Abstract
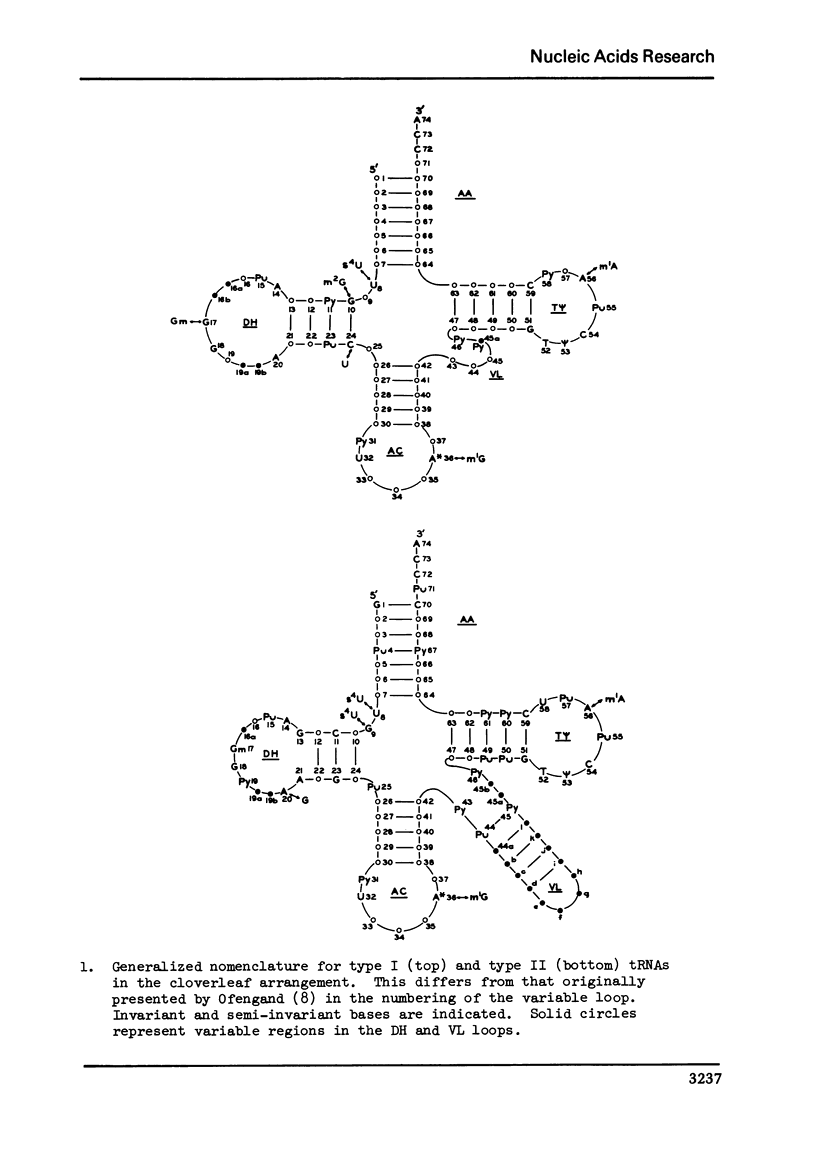
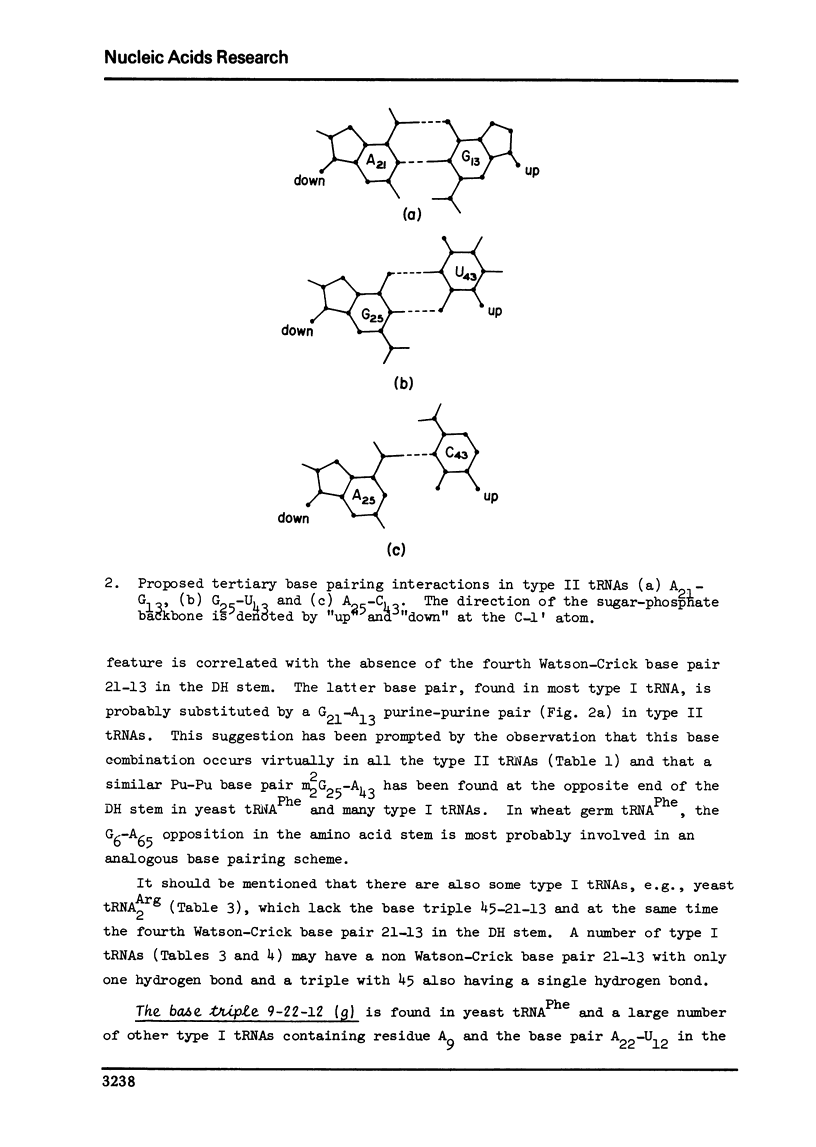
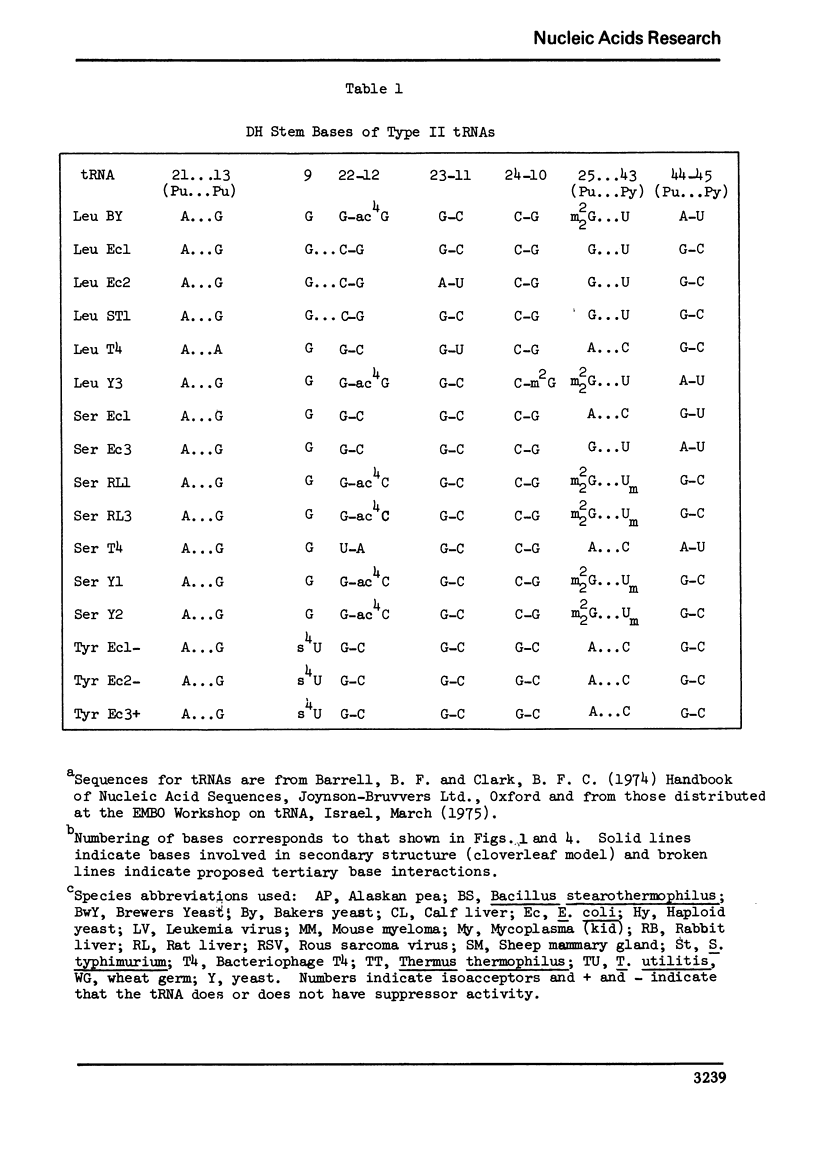
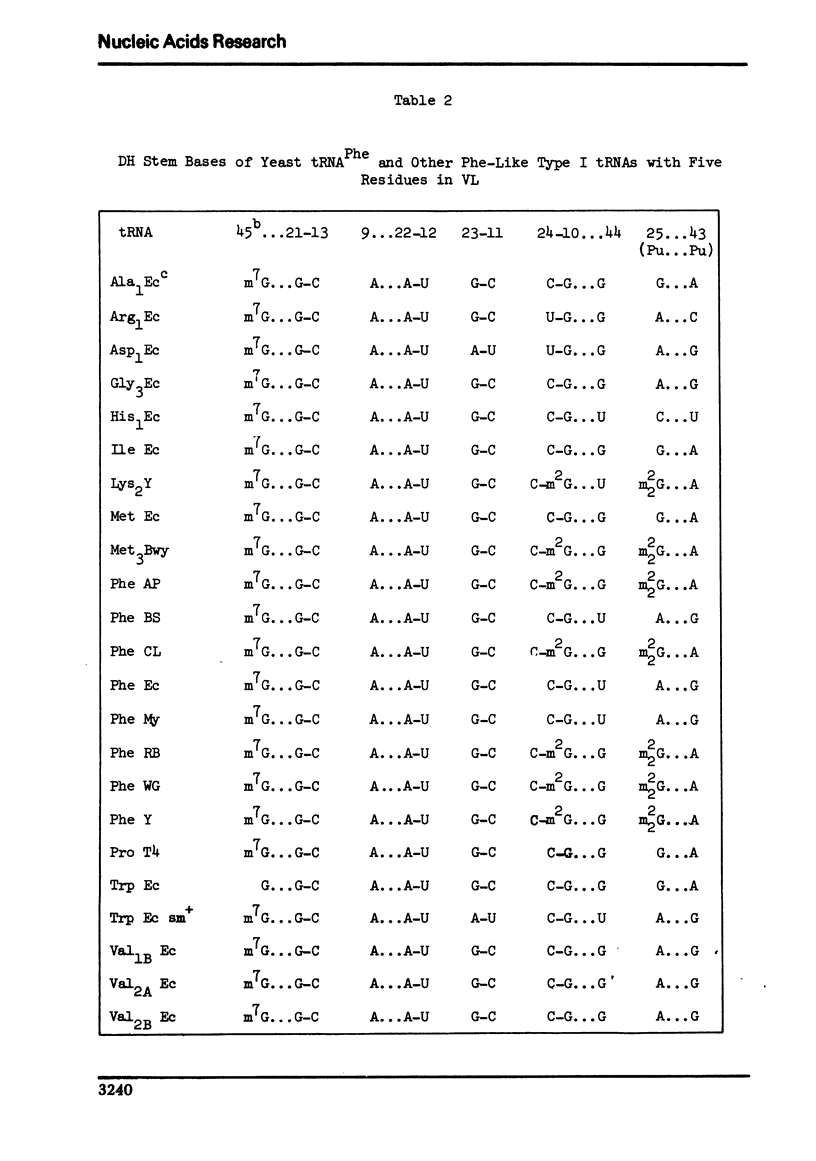
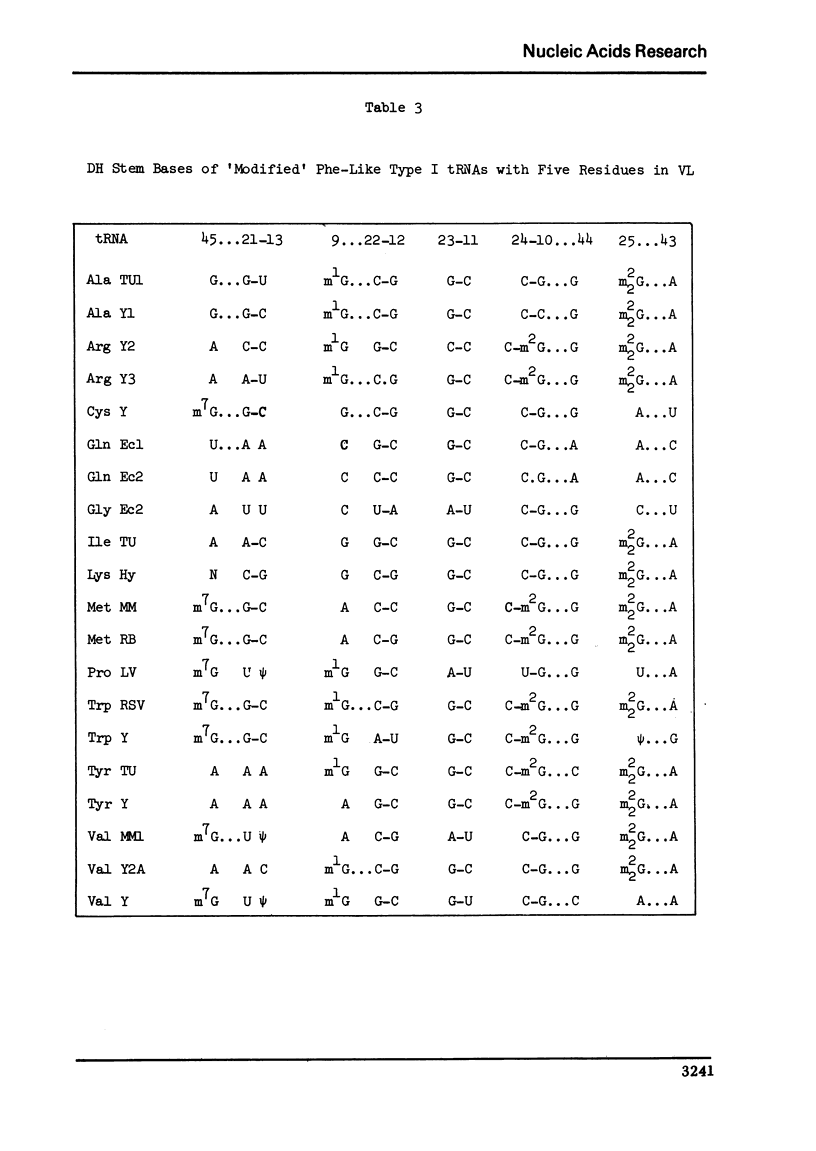
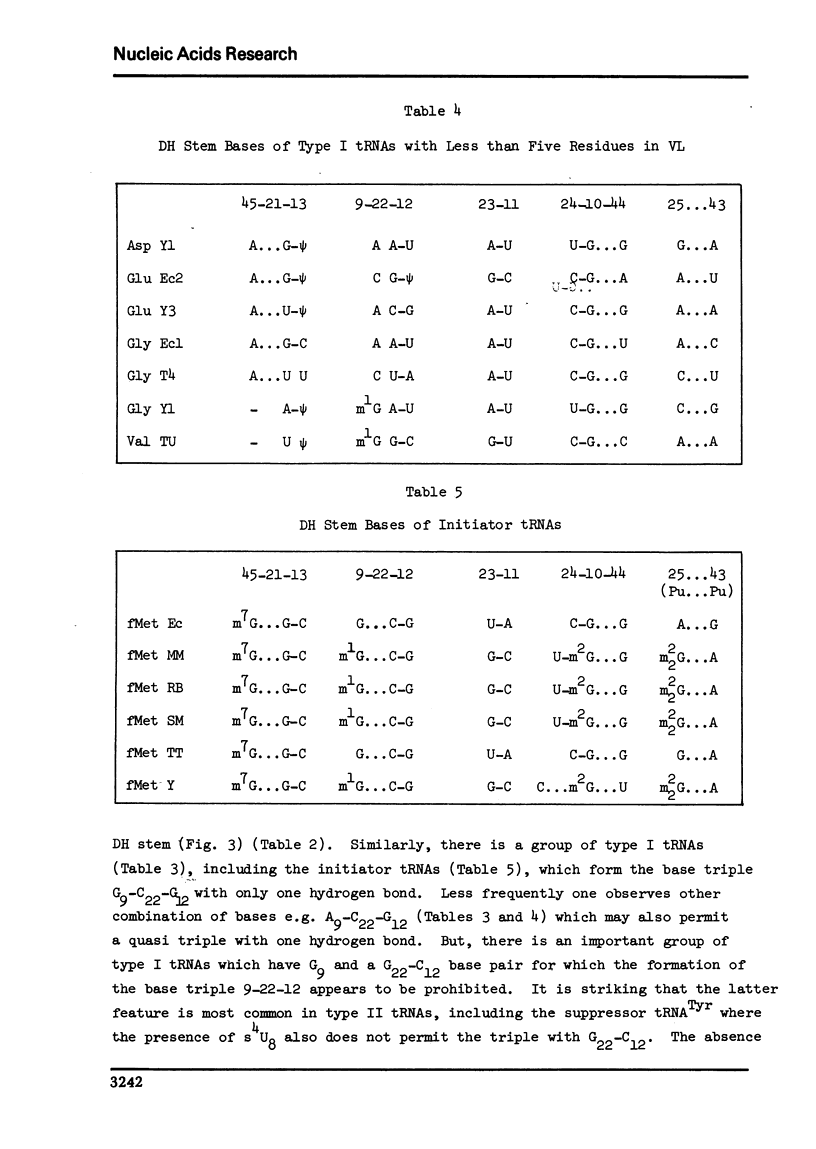
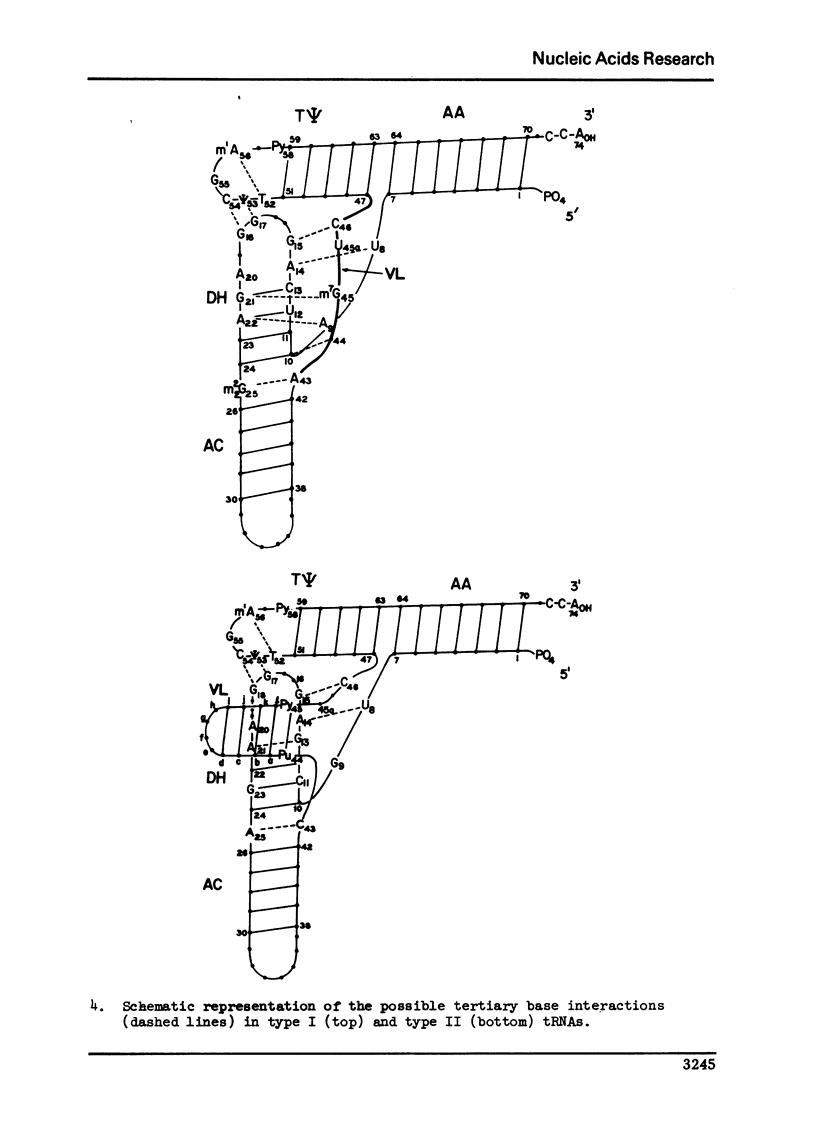
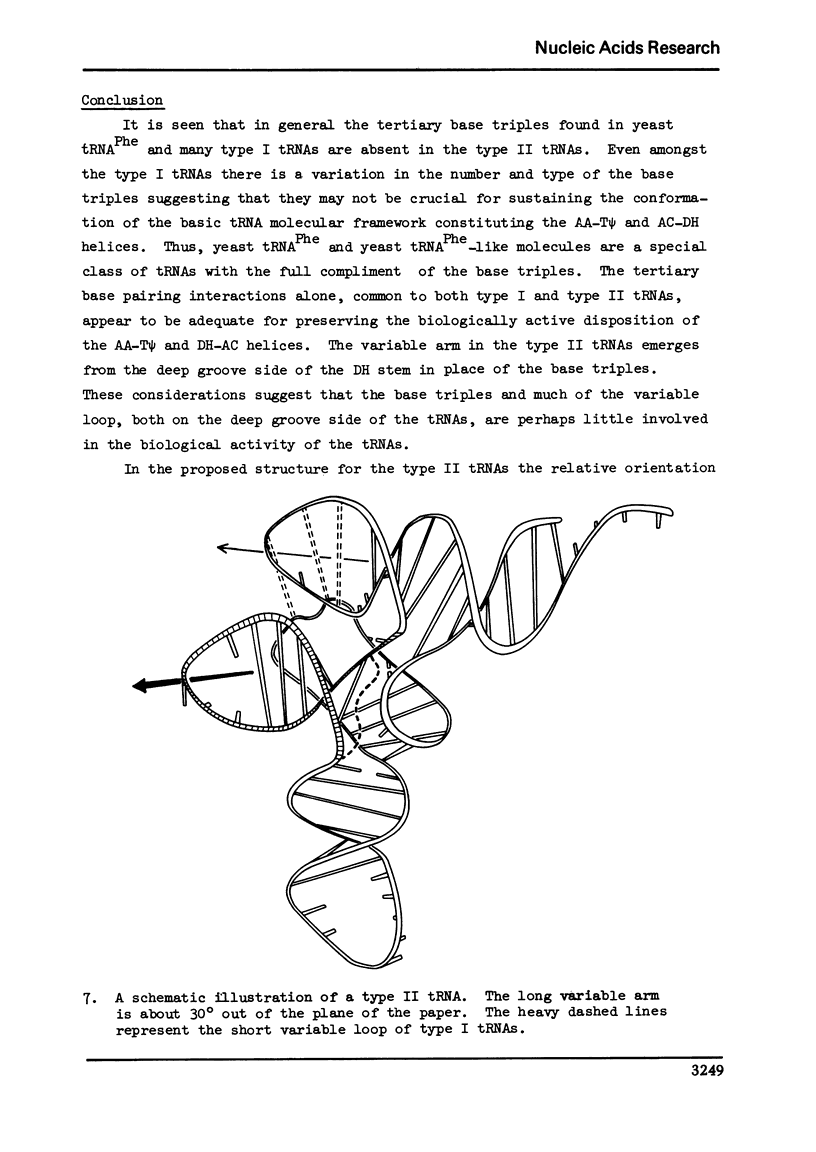
A structure is proposed for the type II tRNA molecules containing the long variable loop and the tertiary base interactions here are compared with type I tRNAs having the short variable loop. The type II tRNAs are similar to the type I tRNAs in their tertiary base pairing interactions but differ from them generally by not having the tertiary base triples. The long