Background: The signal that ensures the specific targeting of β-barrel proteins to either mitochondria or chloroplasts is ill-defined.
Results: Chloroplast β-barrel proteins can be assembled in vitro into the mitochondrial outer membrane.
Conclusion: The mitochondrial import machinery can recognize and process chloroplast β-barrel proteins as substrates.
Significance: Dedicated targeting factors had to evolve in plant cells to prevent mis-sorting of chloroplast β-barrel proteins to mitochondria.
Keywords: Chloroplast, Membrane Proteins, Mitochondria, Mitochondrial Transport, Protein Sorting, β-Barrel Proteins
Abstract
Membrane-embedded β-barrel proteins are found in the outer membranes (OM) of Gram-negative bacteria, mitochondria and chloroplasts. In eukaryotic cells, precursors of these proteins are synthesized in the cytosol and have to be sorted to their corresponding organelle. Currently, the signal that ensures their specific targeting to either mitochondria or chloroplasts is ill-defined. To address this issue, we studied targeting of the chloroplast β-barrel proteins Oep37 and Oep24. We found that both proteins can be integrated in vitro into isolated plant mitochondria. Furthermore, upon their expression in yeast cells Oep37 and Oep24 were exclusively located in the mitochondrial OM. Oep37 partially complemented the growth phenotype of yeast cells lacking Porin, the general metabolite transporter of this membrane. Similarly to mitochondrial β-barrel proteins, Oep37 and Oep24 expressed in yeast cells were assembled into the mitochondrial OM in a pathway dependent on the TOM and TOB complexes. Taken together, this study demonstrates that the central mitochondrial components that mediate the import of yeast β-barrel proteins can deal with precursors of chloroplast β-barrel proteins. This implies that the mitochondrial import machinery does not recognize signals that are unique to mitochondrial β-barrel proteins. Our results further suggest that dedicated targeting factors had to evolve in plant cells to prevent mis-sorting of chloroplast β-barrel proteins to mitochondria.
Introduction
In addition to the outer membrane (OM)3 of Gram-negative bacteria, membrane-embedded β-barrel proteins are found also in the OM of the endosymbiotic organelles, mitochondria and chloroplasts. Compared with the diversity of β-barrel membrane proteins in prokaryotes, the number of organellar OM proteins confirmed to have this structural type is rather limited. In Saccharomyces cerevisiae, bakers' yeast, only five members have been identified: Tom40, Tob55/Sam50, two isoforms of Porin/VDAC, and Mdm10. The number of characterized β-barrel proteins in chloroplast OM is not much higher, although sequence analysis predicted the presence of many such proteins in this membrane (1). Some of those (for example, outer envelope proteins OEP21, OEP24, and OEP37) were proposed to function as pores for small metabolites, and their distinct substrate specificities may point to discrete roles in various metabolic processes (2, 3). The chloroplast OM harbors also Toc75, a β-barrel protein with several isoforms in Arabidopsis thaliana (4). Toc75 (Toc75-III in A. thaliana) forms the protein-conducting pore of the translocase of the OM of chloroplasts (TOC complex) (5, 6).
Newly synthesized mitochondrial β-barrel precursor proteins are devoid of canonical N-terminal presequences or any other characterized linear targeting signal. They are initially recognized by the Tom20 and Tom22 receptor components of the translocase of the outer mitochondrial membrane (TOM complex) before their translocation across the OM via the import pore of this complex. Next, these proteins are relayed to the dedicated complex for topogenesis of outer membrane β-barrel proteins (TOB complex, also known as sorting and assembly machinery), which mediates their assembly into the OM. The known components of the TOB core complex are Tob55/Sam50/Omp85, Tob38/Sam35, and Mas37/Tom37/Sam37. On their way from the TOM to the TOB complex, the β-barrel precursors are exposed to the intermembrane space (IMS) where they interact with the small Tim chaperones (for detailed reviews see Refs. 7–9).
In contrast to our detailed picture on the biogenesis of β-barrel proteins in mitochondria, relatively little is known about their assembly pathways in chloroplasts. Specific signals for targeting of most β-barrel proteins to the chloroplast have not yet been identified. An interesting exception is provided by the unique biogenesis pathway of the precursor of Toc75-III. This precursor protein is synthesized with an N-terminal extension, which functions as a bipartite transit peptide and is processed during maturation (10, 11). The first portion of the targeting signal directs the precursor protein to the chloroplast stroma where it is cleaved by a stromal processing peptidase (10). The second portion functions probably as a stop-transfer segment and was found to be processed by a type I signal peptidase (12). The overall import pathway of Toc75-III seems to support the idea that sorting of β-barrel membrane proteins of chloroplasts occurs in a manner similar to that of mitochondria. The Toc75-III precursor is first completely translocated across the outer envelope by the TOC complex and thus is likely inserted from the inner face into the lipid phase of the OM. Another Toc75 isoform (Toc75-V/OEP80 in A. thaliana) was speculated to be involved in the membrane integration of chloroplast OM β-barrel proteins; however, experimental evidence to support this proposal is still lacking. In any case, this isoform is not a component of the TOC complex (13). Surprisingly, the N-terminal region of Toc75-V is not essential for the targeting, biogenesis, or functionality of the protein suggesting that Toc75-III and Toc75-V do not follow the same targeting pathway (14). Thus, currently Toc75-III is the only known protein in the OM of chloroplasts or mitochondria with a cleavable targeting sequence. Hence, the question regarding how the vast majority of the β-barrel proteins of chloroplasts and all those of mitochondria is targeted to their respective organelle is still an open one.
Bacterial β-barrel proteins can be targeted to mitochondria when expressed in eukaryotic cells suggesting that signals in these proteins are functional in eukaryotic cells for targeting to and assembly in mitochondria (15–18). In addition, previous studies failed to identify a linear sequence that functions as an intracellular targeting signal for mitochondrial β-barrel proteins (9). These findings indicate that the signal for targeting of β-barrel precursors to mitochondria is probably not confined to a single linear sequence but rather involves structural elements unique to membrane-embedded β-barrel proteins. Supporting this assumption is a report that the chloroplast β-barrel protein Oep24 was integrated into the mitochondrial OM upon its expression in yeast cells (19). However, the mechanism of such assembly was not studied, and therefore, it is not clear whether a chloroplast β-barrel can be recognized and processed by the same elements that mediate the biogenesis of mitochondrial β-barrel proteins.
To better understand the specific targeting of these proteins, we expressed the chloroplast β-barrel proteins Oep37 and Oep24 in yeast cells and studied their biogenesis in this system. The proteins were located exclusively in mitochondria where they assembled into the OM. In vitro experiments revealed that both Oep24 and Oep37 were first translocated across the OM by the TOM complex and then integrated into the outer membrane by the TOB complex. Collectively, our results suggest that chloroplast β-barrel proteins can be imported into the OM in a similar pathway to that undertaken by the bona fide mitochondrial β-barrel proteins. Thus, these findings imply the following: (i) the mitochondrial import machinery does not recognize signals that are unique to mitochondrial β-barrel proteins, and (ii) dedicated targeting factors had to evolve in plant cells to avoid mis-targeting of these proteins to mitochondria.
EXPERIMENTAL PROCEDURES
Yeast Strains and Growth Methods
Standard genetic techniques were used for growth and manipulation of yeast strains. The wild-type strains YPH499 and W303α were employed. The tom20Δ, mas37Δ, and tim8Δ/tim13Δ strains were described before (Refs. 17, 20, 21, respectively). The tom70Δ/tom71Δ double-deletion strain is a kind gift of Dr. K. Okamoto (22). For drop-dilution assays, yeast cells were grown in synthetic medium to an A600 of 1.0 and diluted in 5-fold increments, and then 5 μl of each dilution were spotted onto solid media, and growth was monitored for a few days.
Recombinant DNA Techniques
Oep37 and Oep24 were amplified from pea cDNA using standard PCR techniques and subsequently cloned into pGEM4 for cell-free transcription and translation. In addition, Oep37- and Oep24-encoding sequences were cloned into the yeast expression vectors pYX242 or pYX142, respectively. VDAC1 was amplified from A. thaliana cDNA and cloned into pGEM4 for in vitro transcription and translation. ALDH and OE33 constructs for in vitro import experiments were previously described (23, 24).
The constructs for the self-assembly GFP assays were amplified from A. thaliana cDNA using standard PCR techniques and subsequently cloned into the pAVA plasmid (25) containing the fragments for saGFP11 (N-/C-terminal) or saGFP(1–10). Templates for the saGFP(1–10) and saGFP11 fragments were obtained from Dr. G. S. Waldo (Los Alamos, NM).
Mitochondria Isolation and Subcellular Fractionations of Yeast Cells
Mitochondria were isolated from yeast cells grown on galactose-containing medium by differential centrifugation as described (26). For isolation of mitochondria from temperature-sensitive mutants and their parental strains, cells were grown at 24 °C, unless otherwise stated. For highly pure mitochondria, a Percoll gradient purification was performed. For that goal, isolated mitochondria were layered on top of a self-forming gradient (25% Percoll in an SEM buffer (250 mm sucrose, 1 mm EDTA and 10 mm MOPS, pH 7.2)) and centrifuged (80,000 × g, 45 min, 4 °C) (16). Mitochondrial fraction from the lower third of the gradient was collected and resuspended in 30 ml of SEM buffer and reisolated by centrifugation (15,000 × g, 15 min, 4 °C). Microsomes were isolated from yeast cells by differential centrifugation. After obtaining the first mitochondrial pellet, the supernatant was centrifuged (15,000 × g, 15 min, 4 °C) again to avoid contaminations by mitochondrial elements. The post-mitochondrial fraction was subjected to a centrifugation (100,000 × g, 1 h, 4 °C), and the pelleted microsomes were resuspended in SEM buffer.
Biochemical Procedures
Radiolabeled precursor proteins were synthesized in rabbit reticulocyte lysate in the presence of [35S]methionine (PerkinElmer Life Sciences) after in vitro transcription by SP6 polymerase from pGEM4 vector (Promega). Radiolabeled precursor proteins were incubated at 25 °C with isolated yeast mitochondria in an import buffer (250 mm sucrose, 0.25 mg/ml BSA, 80 mm KCl, 5 mm MgCl2, 10 mm MOPS-KOH, 2 mm NADH, 2 mm ATP, pH 7.2). Organelles isolated from mas37Δ and TIM10-1 strains or from their corresponding parental strains were incubated at 37 °C for 15 min before initiating the import reaction.
