Background: Production of insecticidal and nematocidal cyclic peptides is inefficient in transgenic plants.
Results: Efficient cyclization requires cleavage of the N-terminal propeptide from the mature cyclotide domain and a C terminus containing a binding motif.
Conclusion: The cyclization motif has Asn at position P1, a small amino acid at position P1′, and a Leu at position P2′.
Significance: Understanding substrate requirements will help produce cyclotides in transgenic plants.
Keywords: Cysteine Protease, Hydroxyproline, Mass Spectrometry (MS), Peptide Biosynthesis, Peptides, Asparginyl Endoproteinase, Cyclic Peptide, Cyclotide, Kalata B1, Prolylhydroxylase
Abstract
Plant cyclotides are the largest family of gene-encoded cyclic proteins. They act as host defense molecules to protect plants and are promising candidates as insecticidal and nematocidal agents in agriculture. For this promise to be realized a greater understanding of the post-translational processing of these proteins is needed. Cyclotides are cleaved from precursor proteins with subsequent ligation of the N and C termini to form a continuous peptide backbone. This cyclization step is inefficient in transgenic plants and our work aims to shed light on the specificity requirements at the excision sites for cyclic peptide production. Using the prototypic cyclotide kalata B1 (kB1) expressed from the Oak1 gene, MALDI-TOF mass spectrometry was used to examine the cyclization efficiency when mutants of the Oak1 gene were expressed in transgenic Nicotiana benthamiana. Cleavage at the N terminus of the cyclotide domain occurs rapidly with no strict specificity requirements for amino acids at the cleavage site. In contrast, the C-terminal region of the cyclotide domain in the P2, P1, P1′, and P2′ positions is highly conserved and only specific amino acids can occupy these positions. The cyclization reaction requires an Asn at position P1 followed by a small amino acid (Ala, Gly, Ser) at the P1′ position. The P2′ position must be filled by Leu or Ile; in their absence an unusual post-translational modification occurs. Substitution of the P2′ Leu with Ala leads to hydroxylation of the neighboring proline. Through mutational analysis this novel proline hydroxylation motif was determined to be Gly-Ala-Pro-Ser.
Introduction
Cyclotides are a large family of head-to-tail cyclized proteins from plants (1) that function in host defense against insect predation (2, 3), and have exciting potential in agriculture as insecticidal or nematocidal agents and also in drug design applications (4, 5). For this to be realized a greater understanding of the post-translational processing of these proteins is needed. The mechanism of excision and cyclization of cyclotides from their linear precursors involves a fascinating post-translational processing phenomenon that until now has been only partially understood. The joining of the N and C termini occurs via a transpeptidation reaction to create a continuous peptide backbone. This circular structure bestows cyclotides with exceptional structural and proteolytic stability (4, 6).
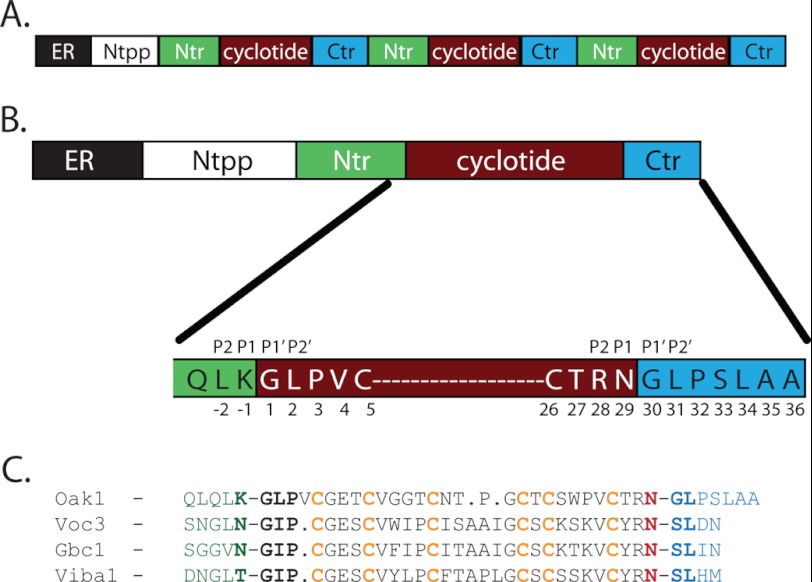
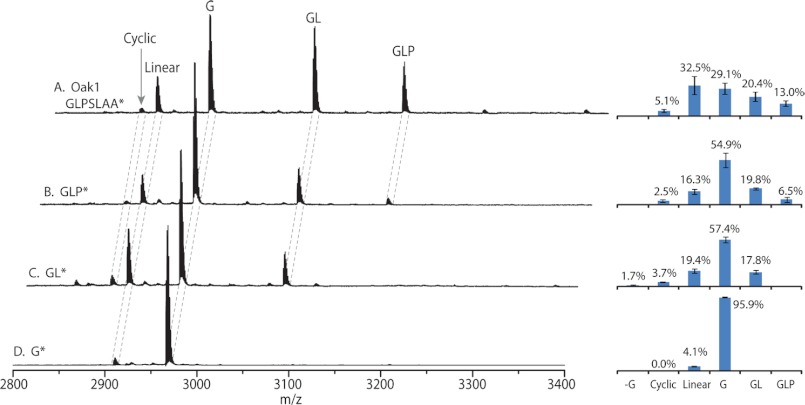
Cyclotides are the largest group of circular peptides containing between 28 and 37 amino acids, and have a well defined three-dimensional structure containing exclusively naturally occurring amino acids (7–9). Cyclotides are characterized by the combination of a cyclic peptide backbone with an embedded cystine knot, collectively called the cyclic cystine knot (10). Cyclotides appear to be cyclized by a protease-mediated biosynthetic pathway, most likely as the result of an asparaginyl endopeptidase (AEP)4 that catalyzes a cleavage reaction as well as the transpeptidation reaction (3, 11, 12). Cyclotides are expressed as larger precursor proteins containing an endoplasmic reticulum (ER) targeting signal, an N-terminal propeptide, an N-terminal repeat (Ntr) region of 16–20 amino acids, and a cyclotide domain. The N-terminal regions both target the cyclotide precursor to the vacuole where the protein is post-translationally processed and cyclized (13). The Ntr and cyclotide domains can be repeated up to three times in some cyclotide precursors and the repeated domains contain a short hydrophobic C-terminal region (Ctr) of 3 to 7 amino acids (Fig. 1A). The prototypic cyclotide kalata B1 (kB1) expressed from the Oak1 gene of Oldenlandia affinis contains a single cyclotide domain (Fig. 1B).
FIGURE 1.
