Background: The chemomechanical properties of myosin-XXI, seemingly the only myosin expressed in Leishmania parasites, are unknown.
Results: Recombinantly expressed full-length myosin-XXI is an active ATPase and, in the presence of calmodulin, moves actin filaments.
Conclusion: Myosin-XXI is a mechanically functional molecular motor.
Significance: Calmodulin-dependent regulation of myosin-XXI might be involved in the various functions of the motor in the parasite lifecycle.
Keywords: Cytoskeleton, Enzyme Kinetics, Leishmania, Molecular Motors, Myosin
Abstract
The genome of the Leishmania parasite contains two classes of myosin. Myosin-XXI, seemingly the only myosin isoform expressed in the protozoan parasite, has been detected in both the promastigote and amastigote stages of the Leishmania life cycle. It has been suggested to perform a variety of functions, including roles in membrane anchorage, but also long-range directed movements of cargo. However, nothing is known about the biochemical or mechanical properties of this motor. Here we designed and expressed various myosin-XXI constructs using a baculovirus expression system. Both full-length (amino acids 1–1051) and minimal motor domain constructs (amino acids 1–800) featured actin-activated ATPase activity. Myosin-XXI was soluble when expressed either with or without calmodulin. In the presence of calcium (pCa 4.1) the full-length motor could bind a single calmodulin at its neck domain (probably amino acids 809–823). Calmodulin binding was required for motility but not for ATPase activity. Once bound, calmodulin remained stably attached independent of calcium concentration (pCa 3–7). In gliding filament assays, myosin-XXI moved actin filaments at ∼15 nm/s, insensitive to both salt (25–1000 mm KCl) and calcium concentrations (pCa 3–7). Calmodulin binding to the neck domain might be involved in regulating the motility of the myosin-XXI motor for its various cellular functions in the different stages of the Leishmania parasite life cycle.
Introduction
Leishmaniasis affects over 12 million people worldwide, with about 2 million newly infected patients each year. The disease is caused by the flagellated protozoan parasite Leishmania and can manifest itself as visceral leishmaniasis, which is potentially fatal, or cutaneous leishmaniasis, which can leave disfiguring mucocutaneous scars (1). The Leishmania parasite has a two-stage life cycle, including a non-motile amastigote stage in mammalian macrophages and a motile promastigote stage in the sand fly alimentary tract (2). The genome of Leishmania donovani contains only two myosin genes, one myosin class IB and one previously assigned to class XXI (3). According to a later classification, Leishmania myosin XXI has been reassigned to class XIII, a kinetoplastide-specific class of myosins (4). Although no expression of myosin-IB has been found in the organism to date, myosin-XXI has been detected in both the promastigote and the amastigote stages of the Leishmania life cycle (5). The motor is preferentially localized to the proximal region of the flagellum but is also found in other flagellar and cell body compartments (6). Myosin-XXI expression depends on both the parasite life cycle (5) and on the growth phase of the parasite. For cultured promastigotes, expression levels were reported to increase almost 4.5-fold from early log phase of growth to stationary phase (6). Katta et al. (6) showed that myosin-XXI is essential for survival of Leishmania promastigotes in culture and that a reduction in expression levels of myosin-XXI results in the loss of endocytosis within the flagellar pocket and impairment of other intracellular trafficking processes. In addition, myosin-XXI heterozygous cells failed to form the paraflagellar rod. The paraflagellar rod is a structure that runs along the length of the flagellum and contains a variety of proteins, including actin, but its functional role remains unclear (7). The detection of only a single myosin isoform in the parasite suggests that this myosin must carry out a variety of functions. Two distinct myosin-XXI populations have been identified. For the membrane-bound population, the tail domain localizes the motor molecules at the base of the flagellum, whereas the detergent-soluble subfraction could be involved in the transport of proteins within the flagellum (5).
The myosin superfamily is composed of 36 classes (3, 4). Myosins consist of a highly conserved motor domain followed by a neck domain of variable length, often including IQ motifs for the binding of light chains of the calmodulin family, and finally a tail domain, which can contain a large variety of motifs (8). Although the motor domain is responsible for the binding to actin and hydrolysis of ATP, it is the tail domain that determines function within the cell by controlling molecular dimerization and motor processivity, motor anchoring to the membrane, and/or selection and transport of specific cargo.
Although myosin-XXI does not contain perfect IQ motifs in the neck domain, there are several less well characterized, degenerative IQ domains present. Subsequent to the converter domain, the proximal tail contains a natural “leucine zipper” motif that is followed by a predicted short coiled-coil domain (MARCOIL) (9) and, finally, near the C terminus, two tandem ubiquitin-associated domains (UBA)2 (Fig. 1A). UBAs are found in a variety of proteins, including members of the ubiquitination pathway, UV excision repair proteins, and certain protein kinases. However, their function is not entirely clear (10). They have been shown to bind ubiquitin and dimerize in a mutually exclusive way (11). They have also been shown to act as a stabilization signal that protects proteins from proteasomal degradation (12). Within the myosin family, UBAs are unique to myosin-XXI, and their role is unknown.
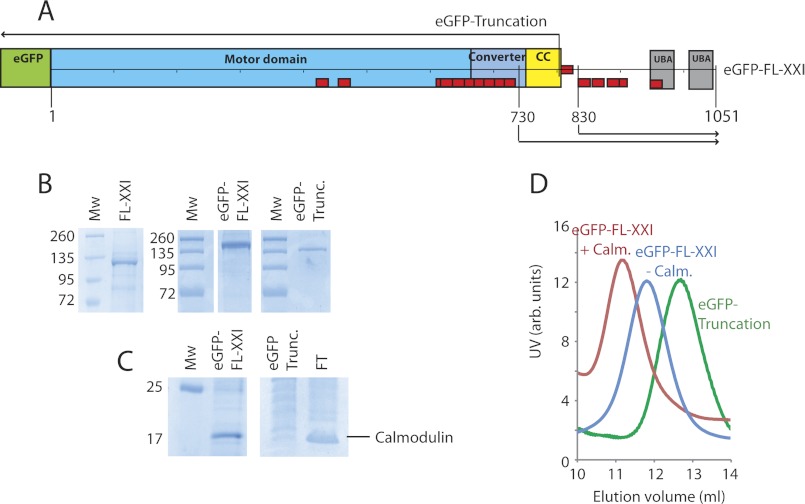
FIGURE 1.
Myosin-XXI constructs and protein purification. A, schematic of eGFP-tagged, full-length myosin-XXI. Other constructs, the eGFP-tagged truncated motor, as well as the 730 and 830 tail constructs are marked. Domains are the motor domain, converter region, predicted coiled coil (CC), and UBA. Red boxes are predicted calmodulin-binding motifs (see Fig. 4A). B, SDS-PAGE gel of full-length and truncated myosin-XXI coexpressed with calmodulin. The gel confirms clean purified protein with very little impurities (size markers in kDa). Mw, molecular weight. C, SDS-PAGE gels show calmodulin bound to the full-length but not to the truncated myosin construct. Calmodulin (untagged) was present in the flow-through (FT) created from the first purification stage where coexpressed proteins are bound to the Ni-Sepharose column. D, Superdextm-200 gel filtration of full-length myosin-XXI expressed in the presence of calmodulin (red), without calmodulin (blue), and of the truncated construct expressed in the presence of calmodulin (green). The elution volume increases with decreasing Stokes radius of the protein complex. All experiments were repeated at least three times for three separate protein purifications. Protein concentrations were between 5–8 μm depending on the preparation.
Controlling expression levels is one mechanism for regulating where and when a myosin is used. However, in a simplified system with only two myosins present in the genome, there clearly must be other mechanisms that allow myosin-XXI to perform its different roles within the cell. The presence of two myosin heavy chain kinases (NCBI accession numbers CBZ38915.1 and CBZ34699.1) and calmodulin in the L. donovani genome suggests other possible regulatory mechanisms.
In this study, we expressed Leishmania full-length myosin-XXI and a truncated minimal motor domain in an Sf21/baculoviral system for biochemical and biophysical analysis. We show that myosin-XXI is an actin-activated ATPase that binds a single calmodulin that is required for motility but not for ATPase activity. EM imaging shows a monomeric molecule that seems to bind cooperatively to actin filament ends.
