Background: The structure of a putative protease from Bacteroides thetaiotaomicron has a unique binding site for a flavin.
Results: The protein tightly binds to lumichrome, riboflavin, FMN, FAD, and flavin derivatives.
Conclusion: The putative protease is a scavenger of flavin and may function as a storage protein in gut bacteria.
Significance: The wealth of information available for cofactors is invaluable to assess protein structures.
Keywords: Flavin, Flavoproteins, Isothermal Titration Calorimetry, Oxidation Reduction, UV Spectroscopy
Abstract
The structure of a putative protease from Bacteroides thetaiotaomicron features an unprecedented binding site for flavin mononucleotide. The flavin isoalloxazine ring is sandwiched between two tryptophan residues in the interface of the dimeric protein. We characterized the recombinant protein with regard to its affinity for naturally occurring flavin derivatives and several chemically modified flavin analogs. Dissociation constants were determined by isothermal titration calorimetry. The protein has high affinity to naturally occurring flavin derivatives, such as riboflavin, FMN, and FAD, as well as lumichrome, a photodegradation product of flavins. Similarly, chemically modified flavin analogs showed high affinity to the protein in the nanomolar range. Replacement of the tryptophan by phenylalanine gave rise to much weaker binding, whereas in the tryptophan to alanine variant, flavin binding was abolished. We propose that the protein is an unspecific scavenger of flavin compounds and may serve as a storage protein in vivo.
Introduction
Structural genomics efforts have led to an increasing number of structures of proteins that are not characterized on a functional biochemical level. In the case of homologous structures, biochemical function may be inferred to some extent and thus provide an initial lead for further functional investigations. In a recent survey of flavin-dependent proteins, we identified a putative protease from Bacteroides thetaiotaomicron, a microbe inhabiting the human gut (1, 2) that presumably binds FMN (3). The structure of the protein was solved to 2 Å resolution by the Midwest Centre for Structural Genomics facility (Protein Data Bank entry 3cne; Fig. 1A) and features a mononuclear metal-binding site, presumably occupied by zinc, although other metals such as cobalt may be the native metal in the protein. In addition, a flavin was found in the protein, which is sandwiched by two tryptophan residues provided by each of the two protomers in the dimeric protein (Fig. 1B). Based on its overall topology, the protein was classified as a member of the DJ-1 superfamily (Pfam DJ-1_PfpI), a heterogenous family of hydrolases and chaperones with many uncharacterized and poorly understood members (4–7). The hydrolase activity found in some members appears to be associated with a catalytic triad formed by a conserved cysteine, histidine, and aspartic acid (5, 8). Other members of the family, such as Hsp31 and YajL from Escherichia coli, were reported to display activity as a holding and covalent chaperone, respectively (9, 10).
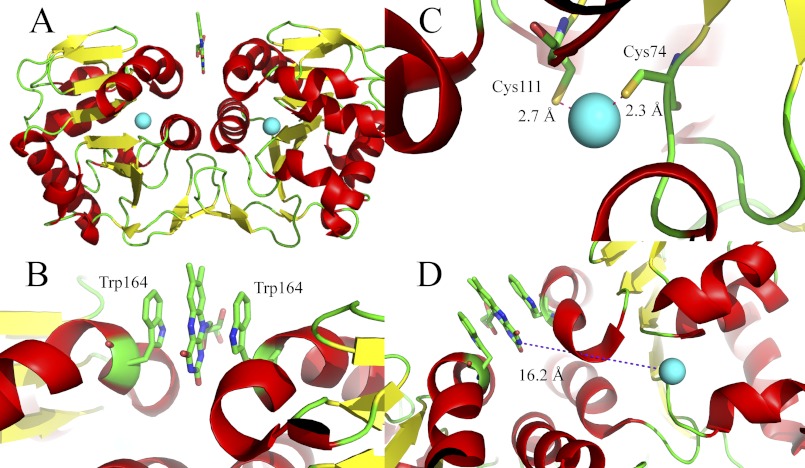
FIGURE 1.
Structural representation of the putative protease from B. thetaiotaomicron (Protein Data Bank entry 3cne). A, overall topology of the dimeric protein in a cartoon representation with α-helices in red and β-strands in yellow. Zinc ions are shown as cyan spheres. The flavin is shown as a stick model. B, flavin-binding site in the dimer interface of the putative protease with the two tryptophan residues (one from each protomer) sandwiching the isoalloxazine ring shown in a stick model. C, zinc ion coordination sphere (one in each of the two protomers). Zinc is shown as a sphere with the two coordinating cysteine residues (Cys74 and Cys111) and two coordinating water molecules in 3.6 and 3.1 Å distance from the zinc ion. D, spatial relationship between the zinc ion and the flavin. The closest atom to the zinc ion is N-3 of the isoalloxazine ring with a distance of 16.2 Å.
The close structural relationship suggests that the zinc ion might be the catalytic center of a hydrolytic activity, although none of the other family members appear to require metal ions for catalytic activity. Furthermore, complexation of the zinc is atypical for zinc-dependent hydrolases with two cysteine-derived thiol groups (Fig. 1C and Ref. 11). In addition, the zinc is liganded by water molecules; however, this interaction varies in the four chains of the asymmetric unit.
In contrast to zinc, the flavin cofactor occurs predominantly in oxidoreductases (∼90% of flavin-dependent enzymes) and not hydrolases. Although the flavin cofactor is also present in other enzyme classes, like transferases, lyases, and isomerases, it has never been reported in a hydrolase (3). The large distance of ≈16 Å between the edge of the flavin cofactor and the zinc argues against a direct cooperation between these two cofactors (Fig. 1D). This intriguing combination of unusual cofactors sparked our interest in the protein. Here, we report the biochemical characterization of the flavin-binding site of the protein and demonstrate that the recombinant protein is capable of binding not only naturally occurring flavin derivatives, such as lumichrome, riboflavin, FMN, and FAD (supplemental Fig. S1A) but also a variety of chemically modified “artificial” flavin analogs, such as iso-riboflavin (6.7-dimethyl-8-nor-riboflavin) and iso-FMN as well as 8-amino- and 1- and 5-deaza-riboflavin (12–14) (supplemental Fig. S1B). Thus it appears that this putative protease from B. thetaiotaomicron (ppBat)2 unspecifically binds tricyclic aromatic systems such as the alloxazine and isoalloxazine ring systems. Furthermore, we showed that replacement of Trp164 (Fig. 1C) by alanine completely abolishes the ability to bind the flavin ring system, whereas the W164F variant retained some affinity. Sequence alignment of ppBat with other related putative proteases from the genus Bacteroides and related bacteria revealed that Trp164 is conserved in the majority of homologs. Therefore, we propose that ppBat may serve as a flavin storage protein in gut bacteria.
EXPERIMENTAL PROCEDURES
Reagents
All of the chemicals were of the highest grade commercially available from Sigma-Aldrich, Fluka (Buchs, Switzerland), or Merck. Ni-NTA-agarose was from GE Healthcare, and Sephadex resin was from Pharmacia Biotech. Flavin analogs were a generous gift from Prof. Sandro Ghisla and the late Prof. Peter Hemmerich (University of Konstanz, Konstanz, Germany).
