Background: Aβ oligomers are major players in Alzheimer disease, but the tools for their detection are not satisfactory.
Results: We developed a SPR-based immunoassay and a test in C. elegans, specifically identifying toxic oligomers.
Conclusion: These methods allow study of the effects of mutations or drugs on Aβ oligomerization.
Significance: The SPR-based immunoassay provides new opportunities for the detection of toxic oligomers in biological samples.
Keywords: Alzheimer Disease, Amyloid, C. elegans, Neurodegenerative Diseases, Protein Misfolding, Surface Plasmon Resonance (SPR)
Abstract
Soluble oligomers of the amyloid-β (Aβ) peptide play a key role in the pathogenesis of Alzheimer's disease, but their elusive nature makes their detection challenging. Here we describe a novel immunoassay based on surface plasmon resonance (SPR) that specifically recognizes biologically active Aβ oligomers. As a capturing agent, we immobilized on the sensor chip the monoclonal antibody 4G8, which targets a central hydrophobic region of Aβ. This SPR assay allows specific recognition of oligomeric intermediates that rapidly appear and disappear during the incubation of synthetic Aβ1–42, discriminating them from monomers and higher order aggregates. The species recognized by SPR generate ionic currents in artificial lipid bilayers and inhibit the physiological pharyngeal contractions in Caenorhabditis elegans, a new method for testing the toxic potential of Aβ oligomers. With these assays we found that the formation of biologically relevant Aβ oligomers is inhibited by epigallocatechin gallate and increased by the A2V mutation, previously reported to induce early onset dementia. The SPR-based immunoassay provides new opportunities for detection of toxic Aβ oligomers in biological samples and could be adapted to study misfolding proteins in other neurodegenerative disorders.
Introduction
Aggregation of β-amyloid (Aβ)3 peptides, eventually leading to brain deposition of amyloid plaques, represents a major and well known hallmark of Alzheimer's disease (AD). Although fibrillar amyloid may contribute to AD pathogenesis, different lines of evidence point to smaller and soluble Aβ assemblies (termed Aβ oligomers) as the main neurotoxic species (1, 2). Aβ oligomers may also represent important biological markers of AD.
The development of reliable tools and sensors to selectively detect the toxic oligomeric species still represents a major need in AD research (3). The recognition that morphologically similar oligomers have very different toxicity points to the existence of subpopulations of biologically active oligomers with specific structural features (4–6). Most of the techniques currently used to visualize Aβ aggregates or to determine their size are not suited for the identification of the toxic oligomers. Conformation- or epitope-specific antibodies may provide a more suitable means for a structure-dependent recognition of specific oligomer subpopulations (5). However, classical immunoassays such as ELISA, Western blot, dot-blot, etc., are time-consuming and require indirect readouts, with long incubation steps and washing procedures that could affect the recognition of transient and unstable species.
Here we describe a novel immunoassay based on surface plasmon resonance (SPR) that specifically detects biologically important oligomers of synthetic Aβ. SPR is a powerful method widely used to study interactions between two macromolecules in real time without labeling the molecules (7, 8). Typically, one of the two interacting partners is immobilized on a sensor chip surface, and the other is flowed through a microfluidic system in contact with the chip surface. Binding is measured in real time as a change of mass at the surface, and the interaction can be characterized in terms of on and off rates (kinetics) and binding strength (affinity).
We immobilized different antibodies on the sensor chip and injected Aβ solutions at different times during the aggregation. We identified an antibody, 4G8, which permitted a direct, real time determination of the binding signal specifically caused by transient oligomers, which could not be obtained with common immunoassays (ELISA or dot-blot).
We then assessed the toxicity of the Aβ oligomers detected by SPR (3) using two new assays suitable to study transient oligomeric species with a relatively short half-life (few hours). We used an electrophysiological assay in model membranes and a behavioral test in the nematode Caenorhabditis elegans, applied here for the first time to test the toxic potential of Aβ oligomers.
For the present studies we used the “depsi-peptide” technique for Aβ synthesis (9, 10), improved in our laboratory (11, 12), which consistently produces seed-free starting solutions. This is a prerequisite for any analysis of the properties of highly aggregating peptides (13).
EXPERIMENTAL PROCEDURES
Anti-Aβ antibodies 4G8 and 6E10 were from Covance; anti-oligomers antibodies A11 and OC (5) were from Invitrogen and Millipore, respectively. Epigallocatechin gallate (EGCG) was a kind gift from INDENA (Milan, Italy).
Aβ Preparation
Depsi-Aβ1–42, depsi-Aβ1–40 (WT), and depsi-Aβ1–40A2V peptides were synthesized as previously described (10, 11). The depsi-peptide is much more soluble than the native one and has a much lower propensity to aggregate, thereby preventing the spontaneous formation of seeds in solution. Aβ peptides were obtained from the corresponding depsi-peptide by a “switching” procedure involving a change in pH (11) and used immediately. The switched solutions were diluted in 10 mm phosphate-buffered solution, containing 150 mm NaCl, pH 7.4 (PBS), and incubated as indicated.
SPR Studies
The SPR apparatus used for the present study (ProteOn XPR36 Protein Interaction Array System; Bio-Rad) has six parallel flow channels that can be used to uniformly immobilize strips of six ligands on the sensor surface. The fluidic system can automatically rotate 90° so that up to six different analytes can be injected, allowing simultaneous monitoring of up to 36 individual molecular interactions in a single run on a single chip (14). The antibodies were immobilized in parallel flow channels of GLC sensor chips (Bio-Rad) using amine coupling chemistry, as previously described (14). Briefly, after surface activation, the antibodies (30 μg/ml in 10 mm acetate buffer, pH 5.0) were injected for 5 min at a flow rate of 30 μl/min, and the remaining activated groups were blocked with ethanolamine, pH 8.0. The final immobilization levels were ∼5000 resonance units (1 RU = 1 pg of protein/mm2), for all the antibodies tested. A “reference” surface was always prepared in parallel using the same immobilization procedure but without addition of the antibody.
After rotation of the microfluidic system, aliquots of Aβ1–42 or Aβ1–40 (WT or A2V) were injected over the immobilized antibodies for 2 min at a flow rate of 30 μl/ml. Dissociation was measured in the following 11 min. The running buffer, also used to dilute the samples, was 10 mm PBS containing 150 mm NaCl and 0.005% Tween 20 (PBST). All of these assays were done at 25 °C. The sensorgrams (time course of the SPR signal in RU) were normalized to a base-line value of 0. The signal observed in the surfaces immobilizing the antibody was corrected by subtracting the nonspecific response observed in the reference surface. The resulting sensorgrams were globally fitted by a two-binding site model (heterogeneous ligand model, ProteOn analysis software) to obtain the corresponding kinetics parameters of the quickly dissociating and the slowly dissociating component (kon and koff) and the equilibrium dissociation constant (KD). Lysates from CL4176 and CL802 C. elegans strains (15, 16) were diluted in PBST to a final protein concentration of 0.05 μg/μl and injected over immobilized 4G8 for 3–5 min at a flow rate of 30 μl/ml.
Size Exclusion Chromatography (SEC)
Synthetic Aβ1–42 (100 μm) was incubated at 25 °C for different times (0, 5, and 24 h). Aliquots were then diluted with 10 mm PBS to a final concentration of 10 μm. SEC was performed using an FPLC apparatus (Biologic DuoFlow FPLC system; Bio-Rad) equipped with a precision column prepacked with: (i) Superdex 75 HR 10/30 (separation range of 3–70 kDa), (ii) Superdex 200 HR 10/30 (10–600 kDa), or (iii) Superose 6 HR 10/30 (5–5000 kDa) (GE Healthcare). The mobile phase (PBS) was set at 0.5 ml/min, and the elution peak was detected at 214-nm UV absorbance. The columns were calibrated using appropriate molecular mass standard proteins (see legend to supplemental Fig. S2). The void volume was determined by blue dextran 2000 (2000 kDa). Lysates from CL4176 C. elegans were fractionated in a Superdex 200 HR 10/30, as described above.
Atomic Force Microscopy (AFM)
Aliquots of synthetic Aβ1–42 were diluted with 10 mm PBS to a final concentration of 10 μm and incubated for 0.5–2 min on a freshly cleaved mica disk. The disk was washed with water and dried under a very gentle nitrogen stream. The sample was then mounted on a Nanoscope V multimode AFM (Veeco/Digital Instruments, Santa Barbara, CA) operating in tapping mode using standard phosphorus-doped silicium probes (Veeco). The scan speed was 1 Hz. For every sample, three pictures from distinct regions were taken. Plane corrected and background removed pictures were obtained with the open source software tool Gwyddion (17).