EXPERIMENTAL PROCEDURES
Plasmids and Generation of Recombinant Baculovirus
Full-length myosin-XXI (3153 bp) cDNA (accession number FJ028724) was chemically synthesized and cloned into pUC57 (Genscript USA, Inc.). For purification, this construct was subcloned into the 6× His-tagged transfer vector pFastBacHbtm (Invitrogen) using standard PCR methods and oligonucleotides XXIFP6 (AGGATCCATGCCGGAGCGTGT-GTCTGTG) and XXIRP4 (AAAAGCGGC-CGCCTAGCTCACCTTGAACAGCATCTTAACGG) via BamHI/NotI sites. An aa 1–800 truncation of myosin-XXI was created using PCR and oligonucleotides XXIFP6 and XXITR6 (AAAAGCGGCCGCCTATTTCTCCGCCGGCACACC). The N terminus of full-length and truncated myosin-XXI was modified by addition of eGFP ligated into the BamHI site of pFastBactHbtm. Recombinant baculovirus DNA was generated by the Bac-to-Bac® method according to the instructions of the manufacturer and transferred into Spodoptera frugiperda (SF21) cells. Viruses were amplified to a P3 stock, and virus titer was determined before infection of cells for protein expression. Xenopus calmodulin was amplified from a P4 stock (gift of J. R. Sellers, NHLBI, National Institutes of Health), and the titer was determined.
Myosin-XXI Protein Expression and Purification
500 ml of SF21 cells (1 × 106 cells/ml) were cultured in Grace's medium supplemented with 10% FCS (heat-inactivated). Normycin and pluronic were coinfected with full-length or truncated myosin-XXI plus calmodulin at a ratio of 2:1 multiplicity of infection. Infected SF21 cells were harvested after 72 h and washed once with PBS. The remaining steps were performed at 4 °C. Cells were resuspended in myosin extraction buffer as described by Margossian and Lowey (13) (300 mm KCl, 150 mm phosphate buffer (pH 6.8), 1 mm DTT, and EDTA-free protease inhibitor tablets (Roche)) and lysed by sonication (sonicator probe Bandelin HD 2070, 5 min total time, 15 s on and 15 s off, at 40% maximal power). The lysate was clarified by centrifugation at 50,000 × g for 20 min. The supernatant was immediately loaded onto a HisTrap FF column (GE Healthcare) on an ÄKTA purifier (liquid chromatography, GE Healthcare), washed with a low-His buffer (50 mm Tris (pH 7.5), 40 mm imidazole, 200 mm NaCl, 1 mm DTT) and eluted using His buffer plus 400 mm imidazole using a step program. Peaks were run on SDS-PAGE gels, and pure fractions were pooled and concentrated to ∼25 μm. This sample was diluted with glycerol 1:1 (v/v) and drop-frozen in liquid nitrogen.
Myosin-XXI Tail and Calmodulin Expression in Escherichia coli
Two tail fragments of myosin-XXI (aa 730–1050 and aa 830–1050) fused with N-terminal mRFP were cloned into pET28a vector using standard PCR and oligonucleotides XXI-730FP (AGAGGCGGCCGCCGGCAAGACGAAGGTGTTCCTCC), XXI-830FP (AGAGGCGGCCGCCGCCGTCGAGGCGGACACGCGCG), and XXI-ENDRP (ACTCGAGCTAGCTCACCTT-GAACAGC). The restriction sites are shown in boldface. Xenopus calmodulin was cloned into the pET28a vector between the HindIII/XhoI sites using PCR and oligonucleotides CamXHindIIIFP (AAAAAGC TTTAATGGCTGACCAACTGACAGAAG) and CamXXhoI-RP (ACTCGAGTCACTTTGCTGT CATCATTTGTAC). Plasmids were transformed into BL21 (DE3) Star E. coli cells (Invitrogen). An overnight starter culture was used to inoculate 2 liters of 2YT medium supplemented with 25 μg/ml kanamycin. The cells were grown at 37 °C until they reached mid-log phase. At this point, 1 mm isopropyl 1-thio-β-d-galactopyranoside was added to the 2YT, and the growth temperature was simultaneously dropped to 27 °C. Growth was continued for a further 4 h before the cells were harvested and resuspended in a low-His buffer containing 20% sucrose. Cells were then stored at −80 °C until further use. Cells were lysed by sonication as described above, and soluble proteins were harvested by centrifugation at 50,000 × g for 20 min. Proteins were then loaded onto a HisTrap FF column and purified as described above. Proteins were concentrated to ∼2–4 mg.ml−1 using Amicon centrifuge concentrators and then frozen in liquid nitrogen and stored at −80 °C.
Calmodulin Stoichiometry
Following the method described by Coluccio (14), purified myosin proteins at known concentrations were boiled for 150 s followed by centrifugation of the samples at 245,000 × g for 20 min to separate supernatant and pellets. The protein present in the supernatant was entirely assignable to calmodulin, as determined by SDS-PAGE, and concentrations were estimated by Bradford assay (15). The portion of the total protein in the original sample attributable to the heavy chain was determined by subtracting the amount of calmodulin from the protein concentration of the original sample. Protein concentrations were converted to molar quantities.
Gel Filtration
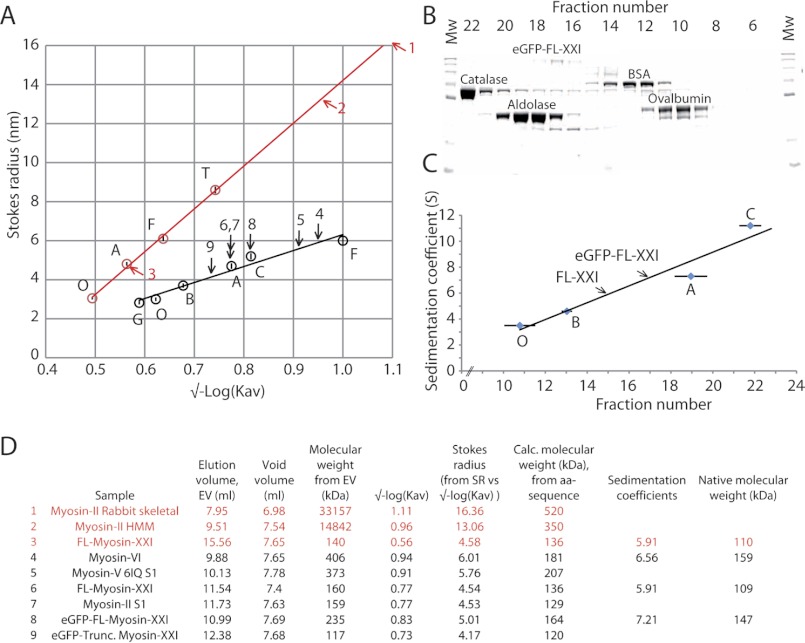
100 μl samples of purified protein were applied to a Superdextm-200 (10/300 GL) analytical column (GE Healthcare) or Superosetm-6 column (10/300 GL) equilibrated in 150 mm NaCl, 50 mm Tris (pH 7.5), 1 mm MgCl2, and 1 mm DTT and controlled using an ÄKTA purifier (see above). The column was calibrated with GFP, chymotrypsin A, ovalbumin, BSA, aldolase, catalase, ferritin, and thyroglobulin standards as described (16) and according to the instructions of the manufacturer (GE Healthcare. The partition coefficient Kav for the standard proteins was calculated as follows (17):
 |
with Ve elution volume of the protein, Ve void volume, and Vc total volume of the column. Because the molecular weight determination of non-globular proteins may not correspond well to the calibration curves established for the globular calibration proteins, we related the Stokes radius Rs of the proteins, rather than their molecular weight, to their partition coefficient Kav and plotted Rs versus SQRT(-log(Kav)) (17).
Sucrose Density Gradients
The 6–20% sucrose gradient contained 500 mm NaCl, 150 mm imidazole (pH 7.4), 10 mm Na2PO42− (pH 7.2), 5 mm MgCl2, and 1 mm EGTA. Samples of purified myosin-XXI (2 mg/ml) together with protein standards of 0.5 mg ovalbumin, 0.25 mg BSA, 0.5 mg aldolase, and 0.5 mg catalase were layered on top of the gradient in a volume of 200 μl. The gradients were centrifuged for 18 h at 4 °C at 38,000 rpm in an SW 40 Ti Beckman rotor. Following centrifugation, 263-μl fractions were dripped from the bottom and run on a 10% SDS-PAGE gel.
