Background: Rabies virus expresses five isoforms of P protein, which undergo nucleocytoplasmic trafficking important to roles as antagonists of interferon-mediated immunity.
Results: P isoform trafficking is mediated by a novel module containing co-regulated overlapping nuclear localization and export signals.
Conclusion: P isoform nuclear localization involves a novel trafficking module.
Significance: A novel molecular switch regulating trafficking, important to a viral interferon antagonist, is identified.
Keywords: Intracellular Trafficking, Nuclear Pore, Nuclear Translocation, Nuclear Transport, Viral Protein
Abstract
Regulated nucleocytoplasmic transport of proteins is central to cellular function and dysfunction during processes such as viral infection. Active protein trafficking into and out of the nucleus is dependent on the presence within cargo proteins of intrinsic specific modular signals for nuclear import (nuclear localization signals, NLSs) and export (nuclear export signals, NESs). Rabies virus (RabV) phospho (P) protein, which is largely responsible for antagonising the host anti-viral response, is expressed as five isoforms (P1–P5). The subcellular trafficking of these isoforms is thought to depend on a balance between the activities of a dominant N-terminal NES (N-NES) and a distinct C-terminal NLS (C-NLS). Specifically, the N-NES-containing isoforms P1 and P2 are cytoplasmic, whereas the shorter P3–P5 isoforms, which lack the N-NES, are believed to be nuclear through the activity of the C-NLS. Here, we show for the first time that RabV P contains an additional strong NLS in the N-terminal region (N-NLS), which, intriguingly, overlaps with the N-NES. This arrangement represents a novel nuclear trafficking module where the N-NLS is inactive in P1 but becomes activated in P3, concomitant with truncation of the N-NES, to become the principal targeting signal conferring nuclear accumulation. Understanding this unique switch arrangement of overlapping, co-regulated NES/NLS sequences is vital to delineating the critical role of RabV P protein in viral infection.
Introduction
The eukaryotic cell nucleus is separated from the cytoplasm by the double-membrane nuclear envelope. All transport across the nuclear envelope occurs through nuclear pore complexes, and active protein transport through the nuclear pore complexes is dependent on nuclear localization signals (NLSs)2 or nuclear export signals (NESs) within the cargo proteins (1–4). This enables the specific regulation of protein access to the nuclear contents including the genomic DNA, an essential mechanism in processes such as development, apoptosis, and immunity (for review, see Refs. 1, 3, 5).
NLS/NES-dependent trafficking is mediated by importin (IMP) proteins, of which there are at least 6 α and 24 β isoforms in humans. IMPs bind directly to NLSs/NESs, either as an IMPα/β1 heterodimer or as an IMPβ homologue alone (6–8). NLSs are commonly short monopartite or bipartite sequences enriched in basic residues (9–11); classical monopartite NLSs such as the SV40 large T-antigen (T-ag) NLS are single short basic sequences that variously conform to the KKXK consensus sequence (where X is any amino acid) (12), whereas bipartite NLSs typically contain two stretches of 4–8 basic residues separated by a 10–12-residue spacer (9, 10). Recent studies have also identified a number of nonclassical bipartite NLSs with longer spacers (up to 30 residues) (9, 13), as well as conformation-dependent NLSs such as that of the signal transducer and activator of transcription 3 (STAT3) (11). NESs are typically 9–15-residue sequences containing a conserved hydrophobic residue motif (7) and are recognized by exportins of the IMPβ superfamily, the best characterized of which is CRM1.
NLSs and NESs are generally functionally independent, modular sequences able to mediate active nuclear import or export of heterologous proteins (12, 14, 15). Thus, the addition/deletion of independent modular NLSs or NESs by mechanisms including alternate mRNA splicing, leaky ribosomal mRNA scanning, and RNA editing, represents a key mechanism by which differential nucleocytoplasmic localization can be conferred on the isoforms of cellular proteins including promyelocytic leukemia (PML) protein, Krüppel-like factor 6, and spastin (16–18), as well as virus-encoded proteins such as RabV P and paramyxovirus P/V/W proteins (19–21); this is thought to play key roles in isoform functional heterogeneity.