A, cyclotides are expressed as multidomain precursor proteins containing five distinct regions (ER, N-terminal propeptide, Ntr, cyclotide, and Ctr), which can contain up to three cyclotide domains. B, the Oak1 precursor from O. affinis with the amino acid sequence displayed for the residues on either side of the cyclotide processing sites. C, alignment of cyclotide sequences from different plant families showing the conservation of the Asn at the C terminus, followed by a small amino acid and a Leu. Residues at the N-terminal side (−1) of the cyclotide domain are not strongly conserved. Oak1 is the precursor of kalata B1 from O. affinis (3, 11), Voc3 is the precursor of cycloviolacin O13 from Viola odorata (25), Gbc1 is from Gloeospermum blakeanum (26), and Viba1 is a precursor from Viola baoshanensis (27).
Genes encoding cyclotide precursors from O. affinis, the most studied cyclotide-bearing plant (6), encode proteins that have a conserved motif that flanks the upstream and downstream processing sites of each cyclotide domain (14). A recent analysis of a large number of cyclotide precursor sequences showed that similar motifs are present in other plant species (15). The conserved motifs that flank the cyclotide domains of the precursor proteins have been proposed to have a crucial role in the cyclization reaction (Fig. 1C) (14, 16).
There are two highly conserved residues near the C terminus of every cyclotide domain. The first is an asparagine or aspartate residue in the P1 position at the C-terminal processing site of the cyclotide domain, which led to the proposal that an AEP could be involved in both the cleavage and cyclization of cyclotides (3, 11, 16, 17). This is consistent with the discovery that cyclotides are targeted to the vacuole which contains AEPs. That is, the vacuole is the site where processing and cyclization occurs (13). These enzymes are common in plants and have a role in processing seed storage proteins, where they cleave peptide bonds adjacent to Asn residues. The second highly conserved residue is a leucine located 2 amino acids downstream of the Asn in the P2′ position. It has been proposed, but not experimentally verified, that this residue is essential for AEP binding and cyclization (16, 18).
Studies of transgenic plants into which modified cyclotide precursor genes are inserted provide a powerful way of probing the role of individual residues in cyclotide processing and cyclization. We recently reported the observation of processing intermediates during the maturation of the O. affinis kalata B1 precursor protein (Oak1) in transgenic Nicotiana benthamiana (16). Substitution of residues surrounding the cleavage sites to produce these intermediates should enable a clearer picture of how cyclotides are cleaved from their precursor proteins and cyclized. The Ntr appears to be cleaved before the cyclotide precursor is cyclized, which suggests the Ctr should be critical in the enzymatic cyclization of the protein. Mutation of this Ctr region should then reveal which residues are essential for cyclization.
Here we focus on the residues at the N and C terminus of the cyclotide domain with particular emphasis on P2, P1, P1′, and P2′ residues at the C terminus of the cyclotide domain to help delineate the residues that are essential for cyclization. Our examination of the in planta protein products resulting from systematic mutation of key residues within the processing site has enabled a detailed understanding of the recognition site required for binding to the enzyme that catalyzes cyclization. During the process of mutating the Ctr of Oak1 a novel proline hydroxylation sequence was discovered that contains the tetrapeptide motif GAPS within the Ctr.
EXPERIMENTAL PROCEDURES
Production of Mutant Oak1 Constructs
The Oak1 mutants were generated from the Oak1 gene (supplemental Fig. S1) using the Phusion site-directed mutagenesis kit (Finnzymes). All constructs were subcloned into pAM9 to incorporate the 35S-cauliflower mosaic virus promoter and terminator sequences (19). The genes were then cloned into the binary vector pBIN19. The pBIN19 expression vectors were transformed into Agrobacterium tumefaciens strain 4404. The Agrobacterium was grown on yeast mannitol plates containing 50 μg/ml of kanamycin and 100 μg/ml of streptomycin.
Transient Expression
Agrobacterium cells were taken from yeast mannitol plates and suspended in a transformation solution containing 10 μm acetosyringone, 10 mm MgCl2 at an A600 of 1.0 and incubated at room temperature for 2 h. The cells were then infiltrated into the abaxial side of the leaves of N. benthamiana plants by forcing the liquid into the tissue using a syringe (16). The full list of constructs expressed in planta is presented in supplemental Fig. S2.
Peptide Analysis of Leaf Tissue
Samples were prepared by grinding N. benthamiana leaves to a powder in liquid N2 before the addition of 50% (v/v) acetonitrile containing 0.1% (v/v) TFA. Samples were centrifuged and the supernatant was de-salted and concentrated using C18 ziptips (Millipore). Samples were mixed 1:1 with a saturated solution of α-cyano-4-hydroxycinnamic acid, spotted onto a sample plate, and air dried. All samples were produced with a minimum of three biological replicates. Mass analysis was performed in positive ion reflector mode on either a 4700 Proteomics Analyzer (AB/Sciex, Foster City, CA) or a Bruker ultraflex III MALDI TOF/TOF mass spectrometer (Bruker AXS GmbH, Karlsruhe, Germany). Two hundred spectra at each of 10 randomly selected positions were accumulated per spot between 1000 and 5000 Da using an MS positive ion reflectron mode acquisition method. Calibration was conducted using a mixture of peptide standards (Bruker Daltonics). Data were acquired and processed with either the accompanying 4000 series Explorer software or Bruker flexAnalysis software.
Relative Quantification of Transiently Expressed Peptides
Spectra were collected and the integrated peak area corresponding to known Oak1 peptides were taken as 100% of the expressed peptides. This enables the amount of cyclic protein to be assessed within the sample as a percentage of the total amount of cyclotide peptides. All mutants were expressed in triplicate within different leaves to give a minimum of three biological replicates. The S.E. ± mean was determined for each peptide mass. This relative quantification method using MALDI-MS was validated against LC-electrospray ionization-MS using an independent t test to assess equality of means (IBM SPSS Statistics, version 19.0) to determine that the datasets produced by both techniques were not significantly different (supplemental Fig. S3). LC-MS could not be used for this study as it has limitations in sensitivity making it difficult to detect cyclic peptides. The final relative levels of cyclic peptide produced by each construct were compared with the WT-Oak1 construct and a paired t test was used to determine whether the levels of cyclic peptide were statistically different (represented as p values).
Purification and Analysis of kB1-GAOS
The kB1-GAOS peptide was purified from plant extracts using HPLC. Proteins were separated on a C8 column in 0.1% TFA and eluted with 0.089% TFA, 80% acetonitrile. Peaks containing the 3236 dalton mass were determined using MALDI-MS and pooled. Amino acid analysis of the 3236 Dalton HPLC peak was performed by the Australian proteome analysis facility (Sydney, Australia) to confirm the presence of hydroxyproline.