Calculation of Native Molecular Weight
The native molecular weight of myosin-XXI was calculated from its Stokes radius measured by gel filtration, and its sedimentation coefficient was determined by sucrose density gradient centrifugation using the following equation as described by Post et al. (18):
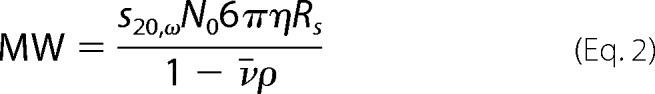 |
with sedimentation coefficient s20,ω, Avogadro's number N0 = 6.02 × 1023, viscosity coefficient η = 1 × 10−2 g/s/cm, Stokes radius Rs, solution density ρ = 1 g/cm3, and partial specific volume υ = 0.72 cm3/g (value for an average soluble protein (18)).
ATPase Assay
Ca2+-actin monomers were converted to Mg2+-actin with 0.2 mm EGTA and 50 mm MgCl2 and then polymerized by dialysis into an assay buffer containing 25 mm KCl, 4 mm MgCl2, 1 mm EGTA, 1 mm dithiothreitol, and 25 mm imidazole (pH 7.4). A 1.1 molar equivalent of phalloidin (Sigma) was used to stabilize actin filaments. ATP (Sigma) was purified by anion exchange chromatography to 99.7% purity, as determined by HPLC. Quantification was performed using a Cary 50 spectrophotometer (Varian) with the extinction coefficients of OD259 of 15,400 M/cm. Steady-state ATPase-activities were measured at 22 (± 0.1) °C in assay buffer supplemented with the NADH-coupled assay components (0.2 mm NADH, 2 mm phosphoenolpyruvate, 3.3 units/ml lactate dehydrogenase, 2.3 units/ml pyruvate kinase) and various actin concentrations (0–30 μm). The final [MgATP] was 2 mm. The myosin-XXI concentration was 100–300 nm. The assay was started by the addition of myosin-XXI. The change in absorption at A340 nm was followed for 5 min. All measurements were repeated at least three times. The kcat and Kactin values were determined by fitting the data to equation 3,
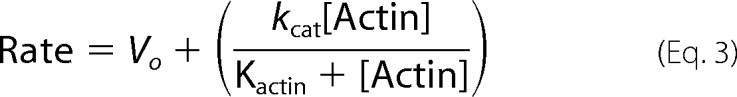 |
where Vo is the basal ATPase activity of myosin-XXI, kcat is the maximum actin-activated ATPase rate, and Kactin is the concentration of actin needed to reach half maximal ATPase activity (i.e. Km for actin).
In Vitro Motility Assay
Rabbit skeletal G-actin was prepared and polymerized with Tetramethyl Rhodamine Isothiocyanate (TRITC)-phalloidin as described previously (19). Motility assays were performed at 22 °C as described by Kron et al. (20). The assay buffer (as described in the previous section) was supplemented with a scavenger system (20 mm DTT, 0.02 mg/ml catalase, 0.1 mg/ml glucose oxidase, 3 mg/ml glucose) and 2 mm ATP. For motility assays at increasing ionic strength or calcium concentrations, the buffer was supplemented with KCl and CaCl2, respectively. Free calcium concentrations were calculated using maxchelator. Accordingly, solutions with pCa 3.1, 6.2, 6.8, and 7.2 where obtained by adding 2, 0.9, 0.7 and 0.5 mm CaCl2 to the assay buffer. For fluorescence imaging, TRITC-phalloidin-labeled actin filaments were excited with a 532-nm laser (5 milliwatt). Images were recorded every 10 s for a total period of 300 s. Only filaments moving continuously for at least 20 frames were included in the data analysis to determine the gliding velocity. Velocities were determined using the kinetic imaging motion analysis software GmimPro.
Electron Microscopy
Myosin-XXI was diluted to 100 nm in a buffer containing 25 mm KCl, 50 mm Tris (pH 7.5), 0.1 mm MgCl2, 0.1 mm DTT. The protein was applied to UV-treated, continuous carbon-coated copper grids and negatively stained with 1% uranyl acetate as described (21). Images were recorded on a JEOL JEM-1011 transmission electron microscope (Lehrstuhl Prof. Joachim Rädler, Physics Department, Ludwig-Maximilians-Universität, Munich, Germany).
RESULTS
Expression and Purification of Full-length Myosin-XXI with Calmodulin
Full-length Leishmania myosin-XXI heavy chain (FL-XXI) and a truncated motor domain (aa 1–800) with and without a N-terminal eGFP tag (Fig. 1A) were cloned into the pFastBacHB vector and coexpressed in the presence and absence of Xenopus calmodulin using a baculoviral/Sf21 system. We used Xenopus calmodulin for coexpression because of its high sequence agreement with calmodulin found in L. donovani (see Fig. 4C). Approximately 5 mg of protein was obtained from 5 × 108 cells, and all constructs were stable with or without calmodulin coexpression, as shown by size exclusion chromatography (SEC) (Fig. 1D). SDS-PAGE indicated that coexpressed calmodulin binds to full-length myosin-XXI (Fig. 1, B and C). Consistent with this, an earlier elution (corresponding to a larger Stokes radius, see below) was found for FL-XXI, with calmodulin-bound FL-XXI compared with FL-XXI expressed in the absence of calmodulin, as shown using SEC (Fig. 1D). Both SDS-PAGE and SEC indicated that the truncated motor domain (aa 1–800) was unable to bind calmodulin when coexpressed in the same way as the full-length protein. This result suggested that calmodulin binds to full-length myosin-XXI downstream of amino acid 800 in the sequence.
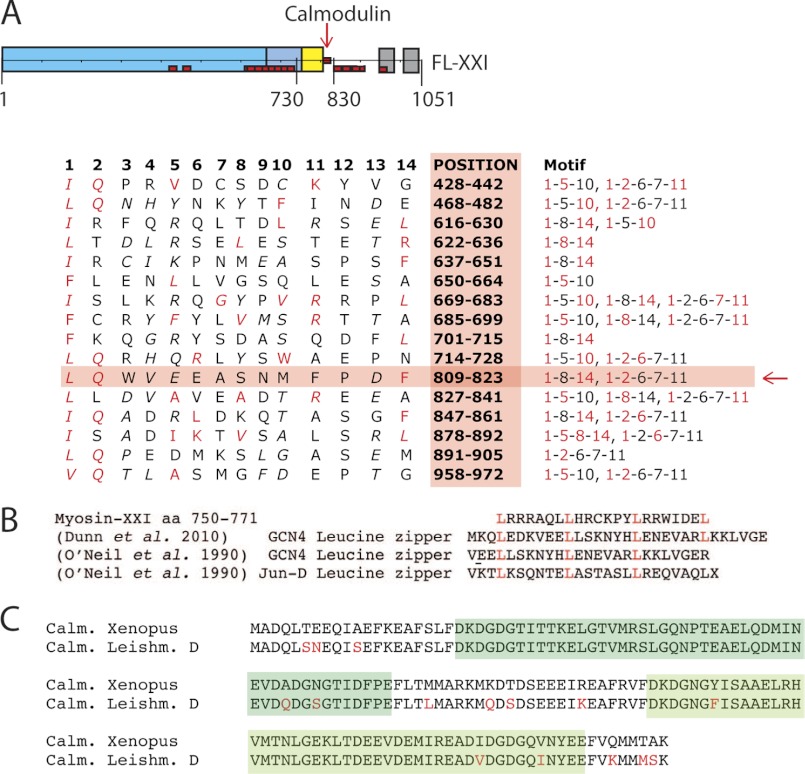
FIGURE 4.
Calmodulin binding site. Sequence analysis of myosin-XXI to identify potential calmodulin binding sites. A, schematic of the FL-XXI construct. Positions of 16 potential binding sites, determined according to the literature (22, 23, 36), are marked in red. The table shows the sequences and positions of the 16 potential calmodulin binding sites for FL-XXI. Red letters indicate residues consistent with calmodulin-binding motifs according to the literature. The red arrow indicates the most probable site from our calmodulin binding studies. B, table showing a selection of known leucine zipper sequences (37, 38) aligned with the predicted leucine zipper of myosin-XXI. C, table showing the high similarity in sequence of calmodulin found in Xenopus (and used in the experiments in this study) and in L. donovani. Marked in green are the two predicted calcium-binding EF-hand domains.