Dynamic Laser Light Scattering (DLS)
Aliquots of synthetic Aβ1–42 were diluted with 10 mm PBS to final concentrations of 1 and 10 μm. DLS measurements were taken on each sample at 25 °C with a homemade apparatus that includes a diode laser (λ = 532 nm), a temperature-controlled cell, and a digital correlator (Brookhaven Instruments Co) (18, 19). Dynamic measurements of the scattered intensity correlation function yielded the translational diffusion coefficient of particles, and then, via the Stokes-Einstein relation, the average hydrodynamic diameter of the particles in solution was calculated. Several runs were performed on each sample to check for data reproducibility. The data were analyzed by the CONTIN method, suitable for polydisperse systems (20).
Native Gels and SDS-PAGE, Western Blot, and Dot-blot Analysis
Synthetic Aβ1–42 (100 μm) was incubated at 25 °C for different times (0–24 h). The aliquots were then diluted with 10 mm PBS to a final concentration of 10 μm, and the Aβ species were evaluated. For native gels, equal amounts of protein were loaded onto 4–16% (w/v) Bis-Tris native gels (Invitrogen) and electrophoresed at 150 V. After blotting, the membranes were probed with 4G8 (1:1000 dilution). NativeMark standards (Invitrogen) were run in parallel. Equal amounts of protein were fractionated by 16% Tris-Tricine SDS-PAGE and, after blotting, were probed with 4G8 (1:1000 dilution). For dot-blot analysis, 50 ng of synthetic Aβ1–42 was spotted onto nitrocellulose membranes (Millipore), blocked with PBST (PBS, pH 7.4, plus 0.1% (v/v) Tween 20), and incubated for 1 h with 4G8 (1:1000 dilution). Anti-mouse IgG peroxidase conjugate (1:2000 dilution; Sigma) was used as a secondary antibody for 4G8. Immunoreactive bands were detected by ECL chemioluminescence and quantified by Quantity One Image Software (Bio-Rad).
Electrophysiological Assay
Single-channel recordings from lipid bilayer were obtained using the Tip-Dip method, as previously described (21). Experiments carried out using only ionic solutions and pure lipid bilayer do not show any ionic flow for more than 1 h of continuous current recordings (n = 5). Aβ samples were made up to a final concentration of 100 nm, alone and in presence of 4G8 or EGCG.
C. elegans Studies
Wild-type N2, the transgenic Aβ muscle expressing strain CL4176 and its control CL802 (15) were obtained from the Caenorhabditis Genetic Center (University of Minnesota). In CL4176, the expression of human Aβ1–42 depends on raising the temperature from 16 to 24 °C and caused progressive worm paralysis (15). All nematode strains were propagated on solid nematode growth medium seeded with Escherichia coli (OP50) for food.
Pharyngeal Pumping Assay
N2 nematodes (L3–L4 larval stage) were collected by washing plates with M9 buffer, transferred to tubes, centrifuged, and washed two times with 5 mm PBS, pH 7.4, to eliminate bacteria. Synthetic Aβ1–42 (100 μm) was incubated at 25 °C, and aliquots were taken at different time points (0, 5, 24 and 48h), diluted to 0.5–10 μm in 5 mm PBS, pH 7.4, and administered to C. elegans (100 worms/100 μl). Control worms were fed vehicle alone. After 2 h of orbital shaking, the worms were transferred on NMG plates seeded with E. coli, and pharyngeal pumping rate was scored after 2 h of recovery by counting the number of times the terminal bulb of the pharynx contracted over a 1-min interval. In some experiments, worms were fed for 2 h with 10 μm Aβ1–42 preincubated or not, for 30 min, with 4G8 antibody (1:500, v/v in 5 mm PBS, pH 7.4) or with 10 μm Aβ1–42 co-incubated with 10 μm EGCG (in 5 mm PBS, pH 7.4) as described above.
Native Oligomers from CL4176 Nematodes
CL4176 and CL802 age-synchronized nematodes were transferred to fresh nematode growth medium plates and, when reaching maturity at 3 days of age, allowed to lay eggs overnight. Isolated hatchlings from the synchronized eggs (day 1) were cultured at 16 °C on fresh NMG plates (35 × 10-mm culture plates, 100 worms/plate) seeded with E. coli. Transgene expression was induced by raising the temperature from 16 to 24 °C for 44 h, at which time ∼60% of animals resulted paralyzed (16). The worms were then collected and homogenized in lysis buffer.
RESULTS
SPR Studies with Synthetic Aβ1–42
Aβ1–42 was dissolved in PBS to a final concentration of 100 μm and incubated at 25 °C in quiescent conditions (22, 23). Aliquots were then taken at different times (from 0 to 72 h), diluted to 1 μm in PBS and flowed over sensor chips on which we had previously immobilized the anti-Aβ antibodies 4G8 and 6E10 or the anti-oligomer antibody A11.
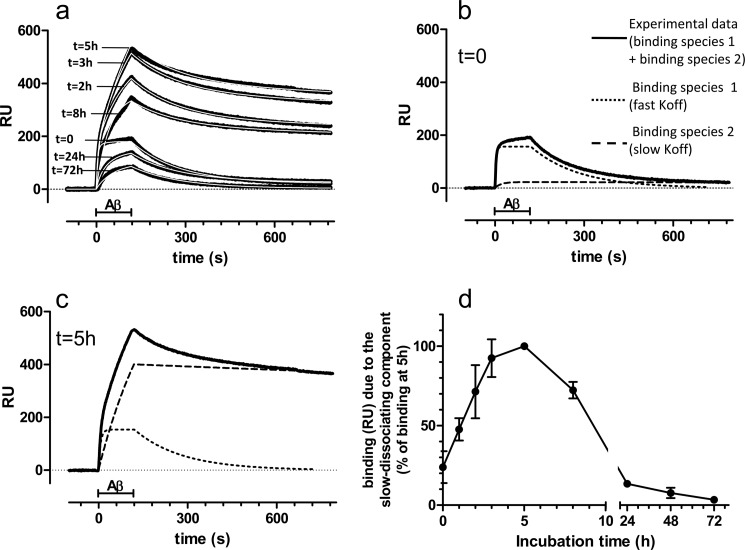
Fig. 1a shows that the binding signal on 4G8 was strikingly affected by the length of incubation of Aβ1–42. In comparison with the signal obtained on injecting the freshly prepared solution (t = 0), a marked increase was seen with solutions analyzed after 2, 3, and 5 h of incubation; with longer incubation (8, 24, and 72 h), the signal progressively declined. The incubation time also affected the shape of the sensorgrams, particularly the dissociation phase: Aβ1–42 incubated for 0, 24, and 72 h dissociated almost completely, whereas only a minor fraction dissociated with Aβ1–42 incubated for 2–8 h. Qualitatively identical results were obtained by diluting Aβ1–42 to 10 μm before the injection in the SPR apparatus (data not shown).
FIGURE 1.
SPR studies. a, synthetic Aβ1–42 (100 μm) was incubated at 25 °C, and samples were taken at different times (from 0 to 72 h), diluted to 1 μm in PBS, and injected over immobilized 4G8 for 2 min (bar), followed by 11 min of dissociation. The figure shows the sensorgrams (time course of the SPR signal expressed in resonance units, RU) obtained in a representative experiment (black lines). Fitting the sensorgrams (white lines) required a model assuming the presence of two binding species with different dissociation rates. b, experimental sensorgram obtained injecting the freshly prepared solution (t = 0, continuous line) is shown together with the theoretical sensorgrams corresponding to the quickly dissociating binding species (dotted lines) and the slowly dissociating binding species (dashed lines). The experimental sensorgram is mainly accounted for by the quickly dissociating species, which bind to 4G8 with a KD of 75 ± 10 nm (n = 5). c, experimental and theoretical sensorgrams obtained injecting the solutions incubated for 5 h. The lines are as in b. Note the increase of the signal caused by the slowly dissociating species (see the complete sets of data in supplemental Fig. S1). d, time course of the slowly dissociating component. Each point is the mean ± S.D. of three or four independent experiments, presented as a percentage of the signal found after 5 h of incubation.
