Background: The headful nuclease is essential for genome processing during viral DNA packaging.
Results: The crystal structure of P22 nuclease reveals a seven-stranded β-sheet core with two magnesium ions poised for catalysis.
Conclusion: P22 headful nuclease is active only in the context of the large terminase.
Significance: The C-terminal headful nuclease is activated by intramolecular cross-talk with the N-terminal ATPase domain.
Keywords: Bacteriophage, Crystal Structure, DNA Viruses, Ribonuclease, X-ray Crystallography, DNA Packaging, Large Terminase, Portal Protein, Small Terminase
Abstract
Packaging of viral genomes into preformed procapsids requires the controlled and synchronized activity of an ATPase and a genome-processing nuclease, both located in the large terminase (L-terminase) subunit. In this paper, we have characterized the structure and regulation of bacteriophage P22 L-terminase (gp2). Limited proteolysis reveals a bipartite organization consisting of an N-terminal ATPase core flexibly connected to a C-terminal nuclease domain. The 2.02 Å crystal structure of P22 headful nuclease obtained by in-drop proteolysis of full-length L-terminase (FL-L-terminase) reveals a central seven-stranded β-sheet core that harbors two magnesium ions. Modeling studies with DNA suggest that the two ions are poised for two-metal ion-dependent catalysis, but the nuclease DNA binding surface is sterically hindered by a loop-helix (L1-α2) motif, which is incompatible with catalysis. Accordingly, the isolated nuclease is completely inactive in vitro, whereas it exhibits endonucleolytic activity in the context of FL-L-terminase. Deleting the autoinhibitory L1-α2 motif (or just the loop L1) restores nuclease activity to a level comparable with FL-L-terminase. Together, these results suggest that the activity of P22 headful nuclease is regulated by intramolecular cross-talk with the N-terminal ATPase domain. This cross-talk allows for precise and controlled cleavage of DNA that is essential for genome packaging.
Introduction
Packaging of viral genomes into preformed procapsids is a nonspontaneous reaction catalyzed in DNA bacteriophages and herpesviruses by a powerful genome-packaging motor (1–3). This machine consists of two packaging proteins known as large and small terminases (L- and S-terminases)2 that assemble onto a dodecamer of portal protein to form a complex of unknown stoichiometry. The packaging motor inserts DNA inside empty procapsids with high precision and efficiency, at translocation rates as high as ∼2,000 bp/s (4). Structural studies on bacteriophage T4 L-terminase have revealed the organization of a prototypical L-terminase subunit (5, 6), thought to be conserved in other DNA viruses. The N terminus of T4 L-terminase contains an ATPase domain that is flexibly connected to a C-terminal nuclease domain. The ATPase burns ATP to power DNA translocation, whereas the nuclease domain introduces cuts in the viral genome to initiate and to end a packaging reaction. In contrast, the S-terminase subunit is thought to bind to a packaging initiation site in preparation for genome packaging and to regulate the ATPase activity associated with genome packaging (1–3).
P22 is a general transducing phage (7) and is often referred to as the representative member of the Podoviridae family of short tailed bacteriophages (8). Genome packaging in P22 has been studied mainly by genetic analysis. In this phage, the substrate for DNA packaging is a repeating polymer (or concatemer) containing up to 10 copies of phage genome (9). Concatemer DNA is produced by rolling circle replication of P22 DNA circularized by homologous recombination (10). DNA packaging initiates at a particular site (pac) located within gene 3 (11–16) that encodes S-terminase. Here the nuclease domain of L-terminase breaks the DNA backbone at several points (known as “series initiation cleavages”), generating a DNA end that is inserted into the procapsid, unidirectionally from the initiation cleavage point (10). The S-terminase subunit is thought to directly recognize the pac site and present it to the packaging motor (12), which, in analogy to T4 (6), likely consists of an oligomer of L-terminase assembled onto the portal vertex. During packaging, an individual unit of P22 genome (∼43 kb) is encapsulated inside an empty procapsid by a “headful packaging” mechanism (10). This process is robust but not perfect, as, in vivo, each round of infection results in ∼2% of newly replicated particles that carry host DNA instead of the viral chromosome (7). Headful packaging indicates a packaging strategy where the length of the DNA encapsulated inside the capsid shell is determined by the interior volume of the mature phage particle. Critical to this packaging mechanism is the headful nuclease of L-terminase that cleaves packaged DNA only when the phage head is full, releasing the remainder of the concatemer to be available for a new round of packaging (10). Consistent with their important role in DNA packaging, the genes encoding L- and S-terminase subunits are essential for P22 viability in vivo (17).
Although the basic principles of DNA packaging have been elucidated, viral genome-packaging motors are poorly characterized. Atomic level structural information is presently available for a catalytically inactive construct of bacteriophage T4 L-terminase (6) and for the isolated L-terminase nuclease domains of SPP1 (18) and human cytomegalovirus (human herpesvirus 5 or HHV-5) (19). While no high resolution structure is available for a L-terminase holoenzyme, a pseudoatomic model of T4 L-terminase was proposed based on a 32 Å asymmetric cryo-electron microscopy reconstruction of T4 procapsid bound to L-terminase (6). In this model, the nuclease domain of L-terminase points away from the portal protein, and its ATPase domain forms a pentameric ring bound directly to the portal vertex (6). Based on these structural data, Sun et al. (6) proposed a model for DNA packaging that postulates the existence of two distinct conformations of L-terminase alternating during packaging to promote DNA translocation. In analogy to a model previously proposed for the ATPase Rad50 (20), the driving force mediating interconversion between relaxed and tensed states is generated by the intramolecular electrostatic contacts between ATPase and nuclease domains of L-terminase, which are connected by a flexible linker (21).
In this paper, we report the structure of bacteriophage P22 headful nuclease refined to a nominal resolution of 2.02 Å. We complement this structure with a biochemical analysis of its activity for the pac-containing gene 3. This work expands the repertoire of viral nuclease structures solved to date and sheds light on several properties governing the activity and regulation of a prototypical headful nuclease.