Expression of Recombinant Protein in E. coli Host Cells
The gene gat1 of B. thetaiotaomicron was cloned into a derivative of pET-28 for isopropyl β-d-thiogalactopyranoside-inducible expression in E. coli BL21 (DE3) host cells (kanamycin resistance). The N terminus of the expressed protein possesses a hexahistidine tag to facilitate purification. Precipitated cells were resuspended in lysis buffer (50 mm NaH2PO4, 300 mm NaCl, 10 mm imidazole, pH 8.0), using 2 ml of buffer/g of wet cells. At this stage lumichrome, riboflavin, FMN, FAD, or modified flavin analogs were added in excess to saturate the protein with the respective ligand. To obtain the apoprotein (protein preparation “as isolated”), no ligands were added. Resuspended cells were disrupted by 0.5-s sonication pulses for 10 min while cooling on ice. The cell debris was removed by centrifugation at 18,000 × g for 30 min at 4 °C. The hexahistidine-tagged protein was purified by Ni-NTA affinity chromatography, loading the supernatant onto a Ni-NTA HisTrapTMHP column (GE Healthcare), previously equilibrated with lysis buffer. After loading of the filtrated lysate, the column was washed with 10 column volumes of wash buffer (50 mm NaH2PO4, 300 mm NaCl, 20 mm imidazole, pH 8.0), and bound protein was recovered with elution buffer (50 mm NaH2PO4, 300 mm NaCl, 150 mm imidazole, pH 8.0). The purity of the eluted fractions was determined by SDS/PAGE. Fractions containing ppBat were pooled, dialyzed against 20 mm Tris/HCl buffer containing 100 mm NaCl (pH 8.0) overnight, and concentrated using Centripreps (Millipore). The final concentration was determined at 280 nm using an ϵ280 of 13,075 m−1 cm−1. The protein was flash-frozen and stored at −20 °C.
Site-directed Mutagenesis
The W164F and W164A variants were prepared using a QuikChange XL-site directed mutagenesis kit (Stratagene) with a slight modification of the protocol. A two-step PCR was carried out, where separate reactions for forward and reverse primer were performed for four cycles prior to another 18 cycles where an equal amount of the forward and reverse reaction were mixed. Following primers were used for the insertion of the mutations: 5′-CTGCACAAGATGAGAATACGATCTTTACAATGTTGCCGAAAGTCATAG-3′and 5′-CTATGACTTTCGGCAACATTGTAAAGATCGTATTCTCATCTTGTGCAG-3′ for W164F and 5′-CTGCACAAGATGAGAATACGATCGCGACAATGTTGCCGAAAGTCATAG-3′ and 5′-CTATGACTTTCGGCAACATTGTCGCGATCGTATTCTCATCTTGTGCAG-3′ for W164A, respectively. After verification of positive transformants by sequencing, expression and purification was achieved as described for the wild-type protein.
UV-visible Absorbance and Fluorescence Spectroscopy
UV-visible absorbance spectra were recorded with a Specord 210 spectrophotometer (Analytik Jena, Jena, Germany). Fluorescence emission spectra were recorded with a Shimadzu RF301 PC spectrofluorophotometer. Difference titrations were carried out at 25 °C in tandem cuvettes by the addition of ppBat to the flavin in the measurement cell and to buffer (20 mm Tris/HCl, 100 mm NaCl, pH 8.0) in the reference cell at 2-min intervals. Fluorescence titrations were performed at 25 °C in 20 mm Tris/HCl, 100 mm NaCl, pH 8.0 buffer. For quenching of the FMN fluorescence, 1.8 ml of 1 μm FMN solution were titrated with 2-μl aliquots of 100 μm protein solution every 2 min under stirring. Emission at 524 nm was measured at an excitation of 467 nm until an end point was reached. The spectra were corrected for dilution as well as for fluorescence of enzyme bound flavin. In case of tryptophan fluorescence, 2 ml of 15 μm protein solution were titrated with 2-μl aliquots of 1.5 mm FMN solution every 2 min (emission at 343 nm and excitation at 282 nm).
Isothermal Titration Calorimetry (ITC)
Dissociation constants for the binding of flavins to ppBat were determined using a VP-ITC system (MicroCal). All of the experiments were performed at 25 °C in 20 mm Tris/HCl, 100 mm NaCl, pH 8.0 buffer, and solutions were degassed immediately before measurements. Titration experiments for FMN, FAD, riboflavin, and lumichrome consisted of 21 injections (7 μl; duration time, 14 s; spacing time, 300 s) of a flavin solution (530 μm) to 1.495 ml ppBat (40 μm). 40 μm enzyme were titrated with iso-FMN (370 μm; 21 × 10 μl; duration time, 10 s; spacing time, 300 s), 5-deaza-riboflavin (200 μm; 25 × 10 μl; duration time, 20 s; spacing time, 300 s), and 25 μm protein were titrated with 8-amino-riboflavin and iso-riboflavin (300 μm; 25 × 7 μl; duration time, 14 s; spacing time, 300 s), respectively. 40 μm of the W164F variant were titrated with 7-μl aliquots of 300 μm riboflavin for 25 times (duration time, 14 s; spacing time, 300 s). One set of sites fitting with Origin version 7.0 (MicroCal) for ITC data analysis was used to obtain dissociation constants.
Determination of Oligomeric State by Size Exclusion Chromatography
Determination of the oligomerization state for the wild-type protein and variants was carried out with a Superdex 200 analytical 10/300 GL column (GE Healthcare) using a ÄKTA explorer system. The calibration curve was prepared according to the manufacturer's protocol. Approximately 1 mg of protein samples as isolated and saturated with FMN, respectively, were dissolved in 20 mm Tris/HCl, 100 mm NaCl, pH 8.0 buffer prior to size exclusion chromatography (flow rate, 0.5 ml/min).
Photoreduction
Photoreduction was carried out as outlined in a previous study (15). The protein was diluted in 20 mm Tris/HCl, pH 8.0, containing 100 mm NaCl, 1 mm EDTA, and 2 μm benzyl viologen. Quartz cuvettes were rendered anaerobic by repeated cycles of evacuation and flushing with nitrogen. Reduction of the flavin cofactor was achieved by irradiation of the sample with a 250-W halogen lamp that was cooled to 15 °C during the time of irradiation. Spectra from 300 to 700 nm were recorded at the same temperature until no further changes were observed. Then the cuvette was opened to air, and the absorbance spectrum was recorded.
Rapid Reaction Studies
Stopped flow measurements were carried out with a Hi-Tech (SF-61DX2) stopped flow device (TgK Scientific Limited, Bradford-on-Avon, UK) positioned in a glove box from Belle Technology (Weymouth, UK) at 25 °C. Reoxidation of FMN free in solution and bound to ppBat was measured by monitoring changes at A451 (ppBat) or A449 (FMN), respectively, with a KinetaScanT diode array detector (MG-6560; TgK Scientific Limited). Reduction of FMN was achieved by the addition of solid sodium dithionite. Reoxidation was then monitored by rapidly mixing the reduced solution with buffer previously equilibrated with air.
HPLC Determination of Flavins and Lumichrome
Cofactors were released by thermal precipitation of ppBat at 95 °C for 10 min, cooling on ice for 5 min, and centrifugation for 5 min. The flavin extracts were loaded onto an Atlantis® dC18 8-μm 4.6 × 250-mm column (Waters) and eluted with a water/acetonitrile multi-step gradient (0–1.5 min, 0–5% acetonitrile; 1.5–20 min, 5–60% acetonitrile; 20–22 min, 95% acetonitrile) at a flow rate of 1 ml/min. The elution was monitored by UV absorption at 370 nm, and the cofactors were identified according to their elution time and UV-visible absorbance spectra.
Zinc Determination
The amount of zinc in ppBat samples was determined by means of inductively coupled plasma optical emission spectrometry.
RESULTS
Expression and Purification of ppBat
Cells transformed with the plasmid carrying the gene of ppBat were cultured, and protein production was induced by isopropyl β-d-thiogalactopyranoside. The majority of the target protein was soluble and purified by a single Ni-NTA affinity chromatography step (the progress of purification is shown in supplemental Fig. S2). The purified protein was obtained with an average yield of 30 mg/g of wet cells (∼140 mg/l bacterial culture).
