Background: The G-box cis-element is enriched in promoters of genes responding to light and to high light.
Results: DTT induces DNA binding activity of bZIP transcription factors by reducing a disulfide bond.
Conclusion: Redox regulation is crucial for DNA binding of the G-group of Arabidopsis bZIP transcription factors.
Significance: Redox-dependent mechanisms modulate the activity of plant bZIPs in response to environmental signals.
Keywords: Cell Biology, Chloroplast, DNA-binding Protein, Redox Regulation, Redox Signaling
Abstract
Plant genes that contain the G-box in their promoters are responsive to a variety of environmental stimuli. Bioinformatics analysis of transcriptome data revealed that the G-box element is significantly enriched in promoters of high light-responsive genes. From nuclear extracts of high light-treated Arabidopsis plants, we identified the AtbZIP16 transcription factor as a component binding to the G-box-containing promoter fragment of light-harvesting chlorophyll a/b-binding protein2.4 (LHCB2.4). AtbZIP16 belongs to the G-group of Arabidopsis basic region leucine zipper (bZIP) type transcription factors. Although AtbZIP16 and its close homologues AtbZIP68 and AtGBF1 bind the G-box, they do not bind the mutated half-sites of the G-box palindrome. In addition, AtbZIP16 interacts with AtbZIP68 and AtGBF1 in the yeast two-hybrid system. A conserved Cys residue was shown to be necessary for redox regulation and enhancement of DNA binding activity in all three proteins. Furthermore, transgenic Arabidopsis lines overexpressing the wild type version of bZIP16 and T-DNA insertion mutants for bZIP68 and GBF1 demonstrated impaired regulation of LHCB2.4 expression. Finally, overexpression lines for the mutated Cys variant of bZIP16 provided support for the biological significance of Cys330 in redox regulation of gene expression. Thus, our results suggest that environmentally induced changes in the redox state regulate the activity of members of the G-group of bZIP transcription factors.
Introduction
Light is not only the primary energy source for plants, but it also provides them with information to modulate a wide range of developmental processes (1, 2). In response to light, a large reorganization of the transcriptional program is initiated to modulate plant growth and development (2). However, sometimes photon fluence exceeds the photon utilization capacity of the chloroplast. Absorption of excess light leads to redox changes in the chloroplast; production of reactive oxygen species such as hydrogen peroxide (H2O2), superoxide (O2˙̄), hydroxyl radicals (OH), and singlet oxygen (1O2); and inactivation of photosynthesis (3). These reactive oxygen species are generated as the consequence of an overreduced electron transport system and consecutive transfer of electrons to oxygen (4). Accumulation of reactive oxygen species causes damage to structural proteins, lipids, DNA, and enzymes important for the function of the chloroplasts (4, 5). To protect themselves against extensive damage, plants have the ability to sense when photon fluence exceeds the photon utilization capacity of the chloroplast and communicate this information to stimulate changes in nuclear and chloroplast gene expression. The redox state of the plastoquinone pool (6, 7), the acceptor availability at photosystem I (8, 9), and accumulation of specific reactive oxygen species (10) are proposed as possible sources for plastid signals involved in communicating the redox changes in the chloroplast to the nucleus (11).
Redox reactions are basic to all cellular processes, and the redox environment of the cell controls the activity of numerous metabolic processes by regulating protein function. The buffering capacity of the cellular redox environment is therefore very critical, and as a result, different mechanisms are used to balance the redox state e.g. by the action of redox pairs such as glutathione, ferredoxin, and thioredoxin; antioxidant systems such as superoxide dismutase, ascorbate peroxidase, and peroxiredoxin; and secondary metabolites such as flavonoids, alkaloids, and carotenoids (12). The DNA binding activity of transcription factors has also been shown to be altered by redox-dependent posttranslational modifications. The plant R2R3 MYB family transcription factor is regulated in a redox-dependent manner. Using the P1 regulator of maize flavonoid biosynthesis, it was shown that two cysteines of the R2R3 domain, Cys49 and Cys53, are essential for DNA binding. Thus, under non-reducing conditions, both cysteines form a disulfide bond that prevents the R2R3 MYB domain from binding to DNA (13, 14). The DNA binding activity of another MYB family transcription factor, AtMYB2, which lacks the first Cys residue, is controlled by an alternative mechanism that involves cysteine S-nitrosylation (15). Thus, both oxidation and S-nitrosylation negatively influence the DNA binding activity of the MYB family transcription factors described (14, 15). Another example of redox regulation of transcription factors is a group of plant homeodomain proteins, HAHR1 and HAHB10, which contain a set of conserved Cys residues. In the oxidized state, the homeodomain transcription factors form intermolecular disulfide bonds resulting in inefficient DNA binding activity. However, under reducing conditions, the DNA binding activity of these proteins is strongly enhanced (16). The localization of transcription factors also can be influenced by redox modifications as was shown for NPR1. The NPR1 monomers are translocated at a low rate to the nucleus, preventing activation of the target genes in the absence of inducing stimulus (17). However, following a pathogen attack, salicylic acid-mediated redox changes lead to translocation of NPR1 to the nucleus where it interacts with several transcription factors (18) such as TGA1 and activates its DNA binding (19). Rap2.4a was the first plant transcription factor shown to be directly regulated by the redox status of the nucleus (20). Rap2.4a is an AP2 domain-containing transcription factor involved in the expression of the chloroplast 2-Cys peroxiredoxin A, and the activity of the Rap2.4a transcription factor was demonstrated to be controlled both by reducing and oxidizing conditions (20). The homodimeric structure of Rap2.4a stabilized by an intermolecular disulfide bond is the active form necessary for DNA binding. Oxidation of the dimer by H2O2 or reduction by DTT strongly reduces its DNA binding affinity. Rap2.4a was proposed to act as a redox sensor and transducer (20).
Our aim was to isolate nuclear components responding to redox changes in the chloroplast following exposure to high light. Using a biochemical approach, we identified the AtbZIP163 protein, a member of the G-group of bZIP transcription factors from Arabidopsis thaliana, as a protein binding to the G-box-containing LHCB2.4 promoter fragment. DTT induces DNA binding activity of AtbZIP16 and two other members of the G-group bZIP transcription factors, bZIP68 and GBF1, by reducing a disulfide bond formed through a conserved cysteine. Transgenic Arabidopsis lines overexpressing the wild type version of bZIP16 and T-DNA insertion mutants for bZIP68 and GBF1 demonstrated impaired regulation of LHCB2.4 expression in response to light. Transgenic lines overexpressing the mutated Cys variant of bZIP16 supported a biological significance for the conserved Cys residue in redox regulation of gene expression. Thus, we propose that a redox-dependent mechanism is necessary to modulate the activity of these transcription factors in response to environmental signals.
EXPERIMENTAL PROCEDURES
Plant Material and Growth Conditions
Seeds of A. thaliana Col-0 wild type were grown on soil at 23 °C (16 h of 100 μmol quanta m−2 s−1 light) and 18 °C (8 h of darkness) at 60% relative humidity. For high light treatment for nuclear preparations, 4-week-old plants were subjected to 3 h of 1000 μmol quanta m−2 s−1 light (metal halide HQI-T 400-watt day light bulbs, Orsam). Seedlings for hypocotyl measurements and LHCB2.4 gene expression were grown on Murashige-Skoog plates without sucrose. The plates were vernalized for 24 h at 4 °C in darkness, placed in 150 μmol m−2 s−1 constant white light for 12 h to induce germination, and then dark-adapted for 24 h prior to 5 days of growth in 10 μmol m−2 s−1 constant white light. Seeds were obtained from The Arabidopsis Information Resource for bzip68 (Salk_147015) and gbf1 (Salk_144534) T-DNA insertion lines (21). Homozygous plants were checked for transcript levels using primers for actin, Act2.1 (5′-GGAAGGATCTGTACGGTAC-3′) and Act2.2 (5′-TGTGAACGATTCCTGGACCT-3′); bZIP68 F (5′-CACCATGGGTAGCAGTGAGATG-3′) and bZIP68 R (5′-CTACGCAACATCCTGACGTGTA-3′); and GBF1 F (5′-CACCATGGGAACGAGCGAAGACAAG-3′) and GBF1 R (5′-TTAATTTGTTCCTTCACCATC-3′).
Nuclear Protein Extraction and DNA Affinity Trapping of DNA-binding Proteins
Using a CelLyticTM PN-Plant nuclei isolation/extraction kit (Sigma), control and 3-h high light-exposed leaves of 4-week-old Arabidopsis plants were used for nuclear protein extraction. The tissue was ground in liquid nitrogen and resuspended in nuclei extraction buffer. The pellets were collected by centrifugation after the tissue mixture was filtered through a nylon net. To solubilize lipid membrane, Triton X-100 was added at 0.3% final concentration, and protein extraction buffer was used to extract nuclear proteins from crude nuclei. The promoter region −1530 to −1674 containing the G-box of LHCB2.4 was used as a probe to trap DNA-binding proteins. To amplify a biotinylated DNA promoter fragment by PCR, Gbox-F (biotin-5′-CTTATTGTCGAGGATGGTCT-3′) and Gbox-R (5′-AGATTCACGTGCCTGAGATA-3′) primers were used. DNA affinity trapping of DNA-binding proteins was performed as described previously (22). 2 mg of beads (Dynabeads® M-280 streptavidin, Invitrogen) were used for DNA immobilization in 2× binding buffer (10 mm Tris-HCl, pH 7.5, 1 mm EDTA, 2 m NaCl). After the beads were resuspended in protein binding buffer (20 mm Tris-HCl, pH 8.0, 1 mm EDTA, 10% glycerol, 100 mm NaCl, 0.05% Triton X-100, 1 mm DTT) and mixed with Arabidopsis nuclear protein extracts, DNA-protein binding was allowed by 15-min incubation at 25 °C. At least three washes with protein binding buffer were performed on the Dynabeads to eliminate unspecific protein binding. DNA-binding proteins were collected in elution buffer (20 mm Tris-HCl, pH 8.0, 1 mm EDTA, 10% glycerol, 1 m NaCl, 0.05% Triton X-100, 1 mm DTT).
Mass Spectrometry
Proteins were incubated in the presence of 0.1 m NH4HCO3 and 10 mm DTT at 95 °C for 15 min and then cooled to room temperature before they were mixed with 8 m urea and incubated for 1 h. After addition of 55 mm iodoacetamide, an alkylation reaction was performed at 37 °C for 30 min in the dark. Urea concentration was reduced to 0.8 m with 50 mm NH4HCO3, and tryptic digestion (enzyme-to-substrate ratio, 1:50) was performed overnight at 37 °C. The resulting peptides were lyophilized, resuspended in 1% TFA, and desalted using a Poros 50 reversed-phase R2 microcolumn (PerSeptive Biosystems). Reversed-phase ultraperformance liquid chromatography using a nanoACQUITY UPLCTM system (Waters, Milford, MA) was used to separate the desalted tryptic peptides prior to MS analysis. A C18 trap column (Symmetry, 180 μm × 20 mm, 5 μm; Waters) was used to concentrate each sample (peptides) and then washed with 2% acetonitrile, 0.1% formic acid at 15 μl/min for 2 min. After elution, samples were separated on a C18 analytical column (75 μm × 100 mm, 1.7 μm; Waters) at 350 nl/min using 0.1% formic acid as solvent A and 0.1% formic acid in acetonitrile as solvent B in a gradient. The gradients used were as follows: linear from 0 to 40% B in 25 min, linear from 40 to 80% B in 1 min, isocratic at 80% B in 1 min, linear from 80 to 5% B in 1 min, and isocratic at 5% B for 7 min. The eluting peptides were sprayed into the mass spectrometer (Q-TOF UltimaTM, Waters) with the capillary voltage set to 2.6 kV and cone voltage set to 40 V. The instrument was operated in data-dependent mode as described (23) without any further changes. For database searching, raw data were converted into peak lists using Protein Lynx Global Server software (V2.2.5). For identification of proteins, a local version of the Mascot search program was used (V2.1.04, Matrix Science Ltd.) using the Arabidopsis protein database from The Arabidopsis Information Resource (TAIR version 9.0; July 19, 2009; 33,410 sequences). Settings used for the database search were trypsin-specific digestion with two missed cleavage allowed, carbamidomethylated cysteine set as a fixed modification, oxidized methionine in variable mode, peptide tolerance of 80 ppm, and fragment tolerance of 0.1 Da. Peptides with Mascot ion scores exceeding the threshold for statistical significance of p < 0.05 were selected and also reprocessed manually to validate their significance.