variable loop, which is comprised of a helical stem and a loop at the end of it, emerges from the deep groove side of the dihydrouridine helix, and is tilted roughly 30° to the plane formed by the amino acid-pseudouridine and anticodon-dihydrouridine helices found in yeast tRNAPhe. The fact that many of the type I tRNAs also lack the full compliment of base triples suggests that the tertiary base pairs may alone suffice to sustain the tRNA fold required for its biological function. The base triples and the variable loop appear to have little functional significance. The base type at position 9 is correlated with the number of base triples and G-C base pairs in the dihydrouridine stem.
Full text
PDF








Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chang S. E., Cashmore A. R., Brown D. M. Selective modification of uridine and guanosine residues in tyrosine transfer ribonucleic acid. J Mol Biol. 1972 Jul 28;68(3):455–464. doi: 10.1016/0022-2836(72)90099-x. [DOI] [PubMed] [Google Scholar]
- Erdmann V. A., Sprinzl M., Pongs O. The involvement of 5S RNA in the binding of tRNA to ribosomes. Biochem Biophys Res Commun. 1973 Oct 1;54(3):942–948. doi: 10.1016/0006-291x(73)90785-7. [DOI] [PubMed] [Google Scholar]
- Kim S. H., Suddath F. L., Quigley G. J., McPherson A., Sussman J. L., Wang A. H., Seeman N. C., Rich A. Three-dimensional tertiary structure of yeast phenylalanine transfer RNA. Science. 