Plant Organelle Isolation and Biochemical Assays Employing These Organelles
Import of the translated 35S-labeled precursor proteins into isolated pea chloroplasts was performed as described before (27). Pea mitochondria for single import assays were isolated according to published procedure (28), and single in vitro import experiments were done as described previously (29). After import (30 min, RT), the organelles were reisolated and subsequently treated with 0.1 m Na2CO3, pH 11.5, with or without the addition of Triton X-100 (1%) and incubated for 30 min on ice. Subsequently, they were centrifuged (100,000 × g, 20 min, 4 °C), and the pellet fractions were analyzed by SDS-PAGE and autoradiography.
Isolation of pea chloroplasts and mitochondria for dual in vitro import was performed according to Rödiger et al. (30). Dual import reactions were done for 30 min at 25 °C as described (31). At the end of the import reactions, the organelles were treated with a final concentration of 120 μg/ml thermolysin or 5 μg/ml proteinase K in dual import buffer supplemented with 50 mm CaCl2 and incubated on ice for 30 or 15 min, respectively. The proteolytic digestion was stopped by 10 mm EDTA, pH 8.0, for thermolysin and 10 mm PMSF for proteinase K. The organelles were repurified and analyzed by SDS-PAGE and autoradiography. For sodium carbonate and Triton X-100 treatment, the organelles were repurified directly after import, subsequently treated with sodium carbonate buffer (0.1 m Na2CO3, pH 11.5, 1 mm EDTA) in the presence or absence of 1% Triton X-100, and incubated on ice for 30 min. The samples were then centrifuged (100,000 × g, 30 min, 4 °C), and the pellet fractions were analyzed by SDS-PAGE and autoradiography. The purity of organelles after dual import was analyzed using one-half of each import reaction sample for immunodecoration with antibodies against psToc75 and atVDAC1 (Agrisera).
Proteolytic digestion of pea chloroplast outer envelope vesicles (isolated according to Ref. 32) was performed as described before (33). The samples were incubated on ice with thermolysin or PK for 30 or 15 min, respectively. Envelope vesicles were recovered by centrifugation (100,000 × g, 30 min, 4 °C) and subsequently analyzed via SDS-PAGE and immunodecoration.
Protoplast Isolation, Transfection, and saGFP Analysis
A. thaliana mesophyll protoplasts were isolated and transfected as described (33). GFP and chloroplast fluorescence was monitored by confocal laser scanning microscopy using a TCS SP5 microscope (Leica) with an HCX PL APO CS 40 × 1.25 NA 1.25 oil objective. Fluorescence was excited and detected as follows: GFP 488/505–525 nm, chlorophyll fluorescence 514/650–750 nm.
RESULTS
Chloroplast β-Barrel Protein Oep37 Is Assembled into the Mitochondrial OM
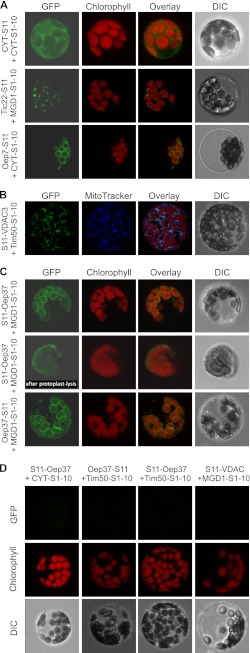
To understand how specific targeting of β-barrel proteins can be achieved in plant cells, we used the model protein Oep37. To study the location of this protein in vivo, we utilized the recently established sa-GFP system where the first 10 β-strands of GFP are fused to one protein while the complementing 11th β-strand is attached to another protein. Only if the two proteins, to which the GFP fragments are fused, are located in the same cellular compartment can a GFP signal be observed (34). As expected, if both fragments were in the cytosol or the IMS of chloroplasts was a GFP signal observed in the corresponding compartment (Fig. 1A). Similarly, a signal was obtained when both parts of the GFP were localized in the mitochondrial IMS (Fig. 1B). Next, strand 11 of GFP was fused to either the N or C terminus of Oep37, and the fusion protein was co-expressed with Mdg1(S1–10) that positions strands 1–10 in between the outer and inner membranes (33). Both combinations resulted in a distinct GFP staining of the chloroplasts (Fig. 1C). Of note, no signal was observed when the fragment containing strands 1–10 was located either in the cytosol or in mitochondria (Fig. 1D). As expected, also the control combination of Mdg1(S1–10) in chloroplasts and S11-VDAC3 in mitochondria did not result in a GFP signal (Fig. 1D). Further control experiments demonstrated that false-positive signal was not observed in other cases where both fragments were not in the same compartment (Fig. 2). Thus, Oep37 is located in vivo solely in chloroplast OM where its two termini are facing the intermembrane space. Interestingly, Oep37 was detected in isolated chloroplasts before (1, 2), but it was predicted to have converse topology (1). Thus, to validate our method, we fused the S11 fragment to the C terminus of the single-span membrane protein Oep7 that was reported to have a Cout orientation (35). As expected, co-expression of this protein with the cytosolic S1–10 fragment gave a GFP signal (Fig. 1A, bottom panel). Thus, the absence of a signal upon co-expression of Oep37-S11 and cytosolic S1–10 is not a technical problem of the used method but rather results from the Nin/Cin topology of the protein.
FIGURE 1.
Chloroplast β-barrel protein Oep37 is exclusively targeted in vivo to chloroplast outer envelopes and possesses an Nin-Cin topology. The indicated constructs were co-transfected into A. thaliana protoplasts that were subsequently analyzed by confocal fluorescence microscopy. The GFP fluorescence (GFP), the autofluorescence of chlorophyll, the overlay of all fluorescence signals, and the differential interference contrast image (DIC) are shown for a representative example. A, two saGFP fragments were targeted to the cytoplasm (CYT-S11 and CYT(1–10), top panel), to the chloroplasts IMS (Tic22-S11 and Mgd1(S1–10), middle panel), or to the outer envelope of chloroplasts (Oep7-S11 and CYT(S1–10), bottom panel). B, two saGFP fragments were targeted to the mitochondrial IMS (S11-VDAC3 and Tim50(S1–10)). A staining of mitochondria with MitoTracker is shown. C, Oep37 either N- or C-terminally fused to saGFP11 (S11-Oep37 or Oep37-S11, respectively) was co-expressed with the chloroplast IMS-located Mgd1(S1–10). The middle panel shows an isolated chloroplast after osmolysis of protoplasts. D, S11-Oep37 fusion proteins were co-expressed with either cytosolically localized S1–10 (CYT(1–10)) or with S1–10 located in the mitochondrial IMS (Tim50(S1–10)). As another control, VDAC3 fused to saGFP11 (S11-VDAC3) was co-expressed with the chloroplast IMS-located Mgd1(S1–10).
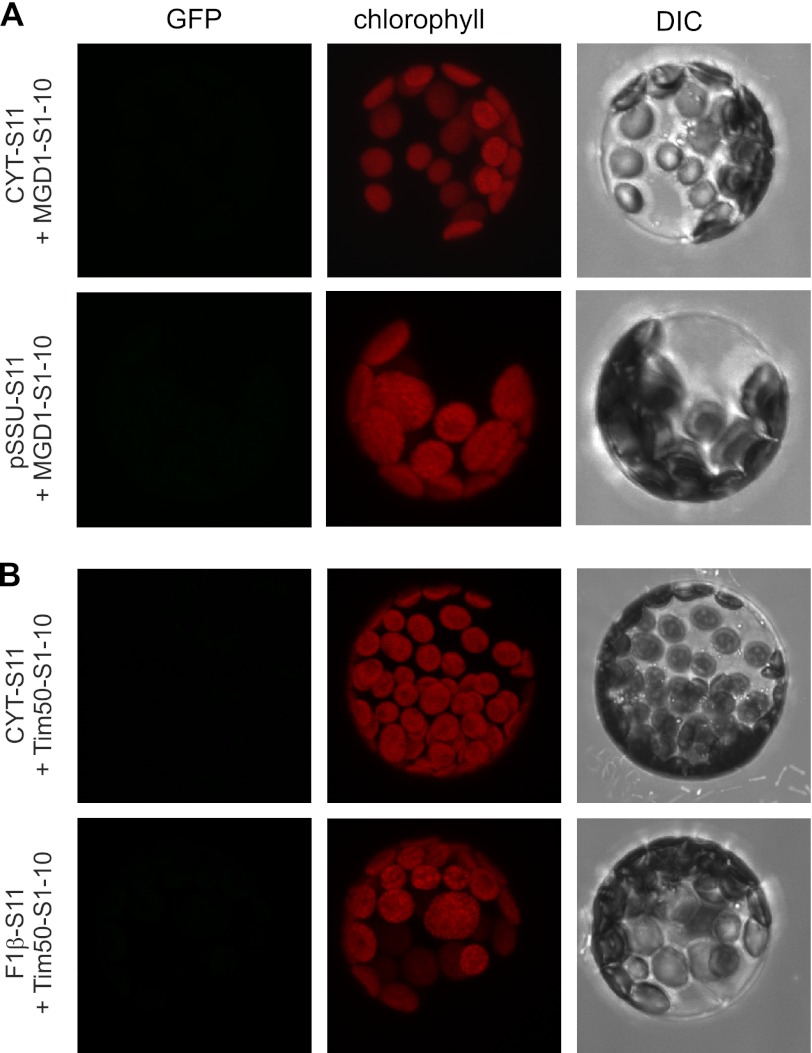
FIGURE 2.
GFP signal is not observed when the two fragments are not in the same compartment. The indicated constructs were co-transfected into A. thaliana protoplasts that were subsequently analyzed by confocal fluorescence microscopy. The GFP fluorescence after GFP assembly (GFP), the autofluorescence of chlorophyll, and the differential interference contrast image (DIC) are shown for a representative example. A, large fragment was targeted to the chloroplast IMS (MGD1(S1–10)), whereas the small one was targeted to either the cytosol (CYT-S11) or the chloroplast stroma (pSSU-S11). B, large fragment was targeted to the mitochondrial IMS (Tim50(S1–10)), whereas the small one was targeted to either the cytosol (CYT-S11) or the mitochondrial matrix (F1β-S11).