Inhibitor Infiltrations
The Oak1 and Oak1 L31A constructs were expressed transiently in N. benthamiana with and without the prolylhydroxylase inhibitor 2,4-pyridinedicarboxylic acid monohydrate (PDCA). A 2 mm solution of the inhibitor was infiltrated into N. benthamiana leaves 12 h after Agrobacterium infiltration and leaves were harvested at 3.5 dpi.
RESULTS
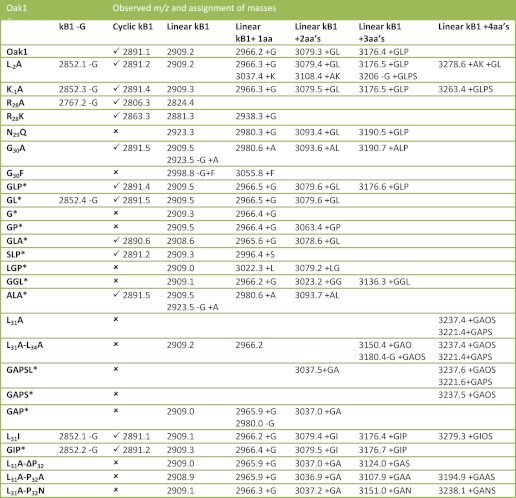
Expression of the Oak1 precursor in transgenic plants produces both cyclic and linear forms of the kalata B1 protein due to processing inefficiencies (16). In this study we have expressed the Oak1 gene and mutants of this gene in N. benthamiana to determine which amino acids within the processing sites of the Oak1 precursor protein are necessary for cyclization to occur. When Oak1 is expressed transiently the largest Oak1 precursor protein intermediate observed via mass spectrometry (MS) was that of the cyclotide domain with the Ctr still attached (Fig. 2A). To determine whether substitution of amino acids within the Oak1 precursor had an effect on the amount of cyclic protein produced a novel method of quantitation was utilized. This method involved taking all MALDI-TOF MS peaks corresponding to the Oak1 precursor, i.e. peptides consisting of the kB1 cyclotide domain with varying lengths of Ctr attached, as 100% of the total Oak1 precursor protein expressed. Using this method it was possible to quantitate the amount of cyclic kB1 produced relative to the linear forms in any one sample (Fig. 2A, right panel). When Oak1 was expressed transiently in N. benthamiana leaves a total of 5.1% (±1.5%) of all peptides expressed were processed to the cyclic form. All of the other expressed peptides were present as linear forms of kalata B1 with varying lengths of Ctr. All peptides produced and their masses are documented in Table 1. The relative amount of each of the peptides produced during expression of the Oak1 mutants is given in Table 2. Analysis of the statistical significance of the levels of cyclic protein as compared with the WT-Oak1 are given in supplemental Fig. S4.
FIGURE 2.
MALDI MS spectra of Oak1 proteins expressed transiently in N. benthamiana. The letters above each peak indicate the amino acids of the Ctr attached to the linear kB1 peptide. A, Oak1 with full-length Ctr (GLPSLAA*) and the proteins produced. B, Oak1 with the Ctr truncated to GLP. C, Oak1 with the Ctr truncated to GL*. D, Oak1 with the Ctr truncated to G*. The right histograms are given showing the average percentage of all of the peptide species for each of the constructs expressed.
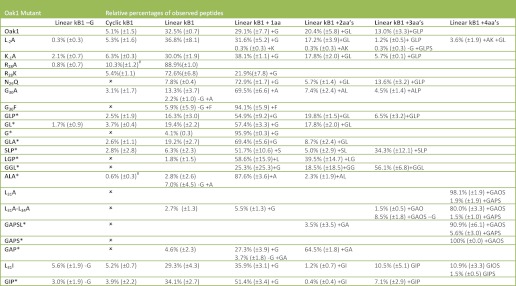
TABLE 1.
Summary of the mutants of Oak1 which were tested for cyclization in N. benthamiana
A tick indicates cyclic peptide was produced. The mass to charge ratio (m/z) is given for each non-cyclic product as well as the corresponding cyclic peptide. −G corresponds to the loss of Gly from the N-terminus of the cyclotide domain and + indicates amino acids present at the C terminus of the cyclotide domain.
* Indicates constructs with a truncated Ctr. These constructs are labeled according to the amino acid sequence of the Ctr.
TABLE 2.
Summary of the relative percentages of each Oak1 peptide produced when mutants of Oak1 were expressed translently in N. benthamiana
The S.E. ± mean is shown in brackets. −G corresponds to the loss of Gly from the N terminus of the cyclotide domain and + indicates amino acids present at the C terminus of the cyclotide domain; # Indicates the level of cyclic protein is significantly different (p < 0.05) to the levels of cyclic protein produced by the Oak1 construct.
Requirements for Cleavage at the N Terminus of the Cyclotide Domain (P1 and P2)
Post-translational processing of the precursor begins with the excision of the region upstream of the cyclotide domain comprising the N-terminal propeptide and Ntr (16). We predict that the Ntr (and regions upstream of it) must be removed from the cyclotide domain before cyclization can occur. The N terminus of the cyclotide domain is liberated after cleavage next to an upstream residue which for Oak1 is a Lys residue (Lys−1). Substitution of Lys−1 with an Ala (K−1A) did not produce significant changes in the levels of cyclic kB1, 6.3% (±0.3%) when expressed transiently in N. benthamiana. Substitution of the residue further upstream at position P2 (Leu−2) with an Ala (L-2A) effected cleavage of the Ntr domain, with normal levels of cyclic protein being produced (5.3% (±1.6)), but a number of peptide products were observed with additional residues at the N terminus (Table 2). This suggests that the Leu residue is necessary for clean cleavage of the Ntr from the cyclotide domain.
Cyclotide Domain C-terminal P1 and P2 Requirements
The last residue of the cyclotide domain before the Ctr is Asn29, this residue is in the P1 position and is necessary for cleavage and cyclization to occur. Substitution of this Asn with an Ala resulted in no cyclic protein being made and mutation to Asp produced minimal cyclic peptide (16). Furthermore, substitution of this residue with a glutamine (N29Q), also failed to produce any cyclic peptide. The P2 residue immediately preceding Asn29 is a highly conserved residue and a search of the cyclotide data base (Cybase) revealed that greater than 50% of documented cyclotides contain an Arg at this position (15). When Arg28 was replaced with Ala (R28A) or Lys (R28K), there was a significant decrease in the amount of detectable protein as determined using MALDI MS. Cyclic protein was still produced, but the level of expression for both of these was significantly lower than observed for the wild-type Oak1 precursor. Quantification suggested an increased ratio of cyclic to linear protein for the R28A mutant with 10.3% (±1.2%) of all protein present being cyclic, but the total amount of Oak1 peptides observed was extremely low. The increase in the relative amount of cyclic protein was statistically significant compared with WT-Oak1 (p = 0.04).