Regulated nuclear trafficking of viral proteins is common even for viruses with entirely cytoplasmic life cycles such as RabV (22–30), as it enables viral modification of host cell functions through interference with intranuclear processes and/or the nucleocytoplasmic trafficking of cellular proteins (31–34). RabV P protein is expressed as five isoforms in infected cells, full-length P1 and the N-terminally truncated P2–P5 (Fig. 1), which perform functions in viral replication processes and antagonism of interferon (IFN)-mediated antiviral responses (19, 29, 35–38). P1 is the most abundantly expressed isoform, presumably enabling essential roles as the viral polymerase (L-protein)-binding co-factor, whereas the “accessory” proteins P2–P5 show a decreasing gradient of expression (19, 29). P isoform nuclear trafficking has been reported to play vital roles in IFN antagonism by enabling cytoplasmic and nuclear targeting of essential factors of the IFN system including STAT1 and 2, to affect their nucleocytoplasmic trafficking/DNA binding, and PML nuclear bodies (29, 35–39). As a result, defects in the trafficking of RabV P correlate with defective viral IFN antagonism and pathogenicity in infected animals (36, 40). Analogous functions have been attributed to isoforms of the vesicular stomatitis virus M protein, and of the P proteins of Nipah and Hendra viruses (21, 24–29, 41). Thus, the expression of multiple isoforms with heterogeneous nucleocytoplasmic localization appears to be an important mechanism whereby viruses with limited genome capacity can optimize their functional proteome to enable efficient evasion of the IFN response (29).
FIGURE 1.
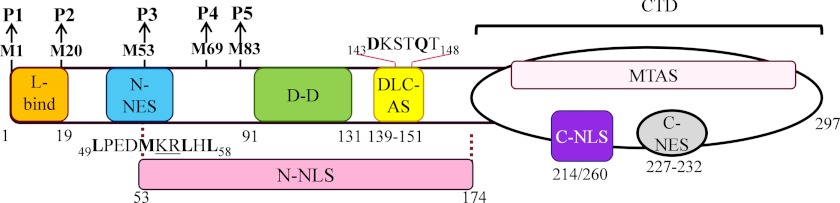
Schematic representation of RabV P protein. The RabV P gene encodes a 297-residue protein (P1) and four N-terminally truncated isoforms (P2–P5), expressed from internal in-frame AUG start codons encoding methionine (M) residues at positions 20, 53, 69, and 83 (19). RabV P interacts with the cellular trafficking machinery through the N-NES (sequence indicated under the protein with key residues in bold) and C-NES, which interact with CRM1, and the C-NLS (the critical residues of which include Lys214/Arg260, which are proximally localized on the CTD surface) (30, 42) and N-NLS (identified in the current study; critical residues of the N-NLS within the N-NES sequence are underlined). RabV P nuclear import is facilitated by interactions with the dynein light chain LC8 via a DLC-AS (sequence indicated above protein with essential residues Asp143/Gln147 in bold) (44, 66). A MTAS, mapped within the C-terminal domain (CTD), also enables association of P3 with the MT network (43). The N-NES is truncated in P3 to remove residues 49–52 and is deleted from P4 and P5, enabling nuclear localization of these isoforms. RabV P also interacts with viral proteins L (via P1–19) and N (not shown) (67, 68), homodimerizes via the dimerization domain (D-D) (58), and interacts with cellular proteins of the IFN system such as STAT 1 and 2, and PML (not shown) (37, 39). The positions of individual domains are indicated by residue numbers beneath the protein.