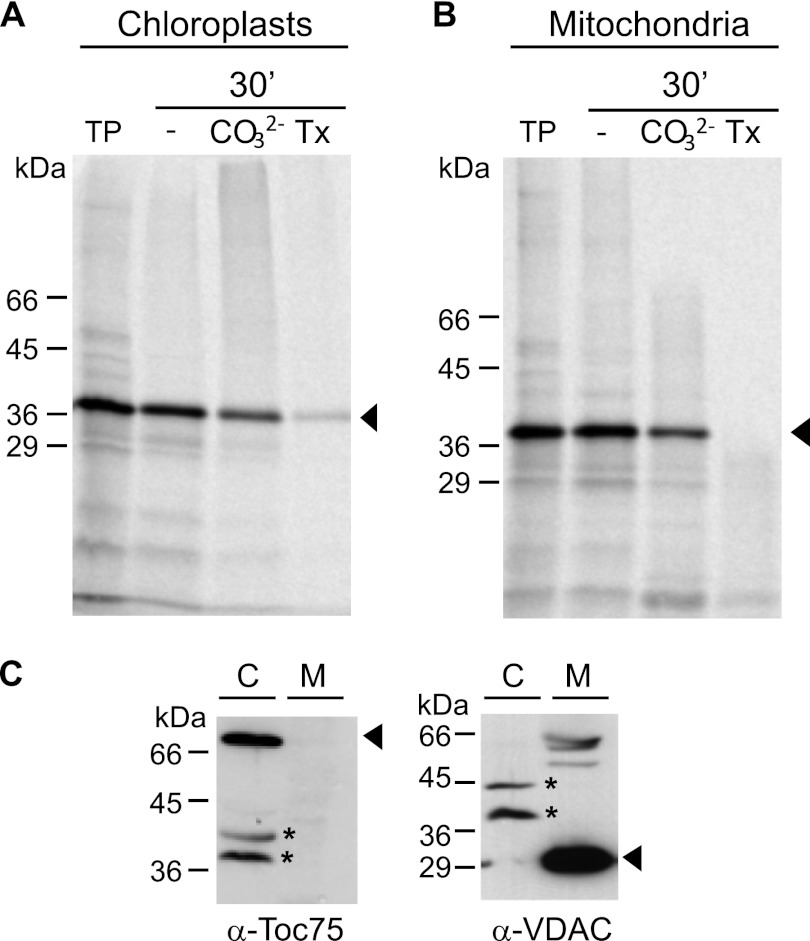
To further study the biogenesis of Oep37, we used a cell-free import system where radiolabeled Oep37 was mixed with either chloroplasts or mitochondria isolated from pea cells. Integration of Oep37 into the target membrane was monitored by alkaline extraction. Surprisingly, the protein was imported into both compartments in a similar efficiency (Fig. 3, A and B). To make sure that the samples used in the in vitro experiments are not cross-contaminated, we analyzed them by immunodecoration with antibodies against marker proteins. Of note, signals corresponding to chloroplast Toc75 and mitochondrial VDAC were observed only in the chloroplasts or mitochondrial samples, respectively (Fig. 3C).
FIGURE 3.
Oep37 can be imported in vitro into chloroplasts and mitochondria. Radiolabeled Oep37 was incubated for 30 min at RT with either isolated pea chloroplasts (A) or isolated mitochondria (B). Membrane insertion was assayed by the resistance to carbonate treatment (CO32−) in the presence or absence of Triton X-100 (Tx). An aliquot (5%) of the translation product (TP) used for each import reaction was loaded as control. The bands corresponding to Oep37 are indicated with an arrowhead. C, purity of the isolated mitochondria (M) or chloroplasts (C) was monitored by analyzing 10 μg of total organellar protein by SDS-PAGE followed by immunodecoration, using organelle-specific antibodies (α-VDAC for mitochondria and α-Toc75 for chloroplasts). The bands corresponding to the marker proteins are indicated with an arrowhead, whereas unspecific bands resulting from cross-reactivity of the antibodies are marked with an asterisk.
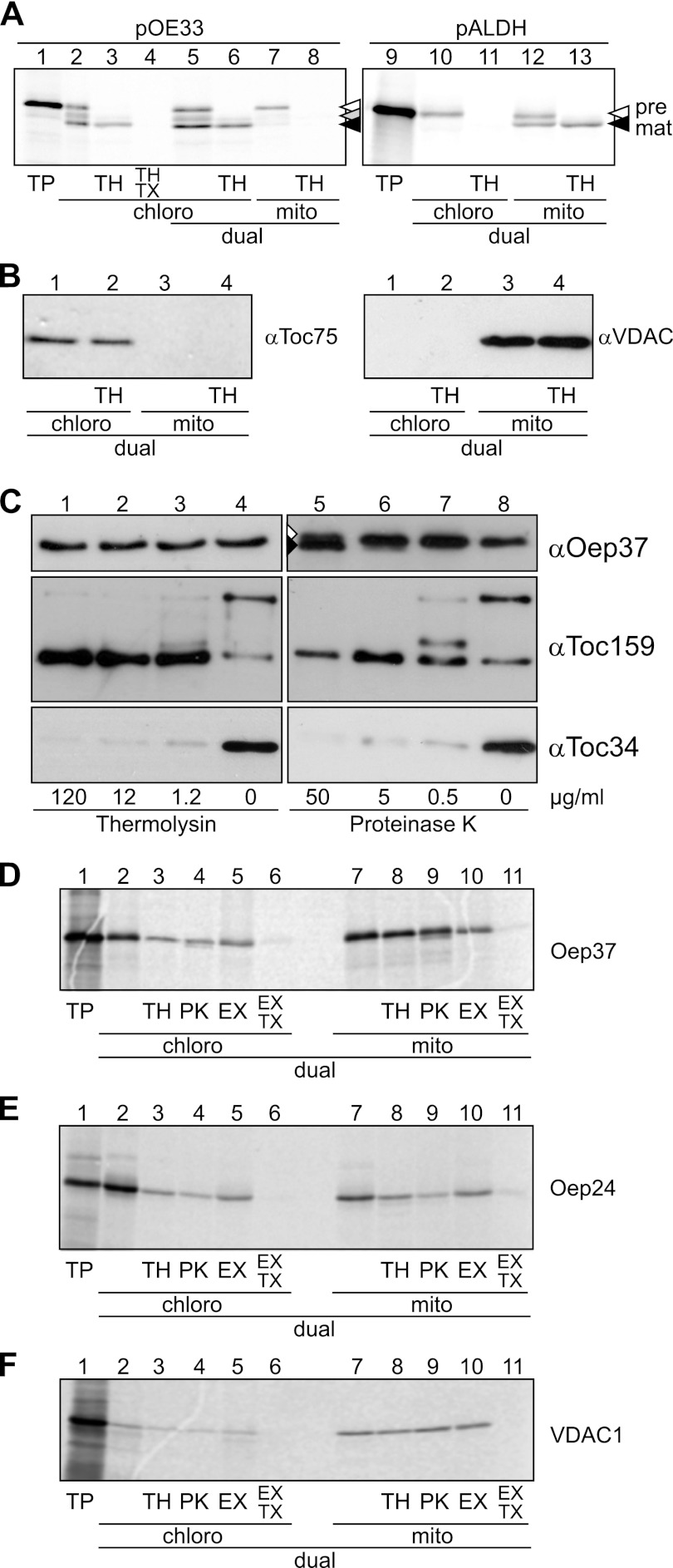
The aforementioned binding experiments were not performed under competitive conditions, and it was often reported that mitochondrial and chloroplast proteins can be imported in vitro into the wrong organelle (36, 37). Therefore, we wanted to exclude the possibility that the rather hydrophobic Oep37 molecules are inserted in vitro into mitochondria simply because this is the only membrane present in the binding reaction. To test the specificity of the binding, we asked whether Oep37 would insert into mitochondria also when competing chloroplasts are present. To this end, we employed a dual system where both isolated chloroplasts and mitochondria are present during the import reaction and are separated only at its end. To check whether the specificity of import is kept under these conditions, we incubated the organelle mixture with radiolabeled precursors of either the chloroplast thylakoid protein pOE33 or the mitochondrial protein pALDH. The results shown in Fig. 4A demonstrate that each precursor protein was imported into its corresponding organelle. Next, we controlled by Western blotting for the purity of the organelles after their separation and found that the chloroplast protein Toc75 was detected only in the chloroplasts fraction, whereas the mitochondrial protein VDAC was found exclusively in the mitochondrial fraction (Fig. 4B). These results validate the dual import system as a specific and reliable assay to monitor the import of precursor proteins.
FIGURE 4.
Oep37 and Oep24 are targeted in vitro in a dual system to the OM of mitochondria and chloroplasts. A, single (lanes 2–4) and dual (lanes 5–13) import of rabbit reticulocyte-translated pOE33 or pALDH into isolated pea chloroplasts (chloro) (lanes 2–6, 10, and 11) and mitochondria (mito) (lanes 7 and 8 and 12 and 13). Translation products (TP, 10%) as input control are shown in lanes 1 and 9. Nonimported proteins were removed by thermolysin (TH, lanes 3, 6, 8, 11, and 13). In addition, 1% (v/v) Triton X-100 was added to one sample (TX, lane 4). Precursor and mature forms of pOE33 and pALDH are indicated by white and black arrowheads, respectively. The stromal intermediate form of pOE33 is indicated with a gray arrowhead. B, purity of the chloroplasts (lanes 1 and 2) and mitochondrial (lanes 3 and 4) fractions after dual import and re-purification of the organelles was assayed by Western blotting with the indicated organelle-specific antibodies. C, chloroplast outer envelope membranes were treated with the indicated concentrations of either thermolysin (lanes 1–4) or proteinase K (lanes 5–8). The membranes were then assayed by Western blotting using the indicated organelle-specific antibodies. Oep37 was partially sensitive to high amounts of proteinase K, and in addition to full-length protein, a slightly smaller degradation product was also observed (lane 5, white and black arrowheads, respectively). D–F, dual import of radiolabeled Oep37 (D), Oep24 (E), and VDAC1 (F) into chloroplasts and mitochondria (lanes 2–11). Translation products (TP, 10%) as input control are shown in lane 1. Nonimported proteins were removed by either thermolysin (TH, lanes 3 and 8) or proteinase K (PK, lanes 4 and 9) treatment. Full integration into the membrane was assayed by carbonate extraction (EX) in the presence or absence of Triton X-100 (TX, lanes 5, 6, 10, and 11).
We aimed to establish an assay to monitor the in vitro import of the chloroplast β-barrel protein Oep37. To this end, we investigated the protease resistance of the endogenous protein in isolated organelles. Oep37 was resistant against the used amounts of both PK and thermolysin. In contrast, as expected, the exposed receptors Toc159 and Toc34 were cleaved under these conditions (Fig. 4C). The protease resistance of Oep37 under these conditions provides further support for the notion that both termini of Oep37 are in the intermembrane space of chloroplasts. The earlier proposal that the N and C termini are exposed to the cytosol was based only on proteolysis of outer envelope membranes with very high concentrations of thermolysin (1). As contrast, our current model is based on improved methodology, namely in vivo data with intact cells and proteolytic assay with reduced proteases concentrations.