Ctr Motif Requirements
Minimum Ctr Motif for Cyclization
The Ctr sequence plays a major role in peptide recognition prior to cyclization. Expressing the wild-type Oak1 precursor in N. benthamiana produced a series of peptides with different length Ctr regions due to the expressed protein being sequentially trimmed by carboxypeptidases. Not only is cyclic protein produced (5.1%), but also linear kB1 (32.5%), linear kB1 with Gly present at the C terminus (29.1%), linear kB1 with Gly-Leu (20.4%), and linear kB1 with Gly-Leu-Pro (13.0%) (Fig. 2A). To determine the minimum Ctr motif required for cyclization to occur efficiently, a series of truncation mutants were produced (supplemental Fig. S2). Expression of a construct where the Ctr was truncated from the native 7 amino acids down to 3 amino acids (GLP*) did not prevent cyclization with 2.5% (±1.9%) cyclic kB1 observed (Fig. 2B). Further truncation of the Ctr to 2 amino acids (GL*) produced 3.7% (±0.4%) cyclic protein (Fig. 2C). The level of cyclic protein in both of these truncation mutants was not significantly different to the WT-Oak1 construct. Shortening the Ctr to a single Gly (G*) resulted in no cyclic peptide formation with >95% of the peptide remaining as the linear +G peptide (Fig. 2D). The relative percentages of all of the peptides expressed are presented as histograms in Fig. 2, and the standard error of these values are presented in Table 2.
Ctr P1′ Requirements
Having determined that the minimum length of the Ctr required for cyclization is 2 amino acids, substitutions were made within the Ctr to further clarify the requirements of this motif. The substitutions encompassed the Ctr P1′, P2′, and P3′ positions, which are predicted to be required for binding to the enzyme that catalyzes cyclization. The Gly at position P1′ (Gly30) was mutated to Ala, Ser, and Phe. Substitution of Gly30 with Ala (G30A), which is a conservative substitution, produced 3.1% (±1.7%) cyclic peptide, whereas mutation to Phe (G30F), a large hydrophobic residue, produced no cyclic protein at all (Table 2). Furthermore, a mutant with a 3-amino acid Ctr was produced where Gly30 was mutated to Ser (G30S). This construct was labeled kB1-SLP* and produced 2.8% (±2.8%) cyclic protein.
Ctr P2′ Requirements
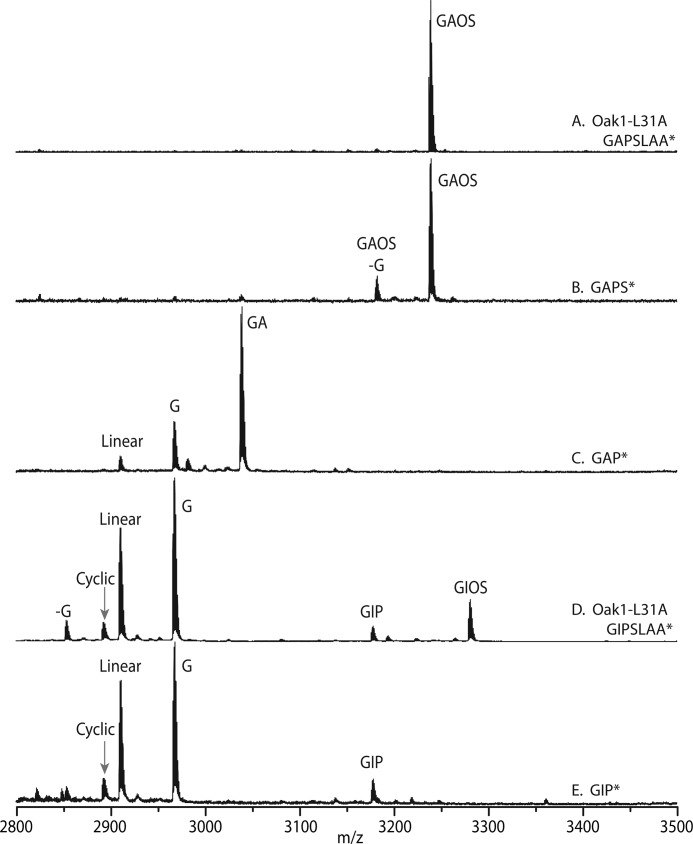
To determine whether the highly conserved Leu residue at position P2′ of the Ctr (Leu31) was necessary for cyclization, Leu was substituted with Ala or Ile. Substitution of Leu to Ala (L31A) resulted in a single linear product with an m/z of 3237.4 (Fig. 3A). This mass did not correlate with any of the predicted linear cyclotides so it was postulated that a post-translational modification of the peptide had occurred. Sequencing of this peptide by LC-MS revealed that the cyclotide domain was unaltered but Ctr contained a hydroxyproline (Hyp) at amino acid 32. The hydroxylated peptide was composed of the cyclotide domain with a 4-amino acid Ctr region, Gly-Ala-Hyp-Ser. Expression of the L31I mutant was interesting as it still produced 5.2% cyclic protein (±0.7%) and also an additional peptide with the post-translational hydroxylation of Pro32 (kB1-Gly-Ile-Hyp-Ser, 3278.4 Da). However, the hydroxylated protein was a minor product making up just 10.9% (±3.3%) of all Oak1 peptides identified.
FIGURE 3.
MALDI MS spectra of Oak1 proteins expressed transiently in N. benthamiana. A, Oak1 L31A. B, Oak1 L31A with the Ctr truncated to GAPS*. C, Oak1 L31A with the Ctr further truncated to GAP*. D, Oak1 L31I. E, Oak1 L31I with the Ctr truncated to GIP*. Hydroxyproline is represented as an O in single letter amino acid code.
Ctr P3′ Requirements
To determine whether a Pro residue is essential at the P3′ position of the Ctr, this amino acid was mutated to Ala (P32A) in a construct containing a 3-amino acid Ctr (kB1-GLA*). The expression of kB1-GLA* produced 2.6% (±1.1%) cyclic kB1 similar to that found for Oak1.
Shuffling of the P1′, P2′, and P3′ Sites
Shuffled variants of the Oak1 Ctr containing only 3 amino acids after the cyclotide domain were produced to further delineate the residues required for cyclization. Expression of mutants where the Leu was moved such as kB1-LGP* and kB1-GGL* or removed such as kB1-GP* resulted in no cyclic protein being produced. Additionally, substitution of both residues either side of the Leu interfered with the cyclization mechanism such that expression of kB1-ALA* only produced 0.6% (±0.3%) cyclic protein (Table 2). This reduction in the amount of cyclic protein produced was statistically significant compared with the relative percentage produced by WT-Oak1 (p = 0.05). These shuffled variants of the Ctr tripeptide motif revealed that Leu or a similar residue at the P2′ position is essential for cyclization.