Characterization of the Isolated Protein
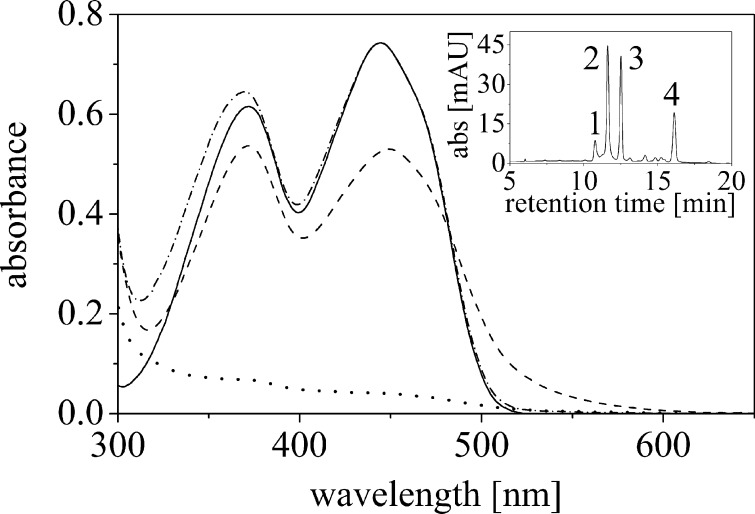
The purified protein was pale yellow and had absorbance maxima at 372 and 449 nm indicative for bound flavin. The absorbance ratio A280/A449 was very high (≈ 45) for a small flavoprotein and suggested that the protein was predominantly present in a flavin-free apo-form (Fig. 2, dotted line). When FMN was added to the extraction buffer, the protein was isolated with a reproducibly lower A280/A449 ratio of 5, indicating that the apoprotein present in the crude extract was reconstituted to the holo-protein (Fig. 2). The extinction coefficient of bound flavin was determined by denaturation of the protein with subsequent determination of the UV-visible absorbance spectrum of the released FMN (ϵfree = 11,500 m−1 cm−1) to ϵbound = 8,960 m−1 cm−1. Using this extinction coefficient, it was estimated that only 8–10% of the isolated protein is present in the holo-form when no flavin was added to the crude extract. The hypochromic effect at both absorbance maxima is accompanied by a long wavelength absorbance extending beyond the typical flavin absorbance (Fig. 2, compare dashed and solid lines). In contrast to the low flavin content of the purified protein, zinc was found in stoichiometric amounts, i.e., one zinc atom/protomer.
FIGURE 2.
UV-visible absorbance spectrum of ppBat as isolated (dotted line), ppBat saturated with FMN (dashed line) and after denaturation (dashed and dotted line). The UV-visible absorbance spectrum of free FMN is shown for comparison (solid line). The inset shows the HPLC analysis of bound ligands. The trace shown was recorded at 370 nm. Peak 1, FAD (retention time = 10.7 min); peak 2, FMN (11.6 min); peak 3, riboflavin (12.5 min); peak 4, lumichrome (16.1 min).
FMN bound to ppBat showed fluorescence excitation and emission maxima similar to that of free flavin; however, fluorescence intensity was ∼47% lower as compared with free FMN. Similarly, FMN binding to ppBat quenched the tryptophan fluorescence emission at 343 nm to ∼28%.
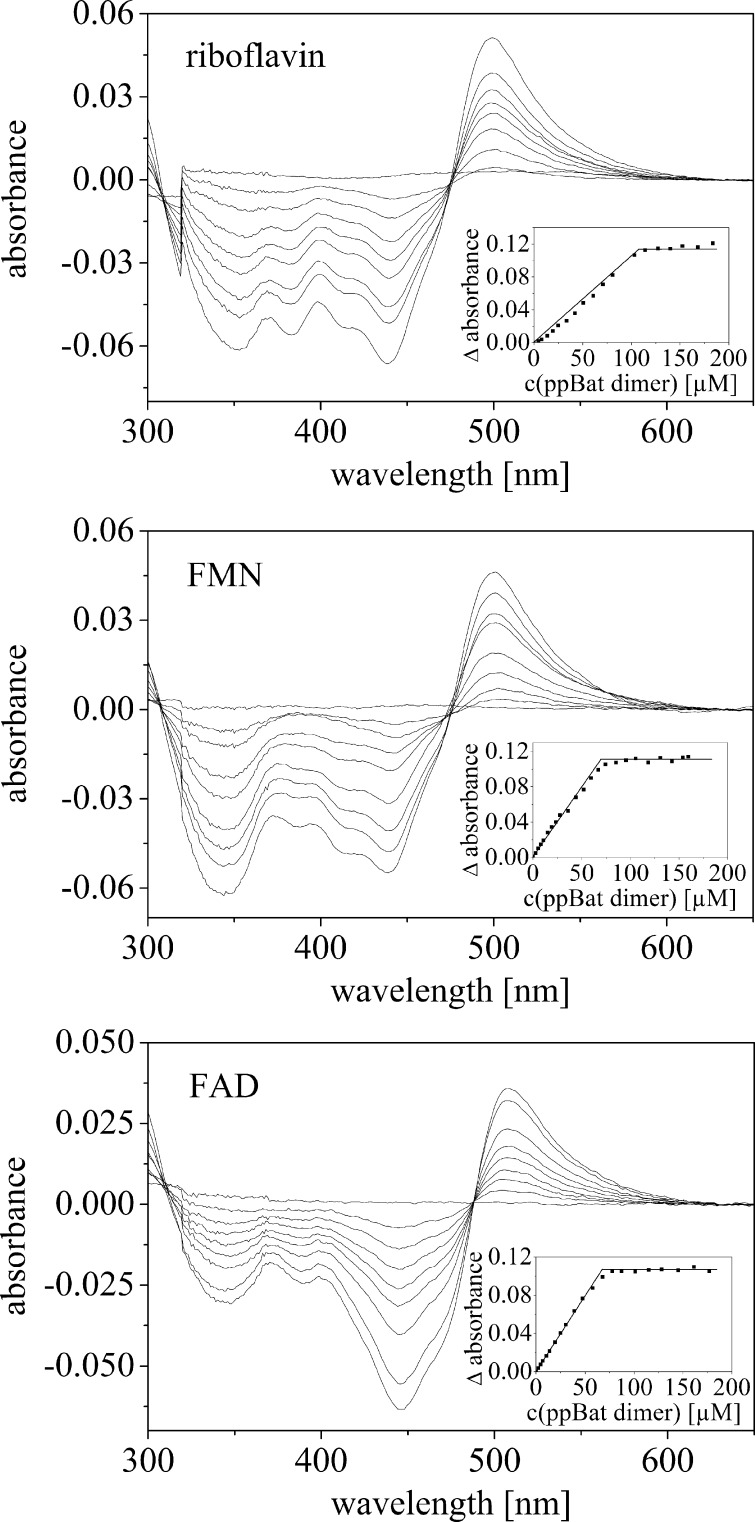
Because the deposited structure of the protein has a truncated N-10 side chain (only four carbon atoms of the side chain were modeled; Fig. 1B), it is not clear which flavin derivative is bound to the protein. Therefore, we also tested riboflavin and FAD as potential binding partners of the recombinant protein. We found that riboflavin and FAD also bind to the protein with spectral changes similar to those seen with FMN. Further analysis by difference UV-visible absorbance spectroscopy revealed that all three flavin compounds bind tightly to the apoprotein with similar spectral perturbations (Fig. 3) and molar extinction coefficients (Table 1). All three flavin derivatives produced a sharp end point at equimolar concentrations of flavin and dimeric protein in agreement with a 1:1 stoichiometry as suggested by the three-dimensional structure.