Using such proteolytic assays and carbonate extraction, we then analyzed the import of Oep37 in the dual import system. In agreement with our results in the single organelle system (Fig. 3), the protein became protected from proteases upon import into both organelles (Fig. 4D). Furthermore, Oep37 was found in the pellet fraction of an alkaline extraction as expected from membrane-embedded proteins. Triton X-100 at concentrations up to 2% is frequently used to wash protein from inclusion bodies that are aggregates of non-native proteins (38). Thus, we applied the previously established principle of Triton X-100 treatment (39) to test whether the protein was indeed inserted into the membrane or just co-sedimented as a nonmembrane-inserted aggregate. Oep37 was not detected in the pellet when we performed the extraction in the presence of the detergent Triton X-100, excluding the possibility of aggregation as a cause for the appearance in the pellet (Fig. 4D). As Oep24 was previously reported to be targeted to mitochondria upon its expression in yeast cells (19), we also imported this protein in the dual system. Similarly to Oep37, Oep24 was integrated into the membrane of both organelles (Fig. 4E). Of note, under these conditions the mitochondrial β-barrel protein VDAC was efficiently imported into mitochondria but only sparsely into chloroplasts (Fig. 4F). The mitochondria and chloroplast fractions were also analyzed after their re-isolation by SDS-PAGE followed by Coomassie staining. This analysis revealed that similar amounts of proteins were contained in each fraction (data not shown), thus excluding the possibility that unequal amounts of proteins affect the import efficiencies. Taken together, our findings suggest that chloroplasts β-barrel proteins can be imported in vitro into isolated mitochondria.
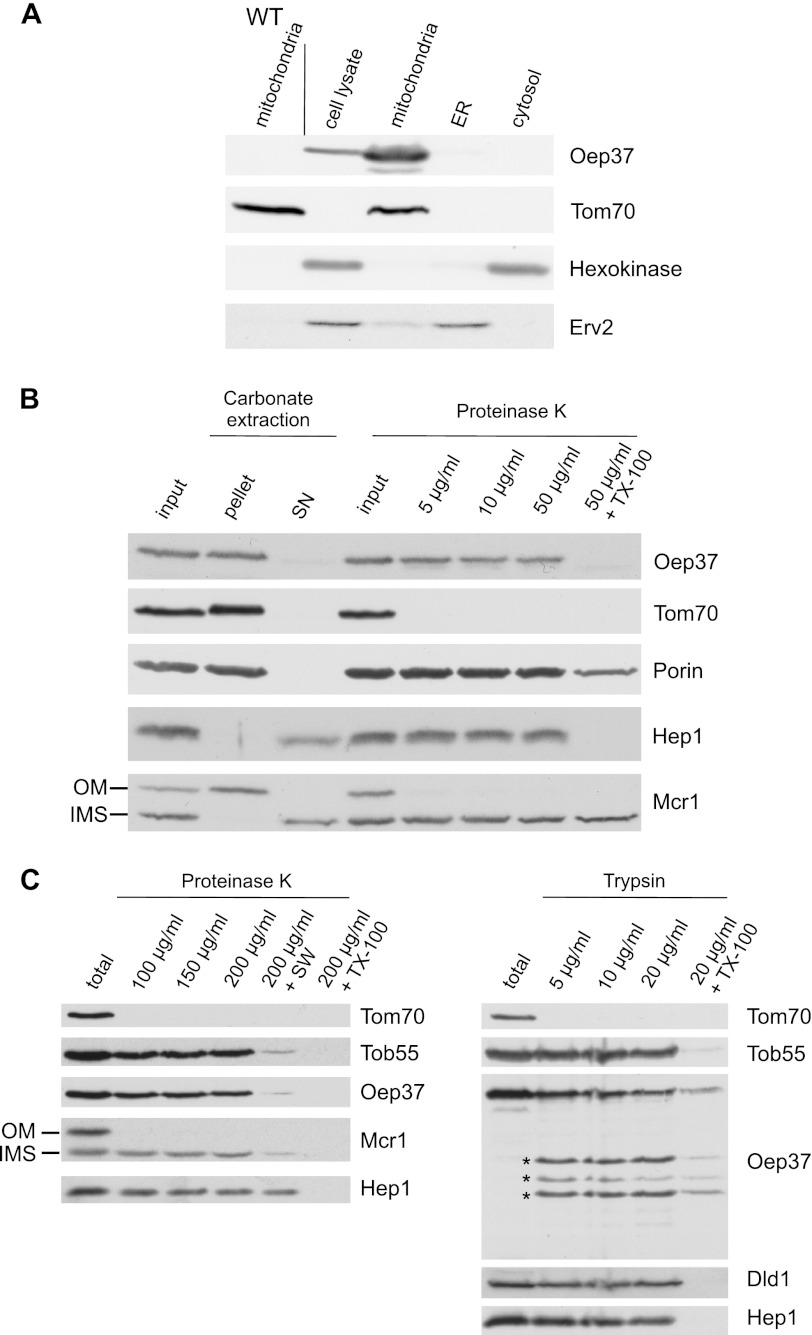
Next, we were interested whether this mitochondrial targeting capacity can also be observed in an in vivo system. We therefore cloned Oep37 into a yeast expression vector and transformed the resulting plasmid into S. cerevisiae cells. Subcellular fractionation of the transformed cells revealed that Oep37 was located exclusively in the mitochondrial fraction (Fig. 5A). As a control for the specificity of the Oep37 antibody, we verified the absence of a signal in mitochondria isolated from a nontransformed strain (Fig. 5A, left lane). Hence, although several different membranes are available for the newly synthesized Oep37 molecules upon their translation in the yeast cytosol, the protein is targeted solely to mitochondria.
FIGURE 5.
Oep37 expressed in yeast cells is assembled into the mitochondrial OM in a native conformation. A, Oep37 is located in mitochondria. Lysates of Oep37-expressing cells and fractions corresponding to mitochondria, endoplasmic reticulum (ER), and cytosol were analyzed by SDS-PAGE and immunodecoration with antibodies against Oep37, the mitochondrial protein Tom70, a marker protein for the cytosol (hexokinase), and the endoplasmic reticulum protein Erv2. Mitochondria isolated from untransformed WT cells were co-analyzed as a control. B, mitochondria isolated from cells expressing Oep37 were analyzed directly by SDS-PAGE (input) or were subjected first to carbonate extraction and then centrifuged to discriminate between membrane proteins in the pellet and soluble proteins in the supernatant (SN). Additional aliquots of mitochondria were left intact or were treated with the indicated amounts of proteinase K. Proteins were analyzed by SDS-PAGE and immunodecorated with antibodies against the indicated proteins as follows: Tom70, an OM protein exposed to the cytosol; Porin, a protein embedded in the OM; Hep1, a mitochondrial soluble matrix protein; Mcr1, a protein with two isoforms, a 34-kDa species exposed on the OM and a 32-kDa soluble one in the IMS. C, mitochondria isolated from cells expressing Oep37 were left intact (total) or were treated with the indicated amounts of proteinase K (left panel) or trypsin (right panel). In one sample, the mitochondria were swelled (+SW) before the treatment with PK. Proteins were analyzed by SDS-PAGE and immunodecorated with antibodies against the indicated proteins. Tob55, a protein embedded in the OM; Dld1, an inner membrane protein exposed to the IMS. Proteolytic fragments are indicated with an asterisk.
To demonstrate that Oep37 was indeed integrated into the outer membrane rather than simply attached on the surface of the organelle, mitochondria were subjected to alkaline extraction. Oep37 was found in the pellet fraction similarly to other membrane-embedded mitochondrial proteins like Tom70 or Porin (Fig. 5B). In contrast, soluble proteins like Hep1 and the IMS isoform of Mcr1 were found in the supernatant fraction (Fig. 5B). Because we observed that native Oep37 is resistant to PK treatment in intact chloroplasts (Fig. 4C), the membrane integration of Oep37 was further analyzed by a similar treatment of intact mitochondria. Similarly to mitochondrial β-barrel proteins like Porin and Tob55, Oep37 was unaffected by addition of external protease to intact mitochondria. Oep37 became accessible to proteinase K only when mitochondrial membranes were solubilized with the detergent, Triton X-100 (Fig. 5, B and C). Similarly, rupturing of the outer membrane by osmotic swelling caused exposure of loops in the IMS to proteinase K and disappearance of the protein signal (Fig. 5C, left panel). As the experiments described so far cannot exclude the possibility that Oep37 is integrated into the inner membrane, we treated intact organelles with higher concentrations of PK or with trypsin as the latter protease cleaves other β-barrel proteins like Tom40. Indeed, upon addition of such elevated amounts of PK or trypsin to intact organelles, we observed the formation of proteolytic fragments (Fig. 5C). Such fragments can be formed only if the protein is embedded into the OM and exposes loops toward the cytosol. The intactness of mitochondria under these conditions is reflected by the resistance of the IMS proteins DLD1 and the IMS isoform of Mcr1 as well as the matrix protein Hep1 to the proteases treatment (Fig. 5C). We conclude that Oep37 is assembled into the mitochondrial OM in a native-like conformation.
Oep37 Can Complement the Absence of Mitochondrial Porin
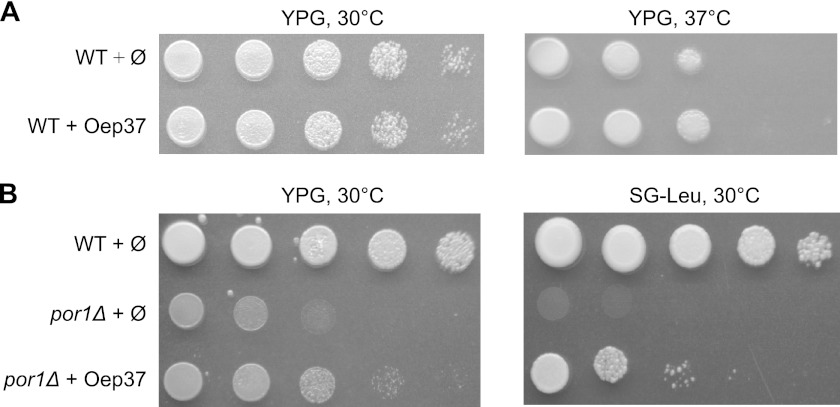
We further investigated whether the expression of Oep37 interferes with crucial functions of mitochondria. To that end, we compared the growth rate of yeast cells expressing the chloroplast protein to those bearing an empty plasmid. The growth of Oep37-expressing cells on a nonfermentable carbon source, where yeast cells require fully functional mitochondria, was comparable with that of control cells (Fig. 6A). Next, we verified that expressing Oep37 in yeast cells did not have any effect on the morphology of the organelle (data not shown). Collectively, it seems that the expression of Oep37 does not obstruct crucial cellular and mitochondrial processes.