RabV P isoform nucleocytoplasmic localization is currently believed to be determined principally by two distinct, independent signals: a dominant CRM1-dependent NES close to the N terminus (N-NES, residues 49–58) and a predicted NLS (C-NLS) in the C-terminal domain (CTD; Fig. 1) (30). P1 and P2 contain the N-NES and are thus cytoplasmic (30). In contrast, P3–P5 can localize in the nucleus, and this is thought to be due to truncation/deletion of the N-NES such that the C-NLS becomes the main “default” targeting signal (Fig. 1) (30). However, the nucleocytoplasmic localization of RabV P isoforms and the NLS function of P with regard to IMP binding have never been quantitatively examined. Using single cell imaging approaches and in vitro molecular interaction analyses, we have examined the trafficking of P protein in detail, finding that the nucleocytoplasmic localization of P isoforms is principally regulated by a novel nuclear trafficking module in which the N-NES overlaps with a newly identified N-terminal NLS (N-NLS) sequence (Fig. 1). The close association of these signals enables their co-regulation by a unique mechanism whereby truncation of the module both inactivates the N-NES and concomitantly activates the N-NLS, producing a coordinated effect on protein trafficking. This work reveals a novel mechanism underlying the efficient regulation of protein nuclear trafficking, and contributes to our understanding of events at the virus-host interface.
EXPERIMENTAL PROCEDURES
Constructs
All P protein constructs were generated using the P gene from RabV CVS11 strain, as described previously (42–44). cDNA encoding P isoforms or truncated derivatives was generated by PCR and cloned in-frame N-terminal to GFP in the pEGFP-N3 vector by BglII-BamHI restriction ligation (42–44) for the expression of GFP-fused protein in mammalian cells. To express untagged proteins, the cDNA was cloned into the pΔEGFP-C1 vector, which we generated by excising GFP from the pEGFP-C1 vector using BspE1 and AgeI. cDNA containing mutations to residues Lys54, Arg55, Asp143, Gln147, Lys139, and Arg144 (positions of the residues in the P protein sequence are shown in superscript) was generated by PCR extension or overlap mutagenesis as described previously (42, 44). Constructs for the expression of His6-tagged GFP fusion proteins in Escherichia coli used the pGFP-RfB vector, and were generated using the GatewayTM Cloning System (Invitrogen) (23, 45).
Cell Culture, Transfection, Drug Treatments, and Immunofluorescence
HeLa cells were routinely cultured in DMEM with 10% FCS (37 °C, 5% CO2). For confocal laser scanning microscopy (CLSM), cells were grown on coverslips to 80–90% confluence before transfection using Lipofectamine 2000 (Invitrogen) and analysis 16–24 h later. For inhibition of CRM1-mediated nuclear export, cells were treated with 2.8 ng/ml leptomycin-B (LMB, a kind gift from M. Yoshida, RIKEN, Japan) for 3 h prior to analysis.
For indirect immunofluorescence of untagged protein, cells were washed with PBS and fixed using 3.7% formaldehyde followed by 90% methanol (43). Cells were then blocked in 1% BSA in PBS before immunostaining with anti-RabV P protein antibody (46) and Alexa Fluor 488-coupled secondary antibody (Invitrogen A11008).
CLSM and Image Analysis
Cells were analyzed using a Nikon Eclipse C1 inverted confocal laser scanning microscope, with an Olympus 60× oil immersion objective (NA 1.4) (for fixed cells) or an Olympus 60× water immersion objective (NA 1.2) and 37 °C heated chamber (for living cells). Analysis of digitized CLSM images was performed using ImageJ software (v1.44p) as described previously (42–44, 47–49) to determine the ratio of nuclear to cytoplasmic fluorescence (Fn/c) for single cells using the formula Fn/c = (Fn − Fb)/(Fc − Fb), where Fn is the nuclear fluorescence, Fc the cytoplasmic fluorescence, and Fb the background fluorescence. The mean Fn/c was calculated for ≥49 cells, and statistical analysis (t test) was performed using GraphPad Instat software as described previously (42).