1974 Aug 2;185(4149):435–440. doi: 10.1126/science.185.4149.435. [DOI] [PubMed] [Google Scholar]
- Kim S. H., Sussman J. L., Suddath F. L., Quigley G. J., McPherson A., Wang A. H., Seeman N. C., RICH A. The general structure of transfer RNA molecules. Proc Natl Acad Sci U S A. 1974 Dec;71(12):4970–4974. doi: 10.1073/pnas.71.12.4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. Symmetry recognition hypothesis model for tRNA binding to aminoacyl-tRNA synthetase. Nature. 1975 Aug 21;256(5519):679–681. doi: 10.1038/256679a0. [DOI] [PubMed] [Google Scholar]
- Klug A., Ladner J., Robertus J. D. The structural geometry of co-ordinated base changes in transfer RNA. J Mol Biol. 1974 Nov 5;89(3):511–516. doi: 10.1016/0022-2836(74)90480-x. [DOI] [PubMed] [Google Scholar]
- Ladner J. E., Jack A., Robertus J. D., Brown R. S., Rhodes D., Clark B. F., Klug A. Atomic co-ordinates for yeast phenylalanine tRNA. Nucleic Acids Res. 1975 Sep;2(9):1629–1637. doi: 10.1093/nar/2.9.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladner J. E., Jack A., Robertus J. D., Brown R. S., Rhodes D., Clark B. F., Klug A. Structure of yeast phenylalanine transfer RNA at 2.5 A resolution. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4414–4418. doi: 10.1073/pnas.72.11.4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt M. Detailed molecular model for transfer ribonucleic acid. Nature. 1969 Nov 22;224(5221):759–763. doi: 10.1038/224759a0. [DOI] [PubMed] [Google Scholar]