FIGURE 6.
Overexpression of Oep37 can partially complement the por1Δ phenotype. A, expression of Oep37 does not interfere with growth on a nonfermentable carbon source. Cells harboring either a plasmid encoding Oep37 or an empty plasmid (Ø) as control were tested by drop dilution assay for their ability to grow on glycerol-containing medium (YPG) at 30 and 37 °C. B, wild-type cells harboring an empty plasmid (Ø) and cells deleted for POR1 (por1Δ) that were transformed with either an empty plasmid (Ø) or a plasmid encoding Oep37 were tested by drop dilution assay for their ability to grow at 30 °C on rich (YPG) or synthetic glycerol-containing medium (SG-Leu).
Oep37 was reported to form a rectifying high conductance channel in artificial membranes (2). Thus, we asked whether this protein can complement the absence of the general solute transporter of the mitochondrial outer membrane, Porin (also called VDAC in higher eukaryotes). Cells lacking Porin hardly grow on a nonfermentable carbon source at elevated temperatures (40). We observed that por1Δ cells expressing Oep37 partially regained their capacity to grow under these conditions (Fig. 6B). This finding suggests that Oep37 can form pores in the mitochondrial outer membrane. Of note, another chloroplast OM protein, Oep24, was previously shown to partially complement Porin deficiency in yeast cells (19). Thus, our current findings indicate that although Oep24 and Oep37 probably have different substrate specificity in chloroplast membranes, they share the ability to complement the function of Porin that serves as the single and general solute transporter in the yeast mitochondrial OM (40).
Oep37 Assembly into Mitochondria Requires the TOM and TOB Complexes
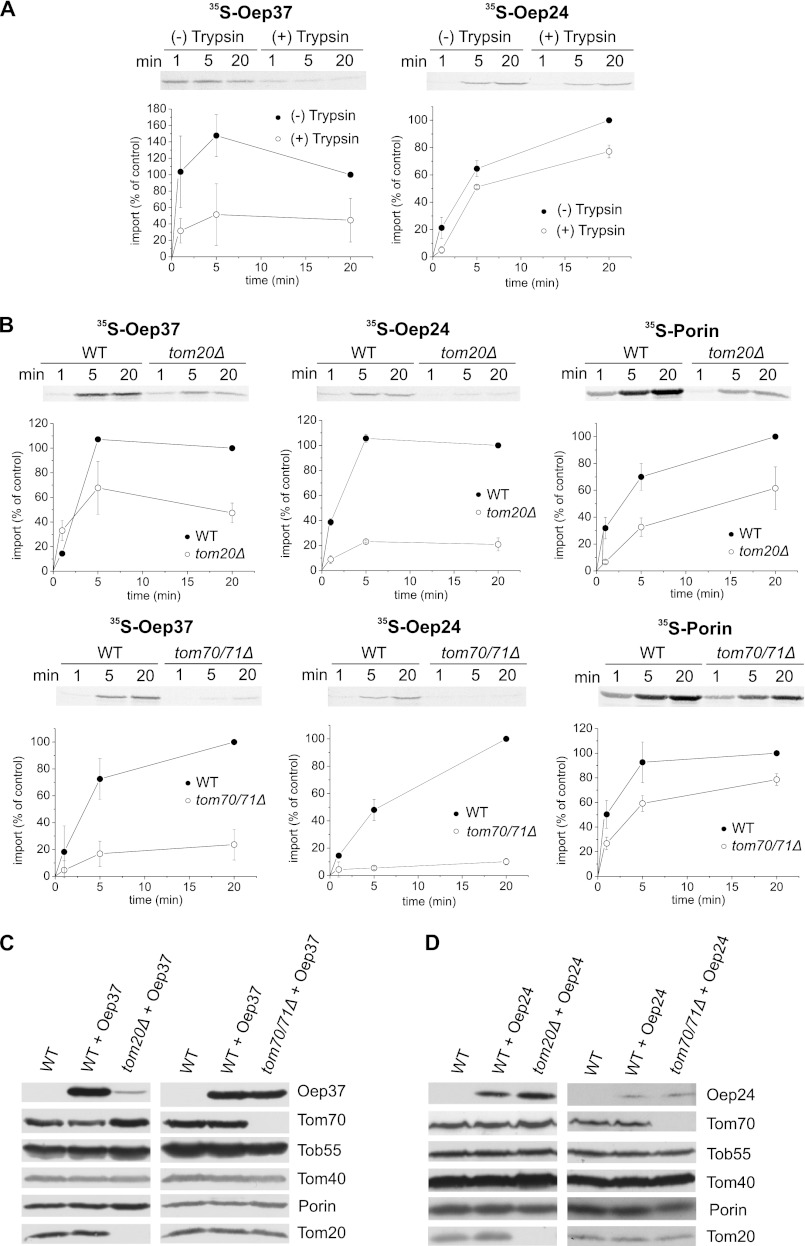
Mitochondrial β-barrel proteins like Tom40, Porin, and Tob55 or bacterial β-barrel proteins expressed in yeast cells are initially recognized by the import receptors Tom20 and Tom70 (16, 17, 20, 41–43). Hence, we used an in vitro import assay to address the importance of these receptors for the mitochondrial assembly of Oep37 and Oep24. Membrane integration of the precursor molecules was analyzed by monitoring those molecules that are proteinase K-resistant. Initially, we observed that the removal of the exposed domains of import receptors by externally added trypsin affected the in vitro import of both proteins into isolated mitochondria (Fig. 7A). To verify the importance of the import receptors, we imported newly synthesized Oep37 or Oep24 molecules into mitochondria isolated from strains lacking either Tom20 or Tom70 and its low abundant paralog Tom71. The import of newly synthesized chloroplast β-barrels and the control mitochondrial β-barrel protein Porin into both types of mutated mitochondria was strongly compromised (Fig. 7B). To further study the importance of the receptors, we transformed plasmids encoding either Oep37 or Oep24 into cells lacking either Tom20 or Tom70/Tom71 and analyzed the steady-state levels in these mutated cells. Crude mitochondria isolated from tom20Δ cells had significantly reduced amounts of Oep37 but wild-type-like levels of Oep24 (Fig. 7, C and D). In contrast, the absence of Tom70/71 caused only a slight reduction or none at all in the observed levels of Oep37 and Oep24, respectively (Fig. 7, C and D). The reduced Tom20 dependence of the in vivo biogenesis of Oep24 might be related to its smaller size. We previously observed that upon their expression in yeast cells the biogenesis of small bacterial β-barrel proteins is Tom20-independent whereas that of their larger counterparts required the presence of this receptor (17). We assume that the higher dependence on Tom70/71 in the in vitro system is due to the use of the reticulocyte lysate in these experiments and the function of Tom70 as an anchor for chaperones present in this lysate. Similar dependence on Tom70 was observed for the import of multispan proteins of the outer and inner mitochondrial membranes (44–47).
FIGURE 7.
Oep37 and Oep24 require the mitochondrial import receptors for their assembly into the OM. A, isolated mitochondria were left intact or pretreated with trypsin followed by re-isolation of the organelles. Next, radiolabeled precursor of either Oep37 or Oep24 was incubated with the trypsin-treated or intact mitochondria for the indicated time periods. At the end of the import reactions, samples were treated with PK, and proteins were analyzed by SDS-PAGE and autoradiography. The insertion of the proteins was quantified by analyzing the PK-protected molecules. The amount of precursor proteins imported into intact mitochondria for 20 min was set to 100%. An autoradiographic representative of three independent repeats and quantification of three independent experiments is presented. B, radiolabeled precursors of Oep37, Oep24, and Porin (as a control) were imported into mitochondria isolated from either tom20Δ or tom70/71Δ and their corresponding WT strains. Imported proteins were analyzed and quantified as described in the legend to A. C and D, mitochondria isolated from nontransformed WT cells and those isolated from either tom20Δ or tom70/71Δ and their corresponding WT strains transformed with either Oep37 (C) or Oep24 (D) encoding plasmid were analyzed by SDS-PAGE and immunodecoration with antibodies against Oep37 or Oep24, respectively. In addition, immunodecoration with antibodies against the indicated mitochondrial proteins was performed.
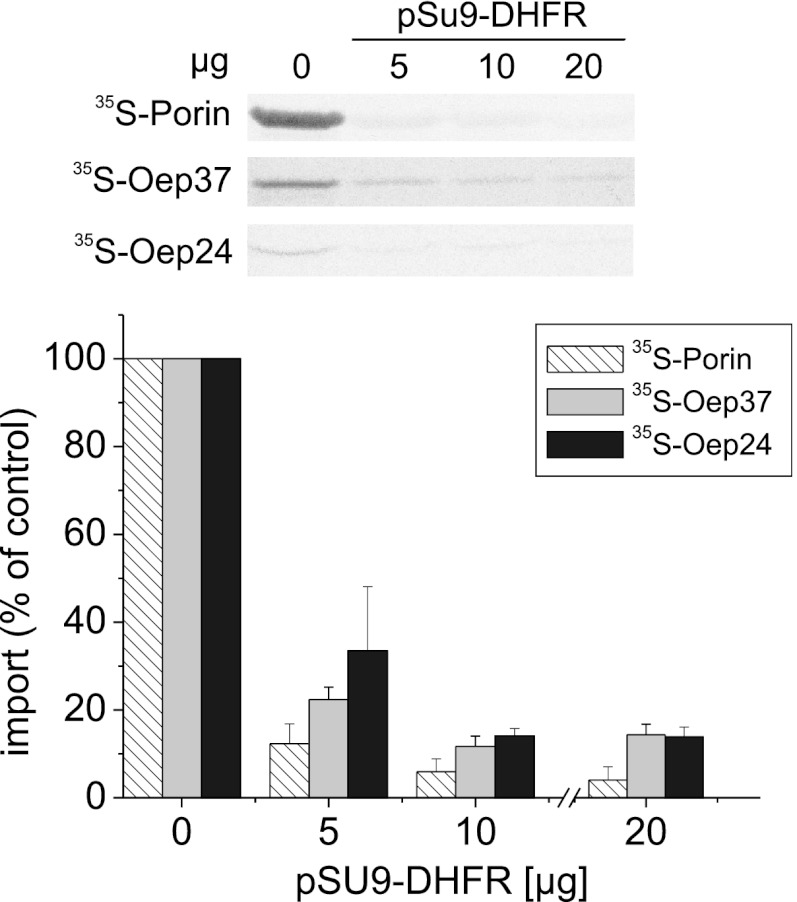
Mitochondrial β-barrel proteins are translocated through the import pore of the TOM complex before their insertion into the mitochondrial OM (7–9). Therefore, we asked whether Oep37 and Oep24 follow a similar pathway. To this end, we added to the in vitro import reaction excess amounts of recombinant matrix-destined precursor (pSu9(1–69)-DHFR), which can block the TOM pore and thus compete with import of other TOM-dependent precursor proteins. This addition resulted in a significant reduction in the membrane integration of Oep37, Oep24, and of Porin as a control (Fig. 8). Hence, it appears that the TOM import pore is used in the membrane-assembly pathway of Oep37 and Oep24.
