., Hitotsumatsu O., McNally E., Pickart C., Ma A. (2004) The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat. Immunol. 5, 1052–1060 [DOI] [PubMed] [Google Scholar]
- 15. Hitotsumatsu O., Ahmad R. C., Tavares R., Wang M., Philpott D., Turer E. E., Lee B. L., Shiffin N., Advincula R., Malynn B. A., Werts C., Ma A. (2008) The ubiquitin-editing enzyme A20 restricts nucleotide-binding oligomerization domain containing 2-triggered signals. Immunity 28, 381–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kumar A., Takada Y., Boriek A. M., Aggarwal B. B. (2004) Nuclear factor-κB. Its role in health and disease. J. Mol. Med. 82, 434–448 [DOI] [PubMed] [Google Scholar]
- 17. Paddison P. J., Cleary M., Silva J. M., Chang K., Sheth N., Sachidanandam R., Hannon G. J. (2004) Cloning of short hairpin RNAs for gene knockdown in mammalian cells. Nat. Methods 1, 163–167 [DOI] [PubMed] [Google Scholar]
- 18. Barton G. M., Kagan J. C. (2009) A cell biological view of Toll-like receptor function. Regulation through compartmentalization. Nat. Rev. Immunol. 9, 535–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sun S. C., Ganchi P. A., Ballard D. W., Greene W. C. (1993) NF-κB controls expression of inhibitor IκBα. Evidence for an inducible autoregulatory pathway. Science 259, 1912–1915 [DOI] [PubMed] [Google Scholar]
- 20. Girardin S. E., Boneca I. G., Viala J., Chamaillard M., Labigne A., Thomas G., Philpott D. J., Sansonetti P. J. (2003) Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J. Biol. Chem. 278, 8869–8872 [DOI] [PubMed] [Google Scholar]
- 21. Colland F., Jacq X., Trouplin V., Mougin C., Groizeleau C., Hamburger A., Meil A., Wojcik J., Legrain P., Gauthier J. M. (2004) Functional proteomics mapping of a human signaling pathway. Genome Res. 14, 1324–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li L., Hailey D. W., Soetandyo N., Li W., Lippincott-Schwartz J., Shu H. B., Ye Y. (2008) Localization of A20 to a lysosome-associated compartment and its role in NFκB signaling. Biochim. Biophys. Acta 1783, 1140–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang S. Q., Kovalenko A., Cantarella G., Wallach D. (2000) Recruitment of the IKK signalosome to the p55 TNF receptor. RIP and A20 bind to NEMO (IKKγ) upon receptor stimulation. Immunity 12, 301–311 [DOI] [PubMed] [Google Scholar]
- 24. Legler D. F., Micheau O., Doucey M. A., Tschopp J., Bron C. (2003) Recruitment of TNF receptor 1 to lipid rafts is essential for TNFα-mediated NF-κB activation. Immunity 18, 655–664 [DOI] [PubMed] [Google Scholar]
- 25. Pobezinskaya Y. L., Kim Y. S., Choksi S., Morgan M. J., Li T., Liu C., Liu Z. (2008) The function of TRADD in signaling through tumor necrosis factor receptor 1 and TRIF-dependent Toll-like receptors. Nat. Immunol. 9, 1047–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gerlach B., Cordier S. M., Schmukle A. C., Emmerich C. H., Rieser E., Haas T. L., Webb A. I., Rickard J. A., Anderton H., Wong W. W., Nachbur U., Gangoda L., Warnken U., Purcell A. W., Silke J., Walczak H. (2011) Linear ubiquitination prevents inflammation and regulates immune signalling. Nature 471, 591–596 [DOI] [PubMed] [Google Scholar]
- 27. Hogue D. L., Kerby L., Ling V. (1999) A mammalian lysosomal membrane protein confers multidrug resistance upon expression in Saccharomyces cerevisiae. J. Biol. Chem. 274, 12877–12882 [DOI] [PubMed] [Google Scholar]
- 28. Li Y., Zhang Q., Tian R., Wang Q., Zhao J. J., Iglehart J. D., Wang Z. C., Richardson A. L. (2011) Lysosomal transmembrane protein LAPTM4B promotes autophagy and tolerance to metabolic stress in cancer cells. Cancer Res. 71, 7481–7489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li Y., Zou L., Li Q., Haibe-Kains B., Tian R., Li Y., Desmedt C., Sotiriou C., Szallasi Z., Iglehart J. D., Richardson A. L., Wang Z. C. (2010) Amplification of LAPTM4B and YWHAZ contributes to chemotherapy resistance and recurrence of breast cancer. Nat. Med. 16, 214–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hsu H., Huang J., Shu H. B., Baichwal V., Goeddel D. V. (1996) TNF-dependent recruitment of the protein kinase RIP to the TNF receptor-1 signaling complex. Immunity 4, 387–396 [DOI] [PubMed] [Google Scholar]
- 31. Seimiya M., O-Wang J., Bahar R., Kawamura K., Wang Y., Saisho H., Tagawa M. (2003) Stage-specific expression of Clast6/E3/LAPTM5 during B cell differentiation. Elevated expression in human B lymphomas. Int. J. Oncol. 22, 301–304 [PubMed] [Google Scholar]
- 32. Harvey K. F., Shearwin-Whyatt L. M., Fotia A., Parton R. G., Kumar S. (2002) N4WBP5, a potential target for ubiquitination by the Nedd4 family of proteins, is a novel Golgi-associated protein. J. Biol. Chem. 277, 9307–9317 [DOI] [PubMed] [Google Scholar]
- 33. Shearwin-Whyatt L. M., Brown D. L., Wylie F. G., Stow J. L., Kumar S. (2004) N4WBP5A (Ndfip2), a Nedd4-interacting protein, localizes to multivesicular bodies and the Golgi, and has a potential role in protein trafficking. J. Cell Sci. 117, 3679–3689 [DOI] [PubMed] [Google Scholar]
- 34. Brudzewsky D., Pedersen A. E., Claesson M. H., Gad M., Kristensen N. N., Lage K., Jensen T., Tommerup N., Larsen L. A., Knudsen S., Tümer Z. (2009) Genome-wide gene expression profiling of SCID mice with T-cell-mediated colitis. Scand. J. Immunol. 69, 437–446 [DOI] [PubMed] [Google Scholar]
- 35. Myles M. H., Dieckgraefe B. K., Criley J. M., Franklin C. L. (2007) Characterization of cecal gene expression in a differentially susceptible mouse model of bacterial-induced inflammatory bowel disease. Inflamm. Bowel. Dis. 13, 822–836 [DOI] [PubMed] [Google Scholar]
- 36. Xavier R. J., Podolsky D. K. (2007) Unravelling the pathogenesis of inflammatory bowel disease. Nature 448, 427–434 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.