FIGURE 3.
UV-visible absorbance difference titration of riboflavin (80 μm, top panel), FMN (61 μm, central panel), and FAD (60 μm, bottom panel) with ppBat. The data points shown in the inset were manually fitted by straight lines to indicate the titration end points.
TABLE 1.
Molar extinction coefficients (ϵ) of flavins and flavin analogs free and bound to ppBat
| Flavin | ϵ (free/bound) | λmax (free/bound) | Reference |
|---|---|---|---|
| m−1 cm−1 | nm | ||
| Lumichrome | 8,200/6,400 | 347/348 | 28 |
| Riboflavin | 12,500/9,800 | 444/449 | 28 |
| iso-Riboflavin | 7,300/5,650 | 448/454 | 29 |
| 1-Deaza-riboflavin | 6,800/6110 | 536/558 | 30 |
| 5-Deaza-riboflavin | 11,500/9,620 | 399/398 | 14 |
| 8-Amino-riboflavin | 42,000/24,570 | 479/487 | 31 |
| 2′-Deoxy-riboflavin | 12,500/9950 | 443/452 | ϵ of riboflavin was used |
| FMN | 12,500/8,960 | 444/449 | 28 and 32 |
| FAD | 11,300/8,500 | 448/449 | 28, 29, and 32 |
| iso-FMN | 7,200/5,600 | 449/454 | 33 |
To identify the ligand(s) bound to ppBat as isolated from E. coli cells, the protein was denatured, and the released compounds were subjected to HPLC analysis. As shown in the inset of Fig. 2, four peaks were observed. Based on the retention times and UV-visible absorbance properties these compounds were identified as FAD (peak 1), FMN (peak 2), riboflavin (peak 3), and lumichrome (peak 4).
Determination of Dissociation Constants
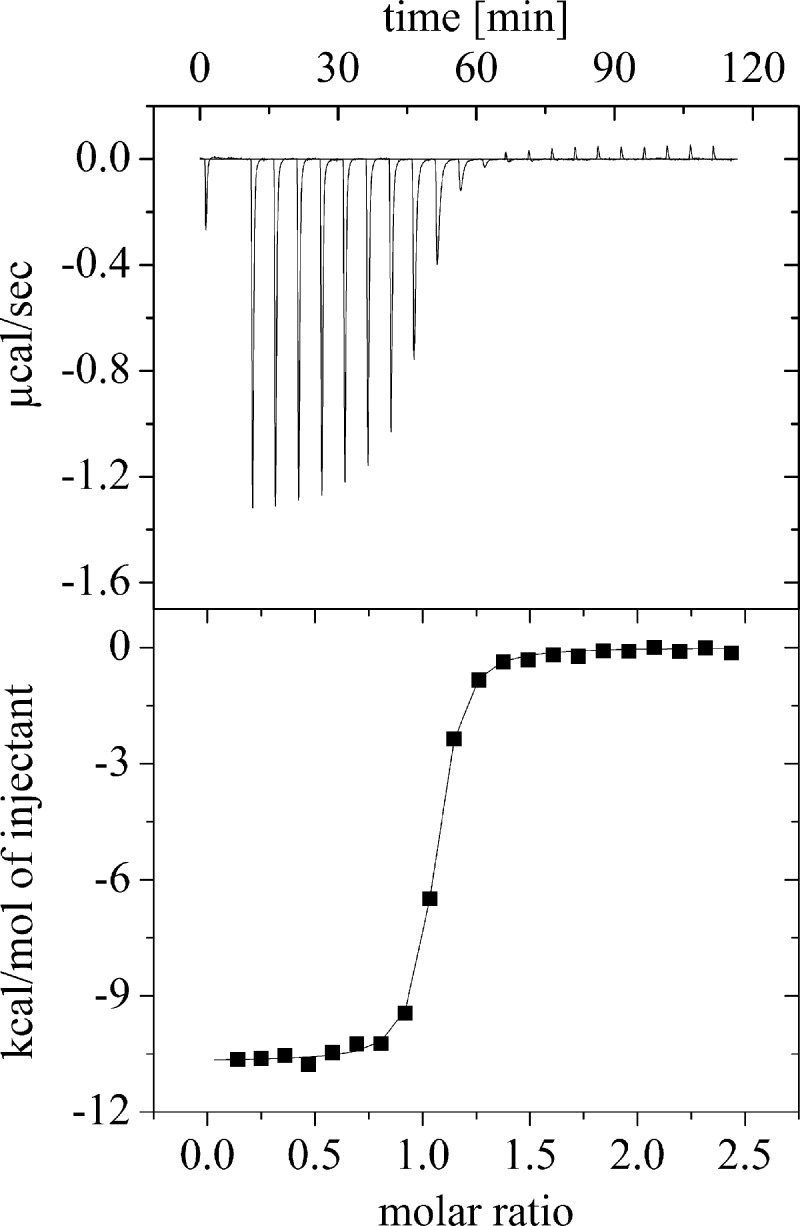
Because of the apparently tight binding of riboflavin, FMN, and FAD to apo-ppBat (Fig. 3), we employed ITC to determine their dissociation constants. As shown in Fig. 4, binding of these compounds to apoprotein is exothermic at 25 °C. The experimental data were fitted with a single site binding model and yielded dissociation constants in the range of 74.3 and 800.6 nm for riboflavin and FAD, respectively (Table 2). Obviously, flavin binding in the dimer interface of ppBat is adversely influenced by the size of the side chain attached to N-10.
FIGURE 4.
Representative example for the determination of a dissociation constant by isothermal titration microcalorimetry. The measurement shows the titration of 40 μm ppBat with riboflavin in 20 mm Tris/HCl, 100 mm NaCl, pH 8.0. at 25 °C.
TABLE 2.
Dissociation constants for binding of lumichrome, riboflavin, FMN and FAD as well as modified flavin analogs to ppBat as determined by ITC
The values given are the averages of three measurements carried out in 20 mm Tris/HCl, 100 mm NaCl, pH 8.0 buffer at 25 °C.