FIGURE 8.
Integration of Oep37 and Oep24 into the mitochondrial OM requires the TOM import pore. Radiolabeled precursors of Oep37, Oep24, and Porin (as a control) were imported into mitochondria in the absence or presence of the indicated amounts of recombinant pSu9-DHFR. Imported proteins were analyzed and quantified as described in the legend to Fig. 7A. The amount of precursor proteins imported into mitochondria without added pSu9-DHFR was set to 100%.
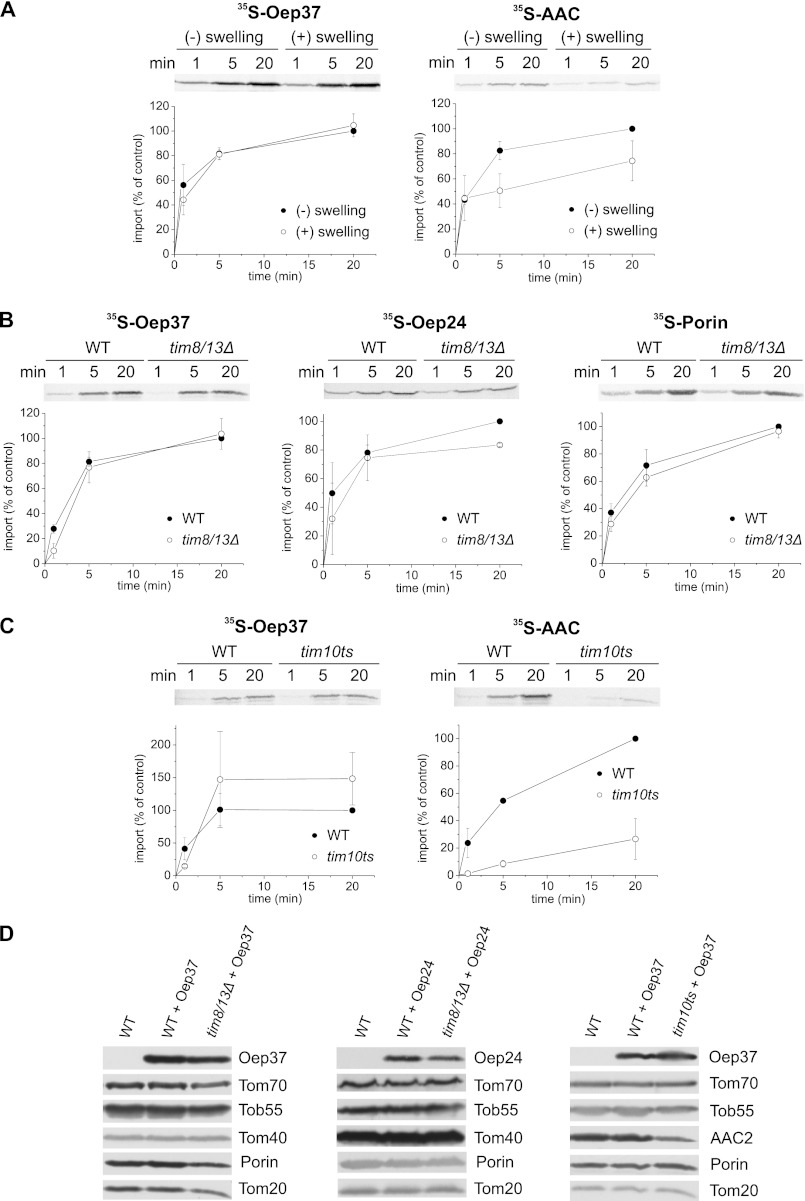
Upon their translocation across the pore of the TOM complex, β-barrel proteins are chaperoned by the small Tim proteins residing in the IMS (17, 20, 48, 49). Therefore, we next investigated whether the chloroplast proteins require these small Tim chaperones for their assembly in the mitochondrial outer membrane. First, we imported Oep37 into mitochondria where the OM was ruptured by osmotic swelling. This treatment results in the release of the small Tims from the IMS that in turn causes a reduction in the assembly efficiency of mitochondrial β-barrel proteins and inner membrane carrier proteins like ADP-ATP carrier (AAC) (Fig. 9A) (20, 49–51). Of note, the membrane integration of Oep37, as monitored by resistance to alkaline extraction, was not compromised in the ruptured organelles, whereas that of AAC was significantly reduced (Fig. 9A). Next, we observed that the capacity of mitochondria isolated from a strain lacking both Tim8 and Tim13 or a strain harboring a temperature-sensitive allele of TIM10 to import in vitro newly synthesized Oep37 molecules was not reduced as compared with that of organelles isolated from wild-type cells (Fig. 9, B and C). Furthermore, Oep37-encoding plasmid was transformed into these mutated cells. Crude mitochondria were isolated from these cells and subjected to SDS-PAGE and immunodecoration. Our results revealed that the observed levels of Oep37 in the mutated cells are very similar to those in the corresponding parental strains (Fig. 9D). The results with Oep24 were slightly different as its import into mitochondria lacking Tim8/13 complex and its levels in cells lacking these proteins were mildly hampered (Fig. 9, B and D). Collectively, these findings suggest that the small Tim proteins play only a minor role, if at all, in the import pathway of Oep24 and Oep37.
FIGURE 9.
Small Tim chaperones have only a minor role in the assembly of Oep37 and Oep24 into the mitochondrial OM. A, rupturing of the outer membrane does not compromise the assembly of Oep37. Radiolabeled precursors of Oep37 and AAC (as a control) were incubated for the indicated time periods with isolated intact mitochondria or with mitochondria that had been subjected to osmotic swelling. After import, mitochondria were pelleted and subjected to alkaline extraction, and the pellet fractions were analyzed by SDS-PAGE followed by autoradiography. The amount of precursor proteins imported into intact mitochondria for 20 min was set to 100%. An autoradiographic representative of three independent repeats and quantification of three independent experiments are presented. B, insertion of Oep37 and Oep24 is hardly affected in mitochondria lacking the Tim8/Tim13 complex. Radiolabeled precursors of Oep37, Oep24, and Porin were imported into mitochondria isolated from either tim8Δ/tim13Δ or its corresponding parental strain. Imported proteins were analyzed and quantified as described in the legends to Fig. 7A. C, insertion of Oep37 is not affected in mitochondria mutated in TIM10. Radiolabeled precursors of Oep37 and AAC were imported into mitochondria isolated from a strain harboring a temperature-sensitive allele of TIM10 (TIM10-1 (58)) or from its corresponding parental strain. Imported proteins were analyzed and quantified as described in the legends to Fig. 7A. D, mitochondria isolated from nontransformed WT cells and those isolated from either tim8/tim13Δ or TIM10-1 and their corresponding parental strains transformed with Oep37- or Oep24-encoding plasmid were analyzed by SDS-PAGE and immunodecoration with antibodies against Oep37 or Oep24, respectively. In addition, immunodecoration with antibodies against the indicated mitochondrial proteins was performed.
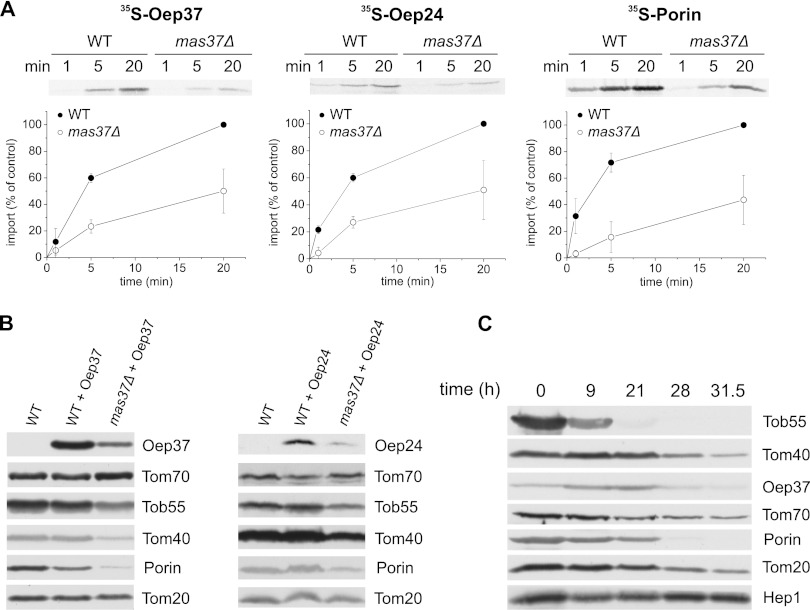
As the TOB complex is absolutely essential for the membrane insertion of mitochondrial β-barrel proteins, we investigated whether Oep37 and Oep24 share this TOB dependence. As anticipated, the in vitro integration of both proteins was heavily compromised in mitochondria lacking Mas37, a peripheral subunit of the TOB complex (Fig. 10A). We further investigated the Mas37 dependence by transforming cells deleted for Mas37 with an Oep37- or Oep24-encoding plasmid. Crude mitochondria were isolated from WT and the mutant cells, and their proteins were analyzed by SDS-PAGE and immunodecoration. In accordance with the in vitro results, this analysis revealed that the steady-state levels of both proteins are strongly reduced in mas37Δ cells (Fig. 10B). To substantiate the dependence on the TOB complex, we expressed Oep37 under the control of the TPI promoter in cells where the essential component Tob55 is under the control of the inducible GAL promoter (52). Growing these cells on glucose results in a gradual reduction in the levels of the essential protein Tob55, which in turn compromises growth of the cells (16, 52). Mitochondria from these Tob55-depleted cells were isolated after growth for various periods on glucose-containing medium, and the levels of mitochondrial proteins were monitored. We also observed in parallel to the gradual reduction of Tob55 a reduction in the other mitochondrial β-barrel protein Tom40 and Porin (Fig. 10C). Importantly, the amounts of Oep37 were also compromised upon the depletion of Tob55 (Fig. 10C). Taken together, our findings demonstrate the involvement of the TOB complex in the membrane assembly of Oep37 and Oep24.