EXPERIMENTAL PROCEDURES
Molecular Cloning and Recombinant Proteins
The gene encoding L-terminase was PCR-amplified from P22 genomic DNA (22) and ligated between BamHI and HindIII restriction sites of expression vectors pMal-2cE (New England Biolabs) and pET-28a (Novagen). ΔC-L-terminase, which contains a stop codon at position 483 and does not express C-terminal residues 483–499, was generated by QuikChange site-directed mutagenesis, using the plasmid pMal-FL-L-terminase as a template. The full-length nuclease (FL-nuclease) domain of P22 L-terminase (residues 289–499) was PCR-amplified from gene 2 and ligated between BamHI and HindIII restriction sites of the expression vector pMal-2cE (New England Biolabs). Deletion constructs nuclease-ΔL1 (that lacks residues 374–382) and nuclease-ΔL1-α2 (that lacks residues 374–400) were generated by long PCR using the plasmid pMal-FL-nuclease as a template. All plasmids were sequenced to confirm the fidelity of the DNA sequence. His-tagged FL-L-terminase (cloned in pET-28a) was expressed in Escherichia coli strain BL21 (DE3/pLysS). Protein expression was induced by adding 0.5 mm isopropyl-β-d-1-thiogalactopyranoside for 16 h at 16 °C. Cell pellets were lysed by sonication in lysis buffer (20 mm Tris-HCl, pH 8.0, 300 mm NaCl, 0.1% (v/v) CHAPS, 5 mm β-mercaptoethanol, 1 mm phenylmethylsulfonyl fluoride. L-terminase was purified by Ni2+ affinity chromatography followed by size exclusion chromatography on a Superdex 200 column (GE Healthcare) equilibrated in 20 mm Tris-HCl, 150 mm NaCl, 5 mm β-mercaptoethanol, 5% (v/v) glycerol, pH 8.0. ΔC-L-terminase and FL-nuclease were expressed in E. coli strain BL21 (DE3/pLysS) fused to an N-terminal maltose-binding protein (MPB). Protein expression was induced by adding 0.1 mm isopropyl-β-d-1-thiogalactopyranoside for 16 h at 16 °C. Deletion constructs nuclease-ΔL1 and nuclease-ΔL1-α2 were expressed (also fused to MBP) in E. coli BL21-AI™ strain (Invitrogen). Protein expression was induced by adding 0.1% arabinose plus 0.1 mm isopropyl-β-d-1-thiogalactopyranoside for 2 h at 30 °C. All MBP-fused proteins were purified over amylose beads without the addition of detergent. After cleavage of MBP with Prescission Protease, the nuclease domain was separated from MBP by heparin chromatography, followed by gel filtration chromatography. Finally, expression, purification, and assembly of P22 nonameric S-terminase (23, 24) and dodecameric portal protein (25, 26) were carried out as described.
Structural Stability
Limited proteolysis was performed by adding chymotrypsin to FL-L-terminase in a ratio ∼1:200 (w/w), as described previously (27). Thermal denaturation was measured by monitoring ellipticity variations at 222 nm while increasing the temperature by 1 °C/min from 4 to 95 °C, as described previously (28). The experiment was carried out in protein simple buffer (3.2 mm Na2HPO4, 0.5 mm KH2PO4, 135 mm NaCl, pH 7.4), using FL-L-terminase at ∼5 μm final concentration. The data were acquired on a Jasco J-715 spectropolarimeter equipped with a temperature-controlling unit at the Kimmel Cancer Center resource facility for x-ray crystallography and molecular characterization.
Crystallization, Data Collection, and Structure Determination
To crystallize P22 nuclease domain, we set up crystallization droplets using ∼10 mg/ml of purified ΔC-L-terminase in the absence of protease inhibitors. In-drop cleavage of ΔC-L-terminase resulted in formation of a stable nuclease core (ΔC-nuclease), which gave microcrystals of ∼30 × 30 × 20 μm3 within 1–2 months, in the presence of 0.2 m magnesium sulfate and 20% w/v PEG 3,350, at pH 6.0. The crystals were cryoprotected by adding ∼27% ethylene glycol to the mother liquor. Several data sets were recorded at the National Synchrotron Light Source Beamline X6a on an ADSC Quantum-270 CCD detector. Diffraction data were integrated and scaled using the HKL-2000 suite (29) and analyzed using CCP4 programs (30) (see Table 1). Crystals belong to space group P21 with four nuclease domains per asymmetric unit (referred to as A, B, C, and D). Assessment of crystallographic data quality with Xtriage (31) revealed that crystals of P22 nuclease suffer from pseudomerohedral twinning, with twinning fraction between 0.32 and 0.47. The structure was determined by molecular replacement with PHASER (31) using the nuclease domain of bacteriophage SPP1 (Protein Data Bank code 2WBN) as a search model. Initial rigid body and positional refinement followed by manual rebuilding in Coot (32) resulted in a nonrandom Rfree of 42% (calculated using ∼2,000 reflections chosen in 20 thin resolution shells). This structure was then subjected to several cycles of manual rebuilding followed by positional and isotropic B-factor refinement in Phenix (31) using 4-fold noncrystallographic symmetry restraints and including a twinning operator (h, −k, and l). Peaks above 3σ in the Fo − Fc difference electron density map were modeled as water molecules. The structure of P22 L-terminase nuclease domain has been refined to Rwork/Rfree of 16.6/19.7%, using all data between 15 and 2.02 Å resolution. The final model contains residues 289–482 for chains A–D, two magnesium atoms (MgA and MgB), a sulfate ion, and 1,112 water molecules. Stereochemistry was checked by PROCHECK (33): the final model has excellent geometry (see Table 1), with 98.8% residues in the most favored regions of the Ramachandran plot, and a root mean square deviation of bond lengths and angles of 0.010 Å and 1.098°, respectively. All ribbon diagrams were generated using PyMol. Topological diagram was generated using PDBsum (34) and structural superimpositions were carried out in Coot (32).
TABLE 1.
Crystallographic data collection and refinement statistics
The numbers in parentheses refer to the statistics for the outer resolution shell (2.09–2.02 Å).
| Data collection statistics | |
| Space group | P21 |
| Unit cell dimensions (Å) | a = 60.92, b = 139.76, c = 61.02 |
| β-Angle (°) | β = 95.10 |
| Resolution range (Å) | 15–2.02 |
| B value from Wilson plot (Å2) | 27.0 |
| Observations (total/unique) | 258,985/60,520 |
| Completeness (%) | 90.8 (55.0) |
| Redundancy | 2.4 (1.9) |
| Rsym (%)a | 7.8 (61.0) |
| <I>/<σ(I)> | 18.4 (2.2) |
| Refinement statistics | |
| Number of reflections (30–2.02Å) | 60,494 |
| Rwork/Rfree (%)b | 16.6/19.7 (31.8/37.9) |
| Number of water molecules | 1,112 |
| B value (Å2) nuclease/Mg/solvent | 39/35/36 |
| Root mean square deviation from ideal bond length (Å) | 0.010 |
| Root mean square deviation from ideal bond angles (°) | 1.097 |
| Ramachandran plot (core/allowed/generously allowed/disallowed, %) | 86.8/12.1/1.2/0.0 |
a Rsym = Σi,h|I(i,h) − <I(h)>|/Σi,h |I(i,h)| where I(i,h) and <I(h)> are the ith and mean measurement of intensity of reflection h.
b The Rfree value was calculated using 2,000 reflections chosen in 20 thin resolution shells.