Myosin-XXI Has Multiple Potential Calmodulin-binding Motifs but Binds Only a Single Calmodulin
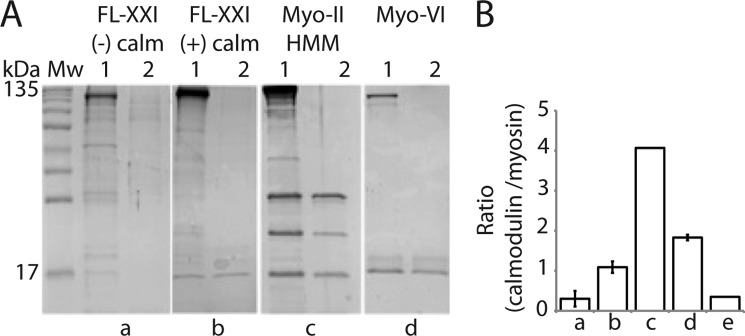
We analyzed the FL-XXI sequence for potential calmodulin-binding positions (22, 23). A total of 16 potential binding sites were mapped (red boxes, Fig. 1A). To determine the number of calmodulins binding to the FL-XXI heavy chain, we performed a calmodulin-binding assay as described by Coluccio (14). In short, samples of known protein concentrations were boiled, subsequently cooled down rapidly on ice, and centrifuged to separate soluble protein in the supernatant from insoluble protein in the pellet. The protein in the supernatant was entirely attributable to calmodulin, which refolds after temperature-dependent denaturation, in contrast to the myosin heavy chain. The heavy chain fraction in the original sample was calculated by subtracting the total amount of calmodulin detected in the supernatant from the starting concentration, and the stoichiometry between calmodulin and myosin heavy chain in the original sample was determined. The assay suggested that only a single calmodulin is bound to FL-XXI and confirmed that no calmodulin is bound to the truncation (Fig. 2, A and B), consistent with a single calmodulin binding downstream of aa 800. To further narrow down calmodulin-binding to potential binding sites, two myosin tail constructs of different lengths were expressed and purified using an E. coli expression system (Fig. 3A). We found that apo-calmodulin (i.e. calcium-free calmodulin) did bind very little or not at all to the tail fragments (Fig. 3, C and D). However, in the presence of calcium pCa 4.1, added calmodulin bound to the 730-tail fragment (Fig. 3E) but not to the 830-tail (C). Only a single calmodulin-binding motif is present between the end of the truncated myosin construct (aa 1–800) and the beginning of the 830 tail fragment (Fig. 4A), indicating that calmodulin probably binds to the motif at aa 809–823. The sequence in this region corresponds, at least in part, to a 1-2-6-7-11 (22) and to a 1-8-14 (23) calmodulin-recognition motif known to be calcium-sensitive.
FIGURE 2.
Calmodulin stoichiometry. A, separation of calmodulin from myosin preparations by heat treatment and analysis by SDS-PAGE as described by Coluccio et al. (14). In panels a–d, lane 1 refers to the sample containing the myosin heavy chain and bound calmodulin before heat treatment. Lane 2 refers to the supernatant after heat treatment and subsequent centrifugation at 245,000 × g for 20 min. Gels are representatives of n experiments: sample FL-XXI expressed without calmodulin (n = 2) (a), FL-XXI co-expressed with calmodulin (n = 4) (b), tissue-purified myosin-II heavy meromyosin (HMM) (n = 1) (c), and chicken full-length myosin-VI coexpressed with calmodulin (n = 2) (d). Note the very faint band at 17 kDa consistent with calmodulin in lane a1. This band probably indicates a small amount of endogenous insect cell calmodulin that has been picked up by the expressed Leishmania myosin heavy chains. B, the histogram shows the ratios of myosin heavy chains to calmodulin (mean ± S.D.). The histogram column (e) shows the truncated myosin-XXI construct (aa 1–800) coexpressed with calmodulin (n = 1).
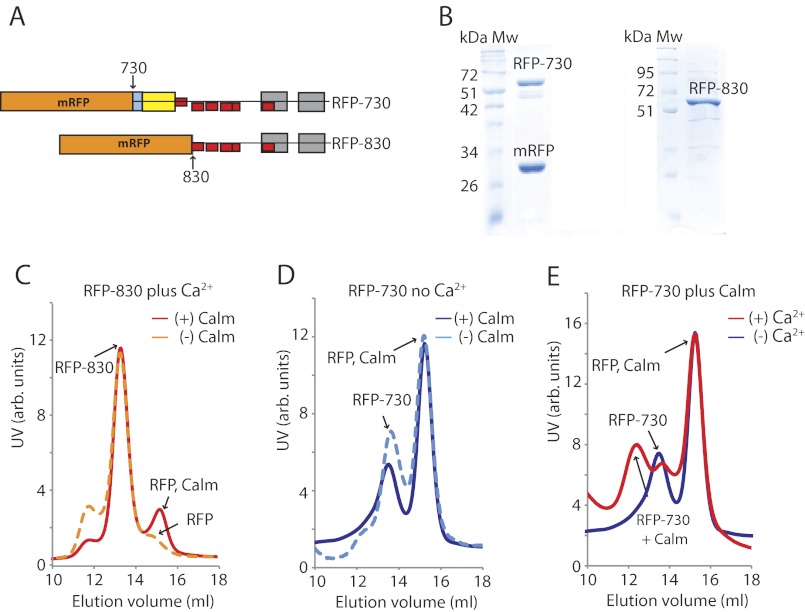
FIGURE 3.
Calmodulin binding to myosin-XXI tail constructs. A, diagram of N-terminally tagged mRFP-myosin-XXI tail constructs (RFP-730 and RFP-830). B, SDS-PAGE of purified RFP-730 and RFP-830. Note that the RFP-730 sample contains a significant amount of mRFP alone because of abortive protein expression of the fusion protein. C–E, size exclusion chromatography of RFP-730 and RFP-830 using a Superdextm-200 column. Traces in red and orange show experiments in the presence of calcium. Traces in blue show experiments in the absence of calcium. Tail constructs (∼1 mg/ml) were mixed for 30 min at 4 °C with calmodulin (∼3-fold molar excess) before loading onto the gel filtration column. Traces are representative of experiments repeated at least three times. C, RFP-830 in the presence of calcium pCa 4.1, without calmodulin (dashed orange line), and with calmodulin (solid red line). Note the calmodulin and copurified abortive RFP-peak at 15 ml elution volume (also in D and E). D, RFP-730 without calcium, without calmodulin (blue dashed line), and with calmodulin (solid blue line). E, RFP-730 in the presence of calmodulin, without calcium (blue line) and with calcium (pCa 4.1, red line). Note the reduced elution volume (about 12 ml) of RFP-730 upon calmodulin binding.
Myosin-XXI Appears to be Monomeric under our Conditions
The section of sequence between the end of the converter domain and the calmodulin-binding motif (aa 754–809) is predicted to form a coiled coil (Fig. 1A). Gel filtration studies suggested, however, that the 730 tail construct (∼68 kDa), which contains this entire section, is monomeric. In these studies, the 730 tail construct eluted at roughly the same volume as the 830 tail construct (∼56 kDa), which does not contain the predicted coiled coil section (Fig. 3, C–E). In fact, the SEC studies showed three clear peaks for both of the constructs. These peaks were consistent with the monomeric 730 and 830 tail constructs on their own, the monomeric tail constructs with calcium-calmodulin-bound, and monomeric red fluorescent protein (∼30 kDa) and calmodulin (∼ 17 kDa) on their own. The presence of some abortive mRFP expression of the mRFP-tail fusion constructs was confirmed by SDS-PAGE (Fig. 3B). Peaks at elution volumes < 12 ml indicating tail fragment dimerization (i.e. > 100 kDa) were not observed. The small peak at ∼12 ml elution volume for the 830 tail might indicate some small fraction of 830 tails with bound calmodulin, possibly partly because of endogenous (calcium)-calmodulin present during protein expression (Fig. 3C). However, the fraction of 830 tails with bound calmodulin was only very small compared with the majority of 730-tails with calcium-calmodulin bound under the same conditions (Fig. 3E). In summary, the SEC studies on the myosin-XXI tails suggested that, at least under the conditions in this study (150 mm NaCl, pCa 3–7), myosin-XXI seems to be monomeric and to bind a single calmodulin between aa 809–823 in the presence of calcium at pCa 4.1. When analyzing the sequence in more detail, the predicted coiled-coil sequence between the end of the converter domain and the calmodulin-binding motif (aa 750–771, Fig. 4B) could, in fact, be made up, at least in part, of a single α-helical (SAH) domain, which does not dimerize (24).