Protein Expression and Purification
Constructs encoding His6-tagged GFP-fused P protein derivatives P54–174 and P1–174 (sequence of P indicated in superscript), and P54–174-(K54N/R55N) (in which Lys54 and Arg55 are mutated to asparagine) were expressed in E. coli strain BL21 pRep4 cells before induction of expression and purification using nickel-nitrilotriacetic acid columns (Qiagen) under denaturing conditions (8 m urea), as described previously (50). Proteins were renatured on the column and eluted with 200 mm imidazole (45), prior to dialysis and concentration using a 30K MWCO column (Amicon). Mouse IMPα2 and IMPβ1 were expressed as GST fusion proteins and purified by affinity chromatography using glutathione-Sepharose (50); biotinylated IMPα2 protein generated as described previously (51), and GFP-T-ag-NLS protein, were kindly donated by Dr. Kylie Wagstaff (Monash University. Australia).
Native Gel Electrophoresis
GFP fusion proteins (2 μm) were incubated with or without IMPα2, IMPβ1, or predimerized IMPα/β (1–9 μm), for 15 min at room temperature and then electrophoresed for 3–5 h at 4 °C in 1× TBE buffer using precast native polyacrylamide (10%) gels (Bio-Rad) (52). The migration of GFP fusion proteins was visualized using a TyphoonTM Trio Variable Mode Imager (GE Life Sciences), and gel images were processed using ImageJ software as described previously (52).
AlphaScreen Binding Assays
GFP-fused P proteins (30 nm final concentration) and 0–60 nm biotinylated IMPα2 or IMPα/β heterodimer were incubated in 384-well plates (PerkinElmer Life Sciences) for 30 min at room temperature. 1 μl of streptavidin-coated acceptor beads (diluted 1:10 in PBS) and 1 μl of 2.5% BSA in PBS were then added to each well (51). After 90 min, 1 μl of nickel chelate-coated donor beads (diluted 1:10 in PBS) was added and incubated for a further 2 h before measurement of AlphaScreen counts using a Fusion-αTM plate reader (PerkinElmer Life Sciences) (51). Triplicate values were averaged and curves plotted using SigmaPlot (51).
RESULTS
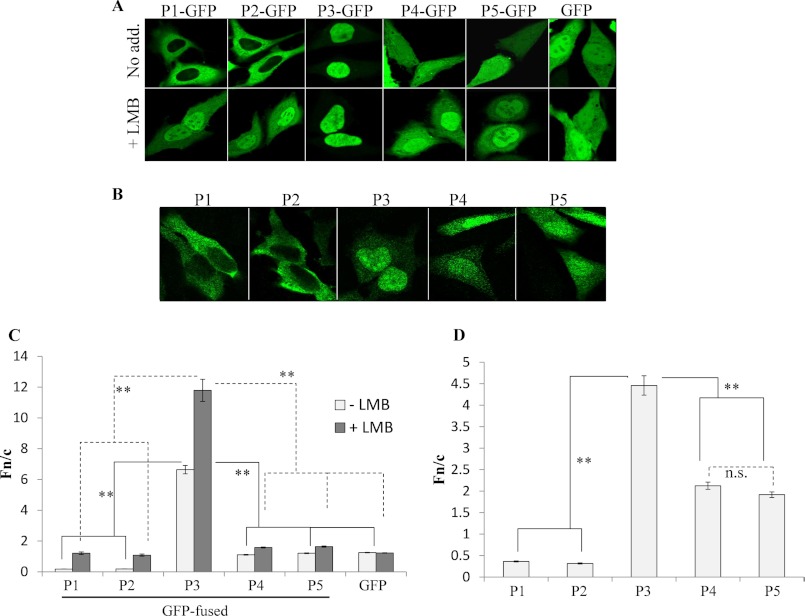
P3 Nuclear Localization Is Significantly Greater Than That of Other Isoforms
Previous research using nonquantitative CLSM analysis of fixed cells indicated that P1 and P2 are cytoplasmic due to the N-NES, whereas P3–P5 are mostly nuclear due to the C-NLS (30). This work also indicated that the level of nuclear localization may differ among P3, P4, and P5, implying that additional sequences or mechanisms contribute to trafficking. To determine quantitatively the nucleocytoplasmic distribution of P1–P5, we transfected HeLa cells to express these proteins fused at their C termini to GFP, enabling analysis of their localization in living cells by CLSM (Fig. 2A). P1-GFP and P2-GFP were clearly excluded from the nucleus, whereas P4-GFP and P5-GFP could enter cell nuclei but showed only moderate nuclear localization. P3-GFP, however, showed clear nuclear accumulation (Fig. 2A). To quantitate the nuclear localization, we determined the nuclear to cytoplasmic fluorescence ratio (Fn/c) as previously (42–44, 47–49) (Fig. 2C), confirming that P1- and P2-GFP were both excluded almost entirely from the nucleus (Fn/c of approximately 0.2), whereas P4- and P5-GFP both exhibited diffuse localization (Fn/c of approximately 1) similar to that of GFP alone. In contrast, the level of accumulation of P3-GFP (Fn/c of approximately 7), was approximately 5-fold higher than that of P4-GFP (Fig. 2C). Comparable results were observed in Vero cells (data not shown).