Site-directed Mutagenesis of Cys Residues of AtbZIP16, AtbZIP68, and GBF1 Proteins
Site-directed mutagenesis was performed through three PCR steps as described previously in Montemartini et al. (24). To generate the different mutants, two PCR products that overlap in the sequence containing the same mutation were synthesized in two separate PCRs. In the first PCR, a mutation in the reverse primer was used, and in the second PCR, the same mutation was included in the forward primer. The two PCR products were separated on an agarose gel, and the DNA was excised and purified with a gel extraction kit (E.Z.N.A.). The purified DNA (PCR1 and PCR2) served as a template for a third PCR performed with the forward and reverse primers used to amplify wild type proteins (Table 1). The presence of the desired mutations was confirmed by sequencing the entire genes.
TABLE 1.
Primers used to generate AtbZIP16, GBF1, and AtbZIP68 wild type and mutant proteins
| Protein | Primer | Sequence |
|---|---|---|
| AtbZIP16-WT | bZIP-F1 | 5′-CACCATGGCTAGCAATGAGATGGA-3′ |
| bZIP-R | 5′-TCACGTTGAGTCTTTGTATGAATC-3′ | |
| GBF1-WT | GBF1-F | 5′-CACCATGGGAACGAGCGAAGACAAG-3′ |
| GBF1-R | 5′-TTAATTTGTTCCTTCACCATC-3′ | |
| AtbZIP68-WT | BamHI-bZIP68-F | 5′-AAAAGGATCCATGGGTAGCAGTGAGA-3′ |
| XhoI-bZIP68-R | 5′-AAAACTCGAGCTACGCAACATCCTGA-3′ | |
| AtbZIP16-C1 | bZIP-C1-R | 5′-CCAGCTCATCCAATTCGGCCTGT-3′ |
| bZIP-C1-F | 5′-ACAGGCCGAATTGGATGAGCTGG-3′ | |
| AtbZIP16-C2 | bZIP-C2-R | 5′-TAAGCTCTTCCAATTGGCTTTTG-3′ |
| bZIP-C2-F | 5′-CAAAAGCCAATTGGAAGAGCTTA-3 | |
| GBF1-C1 | GBF1-C1-R | 5′-GAAGTTGTTCCAATTCGGCCTGC-3′ |
| GBF1-C1-F | 5′-GCAGGCCGAATTGGAACAACTTC-3′ | |
| GBF1-C2 | GBF1-C2-R | 5′-TGAGCTTATCCAATTCGCTTGAG-3′ |
| GBF1-C2-F | 5′-CTCAAGCGAATTGGATAAGCTCA-3′ | |
| AtbZIP68-C1 | bZIP68-C1-R | 5′-CACTTTTAGACAAAGCTCCATTA-3′ |
| bZIP68-C1-F | 5′-TAATGGAGCTTTGTCTAAAAGTG-3′ | |
| AtbZIP68-C2 | bZIP68-C2-R | 5′-CTAGCTCATCCAACTCCGCCTGT-3′ |
| bZIP68-C2-F | 5′-ACAGGCGGAGTTGGATGAGCTAG-3′ | |
| AtbZIP16BR | bZIP-F1 | 5′-CACCATGGCTAGCAATGAGATGGA-3′ |
| bZIP16basic-R | 5′-TCACTCATCACATTCGGCCTGTTT-3′ |
Cloning, Expression, and Purification of Recombinant Proteins
The PCR products of AtbZIP16-WT, AtbZIP16-C1, AtbZIP16-C2, AtbZIP16-C1C2, GBF1-WT, GBF1-C1, and GBF1-C2 were cloned into the pET100D TOPO vector according to the manufacturer's instructions (Invitrogen). PCR-amplified cDNAs of AtbZIP68-WT, AtbZIP68-C1, and AtbZIP68-C2 were cloned into BamHI/XhoI sites of pET32a (+) vector (Novagen). The resulting plasmids were transformed into Escherichia coli BL21 (DE3) Star. After 5–16 h of induction with 2 mm isopropyl 1-thio-β-d-galactopyranoside, expressed proteins were affinity-purified on Ni2+-nitrilotriacetic acid-agarose resin (Qiagen).
Electrophoretic Mobility Shift Assays
The promoter region −1530 to −1674 upstream of the ATG start codon of LHCB2.4 gene was PCR-amplified to generate a 144-bp fragment containing the G-box using the primers Gbox-F (5′-CTTATTGTCGAGGATGGTCT-3′) and Gbox-R (5′-AGATTCACGTGCCTGAGATA-3′). To generate the G-box cis-element and its mutant variants, forward primers, namely Gboxcis-F (5′-TCAACTGACACGTGGCATAAC-3′), GboxcisM1-F (5′-TCAACTGAACAGTGGCATAAC-3′), and Gboxcis-M2-F (5′-TCAACTGACACACAGCATAAC-3′), were annealed with their complementary oligonucleotides at room temperature after they were incubated at 70 °C for 5 min. The bold letters show the Gbox and its modified variants. DNA probes were labeled with biotin-14-dCTP at their 3′-end in 50 μl of labeling reaction containing 1× terminal deoxynucleotidyltransferase (TdT) reaction buffer, 100 nm unlabeled DNA, 0.5 μm biotin-14-dCTP (Invitrogen), and 0.2 unit/μl TdT (Invitrogen). After a 30-min incubation at 37 °C, reactions were stopped by addition of 5 mm EDTA, and TdT was extracted with an equal volume of chloroform/isoamyl alcohol. DNA-protein interactions and biotin detection were performed using a LightShift chemiluminescent electrophoretic mobility shift assay (EMSA) kit (Pierce) and chemiluminescent nucleic acid detection module (Pierce), respectively, according to the supplier's instructions.
Subcellular Localization of AtbZIP16-WT
Isolation and transfection of A. thaliana mesophyll protoplasts was performed as described (25). AtbZIP16-WT full-length coding sequence lacking the stop codon was amplified by PCR using the primers BamHI-AtbZIP16-F (5′-AAAAGGATCCATGGCTAGCAATGAGATGG-3′) and KpnI-AtbZIP16-R (5′-AAAAGGTACCCGTTGAGTCTTTGTATGAAT-3′). The amplified product was fused to the N terminus of CFP in the 35S-CFP vector to generate the construct AtbZIP16-CFP. AtbZIP16-CFP and the nuclear marker ABI5-YFP (20) were co-transfected in the protoplasts and incubated for 16 h in the dark. Expression in protoplasts was analyzed by confocal microscopy using an SP2 confocal laser scanning microscope (Leica). All micrographs were taken with ×63 water immersion objective with a numerical aperture of 0.75. Images were taken at 433 and 514 nm specific for CFP and YFP, respectively. Chlorophyll was excited with a 543 nm laser and detected using a 560–700-nm filter.
Yeast Two-hybrid Assays
cDNA of the AtbZIP16 gene was cloned into pLexA-N vector to generate a fusion protein with LexA DNA binding domain (bait). AtbZIP68 and AtGBF1 full-length cDNAs were cloned into pGADHA vector to generate fusions of the prey protein with the GAL4 activation domain (prey). Bait and prey vectors were co-transformed into NMY51, and the transformants were selected on selective medium lacking tryptophan, leucine, histidine, and adenine (SD/-Trp-Leu-His-Ade). The competitive inhibitor 3-aminotriazole was used to suppress the leaky expression of the HIS gene when needed. The yeast two-hybrid assay was performed with the DUALhybrid system (Dualsystems Biotech) according to the manufacturer's instructions. A β-galactosidase overlay assay to detect LacZ activation was performed with an overlay buffer containing 0.5 m potassium phosphate, pH 7.0, 6% N,N-dimethylformamide, 0.1% SDS, 50 μl/100 ml β-mercaptoethanol, 5 mg/ml low melting agarose, and 0.05% X-gal (Fermentas). Overlay buffer was spread on top of the cells, and the yeast plates were incubated at 30 °C until blue color developed. The primers for AtbZIP16 used in the bait constructs are as follows: EcoRI-bZIP16-F (5′-AAAAGAATTCATGGCTAGCAATGAGATG-3′) and KpnI-bZIP16-R (5′-AAAAGGTACCACTCGTTGAGTCTTTGTATGAAT-3′). The primers used to generate cDNAs for prey cloning are as follows: BamHI-bZIP68-F (5′-AAAAGGATCCATGGGTAGCAGTGAGA-3′) and XhoI-bZIP68-R (5′-AAAACTCGAGCTACGCAACATCCTGA-3′) for AtbZIP68 and BamHI-GBF1-F (5′-AAAAGGATCCATGGGAACGAGCGAAGAC-3′) and XhoI-GBF1-R (5′-AAAACTCGAGTTAATTTGTTCCTTCACC-3′) for AtGBF1.
Midpoint Redox Potential
The recombinant AtbZIP16-WT and its mutant variants, AtbZIP16-C1 and AtbZIP16-C2, were used for redox midpoint potential measurements as described in Hirasawa et al. (26). Samples contained 100 mm HEPES buffer, pH 7.0, 100 μg of recombinant protein, and 2 mm total DTT (different ratios of reduced:oxidized DTT) and were incubated at ambient temperature. After a 3-h incubation, monobromobimane (Sigma) was added at a final concentration of 10 mm. Proteins were precipitated with 20% trichloroacetic acid after a 30-min incubation in the dark. Protein pellets were collected by centrifugation at 13,000 rpm and then washed with 1% trichloroacetic acid before they were resuspended in 250 μl of buffer containing 100 mm Tris-HCl, pH 8.0 and 1% SDS. Fluorescence was measured using a Spectra Max Gemini plate reader (Molecular Devices) with excitation at 380 nm and emission at 450 nm. All Em value calculations were based on a value of −330 mV for the Em of DTT at pH 7.0. The data were fitted to the Nernst equation with n = 1 or with n not fixed. Best fit values for Em were determined by fitting titration data to the Nernst equation using Grafit 5.0 software.
Homology Modeling of bZIP Domain
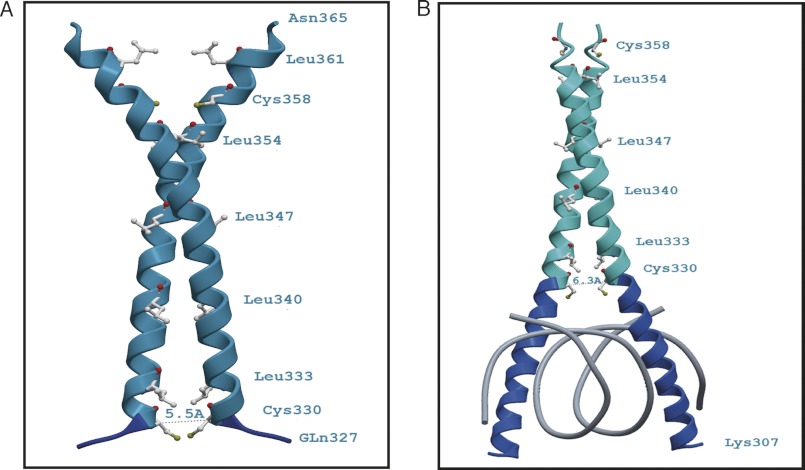
Modeling was performed using the SWISS-MODEL and the LOMETS servers (27, 28). Two peptide sequence variants of the AtbZIP16 bZIP domain were used to identify structures with sufficient similarity to perform homology modeling. A Blast search with the sequence variant Arg304–Asn365 (62 amino acids) was performed against the Protein Data Bank (29) to identify structural templates. The highest ranking match with a sequence identity of 41% over 49 aligned amino acids was obtained to the structure of mouse CREB341 bound to DNA (Protein Data Bank code 1DH3). The AtbZIP16 fragment Arg304–Asn365 was submitted to the SWISS-MODEL server together with the coordinates of a mouse CREB341 monomer (Protein Data Bank code 1DH3_A) as the structural template. After generating the homology model, its quality was scrutinized. The final model contains AtbZIP16 amino acids Lys307–Glu359 (53 amino acids). The likely AtbZIP16 “dimer” was constructed by duplicating the monomer model coordinates and superimposing each of the duplicates onto the structure of the mouse CREB341 dimer (Protein Data Bank code 1DH3). The second, shorter sequence variant Gln327–Asn365 (39 amino acids), which lacks the basic region (BR), did not yield any matches in a Blast search against the Protein Data Bank. However, the LOMETS server identified the synthetic leucine zipper structure with Protein Data Bank code 3HE4_A as the best structural template. For the shorter model of the AtbZIP16, bZIP16 was superimposed onto the structure of the mouse CREB341 dimer. To confirm the correctness of the AtbZIP16 homology models, the longer model also was superimposed onto the DNA-bound GCN4 basic Leu zipper structure (Protein Data Bank code 1YSA), and the shorter AtbZIP16 model was overlaid with the GCN4 Leu zipper structure (Protein Data Bank code 2ZTA), which lacks the basic region. In all cases, Swiss-PdbViewer 4.0.1 (30) was used to superimpose the coordinates of the three-dimensional structures and the homology models using the “Magic Fit” routine followed by the “Iterative Magic Fit” option, which minimizes the root mean square distance between corresponding atom positions. The ribbon diagrams of Fig. 8, A and B, were generated using the ICM-Browser, which was also used to determine the distances between the Cys330 residues of the bZIP16 monomers.