In Vitro Nuclease Assay
The coding region of P22 gene 3 (486 bp) was PCR-amplified from a P22 genomic DNA and purified using a QIAquick gel extraction kit (Qiagen). In vitro nuclease assay was carried out by adding 1 μm FL-L-terminase or FL-nuclease to 100 ng of gp3-DNA, in a 100-μl reaction volume containing 10 mm Tris-HCl, pH 7.5, 50 mm NaCl, 1 mm MgCl2. The samples were incubated at 37 °C for up to 60 min, and aliquots of 20 μl were taken after 10, 20, 30, and 60 min. Nuclease activity was stopped by adding 50 mm EDTA, and samples were analyzed by electrophoresis on a 1.2% (w/v) agarose gel run at 100 V for ∼45 min, followed by ethidium bromide staining. Additional experiments were carried out in the presence of 1 μm nonameric S-terminase (with and without 1 mm ATP) and dodecameric portal protein. In all cases, bands corresponding to the gp3-DNA were quantified using ImageJ (35), and the percentage of gp3-DNA remaining after digestion was plotted against the time of reaction using the program Origin.
RESULTS
Domain Organization
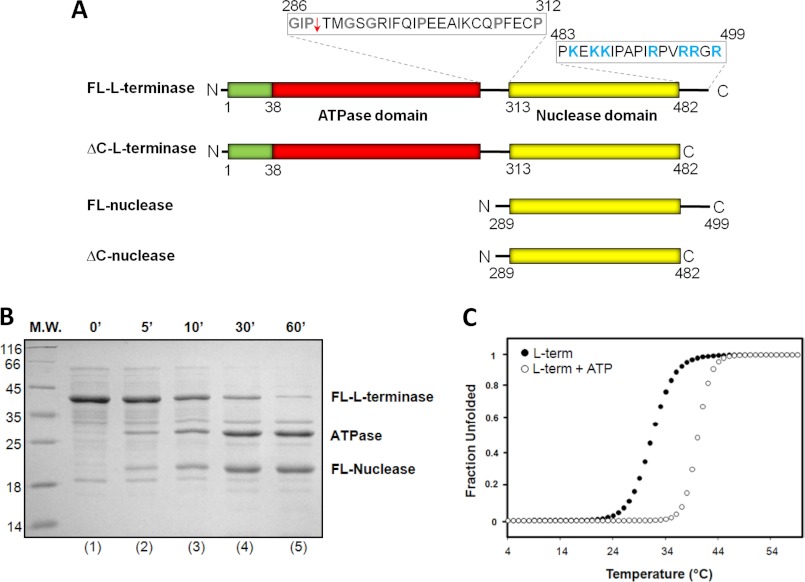
The L-terminase subunit of bacteriophage P22 consists of 499 residues and presents a bipartite organization. Analogous to T4 (6), the N terminus of L-terminase contains a Walker A motif, characteristic of an ATPase (36), followed at its C terminus by a nuclease domain (Fig. 1A). A linker rich in prolines and glycines connects the two domains (Fig. 1A). The first 38 residues of P22 L-terminase also contain a putative coiled coil motif, as predicted by the software Coils (37). In the test tube, purified P22 L-terminase quickly aggregates under physiological conditions, but it can be solubilized by addition of mild detergents (38). Removing the highly basic C-terminal tail spanning residues 483–499 (Fig. 1A) yielded a soluble and monodisperse protein (referred to as ΔC-L-terminase) that could be purified without detergents from bacterial extracts. To probe the flexibility of P22 L-terminase, we subjected the protein to limited proteolysis in the presence of chymotrypsin (Fig. 1B). This treatment readily yielded two stable domains that subsequent mass spectrometry analysis and N-terminal sequencing mapped to the N-terminal ATPase and C-terminal nuclease domains, respectively (Fig. 1A). Thus, in analogy to T4 L-terminase (21), the L-terminase subunit of bacteriophage P22 also contains two folded domains linked by a flexible loop that is readily cleaved by proteases in solution.
FIGURE 1.
Structural stability of P22 L-terminase subunit. A, schematic diagram of P22 L-terminase indicating the presence of an N-terminal ATPase domain connected to a C-terminal nuclease domain by a flexible linker. Amino acid sequences for the interdomain linker (residues 286–312) and C-terminal basic tail (residues 483–499) are also indicated, with flexible and basic residues colored in gray and blue, respectively. The site of proteolytic cleavage in the linker is indicated by a red arrow. B, time course of limited proteolysis of purified L-terminase in the presence of chymotrypsin. M.W., molecular mass. C, structural stability of FL-L-terminase against thermal denaturation monitored by measuring changes in the ellipticity intensity at 222 nm as a function of temperature. The fraction unfolded of FL-L-terminase was plotted against the temperature, revealing an apTm of 31.3 and 42 °C, in the presence and absence of 1 mm ATP, respectively. term, terminase.
To assess the structural stability of L-terminase, we studied its thermal unfolding using CD. Because purified L-terminase displays ∼60–70% helical content in solution (data not shown), denaturation curves were measured by monitoring variation in ellipticity at 222 nm as a function of the temperature between 4 and 95 °C. Under these conditions, L-terminase unfolded irreversibly as a single entity with an apparent melting temperature (apTm) of 31.3 °C (Fig. 1C). The shape of the native-to-denatured unfolding transition suggested a cooperative denaturation process, but the observed apTm was lower than that of most globular proteins (39) and indicative of intrinsic instability. To rule out the possibility that such a low apTm was caused by partial misfolding caused by detergent solubilization, we also repeated the thermal unfolding using ΔC-L-terminase purified under native conditions. This construct yielded identical apTm, confirming that neither the use of detergent nor the highly basic C-terminal tail (Fig. 1A) affects L-terminase overall stability. Instead, the addition of ∼1 mm ATP increased the apTm to ∼42 °C (Fig. 1C), suggesting that nucleotide binding to the ATPase domain is crucial to enhance the overall stability of the protein. This result agrees well with the observation made by Poteete and Botstein (40) that ATP, and not GTP, stabilizes L-terminase in partially purified extracts of P22 strain carrying amber mutations in genes 5 and 8.