None of the sensorgrams could be adequately fitted by the equation modeling a simple bimolecular interaction but required a complex model. All the curves could be fitted well (Fig. 1a, white lines) by assuming there were two binding species (Fig. 1, b and c, and supplemental Fig. S1) with markedly different dissociation rate constants (koff): one (dotted lines) dissociates relatively fast (koff: 6.3 × 10−3 s−1) and the other (dashed lines) had a >50-fold slower rate (koff: 1.1 × 10−4 s−1), approaching pseudoirreversible binding.
The quickly dissociating binding species predominant in the freshly prepared Aβ1–42 solution (t = 0; Fig. 1b) likely correspond to monomers. Thus, SEC analysis at t = 0 showed a single peak at an elution time corresponding to an apparent molecular mass of 12–13 kDa (supplemental Fig. S2, a, d, and e, green traces). It has been shown that this elution position corresponds to Aβ1–42 monomers, as determined by multiangle laser light scattering coupled to SEC (24). SPR binding caused by the quickly dissociating binding species (monomers) decreased with the incubation time (supplemental Fig. S1, a–f, blue dotted lines), indicating evolution toward other molecular species.
Slowly dissociating species made a negligible contribution in freshly prepared Aβ1–42 solutions (t = 0; Fig. 1b) and in solutions incubated for 24–72 h (supplemental Fig. S1, e and f), but it was up to 20 times higher at 2–8 h of incubation (Fig. 1c and Fig. S1, b–d). Fig. 1d illustrates the binding signal caused by these species in relation to Aβ1–42 incubation, with a bell-shaped kinetics and a peak at t = 5 h. SEC analysis at this time (Fig. S2, a, d, and e, red traces) showed the appearance of a peak in the void volume. Dot-blot analysis of the void volume with either 4G8 and 6E10 confirmed a marked increase of signal at t = 5 h (data not shown). SPR analysis on the fraction corresponding to the void volume showed that it contained the slowly dissociating species only (supplemental Fig. S2b), whereas the fraction corresponding to the monomeric peak contained the quickly dissociating species only (supplemental Fig. S2c). AFM at t = 5 h (supplemental Fig. S3b) confirmed the appearance of aggregates, and analysis of the void volume (supplemental Fig. S3, d–f) highlighted the presence of species with different morphologies, including globular aggregates and short protofibrils. DLS analysis at t = 5 h (supplemental Fig. S4) indicated that most of the aggregates have a hydrodynamic diameter of 10–30 nm, although larger species were also observed. DLS volume distributions were similar when diluting Aβ1–42 to 1 μm (i.e., the concentration used for SPR studies) or to 10 μm (i.e., the concentrations needed for AFM or SEC) (supplemental Fig. S4). Native gel confirmed the appearance, at t = 5 h, of high order aggregates (supplemental Fig. S5a), which were SDS-labile and migrated on SDS-PAGE as monomers and dimers (supplemental Fig. S5b).
The SPR signal caused by the slowly dissociating species decreased with an incubation time of ≥8 h until negligible values after 24–72 h of incubation (Fig. 1d). This finding is likely due to the concomitant evolution of Aβ1–42 toward different aggregated species, losing the property to bind immobilized 4G8. In agreement, SEC analysis showed that the peak in the void volume markedly decreased after 24 h (supplemental Fig. S2, a, d, and e, blue traces), suggesting that the aggregates formed at t = 24 h do not enter the columns. At this time, DLS analysis showed bigger species, the majority of which had a hydrodynamic diameter of ∼500 nm (supplemental Fig. S4, lower panels). Larger aggregates were also detected by AFM (supplemental Fig. S3c) and native gel (supplemental Fig. S5a). Western blot analysis, carried out under denaturing conditions, showed a decrease of monomers, an increase of dimers, and the appearance of higher molecular mass SDS-stable species (supplemental Fig. S5b). Dot-blot analysis showed that 4G8 recognized in a similar way the assemblies present at t = 5 h and those formed at t = 24 h (supplemental Fig. S5, c and d), indicating that this technique cannot be used to characterize transient oligomeric species. In summary, the SPR-based immunoassay specifically detects a transient subpopulation of Aβ1–42 oligomers, characterized by a pseudoirreversible binding to immobilized 4G8.
SPR studies using 6E10 as a capturing antibody showed low dissociation rates for both Aβ1–42 monomers and oligomers (supplemental Fig. S6). Therefore, the contribution of monomers and oligomers could not be investigated with this antibody. We also did not detect any binding of Aβ1–42 (tested at concentrations up to 10 μm) on immobilized A11 (not shown).
Another set of SPR studies was designed to investigate the direct binding of A11 and OC antibodies (5) to the Aβ1–42 oligomers, which had been previously captured by immobilized 4G8. We found a concentration-dependent binding of OC (supplemental Fig. S7), indicating that the 4G8-binding oligomers are also OC-positive. No reliable data could be obtained with A11 because we found a very high nonspecific binding to immobilized 4G8, even in the absence of captured oligomers.
We then used the 4G8-based SPR immunoassay to examine how the incubation conditions affects the kinetics of appearance/disappearance of Aβ1–42 oligomers. Supplemental Fig. S8 shows that raising the temperature from 25 to 37 °C shifted the peak time from 5 to 1 h, indicating much faster oligomer formation. The disappearance of the oligomers was also faster and was almost complete after 8 h of incubation. In addition, the peak time was affected by the peptide concentration, being 1, 3, and 5 h at 100, 50, and 25 μm Aβ1–42, respectively. The maximum resonance signal was also affected, with lower concentrations producing fewer oligomers. Finally, the rate of disappearance of the oligomers was directly proportional to the concentration of Aβ1–42 (supplemental Fig. S8). In summary, higher temperatures or higher concentrations of Aβ1–42 resulted in faster oligomerization and faster evolution toward species not recognized by 4G8.
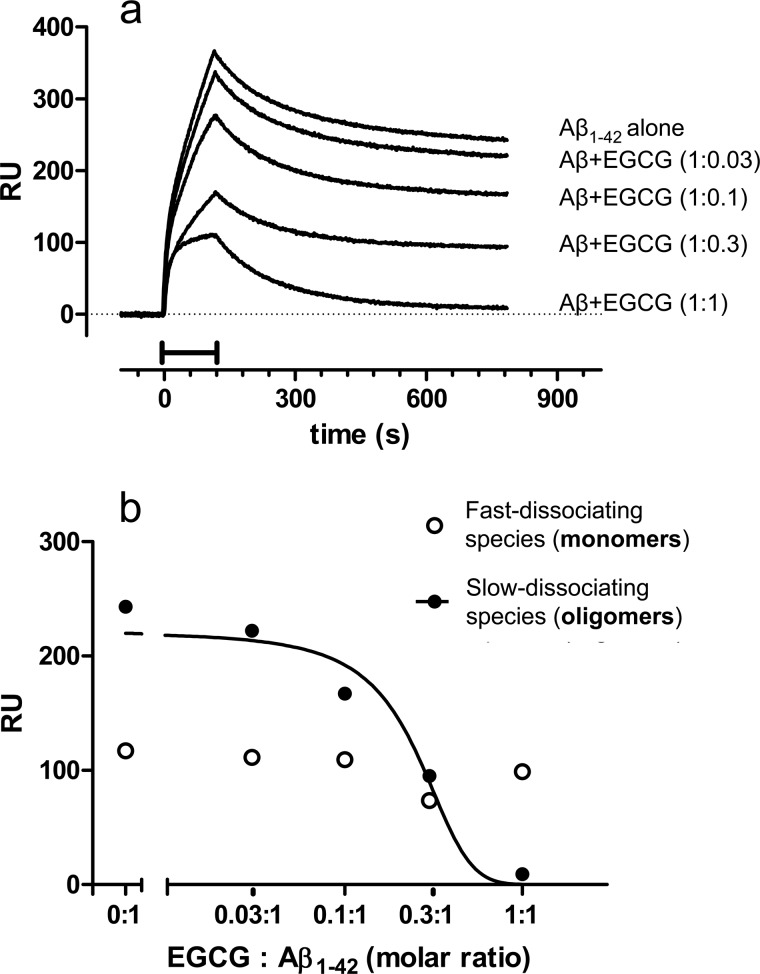
To investigate whether the SPR-based immunoassay could be applied to study molecules with potential anti-oligomeric effect, we tested the small polyphenolic green tea constituent (−)-EGCG, whose effects on Aβ aggregation and fibrillogenesis have been reported (25, 26). We incubated Aβ1–42 (100 μm) for 5 h with or without 3–100 μm EGCG. The samples were then diluted in PBS and flowed over immobilized 4G8. Preincubation with EGCG dose-dependently reduced Aβ1–42 binding to 4G8 (Fig. 2a). The reduction was specifically due to the slowly dissociating component (Fig. 2b), which completely disappeared with equimolar concentrations of EGCG. The binding of preformed Aβ1–42 oligomers to 4G8 was not affected by co-injection of equimolar concentrations of EGCG (data not shown), suggesting that EGCG inhibits the oligomer formation. Thus, the SPR assay can be used to monitor the effect of potential anti-oligomeric compounds.