| Flavin ligand | Kd |
|---|---|
| nm | |
| Lumichrome | 272 ± 61 |
| Riboflavin | 74 ± 5 |
| 5-Deaza-riboflavin | 84 ± 5 |
| iso-Riboflavin | 96 ± 2 |
| 8-Amino-riboflavin | 227 ± 26 |
| 2′-Deoxy-riboflavin | 117 ± 3 |
| FMN | 487 ± 19 |
| iso-FMN | 295 ± 21 |
| FAD | 801 ± 82 |
Binding of Chemically Modified Flavins
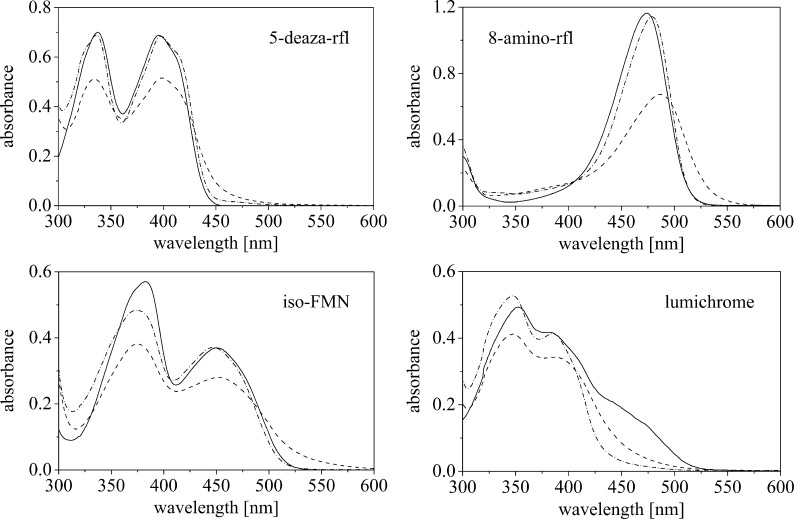
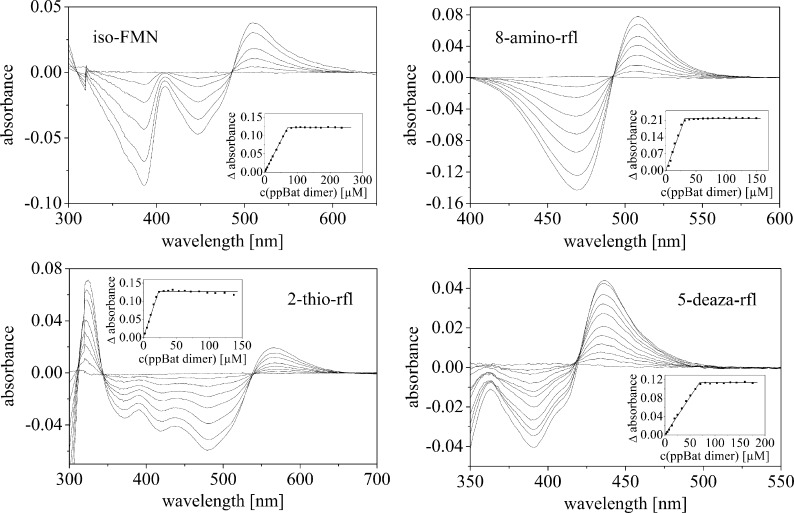
In addition to the naturally occurring flavin compounds, ppBat also hosted a variety of derivatized riboflavin and FMN compounds, such as iso-riboflavin and iso-FMN (6.7-dimethylisoalloxazine), 2-thio-, 8-amino-, 1-, and 5-deaza-riboflavin, as well as 5-deaza-lumiflavin and lumichrome. Lumazine did not bind to ppBat. UV-visible absorbance spectra of four selected examples are shown in Fig. 5 (see also supplemental Figs. S3 and S4). To determine the stoichiometry and affinity of these flavin analogs, difference titrations were performed with an UV-visible absorbance spectrophotometer. As shown in Fig. 6, we found tight binding with all of the tested flavin analogs and a stoichiometry of 1:1 (flavin:dimer) for complex formation with ppBat. The binding of five flavin analogs was further examined by ITC. The three riboflavin analogs, 5-deaza-riboflavin, iso-riboflavin, and 8-amino-riboflavin showed dissociation constants similar to riboflavin and iso-FMN bound even tighter than FMN (Table 2). Interestingly, 2′-deoxy-riboflavin lacking the hydroxyl group at the 2′-position of the ribityl side chain had only a marginally higher dissociation constant (117.4 versus 74.3 nm; see also supplemental Fig. S5), suggesting that the carbonyl oxygen of Asn161 forms only a weak hydrogen bond to the C2′-OH group (Table 2).
FIGURE 5.
UV-visible absorbance spectra of ppBat with three different flavin analogs and lumichrome. The dashed lines represent spectra of ppBat saturated with the ligand, dashed and dotted lines were recorded after denaturation of the protein, and solid lines are the spectra of the free chromophore.
FIGURE 6.
UV/Vis absorbance difference titration of iso-FMN (76 μm), 8-amino-riboflavin (rfl) (17 μm), 2-thio-riboflavin (32 μm), and 5-deaza-riboflavin (48 μm).
Properties of Variants W164F and W164A
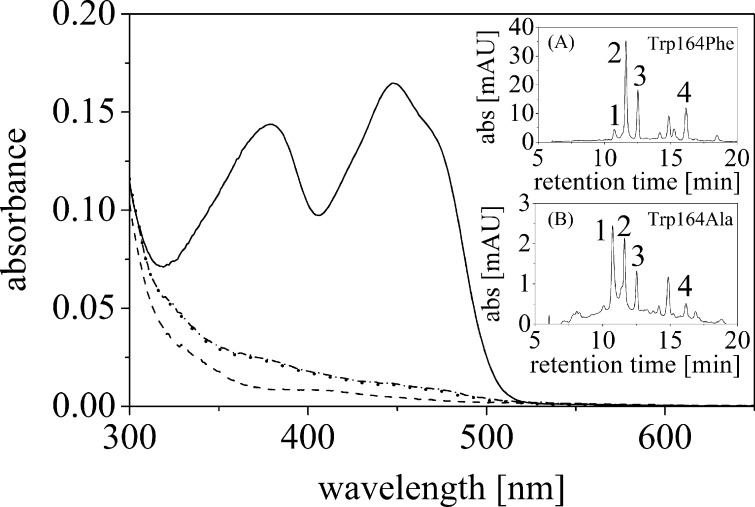
To investigate the importance of the indole side chain of Trp164 for dimerization of the protein and for flavin binding, two single replacement variants were generated by site-directed mutagenesis: W164F and W164A. Similar to wild-type ppBat, both variants were found to form dimers in solution as judged by size exclusion chromatography (supplemental Fig. S6). In the presence of FMN in the crude extract, the W164F variant retained the ability to bind FMN, whereas the W164A variant was essentially devoid of FMN (Fig. 7). In the absence of FMN in the crude extract, only trace amounts of flavin could be detected in both variants. Detailed HPLC analysis of ligands released upon denaturation of the protein variants revealed that small amounts of FAD, FMN, riboflavin, and lumichrome were bound (Fig. 7, insets). For the W164F variant, the dissociation constant of riboflavin was determined by ITC to 1.19 ± 0.15 mm, i.e., 16 times higher than for wild-type ppBat.
FIGURE 7.
UV-visible absorbance spectra of the W164F and W164A variants. The solid and dashed lines represent the absorbance spectra of the W164F and W164A variant, respectively, purified with FMN added to the crude extract. The dotted and dashed and dotted lines were obtained in the absence of additional FMN (“as isolated”). The insets show the HPLC analysis of bound ligands for the two variants as isolated. The traces show the absorbance at 370 nm. Peak 1, FAD; peak 2, FMN; peak 3, riboflavin; peak 4, lumichrome.
Recombinant ppBat loaded with FMN was found to be present as a dimer in solution using analytical size exclusion chromatography. Both tryptophan variants yielded a virtually identical peak, indicating that neither of the two amino acid substitutions affected dimerization of ppBat (supplemental Fig. S6).