Characterization of the Proline Hydroxylation Motif
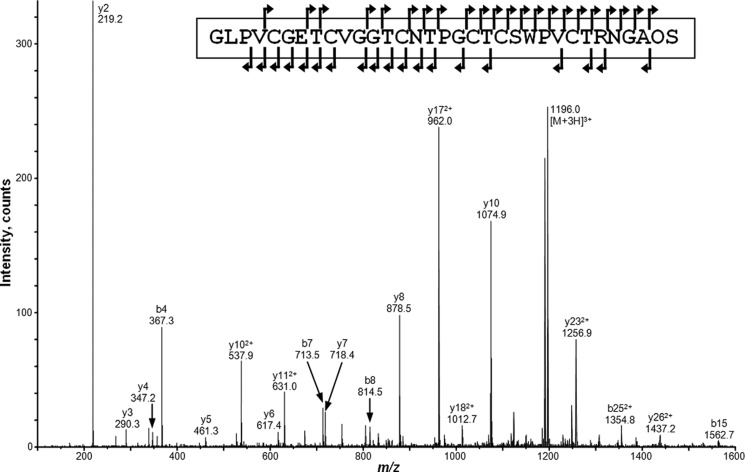
As mentioned previously substitution of Leu with Ala at position 31 resulted in accumulation of a single peptide with an unexpected mass of 3237.4 as a result of hydroxylation of Pro32. MS/MS sequencing of this mass revealed the amino acid sequence to be the kalata B1 + GAOS peptide (Fig. 4). Amino acid analysis further confirmed that a Hyp residue was present in this peptide. The proline at position 32 was found to be essential for the post-translational modification event as deletion of this residue or mutation to an Ala or Asn (double mutants L31A-ΔP32, L31A-P32A, and L31A-P32N) resulted in proteolytic trimming of the Ctr over time with no cyclic protein formation nor any post-translational hydroxylation events.
FIGURE 4.
MS/MS spectrum of the reduced and alkylated form of the Oak1 L31A product with a precursor ion observed at m/z 1196.03+ corresponding to a mass of 3585 Da. Sequence analysis revealed that the peptide sequence was GLPVCGETCVGGTCNTPGCTCSWPVCTRNGAOS, where O = Hyp. Singly charged ions present are labeled.
Determination of the Minimum Motif for Hydroxylation
To determine the minimum motif for hydroxylation of Pro32, a series of Oak1 L31A truncation mutants were made that corresponded to sequences kB1-GAPSL*, kB1-GAPS*, and kB1-GAP*. The GAPSL* and GAPS* mutants produced only a single mass (3237.4) corresponding to Gly-Ala-Hyp-Ser motif at the Ctr (Fig. 3B). Further truncation to GAP* did not produce any masses that corresponded with the presence of Hyp (Fig. 3C). Truncation of the L31I mutant to a Ctr of only 3 amino acids (kB1-GIP*) also no longer produced Hyp, but still produced cyclic peptide (Fig. 3E).
Prevention of Carboxypeptidase Activity by Hyp
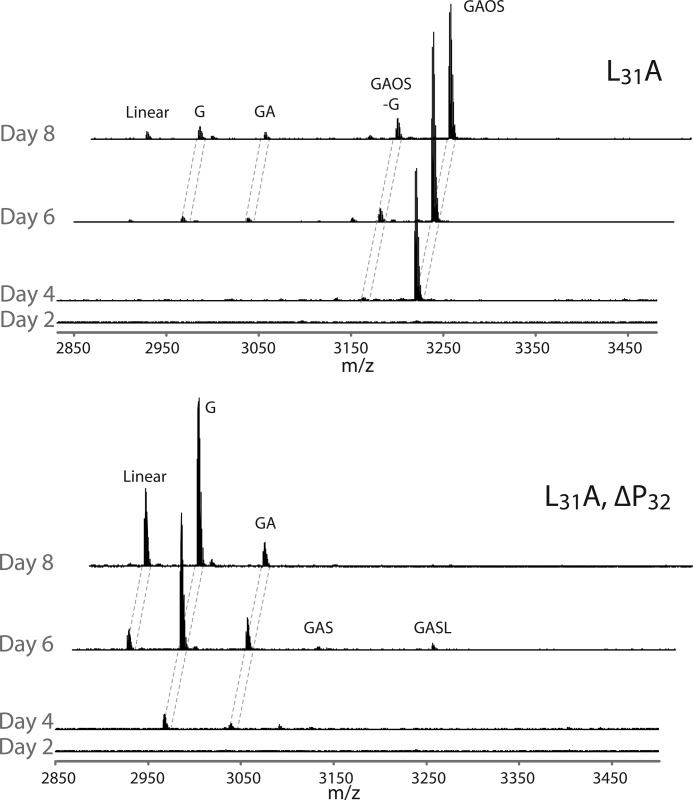
A time course was undertaken to examine the products resulting from expression of both L31A and the double mutant L31A-ΔPro32 in N. benthamiana over a period of 8 days. It was apparent that biosynthesis and trimming differed greatly for these two constructs (Fig. 5). The L31A mutant produced a single protein product of m/z 3237, which was the kalata B1-GAOS peptide. This hydroxylated peptide was stable over a longer period of time and was not readily trimmed from the C terminus. The L31A-ΔPro32 double mutant was trimmed via proteolysis yielding linear kB1 and kB1 + G as the major products at day 8. The hydroxylation of the proline prevented the action of carboxypeptidases resulting in a long-lived product.
FIGURE 5.
Time course for the expression and processing of two Oak1 mutants over 8 days in N. benthamiana. Top, L31A yields kB1 with a truncated and hydroxylated Ctr with m/z 3237, this peptide is resistant to proteolytic breakdown. Bottom, L31A,ΔPro32 is trimmed over 8 days and produces linear kB1 and kB1 + G as the predominant products by day 8.