Atomic Structure of P22 Headful Nuclease
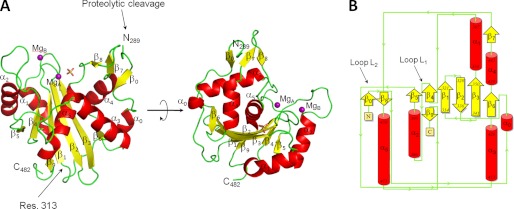
Consistent with its structural flexibility, both FL- and ΔC-L-terminase (Fig. 1A) failed to crystallize as full-length proteins. In a search for crystallization conditions, we found that omitting protease inhibitors during crystallization resulted in slow degradation of L-terminase in the crystallization drop, with subsequent formation of a stable nuclease core that crystallized within ∼1–2 months. Although these crystals were only 30 μm in the longest dimension, diffraction data to a nominal resolution of 2.02 Å were collected using a 50 × 50 μm2 sized x-ray beam. We determined the crystal structure of P22 L-terminase nuclease domain by molecular replacement using the nuclease domain of bacteriophage SPP1 as a search model (18). Although the sequence identity between these two nuclease domains is only 16%, this model was sufficient to yield an initial set of phases that were dramatically improved using the 4-fold noncrystallographic symmetry present in crystal. The polypeptide chain of P22 L-terminase nuclease was unambiguously traced between residues 289 and 482, and the final model was refined to Rwork/Rfree of 16.6/19.7%, at a nominal resolution of 2.02 Å (Table 1). The three-dimensional structure of P22 L-terminase nuclease domain is illustrated in Fig. 2A. The proteolytic cleavage that frees the nuclease domain from the N-terminal ATPase occurs between residues 288 and 289 of the Gly/Pro-rich linker connecting the two domains (indicated by an arrow in Fig. 1A). This linker (residues 289–313) is clearly visible in the electron density and contains a short β-strand (β0) and a 3/10 α-helix (α0) (Fig. 2). It wraps around the other surface of the nuclease core that starts at position 313 (Fig. 2A). The headful nuclease folds into a roughly globular α/β structure, with average dimensions 65 × 45 × 40 Å, that resembles the classical nuclease domain of RNase H1 (41). The tertiary structure of P22 nuclease is built by a central seven-stranded β-sheet sandwiched between two clusters of α-helices (Fig. 2B). Three of the central β-strands, β1–β3, are highly bent and curve around helix α5. Two additional sets of helices (α1-α2 and α3-α4) surround the central core making contacts with the solvent. Overall, P22 headful nuclease is very similar to the nuclease domain of SPP1 (18), T4 (6), and HHV-5 (19). Despite the low sequence identity (16% with SPP1, 14% with T4, 12% with the T4-like phage RB49, and 12% with HHV-5), the four viral nucleases are structurally superimposable. P22 and SPP1 have the greatest structural similarities, with root mean square deviation of ∼2.2 Å, whereas HHV-5 and T4 are more distantly related (root mean square deviation of 2.5 and 2.9 Å, respectively).
FIGURE 2.
Crystal structure of P22 l-terminase nuclease domain at 2.02 Å resolution. A, ribbon diagram of P22 headful nuclease colored by secondary structure elements with α-helices, β-strands, and loops in red, yellow, and green, respectively. The site of proteolytic cleavage (residue 289) and the beginning of the nuclease domain (residue 313) are indicated. B, topological diagram of P22 nuclease (color coded as in A) showing all secondary structure elements identified in the crystal structure. Loops L1 and L2 are also indicated by arrows.
Organization of the Active Site
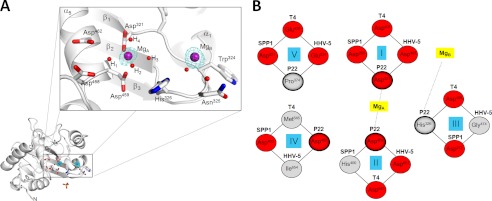
The active site of P22 headful nuclease lies within the discrete acidic pocket formed at the interface between strands β1-β3 and helix α5 (Fig. 2). Two magnesium ions (named MgA and MgB) were identified in the electron density as ∼10 σ peaks of positive density (Fig. 3A). The two ions have similar B-factors (∼35 Å2) but different chemistry of coordination to the nuclease active site. MgA is octahedrally coordinated to Asp459/Asp321 and four (clearly visible) water molecules (named H1–H4 in Fig. 3A). MgA lies in the active site of P22 nuclease at a position equivalent to the magnesium seen in the high resolution structure of T4/RB49 nuclease (6) or the manganese ion visualized in SPP1 (18) and HHV-5 nucleases (19). The second site, MgB, is surrounded by four less defined water molecules (of which only one was included in the final model), likely consistent with a tetrahedral coordination (42) (Fig. 3A). MgB sits nicely in a pocket formed by the main chain of Gly323-Trp324-Asn325-His326; water molecules coordinating MgB hydrogen bound main chain atoms as well as the side chains of Asn325 and His326 (Fig. 3A). In contrast to the first metal ion, MgB is missing in SPP1 (18) and T4/RB49 (6) and occupies a different position in HHV-5 nuclease (19). To account for such differences, we superimposed the four nuclease domains of P22, SPP1, T4/RB49, and HHV-5 and focused on five active site residues responsible for nuclease activity (indicated as sites I–V in Fig. 3B). This structural alignment revealed that Asp321 (site I in Fig. 3B) is the only residue universally conserved in all four nucleases, whereas the position and frequency of occurrence of other residues vary in these viruses. For instance, P22 nuclease has a proline at position 374 (site V in Fig. 3B) that is occupied by an acidic residue (Glu/Asp) in all the other nucleases. The lack of an acidic residue at this position of P22 is likely compensated by Asp462 (site IV in Fig. 3B), which is replaced by Met545/Ile654 in T4 and HHV-5, respectively. Likewise, the position of the catalytic pair His400/Asp272 (sites II and III in Fig. 3B) in SPP1 (18) is inverted in P22, which contains Asp459/His326. Thus, the superimposable RNase H1 fold of viral headful nucleases defines a nonidentical arrangement of metal ions and active site residues, likely reflecting an evolutionary adaptation for recognizing different viral DNA sequences.
FIGURE 3.
Architecture of P22 nuclease active site. A, left panel, ribbon diagram of P22 nuclease domain highlighting only active site residues and metal ions. In the right panel is a magnified view of the active site, which includes essential residues involved in catalysis, two magnesium ions (MgA and MgB) colored in purple, and several water molecules (small red spheres). The Fo − Fc electron density map (in cyan) overlaid to the magnesium sites is contoured at 7 σ above background and was computed at 2.02 Å resolution by omitting the magnesium atoms from the final model. B, comparison of five active site residues from the crystal structure of the L-terminase nuclease of P22, SPP1 (18), T4 (6), and HHV-5 (19). Acidic residues are colored in red, and nonacidic amino acids are in gray. The two metal ions are also shown in yellow.