FIGURE 2.
P3 is more nuclear than other P isoforms due to differing nuclear import. A and B, HeLa cells transfected to express the indicated proteins were treated with or without LMB (2.8 ng/ml, 3 h) prior to imaging of living cells by CLSM (A), or fixation and immunostaining of cells with anti-P antibody for CLSM analysis (B). C and D, images such as those shown in A and B were analyzed to determine the ratio of nuclear to cytoplasmic fluorescence (Fn/c) as described previously (36, 42–44, 47, 48). Data are shown as mean Fn/c ± S.E. (error bars), n ≥ 52 from ≥ 2 separate assays. Statistical analysis used Student's t test (unpaired) (**, p < 0.0001; n.s., not significant).
We also examined the localization of untagged P protein isoforms in transfected HeLa cells by fixation and immunostaining with anti-P antibody before quantitative CLSM analysis (Fig. 2, B and D). This confirmed the observations from live-cell analyses, showing that P3 accumulates in the nucleus to a much greater extent than the other isoforms (Fig. 2D).
P3–P5 each lack the N-NES and contain the C-NLS that is predicted to mediate nuclear localization of these isoforms (30), as well as a CRM1-dependent NES in the CTD (C-NES), which can modulate nuclear localization by the C-NLS (42) (Fig. 1). Thus, the differential nuclear trafficking of P3–P5 could indicate that the C-NLS is a relatively weak signal and that P3 contains an additional, stronger NLS, or that the activity of the C-NES is increased in P4 and P5 compared with P3. To test this, we treated cells expressing GFP-fused P isoforms with or without the CRM1 inhibitor LMB before analysis by CLSM (42) (Fig. 2A). LMB treatment significantly increased the nuclear localization of all GFP-fused P isoforms but not that of GFP alone, confirming that the localization of all P isoforms is regulated by CRM1, due to the N-NES and/or C-NES (Fig. 2, A and C) (42). However, even in LMB-treated cells, P3-GFP nuclear accumulation remained much higher (>7-fold) than that of P4- or P5-GFP (Fig. 2C). Intriguingly, the Fn/c for P3-GFP was also approximately 10-fold that for P1-GFP, even though P1 contains the entire P3 sequence. Thus, it appeared that P3 contains a previously uncharacterized NLS that is inactive in P1 due to the presence of P protein residues 1–52, and absent or inactivated in P4 and P5 due to the deletion of the residues 53–68 (Fig. 1).