Infiltration of a Prolylhydroxylase Inhibitor
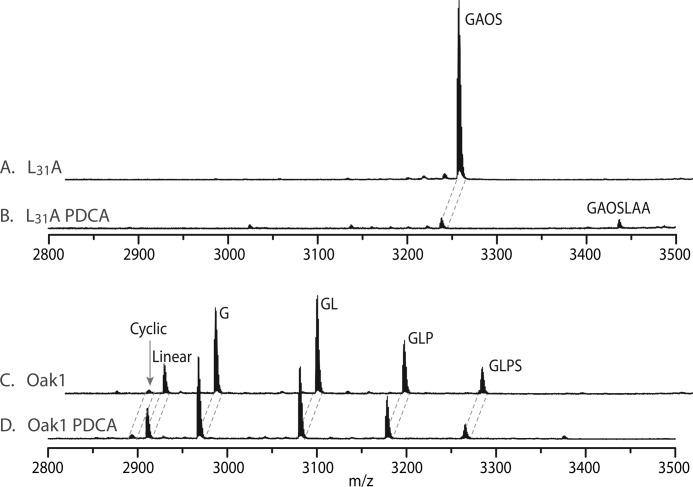
Transient expression of L31A in N. benthamiana with and without the prolylhydroxylase inhibitor PDCA produced a marked difference in the peptides produced. Without PDCA, expression of L31A resulted in production of the hydroxylated kB1-GAOS peptide (Fig. 6A), but in the presence of PDCA production of this peptide was almost completely abolished (Fig. 6B). Interestingly, a small peak coinciding with the mass expected for kB1 + GAOSLAA (m/z 3435.9) was found in the L31A PDCA sample, although there was insufficient peptide for sequencing. Expression of the WT-Oak1 precursor with and without PDCA did not affect expression levels or processing producing near identical spectra (Fig. 6, C and D). The sharp reduction in the amount of kalata B1 peptide produced with the PDCA-treated L31A construct was unexpected, but may be due to misprocessing in the ER and resultant degradation of the protein product.
FIGURE 6.
MALDI MS spectra of transiently expressed Oak1 L31A and Oak1 with and without the prolylhydroxylase inhibitor PDCA. A, Oak1 L31A. B, Oak1 L31A in the presence of PDCA. C, Oak1. D, Oak1 in the presence of PDCA.
DISCUSSION
The processing of cyclotides in planta is an interesting phenomenon with multiple post-translational processing steps needed to produce the final cyclic peptide. For these processing events to occur, specific residues are required for enzyme recognition and binding. The post-translational processing events that produce cyclic protein appear to start with the cleavage of the N-terminal propeptide and Ntr from the precursor sequence. When Oak1 is expressed in N. benthamiana, a plant that does not naturally produce cyclotides, it is processed to cyclic protein, linear cyclotide domain alone, or the cyclotide domain with varying lengths of the Ctr attached.
Cleavage of the Ntr from the cyclotide domain of Oak1 occurs at a lysine residue (Lys−1) at the P1 position immediately upstream of the cyclotide domain. Substitution of this residue significantly increased the relative amount of cyclic to linear protein product and substitution of the Leu residue in position P2 further upstream had a greater impact as it resulted in inefficient cleavage of the Ntr producing linear cyclotide with additional residues still present at the N terminus.
The nature of the residue in the P1 position immediately upstream of the cyclotide domain is probably not important; it can be a Lys, as in the O. affinis cyclotides, or an Asn as occurs in other cyclotide subfamilies, as long as the Ntr is surface-exposed and accessible to proteases and cleavage at this junction prior to the cleavage at the Ctr that results in cyclization. This is supported by the recent discovery of cyclotides in the Fabaceae family where the cyclotide domain is directly preceded by the ER signal peptide and the Ntr is absent (20). The cleavage of this signal peptide in the ER would free up the N terminus of the cyclotide, which presumably remains free until the cyclotide precursor reaches the compartment where cyclization occurs.
Although the residues upstream of the cyclotide domain in positions P2 and P1 are not conserved, the first two residues of the cyclotide domain itself in positions P1′ and P2′ (Gly-Leu) are conserved and these residues appear necessary for cyclization. Substitution of either of these residues resulted in little or no cyclic protein being formed during transient expression of the Oak1 construct (16). These residues are similar across species, with a common theme of a small residue followed by a larger hydrophobic residue (Leu, Ile, or Val).
The residues at the C terminus of the cyclotide domain in positions P1 and P2 are essential for cyclization. Asn29 in position P1 prior to the Ctr is necessary for cleavage and cyclization to occur with mutation of this residue producing no cyclic protein. The residue at position P2 in the cyclotide domain is not completely conserved, but is generally an Arg. Substitution of this residue did not reduce the relative percentage of cyclic protein produced, but severely reduced the total amount of kalata B1 peptide produced such as to be difficult to detect with MALDI MS. The reduction in peptide levels is likely due to misfolding of the precursor protein resulting in degradation or some other event preventing exit of the protein from the ER.
The fact that Arg28 is not required for cyclization is interesting and allows an earlier proposed self-cyclization mechanism to be discounted. We initially speculated that Arg could act as an electron withdrawing group to activate an intein-type processing event involving the autocatalytic removal of residues on the C-terminal side of Asn29 via the formation of a succinimide/lactone intermediate, with ligation of the free termini to the amide from the N-terminal Gly, facilitated by the activated Asn residue (3, 11). For this process to yield mature cyclotide, removal of the N-terminal prodomain must precede the ligation event.
Ctr Motif Requirements
From our hypothesis that the Ctr is essential for cyclization the residues required in this tail region were studied to determine not only the minimum motif for cyclization, but also the type of amino acids permitted at each position relative to the cleavage site. Truncation mutants of the Ctr revealed that a minimum of 2 amino acids are required after Asn29 for cyclization to occur. Trimming of the Ctr beyond the Leu residue in the P2′ position results in complete abolishment of all cyclic peptide production.
The P1′ amino acid within the Ctr was substituted with various amino acids and from this it was obvious that small amino acids are tolerated and a large hydrophobic amino acid was capable of completely abolishing cyclic peptide production. This finding is strengthened by the high conservation of a small residue in this position in cyclotide precursors from the majority of species (15).
The importance of the P2′ residue within the Ctr was exemplified with the truncation mutants that showed that Leu is essential for cyclization and this is not entirely surprising as a Leu in this position is completely conserved across all cyclic peptides found in all species except the Fabaceae, which contain an Ile (20). Cleavage and cyclization occurs 2 amino acids from this leucine. It is proposed that this Leu residue binds within a hydrophobic pocket of the asparaginyl endopeptidase acting to hold the Ctr and position the Asn in an optimal conformation for cleavage and ligation of the cyclotide domain. Substitution of Leu31 with isoleucine (L31I), which is a conservative change, still produced cyclic protein in N. benthamiana with equal efficiency to the wild-type Oak1 gene.
The enzyme that carries out the cyclization reaction, which is most likely an AEP, requires a small residue in the P1′ position and a larger hydrophobic amino acid in the P2′ position. These are not completely novel requirements with the P1′ residue often required to be a small amino acid in the case of naturally occurring protease inhibitors due to steric constraints in binding to a protease. The coagulation proteases such as plasmin and kallikrein in mammals prefer a hydrophobic residue such as Leu to bind in the S2′ binding pocket (21).
Although changing Leu31 in the P2′ position to a similar amino acid had little effect on cyclization, substitution of this residue with Ala (L31A) resulted in production of a single post-translationally modified peptide of 3236.4 Da. Sequencing revealed that this peptide was kalata B1 with a truncated and modified form of the Ctr containing the amino acid sequence Gly-Ala-Hyp-Ser. No other processing intermediates were produced with this construct, suggesting that the enzyme that hydroxylates the proline does so with high efficiency.