The DNA Binding Surface
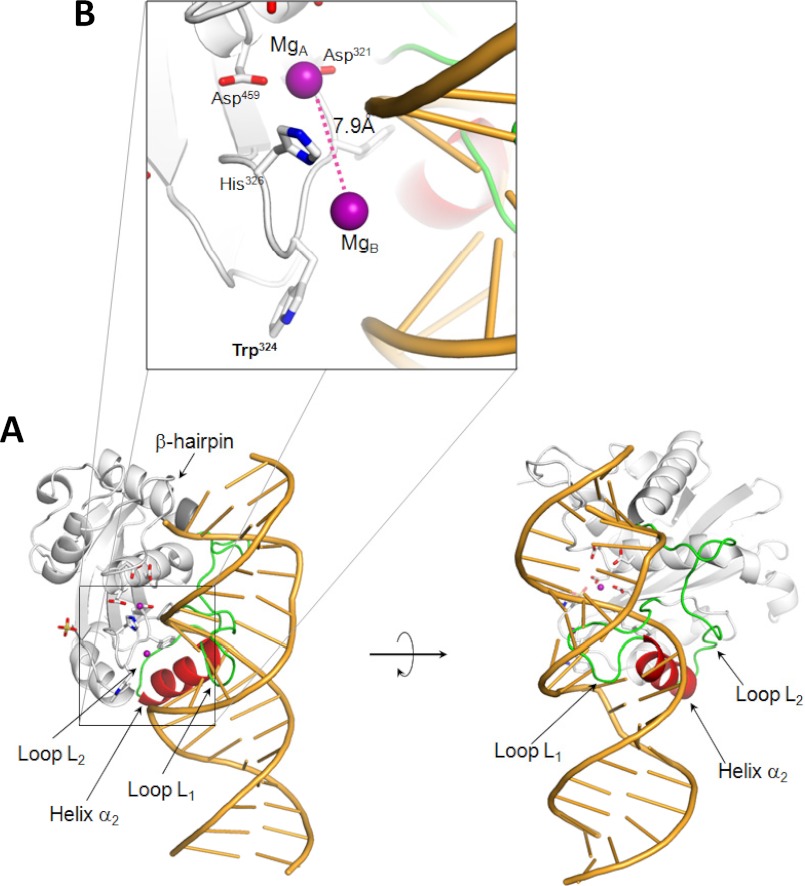
The surface of P22 nuclease is mainly negatively charged, consistent with an isoelectric point of 5.7. To identify residues involved in DNA binding, we superimposed the structure of P22 nuclease on that of RNase H1 determined in complex with a DNA:RNA hybrid (41) (Fig. 4A). This structural alignment allowed us to predict the position of DNA bound to P22 nuclease and indicated three important aspects of this interaction. First, several basic residues in L-terminase (e.g., Arg447, Lys452, Lys455, Arg465, Lys382, etc.) face the surface of DNA and are good candidates to mediate DNA-protein interaction. Some of these residues are conserved in other viral L-terminases; for instance, Arg447, Lys452, and Arg465 in P22 are replaced by Gln383, Arg393, and Lys529 in SPP1 and Arg447 and Lys452 are replaced by Lys529 and Arg406 in T4. This suggests that DNA binds in the vicinity of the active site by a combination of shape complementarity and charge-charge interactions. Second, the structural alignment locates the DNA backbone exactly in between the two active site magnesium ions (Fig. 4B), providing strong structural evidence in support of the two-metal ion-dependent catalysis model (42). However, the distance between these two Mg atoms in our model is 7.9 Å, greater than the ∼4 Å expected for two-metal ion catalysis (42). Third, a tryptophan residue in L-terminase nuclease domain, Trp324 protrudes on the surface of the turn connecting strands β1-β2 (Figs. 3A and 4B). This residue, located in close proximity of the DNA minor groove, may engage in contacts with nucleobases.
FIGURE 4.
Modeling the DNA binding surface of P22 headful nuclease. A, model of P22 nuclease domain (in gray) bound to a DNA-RNA hybrid (in orange). The nucleic acid model was obtained by superimposing the structure of the human RNase H1·DNA-RNA complex (Protein Data Bank code 2QKB) to that of P22 nuclease and removing the atoms for the RNase H1. In this model, helix α2 (in red) and the loop L1 (in green) adopt a conformation incompatible with DNA binding. B, magnified view of the active site highlighting the position and distance (∼7.9Å) of the two metal ions (purple sphere) symmetrically located in the vicinity of the DNA phosphate backbone.
The structural alignment also suggested that the positions of the Pro374–Gly385 loop L1 and helix α2 (residues 386–395) (Fig. 2B) are incompatible with DNA binding, because the loop-helix (L1-α2) motif would sterically obstruct DNA binding (Fig. 4A). Interestingly, loop L1, which links helix α2 to strand β4 (Fig. 2B) is dramatically longer in P22 than in other viral nucleases (12 versus 2–3 residues in SPP1/T4/HHV-5). Furthermore, this loop is poorly visible in our structure (with average B-factor of ∼90 Å2 versus ∼27 Å2 for the rest of the protein) and adopts slightly different conformations in all analyzed crystal forms of P22 L-terminase (data not shown). On the opposite side, helix α2 is connected to helix α3 by a long, mainly random coil linker, L2, that spans residues Gly396–Val412 (Figs. 2B and 4A). Given the loose association of helix α2 with the rest of the nuclease core, it is possible that a conformational change in this region allows for efficient binding to DNA and enhances nuclease activity.
The Nuclease Domain Is Active Only in the Context of FL-L-terminase
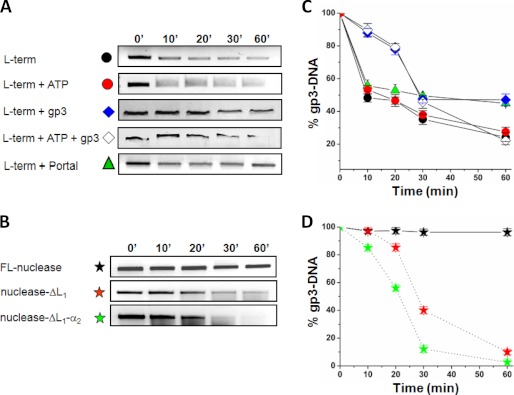
We used an in vitro nuclease assay to characterize the endonucleolytic activity of P22 L-terminase. As substrate for L-terminase, we used P22 gene 3 (gp3-DNA) that contains a pac site between nucleotides 265–286 (11). This is the site where most L-terminase cuts occur in vivo, usually concentrated in 20-bp intervals (12). The nuclease reaction was carried out by incubating homogeneously purified FL-L-terminase or FL-nuclease with the gp3-DNA for up to 60 min. At time intervals, the samples were removed, the nuclease was inactivated, and samples were subjected to separation by electrophoresis on agarose gels (Fig. 5, A and B). Quantification of gp3-DNA bands suggested that FL-L-terminase is catalytically active and degrades (both in the presence and absence of ATP) ∼70–75% of gp3-DNA over the course of 60 min (Fig. 5C). Instead, the isolated nuclease domain incubated with gp3-DNA under identical experimental conditions was essentially inactive (Fig. 5D). This result is consistent with previous reports that the nuclease domain of SPP1 L-terminase is not active when expressed in isolation (18), and deletion of the ATPase domain of T4 L-terminase reduces nuclease activity by ∼13-fold (41). To test the putative inhibitory role of loop L1 and helix α2 (Fig. 4A) on nuclease activity, we generated two deletion constructs of FL-nuclease that lack either loop L1 (nuclease-ΔL1) or loop L1 plus helix α2 (nuclease-ΔL1-α2). Strikingly, both mutants efficiently digested gp3-DNA over 60 min of reaction (Fig. 5B), displaying comparable endonucleolytic activity as FL-L-terminase (Fig. 5, C and D). Interestingly, although nuclease-ΔL1-α2 had the highest activity (digesting >95% of substrate in 60 min), the construct lacking only loop L1, (nuclease-ΔL1) also displayed nearly fully restored nuclease activity, digesting ∼90% of gp3-DNA within 60 min of reaction (Fig. 5D). Thus, the loop L1 is the main determinant for intramolecular autoinhibition of P22 headful nuclease.