SEC Suggested That, Compared with Other Classes of Myosin, Myosin-XXI Has a Compact Conformation in Vitro, Similar to Skeletal Myosin II S1
When using SEC to determine the hydrodynamic properties of proteins, the elution volume can be related directly to the protein molecular weight if the proteins are globular in shape (Fig. 5). The elongated shape of myosins, however, causes many of them to run through the SEC column much quicker than their molecular weights would predict, indicating relatively high Stokes radii (Fig. 5A, see, for example, skeletal muscle myosin and the myosin-II HMM subfragment). To identify possible oligomerization states of myosin-XXI, we used two SEC columns with different ranges of separable Stokes radii (Superosetm-6 and Superdextm-200). To relate the measured Stokes radii to molecular weight, the sedimentation coefficient of the proteins was measured to take into account the molecule shapes deviating from globular (16). We used sucrose density gradient centrifugation to establish the sedimentation coefficient of standard proteins and of myosin-XXI (Fig. 5B). This, combined with the Stokes radius, allowed us to determine a “native” molecular weight for FL-XXI, both with and without the eGFP tag (Fig. 5C). At 4 °C and in our buffer conditions, FL-XXI was monomeric and eluted from the SEC columns as a compact globular-like protein. In fact, FL-XXI (molecular weight calculated from amino acid sequence ∼119 kDa plus 17 kDa with bound calmodulin) eluted at about the same volume as skeletal myosin-II S1 (∼110 kDa including both light chains, Fig. 5D). Calmodulin binding increased the Stokes radius of FL-XXI as expected (Fig. 1D). The native molecular mass of ∼110 kDa estimated in these experiments was somewhat smaller than the theoretical value of 119 kDa plus 17 kDa for a single bound calmodulin (Fig. 5D). We validated our approach by determining the molecular weight of myosin-VI (Fig. 5, A and D), which is known to adopt a compact conformation (16). The discrepancy between the calculated molecular mass on the basis of amino acid sequence (myosin-VI, ∼181 kDa; FL-XXI, ∼136 kDa) and our estimate on the basis of SEC and sucrose gradients (myosin-VI, ∼159 kDa; FL-XXI, ∼110 kDa) was within the error of these techniques. The slight underestimation of the true size of FL-XXI might be due to a more tightly packed conformation in our conditions.
FIGURE 5.
Hydrodynamic properties of myosin-XXI. The hydrodynamic properties of the myosin were determined using size exclusion chromatography and sucrose density gradients. A, calibration curves for the Superdex™-200 (black) and Superose™-6 (red) columns were generated using standard proteins of known Stokes radii. Stokes radii were plotted versus √-Log(Kav) (see “Experimental Procedures”). Arrows mark the Stokes radius of different myosin constructs (see D for index). All experiments were performed at least three times. Standard proteins used were GFP G (∼27 kDa), ovalbumin O (∼45 kDa), BSA B (∼66 kDa), aldolase A (∼160 kDa), catalase C (∼240 kDa), ferritin F (∼450 kDa), and thyroglobulin T (∼660 kDa). B, ultracentrifugation of eGFP-FL-XXI and standard proteins on a sucrose density gradient were used to determine the sedimentation coefficients. SDS-PAGE shows the fractions from a typical 6–20% sucrose gradient. Note that catalase and aldolase are tetramers but run as monomers on SDS-PAGE. Molecular weight (Mw) markers are shown at both ends of the gel. C, calibration curve of standard proteins with known sedimentation coefficients (mean ± S.D.). Tagged and untagged FL-myosin-XXI are marked by arrows. Each sample was repeated at least three times, and markers were run at least 10 times. Shown are the sedimentation coefficients of standard proteins catalase 11.2 S, aldolase 7.3 S, BSA 4.6 S, and ovalbumin 3.55 S. eGFP-FL-XXI 7.21 S and untagged FL-XXI 5.91 S. All experiments were performed at 4 °C. D, table summarizing the data above (data from Superdex™200 in black, from Superose™ 6 in red) and shows the predicted and calculated molecular weights of myosin-XXI. This method was verified by reproducing the known Stokes radius for myosin-VI (16).
The Myosin-XXI Tail Does Not Inhibit ATPase Activity
We determined the steady-state actin-activated ATPase activity for full-length and truncated myosin-XXI using the NADH-coupled assay (Fig. 6) (25). The FL-XXI used here was expressed in the presence of calmodulin. In the absence of actin, the basal ATPase rate of FL-XXI was ∼0.04 s. The ATPase activity was activated by actin about 15-fold to a maximum rate of 0.5 s with a calculated Kactin of ∼12 μm (Fig. 6A). The truncated construct had a slightly higher basal ATPase rate (∼ 0.05 s). The actin activation of the ATPase activity was slightly lower, with a maximum rate of ∼ 0.3 s. Kactin was reduced to ∼ 2.8 μm (Fig. 6B).
FIGURE 6.
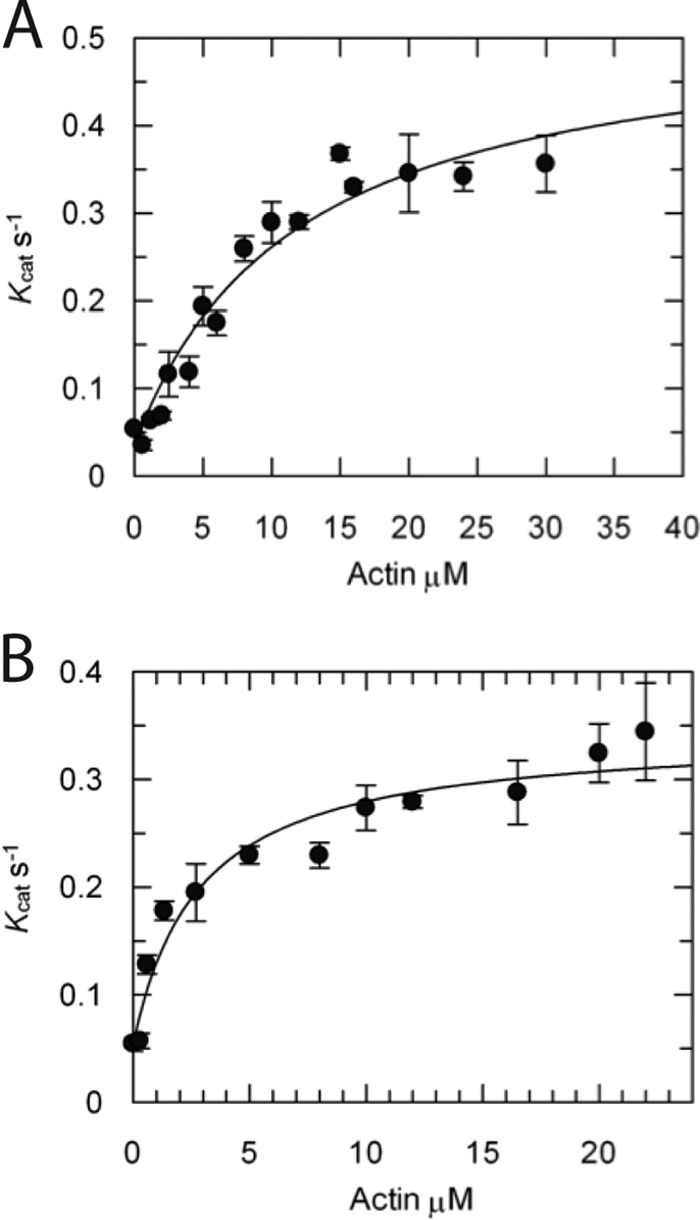
ATPase activity. Steady-state actin-activated ATPase activity was determined for A, eGFP-FL-XXI expressed in the presence of calmodulin, and B, eGFP-Truncation using an NADH-coupled assay (see “Experimental Procedures”). All experiments were repeated using three different protein purification preps. Rates are mean values ± S.D. (n = 3). Rates were fitted according to Michaelis-Menten kinetics (equation 1, see “Experimental Procedures”).