FIGURE 10.
TOB complex is crucial for the mitochondrial integration of Oep37 and Oep24. A, radiolabeled precursors of Oep37, Oep24, and Porin were imported into mitochondria isolated from either WT or mas37Δ strains. Imported proteins were analyzed and quantified as described in the legends to Fig. 7A. B, mitochondria isolated from nontransformed WT cells and those isolated from mas37Δ and their corresponding wild-type cells transformed with either Oep37- or Oep24-encoding plasmid were analyzed by SDS-PAGE and immunodecoration with antibodies against Oep37 or Oep24, respectively. In addition, immunodecoration with antibodies against the indicated mitochondrial proteins was performed. C, Oep37 was transformed into cells expressing Tob55 under the control of the GAL10 promoter. Cells were harvested at the indicated time points after a shift from galactose- to glucose-containing medium. Crude mitochondria were isolated, and proteins were analyzed by SDS-PAGE and immunodecoration with antibodies against Oep37 and the indicated mitochondrial proteins. Tob55, Tom40, and Porin are β-barrel proteins.
DISCUSSION
β-Barrel proteins in modern endosymbiotic organelles evolved most probably from structurally similar proteins in their corresponding ancestors (53, 54). Furthermore, detailed studies on the biogenesis of these proteins in bacteria and mitochondria demonstrated that the central protein component and the basic mechanism in the biogenesis pathway are conserved (9, 55). Accordingly, we and others could previously show that signals in bacterial β-barrel proteins are functional in eukaryotic cells for targeting to and assembly in mitochondria (16, 18). However, it is not clear whether this similarity between two β-barrel containing systems, mitochondria and bacteria, can be extrapolated to the biogenesis of β-barrel proteins in the third membrane that contains such structures, namely the OM of chloroplasts. Thus, in this study we addressed the question whether signals in chloroplast β-barrel proteins can be recognized and processed by the mitochondrial import machinery.
To that goal, the chloroplast β-barrel proteins Oep37 and Oep24 were expressed in yeast cells. Our current results demonstrate that the proteins were assembled into the yeast mitochondrial OM in a process that required the TOM complex and the TOB machinery. Thus, their assembly pathway was similar to the one taken by the bona fide mitochondrial β-barrel proteins. Of note, although Oep37 and Oep24 do not share sequence similarity with endogenous mitochondrial β-barrels, they appear to bear the signals required for recognition by the aforementioned fungal mitochondrial import components. Although the import receptors of plant mitochondria are somewhat altered in comparison with their counterparts in fungal cells, they recognize a similar set of substrate proteins (56). We believe that the plant TOM receptors can also recognize mitochondrial β-barrel proteins. However, because we did not address the recognition of Oep37 by plant TOM receptors, we cannot exclude the unlikely possibility that the in vitro import into plant mitochondria was mediated by other mitochondrial surface proteins.
Our findings underscore the importance of structural elements rather than mitochondrial specific sequences for the biogenesis of β-barrel proteins in mitochondria. They also might imply that the yet to be identified machinery that assembles β-barrel proteins into the chloroplast OM uses similar signals as the mitochondrial counterpart. Collectively, it seems that the evolutionary relations of mitochondrial and chloroplast β-barrel proteins to their bacterial ancestral proteins ensured a certain degree of similarity also among β-barrel proteins from both endosymbiotic organelles. Considering the evolutionary link, it is also tempting to speculate that the principles of β-barrel biogenesis have been conserved from a cyanobacterium to chloroplasts.
An interesting open question is how the precursors of eukaryotic β-barrel proteins are targeted from the cytosol to their target membrane. The signals that facilitate the specific targeting of such precursors to either mitochondria or chloroplasts are not yet characterized. So far, a linear well defined sequence that can function as an intracellular targeting signal was not identified. The only exception is the chloroplast Toc75-III where an N-terminal extension functions as a targeting sequence (10). Thus, it can be assumed that the mitochondrial and chloroplast protein import machineries recognize β-barrel-related structural elements (9, 57). However, this assumption raises the question whether such elements in chloroplast β-barrel proteins are distinct from those in β-barrel proteins destined to mitochondria. Our current results suggest that this is probably not the case as both Oep37 and Oep24 were imported in vitro into plant mitochondria and both in vivo and in vitro into yeast mitochondria.
Conversely, the mitochondrial VDAC protein was imported in vivo and in vitro only to plant mitochondria. One possibility to explain these observations is the difference in the availability of both organelles in plant cells and especially in mesophyll cells. Chloroplasts are predominant in these cells and expose a much larger surface as compared with mitochondria. Hence, the former organelles must ensure that only the correct proteins are inserted. Thus, a bouncing off mechanism in chloroplasts seems to exist. Because of the difference in their surfaces, the likelihood that chloroplast proteins are targeted to mitochondria by free diffusion is rather low, thus mitochondria did not evolve such a mechanism. An additional potential explanation to the absence of import of VDAC into chloroplasts might be its weak affinity to the import receptors of the chloroplasts. As currently it is not clear which proteins recognize β-barrel substrates on the surface of chloroplasts, a detailed study on such recognition depends on future studies that will shed light on this issue.
The aforementioned mechanisms are probably not sufficient to ensure specific targeting. Another potential quality control measure could be the degradation of mis-targeted chloroplast β-barrel proteins by mitochondrial proteases. In any case, it appears that plant cells should not allow those β-barrel proteins destined to chloroplasts any contact with mitochondria as this latter organelle can serve as a default target to all β-barrel proteins. This idea is in line with the common assumption that chloroplasts were integrated into cells that already contained mitochondria. Thus, whereas the mitochondrial β-barrels could follow a general default pathway, the recently arrived chloroplast ones had to develop a mechanism to avoid this destination. Part of such an evading pathway of the chloroplast-destined β-barrels could involve recognition by dedicated factors already in the cytosol. A challenge for future studies will be to identify such putative factors and to understand how they can recognize specifically chloroplast β-barrel proteins.
Acknowledgments
We thank K. Rehn, G. Englich, and E. Kracker for technical support and G. S. Waldo (Los Alamos National Laboratory, Los Alamos, NM) for providing templates for the self-assembling GFP.
This work was supported by the Deutsche Forschungsgemeinschaft Grants RA 1048/4-1 and SFB766/TP B11 (to D. R.) and SFB807/P17 (to E. S.).
- OM
- outer membrane
- IMS
- intermembrane space
- sa
- self-assembly
- AAC
- ADP-ATP carrier
- ALDH
- aldehyde dehydrogenase.
REFERENCES
- 1. Schleiff E., Eichacker L. A., Eckart K., Becker T., Mirus O., Stahl T., Soll J. (2003) Prediction of the plant β-barrel proteome. A case study of the chloroplast outer envelope. Protein Sci. 12, 748–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Goetze T. A., Philippar K., Ilkavets I., Soll J., Wagner R. (2006) OEP37 is a new member of the chloroplast outer membrane ion channels. J. Biol. Chem. 281, 17989–17998 [DOI] [PubMed] [Google Scholar]
- 3. Hemmler R., Becker T., Schleiff E., Bölter B., Stahl T., Soll J., Götze T. A., Braams S., Wagner R. (2006) Molecular properties of Oep21, an ATP-regulated anion-selective solute channel from the outer chloroplast membrane. J. Biol. Chem. 281, 12020–12029 [DOI] [PubMed] [Google Scholar]
- 4. Schleiff E., Maier U. G., Becker T. (2011) Omp85 in eukaryotic systems. One protein family with distinct functions. Biol. Chem. 392, 21–27 [DOI] [PubMed] [Google Scholar]
- 5. Soll J., Schleiff E. (2004) Protein import into chloroplasts. Nat. Rev. Mol. Cell Biol. 5, 198–208 [DOI] [PubMed] [Google Scholar]
- 6. Schleiff E., Becker T. (2011) Common ground for protein translocation. Access control for mitochondria and chloroplasts. Nat. Rev. Mol. Cell Biol. 12, 48–59 [DOI] [PubMed] [Google Scholar]
- 7. Pfanner N., Wiedemann N., Meisinger C., Lithgow T. (2004) Assembling the mitochondrial outer membrane. Nat. Struct. Mol. Biol. 11, 1044–1048 [DOI] [PubMed] [Google Scholar]
- 8. Endo T., Yamano K. (2009) Multiple pathways for mitochondrial protein traffic. Biol. Chem. 390, 723–730 [DOI] [PubMed] [Google Scholar]
- 9. Walther D. M., Rapaport D., Tommassen J. (2009) Biogenesis of β-barrel membrane proteins in bacteria and eukaryotes. Evolutionary conservation and divergence. Cell. Mol. Life Sci. 66, 2789–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tranel P. J., Keegstra K. (1996) A novel, bipartite transit peptide targets OEP75 to the outer membrane of the chloroplastic envelope. Plant Cell 8, 2093–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schleiff E., Klösgen R. B. (2001) Without a little help from 'my' friends. Direct insertion of proteins into chloroplast membranes? Biochim. Biophys. Acta 1541, 22–33 [DOI] [PubMed] [Google Scholar]