The L31I mutant, while producing cyclic protein, also produced a mass that matched the post-translational hydroxylation of Pro32 with the sequence kB1 + Gly-Ile-Hyp-Ser (3278.4 Da). The hydroxylated protein was a minor product, but it is interesting to note that such a conservative change could produce this hydroxylation event.
The P3′ position of the Ctr had little effect on cyclization efficiency. Shortening the Ctr past this residue did not prevent cyclization and mutation also had little effect. This suggests that the major determinant of specificity are the P1, P2, P1′, and P2′ residues. The amino acids within the Ctr are highly conserved across species and expression of mutants where Leu31 was removed or moved resulted in no cyclic protein being produced. It was also interesting to note that substitution of both residues on either side of the Leu interfered with cyclization.
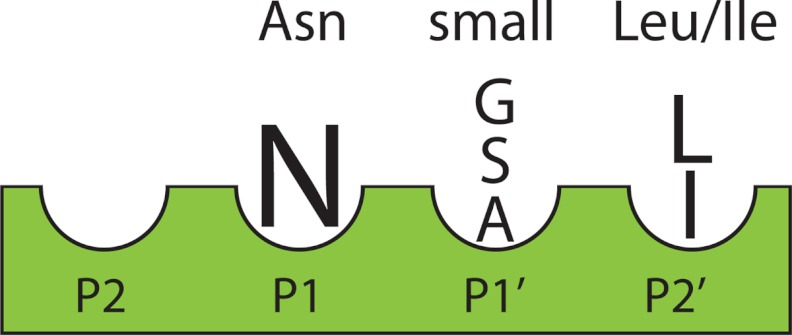
The mutants expressed in this work reveal a cyclization motif that can be modified within specific limits. The P2 residue could not be modified due to its effect on protein levels, but not due to cyclization constraints. The P1 position requires an Asn although an Asp can function inefficiently at this position. P1′ requires a small amino acid and P2′ requires Leu or Ile for efficient cyclization to occur. These mutants all reinforce the need for an Asn-small aa-Leu, at the C terminus for efficient cyclization to occur (Fig. 7).
FIGURE 7.
The specificity requirements of the C-terminal region of the Oak1 cyclotide precursor for production of cyclic kalata B1.
The Proline Hydroxylation Motif
The modification of the P2′ residue had major implications producing post-translationally modified protein products. Mutation to Ala resulted in the production of kalata B1 with a Ctr containing the sequence GAOS. The hydroxylation of this protein appears unusual as it does not coincide with a known hydroxylation motif, although these are poorly characterized in plants (22).
Truncation of the Ctr revealed that the minimum motif for hydroxylation of Pro32 was GAPS. Removal of the C-terminal Ser residue prevented hydroxylation of the proline and resulted in proteolytic trimming of the Ctr. Deletion or substitution of the proline residue in the Ctr negates any post-translational modification of the expressed protein and resulted in proteolytic trimming of the Ctr over time. A time course for the processing of the L31A and L31A-ΔP32 mutants showed that the L31A mutant produces only the kalata-GAOS product, which is quite resistant to breakdown, although the L31A-ΔP32 double mutant was simply trimmed via proteolysis.
Cyclotides are processed inside the ER where protein-disulfide isomerase helps with the formation of disulfide bonds within the cysteine knot motif (23). Protein-disulfide isomerases are thought to exist in complex with prolylhydroxylases in the ER so the cyclotide precursor would be in close proximity to these proteins when being folded (24). This could account for the high efficiency of the hydroxylation reaction in the L31A mutant.
Although the hydroxylation reaction is catalyzed by a prolylhydroxylase, it is more difficult to speculate on the enzyme responsible for cleaving the Ctr to produce the single product kB1-GAOS. Some clues are given when the L31A mutant was infiltrated with the prolylhydroxylase inhibitor. This resulted in very little cyclotide protein being produced but the masses that were found coincided with kB1-GAOS as well as kB1-GAOSLAA. This suggests that the prolylhydroxylase is acting to hydroxylate the protein prior to cleavage of the last 3 amino acids of the Ctr (LAA). The fact that this cleavage reaction is inhibited by a prolylhydroxylase inhibitor also points to the prolylhydroxylase being responsible for hydroxylation of the proline before the cleavage reaction.
Conclusions
From this work we can conclude that for cyclization to occur the N-terminal propeptide must be cleaved to free up the N terminus of the cyclotide domain. Also the Ctr is necessary for cyclization with a specific amino acid sequence required for this reaction to take place. This motif requires an Asn at position P1, followed by a small amino acid in the P1′ position and a Leu in the P2′ position. The finding that a single amino acid substitution in the Oak1 C-terminal cyclization motif can produce a post-translational hydroxylation event suggests that there may be other hydroxylated cyclotides present in nature.
The fact that noncyclotide producing plants can make the cyclic form of these peptides when expressed recombinantly points to the ability of enzymes, which normally have other functions, to carry out the unique cyclization process in all plants. The AEPs responsible for this process are predicted to be very similar in both cyclotide and noncyclotide producing plants. The fact that the cyclization process is very efficient in the cyclotide producing plants may suggest that some optimization of these enzymes or the cyclotide precursor has occurred over time to enhance the levels of cyclic versus linear peptide.
Supplementary Material
Acknowledgments
Bruce McGinness for growing all of the plants used in this study, James McKenna for statistical analysis, and Dr. Gert Talbo for advice on MALDI MS analysis.
This work was supported by Grant DP0984390 from the Australian Research Council.

This article contains supplemental Figs. S1–S4.
- AEP
- asparaginyl endopeptidase
- Hyp
- hydroxyproline
- Ntr
- N-terminal repeat
- Ctr
- C-terminal repeat
- Oak1
- O. affinis kalata B1 precursor gene
- Oak1
- O. affinis kalata B1 precursor protein
- kB1
- kalata B1
- PCDA
- 2,4-pyridinedicarboxylic acid monohydrate
- ER
- endoplasmic reticulum.