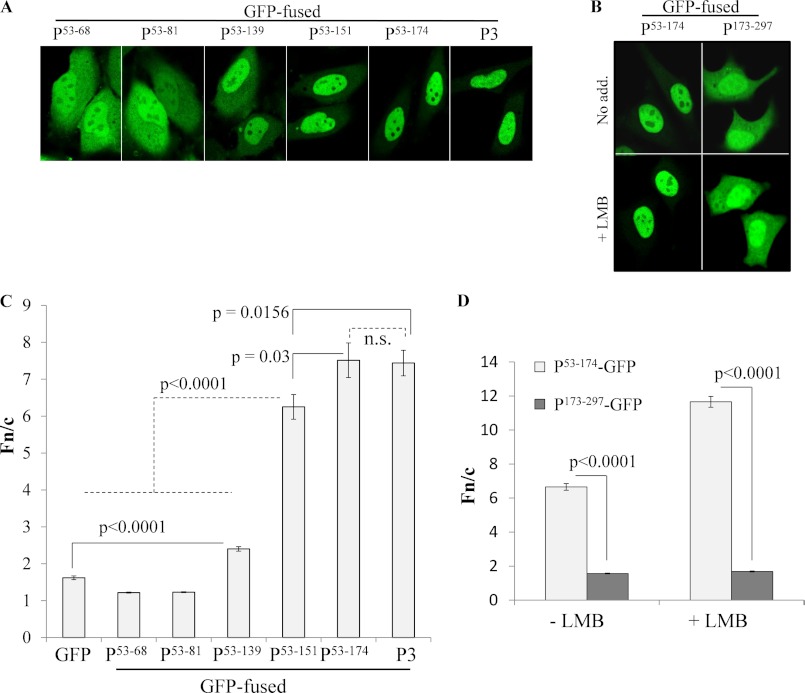
P3 Contains a NLS in Its N-terminal Region
To determine the minimal region of P3 sufficient to account for its increased nuclear localization compared with other isoforms, we generated several C-terminally truncated versions of P3 fused to GFP and determined their subcellular localization in living HeLa cells as above (Fig. 3A).
FIGURE 3.
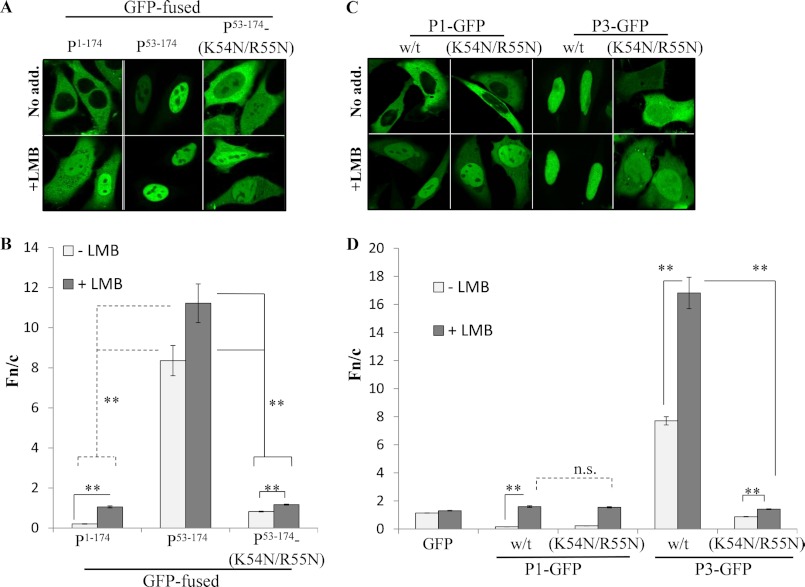
P53–174 is necessary and sufficient to confer nuclear accumulation equivalent to that of P3. A and B, HeLa cells were transfected to express the indicated proteins. C and D, live cells, treated with or without LMB, were analyzed by CLSM to calculate the Fn/c (mean ± S.E. (error bars), n ≥ 49 from 3 separate assays), as described in the legend to Fig. 2. Statistical analysis was according to the legend to Fig. 2 with p values indicated (n.s., not significant).
P53–68-GFP and P53–81-GFP (encoding the P protein residues indicated in superscript) correspond to the regions deleted from P4 and P5 respectively compared with P3. Both regions contain several basic residues that could form part of a NLS (Lys54, Arg55, Lys62, Arg70, Arg77), and NLSmapper online prediction software (53) indicated that P53–81 may contain a bipartite NLS (data not shown). However, both regions failed to confer nuclear accumulation on GFP (Fig. 3, A and C; Fn/c of approximately 1), indicating that additional sequence is required. The minimal region able to confer nuclear accumulation on GFP was within P53–139, which showed an Fn/c value of approximately 2.5 (Fig. 3, A and C); this level of nuclear accumulation, however, was not as high as that of P3.