FIGURE 2.
Effect of (−)-EGCG. Synthetic Aβ1–42 (100 μm) was incubated at 25 °C in the absence or presence of different concentrations of EGCG (3, 10, 30, and 100 μm). Samples were taken after 5 h of incubation, diluted 100-fold, and injected over immobilized 4G8 for 2 min (bar), followed by 11 min of dissociation. a, shows the sensorgrams (time course of the SPR signal expressed in resonance units, RU), which could be dissected for the presence of quickly dissociating species (monomers) and slowly dissociating species (oligomers) (see also Fig. 1). b, shows the effects of EGCG on the SPR binding signal caused by each of these components, highlighting a selective inhibitory effect on the oligomeric species.
SPR Studies with Native Aβ1–42
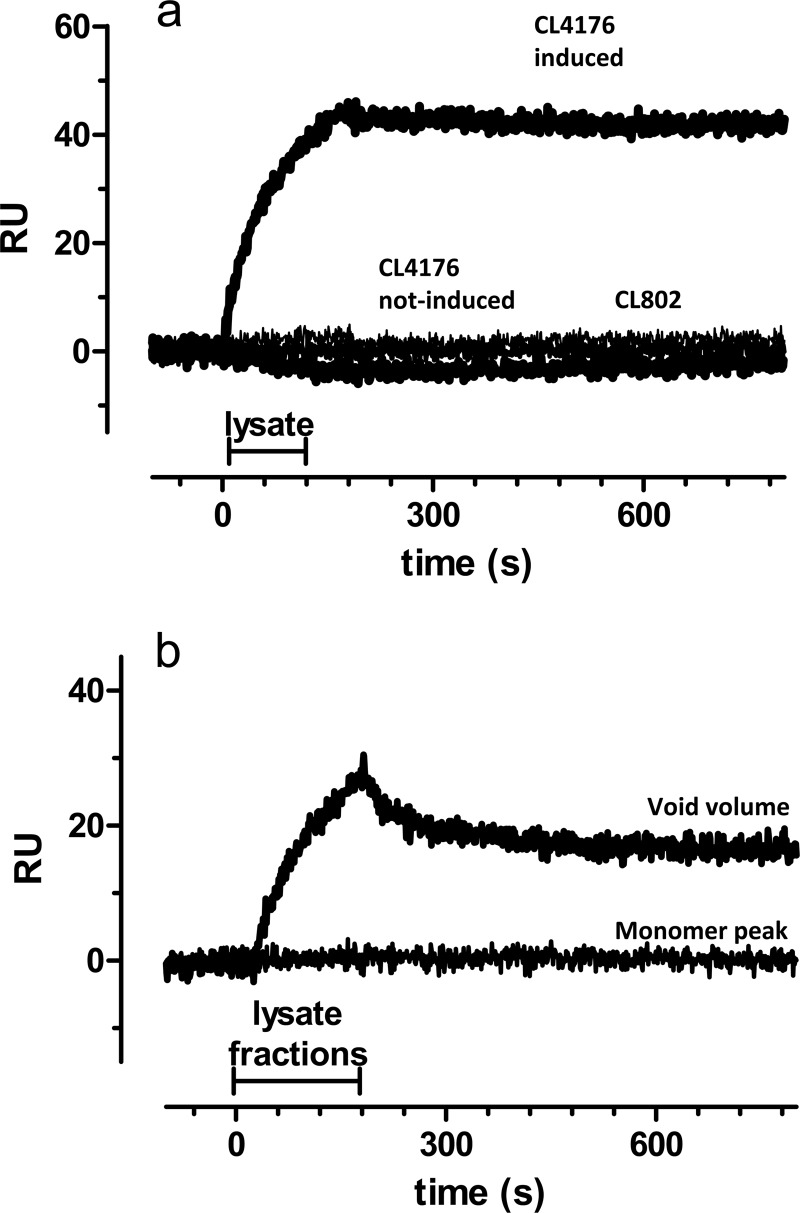
Next we tested whether the SPR-based immunoassay could detect natural (i.e., not synthetic) Aβ1–42 oligomers. We employed the transgenic CL4176 strain of the nematode C. elegans engineered to express human Aβ1–42 peptide in body wall muscles when the temperature is raised (15). As previously reported, ∼60% of the worms were paralyzed 44 h after temperature induction, in parallel with the deposition of oligomeric Aβ assemblies in the body wall muscle cells (16). Noninduced CL4176 and CL802 nematodes, which do not express the Aβ1–42 transgene, were used as controls. SPR analysis showed a clear binding signal only with lysates from induced CL4176 worms (Fig. 3a). The shape of the sensorgram, particularly the slow dissociation rate, suggests that the binding signal is due to the Aβ1–42 oligomer species. To substantiate this finding, lysates from CL4176 worms were eluted by SEC, and fractions were collected, in particular at time points corresponding to the elution of “monomers” and “oligomers” (void volume). The binding signal was only present when injecting the oligomeric fraction (Fig. 3b), consistent with the results obtained with the whole lysate. It is likely that monomers are not detectable under these conditions for sensitivity reasons (see discussion). These data indicate that the SPR assay can be used to detect the presence of native oligomers.
FIGURE 3.
SPR studies with native Aβ1–42 from C. elegans. a, lysates obtained from CL4176 transgenic C. elegans strain 44 h after the temperature rise (i.e., after induction of oligomeric Aβ1–42 expression) were injected onto immobilized 4G8. Lysates from uninduced CL4176 and CL802 nematodes, which do not express the Aβ1–42 transgene, were used as control. This experiment was carried out twice with similar results. b, lysates from induced CL4176 worms (i.e., 44 h after the temperature rise) were eluted by SEC using a S200 column. Fractions collected at time points corresponding to the elution of monomers and oligomers (void volume) were injected onto immobilized 4G8.
Biological Effects of Synthetic Aβ1–42
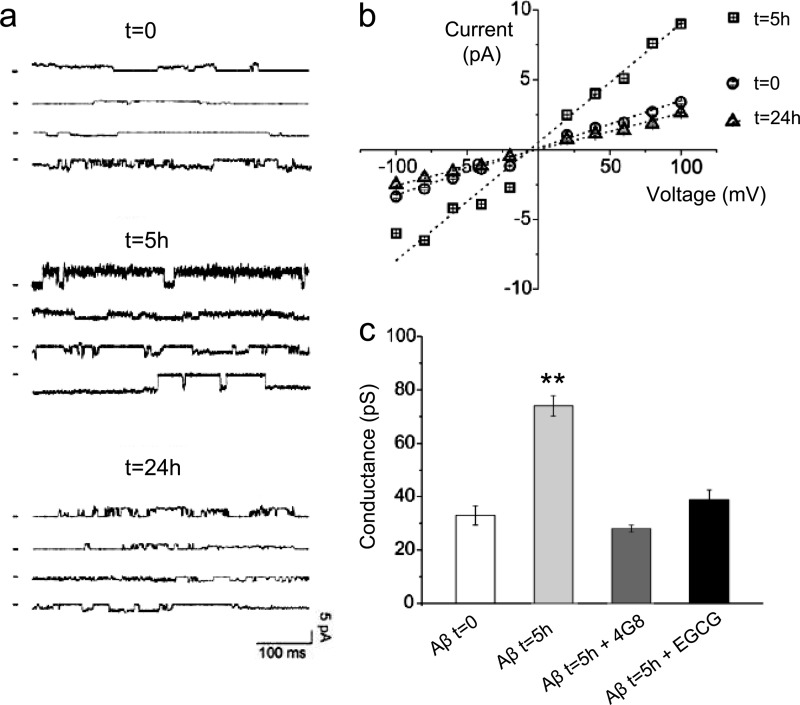
To examine the biological effects of the short-lived oligomers detected by SPR, we used in vitro and in vivo tests. It has been reported that soluble oligomers from several types of amyloid boosted lipid bilayer conductance regardless of the sequence, whereas fibrils and monomeric species did not (27). Electrophysiological Tip-Dip experiments (21) were used to measure the effects of the different Aβ1–42 species on the ionic conductance of a lipid bilayer. Samples of synthetic Aβ1–42 were analyzed at t = 0 (monomers), t = 5 h (4G8-binding oligomers), and t = 24 h (when 4G8-binding oligomers are no longer detected). Pure lipid bilayer exposed to vehicle did not show any ionic current. The Aβ1–42 solution incubated for 5 h had more effect on ion conductivity than solutions incubated for 0 and 24 h (Fig. 4a). Analysis of the corresponding current/voltage relationships (Fig. 4b) showed conductance values of 33 ± 3.6, 82 ± 5.0, and 26 ± 2.5 pS at 0, 5, and 24 h of incubation, respectively. The oligomer-associated increase of conductance was completely abolished by 4G8 or EGCG (Fig. 4c). These data indicate that the 4G8-binding oligomeric species specifically affect membrane permeability.