REFERENCES
- 1. Craik D. J., Daly N. L., Bond T., Waine C. (1999) Plant cyclotides. A unique family of cyclic and knotted proteins that defines the cyclic cystine knot structural motif. J. Mol. Biol. 294, 1327–1336 [DOI] [PubMed] [Google Scholar]
- 2. Gruber C. W., Cemazar M., Anderson M. A., Craik D. J. (2007) Insecticidal plant cyclotides and related cystine knot toxins. Toxicon 49, 561–575 [DOI] [PubMed] [Google Scholar]
- 3. Jennings C., West J., Waine C., Craik D., Anderson M. (2001) Biosynthesis and insecticidal properties of plant cyclotides. The cyclic knotted proteins from Oldenlandia affinis. Proc. Natl. Acad. Sci. U.S.A. 98, 10614–10619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Colgrave M. L., Craik D. J. (2004) Thermal, chemical, and enzymatic stability of the cyclotide kalata B1. The importance of the cyclic cystine knot. Biochemistry 43, 5965–5975 [DOI] [PubMed] [Google Scholar]
- 5. Craik D. J., Simonsen S., Daly N. L. (2002) The cyclotides. Novel macrocyclic peptides as scaffolds in drug design. Curr. Opin. Drug Discov. Devel. 5, 251–260 [PubMed] [Google Scholar]
- 6. Grain L. (1973) Isolation of oxytocic peptides from Oldenlandia affinis by solvent extraction of tetraphenylborate complexes and chromatography on Sephadex LH-20. Lloydia 36, 207–208 [PubMed] [Google Scholar]
- 7. Craik D. J., Daly N. L., Mulvenna J., Plan M. R., Trabi M. (2004) Discovery, structure, and biological activities of the cyclotides. Curr. Protein Pept. Sci. 5, 297–315 [DOI] [PubMed] [Google Scholar]
- 8. Craik D. J., Daly N. L., Saska I., Trabi M., Rosengren K. J. (2003) Structures of naturally occurring circular proteins from bacteria. J. Bacteriol. 185, 4011–4021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tan N. H., Zhou J. (2006) Plant cyclopeptides. Chem. Rev. 106, 840–895 [DOI] [PubMed] [Google Scholar]
- 10. Craik D. J., Anderson M. A., Barry D. G., Clark R. J., Daly N. L., Jennings C. V., Mulvenna J. (2001) Lett. Peptide Sci. 8, 119–128 [Google Scholar]
- 11. Mulvenna J. P., Foley F. M., Craik D. J. (2005) Discovery, structural determination, and putative processing of the precursor protein that produces the cyclic trypsin inhibitor sunflower trypsin inhibitor 1. J. Biol. Chem. 280, 32245–32253 [DOI] [PubMed] [Google Scholar]
- 12. Mylne J. S., Colgrave M. L., Daly N. L., Chanson A. H., Elliott A. G., McCallum E. J., Jones A., Craik D. J. (2011) Albumins and their processing machinery are hijacked for cyclic peptides in sunflower. Nat. Chem. Biol. 7, 257–259 [DOI] [PubMed] [Google Scholar]
- 13. Conlan B. F., Gillon A. D., Barbeta B. L., Anderson M. A. (2011) Subcellular targeting and biosynthesis of cyclotides in plant cells. Am. J. Bot. 98, 2018–2026 [DOI] [PubMed] [Google Scholar]
- 14. Conlan B. F., Anderson M. A. (2011) Circular microproteins and mechanisms of cyclization. Curr. Pharm. Des. 17, 4318–4328 [DOI] [PubMed] [Google Scholar]
- 15. Kaas Q., Craik D. J. (2010) Analysis and classification of circular proteins in CyBase. Biopolymers 94, 584–591 [DOI] [PubMed] [Google Scholar]
- 16. Gillon A. D., Saska I., Jennings C. V., Guarino R. F., Craik D. J., Anderson M. A. (2008) Biosynthesis of circular proteins in plants. Plant J. 53, 505–515 [DOI] [PubMed] [Google Scholar]
- 17. Saska I., Gillon A. D., Hatsugai N., Dietzgen R. G., Hara-Nishimura I., Anderson M. A., Craik D. J. (2007) An asparaginyl endopeptidase mediates in vivo protein backbone cyclization. J. Biol. Chem. 282, 29721–29728 [DOI] [PubMed] [Google Scholar]
- 18. Conlan B. F., Gillon A. D., Craik D. J., Anderson M. A. (2010) Circular proteins and mechanisms of cyclization. Biopolymers 94, 573–583 [DOI] [PubMed] [Google Scholar]
- 19. Anderson M. A., Van Heeswijck R., West J., Bateman K., Lee M., Christeller J. T., McDonald G., Heath R. L. (1997) Proteinase inhibitors from Nicotiana alata enhance plant resistance to insect pests. J. Insect. Physiol. 43, 833–842 [DOI] [PubMed] [Google Scholar]
- 20. Poth A. G., Colgrave M. L., Lyons R. E., Daly N. L., Craik D. J. (2011) Discovery of an unusual biosynthetic origin for circular proteins in legumes. Proc. Natl. Acad. Sci. U.S.A. 108, 10127–10132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bajaj M. S., Ogueli G. I., Kumar Y., Vadivel K., Lawson G., Shanker S., Schmidt A. E., Bajaj S. P. (2011) Engineering kunitz domain 1 (KD1) of human tissue factor pathway inhibitor-2 to selectively inhibit fibrinolysis. Properties of KD1-L17R variant. J. Biol. Chem. 286, 4329–4340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shimizu M., Igasaki T., Yamada M., Yuasa K., Hasegawa J., Kato T., Tsukagoshi H., Nakamura K., Fukuda H., Matsuoka K. (2005) Experimental determination of proline hydroxylation and hydroxyproline arabinogalactosylation motifs in secretory proteins. Plant J. 42, 877–889 [DOI] [PubMed] [Google Scholar]
- 23. Gruber C. W., Cemazar M., Clark R. J., Horibe T., Renda R. F., Anderson M. A., Craik D. J. (2007) A novel plant protein-disulfide isomerase involved in the oxidative folding of cystine knot defense proteins. J. Biol. Chem. 282, 20435–20446 [DOI] [PubMed] [Google Scholar]
- 24. Yuasa K., Toyooka K., Fukuda H., Matsuoka K. (2005) Membrane-anchored prolyl hydroxylase with an export signal from the endoplasmic reticulum. The Plant J: for Cell Mol. Biol. 41, 81–94 [DOI] [PubMed] [Google Scholar]
- 25. Dutton J. L., Renda R. F., Waine C., Clark R. J., Daly N. L., Jennings C. V., Anderson M. A., Craik D. J. (2004) Conserved structural and sequence elements implicated in the processing of gene-encoded circular proteins. J. Biol. Chem. 279, 46858–46867 [DOI] [PubMed] [Google Scholar]
- 26. Burman R., Gruber C. W., Rizzardi K., Herrmann A., Craik D. J., Gupta M. P., Göransson U. (2010) Cyclotide proteins and precursors from the genus Gloeospermum. Filling a blank spot in the cyclotide map of Violaceae. Phytochemistry 71, 13–20 [DOI] [PubMed] [Google Scholar]
- 27. Zhang J., Liao B., Craik D. J., Li J. T., Hu M., Shu W. S. (2009) Identification of two suites of cyclotide precursor genes from metallophyte Viola baoshanensis. cDNA sequence variation, alternative RNA splicing, and potential cyclotide diversity. Gene 431, 23–32 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.