P53–151 and P53–174 both contain the RabV P dynein light chain-association sequence (DLC-AS) (see Fig. 1), which facilitates NLS-mediated nuclear import (44, 49). P53–151-GFP accumulated in the nucleus to a significantly (p < 0.0001) higher extent (Fn/c of approximately 6) than P53–139-GFP, but to a significantly (p = 0.0156) lower extent than P3-GFP (Fn/c > 7) (Fig. 3, A and C). In contrast, P53–174-GFP (Fn/c of 7.5) accumulated in the nucleus to levels completely in keeping with those of P3-GFP (Fig. 3, A and C), indicating that P53–174 contains the sequence necessary and sufficient to mediate full nuclear localization of P3. This sequence was named the N-terminal NLS (N-NLS).
Importantly, P53–174 lacks the C-NLS/C-NES-containing CTD (Fig. 1), indicating that the predicted C-NLS makes little or no contribution to P3 nuclear accumulation. To test this, we compared the nuclear accumulation of P53–174-GFP with that of the CTD (P173–297-GFP), in cells treated with or without LMB (Fig. 3, B and D). P53–174 conferred significantly (p < 0.0001) higher nuclear accumulation (4-fold) on GFP than did the CTD. Further, this difference was almost 7-fold in LMB-treated cells (Fig. 3, B and D), indicating that the N-terminal region/N-NLS makes a greater contribution to the nuclear import of P3 than the CTD/C-NLS.
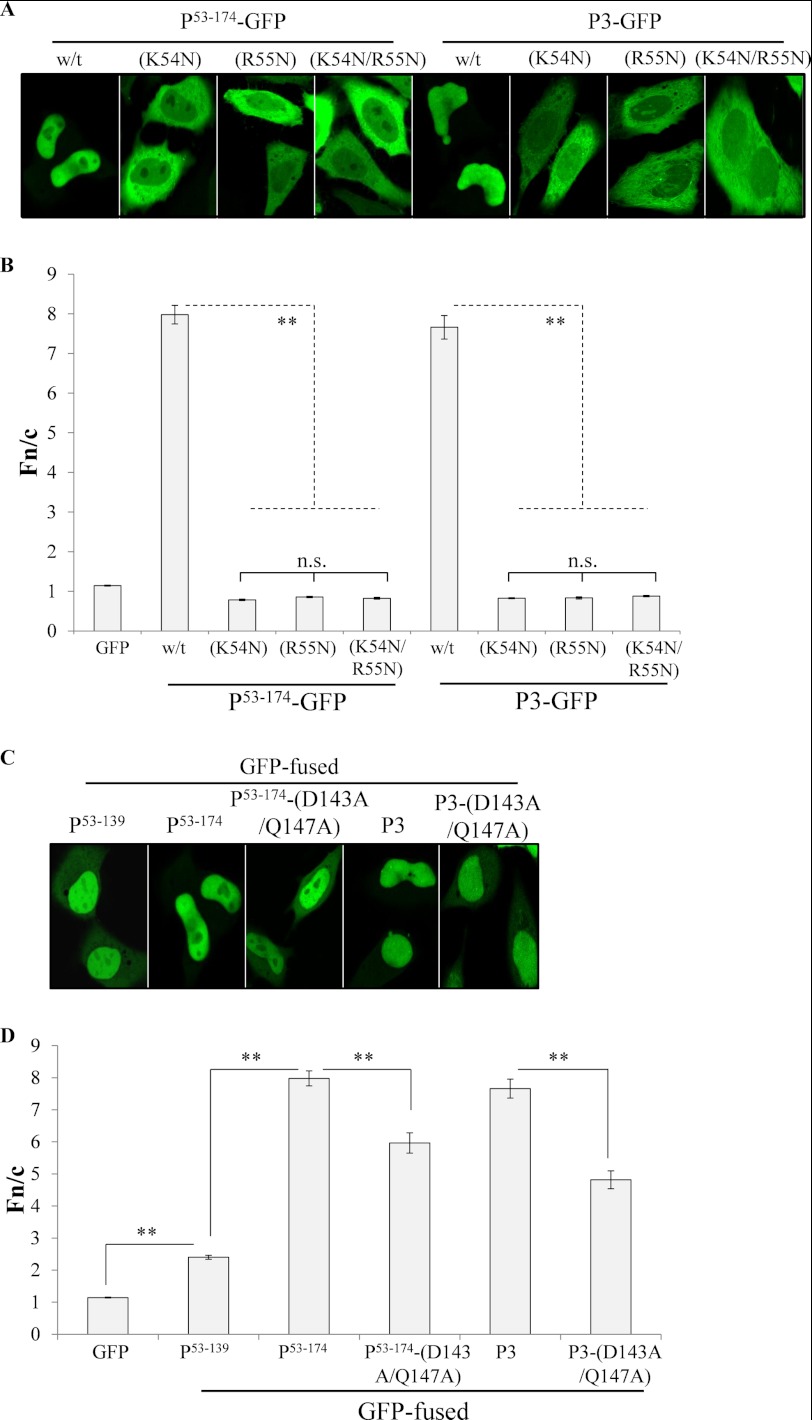
Identification of Residues Important for P3 Nuclear Accumulation
Because P3 accumulates in the nucleus to a significantly higher extent than P4 (see above), P53–68 is likely to contain key residues of the N-NLS. As NLSs commonly contain clustered basic residues, we tested the role of Lys54 and Arg55 in P3 nuclear accumulation by generating constructs for the expression of GFP-fused P protein derivatives in which these residues were mutated to asparagine: P3-(K54N/R55N)-GFP and P53–174-(K54N/R55N)-GFP, in which both residues are mutated, and P3-(K54N)-GFP, P53–174-(K54N)-GFP, P3-(R55N)-GFP and P53–174-(R55N)-GFP, which contain single mutations. CLSM analysis of live HeLa cells expressing these