FIGURE 4.
Membrane conductance changes induced by synthetic Aβ1–42 species. a, representative current traces obtained after applying synthetic Aβ1–42 solutions to an artificial lipid bilayer. Synthetic Aβ1–42 (100 μm) was incubated at 25 °C and sampled at different times (t = 0, 5, and 24 h), and then diluted to 100 nm in 144 mm NaCl, 1.8 mm CaCl2, 1.2 mm MgCl2, 10 mm Hepes, pH 5.0, and applied to a lipid bilayer. Current traces were recorded at different potentials (+80, +40 mV, −40 mV, and −80 mV). b, the current/voltage relationship from a complete set of Tip-Dip currents. c, representative histogram comparing the conductance using freshly prepared Aβ1–42 (t = 0, monomers) and after 5 h of incubation (t = 5 h), with the latter preincubated or not with 4G8 (1:500 v/v) or EGCG (equimolar). The values are the means ± S.D. (n = 5). **, p < 0.01 versus all the other groups (Bonferroni's multiple comparison test following one-way ANOVA). 4G8 or EGCG alone had no effect on ionic currents, nor they did induce lipid bilayer ruptures (not shown).
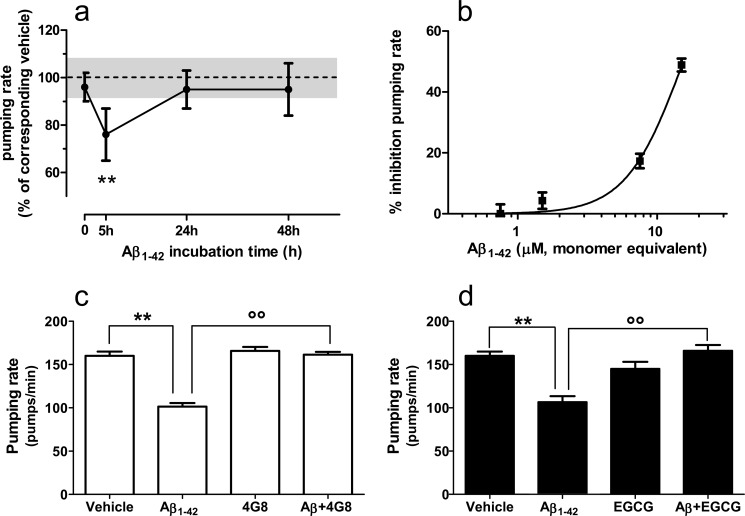
Next we tested the effect of Aβ1–42 on the pharyngeal pumping rate in C. elegans. This is the rhythmic contraction and relaxation of the pharyngeal muscle responsible for the ingestion and transport of food from the mouth to the intestine and is rapidly reduced by sublethal doses of chemical stressors (28). N2 ancestral nematodes were fed with Aβ1–42 taken after 0, 5, 24, or 48 h incubation. Two hours later the worms were transferred onto fresh nematode growth medium agar seeded with E. coli, and the pumping rate was measured. The pumping rate was significantly impaired only in nematodes fed with the Aβ1–42 solution incubated for 5 h, corresponding to the peak of 4G8-binding Aβ1–42 oligomers (Fig. 5a). The solutions containing monomers only (t = 0) or lacking 4G8-reactive oligomers (t = 24–48 h) had no effect. The effect of Aβ1–42 oligomers was dose-dependent (Fig. 5b) and reversible, with the pumping rate returning to base line 16 h after exposure to Aβ1–42 (data not shown). Preincubation with 4G8 antagonized the effect on the pumping rate (Fig. 5c), as did EGCG (Fig. 5d), suggesting that it was due to the 4G8-reactive oligomeric forms recognized by SPR.
FIGURE 5.
Effects of Aβ1–42 species on pharyngeal pumping rate in C. elegans. a, synthetic Aβ1–42 (100 μm) was incubated at 25 °C and sampled at different times (t = 0, 5, and 24 h), diluted to 10 μm in PBS, and administered to N2 worms for 2 h. Control worms were fed vehicle alone (dotted line). Only the samples preincubated for 5 h (i.e., those containing the 4G8-binding Aβ1–42 oligomers) markedly inhibited the pumping rate. The values are the means ± S.E. (n = 20), expressed as a percentage of the corresponding control. **, p < 0.01 versus t = 0, Bonferroni's test after two-way ANOVA. b, dose-response effect of Aβ1–42 oligomers on pharyngeal pumping. Worms were fed Aβ1–42 (0.5–10 μm) previously incubated for 5 h. The values are the means ± S.E., expressed as percentages of control worms fed vehicle. c, the inhibitory effect of Aβ1–42 oligomers on pumping rate was antagonized by 4G8. Aβ1–42 was preincubated for 5 h, added with 4G8 (1:500, v/v in vehicle), and incubated for further 30 min before administration to worms. Control worms were fed vehicle or 4G8 alone. The values are the means ± S.E. (n = 16). Two-way ANOVA showed a significant interaction between Aβ1–42 and 4G8 (p < 0.01). **, p < 0.001 versus vehicle; °°, p < 0.001 versus Aβ1–42 (Bonferroni's test). d, EGCG antagonized the inhibitory effect of Aβ1–42 oligomers on pharyngeal pumping. Aβ1–42 (100 μm) was incubated at 25 °C with or without EGCG (100 μm). After 5 h, the samples were diluted 10 times and administered to worms. Control worms were fed vehicle or 10 μm EGCG alone. The values are the means ± S.E. (n = 16). Two-way ANOVA showed a significant interaction between Aβ1–42 and 4G8 (p < 0.01). **, p < 0.001 versus vehicle; °°, p < 0.001 versus of Aβ1–42 (Bonferroni's test).
Studies with Wild-type and A2V-mutated Aβ1–40
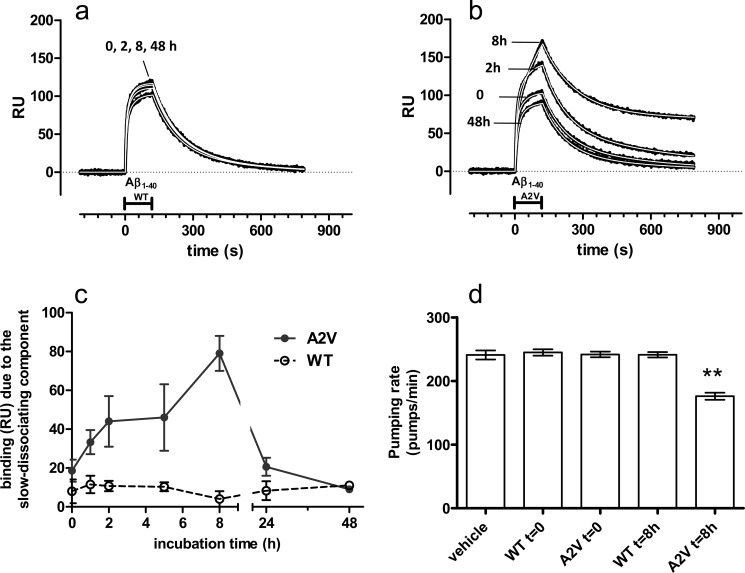
Finally, we applied the SPR-based immunoassay and the pumping rate test to study the A2V mutated form of Aβ1–40, previously found to be more amyloidogenic and more cytotoxic than the WT peptide (19). This peptide was selected on the basis of the neuropathological studies conducted on the brain of the homozygous proband, which indicated that the deposition of Aβ1–40 species were overrepresented and that the effects of A2V genetic mutation might be more pronounced on Aβ1–40 than on Aβ1–42 (29). During incubation at 37 °C in quiescent conditions, Aβ1–40 A2V formed transient 4G8-binding species, whereas Aβ1–40 WT did not (Fig. 6, a and b). The maximum binding signal was found after 8 h of incubation (Fig. 6c). Studies in C. elegans at this time point indicated a significant decrease in the pumping rate with Aβ1–40A2V but not Aβ1–40WT (Fig. 6d).