- Quigley G. J., Seeman N. C., Wang A. H., Suddath F. L., Rich A. Yeast phenylalanine transfer RNA: atomic coordinates and torsion angles. Nucleic Acids Res. 1975 Dec;2(12):2329–2341. doi: 10.1093/nar/2.12.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley G. J., Wang A. H., Seeman N. C., Suddath F. L., Rich A., Sussman J. L., Kim S. H. Hydrogen bonding in yeast phenylalanine transfer RNA. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4866–4870. doi: 10.1073/pnas.72.12.4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich A. Transfer RNA and protein synthesis. Biochimie. 1974;56(11-12):1441–1449. doi: 10.1016/s0300-9084(75)80265-3. [DOI] [PubMed] [Google Scholar]
- Richter D., Erdmann V. A., Sprinzl M. Specific recognition of GTpsiC loop (loop IV) of tRNA by 50S ribosomal subunits from E. coli. Nat New Biol. 1973 Dec 5;246(153):132–135. doi: 10.1038/newbio246132a0. [DOI] [PubMed] [Google Scholar]
- Robertus J. D., Ladner J. E., Finch J. T., Rhodes D., Brown R. S., Clark B. F., Klug A. Structure of yeast phenylalanine tRNA at 3 A resolution. Nature. 1974 Aug 16;250(467):546–551. doi: 10.1038/250546a0. [DOI] [PubMed] [Google Scholar]
- Sigler P. B. An analysis of the structure of tRNA. Annu Rev Biophys Bioeng. 1975;4(00):477–527. doi: 10.1146/annurev.bb.04.060175.002401. [DOI] [PubMed] [Google Scholar]
- Stout C. D., Mizuno H., Rubin J., Brennan T., Rao S. T., Sundaralingam M. Atomic coordinates and molecular conformation of yeast phenylalanyl tRNA. An independent investigation. Nucleic Acids Res. 1976 Apr;3(4):1111–1123. doi: 10.1093/nar/3.4.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman J. L., Kim S. H. Idealized atomic coordinates of yeast phenylalanine transfer RNA. Biochem Biophys Res Commun. 1976 Jan 12;68(1):89–96. doi: 10.1016/0006-291x(76)90014-0. [DOI] [PubMed] [Google Scholar]
- Uhlenbeck O. C. Complementary oligonucleotide binding to transfer RNA. J Mol Biol. 1972 Mar 14;65(1):25–41. doi: 10.1016/0022-2836(72)90489-5. [DOI] [PubMed] [Google Scholar]