FIGURE 5.
In vitro nuclease assay. A, time course of nuclease digestion obtained by incubating gp3-DNA with 1 μm of FL-L-terminase, FL-L-terminase + ATP, FL-L-terminase + nonameric S-terminase (referred to as gp3) with and without ATP and FL-L-terminase + dodecameric portal protein. B, time course of nuclease digestion obtained by incubating gp3-DNA with 1 μm of purified FL-nuclease, nuclease-ΔL1, and nuclease-ΔL1-α2. In both panels, samples were separated on a 1.2% agarose gel followed by ethidium bromide staining. The abnormal migration of gp3-DNA in the presence of gp3 in A is caused by S-terminase tight binding to DNA and was also observed in control experiments where L-terminase was omitted (24). C and D, quantification of agarose bands in A and B. The percentage of gp3-DNA left on the gel is plotted as a function of time. The error bars are based on three independent repeats. term, terminase.
Using the same in vitro assay, we also tested the effect of the nonameric S-terminase (23, 43) and dodecameric portal protein (26) on the nuclease activity of FL-L-terminase (Fig. 5A). In the presence and absence of ATP, a stoichiometric quantity of S-terminase significantly inhibited nuclease activity within the first 20 min of reaction, resulting in a 40% higher recovery of gp3-DNA (Fig. 5, A and C). However, over 30–60 min in the presence of ATP this inhibitory effect was completely lost, whereas it was partially retained in the absence of ATP, where twice as much gp3-DNA remained on gel after 60 min (∼50%), as compared with its counterpart with ATP (Fig. 5C). Finally, consistent with the observation that series initiation cleavages occur in the absence of procapsids in P22 extracts (10), the addition of a stoichiometric quantity of P22 dodecameric portal protein (which is known to bind to L-terminase (44)), did not dramatically affect nuclease activity (Fig. 5, A and C). Thus, the headful nuclease of bacteriophage P22 is inactive when in isolation and active in the context of the FL-L-terminase (or when the autoinhibitory L1-α2 motif is removed), and its activity is partially modulated by the S-terminase subunit and ATP.
DISCUSSION
Two endonucleolytic cuts are required to sustain efficient packaging of concatameric genomes in viruses that package DNA by a headful mechanism. A first initiation cut allows insertion of a linear end of the genome into the procapsid, and a final headful cut terminates packaging into a head, thereby releasing the remainder of the genome for new rounds of packaging. Both cuts are catalyzed by the nuclease domain of L-terminase, which represents the catalytic core of a viral genome-packaging motor (1–3).
Conservation and Specialization of Viral Headful Nucleases
The 2.02 Å crystal structure of P22 L-terminase nuclease domain reveals the detailed architecture of a headful nuclease, related in fold and catalytic mechanism to the RNase H1 family of retroviral integrase superfamily (45). Despite minimal sequence identity (below 16%), P22 nuclease domain is remarkably similar to those previously observed in SPP1 (18), T4 (6), and HHV-5 (19). The identification of two magnesium ions in the active site strengthens the idea that P22 headful packaging nuclease uses a two-metal ion-dependent catalysis to hydrolyze the phosphate ester backbone of DNA (42). By superimposing the active sites of the four viral nucleases solved crystallographically, we determined that Asp321 is the only invariant active site residue. The positions of other active site residues and the exact number and location of metal ions vary in different nucleases (Fig. 3B), possibly underscoring a different degree of DNA sequence specificity. For instance, in pac phages like P22 (10) and SPP1 (46, 47), the nuclease first cleaves within a pac site and then cleaves a second time after ∼102–110% genome length is packaged. Because the first cut always occurs at a pac site, the headful nuclease is likely to have (weak) sequence specificity in vivo. In support of this hypothesis, Casjens et al. (12) observed that P22 headful nuclease cleavages are not random but are concentrated at 20-bp intervals across ∼120 bp of the gp3-DNA. Intriguingly, the structure of P22 nuclease described in this paper reveals a tryptophan residue, Trp324 (Tyr269 in SPP1) in close proximity to the putative DNA binding surface (Fig. 4B), at a position where the tryptophan indole could make specific contacts with DNA nucleobases. Because a tryptophyl residue can intercalate specifically with DNA (48, 49), Trp324 is a potential determinant for sequence specificity. In contrast, in other phages like T4 (that has Glu404 in lieu of Trp324), although a pac site has been reported (50), the first cut does not strictly occur in the proximity of a unique pac sequence (51). Regardless of differences in the position of the first cut site, all viral nucleases make a second headful cut after one equivalent of genome length has been encapsulated. Thus, we propose that only the first initiation cut in DNA packaging is biased by the DNA sequence and reflects the degree of sequence specificity intrinsic to different L-terminases. The second headful cut, instead, is mechanically determined exclusively by the volume of the procapsid and by how much DNA can be efficiently packaged inside.
Regulation of Nuclease Activity by Interdomain Cross-talk
Several lines of evidence presented in this paper support the idea that P22 L-terminase is regulated by interdomain cross-talk between its N-terminal ATPase and C-terminal nuclease domains. First, the two domains are linked together by a protease-sensitive linker (Fig. 1, A and B) but do not function as separate entities (e.g. “beads on a string”). The addition of ATP results in an overall stabilization of L-terminase by ∼11 °C (Fig. 1C) and renders a protein that is unstable at physiological temperature (apTm = ∼31 °C) into a stable enzyme (apTm = ∼42 °C). The energy of stabilization is likely provided by intramolecular contacts between the two domains within the tertiary structure of L-terminase, as proposed by Sun et al. (6) for T4 packaging motor. Second, the nuclease domain of P22 L-terminase was inactive when activity was measured using the isolated nuclease domain. It is unlikely that protein misfolding explains the lack of enzymatic activity, because this domain could be readily crystallized, and most of its structure is well folded and visible in the electron density (Fig. 2A). Instead, modeling studies suggested the isolated nuclease adopts an inactive conformation, with the DNA binding surface autoinhibited by the flexible loop L1 and helix α2 (Fig. 4A). In this autoinhibited state, the two active site magnesium ions are ∼7.9 Å far apart (Fig. 4B), approximately twice the distance required for two-metal ion-dependent catalysis (∼4 Å) (42). The hypothesis of an inactive conformation of the headful nuclease was experimentally tested and successfully validated by deleting the L1-α2 motif or just the loop L1, which yielded two gain of function mutants displaying wild type endonucleolytic activity. Based on these results, we hypothesize that binding of both ATPase and nuclease domains to a pac sequence induces a conformational change in the tertiary structure of FL-L-terminase that brings the two metal ions closer to each other (∼4 Å) in a catalytically active conformation. This conformational change would move helix α2 away from the crystallographic conformation by swinging it counter-clockwise onto the surface of the nuclease core, to which it is flexibly connected by loops L1 and L2 (Fig. 4A). A similar regulation by interdomain cross-talk was previously hypothesized for T4 (52) and SPP1 (18). However, in both of these cases, the authors identified a β-hairpin in the headful nuclease (β7-β8 in P22 nuclease; Fig. 4A), thought to mediate interdomain cross-talk. In P22, this β-hairpin does not appear to be a major determinant for autoinhibition, because unlike its 14 residue length in SPP1, in P22 the β-hairpin is only eight residues long and is predicted to clash minimally with DNA (indicated by an arrow in Fig. 4A). Finally, our results shed light on the role of P22 nonameric S-terminase in inhibiting nuclease activity. At a time scale comparable with that of DNA packaging (1–20 min), S-terminase inhibited nuclease activity, both in the presence and absence of ATP. However, at longer times of incubation, the inhibition was lost in the presence of ATP but persisted in the absence of ATP (Fig. 5C). The molecular basis for this time-dependent inhibition of nuclease activity is unclear. In P22, as in other viruses (3), S-terminase is also involved in stimulating the ATPase activity of L-terminase (24). The inhibitory effect of S-terminase on nuclease activity may reflect conformational changes in the ATPase active site induced by S-terminase that disrupt interdomain cross-talk with the nuclease domain, thereby preventing its activation. In summary, the structure of P22 headful nuclease that we present in this paper provides new clues to dissect the complex chemistry of P22 genome-packaging motor. Further structural analysis of the L-terminase in complex with the S-terminase will be instrumental to fully understand the molecular nature of the intramolecular cross-talk between ATPase and nuclease domains probed biochemically and predicted by our structure.