FIGURE 6.
Effect of A2V mutation of Aβ1–40 on the kinetics of 4G8-binding oligomers and on pharyngeal pumping rate in C. elegans. a and b, synthetic Aβ1–40 WT (a) or A2V (b) (100 μm) were incubated at 37 °C, and samples were taken at different times (from 0 to 48 h), diluted to 3 μm in PBS, and injected over immobilized 4G8 for 2 min (bar), followed by 11 min of dissociation. The figure shows the sensorgrams (time course of the SPR signal expressed in resonance units, RU) (black lines) obtained in a representative experiment. Sensorgrams were fitted as described for Fig. 1 and supplemental Fig. S1, dissecting the experimental curve into its quickly and slowly dissociating components (koff: 8.0 ± 0.2 × 10−3 s−1 and 6.0 ± 1.6 × 10−4 s−1, respectively). c, time course of the slowly dissociating components. Each point is the mean ± S.E. of three independent experiments. d, worms were fed for 2 h Aβ1–40, WT, or A2V (50 μm), freshly prepared (t = 0) or preincubated at 37 °C for 8 h. Control worms were fed vehicle alone. The values are the means ± S.E. (n = 20 worms). **, p < 0.01 versus all the other groups (Bonferroni's test after one-way ANOVA).
DISCUSSION
We describe here a new SPR-based immunoassay that selectively recognizes a subpopulation of biologically active Aβ oligomers. The method exploits the fact that these soluble aggregated species bind immobilized 4G8 in a pseudoirreversible manner (very slow dissociation rates), whereas monomers bind 4G8 with significantly faster dissociation rates. The possibility of accurately determining the binding constants is the key feature that allows the use of 4G8 in the SPR immunoassay to distinguish between different Aβ species.
SPR studies were also done with the anti-Aβ antibody 6E10, which, however, proved unsuitable for distinguishing monomers from oligomers because both species bound the antibody with strong affinities and very slow dissociation rates (30). It might be interesting to test other antibodies, particularly those raised using Aβ oligomers as antigens (31, 32). Recently, SPR data have been reported employing an “oligomer-specific” IgM anti-Aβ antibody (OMAB) (33). In that study, the binding of Aβ1–40/2 monomers/dimers to OMAB had fast off rate kinetics, whereas binding of SEC-enriched Aβ1–40/2 oligomers resulted in a strong complex with an exceptionally slow koff. OMAB might thus serve as another capturing antibody useful for recognition of oligomers by SPR. It will be interesting to see whether 4G8 and OMAB recognize the same species.
In principle the SPR assay might employ conformation-dependent antibodies that recognize oligomers of different proteins or peptides, such as A11 or OC (5, 34). A reliable immobilization of OC requires a purified antibody, which is currently not available. The immobilization of A11 did not result in any binding signal. Whether this antibody is unable to recognize transient Aβ1–42 oligomers or whether its activity is abolished by the immobilization on the SPR sensor chip remains to be established.
The aggregated species that bind immobilized 4G8 in a pseudoirreversible manner rapidly appear and disappear during the incubation of synthetic Aβ1–42. With 100 μm peptide at 25 °C, the oligomer-dependent SPR signal was maximal after 5 h of incubation decreasing thereafter to negligible values at t ≥ 24 h. Analysis of the solution incubated for 5 h (by SEC, DLS, AFM, and gel electrophoresis) showed the presence of a heterogeneous population of SDS-labile aggregates, including globular species and short protofibrils, with a main hydrodynamic diameter of 10–30 nm and eluting in the void volume even with SEC columns suitable for high molecular mass molecules. However, the morphology of Aβ aggregates, as well as their interaction with column matrix, can significantly alter migration times and may result in overestimated dimensions (24, 35). The solution incubated for 24 h mainly contained greater and SDS-stable species, which do not even enter in the SEC columns. Thus, the 4G8-based SPR immunoassay selectively recognized a specific population of soluble aggregates. This population is also recognized by OC, a conformation-specific antibody proposed to selectively target fibrillar oligomers (5), as suggested by an SPR assay in which OC was flowed onto 4G8-captured oligomers. This “sandwich” SPR format (supplemental Fig. S7) could be applied for identifying interactors of captured oligomers.
When tested in a dot-blot assay, 4G8 recognized the oligomeric assemblies present at t = 5 h and t = 24 h in a similar way, in line with published evidence (36). There are two possible explanations for the discrepancy between the dot-blot and SPR results: (i) it is possible that steric hindrance or specific features of the SPR technology do not allow detection of the species at t = 24 h, for example because of impaired diffusion of large aggregates in the microfluidic channels (37); and (ii) the fact that SPR measures the binding events in a dynamic manner (time scale of seconds) could provide a much higher sensitivity to discriminate between species with different affinity values, which cannot be achieved by dot-blot in which long incubation steps may level out differences in affinity.
The pseudoirreversible binding observed by SPR at t = 5 h suggests multivalent interactions between different epitopes on a single oligomeric assembly and different immobilized 4G8 antibodies (38). The affinity of the oligomers for 4G8 could not be determined because of the lack of information about their actual concentration, a consequence of the uncertainty about the precise molecular mass. However, assuming (from SEC data) that after 5 h of incubation ∼40% of monomers have assembled into oligomers and assuming a mass ranging from 90 to 400 kDa (39), it follows that the concentration of oligomers is in the low nanomolar range (4–20 nm) and their affinity for immobilized 4G8 is exceptionally high (KD<1 nm). The fact that monomers have a much lower affinity (KD = 75 nm) and mass could explain why monomers are recognized with a much lower sensitivity than oligomers. Thus, the data in supplemental Fig. S2 (a–c) show that the SPR signal found using monomeric fractions was much lower than found with oligomeric fractions, although the SEC peaks were similar.
The formation of 4G8-binding oligomeric species was completely prevented by the co-incubation of Aβ1–42 in the presence of equimolar concentrations of EGCG, a prototypical inhibitor of oligomerization (25, 26). A previous study showed that EGCG prevented the conversion of Aβ1–42 into toxic intermediates and favored the formation of unstructured nontoxic oligomers (25). Moreover, we found that the A2V mutation, which enhances the aggregation and fibrillogenic properties of Aβ and results in early onset dementia (19), boosts the formation of Aβ1–40 transient oligomeric species recognized by 4G8.
4G8 recognizes a central region of Aβ (amino acids 17–21, LVFFA), which is believed to be implicated in the hydrophobic interactions underlying the formation and elongation of amyloid fibrils (40) but not in Aβ oligomerization (41). Our SPR data indicate that the short-lived Aβ1–42 oligomers expose this 4G8-binding hydrophobic sequence. Very recent data (4, 6) suggest that the exposure of hydrophobic motifs is the main conformational determinant of oligomer toxicity, whereas size and secondary structures are probably less important (6).
Thus, the 4G8-based SPR immunoassay has the potential to specifically recognize the toxic oligomeric species of Aβ1–42. To demonstrate this, we used two assays that require short term exposure to Aβ1–42 solutions, a pivotal condition for studying the biological effects of short-lived species.
Several reports suggest a direct interaction between Aβ and the plasma membrane as one possible mechanism of toxicity (21, 42–45). Electrophysiological experiments employed the Tip-Dip technique, which accurately studies single ion permeability in artificial lipid bilayers (21). The results showed that Aβ1–42 solutions preincubated for 5 h (i.e., those containing 4G8-binding oligomers) had a much greater effect than freshly prepared solutions (monomers) or solutions preincubated for 24 h (lacking 4G8-binding oligomers). 4G8 and EGCG counteracted the oligomer-induced increase in conductance but had no effect on the conductance associated with monomers.