FIGURE 8.
Ribbon diagrams of the theoretical three-dimensional structures (comparative models) of the bZIP domain of AtbZIP16. The figures show the proximity of the cysteine residues at position 330 of the AtbZIP16 sequence without the basic DNA binding region (A) and when bound to DNA (B). A, a model based on amino acids Gln327–Asn365 built onto a DNA-free template (Protein Data Bank code 3HE4_A). The position of Cys330 is indicated, and the distance between the two corresponding Cys330 Cα atoms was determined to be roughly 5.5 Å. At this Cα-Cα distance, the sulfur atoms (Sγ) of the cysteine residues would be sufficiently close to form a disulfide bond. The S-S separation in the model is about 1.5 Å. B, a homology model of residues Lys307–Glu359 built onto the DNA-bound structure of CREB from mouse (Protein Data Bank code 1DH3_A). In this case, the distance between the Cys330 Cα atoms is about 6.3 Å, which would be too far to allow a disulfide bond to form. Dark blue depicts the basic region, and cyan marks the leucine zipper part of AtbZIP16. Amino acids are depicted in gray with the sulfur atoms (Sγ) in yellow and carbonyl oxygen atoms in red.
Overexpression of bZIP16 Wild Type and Mutant Variants in Arabidopsis Transgenic Lines
To generate 35S:bZIP16-WT (WTOX), 35S:bZIP16-C1 (C1OX) and 35S:bZIP16-C1C2 (C1C2OX) overexpresser lines, the PCR products of the full-length coding sequence of bZIP16 wild type or mutant variants was cloned first into the GATEWAY entry vector pDONR207 (Invitrogen) and recombined with pH2GW7.0 destination binary vector. Recombinations were performed using the GATEWAY LR Clonase enzyme mixture according to the manufacturer's instructions (Invitrogen). Wild type Arabidopsis plants were transformed with Agrobacterium strain GV3101 harboring the respective constructs using the floral dip method (31). Transgenic plants (T1) were screened on Murashige-Skoog plates containing 25 μg of hygromycin. Segregation analysis of hygromycin-resistant versus -sensitive ratios (3:1) was performed to select individual lines with single T-DNA insertion, and homozygous transgenic plants were selected for further analysis.
RNA Isolation, cDNA Synthesis, and Real Time PCR
To isolate total RNA, a plant RNA minikit (E.Z.N.A.) was used according to the supplier's instructions. An iScript cDNA synthesis kit (Bio-Rad) was used for cDNA synthesis according to the manufacturer's instructions. Quantitative PCR was performed in 20-μl reactions containing 2 μl of cDNA (1:10 dilution) using iQSYBR Green Supermix (Bio-Rad), and the amplification was performed using a CFX96 Real-Time System (C1000 Thermal Cycler, Bio-Rad). All reactions were performed in triplicates, and the relative transcript abundance of each tested gene was normalized to the expression level of ubiquitin as described (32). The primers used in the quantitative PCR were as follows: Ubiquitin-like protein F (5′-CTGTTCACGGAACCCAATTC-3′) and Ubiquitin like protein R (5′-GGAAAAAGGTCTGACCGACA-3′), LHCB2.4 F (5′-GGAAAAAGGTCTGACCGACA-3′) and LHCB2.4 R (5′-GGAAAAAGGTCTGACCGACA-3′), and bZIP16 F (5′-CGAAGAAAACACAAATCTC-3′) and bZIP16 R (5′-GAGTCTTTGTATGAATCAACCTTTCT-3′). The data were analyzed using LinRegPCR software.
RESULTS
AtbZIP16 Transcription Factor Binds G-box Containing LHCB2.4 Promoter
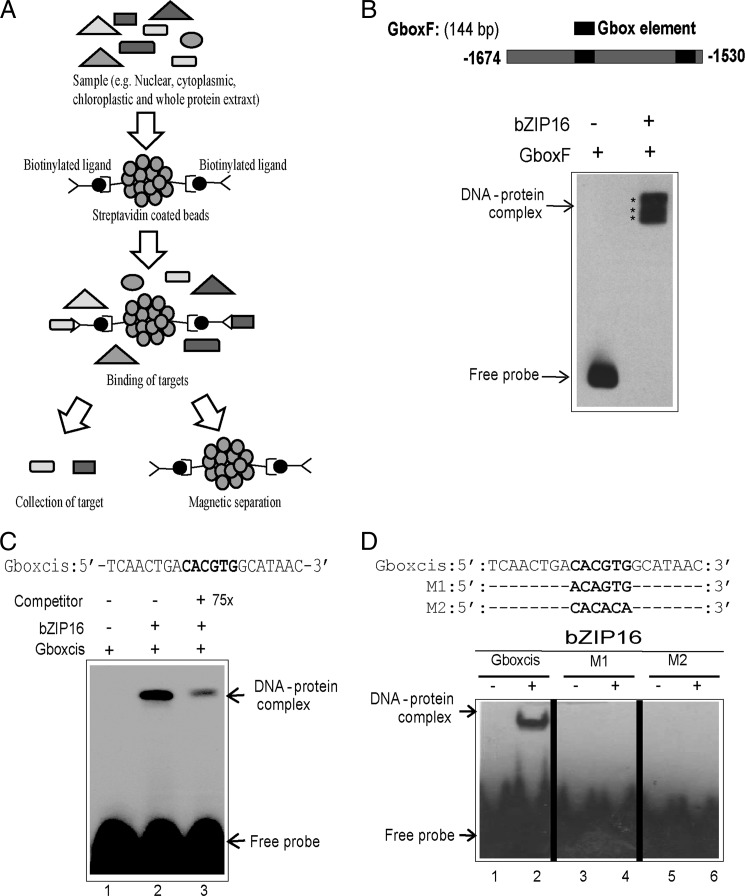
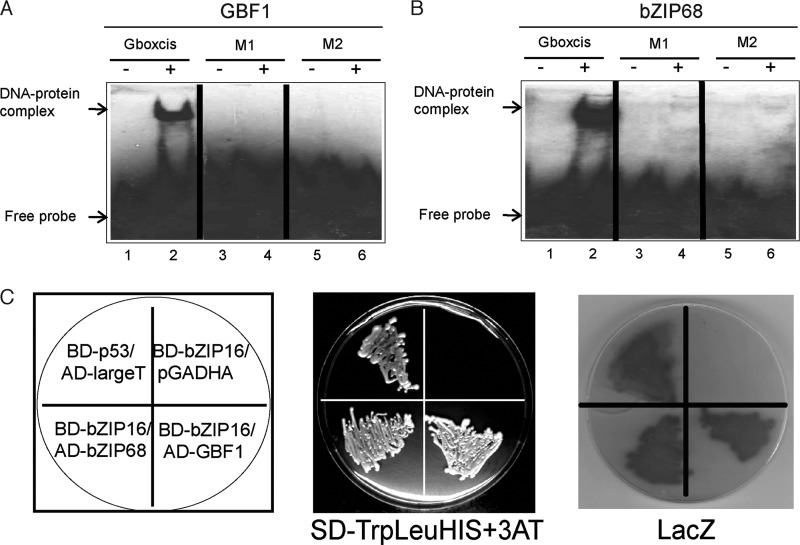
The G-box (CACGTG) cis-element was shown to be enriched in the promoter sequences of genes responding to high light or redox changes in the chloroplast (33). This element was significantly enriched both in the promoters of genes induced and in the promoters of genes repressed by high light, suggesting interaction with both activators and repressors of gene expression (33). To isolate transcription factors responding to changes in light intensity/redox changes and interacting with the G-box element, a 144-bp G-box-containing LHCB2.4 promoter fragment was used. The LHCB2.4 gene (At3g22840) belongs to the LHC supergene family and is representative for genes responding to light and for genes repressed in response to redox changes (33). We used a biochemical DNA affinity trapping approach to isolate transcription factors. This approach was previously proven successful to isolate transcription factors from simple bacterial nuclear protein mixtures (34) and from complex mixtures from plants (35). Nuclear proteins prepared from wild type plants exposed to control and high light conditions were incubated together with the biotin-labeled DNA fragment after it was immobilized on magnetic beads coated with streptavidin. Using this affinity approach, a protein from high light-treated plants was identified to bind the DNA fragment containing the G-box element (Fig. 1A). Q-TOF mass spectroscopic analysis identified the protein as AtbZIP16 (At2g35530) (supplemental Fig. S1). The Arabidopsis bZIP family is subdivided into 10 groups according to sequence similarities of their basic region, conserved motifs, and additional features such as the size of the leucine zipper. The high similarity among the proteins in each group suggests that members of a given group probably bind similar cis-elements (36). AtbZIP16, named according to Jakoby et al. (36), was clustered in group G together with the putative AtbZIP68, GBF1, GBF2, and GBF3 proteins (36). The three GBF proteins have been demonstrated to specifically recognize and bind to the G-box cis-element (37).
FIGURE 1.
DNA affinity trapping of AtbZIP16 using LHCB2.4 promoter. A, biotinylated ligands were immobilized on streptavidin-coated beads and mixed with samples to allow binding. Magnetic separation was used, and the target proteins were collected for subsequent analysis. B, EMSA verified binding of recombinant AtbZIP16 protein to the 144-bp fragment of the LHCB2.4 promoter containing the G-box (positions −1530 to −1674). C, EMSA with complementary synthetic oligonucleotides representing the G-box element and recombinant AtbZIP16 protein. A competition assay was performed by adding unlabeled G-box probe at 75× excess. D, sequence of complementary synthetic oligonucleotides representing the G-box and its mutagenized half-sites used with AtbZIP16 in EMSA. The biotin-labeled DNA probes were incubated in the presence (+) or absence (−) of 4 μg of recombinant AtbZIP16 protein, and the DNA-protein complexes were separated from free DNA by non-denaturing polyacrylamide gel electrophoresis. The signals were detected with a chemiluminescent nucleic acid detection method. Positions of free DNA and protein-DNA complexes are indicated by an arrow.