Gliding Filament Assays Showed That Calmodulin Binding Is Necessary for the Translocation of Actin Filaments and That the Sliding Velocity Is Insensitive to Salt (25–1000 mm) and Free Calcium (pCa 3–7)
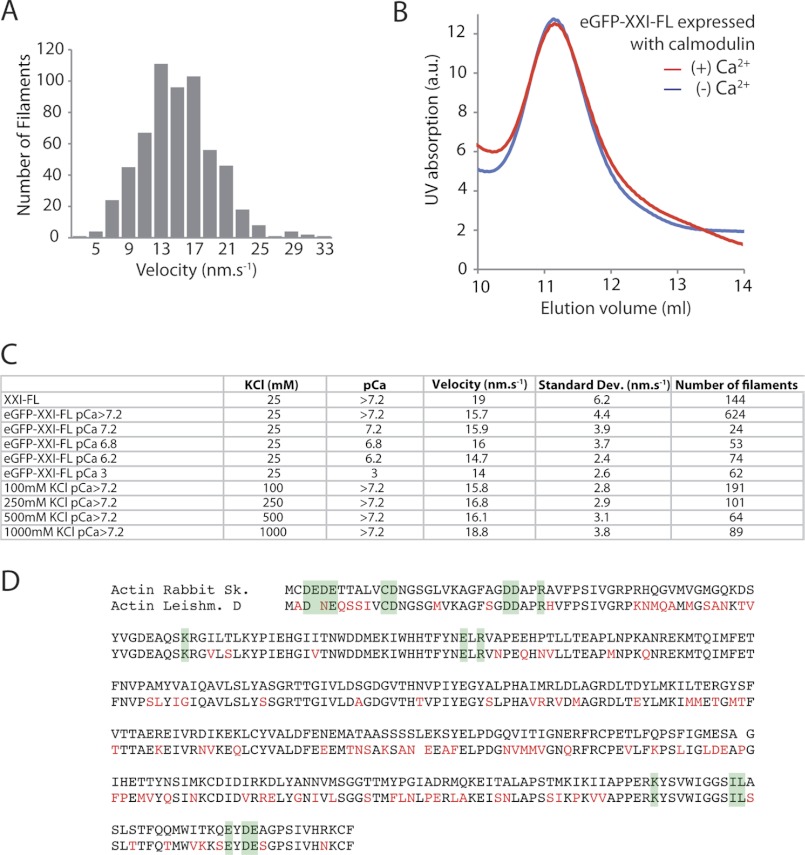
Myosin-XXI actively transported rhodamine-phalloidin-labeled actin filaments in the sliding filament assay (20) with a mean velocity of ∼15 nm/s at 22 °C (Fig. 7, A and C). The truncated and the FL-XXI expressed in the absence of calmodulin did bind to actin in an ATP-dependent fashion, but neither construct generated movement in this assay. The eGFP tag did not affect the velocity of myosin-XXI significantly, and FL-XXI was insensitive to both a wide range of free calcium concentrations and ionic strength (Fig. 7C). This data suggested that once bound, calmodulin could not be displaced by high calcium concentrations. This was confirmed in a SEC experiment where the reduction in calcium concentration did not affect the elution volume of the complex, consistent with calmodulin not detaching from the myosin heavy chain (Fig. 7B).
FIGURE 7.
Gliding filament assay. A, the distribution of velocities of rhodamine-phalloidin-labeled actin filaments gliding over eGFP-FL-XXI. Myosin was deposited on nitrocellulose surfaces at 200 μg/ml. B, SEC (Superdex-200) of FL-XXI coexpressed with calmodulin in the absence and presence of calcium pCa 4.1. Experiments were repeated a minimum of three times from three separate protein purifications. C, comparison of the mean velocity obtained with myosin-XXI at different ionic strengths and calcium concentrations. All experiments were carried out at 22 °C. Data were collected from at least three separate flow cells for each condition. D, multiple sequence alignment of rabbit skeletal and L. donovani actin. Differences are marked in red. Residues on the surface of actin involved in myosin binding according to Kabsch et al. (35) are highlighted in green.
A Compact Conformation of Monomeric Myosin-XXI Was Confirmed by Electron Microscopy
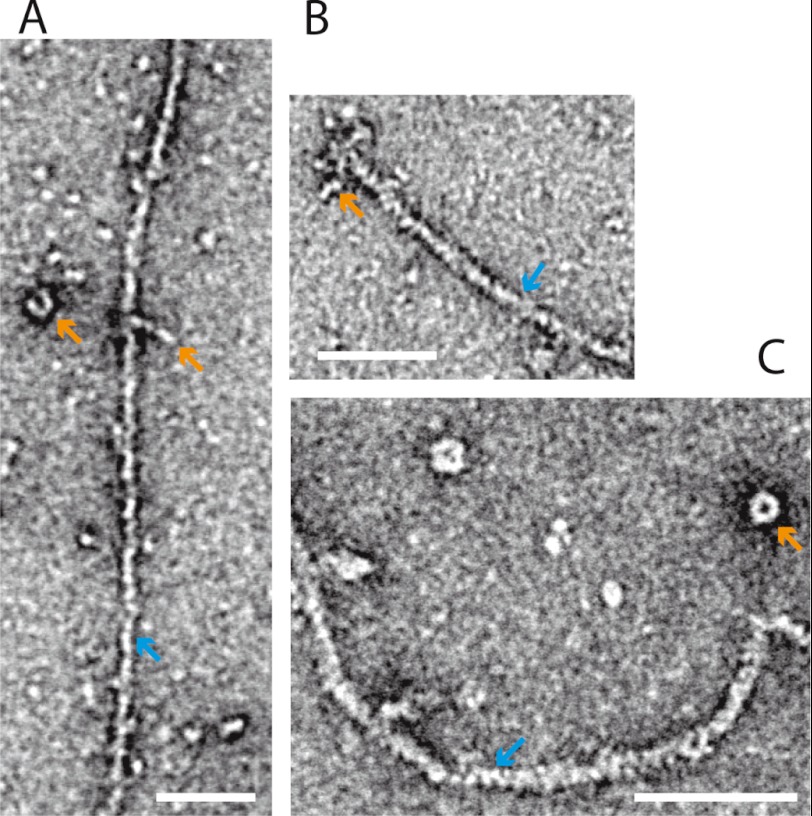
We have used transmission electron microscopy to study the conformation of negatively stained myosin-XXI in the absence and presence of actin (Fig. 8). Detached from actin, the myosin molecules often adopted a compact, sometimes ring-like conformation (orange arrows Fig. 8, A and C). When bound to actin, myosins were often found accumulated at the end of the actin filaments (Fig. 8B). Further electron microscopic studies are underway to characterize the different conformations of myosin-XXI in the presence and absence of actin in more detail.
FIGURE 8.
Electron microscopy. Negative stain electron microscopy of eGFP-FL-XXI bound to F-actin (see “Experimental Procedures”). A, single myosin-XXI molecules (orange arrow) are bound to an actin filament (blue arrow). When attached to the surface on their own they often adopted a compact structure (orange arrow). B, several myosin-XXI molecules are attached to the end of an actin filament. C, myosin-XXI molecules form a compact, ring-like structure when detached from actin. Scale bars = 50 nm.
DISCUSSION
We have successfully expressed full-length myosin-XXI as well as a truncated motor domain construct to investigate the structural and chemomechanical properties of this novel myosin motor found in Leishmania parasites. Sequence analysis of the protein suggested 16 potential calmodulin binding motifs on the myosin heavy chain. However, the stoichiometry between myosin-XXI heavy chain and bound light chains, combined with the calmodulin-binding studies to different tail fragments, indicated that myosin-XXI binds only a single calmodulin to a calmodulin-binding motif at aa 809–823 in the heavy chain sequence.