In vivo we used C. elegans, an attractive model for several studies, including toxicological investigations. The rhythmic contraction and relaxation of the C. elegans pharynx is fundamental for the worm's feeding and is affected by a variety of chemical stressors, such as alcohol, heavy metals, and sulfhydryl-reactive compounds, which induce the production of cellular stress proteins (28). Stress-induced inhibition of feeding was in fact suggested as an important survival mechanism that limits the intake of toxic solutes. We found, for the first time, that Aβ1–42 oligomers, but not monomers or larger aggregates, act as “stressors” in C. elegans, significantly inhibiting their pharyngeal pumping. This innovative in vivo model may serve as a rapid and convenient marker of the toxic potential of Aβ oligomers, as also suggested by the significant effects found with the toxic Aβ1–40A2V mutant. The correspondence between inhibition of the pumping rate in C. elegans and the peculiar binding signal in SPR studies (the pseudoirreversible binding to 4G8) is also noteworthy; this was true when using Aβ1–42 after different incubation times, when comparing Aβ1–40WT with Aβ1–40A2V, and when incubating with EGCG. These findings strongly suggest that the oligomers recognized by 4G8 in the SPR immunoassays are the same as those that are toxic in the C. elegans pharyngeal pumping test. In fact, 4G8 antagonized the effect of Aβ1–42 oligomers on pharyngeal behavior, emerging as an antibody recognizing and antagonizing the toxic Aβ oligomers. Consistently, we previously found that 4G8 also antagonizes the memory impairment induced by intracerebroventricular injection of synthetic Aβ1–42 oligomers in mice (22).
The present study also shows that the SPR immunoassay can recognize natural oligomers formed in transgenic C. elegans expressing Aβ1–42. The ability of this assay to detect native toxic oligomers in other biological samples, including those from human AD patients, is under investigation.
Supplementary Material
Acknowledgments
We thank Margherita Romeo for technical assistance with C. elegans experiments, Stefano Stabilini for electrophysiological TipDip studies, and Flamma SpA Bergamo for the kind gift of FMOC amino acids.
This work was supported in part by Cariplo Foundation Project 2009-2543, Telethon Foundation Project GGP10120, MIUR Grant RPAB11FRE9 (to F. T.), Italian Ministry of Health Grant PS 2007.39 (to F. T.), Italian Ministry of University and Scientific Research Grant 20078RWJBN (to M. M.), and Alzheimer's Association Project NIRG-10-171655, and by the Mario Negri Foundation and Banca Intesa.

This article contains supplemental Figs. S1–S8.
- Aβ
- amyloid-β
- AFM
- atomic force microscopy
- DLS
- dynamic laser light scattering
- EGCG
- epigallocathechin gallate
- SEC
- size exclusion chromatography
- SPR
- surface plasmon resonance
- AD
- Alzheimer disease
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- OMAB
- oligomer-specific IgM anti-Aβ antibody
- ANOVA
- analysis of variance.
REFERENCES
- 1. Lambert M. P., Barlow A. K., Chromy B. A., Edwards C., Freed R., Liosatos M., Morgan T. E., Rozovsky I., Trommer B., Viola K. L., Wals P., Zhang C., Finch C. E., Krafft G. A., Klein W. L. (1998) Diffusible, nonfibrillar ligands derived from Aβ1–42 are potent central nervous system neurotoxins. Proc. Natl. Acad. Sci. U.S.A. 95, 6448–6453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Walsh D. M., Selkoe D. J. (2007) A β oligomers. A decade of discovery. J. Neurochem. 101, 1172–1184 [DOI] [PubMed] [Google Scholar]
- 3. Holtzman D. M., Goate A., Kelly J., Sperling R. (2011) Mapping the road forward in Alzheimer's disease. Sci. Transl. Med. 3, 114–148 [DOI] [PubMed] [Google Scholar]
- 4. Campioni S., Mannini B., Zampagni M., Pensalfini A., Parrini C., Evangelisti E., Relini A., Stefani M., Dobson C. M., Cecchi C., Chiti F. (2010) A causative link between the structure of aberrant protein oligomers and their toxicity. Nat. Chem. Biol. 6, 140–147 [DOI] [PubMed] [Google Scholar]
- 5. Glabe C. G. (2008) Structural classification of toxic amyloid oligomers. J. Biol. Chem. 283, 29639–29643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ladiwala A. R., Litt J., Kane R. S., Aucoin D. S., Smith S. O., Ranjan S., Davis J., Vannostrand W. E., Tessier P. M. (2012) Conformational differences between two amyloid β oligomers of similar size and dissimilar toxicity. J. Biol. Chem. DOI 10.1074/jbc.M111.329763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cooper M. A. (2002) Optical biosensors in drug discovery. Nat. Rev. Drug Discov. 1, 515–528 [DOI] [PubMed] [Google Scholar]
- 8. Rich R. L., Myszka D. G. (2007) Higher-throughput, label-free, real-time molecular interaction analysis. Anal. Biochem. 361, 1–6 [DOI] [PubMed] [Google Scholar]
- 9. Sohma Y., Hayashi Y., Kimura M., Chiyomori Y., Taniguchi A., Sasaki M., Kimura T., Kiso Y. (2005) The “O-acyl isopeptide method” for the synthesis of difficult sequence-containing peptides. Application to the synthesis of Alzheimer's disease-related amyloid β peptide (Aβ) 1–42. J. Pept. Sci. 11, 441–451 [DOI] [PubMed] [Google Scholar]
- 10. Taniguchi A., Sohma Y., Hirayama Y., Mukai H., Kimura T., Hayashi Y., Matsuzaki K., Kiso Y. (2009) “Click peptide.” pH-triggered in situ production and aggregation of monomer Aβ1–42. Chembiochem. 10, 710–715 [DOI] [PubMed] [Google Scholar]
- 11. Beeg M., Stravalaci M., Bastone A., Salmona M., Gobbi M. (2011) A modified protocol to prepare seed-free starting solutions of amyloid-β Aβ1–40 and Aβ1–42 from the corresponding depsipeptides. Anal. Biochem. 411, 297–299 [DOI] [PubMed] [Google Scholar]
- 12. Stravalaci M., Beeg M., Salmona M., Gobbi M. (2011) Use of surface plasmon resonance to study the elongation kinetics and the binding properties of the highly amyloidogenic Aβ1–42 peptide, synthesized by depsi-peptide technique. Biosens Bioelectron. 26, 2772–2775 [DOI] [PubMed] [Google Scholar]
- 13. Anonymous (2011) State of aggregation (Editorial). Nat. Neurosci. 14, 399. [DOI] [PubMed] [Google Scholar]
- 14. Bravman T., Bronner V., Lavie K., Notcovich A., Papalia G. A., Myszka D. G. (2006) Exploring “one-shot” kinetics and small molecule analysis using the ProteOn XPR36 array biosensor. Anal. Biochem. 358, 281–288 [DOI] [PubMed] [Google Scholar]
- 15. Link C. D. (2005) Invertebrate models of Alzheimer's disease. Genes Brain Behav. 4, 147–156 [DOI] [PubMed] [Google Scholar]
- 16. Diomede L., Cassata G., Fiordaliso F., Salio M., Ami D., Natalello A., Doglia S. M., De Luigi A., Salmona M. (2010) Tetracycline and its analogues protect Caenorhabditis elegans from β amyloid-induced toxicity by targeting oligomers. Neurobiol. Dis. 40, 424–431 [DOI] [PubMed] [Google Scholar]
- 17. Nečas D., Klapetek P. (2012) Gwyddion. An open-source software for SPM data analysis. Central Eur. J. Physics 10, 181–188 [Google Scholar]
- 18. Lago P., Rovati L., Cantù L., Corti M. A. (1993) Quasielastic light scattering detector for chromatographic analysis. Rev. Sci. Instrum. 64, 1797–1802 [Google Scholar]