EMSA Confirms Binding of AtbZIP16 to the G-box
EMSA was performed to confirm the interaction between AtbZIP16 and the 144-bp promoter fragment used in the affinity trapping assay. The full-length coding sequence of AtbZIP16 was expressed in E. coli as a His-tagged recombinant protein (supplemental Fig. S2). Binding of AtbZIP16 to the 144-bp G-box-containing DNA fragment (Fig. 1B) was confirmed with EMSA (Fig. 1B). The observed multiple shifts resembling the DNA-protein complex are most likely caused either by binding of bZIP16 protein to both of the binding sites within the 144-bp target DNA (Fig. 1B) or by the DNA binding to multiple forms of the bZIP16 protein such as monomers, dimers, etc. (Fig. 1B). To assess the specificity of the interaction between AtbZIP16 and its putative G-box target sequence, EMSA was performed again using 21-bp oligonucleotides containing the G-box element of the Arabidopsis LHCB2.4 promoter as a probe (Gboxcis) (Fig. 1C). Recombinant AtbZIP16 fusion protein demonstrated clear binding to the 21-bp DNA fragment (Fig. 1C). Moreover, 75× excess unlabeled G-box fragment was able to compete, although not completely, for the binding activity of AtbZIP16 (Fig. 1C), and complete competition was revealed when 100× excess unlabeled G-box fragment was used (supplemental Fig. S3). The G-box cis-element is a palindromic DNA motif that is composed of two identical half-sites (C−3A−2C−1G+1T+2G+3). Here we have numbered the base pairs as −3 to +3 (starting from 5′ to 3′) of the G-box. Binding activity of recombinant AtbZIP16 protein was abolished when either the first (M1; −3 to −1) or the second half-site (M2; +1 to +3) was mutated (Fig. 1D). Taken together, these results indicate high specificity of the interaction between AtbZIP16 and the G-box core sequence.
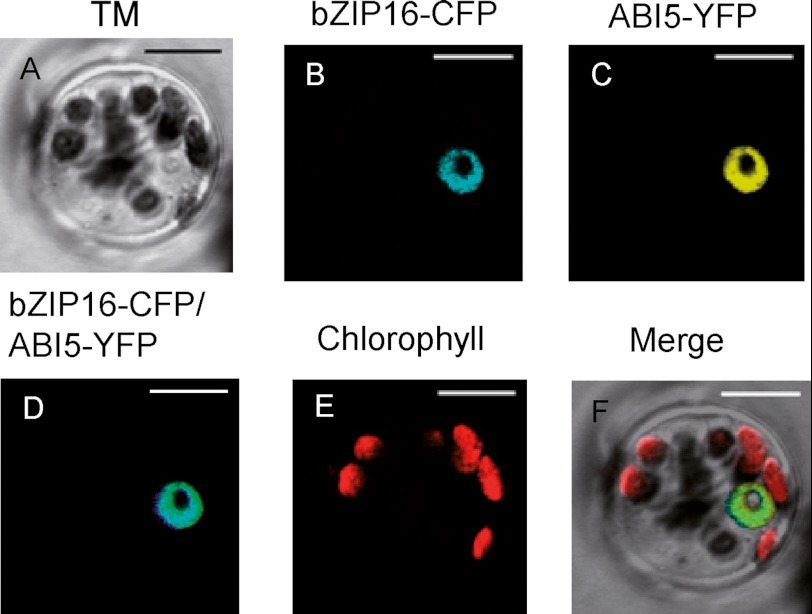
Subcellular Localization of AtbZIP16
Sequence analysis of AtbZIP16 protein revealed the presence of KRQRRKQSNRESARRSR amino acid sequence indicating a putative bipartite nuclear localization sequence. The nuclear localization sequence overlaps with the basic region of the bZIP DNA binding domain. We analyzed the intracellular distribution of AtbZIP16-CFP fusion protein in transiently transformed Arabidopsis mesophyll protoplasts. The CFP fusion plasmid was transfected into protoplasts, and CFP localization was examined by confocal microscopy following overnight expression in the dark. The AtbZIP16-CFP fusion protein was exclusively localized to the nucleus (Fig. 2). As a nuclear marker, we used ABI5-YFP fusion protein (20), which exclusively co-localized with AtbZIP16 in the nucleus (Fig. 2).
FIGURE 2.
Subcellular localization of bZIP16 fusion protein. AtbZIP16-CFP and ABI5-YFP were co-transfected into A. thaliana mesophyll protoplasts. Confocal images are shown as follows: A, bright field transmission; B, AtbZIP16-CFP fluorescence; C, ABI5-YFP fluorescence; D, AtbZIP16-CFP/ABI5-YFP overlay; E, chlorophyll autofluorescence; F, the merged image of A–E. Bar length, 10 μm. TM, transmitted light image.
GBF1 and AtbZIP68 Proteins Bind G-box and Interact with AtbZIP16
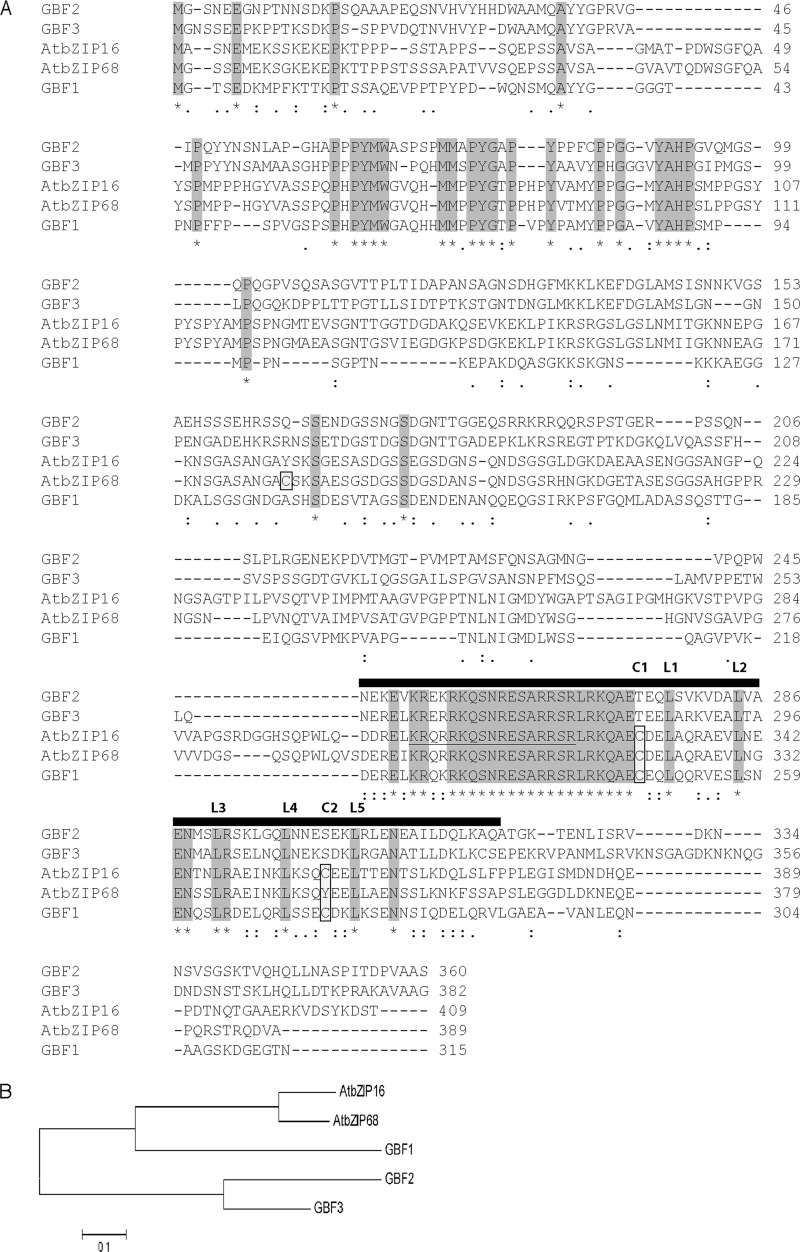
The cluster of group G contains in addition to AtbZIP16 the putative AtbZIP68, GBF1, GBF2, and GBF3 proteins (36). The bZIP domain shares high similarity (more than 85%) among all members of the G-group (Fig. 3A). Moreover, AtbZIP16 shares 78% overall similarity with AtbZIP68, 48% with GBF1, and less than 40% with GBF2 and GBF3 (Fig. 3B). The high sequence similarity among AtbZIP16, AtbZIP68, and GBF1 suggests that these proteins might bind the same cis-element. To test this assumption, recombinant AtbZIP68 and AtGBF1 proteins were expressed in E. coli (supplemental Fig. S2) and incubated with the 21-bp oligonucleotides containing the G-box element of the Arabidopsis LHCB2.4 promoter (Gboxcis). The two proteins interacted with Gboxcis oligonucleotides (Fig. 4, A and B). GBF1 has been isolated from an Arabidopsis cDNA expression library based on its DNA binding activity to a synthetic oligonucleotide from the tomato RBCS-3A G-box-like element G-3A (37). GBF1 has also been shown to interact strongly with the G-box-containing promoter of Arabidopsis RBCS-1A (38). Like AtbZIP16, binding activity of AtbZIP68 and AtGBF1 recombinant proteins was abolished when they were incubated with M1 and M2 mutagenized half-sites of oligonucleotides containing the G-box element of Arabidopsis LHCB2.4 promoter (Fig. 4, A and B).
FIGURE 3.
Sequence comparison of the G-group of the bZIP family in A. thaliana. A, alignment of the predicted amino acid sequence of bZIP16 (At2g35530) with bZIP68 (At1g32150), GBF1 (At4g36730), GBF2 (At4g01120), and GBF3 (At2g46270), which comprise the G-group of bZIP family. The asterisks (*) and the gray shading indicate fully conserved amino acid residues, : indicates general similarities, similarity among two to four is represented by., and dashes (-) indicate gaps introduced to maximize alignment. Cys residues are indicated by boxes and light gray shading, and the nuclear localization sequence of bZIP16 is underlined. The black rectangle indicates the location of the bZIP domain. B, the phylogenetic tree was produced using Mega5.0 software. The scale bar represents substitutions per site.
FIGURE 4.
DNA binding of AtbZIP68 and AtGBF1 and interaction among bZIP16, AtbZIP68, and AtGBF1. Complementary synthetic oligonucleotides representing the G-box and its mutagenized half-sites were labeled with biotin and incubated in the presence (+) or the absence (−) of AtGBF1 (A) or AtbZIP68 (B). Binding reactions were separated by 6% native PAGE. The signals were detected with a chemiluminescent nucleic acid detection module, and positions of free DNA and protein-DNA complexes are indicated by arrows. C, interaction among AtbZIP16, AtbZIP68, and AtGBF1 in a yeast-two hybrid system. The full-length coding sequence of AtbZIP16 was fused to GAL4 binding domain in pLexA-N vector to generate the bait BD-bZIP16. AtbZIP68 or AtGBF1 was fused to the GAL4 activation domain in pGADHA vector to produce the prey clones, respectively. Yeast strain NMY51 was co-transformed with bZIP16 bait and pGADHA empty prey vector (negative control), bZIP16 bait and bZIP68 prey, bZIP16 bait and GBF1 prey, and p53 bait and LargeT prey (positive control). Interaction is indicated by activation of HIS3 reporter gene and LacZ activation. Growth due to the activation of HIS3 reporter gene was examined in the presence of 3-aminotriazole (3AT).
The bZIP transcription factors are dimeric proteins. We therefore tested interaction among AbZIP16, AtbZIP68, and AtGBF1 using the yeast two-hybrid assay. We expressed AtbZIP16 as a fusion to the GAL4 DNA binding domain in pLexA-N vector (BD-bZIP16) and AtbZIP68 and AtGBF1 as fusions to the GAL4 activation domain in pGADHA vector (AD-bZIP68 and AD-GBF1), respectively. We introduced them into yeast strain NMY51 containing HIS and/or LacZ genes under the control of GAL4 binding sites (Fig. 4C). Transformants were grown on selective medium lacking the nutritional selective markers Trp, Leu, and His. Leaky expression of the HIS gene due to the self-activating capacity of bZIP16 was eliminated with 3-aminotriazole. Like BD-p53 interaction with AD-LargeT (positive control), BD-bZIP16 bait was able to interact with the preys AD-bZIP68 and AD-GBF1 but not with the empty AD vector (Fig. 4C). The interactions were confirmed in β-galactosidase overlay assays demonstrating LacZ activation. Similarly to the positive control (p53 interaction with LargeT), strong interaction between AtbZIP16 and its homologues AtbZIP68 and GBF1 was demonstrated by the LacZ reporter activity (Fig. 4C). AtbZIP16 was also shown previously to form heterodimers with AtbZIP68, GBF1, and the other members of the G-group in the presence of DNA (39).