Comparing the sequence of calmodulin and calmodulin-like proteins in the L. donovani genome, we found a high similarity to Xenopus calmodulin (Fig. 4C), which is why we used Xenopus calmodulin for coexpression with the Leishmania myosin-XXI heavy chain. In addition, Leishmania calmodulin has only a small number of differences (12 amino acids) compared with Drosophila calmodulin, which has been shown to change conformation upon calcium-binding, to expose the hydrophobic patches required for typical target binding (26). The sequences of Drosophila and Xenopus calmodulin, on the other hand, are essentially identical. They differ only in two amino acids (aa 144 and 148). Furthermore, many of the differences in the calmodulin sequence between Leishmania on the one hand and Drosophila or Xenopus on the other are conservative (e.g. isoleucine for valine, Fig. 4C). The calcium-binding EF-hand domains in the Leishmania sequence (aa 21–68 and aa 94–141) and in the Drosophila or Xenopus sequences are almost identical (Fig. 4C, green sections). None of the very small differences in sequence (five amino acids) in these sections are expected to affect the ability of the protein to bind calcium. This is entirely consistent with our finding that Xenopus calmodulin was found to bind to Leishmania myosin heavy chain in a calcium-dependent manner but would not detach from the target sequence once bound to it, neither by changes in ionic strength nor by changes in calcium concentration (26–28). The target sequence (aa 809–823) on the myosin heavy chain that we identified for calmodulin binding is somewhat unusual in the sense that it contains a lot of negative charge. It is also interesting to note that this calmodulin binding motif is not the strongest motif predicted from sequence alignments (Fig. 4A). However, it does have bulky hydrophobic residues at the appropriate positions, such as positions 1–4, 10–11, and 14, as shown in Fig. 4A. It is therefore expected that this sequence will bind calmodulin and that calmodulin-binding would be calcium-dependent, as described in the literature (26–28) and as observed in our study.
In the sequence, the calmodulin-binding motif is preceded by a section identified as a natural leucine zipper motif (Fig. 4B), indicating the possibility of dimerization at this part of the structure. However, the size exclusion studies suggested that, at least in the conditions applied in this study, myosin-XXI as well as the tail fragments were monomeric. Closer inspection of the sequence between the end of the converter (aa 750) and the calmodulin-binding motif at aa 809–823, revealed that this section would also be consistent with an SAH domain (24). In myosin-X (24) and myosin-VI (29), such domains have been suggested to contribute to the lever arm that amplifies small conformational changes in the catalytic domain of the motor to produce force and movement (30). In the in vitro motility assays, monomeric full-length myosin-XXI with calmodulin bound was mechanically functional and able to move actin filaments. This is consistent with a mechanically stable domain (such as a SAH domain perhaps) bridging the gap between the end of the converter and the calmodulin-binding lever arm region of the motor molecule. If that bridging section were flexible, it would be hard to envisage how conformational changes in the catalytic domain could be transduced into productive movement at the end of the lever arm. On the other hand, however, the motor truncated at the end of the predicted SAH domain showed nearly unchanged ATPase activity compared with the full-length molecule but was not mechanically functional. This would suggest that the predicted SAH domain on its own (that means in the absence of the following calmodulin-binding region) is unable to form a mechanically functional lever arm and that both the predicted SAH domain and the subsequent calmodulin-binding region are necessary to form a functional lever arm structure. However, at this stage we cannot exclude the possibility that the truncated motor simply bound to the nitrocellulose surface in an unfavorable way so that lever arm movement was impaired for that reason. High-resolution structural studies are currently underway to characterize these domains of the myosin molecule in detail.
The SEC experiments and EM studies suggested that, at least in the experimental conditions here, myosin-XXI is monomeric and has a compact structure with a Stokes radius comparable with muscle myosin-II S1, which has a similar molecular weight. The Stokes radius of myosin-XXI was slightly larger when calmodulin was bound, consistent with calmodulin stabilizing the extended α-helical target sequence that forms a functional lever arm in other myosins (30). The EM studies also suggested that myosin-XXI does bind along the length of actin filaments but tends to accumulate at the filament ends (Fig. 8B). Further experiments are underway to investigate this observation in more detail. In the absence of actin, myosin-XXI frequently seemed to form compact, ring-like structures possibly composed of more than one myosin-XXI molecule. Again, further studies are currently underway to characterize the conditions for ring formation in more detail.
The full-length and truncated Leishmania myosin constructs showed similar actin-activated ATPase activity. We therefore conclude that calmodulin binding and the absence of the tail in the truncation has little effect on the ATPase activity. The rates were similar to smooth muscle myosin with phosphorylated regulatory light chain (31). Interestingly, the sliding velocity was 50-fold lower compared with smooth muscle myosin. This could be due to unfavorable binding of myosin-XXI, which was adsorbed non-specifically to the nitrocellulose surface. However, under the same binding conditions, 20- to 200-fold higher sliding velocities were observed for smooth muscle myosin S1 (32) and skeletal muscle myosin S1 (33, 34), respectively. If not because of unfavorable binding, the low mechanical speed could also point toward a function as an anchor or tether for this monomeric form of Leishmania myosin-XXI that does not require high speed of movement. Further studies are in progress to immobilize full-length and truncated myosin-XXI motor molecules in a more specific fashion on the surface to address this issue.
For both the solution kinetics studies and in vitro motility assays, we used actin tissue-purified from rabbit skeletal muscle. This can be obtained more readily than the Leishmania-specific actin, which would have to be recombinantly expressed. Intriguingly, Leishmania actin and rabbit skeletal actin share only about 70% sequence identity and 87% similarity as determined by multiple sequence alignment (Macvector). However, residues on the surface of actin monomers assumed to be involved in myosin binding (35) are highly conserved (Fig. 7D). This suggests that rabbit skeletal actin might interact with myosin-XXI in a similar fashion compared with endogenous Leishmania actin. In addition this approach enables us to compare the functional assays on myosin-XXI to other unconventional myosins from different species, which have also been carried out with actin purified from rabbit skeletal muscle.
To summarize, we found that myosin-XXI, to date the only myosin motor protein detected in Leishmania parasites, can be expressed recombinantly using a baculovirus expression system. Under our conditions, this myosin is a monomeric, mechanically functional molecular motor that binds a single calmodulin and moves actin filaments at a very low speed. This would be consistent with the motor acting, at least in its monomeric form, as a tether or anchor in the parasite, possibly involved in endocytic processes within the flagellar pocket, possibly also in other intracellular trafficking processes and in the formation of the paraflagellar rod structure (6). Intriguingly, sequence analysis indicates several coiled-coil sections in the tail domain of this myosin, not to mention the ubiquitin binding domains the regulatory functions of which remain completely unclear. It is therefore possible that in different conditions myosin-XXI might well dimerize or even oligomerize to perform the variety of functions it is expected to be involved in, as it seems to be the only myosin motor expressed in the Leishmania parasite.
Acknowledgments
We thank Joachim Rädler at the Physics Department of Ludwig-Maximilians-Universität, Munich, Germany, for letting us use TEM for this study and Steve Martin at The National Institute for Medical Research London, UK, for scientific discussions.
This work was funded by the Deutsche Forschungsgemeinschaft SFB 863, by the Friedrich-Baur Stiftung, and by the Munich Centre for Nanoscience (CeNS). This work was also supported by an EMBO long-term fellowship (to C. P. T.).
- UBA
- ubiquitin-associated domain
- aa
- amino acid(s)
- SEC
- size exclusion chromatography
- FL-XXI
- full-length Leishmania myosin-XXI heavy chain
- SAH
- single α-helical.