Photoreduction and Reoxidation
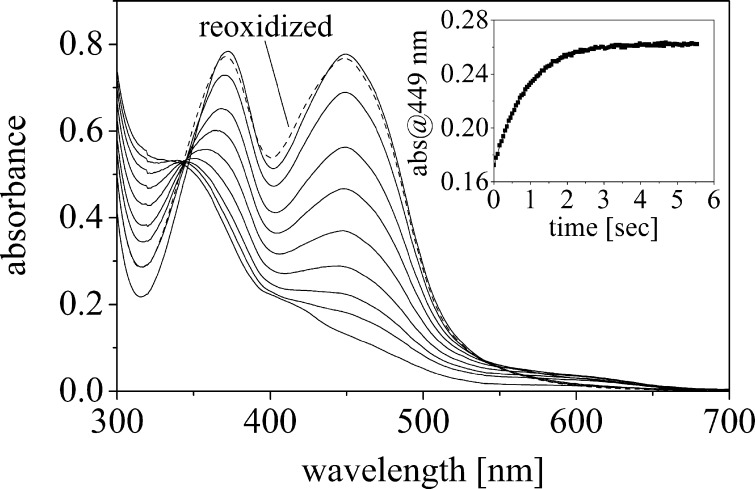
Wild-type ppBat reconstituted with FMN was subjected to photoreduction under anoxic conditions. The course of the reduction is shown in Fig. 8. The spectral changes upon photoreduction featured a loss of absorption at 449 nm and an isosbestic point at 345 nm. The final spectrum is characteristic for the fully reduced dihydroquinone form of the flavin. During reduction, spectral hallmarks of a flavin radical species, like an increased absorbance at 370 nm or absorbance above 550 nm, were absent, indicating that no radical species is thermodynamically stabilized during reduction. The fully reduced dihydroflavin could be readily reoxidized by allowing access of oxygen to the cuvette (Fig. 8). The rate of reoxidation was determined in a stopped flow apparatus by rapid mixing of sodium dithionite-reduced ppBat with air-equilibrated buffer (Fig. 8, inset). This yielded a kobs = 1.1 ± 0.02 s−1 (average of five independent measurements). Using the same experimental set-up, the reoxidation of free FMN yielded a kobs = 6.7 ± 0.09 s−1, i.e., ∼6 times faster than for FMN bound to ppBat (all measurements were performed at 25 °C).
FIGURE 8.
Anaerobic photoreduction of ppBat in the presence of EDTA and benzylviologen shows that the FMN moiety can be directly reduced to its fully reduced form in a two-electron reduction process (isosbestic point at 343 nm) without the formation of either the red or the blue semiquinone radical. Absorbance spectra were recorded prior to illumination (top, solid line) and after 5, 7, 9, 11, 14, 21, 33, and 103 min, respectively (from top to bottom). The spectrum after reoxidation with dioxygen is shown as a dashed line. The inset shows reoxidation in the stopped flow apparatus by molecular oxygen.
DISCUSSION
Flavins are widely used coenzymes for reduction-oxidation processes, with well over 300 different enzymatic reactions described in the literature (3, 16–18). In addition to their role as redox catalysts, flavins are also found in some none redox enzymes, such as isomerases, lyases, and transferases (3, 19). However, the occurrence of a flavin (purportedly FMN) in a ppBat was a novelty because hydrolases have never been associated with this cofactor. The discovery that ppBat features a flavin prompted us to investigate the properties of the flavin-binding site in this unusual family of proteases/chaperones (Fig. 1). At first, the low flavin content of isolated ppBat was surprising in view of the fact that the deposited crystal structure clearly showed the presence of flavin bound between two tryptophans in the dimer interface of the protein (Fig. 1, A and B). The addition of a flavin, for example FMN, increased the flavin content close to one flavin per protein dimer, as could be demonstrated by titration experiments (see Figs. 3 and 6). According to the information given by the authors of the crystal structure, no flavin (neither riboflavin, FMN, nor FAD) was added in the crystallization trials, and hence it follows that the population of ppBat with flavin bound (∼10%) preferentially crystallized, i.e., was selected by the applied conditions. Because our analysis of ppBat isolated without flavin addition has revealed the presence of several chromophores (lumichrome, riboflavin, FMN, and FAD), the nature of the bound species obviously does not matter with regard to the propensity to generate crystals. The deposited crystal structure is also lacking information on the exact nature of the side chain because only the four carbon atoms proximal to N-10 were modeled. Therefore the structure cannot distinguish between riboflavin, FMN, and FAD. In the present study, we have shown that lumichrome as well as riboflavin, FMN, and FAD all bind to ppBat with dissociation constants in the nanomolar range (Table 2).
The UV-visible absorbance spectra of all flavins bound to ppBat showed pronounced hypochromicity and long wavelength absorbance indicative of a charge-transfer complex (Table 1). This is in contrast to the rather modest quenching of both the flavin and the tryptophan fluorescence emission. Interestingly, virtually complete fluorescence quenching was observed for both fluorophores in model compounds, where stacking interactions between the isoalloxazine and indole ring were assumed (20). Apparently, aromatic stacking interactions do not necessarily lead to complete fluorescence quenching.
A salient feature of flavoproteins is the stabilization of either the red (anionic) or blue (neutral) flavin radical (semiquinone) (12). In the case of ppBat, reduction of bound flavin to the hydroquinone proceeded without the observable occurrence of a radical species (Fig. 7). Similarly, reoxidation of the reduced flavin yielded the oxidized form without any observable radical intermediate. Interestingly, the observed rate of reoxidation was approximately six times slower than for free flavin, indicating that the mode of flavin binding in ppBat protects the flavin from reoxidation. This is in keeping with the current understanding of the control of oxygen reactivity by flavoenzymes. In recent years, it has emerged that enhanced oxygen reactivity is effected by an oxygen binding site near the C-4a position of the isoalloxazine ring where a superoxide anion is stabilized by a nearby positive charge, e.g., a lysine side chain (21, 22). The flavin-binding site in ppBat neither provides interactions necessary to stabilize a flavin radical nor a superoxide anion near the C-4a position. Thus the observed properties are in full agreement with the flavin binding mode.
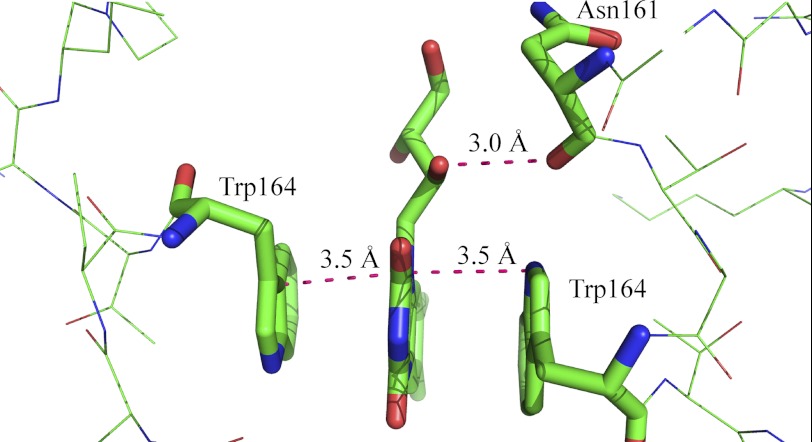
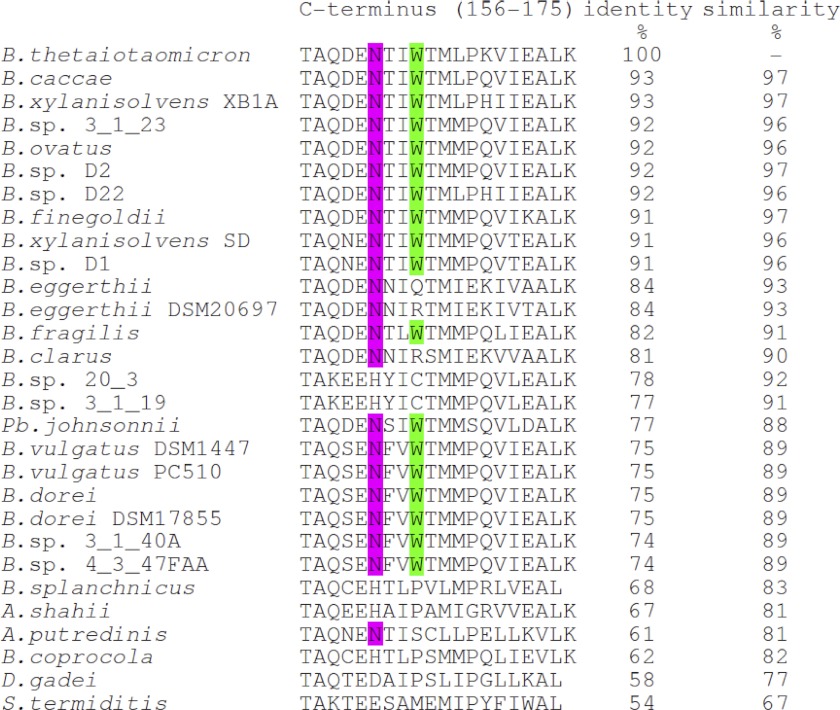
Our studies have clearly shown that Trp164 is crucial for binding of flavins to the dimer interface of ppBat. In contrast to the aromatic stacking interaction, the assumed contact between the C2′-hydroxyl group of the ribityl side chain and the main chain carbonyl of Asn161 contributes very little (1.14 kJ/mol) to the binding energy (Fig. 9). On the other hand, lumichrome, which features an alloxazine rather than an isoalloxazine π-electron system and has no side chain at N-10, binds weaker than riboflavin (∼6 kJ/mol) and 2′-deoxy-riboflavin (∼4.86 kJ/mol), suggesting that the side chain favorably affects binding to ppBat. Alignment of 29 sequences with at least 50% overall sequence identity showed that Trp164 and Asn161 are conserved in 18 and 22 proteins, respectively (Fig. 10). On the other hand, the two cysteines that complex the zinc ion are present in all of the 29 proteins. This suggests that the possibly catalytic function of the protein is related to the presence of the zinc center, and flavin binding may have evolved as an additional asset in a subset of homologous proteins.