- 19. Di Fede G., Catania M., Morbin M., Rossi G., Suardi S., Mazzoleni G., Merlin M., Giovagnoli A. R., Prioni S., Erbetta A., Falcone C., Gobbi M., Colombo L., Bastone A., Beeg M., Manzoni C., Francescucci B., Spagnoli A., Cantù L., Del Favero E., Levy E., Salmona M., Tagliavini F. (2009) A recessive mutation in the APP gene with dominant-negative effect on amyloidogenesis. Science 323, 1473–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Provencher S. W. (1982) A constrained regularization method for inverting data represented by linear algebraic or integral equations. Comput. Phys. Comm. 27, 213–227 [Google Scholar]
- 21. Paulis D., Maras B., Schinina M. E., Francesco L., Principe S., Galeno R., Abdel-Haq H., Cardone F., Florio T., Pocchiari M., Mazzanti M. (2011) The pathological prion protein forms ionic conductance in lipid bilayer. Neurochem. Int. 59, 168–174 [DOI] [PubMed] [Google Scholar]
- 22. Balducci C., Beeg M., Stravalaci M., Bastone A., Sclip A., Biasini E., Tapella L., Colombo L., Manzoni C., Borsello T., Chiesa R., Gobbi M., Salmona M., Forloni G. (2010) Synthetic amyloid-β oligomers impair long-term memory independently of cellular prion protein. Proc. Natl. Acad. Sci. U.S.A. 107, 2295–2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Laurén J., Gimbel D. A., Nygaard H. B., Gilbert J. W., Strittmatter S. M. (2009) Cellular prion protein mediates impairment of synaptic plasticity by amyloid-β oligomers. Nature 457, 1128–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hepler R. W., Grimm K. M., Nahas D. D., Breese R., Dodson E. C., Acton P., Keller P. M., Yeager M., Wang H., Shughrue P., Kinney G., Joyce J. G. (2006) Solution state characterization of amyloid β-derived diffusible ligands. Biochemistry 45, 15157–15167 [DOI] [PubMed] [Google Scholar]
- 25. Ehrnhoefer D. E., Bieschke J., Boeddrich A., Herbst M., Masino L., Lurz R., Engemann S., Pastore A., Wanker E. E. (2008) EGCG redirects amyloidogenic polypeptides into unstructured, off-pathway oligomers. Nat. Struct. Mol. Biol. 15, 558–566 [DOI] [PubMed] [Google Scholar]
- 26. Bieschke J., Russ J., Friedrich R. P., Ehrnhoefer D. E., Wobst H., Neugebauer K., Wanker E. E. (2010) EGCG remodels mature alpha-synuclein and amyloid-β fibrils and reduces cellular toxicity. Proc. Natl. Acad. Sci. U.S.A. 107, 7710–7715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kayed R., Sokolov Y., Edmonds B., McIntire T. M., Milton S. C., Hall J. E., Glabe C. G. (2004) Permeabilization of lipid bilayers is a common conformation-dependent activity of soluble amyloid oligomers in protein misfolding diseases. J. Biol. Chem. 279, 46363–46366 [DOI] [PubMed] [Google Scholar]
- 28. Jones D., Candido E. P. (1999) Feeding is inhibited by sublethal concentrations of toxicants and by heat stress in the nematode Caenorhabditis elegans. Relationship to the cellular stress response. J. Exp. Zool 284, 147–157 [DOI] [PubMed] [Google Scholar]
- 29. Giaccone G., Morbin M., Moda F., Botta M., Mazzoleni G., Uggetti A., Catania M., Moro M. L., Redaelli V., Spagnoli A., Rossi R. S., Salmona M., Di Fede G., Tagliavini F. (2010) Neuropathology of the recessive A673V APP mutation. Alzheimer disease with distinctive features. Acta Neuropathol. 120, 803–812 [DOI] [PubMed] [Google Scholar]
- 30. Ramakrishnan M., Kandimalla K. K., Wengenack T. M., Howell K. G., Poduslo J. F. (2009) Surface plasmon resonance binding kinetics of Alzheimer's disease amyloid β peptide-capturing and plaque-binding monoclonal antibodies. Biochemistry 48, 10405–10415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Barghorn S., Nimmrich V., Striebinger A., Krantz C., Keller P., Janson B., Bahr M., Schmidt M., Bitner R. S., Harlan J., Barlow E., Ebert U., Hillen H. (2005) Globular amyloid β-peptide oligomer. A homogenous and stable neuropathological protein in Alzheimer's disease. J. Neurochem. 95, 834–847 [DOI] [PubMed] [Google Scholar]
- 32. Lambert M. P., Velasco P. T., Chang L., Viola K. L., Fernandez S., Lacor P. N., Khuon D., Gong Y., Bigio E. H., Shaw P., De Felice F. G., Krafft G. A., Klein W. L. (2007) Monoclonal antibodies that target pathological assemblies of Aβ. J. Neurochem. 100, 23–35 [DOI] [PubMed] [Google Scholar]
- 33. Lindhagen-Persson M., Brännström K., Vestling M., Steinitz M., Olofsson A. (2010) Amyloid-β oligomer specificity mediated by the IgM isotype. Implications for a specific protective mechanism exerted by endogenous auto-antibodies. PLoS One 5, e13928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kayed R. (2003) Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science 300, 486–489 [DOI] [PubMed] [Google Scholar]
- 35. Benilova I., Karran E., De Strooper B. (2012) The toxic Aβ oligomer and Alzheimer's disease. An emperor in need of clothes. Nat. Neurosci. 15, 349–357 [DOI] [PubMed] [Google Scholar]
- 36. Perchiacca J. M., Ladiwala A. R., Bhattacharya M., Tessier P. M. (2012) Structure-based design of conformation- and sequence-specific antibodies against amyloid β. Proc. Natl. Acad. Sci. U.S.A. 109, 84–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Štěpánek J., Vaisocherová H., Piliarik M., Homola J. (2006) Molecular interactions in SPR sensors, in Surface Plasmon Resonance Based Sensors, pp. 69–91, Springer, Berlin, Germany [Google Scholar]
- 38. Hong S., Leroueil P. R., Majoros I. J., Orr B. G., Baker J. R., Jr., Banaszak Holl M. M. (2007) The binding avidity of a nanoparticle-based multivalent targeted drug delivery platform. Chem. Biol. 14, 107–115 [DOI] [PubMed] [Google Scholar]
- 39. Freir D. B., Nicoll A. J., Klyubin I., Panico S., Mc Donald J. M., Risse E., Asante E. A., Farrow M. A., Sessions R. B., Saibil H. R., Clarke A. R., Rowan M. J., Walsh D. M., Collinge J. (2011) Interaction between prion protein and toxic amyloid β assemblies can be therapeutically targeted at multiple sites. Nat. Commun. 2, 336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tjernberg L. O., Callaway D. J., Tjernberg A., Hahne S., Lilliehöök C., Terenius L., Thyberg J., Nordstedt C. (1999) A molecular model of Alzheimer amyloid β-peptide fibril formation. J. Biol. Chem. 274, 12619–12625 [DOI] [PubMed] [Google Scholar]
- 41. Matsumura S., Shinoda K., Yamada M., Yokojima S., Inoue M., Ohnishi T., Shimada T., Kikuchi K., Masui D., Hashimoto S., Sato M., Ito A., Akioka M., Takagi S., Nakamura Y., Nemoto K., Hasegawa Y., Takamoto H., Inoue H., Nakamura S., Nabeshima Y., Teplow D. B., Kinjo M., Hoshi M. (2011) Two distinct amyloid β-protein (Aβ) assembly pathways leading to oligomers and fibrils identified by combined fluorescence correlation spectroscopy, morphology, and toxicity analyses. J. Biol. Chem. 286, 11555–11562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kagan B. L., Azimov R., Azimova R. (2004) Amyloid peptide channels. J. Membr. Biol. 202, 1–10 [DOI] [PubMed] [Google Scholar]
- 43. Kawahara M., Ohtsuka I., Yokoyama S., Kato-Negishi M., Sadakane Y. (2011) Membrane incorporation, channel formation, and disruption of calcium homeostasis by Alzheimer's β-amyloid protein. Int. J. Alzheimers Dis. 2011, 304583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Quist A., Doudevski I., Lin H., Azimova R., Ng D., Frangione B., Kagan B., Ghiso J., Lal R. (2005) Amyloid ion channels. A common structural link for protein-misfolding disease. Proc. Natl. Acad. Sci. U.S.A. 102, 10427–10432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shirwany N. A., Payette D., Xie J., Guo Q. (2007) The amyloid β ion channel hypothesis of Alzheimer's disease. Neuropsychiatr. Dis. Treat. 3, 597–612 [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.