Acknowledgments
We are grateful to Vivian Stojanoff and staff at National Synchrotron Light Source Beamline X6A for beamline time and assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants 1R56 AI095974-01 and 1R01GM100888-01A1 (to G. C.).
The atomic coordinates and structure factors (code 4DKW) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- L-terminase
- large terminase
- S-terminase
- small terminase
- FL
- full-length
- HHV-5
- human herpesvirus 5
- MBP
- maltose-binding protein.
REFERENCES
- 1. Casjens S. R. (2011) The DNA packaging nanomotor of tailed bacteriophages. Nat. Rev. Microbiol. 9, 647–657 [DOI] [PubMed] [Google Scholar]
- 2. Sun S., Rao V. B., Rossmann M. G. (2010) Genome packaging in viruses. Curr. Opin Struct. Biol. 20, 114–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rao V. B., Feiss M. (2008) The bacteriophage DNA packaging motor. Annu. Rev. Genet. 42, 647–681 [DOI] [PubMed] [Google Scholar]
- 4. Fuller D. N., Raymer D. M., Kottadiel V. I., Rao V. B., Smith D. E. (2007) Single phage T4 DNA packaging motors exhibit large force generation, high velocity, and dynamic variability. Proc. Natl. Acad. Sci. U.S.A. 104, 16868–16873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sun S., Kondabagil K., Gentz P. M., Rossmann M. G., Rao V. B. (2007) The structure of the ATPase that powers DNA packaging into bacteriophage T4 procapsids. Mol. Cell 25, 943–949 [DOI] [PubMed] [Google Scholar]
- 6. Sun S., Kondabagil K., Draper B., Alam T. I., Bowman V. D., Zhang Z., Hegde S., Fokine A., Rossmann M. G., Rao V. B. (2008) The structure of the phage T4 DNA packaging motor suggests a mechanism dependent on electrostatic forces. Cell 135, 1251–1262 [DOI] [PubMed] [Google Scholar]
- 7. Ebel-Tsipis J., Botstein D., Fox M. S. (1972) Generalized transduction by phage P22 in Salmonella typhimurium. I. Molecular origin of transducing DNA. J. Mol. Biol. 71, 433–448 [DOI] [PubMed] [Google Scholar]
- 8. Teschke C. M., Parent K. N. (2010) “Let the phage do the work.” Using the phage P22 coat protein structures as a framework to understand its folding and assembly mutants. Virology 401, 119–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Botstein D., Levine M. (1968) Intermediates in the synthesis of phage P22 DNA. Cold Spring Harb. Symp. Quant. Biol. 33, 659–667 [DOI] [PubMed] [Google Scholar]
- 10. Casjens S., Weigele P. (2005) in Viral Genome Packaging Machines: Genetics, Structure and Mechanism (Catalano C., ed.) pp. 80–88, Landes Publishing, Georgetown, TX [Google Scholar]
- 11. Wu H., Sampson L., Parr R., Casjens S. (2002) The DNA site utilized by bacteriophage P22 for initiation of DNA packaging. Mol. Microbiol. 45, 1631–1646 [DOI] [PubMed] [Google Scholar]
- 12. Casjens S., Sampson L., Randall S., Eppler K., Wu H., Petri J. B., Schmieger H. (1992) Molecular genetic analysis of bacteriophage P22 gene 3 product, a protein involved in the initiation of headful DNA packaging. J. Mol. Biol. 227, 1086–1099 [DOI] [PubMed] [Google Scholar]
- 13. Casjens S., Huang W. M., Hayden M., Parr R. (1987) Initiation of bacteriophage P22 DNA packaging series. Analysis of a mutant that alters the DNA target specificity of the packaging apparatus. J. Mol. Biol. 194, 411–422 [DOI] [PubMed] [Google Scholar]
- 14. Laski F., Jackson E. N. (1982) Maturation cleavage of bacteriophage P22 DNA in the absence of DNA packaging. J. Mol. Biol. 154, 565–579 [DOI] [PubMed] [Google Scholar]
- 15. Casjens S., King J. (1974) P22 morphogenesis. I. Catalytic scaffolding protein in capsid assembly. J. Supramol. Struct. 2, 202–224 [DOI] [PubMed] [Google Scholar]
- 16. Raj A. S., Raj A. Y., Schmieger H. (1974) Phage genes involved in the formation generalized transducing particles in Salmonella. Phage P22. Mol. Gen. Genet. 135, 175–184 [DOI] [PubMed] [Google Scholar]
- 17. Botstein D., Waddell C. H., King J. (1973) Mechanism of head assembly and DNA encapsulation in Salmonella phage p22. I. Genes, proteins, structures and DNA maturation. J. Mol. Biol. 80, 669–695 [DOI] [PubMed] [Google Scholar]
- 18. Smits C., Chechik M., Kovalevskiy O. V., Shevtsov M. B., Foster A. W., Alonso J. C., Antson A. A. (2009) Structural basis for the nuclease activity of a bacteriophage large terminase. EMBO Rep. 10, 592–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nadal M., Mas P. J., Blanco A. G., Arnan C., Solà M., Hart D. J., Coll M. (2010) Structure and inhibition of herpesvirus DNA packaging terminase nuclease domain. Proc. Natl. Acad. Sci. U.S.A. 107, 16078–16083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hopfner K. P., Karcher A., Shin D. S., Craig L., Arthur L. M., Carney J. P., Tainer J. A. (2000) Structural biology of Rad50 ATPase. ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell 101, 789–800 [DOI] [PubMed] [Google Scholar]
- 21. Kanamaru S., Kondabagil K., Rossmann M. G., Rao V. B. (2004) The functional domains of bacteriophage t4 terminase. J. Biol. Chem. 279, 40795–40801 [DOI] [PubMed] [Google Scholar]
- 22. Pedulla M. L., Ford M. E., Karthikeyan T., Houtz J. M., Hendrix R. W., Hatfull G. F., Poteete A. R., Gilcrease E. B., Winn-Stapley D. A., Casjens S. R. (2003) Corrected sequence of the bacteriophage p22 genome. J. Bacteriol. 185, 1475–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Roy A., Bhardwaj A., Cingolani G. (2011) Crystallization of the nonameric small terminase subunit of bacteriophage P22. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 67, 104–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Roy A., Bhardwaj A., Datta P., Lander G. C., Cingolani G. (2011) Small terminase couples viral DNA binding to genome-packaging ATPase activity. Structure http://dx.doi.org/10.1016/j.str.2012.05.01 [DOI] [PMC free article] [PubMed]