FIGURE 9.
Close-up of the flavin-binding site of ppBat. The two tryptophans are almost equidistant to the plane of the isoalloxazine ring system (orange lines). The main chain carbonyl group of Asn161 engages in a hydrogen bond (magenta) to C′2-OH of the ribityl side chain. The distances are given in Ångstroms. The graphic was generated using PyMOL.
FIGURE 10.
Alignment of the last 20 amino acids of the C terminus of ppBat (amino acids 156–175) and related proteins from Bacteroides (B.), Parabacteroides (Pb.), Alistipes (A.), Dysgomonas (D.), and Sebaldella (S.). The tryptophan at position 164 is highlighted in green, and the asparagine 161 is highlighted in magenta. The percentage identity and similarity are given for the full-length proteins (175 amino acids). Two cysteines (Cys74 and Cys111; see Fig. 1C) are conserved in all sequences shown in this figure. Additionally, Asp76, His131, and Glu160, which are between 5 and 6 Å from the metal-binding site, are conserved in all sequences but Bacteroides vulgatus.
The binding mode in ppBat also rationalizes our findings that the protein is rather insensitive to the N-10 side chain carried by the isoalloxazine ring. Most flavoenzymes exhibit pronounced preference for either FMN or FAD and riboflavin is not used at all except in one case (3). In addition, flavins with modifications in the isoalloxazine ring have lower affinity because of adverse effects on specific interactions with amino acids in the flavin-binding pocket. As demonstrated by our affinity measurements, binding of modified flavins is in the same range as for the naturally occurring flavins and mainly governed by the side chain (i.e., iso-riboflavin and 5-deaza-riboflavin have similar affinities than riboflavin, and iso-FMN is similar to FMN). Therefore it appears that ppBat does not utilize the bound flavin for catalysis. Likewise, because flavin binding is also not required to maintain the dimeric structure of ppBat, we conclude that flavin binding to ppBat does not concern the potential catalytic function of the protein. On the other hand the strong binding of flavins, in particular of riboflavin, suggests that ppBat may play a role as a storage protein for gut bacteria.
A related flavin binding mode was recently discovered in a family of archaeal and bacterial proteins. These so-called dodecins consist of 12 protomers of 68–71 amino acids with six flavin-binding sites (23). As in ppBat, two tryptophans sandwich the isoalloxazine or alloxazine (lumichrome) ring system, although each binding site accommodates two ligands instead of one. In addition, other amino acids engage in hydrogen bond interactions to the flavin ligand, for example an arginine side chain forms hydrogen bonds to C-4=O of the isoalloxazine ring (23–25). In the case of ppBat, the only contacts are between the tryptophans of the two protomers and the isoalloxazine ring and a (weak) hydrogen bond between the carbonyl group of Asn161 of one protomer and the C′2-hydroxyl group of the ribityl side chain. Contacts between the pyrimidine ring and ppBat are not observed in the structure, and hence the affinity of flavins to ppBat is mainly governed by the interaction with the indole side chains of the two tryptophans. Therefore it is not surprising that replacement of the tryptophans by alanine and phenylalanine has marked effects on the binding affinity. In the case of alanine, binding was virtually abolished, whereas the W164F exchange has retained some affinity (Fig. 7). In contrast, replacement of Trp38 in dodecin from Thermophilus has apparently no consequence on the propensity to bind flavins because several other interactions still ensure stable complex formation (see Protein Data Bank entry 2vyx (23)). This protein also binds coenzyme A, and it appears that the dodacamer binds a mixture of cofactors in vivo (23, 25). Therefore dodecins appear to serve as cofactor storage proteins and may also be important as scavengers of potentially harmful cofactor degradation products, such as lumichrome, which is produced in the course of photodegradation of riboflavin in vivo (23, 25, 26). The dissociation constants determined for dodecin from Thermophilus are similar to those found for ppBat and support the notion that ppBat may also serve as a storage protein in gut bacteria in vivo. A BLASTp search using the dodecin sequence of Thermus thermophilus against the B. thetaiotaomicron genome failed to retrieve a dodecin homolog.
The distance between the plane of the indole ring of the two tryptophans and the isoalloxazine in ppBat is ∼3.5–3.7 Å (Fig. 9). A similar distance for π-stacking interaction of a tryptophan and an isoalloxazine ring was seen in the dodecins (3.4 to 3.8 Å) and was also found in the case of FMN adenylyltransferase (27). Apparently, this distance is optimal for π-stacking interactions, although the relative orientation of the interacting aromatic ring systems differs in the above mentioned proteins. Because the main contribution for flavin binding stems from the π-stacking interaction with the two tryptophan residues, we assume that the asymmetric isoalloxazine binds in two possible orientations in the symmetric binding site (2-fold axis) provided by the two protomers of the dimeric protein. The electron density of Protein Data Bank entry 3cne is consistent with 50% occupancy of the two possible orientations of the tricyclic ring system.
Supplementary Material
Acknowledgments
We thank Dr. Michael K. Uhl for help with the structural representations and Prof. Albin Hermetter and Claudia Ramprecht MSc for help with fluorescence measurements.
This work was supported by funds from Graz University of Technology (to P. M.).
This work is dedicated to Prof. Sandro Ghisla, a pioneer in the field of chemically modified flavins, on the occasion of his 70th birthday.

This article contains supplemental Figs. S1–S6.
- ppBat
- putative protease from B. thetaiotaomicron
- ITC
- isothermal titration calorimetry
- Ni-NTA
- nickel-nitrilotriacetic acid.