Redox Regulation of AtbZIP16
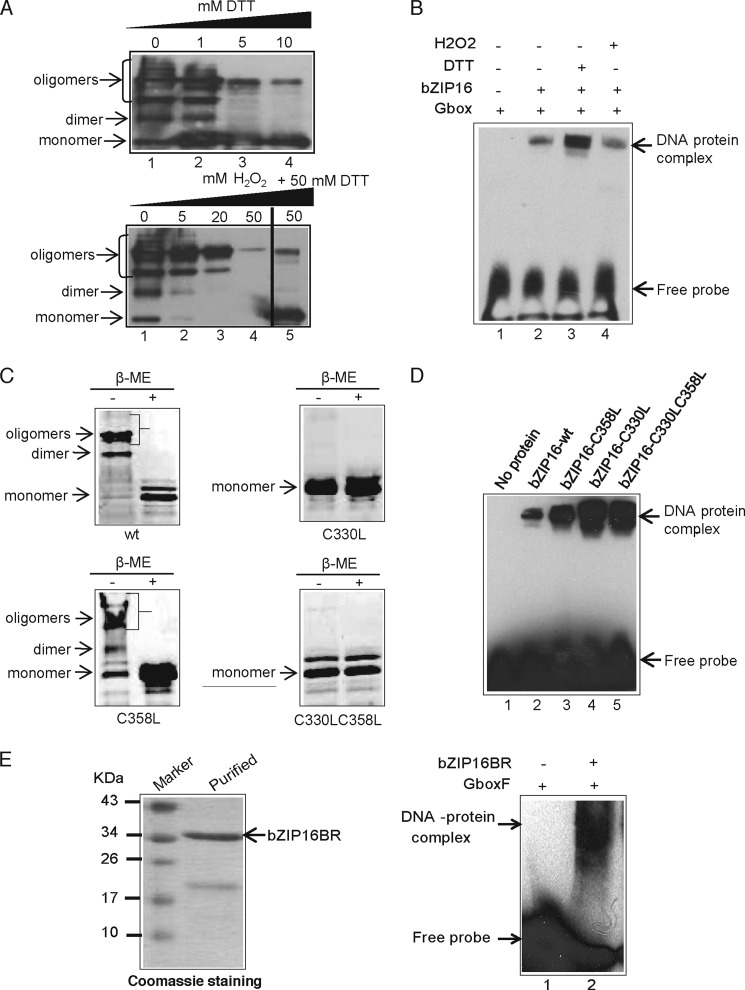
AtbZIP16 protein contains two cysteine residues at positions 330 and 358 within its bZIP domain (Fig. 3A), suggesting the possibility to form disulfide bonds. To investigate this possibility, we performed denaturing polyacrylamide gels (SDS-PAGE) with AtbZIP16 protein subjected to different reducing or oxidizing conditions. Under non-reducing conditions, several bands were detected in the regions corresponding to monomeric, dimeric, and oligomeric forms of AtbZIP16 (Fig. 5A). Increasing concentrations of DTT reduced the oligomers and the dimers (Fig. 5A) and led to complete conversion to monomers (supplemental Fig. S4). In contrast, addition of increasing concentrations of H2O2 resulted in a gradual loss of the different detected forms, suggesting formation of very high molecular mass oligomers that were beyond the migration capacity of the gel and/or the transfer limit of the membrane (Fig. 5A). Restoring reducing conditions by addition of a reducing agent should reverse the effect of H2O2 and break up the high molecular mass oligomers. AtbZIP16 was incubated in the presence of 50 mm H2O2 for 10 min before the addition of the reductant DTT, which resulted in high amounts of AtbZIP16 monomer, demonstrating that H2O2 formed reversible high molecular weight complexes (Fig. 5A).
FIGURE 5.
Redox regulation of AtbZIP16 and its DNA binding activity. A, the quaternary structure of AtbZIP16 protein was analyzed under reducing and oxidizing conditions by incubating AtbZIP16 in the presence of either 0–10 mm DTT or 0–50 mm H2O2. Proteins were detected using anti-His antibody, and positions of monomers, dimers, and oligomers are indicated by arrows. B, the effect of DTT and H2O2 on DNA binding activity of AtbZIP16 was analyzed by EMSA. Binding reactions were carried out in the absence or presence of AtbZIP16, 10 mm DTT, and 10 mm H2O2. Positions of free DNA and protein-DNA complexes are indicated by arrows. C, intermolecular disulfide bond formation in AtbZIP16. AtbZIP16 protein or its mutant variants were either reduced with β-mercaptoethanol (β-ME) or left in their unreduced forms before loading for non-reducing SDS-PAGE. Locations of the monomers, dimers, and oligomers are indicated by arrows for AtbZIP16-WT protein, AtbZIP16 containing a Cys to Leu mutation at position 330, AtbZIP16 containing a Cys to Leu mutation at position 358, and AtbZIP16 containing Cys to Leu mutations at positions 330 and 358. D, binding of AtbZIP16-WT protein and its mutant variants to the G-box element. EMSA was performed using the biotin-labeled G-box element without protein or with AtbZIP16-WT, AtbZIP16 containing a Cys to Leu mutation at position 358, AtbZIP16 containing a Cys to Leu mutation at position 330, and AtbZIP16 containing Cys to Leu mutations at positions 330 and 358. Positions of free DNA and protein-DNA complexes are indicated by arrows. E, Coomassie staining of purified His-tagged bZIP16 protein containing only the basic region and lacking the leucine zipper (bZIP16BR). The band corresponding to the respective protein is indicated by an arrow. bZIP16BR was analyzed for the formation of DNA-protein complex with G-box by EMSA. Binding reactions contained the biotin-labeled G-box element in the absence or presence of AtbZIP16BR. Positions of free DNA and protein-DNA complexes are indicated by arrows.
Redox Regulation of DNA Binding Activity of AtbZIP16
The DNA binding activity of plant transcription factors has been shown to be directly (14, 16, 20) or indirectly (19) regulated by the redox environment. To determine whether the DNA binding activity of AtbZIP16 to the G-box is regulated by changes in the redox state, we treated binding reactions with fixed amounts of DTT and H2O2 before determining DNA binding activity by EMSA. As shown in Fig. 5B, the binding activity of AtbZIP16 was greatly enhanced following DTT treatment compared with that obtained under control conditions or H2O2 treatment (Fig. 5B), suggesting that cysteines in the reduced state are required for efficient DNA binding.
We performed cysteine mutagenesis to probe the importance of these residues for disulfide formation in AtbZIP16. As shown in Fig. 5C, mutant proteins with C330L and C330L/C358L but not C358L alone were converted into a monomer under non-reducing conditions (in the absence of β-mercaptoethanol). Thus, the redox-regulated intermolecular disulfide formation is specifically targeted to the cysteine residue at position 330. Furthermore, the DNA binding activity was greatly enhanced in both the C330L and C330L/C358L mutant variants compared with the wild type protein (Fig. 5D). The C358L mutant protein resulted in a slight increase in DNA binding activity (Fig. 5D). Taken together, these results greatly support the importance of the Cys330 disulfide bridge for redox regulation of AtbZIP16 DNA binding activity. The different behavior of the single mutants C330L and C358L may reflect differences in redox potential of the respective cysteines.
Monomer and dimer pathways of DNA binding have been proposed for bZIP and other transcription factors where the monomeric forms can recognize and bind the DNA. These monomers then dimerize while bound to the DNA (40–42). To test the possibility that bZIP16 could bind its DNA target sequence as a monomer, EMSA was performed using the G-box element and recombinant bZIP16 protein containing the basic region but lacking the leucine zipper, bZIP16BR (Fig. 5E). The truncated form of bZIP16 was able to bind the DNA as a monomer (Fig. 5E). Specificity might be increased by the monomer DNA binding pathway, which prevent trapping of transcription factors at nonspecific DNA sites (40–42).
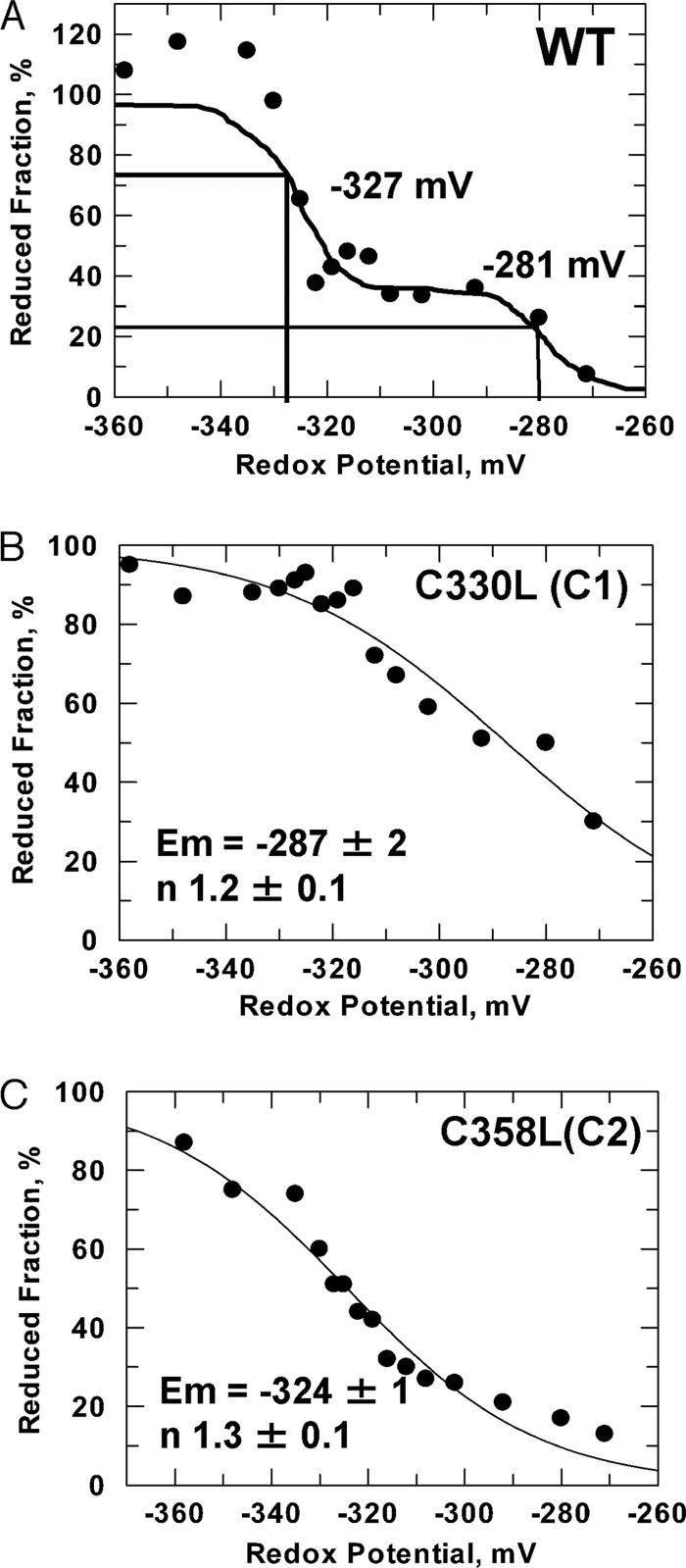
Midpoint Redox Potential of AtbZIP16
A shift of the redox potential to oxidative conditions may induce the formation of intermolecular disulfide bonds between vicinal cysteine residues of protein subunits. Cys330 is responsible for the intermolecular disulfide bond formation in AtbZIP16 and likely stimulates its DNA binding activity (Fig. 5, C and D). The redox state of AtbZIP16-WT and its mutant variants AtbZIP16-C1 (lacking Cys330) and AtbZIP16-C2 (lacking Cys358) was titrated in the presence of varying ratios of DTTreduced/DTToxidized to adjust defined thiol redox potentials. After labeling with monobromobimane, the redox state was determined in a fluorometric assay. Titration of the redox state of AtbZIP16-WT revealed two midpoint redox potentials of −281 and −327 mV, respectively (Fig. 6A), indicating a redox regulatory role for both cysteines. To examine the effect of the cysteine mutations on the Em value of AtbZIP16, additional titrations were performed (Fig. 6, B and C). Redox titration experiments with the two Cys mutants (C1 and C2) yielded Em values similar to those obtained with the wild type protein (for C1, Em = −287 ± 2 mV and for C2, Em = −324 ± 1 mV). The C330L (C1) mutant titration characterizes the properties of the Cys358, whereas the C358L (C2) mutant titration characterizes the properties of Cys330 (C1). The Cys330 residue, which is responsible for disulfide formation, has a midpoint redox potential of −324 mV, which is within the range of values (−290 to −330 mV) reported for other redox-active proteins (26). In addition, the second cysteine of AtbZIP16, Cys358, has a midpoint redox potential of −287 mV, indicating that it could also play a redox regulatory role possibly through glutathionylation. Taken together, AtbZIP16 conformation and activity respond to changes in the intracellular redox environment similarly to other plant proteins with established roles in redox-regulated systems (26).