REFERENCES
- 1. Desjeux P. (2004) Leishmaniasis. Current situation and new perspectives. Comp. Immunol. Microbiol. Infect. Dis. 27, 305–318 [DOI] [PubMed] [Google Scholar]
- 2. Sacks D., Kamhawi S. (2001) Molecular aspects of parasite-vector and vector-host interactions in leishmaniasis. Annu. Rev. Microbiol. 55, 453–483 [DOI] [PubMed] [Google Scholar]
- 3. Foth B. J., Goedecke M. C., Soldati D. (2006) New insights into myosin evolution and classification. Proc. Natl. Acad. Sci. U.S.A. 103, 3681–3686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Odronitz F., Kollmar M. (2007) Drawing the tree of eukaryotic life based on the analysis of 2,269 manually annotated myosins from 328 species. Genome Biol. 8, R196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Katta S. S., Sahasrabuddhe A. A., Gupta C. M. (2009) Flagellar localization of a novel isoform of myosin, myosin XXI, in Leishmania. Mol. Biochem. Parasitol. 164, 105–110 [DOI] [PubMed] [Google Scholar]
- 6. Katta S. S., Tammana T. V., Sahasrabuddhe A. A., Bajpai V. K., Gupta C. M. (2010) Trafficking activity of myosin XXI is required in assembly of Leishmania flagellum. J. Cell Sci. 123, 2035–2044 [DOI] [PubMed] [Google Scholar]
- 7. Santrich C., Moore L., Sherwin T., Bastin P., Brokaw C., Gull K., LeBowitz J. H. (1997) A motility function for the paraflagellar rod of Leishmania parasites revealed by PFR-2 gene knockouts. Mol. Biochem. Parasitol. 90, 95–109 [DOI] [PubMed] [Google Scholar]
- 8. Krendel M., Mooseker M. S. (2005) Myosins. Tails (and heads) of functional diversity. Physiology 20, 239–251 [DOI] [PubMed] [Google Scholar]
- 9. Delorenzi M., Speed T. (2002) An HMM model for coiled-coil domains and a comparison with PSSM-based predictions. Bioinformatics 18, 617–625 [DOI] [PubMed] [Google Scholar]
- 10. Dikic I., Wakatsuki S., Walters K. J. (2009) Ubiquitin-binding domains. From structures to functions. Nat. Rev. Mol. Cell Biol. 10, 659–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Long J., Garner T. P., Pandya M. J., Craven C. J., Chen P., Shaw B., Williamson M. P., Layfield R., Searle M. S. (2010) Dimerisation of the UBA domain of p62 inhibits ubiquitin binding and regulates NF-κB signalling. J. Mol. Biol. 396, 178–194 [DOI] [PubMed] [Google Scholar]
- 12. Heinen C., Acs K., Hoogstraten D., Dantuma N. P. (2011) C-terminal UBA domains protect ubiquitin receptors by preventing initiation of protein degradation. Nat. Commun. 2, 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Margossian S. S., Lowey S. (1982) Preparation of myosin and its subfragments from rabbit skeletal muscle. Methods Enzymol. 85, 55–71 [DOI] [PubMed] [Google Scholar]
- 14. Coluccio L. M. (1994) Differential calmodulin binding to three myosin-1 isoforms from liver. J. Cell Sci. 107, 2279–2284 [DOI] [PubMed] [Google Scholar]
- 15. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 16. Lister I., Schmitz S., Walker M., Trinick J., Buss F., Veigel C., Kendrick-Jones J. (2004) A monomeric myosin VI with a large working stroke. EMBO J. 23, 1729–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ohno H., Blackwell J., Jamieson A. M., Carrino D. A., Caplan A. I. (1986) Calibration of the relative molecular mass of proteoglycan subunit by column chromatography on Sepharose CL-2B. Biochem. J. 235, 553–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Post P. L., Tyska M. J., O'Connell C. B., Johung K., Hayward A., Mooseker M. S. (2002) Myosin-IXb is a single-headed and processive motor. J. Biol. Chem. 277, 11679–11683 [DOI] [PubMed] [Google Scholar]
- 19. Pardee J. D., Spudich J. A. (1982) Purification of muscle actin. Methods Enzymol. 85, 164–181 [DOI] [PubMed] [Google Scholar]
- 20. Kron S. J., Toyoshima Y. Y., Uyeda T. Q., Spudich J. A. (1991) Assays for actin sliding movement over myosin-coated surfaces. Methods Enzymol. 196, 399–416 [DOI] [PubMed] [Google Scholar]
- 21. Walker M. L., Burgess S. A., Sellers J. R., Wang F., Hammer J. A., 3rd, Trinick J., Knight P. J. (2000) Two-headed binding of a processive myosin to F-actin. Nature 405, 804–807 [DOI] [PubMed] [Google Scholar]
- 22. Cheney R. E., Mooseker M. S. (1992) Unconventional myosins. Curr. Opin. Cell Biol. 4, 27–35 [DOI] [PubMed] [Google Scholar]
- 23. Rhoads A. R., Friedberg F. (1997) Sequence motifs for calmodulin recognition. FASEB J. 11, 331–340 [DOI] [PubMed] [Google Scholar]
- 24. Knight P. J., Thirumurugan K., Xu Y., Wang F., Kalverda A. P., Stafford W. F., 3rd, Sellers J. R., Peckham M. (2005) The predicted coiled-coil domain of myosin 10 forms a novel elongated domain that lengthens the head. J. Biol. Chem. 280, 34702–34708 [DOI] [PubMed] [Google Scholar]
- 25. De La Cruz E. M., Ostap E. M. (2009) in Methods in Enzymology: Biothermodynamics, Vol. 455, Part A (Johnson M. L., Holt J. M., Ackers G. K., eds), pp 157–192, Elsevier Academic Press Inc, San Diego [Google Scholar]
- 26. Wang B., Martin S. R., Newman R. A., Hamilton S. L., Shea M. A., Bayley P. M., Beckingham K. M. (2004) Biochemical properties of V91G calmodulin. A calmodulin point mutation that deregulates muscle contraction in Drosophila. Protein Sci. 13, 3285–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Martin S. R., Bayley P. M. (2004) Calmodulin bridging of IQ motifs in myosin-V. FEBS Lett. 567, 166–170 [DOI] [PubMed] [Google Scholar]
- 28. Lewit-Bentley A., Réty S. (2000) EF-hand calcium-binding proteins. Curr. Opin. Struct. Biol. 10, 637–643 [DOI] [PubMed] [Google Scholar]
- 29. Spink B. J., Sivaramakrishnan S., Lipfert J., Doniach S., Spudich J. A. (2008) Long single α-helical tail domains bridge the gap between structure and function of myosin VI. Nat. Struct. Mol. Biol. 15, 591–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rayment I., Rypniewski W. R., Schmidt-Bäse K., Smith R., Tomchick D. R., Benning M. M., Winkelmann D. A., Wesenberg G., Holden H. M. (1993) Three-dimensional structure of myosin subfragment-1. A molecular motor. Science 261, 50–58 [DOI] [PubMed] [Google Scholar]
- 31. Trybus K. M., Waller G. S., Chatman T. A. (1994) Coupling of ATPase activity and motility in smooth muscle myosin is mediated by the regulatory light chain. J. Cell Biol. 124, 963–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cremo C. R., Sellers J. R., Facemyer K. C. (1995) Two heads are required for phosphorylation-dependent regulation of smooth muscle myosin. J. Biol. Chem. 270, 2171–2175 [DOI] [PubMed] [Google Scholar]
- 33. Molloy J. E., Kendrick-Jones J., Veigel C., Tregear R. T. (2000) An unexpectedly large working stroke from chymotryptic fragments of myosin II. FEBS Lett. 480, 293–297 [DOI] [PubMed] [Google Scholar]
- 34. Uyeda T. Q., Abramson P. D., Spudich J. A. (1996) The neck region of the myosin motor domain acts as a lever arm to generate movement. Proc. Natl. Acad. Sci. U.S.A. 93, 4459–4464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kabsch W., Mannherz H. G., Suck D., Pai E. F., Holmes K. C. (1990) Atomic structure of the actin:DNase I complex. Nature 347, 37–44 [DOI] [PubMed] [Google Scholar]
- 36. Osawa M., Tokumitsu H., Swindells M. B., Kurihara H., Orita M., Shibanuma T., Furuya T., Ikura M. (1999) A novel target recognition revealed by calmodulin in complex with Ca2+-calmodulin-dependent kinase kinase. Nat. Struct. Biol. 6, 819–824 [DOI] [PubMed] [Google Scholar]
- 37. Dunn A. R., Chuan P., Bryant Z., Spudich J. A. (2010) Contribution of the myosin VI tail domain to processive stepping and intramolecular tension sensing. Proc. Natl. Acad. Sci. U.S.A. 107, 7746–7750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. O'Neil K. T., Hoess R. H., DeGrado W. F. (1990) Design of DNA-binding peptides based on the leucine zipper motif. Science 249, 774–778 [DOI] [PubMed] [Google Scholar]