REFERENCES
- 1. Comstock L. E., Coyne M. J. (2003) Bacteroides thetaiotaomicron. A dynamic, niche-adapted human symbiont. Bioessays 25, 926–929 [DOI] [PubMed] [Google Scholar]
- 2. Mahowald M. A., Rey F. E., Seedorf H., Turnbaugh P. J., Fulton R. S., Wollam A., Shah N., Wang C., Magrini V., Wilson R. K., Cantarel B. L., Coutinho P. M., Henrissat B., Crock L. W., Russell A., Verberkmoes N. C., Hettich R. L., Gordon J. I. (2009) Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc. Natl. Acad. Sci. U.S.A. 106, 5859–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Macheroux P., Kappes B., Ealick S. E. (2011) Flavogenomics: A genomic and structural view of flavin-dependent proteins. FEBS J. 278, 2625–2634 [DOI] [PubMed] [Google Scholar]
- 4. Honbou K., Suzuki N. N., Horiuchi M., Niki T., Taira T., Ariga H., Inagaki F. (2003) The crystal structure of DJ-1, a protein related to male fertility and Parkinson's disease. J. Biol. Chem. 278, 31380–31384 [DOI] [PubMed] [Google Scholar]
- 5. Quigley P. M., Korotkov K., Baneyx F., Hol W. G. (2003) The 1.6-Å crystal structure of the class of chaperones represented by Escherichia coli Hsp31 reveals a putative catalyitc cleft. Proc. Natl. Acad. Sci. U.S.A. 100, 3137–3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tao X., Tong L. (2003) Crystal structure of human DJ-1, a protein associated with early onset Parkinson's disease. J. Biol. Chem. 278, 31372–31379 [DOI] [PubMed] [Google Scholar]
- 7. Wei Y., Ringe D., Wilson M. A., Ondrechen M. J. (2007) Identification of functional subclasses in the DJ-1 superfamily proteins. PLoS Comput. Biol. 3, e10. [DOI] [PubMed] [Google Scholar]
- 8. Malki A., Caldas T., Abdallah J., Kern R., Eckey V., Kim S. J., Cha S. S., Mori H., Richarme G. (2005) Peptidase activity of the Escherichia coli Hsp31 chaperone. J. Biol. Chem. 280, 14420–14426 [DOI] [PubMed] [Google Scholar]
- 9. Le H. T., Gautier V., Kthiri F., Malki A., Messaoudi N., Mihoub M., Landoulsi A., An Y. J., Cha S. S., Richarme G. (2012) YajL, prokaryotic homolog of parkinsonism-associated protein DJ-1, functions as a covalent chaperone for thiol proteome. J. Biol. Chem. 287, 5861–5870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mujacic M., Bader M. W., Baneyx F. (2004) Escherichia coli Hsp31 functions as a holding chaperone that cooperates with the DnaK-DnaJ-GrpE system in the management of protein misfolding under severe stress conditions. Mol. Microbiol. 51, 849–859 [DOI] [PubMed] [Google Scholar]
- 11. Parkin G. (2004) Synthetic analogues relevant to the structure and function of zinc enzymes. Chem. Rev. 104, 699–767 [DOI] [PubMed] [Google Scholar]
- 12. Ghisla S., Massey V. (1986) New flavins for old. Artificial flavins as active site probes of flavoproteins. Biochem. J. 239, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Matsui K., Kasai S. (1974) Effect of 8-amino-8-dimethyl-d-araboflavin, 8-amino-8-dimethyl-d-riboflavin and 8-hydroxy-8-demethyl-d-riboflavin on mice. J. Nutr. Sci. Vitam. 20, 411–420 [DOI] [PubMed] [Google Scholar]
- 14. Spencer R., Fisher J., Walsh C. (1976) Preparation, characterization, and chemical properties of the flavin coenzyme analogues 5-deazariboflavin, 5-deazariboflavin 5′-phosphate, and 5-deazariboflavin 5′-diphosphate, 5′-5′-adenosine ester. Biochemistry 15, 1043–1053 [DOI] [PubMed] [Google Scholar]
- 15. Massey V., Hemmerich P. (1978) Photoreduction of flavoproteins and other biological compounds catalyzed by deazaflavins. Biochemistry 17, 9–16 [DOI] [PubMed] [Google Scholar]
- 16. Fagan R. L., Palfey B. A. (2010) Flavin-dependent enzymes. In Comprehensive Natural Products II ((Begley T. P., ed) Elsevier Science Publishing Co., Inc., New York [Google Scholar]
- 17. Joosten V., van Berkel W. J. (2007) Flavoenzymes. Curr. Opin. Chem. Biol. 11, 195–202 [DOI] [PubMed] [Google Scholar]
- 18. Mansoorabadi S. O., Thibodeaux C. J., Liu H. W. (2007) The diverse roles of flavin coenzymes: Nature's most versatile thespians. J. Org. Chem. 72, 6329–6342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bornemann S. (2002) Flavoenzymes that catalyse reactions with no net redox change. Nat. Prod. Rep. 19, 761–772 [DOI] [PubMed] [Google Scholar]
- 20. McCormick D. (1977) Interactions of flavins with amino acid residues. Assessments from spectral and photochemical studies. Photochem. Photobiol. 26, 169–182 [DOI] [PubMed] [Google Scholar]
- 21. Kommoju P. R., Chen Z. W., Bruckner R. C., Mathews F. S., Jorns M. S. (2011) Probing oxygen activation sites in two flavoprotein oxidases using chloride as an oxygen surrogate. Biochemistry 50, 5521–5534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McDonald C. A., Fagan R. L., Collard F., Monnier V. M., Palfey B. A. (2011) Oxygen reactivity in flavoenzymes. Context matters. J. Am. Chem. Soc. 133, 16809–16811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Meissner B., Schleicher E., Weber S., Essen L. O. (2007) The dodecin from Thermus thermophilus, a bifunctional cofactor storage protein. J. Biol. Chem. 282, 33142–33154 [DOI] [PubMed] [Google Scholar]
- 24. Grininger M., Seiler F., Zeth K., Oesterhelt D. (2006) Dodecin sequesters FAD in closed conformation from the aqueous solution. J. Mol. Biol. 364, 561–566 [DOI] [PubMed] [Google Scholar]
- 25. Grininger M., Staudt H., Johansson P., Wachtveitl J., Oesterhelt D. (2009) Dodecin is the key player in flavin homeostasis of archaea. J. Biol. Chem. 284, 13068–13076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cairns W. L., Metzler D. E. (1971) Photochemical degradation of flavins. VI. A new photoproduct and its use in studying the photolytic mechanism. J. Am. Chem. Soc. 93, 2772–2777 [DOI] [PubMed] [Google Scholar]
- 27. Huerta C., Borek D., Machius M., Grishin N. V., Zhang H. (2009) Structure and mechanism of a eukaryotic FMN adenylyltransferase. J. Mol. Biol. 389, 388–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Müller F., Ghisla S., Bacher A. (1988) Vitamin B2 und natürliche Flavine. In Vitamine II (Isler O., Brubacher G., Ghisla S., Kräutler B., eds) Georg Thieme, New York [Google Scholar]
- 29. Whitby L. G. (1954) Transglucosidation reactions with flavins. Biochem. J. 57, 390–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Spencer R., Fisher J., Walsh C. (1977) One- and two-electron redox chemistry of 1-carba-1-deazariboflavin. Biochemistry 16, 3586–3594 [DOI] [PubMed] [Google Scholar]
- 31. Fitzpatrick P. F., Ghisla S., Massey V. (1985) 8-Azidoflavins as photoaffinity labels for flavoproteins. J. Biol. Chem. 260, 8483–8491 [PubMed] [Google Scholar]
- 32. Macheroux P. (1999) UV-visible spectroscopy as a tool to study flavoproteins. Methods Mol. Biol. 131, 1–7 [DOI] [PubMed] [Google Scholar]
- 33. Choong Y. S., Massey V. (1981) Studies on lactate oxidase substituted with synthetic flavins, iso-FMN lactate oxidase. J. Biol. Chem. 256, 8671–8678 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.