FIGURE 6.
Midpoint redox potential of AtbZIP16 and its mutant variants. The ambient redox potential, adjusted by defined ratios of DTToxidized/DTTreduced, ranged from −260 to −360 mV. After labeling the reduced protein thiols with monobromobimane, the fluorescence was quantified in each sample. The redox potential of AtbZIP16-WT (A), AtbZIP16-C1 (C330L) (B), and AtbZIP16-C2 (C358L) (C) was assessed using the Nernst equation (for details, see “Experimental Procedures”).
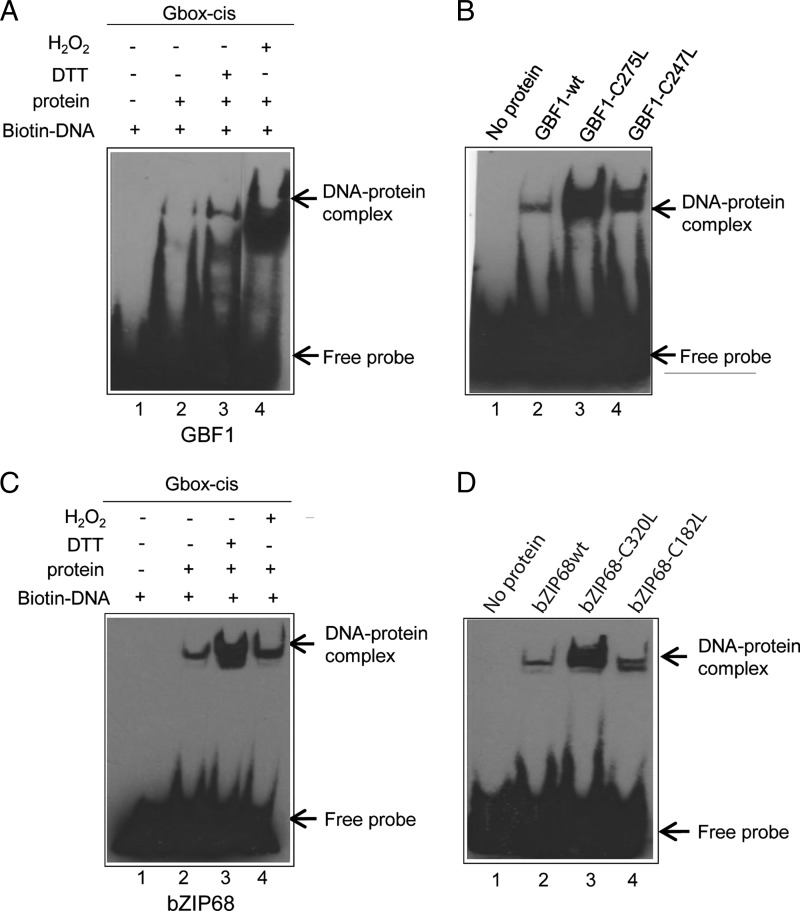
Redox-dependent DNA Binding Activity of AtbZIP68 and AtGBF1
Analysis of the conserved cysteines in the other members of the G-group of bZIP transcription factors revealed that C1 and C2 of AtbZIP16 are also conserved in AtGBF1. C2 of AtbZIP68 resembles C1 in AtbZIP16 and AtGBF1. In AtbZIP68, C1 is located at position 182 in the N terminus and upstream of the bZIP domain (Fig. 3A). The other members, AtGBF2 and AtGBF3, contain threonine and serine instead of cysteines at the respective positions (Fig. 3A). To determine whether the DNA binding activity of AtbZIP68 and AtGBF1 also is subjected to redox-dependent modulation, we expressed these proteins in E. coli and performed studies by EMSA similar to those described above for AtbZIP16. Similarly to AtbZIP16, DTT greatly enhanced DNA binding of both AtbZIP68 and AtGBF1 (Fig. 7, A and C), suggesting involvement of the cysteines in redox-dependent regulation. The role of these cysteines was studied using the mutant proteins AtbZIP68-C1 (C182L), AtbZIP68-C2 (C320L), AtGBF1-C1 (C247L), and AtGBF1-C2 (C275L) in which the cysteines were exchanged for leucines. The AtbZIP68-C2 and AtGBF1-C1 mutant forms revealed significantly increased DNA binding activity (Fig. 7, B and D) consistent with the result of DTT-induced DNA binding activity. AtbZIP68-C1 and AtGBF1-C2 mutant proteins resulted in a slight increase in DNA binding activity (Fig. 7, B and D). These results indicate that the behavior of AtbZIP68 and AtGBF1 resembles that observed for AtbZIP16 (Fig. 5).
FIGURE 7.
Redox modulation of AtbZIP68 and AtGBF1 binding affinities to the G-box. A, the effect of DTT and H2O2 on DNA binding activity of GBF1 was tested by EMSA. EMSA was performed using biotin-labeled complementary synthetic oligonucleotides representing the G-box element without protein or with GBF1-WT, 10 mm DTT, and 10 mm H2O2. B, binding of GBF1-WT protein and its mutant variants to the G-box element. Binding reactions in the absence of GBF1 protein or with GBF1-WT, GBF1 containing a Cys to Leu mutation at position 275, or GBF1 containing a Cys to Leu mutation at position 247. C, the effect of DTT and H2O2 on DNA binding activity of bZIP68 was analyzed by EMSA. Binding reactions in the absence or presence of bZIP68 protein, 10 mm DTT, and 10 mm H2O2. D, binding of bZIP68-WT protein and its mutant variants to the G-box element. EMSA was performed using the biotin-labeled G-box element without protein or with bZIP68-WT, bZIP86 containing a Cys to Leu mutation at position 320, or bZIP68 containing a Cys to Leu mutation at position 182. Biotin-labeled probes were detected with a chemiluminescent nucleic acid detection module, and positions of free DNA and protein-DNA complexes are indicated by arrows.
Modeling of the bZIP Domain of AtbZIP16
Comparative modeling produced three-dimensional models of the free and DNA-bound bZIP domain of AtbZIP16 (Fig. 8). The longer variant comprising the basic DNA binding region (residues Arg304–Asn365) was modeled onto the DNA-bound cAMP-response element-binding (CREB341) bZIP protein from mouse (Protein Data Bank code 1DH3_A). The shorter variant lacking the basic region (residues Gln327–Asn365) was modeled onto the structure of a synthetic Leu zipper (Protein Data Bank code 3HE4_A). The predicted AtbZIP16 structures show that Cys330 (C1) is positioned at the transition between the zipper and the basic region just outside the direct DNA contact sites of the BR. In the DNA-free bZIP form, the Cys330 (C1) residues of the respective monomers are located in close proximity of each other. The distance between the two corresponding Cys330 Cα atoms is estimated to 5.5 Å. The sulfur atoms of the cysteine residues would then be sufficiently close to allow disulfide bond formation. The Sγ to Sγ atom separation (S-S distance) in this model is estimated to 1.5 Å, which is slightly closer than the most commonly observed Sγ-Sγ distance of 2.02 Å in disulfide bond-forming proteins (Fig. 8A) (43). However, upon DNA binding, the separation between the Cys330 residues on the two monomers is predicted to increase to about 6.3 Å, which will most likely not allow formation of the disulfide bond (Fig. 8B).
Transgenic Lines Overexpressing bZIP16 Wild Type and Mutated Cys Variants and T-DNA Insertion Mutants for bZIP68 and GBF1
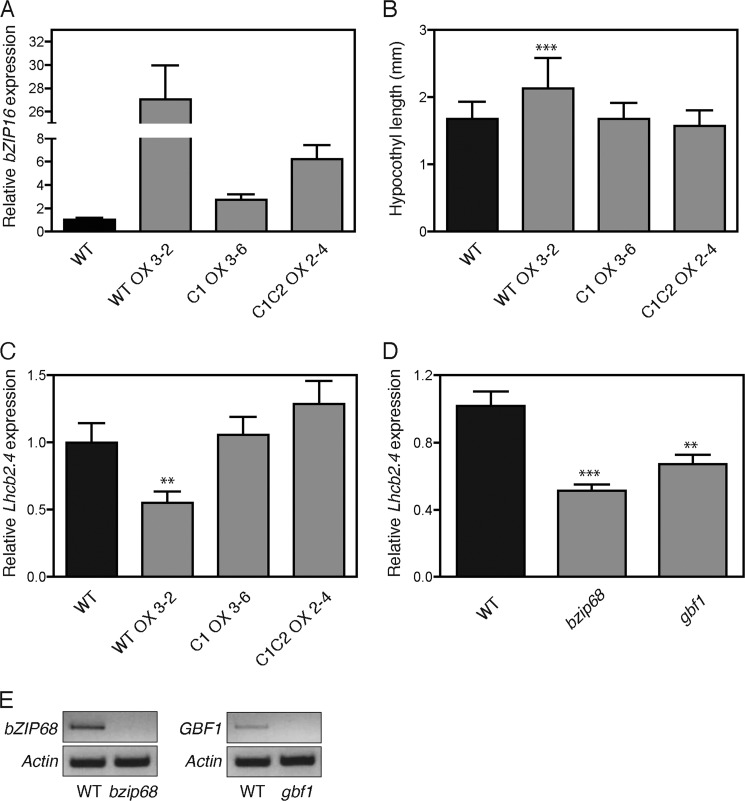
To test whether the identified interaction between the G-box element and bZIP16 and the Cys330-mediated redox regulation of bZIP activity are biologically significant, we generated overexpression lines of wild type bZIP16 (bZIP16-WTOX) and bZIP16 Cys mutants (bZIP16-C1OX and bZIP16-C1C2OX). Homozygote lines carrying only single T-DNA insertions were identified and used for the experiments. The bZIP16 transcript levels were significantly higher compared with wild type in all lines produced (Fig. 9A). GBF1 was shown previously to play a role during the early light response in Arabidopsis (44), and we therefore monitored the growth of 5-day-old seedlings of the overexpression lines in constant white light. Examination of the hypocotyl length revealed that the bZIP16-WTOX line displayed a significant reduction of inhibition of hypocotyl elongation in response to white light (Fig. 9B). In contrast, the overexpression lines with the cysteine mutations bZIP16-C1OX and bZIP16-C1C2OX displayed hypocotyl length similar to that of the wild type. These results indicate a biological significance and a functional involvement of the redox status of Cys330 in planta (Fig. 9B). Furthermore, we investigated whether bZIP16 plays a role in light-regulated gene expression of LHCB2.4 in planta and whether Cys330 is important for such regulation. Expression of LHCB2.4 gene was investigated in 5-day-old seedlings of the bZIP16-WTOX, bZIP16-C1OX, and bZIP16-C1C2OX lines grown in constant white light and compared with wild type (Fig. 9C). The expression level of LHCB2.4 was significantly lower in the bZIP16-WTOX line compared with wild type, suggesting that bZIP16 acts as a repressor of LHCB2.4. Similarly to the hypocotyl elongation, the bZIP16-C1OX and bZIP16-C1C2OX lines displayed wild type levels of the LHCB2.4 transcript (Fig. 9C). Taken together, the results provide biological relevance for the interaction between bZIP16 and the G-box and support the proposed regulatory role of Cys330.
FIGURE 9.