- 12. Inoue K., Baldwin A. J., Shipman R. L., Matsui K., Theg S. M., Ohme-Takagi M. (2005) Complete maturation of the plastid protein translocation channel requires a type I signal peptidase. J. Cell Biol. 171, 425–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Eckart K., Eichacker L., Sohrt K., Schleiff E., Heins L., Soll J. (2002) A Toc75-like protein import channel is abundant in chloroplasts. EMBO Rep. 3, 557–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Patel R., Hsu S. C., Bédard J., Inoue K., Jarvis P. (2008) The Omp85-related chloroplast outer envelope protein OEP80 is essential for viability in Arabidopsis. Plant Physiol. 148, 235–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Müller A., Rassow J., Grimm J., Machuy N., Meyer T. F., Rudel T. (2002) VDAC and the bacterial Porin PorB of Neisseria gonorrhoeae share mitochondrial import pathways. EMBO J. 21, 1916–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Walther D. M., Papic D., Bos M. P., Tommassen J., Rapaport D. (2009) Signals in bacterial β-barrel proteins are functional in eukaryotic cells for targeting to and assembly in mitochondria. Proc. Natl. Acad. Sci. U.S.A. 106, 2531–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Müller J. E., Papic D., Ulrich T., Grin I., Schütz M., Oberhettinger P., Tommassen J., Linke D., Dimmer K. S., Autenrieth I. B., Rapaport D. (2011) Mitochondria can recognize and assemble fragments of a β-barrel structure. Mol. Biol. Cell 22, 1638–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kozjak-Pavlovic V., Ott C., Götz M., Rudel T. (2011) Neisserial Omp85 protein is selectively recognized and assembled into functional complexes in the outer membrane of human mitochondria. J. Biol. Chem. 286, 27019–27026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Röhl T., Motzkus M., Soll J. (1999) The outer envelope protein OEP24 from pea chloroplasts can functionally replace the mitochondrial VDAC in yeast. FEBS Lett. 460, 491–494 [DOI] [PubMed] [Google Scholar]
- 20. Habib S. J., Waizenegger T., Lech M., Neupert W., Rapaport D. (2005) Assembly of the TOB complex of mitochondria. J. Biol. Chem. 280, 6434–6440 [DOI] [PubMed] [Google Scholar]
- 21. Paschen S. A., Rothbauer U., Káldi K., Bauer M. F., Neupert W., Brunner M. (2000) The role of the TIM8–13 complex in the import of Tim23 into mitochondria. EMBO J. 19, 6392–6400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kondo-Okamoto N., Shaw J. M., Okamoto K. (2008) Tetratricopeptide repeat proteins Tom70 and Tom71 mediate yeast mitochondrial morphogenesis. EMBO Rep. 9, 63–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schleiff E., Heard T. S., Weiner H. (1999) Positively charged residues, the helical conformation and the structural flexibility of the leader sequence of pALDH are important for recognition by hTom20. FEBS Lett. 461, 9–12 [DOI] [PubMed] [Google Scholar]
- 24. Kirwin P. M., Meadows J. W., Shackleton J. B., Musgrove J. E., Elderfield P. D., Mould R., Hay N. A., Robinson C. (1989) ATP-dependent import of a lumenal protein by isolated thylakoid vesicles. EMBO J. 8, 2251–2255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. von Arnim A. G., Deng X. W., Stacey M. G. (1998) Cloning vectors for the expression of green fluorescent protein fusion proteins in transgenic plants. Gene 221, 35–43 [DOI] [PubMed] [Google Scholar]
- 26. Daum G., Böhni P. C., Schatz G. (1982) Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J. Biol. Chem. 257, 13028–13033 [PubMed] [Google Scholar]
- 27. Bionda T., Tillmann B., Simm S., Beilstein K., Ruprecht M., Schleiff E. (2010) Chloroplast import signals. The length requirement for translocation in vitro and in vivo. J. Mol. Biol. 402, 510–523 [DOI] [PubMed] [Google Scholar]
- 28. Day D. A., Neuburger M., Douce R. (1985) Biochemical characterization of chlorophyll-free mitochondria from pea leaves. Aust. J. Plant Physiol. 12, 219–228 [Google Scholar]
- 29. Hausmann N., Werhahn W., Huchzermeyer B., Braun H. P., Papenbrock J. (2003) How to document the purity of mitochondria prepared from green tissue of pea, tobacco, and Arabidopsis thaliana. Phyton 43, 215–229 [Google Scholar]
- 30. Rödiger A., Baudisch B., Klösgen R. B. (2010) Simultaneous isolation of intact mitochondria and chloroplasts from a single pulping of plant tissue. J. Plant Physiol. 167, 620–624 [DOI] [PubMed] [Google Scholar]
- 31. Rudhe C., Chew O., Whelan J., Glaser E. (2002) A novel in vitro system for simultaneous import of precursor proteins into mitochondria and chloroplasts. Plant J. 30, 213–220 [DOI] [PubMed] [Google Scholar]
- 32. Schleiff E., Soll J., Küchler M., Kühlbrandt W., Harrer R. (2003) Characterization of the translocon of the outer envelope of chloroplasts. J. Cell Biol. 160, 541–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sommer M. S., Daum B., Gross L. E., Weis B. L., Mirus O., Abram L., Maier U. G., Kühlbrandt W., Schleiff E. (2011) Chloroplast Omp85 proteins change orientation during evolution. Proc. Natl. Acad. Sci. U.S.A. 108, 13841–13846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Machettira A. B., Gross L. E., Sommer M. S., Weis B. L., Englich G., Tripp J., Schleiff E. (2011) The localization of Tic20 proteins in Arabidopsis thaliana is not restricted to the inner envelope membrane of chloroplasts. Plant Mol. Biol. 77, 381–390 [DOI] [PubMed] [Google Scholar]
- 35. Schleiff E., Tien R., Salomon M., Soll J. (2001) Lipid composition of outer leaflet of chloroplast outer envelope determines topology of OEP7. Mol. Biol. Cell 12, 4090–4102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hurt E. C., Soltanifar N., Goldschmidt-Clermont M., Rochaix J. D., Schatz G. (1986) The cleavable presequence of an imported chloroplast protein directs attached polypeptides into yeast mitochondria. EMBO J. 5, 1343–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Brink S., Flügge U. I., Chaumont F., Boutry M., Emmermann M., Schmitz U., Becker K., Pfanner N. (1994) Preproteins of chloroplast envelope inner membrane contain targeting information for receptor-dependent import into fungal mitochondria. J. Biol. Chem. 269, 16478–16485 [PubMed] [Google Scholar]
- 38. Rudolph R., Lilie H. (1996) In vitro folding of inclusion body proteins. FASEB J. 10, 49–56 [PubMed] [Google Scholar]
- 39. Lee Y. J., Kim D. H., Kim Y. W., Hwang I. (2001) Identification of a signal that distinguishes between the chloroplast outer envelope membrane and the endomembrane system in vivo. Plant Cell 13, 2175–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Blachly-Dyson E., Song J., Wolfgang W. J., Colombini M., Forte M. (1997) Multicopy suppressors of phenotypes resulting from the absence of yeast VDAC encode a VDAC-like protein. Mol. Cell. Biol. 17, 5727–5738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rapaport D., Neupert W. (1999) Biogenesis of Tom40, core component of the TOM complex of mitochondria. J. Cell Biol. 146, 321–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Krimmer T., Rapaport D., Ryan M. T., Meisinger C., Kassenbrock C. K., Blachly-Dyson E., Forte M., Douglas M. G., Neupert W., Nargang F. E., Pfanner N. (2001) Biogenesis of the major mitochondrial outer membrane protein Porin involves a complex import pathway via receptors and the general import pore. J. Cell Biol. 152, 289–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yamano K., Yatsukawa Y., Esaki M., Hobbs A. E., Jensen R. E., Endo T. (2008) Tom20 and Tom22 share the common signal recognition pathway in mitochondrial protein import. J. Biol. Chem. 283, 3799–3807 [DOI] [PubMed] [Google Scholar]
- 44. Wiedemann N., Pfanner N., Ryan M. T. (2001) The three modules of ADP/ATP carrier cooperate in receptor recruitment and translocation into mitochondria. EMBO J. 20, 951–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Young J. C., Hoogenraad N. J., Hartl F. U. (2003) Molecular chaperones Hsp90 and Hsp70 Deliver preproteins to the mitochondrial import receptor Tom70. Cell 112, 41–50 [DOI] [PubMed] [Google Scholar]
- 46. Becker T., Wenz L. S., Krüger V., Lehmann W., Müller J. M., Goroncy L., Zufall N., Lithgow T., Guiard B., Chacinska A., Wagner R., Meisinger C., Pfanner N. (2011) The mitochondrial import protein Mim1 promotes biogenesis of multispanning outer membrane proteins. J. Cell Biol. 194, 387–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Papic D., Krumpe K., Dukanovic J., Dimmer K. S., Rapaport D. (2011) Multispan mitochondrial outer membrane protein Ugo1 follows a unique Mim1-dependent import pathway. J. Cell Biol. 194, 397–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hoppins S. C., Nargang F. E. (2004) The Tim8-Tim13 complex of Neurospora crassa functions in the assembly of proteins into both mitochondrial membranes. J. Biol. Chem. 279, 12396–12405 [DOI] [PubMed] [Google Scholar]
- 49. Wiedemann N., Frazier A. E., Pfanner N. (2004) The protein import machinery of mitochondria. J. Biol. Chem. 279, 14473–14476 [DOI] [PubMed] [Google Scholar]
- 50. Smith M., Hicks S., Baker K., McCauley R. (1994) Rupture of the mitochondrial outer membrane impairs Porin assembly. J. Biol. Chem. 269, 28460–28464 [PubMed] [Google Scholar]
- 51. Kübrich M., Rassow J., Voos W., Pfanner N., Hönlinger A. (1998) The import route of ADP/ATP carrier into mitochondria separates from the general import pathway of cleavable preproteins at the trans side of the outer membrane. J. Biol. Chem. 273, 16374–16381 [DOI] [PubMed] [Google Scholar]
- 52. Paschen S. A., Waizenegger T., Stan T., Preuss M., Cyrklaff M., Hell K., Rapaport D., Neupert W. (2003) Evolutionary conservation of biogenesis of β-barrel membrane proteins. Nature 426, 862–866 [DOI] [PubMed] [Google Scholar]
- 53. Schleiff E., Soll J. (2005) Membrane protein insertion. Mixing eukaryotic and prokaryotic concepts. EMBO Rep. 6, 1023–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Alcock F., Clements A., Webb C., Lithgow T. (2010) Evolution. Tinkering inside the organelle. Science 327, 649–650 [DOI] [PubMed] [Google Scholar]
- 55. Gentle I., Gabriel K., Beech P., Waller R., Lithgow T. (2004) The Omp85 family of proteins is essential for outer membrane biogenesis in mitochondria and bacteria. J. Cell Biol. 164, 19–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Duncan O., Murcha M. W., Whelan J. (2012) Unique components of the plant mitochondrial protein import apparatus. Biochim. Biophys. Acta, in press [DOI] [PubMed] [Google Scholar]
- 57. Rapaport D. (2003) How to find the right organelle. Targeting signals in mitochondrial outer membrane proteins. EMBO Rep. 4, 948–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Koehler C. M., Jarosch E., Tokatlidis K., Schmid K., Schweyen R. J., Schatz G. (1998) Import of mitochondrial carriers mediated by essential proteins of the intermembrane space. Science 279, 369–373 [DOI] [PubMed] [Google Scholar]