- 25. Lorenzen K., Olia A. S., Uetrecht C., Cingolani G., Heck A. J. (2008) Determination of stoichiometry and conformational changes in the first step of the P22 tail assembly. J. Mol. Biol. 379, 385–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Olia A. S., Prevelige P. E., Jr., Johnson J. E., Cingolani G. (2011) Three-dimensional structure of a viral genome-delivery portal vertex. Nat. Struct. Mol. Biol 18, 597–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cingolani G., Lashuel H. A., Gerace L., Müller C. W. (2000) Nuclear import factors importin α and importin β undergo mutually induced conformational changes upon association. FEBS Lett. 484, 291–298 [DOI] [PubMed] [Google Scholar]
- 28. Bhardwaj A., Olia A. S., Walker-Kopp N., Cingolani G. (2007) Domain organization and polarity of tail needle GP26 in the portal vertex structure of bacteriophage P22. J. Mol. Biol. 371, 374–387 [DOI] [PubMed] [Google Scholar]
- 29. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 30. Collaborative Computational Project, Number 4 (1994) The CCP4 suite. Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 31. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) PHENIX. Building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 32. Emsley P., Cowtan K. (2004) Coot. Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 33. Bullough P. A., Hughson F. M., Skehel J. J., Wiley D. C. (1994) Structure of influenza haemagglutinin at the pH of membrane fusion. Nature 371, 37–43 [DOI] [PubMed] [Google Scholar]
- 34. Laskowski R. A. (2009) PDBsum new things. Nucleic Acids Res. 37, D355–D359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Abramoff M. D., Magelhaes P. J., Ram S. J. (2004) Image processing with ImageJ. Biophotonics Int. 11, 36–42 [Google Scholar]
- 36. Mitchell M. S., Rao V. B. (2004) Novel and deviant Walker A ATP-binding motifs in bacteriophage large terminase-DNA packaging proteins. Virology 321, 217–221 [DOI] [PubMed] [Google Scholar]
- 37. Lupas A., Van Dyke M., Stock J. (1991) Predicting coiled coils from protein sequences. Science 252, 1162–1164 [DOI] [PubMed] [Google Scholar]
- 38. Nemecek D., Gilcrease E. B., Kang S., Prevelige P. E., Jr., Casjens S., Thomas G. J., Jr. (2007) Subunit conformations and assembly states of a DNA-translocating motor. The terminase of bacteriophage P22. J. Mol. Biol. 374, 817–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pace C. N., Scholtz J. M. (1997) in Protein Structure: A Practical Approach (Creighton T. E., ed) pp. 299–321, IRL Press, Oxford, U.K [Google Scholar]
- 40. Poteete A. R., Botstein D. (1979) Purification and properties of proteins essential to DNA encapsulation by phage P22. Virology 95, 565–573 [DOI] [PubMed] [Google Scholar]
- 41. Nowotny M., Gaidamakov S. A., Ghirlando R., Cerritelli S. M., Crouch R. J., Yang W. (2007) Structure of human RNase H1 complexed with an RNA/DNA hybrid. Insight into HIV reverse transcription. Mol. Cell 28, 264–276 [DOI] [PubMed] [Google Scholar]
- 42. Yang W. (2011) Nucleases. Diversity of structure, function and mechanism. Q. Rev. Biophys. 44, 1–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nemecek D., Lander G. C., Johnson J. E., Casjens S. R., Thomas G. J., Jr. (2008) Assembly architecture and DNA binding of the bacteriophage P22 terminase small subunit. J. Mol. Biol. 383, 494–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Moore S. D., Prevelige P. E., Jr. (2002) Bacteriophage p22 portal vertex formation in vivo. J. Mol. Biol. 315, 975–994 [DOI] [PubMed] [Google Scholar]
- 45. Nowotny M. (2009) Retroviral integrase superfamily. The structural perspective. EMBO Rep. 10, 144–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bravo A., Alonso J. C., Trautner T. A. (1990) Functional analysis of the Bacillus subtilis bacteriophage SPP1 pac site. Nucleic Acids Res. 18, 2881–2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Camacho A. G., Gual A., Lurz R., Tavares P., Alonso J. C. (2003) Bacillus subtilis bacteriophage SPP1 DNA packaging motor requires terminase and portal proteins. J. Biol. Chem. 278, 23251–23259 [DOI] [PubMed] [Google Scholar]
- 48. Rajeswari M. R., Montenay-Garestier T., Hélène C. (1987) Does tryptophan intercalate in DNA? A comparative study of peptide binding to alternating and nonalternating A.T sequences. Biochemistry 26, 6825–6831 [DOI] [PubMed] [Google Scholar]
- 49. Jain A. A., Rajeswari M. R. (2003) Binding studies on peptide-oligonucleotide complex. Intercalation of tryptophan in GC-rich region of c-myc gene. Biochim. Biophys. Acta 1622, 73–81 [DOI] [PubMed] [Google Scholar]
- 50. Lin H., Black L. W. (1998) DNA requirements in vivo for phage T4 packaging. Virology 242, 118–127 [DOI] [PubMed] [Google Scholar]
- 51. Rao V. B., Black L. W. (2005) in Viral Genome Packaging Machines: Genetics, Structure and Mechanism (Catalano C., ed) pp. 40–58, Landes Publishing, Georgetown, TX [Google Scholar]
- 52. Ghosh-Kumar M., Alam T. I., Draper B., Stack J. D., Rao V. B. (2011) Regulation by interdomain communication of a headful packaging nuclease from bacteriophage T4. Nucleic Acids Res. 39, 2742–2755 [DOI] [PMC free article] [PubMed] [Google Scholar]