Characterization of bZIP16 overexpression lines and bzip68 and gbf1 T-DNA insertion mutants. A, real time analysis of bZIP16 expression in the overexpression lines of wild type bZIP16 (bZIP16-WTOX) and bZIP16 Cys mutants (bZIP16-C1OX and bZIP16-C1C2OX). B, 5-day-old seedlings of wild type and the overexpression lines were used to measure the hypocotyl lengths. The seedlings were grown in 10 μmol quanta m−2 s−1 constant white light, and 90–100 seedlings were measured for each genotype. C and D, real time RT-PCR analysis of LHCB2.4 (At3g27690) transcript levels in response to white light in 5-day-old seedlings of WT; bZIP16-WTOX, bZIP16-C1OX, and bZIP16-C1C2OX (C); and the T-DNA insertion mutants bzip68 and gbf1 (D). The gene expression was normalized to the expression level of At4g36800, which encodes a ubiquitin-protein ligase-like protein. The mean ± S.E. (error bars) of at least three biological replicates is shown. The expression was significantly different from wild type in the transgenic lines as demonstrated by Student's t test: **, p < 0.01 and ***, p < 0.001. E, semi-quantitative PCR analysis of transcript levels in the bzip68 and gbf1 T-DNA insertion lines using gene-specific primers. Actin (At5g09810) was used as a reference gene.
In addition to the study of the bZIP16 overexpresser lines, we also investigated LHCB2.4 expression in T-DNA insertion mutants of the other two G-group members, bZIP68 and GBF1. Eliminating the bZIP68 and GBF1 proteins resulted in a decrease of LHCB2.4 transcript compared with wild type, suggesting repression of the light-regulated gene expression (Fig. 9, D and E). Surprisingly, this regulation was similar to the overexpression of the bZIP16 protein in bZIP16-WTOX (Fig. 9C), implying that in contrast to bZIP16 bZIP68 and GBF1 function as activators of LHCB2.4 expression. Furthermore, this deregulation of gene expression suggests that despite the high similarity among the three G-group members bZIP16, bZIP68, and GBF1 their function is not redundant.
DISCUSSION
Exposure to redox changes induced by exposure to excess light results in dramatic changes in gene expression (6, 33). Analysis of the 500-bp promoter sequences of genes responding to exposure to high light or redox changes in the chloroplast revealed that the G-box was enriched in the promoters of genes repressed by excess light such as LHCB2.4 (At3g27690) (33). Using a biochemical approach, we identified AtbZIP16 as a transcriptional regulator involved in the light- and/or redox-triggered regulation of LHCB2.4 expression in Arabidopsis.
The A. thaliana genome encodes at least 75 predicted bZIP transcription factors (36, 45, 46), which are clustered into 10 subgroups (A–I and S) (36). Plant bZIP transcription factors have been shown to bind the G-box, C-box, and A-box elements, all containing the functional ACGT core (47). AtbZIP16 belongs to the G-group and clusters with the AtbZIP68, AtGBF1, AtGBF2, and AtGBF3 proteins. AtGBF1, AtGBF2, and AtGBF3 have been shown to bind the G-box element (37). We further demonstrated that AtbZIP16, AtbZIP68, and AtGBF1 recognize and specifically bind to the oligonucleotide sequences containing the CACGTG G-box derived from LHCB2.4 promoter. Site-directed mutations of the ACGT core abolished binding of AtbZIP16, AtbZIP68, and AtGBF1 to DNA.
Regulatory proteins and transcription factors are subject to posttranslational modifications necessary to modulate their activities. Dithiol/disulfide exchange is a key component for fast responses to changes in the redox environment and for modifying the activity of the protein (48). The EMSA for AtbZIP16, AtbZIP68, and AtGBF1 clearly demonstrated that binding to the DNA target is significantly affected by redox conditions and that DNA binding is enhanced under reducing conditions (Figs. 5 and 7). Analysis of Cys330, Cys247, and Cys320 residues in the basic region of AtbZIP16, AtGBF1, and AtbZIP68, respectively, demonstrated that these residues account for the observed changes in DNA binding activities of the respective proteins. Furthermore, our data suggest that Cys330 in bZIP16 is essential both for regulation of DNA binding activity (Fig. 5D) and for disulfide bond formation (Fig. 5C). The midpoint redox potential for Cys330 was determined to be −324 mV, which is biologically relevant and falls within the range of values (−290 to −330 mV) reported for other redox-active proteins, including thioredoxins, ferredoxin-thioredoxin reductase, and thioredoxin-regulated proteins (26). In addition, the midpoint redox potential for the second cysteine of bZIP16, Cys358 (C2), was determined to −287 mV, although it does not form intra- or intermolecular disulfide bonds (Fig. 5). This redox potential is more negative than that reported for glutathione (−240 mV; Ref. 49), suggesting that this cysteine is prone to redox regulation possibly through glutathionylation or mixed disulfide bonds. The midpoint redox potential of the redox-regulated AP2 domain-containing transcription factor Rap2.4a was determined to be −269 mV. Moderate oxidation of the glutathione pool was suggested to activate Rap2.4a-dependent gene expression, whereas stronger deviations from the normal cellular redox states inactivate Rap2.4a by aggregate formation (20).
Structural analysis of bZIP transcription factors revealed that these proteins bind DNA as dimers formed by the interaction of two α-helical stretches, which consist of 7 amino acid repeats per DNA turn (50). Under reducing and non-reducing conditions, the DNA-protein complexes formed between AtbZIP16 and its target DNA migrated similarly in the EMSA, suggesting that the homodimer of the protein is still formed and bound to the DNA. DTT induced monomerization of the AtbZIP16 proteins in vitro, and DTT was shown to stimulate DNA binding activity (Fig. 5). The conventional model for DNA binding of bZIP transcription factors proposes that dimerization of the leucine zipper protein is a prerequisite to specific recognition and DNA binding (51). However, more recent reports suggest that there are two possible pathways for DNA binding of bZIP transcription factors. In pathway I (dimer pathway), bZIP dimer formation precedes DNA binding, whereas pathway II (monomer pathway) involves the initial formation of the bZIP monomer-DNA complex, which then binds a second bZIP monomer in a subsequent step (40–42, 52). DTT stimulates monomerization, which might improve DNA binding activity of AtbZIP16 through pathway II where the monomer formed by the disruption of disulfide bond binds DNA, the monomer-DNA complex binds a second AtbZIP16 monomer, and subsequent dimerization takes place through the leucine zippers of the proteins. We show that the truncated form of AtbZIP16 containing only the BR of the DNA binding domain has DNA binding activity (Fig. 5E). The DNA binding of this truncated variant of AtbZIP16, which is unable to form a dimer via the leucine zipper, suggests that AtbZIP16 indeed can bind DNA as a monomer. Thus, it is likely that DNA binding of AtbZIP16 occurs via pathway II. Furthermore, it has been proposed that the monomer pathway not only allows for rapid identification of a specific DNA site in response to cellular stimuli and faster assembly of the bZIP dimer-DNA complex, but it could also provide an efficient means for discriminating between specific and nonspecific DNA target sites (42). We hypothesize that the monomeric form of AtbZIP16 binds to the DNA before the formation of the functional dimer to enhance the response to changes in the environment such as changing light conditions.
A model of the bZIP domain from AtbZIP16 was generated using the SWISS-MODEL and LOMETS software. From this model, the theoretical structural position of the regulatory Cys330 was assigned to a critical location within the basic region but just outside the direct contact site between the BR and the DNA (Fig. 8). In the free bZIP form, the position of Cys330 in each monomer and the distance between them allows for formation of a disulfide bond (Fig. 8A). Thus, the redox status of Cys330 could potentially be critical for DNA binding. If a disulfide bridge is formed between the Cys330 residues of the two bZIP16 monomers, the configuration of the zipper may not be open or flexible enough to allow DNA binding. Furthermore, upon DNA binding, the distance between the two Cys330 residues increases significantly and may not allow formation of the disulfide bond (Fig. 8B). Thus, the theoretical model supports the experimental evidence for a stimulation of DNA binding activity under reducing conditions (Figs. 5 and 7).
When the early light response was investigated in the bZIP16-WTOX lines, impaired induction of LHCB2.4 expression compared with wild type was observed. In the bZIP16-WTOX line, the levels of LHCB2.4 transcript was significantly reduced compared with wild type, suggesting that bZIP16 acts as a repressor of LHCB2.4. Expression of LHCB2.4 is strongly repressed in response to redox changes in the chloroplast (7), and bZIP16 possibly plays a role in mediating this repression. Interestingly, when the mutated forms of bZIP16 (OXC1 and OXC1C2) were overexpressed, no effect on LHCB2.4 expression was observed. This suggests that redox regulation of Cys330 is important for the activity of bZIP16. In support of the gene expression data and of the proposed role for bZIP16 as a repressor of photosynthetic gene expression, the bZIP16-WTOX line demonstrated elongated hypocotyls compared with wild type in response to light. This phenotype was observed neither in the OXC1 nor in the OXC1C2 line (Fig. 9). The T-DNA insertion lines for bZIP68 and GBF1 also demonstrated reduced LHCB2.4 expression levels in response to light compared with wild type, suggesting a role as activators for bZIP68 and GBF1. GBF1 has been shown to function as a transcriptional repressor of RBCS and CAT2 but as an activator of LHCs (21, 44). In addition, gbf1 mutants have also been shown to exhibit elongated hypocotyls in response to white and blue light (44). It is clear that AtbZIP16, AtbZIP68, and AtGBF1 form homo- and heterodimers (Fig. 4) (39). Heterodimer formation increases the diversity of functional G-box binding combinations, and the monomeric form of AtbZIP16 possibly binds to the DNA, and depending on the recruited partner, a different output is generated. DNA binding affinity and specificity, transactivation potential, and overall cell physiological function have been suggested to be altered by heterodimerization (53). Thus, our results suggest that combinatorial interactions among bZIP16, bZIP68, and GBF1 play a role in generating light signaling outputs and that these are possibly regulated by redox modifications to the proteins via the conserved Cys residues. The formation of bZIP homo- or heterodimers offers flexibility to a regulatory system, enabling a redox-controlled response to changes in the environment.
In conclusion, changes in intracellular redox potential triggers changes in the activity of many proteins (12). Thus, redox regulation of G-box-binding transcription factors offers a fundamental and mechanistic link between photosynthesis-dependent changes in the redox environment and regulation of nuclearly encoded photosynthesis genes.
Supplementary Material
Acknowledgments
We thank Dr. Thorsten Seidel Bielefeld University for the ABI5:YFP construct and Dr. Ulrike Zentgraf for the gbf1 mutant seeds.
This work was supported in part by grants from the Swedish research foundation Vetenskapsrådet and a Foundation for Strategic Research FFL2 grant (to Å. S.).

This article contains supplemental Figs. S1–S4.
- bZIP
- basic region leucine zipper
- LHCB2.4
- light-harvesting chlorophyll a/b-binding protein2.4
- CFP
- cyan fluorescent protein
- LHC
- light-harvesting complex
- GBF
- G-box binding factor
- CREB
- cAMP-response element-binding protein
- BR
- basic region.
REFERENCES
- 1. Fankhauser C., Staiger D. (2002) Photoreceptors in Arabidopsis thaliana: light perception, signal transduction and entrainment of the endogenous clock. Planta 216, 1–16 [DOI] [PubMed] [Google Scholar]
- 2. Chen M., Chory J., Fankhauser C. (2004) Light signal transduction in higher plants. Annu. Rev. Genet. 38, 87–117 [DOI] [PubMed] [Google Scholar]
- 3. Niyogi K. K. (1999) Photoprotection revisited: genetic and molecular approaches. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 333–359 [DOI] [PubMed] [Google Scholar]
- 4. Asada K. (2006) Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol. 141, 391–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Foyer C. H., Descourvieres P., Kunert K. J. (1994) Protection against oxygen radicals: an important defence mechanism studied in transgenic plants. Plant Cell Environ. 17, 507–523 [Google Scholar]
- 6. Bräutigam K., Dietzel L., Kleine T., Ströher E., Wormuth D., Dietz K. J., Radke D., Wirtz M., Hell R., Dörmann P., Nunes-Nesi A., Schauer N., Fernie A. R., Oliver S. N., Geigenberger P., Leister D., Pfannschmidt T. (2009) Dynamic plastid redox signals integrate gene expression and metabolism to induce distinct metabolic states in photosynthetic acclimation in Arabidopsis. Plant Cell 21, 2715–2732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kindgren P., Kremnev D., Blanco N. E., de Dios Barajas Lopez J., Fernandez A. P., Tellgren-Roth C., Small I., Strand Å. (2012) The plastid redox insensitive 2 mutant of Arabidopsis is impaired in PEP activity and high light-dependent plastid redox signalling to the nucleus. Plant J. 70, 279–291 [DOI] [PubMed] [Google Scholar]
- 8. Piippo M., Allahverdiyeva Y., Paakkarinen V., Suoranta U. M., Battchikova N., Aro E. M. (2006) Chloroplast-mediated regulation of nuclear genes in Arabidopsis thaliana in the absence of light stress. Physiol. Genomics 25, 142–152 [DOI] [PubMed] [Google Scholar]
- 9. Baier M., Stroher E., Dietz K. J. (2004) The acceptor availability at photosystem I and ABA control nuclear expression of 2-Cys peroxiredoxin-A in Arabidopsis thaliana. Plant Cell Physiol. 45, 997–1006 [DOI] [PubMed] [Google Scholar]
- 10. Rossel J. B., Wilson I. W., Pogson B. J. (2002) Global changes in gene expression in response to high light in Arabidopsis. Plant Physiol. 130, 1109–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fernandez A. P., Strand Å. (2008) Retrograde signaling and plant stress: plastid signals initiate cellular stress responses. Curr. Opin. Plant Biol. 11, 509–513 [DOI] [PubMed] [Google Scholar]
- 12. Dietz K. J. (2003) Redox control, redox signaling, and redox homeostasis in plant cells. Int. Rev. Cytol. 228, 141–193 [DOI] [PubMed] [Google Scholar]
- 13. Hernandez J. M., Heine G. F., Irani N. G., Feller A., Kim M. G., Matulnik T., Chandler V. L., Grotewold E. (2004) Different mechanisms participate in the R-dependent activity of the R2R3 MYB transcription factor C1. J. Biol. Chem. 279, 48205–48213 [DOI] [PubMed] [Google Scholar]
- 14. Heine G. F., Hernandez J. M., Grotewold E. (2004) Two cysteines in plant R2R3 MYB domains participate in redox-dependent DNA binding. J. Biol. Chem. 279, 37878–37885 [DOI] [PubMed] [Google Scholar]
- 15. Serpa V., Vernal J., Lamattina L., Grotewold E., Cassia R., Terenzi H. (2007) Inhibition of AtMYB2 DNA-binding by nitric oxide involves cysteine S-nitrosylation. Biochem. Biophys. Res. Commun. 361, 1048–1053 [DOI] [PubMed] [Google Scholar]
- 16. Tron A. E., Bertoncini C. W., Chan R. L., Gonzalez D. H. (2002) Redox regulation of plant homeodomain transcription factors. J. Biol. Chem. 277, 34800–34807 [DOI] [PubMed] [Google Scholar]
- 17. Spoel S. H., Tada Y., Loake G. J. (2010) Post-translational protein modification as a tool for transcription reprogramming. New Phytol. 186, 333–339 [DOI] [PubMed] [Google Scholar]
- 18. Tada Y., Spoel S. H., Pajerowska-Mukhtar K., Mou Z., Song J., Wang C., Zuo J., Dong X. (2008) Plant immunity requires conformational changes [corrected] of NPR1 via S-nitrosylation and thioredoxins. Science 321, 952–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Després C., Chubak C., Rochon A., Clark R., Bethune T., Desveaux D., Fobert P. R. (2003) The Arabidopsis NPR1 disease resistance protein is a novel cofactor that confers redox regulation of DNA binding activity to the basic domain/leucine zipper transcription factor TGA1. Plant Cell 15, 2181–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shaikhali J., Heiber I., Seidel T., Stroher E., Hiltscher H., Birkmann S., Dietz K. J., Baier M. (2008) The redox-sensitive transcription factor Rap2.4a controls nuclear expression of 2-Cys peroxiredoxin A and other chloroplast antioxidant enzymes. BMC Plant Biol. 8, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Smykowski A., Zimmermann P., Zentgraf U. (2010) G-Box binding factor1 reduces CATALASE2 expression and regulates the onset of leaf senescence in Arabidopsis. Plant Physiol. 153, 1321–1331 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22. Gabrielsen O. S., Hornes E., Korsnes L., Ruet A., Oyen T. B. (1989) Magnetic DNA affinity purification of yeast transcription factor tau—a new purification principle for the ultrarapid isolation of near homogeneous factor. Nucleic Acids Res. 17, 6253–6267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Srivastava V., Srivastava M. K., Chibani K., Nilsson R., Rouhier N., Melzer M., Wingsle G. (2009) Alternative splicing studies of the reactive oxygen species gene network in Populus reveal two isoforms of high-isoelectric-point superoxide dismutase. Plant Physiol. 149, 1848–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Montemartini M., Kalisz H. M., Kiess M., Nogoceke E., Singh M., Steinert P., Flohé L. (1998) Sequence, heterologous expression and functional characterization of a novel tryparedoxin from Crithidia fasciculata. Biol. Chem. 379, 1137–1142 [DOI] [PubMed] [Google Scholar]
- 25. Zhai Z., Sooksa-nguan T., Vatamaniuk O. K. (2009) Establishing RNA interference as a reverse-genetic approach for gene functional analysis in protoplasts. Plant Physiol. 149, 642–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hirasawa M., Schürmann P., Jacquot J. P., Manieri W., Jacquot P., Keryer E., Hartman F. C., Knaff D. B. (1999) Oxidation-reduction properties of chloroplast thioredoxins, ferredoxin:thioredoxin reductase, and thioredoxin f-regulated enzymes. Biochemistry 38, 5200–5205 [DOI] [PubMed] [Google Scholar]
- 27. Arnold K., Bordoli L., Kopp J., Schwede T. (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 28. Wu S., Zhang Y. (2007) LOMETS: a local meta-threading-server for protein structure prediction. Nucleic Acids Res. 35, 3375–3382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., Shindyalov I. N., Bourne P. E. (2000) The Protein Data Bank. Nucleic Acids Res. 28, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Guex N., Peitsch M. C. (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18, 2714–2723 [DOI] [PubMed] [Google Scholar]
- 31. Clough S. J., Bent A. F. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 [DOI] [PubMed] [Google Scholar]
- 32. Kindgren P., Norén L., Barajas López J. D., Shaikhali J., Strand Å. (December 26, 2011) Interplay between HEAT SHOCK PROTEIN 90 and HY5 controls PhANG expression in response to the GUN5 plastid signal. Mol. Plant 10.1093/mp/ssr112 [DOI] [PubMed] [Google Scholar]
- 33. Kleine T., Kindgren P., Benedict C., Hendrickson L., Strand Å. (2007) Genome-wide gene expression analysis reveals a critical role for CRYPTOCHROME1 in the response of Arabidopsis to high irradiance. Plant Physiol. 144, 1391–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rey D. A., Puhler A., Kalinowski J. (2003) The putative transcriptional repressor McbR, member of the TetR-family, is involved in the regulation of the metabolic network directing the synthesis of sulfur containing amino acids in Corynebacterium glutamicum. J. Biotechnol. 103, 51–65 [DOI] [PubMed] [Google Scholar]
- 35. Shaikhali J., Barajas-Lopéz J., Ötvös K., Sánchez Garcia A., Srivastava V., Wingsle G., Bako L., Strand Å. (2012) The CRYPTOCHROME1-dependent response to excess light is mediated through the transcriptional activators ZINC FINGER PROTEIN EXPRESSED IN INFLORESCENCE MERISTEM LIKE1 and 2 in Arabidopsis thaliana. Plant Cell, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jakoby M., Weisshaar B., Dröge-Laser W., Vicente-Carbajosa J., Tiedemann J., Kroj T., Parcy F.; bZIP Research Group (2002) bZIP transcription factors in Arabidopsis. Trends Plant Sci. 7, 106–111 [DOI] [PubMed] [Google Scholar]
- 37. Schindler U., Terzaghi W., Beckmann H., Kadesch T., Cashmore A. R. (1992) DNA binding site preferences and transcriptional activation properties of the Arabidopsis transcription factor GBF1. EMBO J. 11, 1275–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schindler U., Menkens A. E., Beckmann H., Ecker J. R., Cashmore A. R. (1992) Heterodimerization between light-regulated and ubiquitously expressed Arabidopsis GBF bZIP proteins. EMBO J. 11, 1261–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shen H., Cao K., Wang X. (2008) AtbZIP16 and AtbZIP68, two new members of GBFs, can interact with other G group bZIPs in Arabidopsis thaliana. BMB Rep. 41, 132–138 [DOI] [PubMed] [Google Scholar]
- 40. Metallo S. J., Schepartz A. (1997) Certain bZIP peptides bind DNA sequentially as monomers and dimerize on the DNA. Nat. Struct. Biol. 4, 115–117 [DOI] [PubMed] [Google Scholar]
- 41. Berger C., Piubelli L., Haditsch U., Bosshard H. R. (1998) Diffusion-controlled DNA recognition by an unfolded, monomeric bZIP transcription factor. FEBS Lett. 425, 14–18 [DOI] [PubMed] [Google Scholar]
- 42. Kohler J. J., Metallo S. J., Schneider T. L., Schepartz A. (1999) DNA specificity enhanced by sequential binding of protein monomers. Proc. Natl. Acad. Sci. U.S.A. 96, 11735–11739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Petersen M. T., Jonson P. H., Petersen S. B. (1999) Amino acid neighbours and detailed conformational analysis of cysteines in proteins. Protein Eng. 12, 535–548 [DOI] [PubMed] [Google Scholar]
- 44. Mallappa C., Yadav V., Negi P., Chattopadhyay S. (2006) A basic leucine zipper transcription factor, G-box-binding factor 1, regulates blue light-mediated photomorphogenic growth in Arabidopsis. J. Biol. Chem. 281, 22190–22199 [DOI] [PubMed] [Google Scholar]
- 45. Deppmann C. D., Acharya A., Rishi V., Wobbes B., Smeekens S., Taparowsky E. J., Vinson C. (2004) Dimerization specificity of all 67 B-ZIP motifs in Arabidopsis thaliana: a comparison to Homo sapiens B-ZIP motifs. Nucleic Acids Res. 32, 3435–3445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vincentz M., Bandeira-Kobarg C., Gauer L., Schlögl P., Leite A. (2003) Evolutionary pattern of angiosperm bZIP factors homologous to the maize Opaque2 regulatory protein. J. Mol. Evol. 56, 105–116 [DOI] [PubMed] [Google Scholar]
- 47. Foster R., Izawa T., Chua N. H. (1994) Plant bZIP proteins gather at ACGT elements. FASEB J. 8, 192–200 [DOI] [PubMed] [Google Scholar]
- 48. Amoutzias G. D., Bornberg-Bauer E., Oliver S. G., Robertson D. L. (2006) Reduction/oxidation-phosphorylation control of DNA binding in the bZIP dimerization network. BMC Genomics 7, 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Diaz Vivancos P., Wolff T., Markovic J., Pallardó F. V., Foyer C. H. (2010) A nuclear glutathione cycle within the cell cycle. Biochem. J. 431, 169–178 [DOI] [PubMed] [Google Scholar]
- 50. Ellenberger T. E., Brandl C. J., Struhl K., Harrison S. C. (1992) The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted alpha helices: crystal structure of the protein-DNA complex. Cell 71, 1223–1237 [DOI] [PubMed] [Google Scholar]
- 51. Gentz R., Rauscher F. J., 3rd, Abate C., Curran T. (1989) Parallel association of Fos and Jun leucine zippers juxtaposes DNA binding domains. Science 243, 1695–1699 [DOI] [PubMed] [Google Scholar]
- 52. Cranz S., Berger C., Baici A., Jelesarov I., Bosshard H. R. (2004) Monomeric and dimeric bZIP transcription factor GCN4 bind at the same rate to their target DNA site. Biochemistry 43, 718–727 [DOI] [PubMed] [Google Scholar]
- 53. Näär A. M., Lemon B. D., Tjian R. (2001) Transcriptional coactivator complexes. Annu. Rev. Biochem. 70, 475–501 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.