proteins (Fig. 4, A and B) indicated that both residues are important for nuclear localization of P3 and P53–174, as mutation of either residue individually or their combined mutation, resulted in a significant decrease in nuclear accumulation compared with wild-type (WT) protein (Fig. 4B). No significant difference in Fn/c was found between proteins containing either the K54N or R55N single mutations, or the K54N/R55N double mutation (Fn/c values of approximately 0.8 in all cases), indicating that both residues are critical to the N-NLS. Similar effects were observed for constructs in which residues Lys54 and Arg55 were deleted (data not shown), and a similar decrease in nuclear accumulation was observed in fixed, immunostained cells transfected with untagged P3-(K54N/R55N) compared with WT P3 (Fn/c of approximately 2 and 5 respectively; images not shown). Thus, Lys54/Arg55 are essential to N-NLS function, and their proximity to the N terminus may account for the quantitatively lower levels of nuclear accumulation observed in previous studies using P3 modified at its N terminus (42, 43).
FIGURE 4.
P3 nuclear accumulation is critically dependent on residues Lys54 and Arg55 and is facilitated by the DLC-AS. A and C, HeLa cells transfected to express the indicated proteins were analyzed live by CLSM. B and D, images such as those shown were analyzed to determine the Fn/c (mean ± S.E. (error bars), n ≥ 57 from three separate assays) as described in the legend to Fig. 2. w/t, wild-type. Statistical analysis was according to the legend to Fig. 2.
To investigate directly the potential role of the DLC-AS in facilitating N-NLS-mediated nuclear import, we converted the essential DLC-AS residues Asp143 and Gln147 to alanine in P3 and P53–174 (P3-(D143A/Q147A)-GFP and P53–174-(D143A/Q147A)-GFP, respectively), which has been shown to inhibit DLC-AS-facilitated nuclear import (44, 54). The mutations resulted in a significant decrease in nuclear accumulation of GFP-fused P3 and P53–174 proteins (Fig. 4, C and D). However, they did not reduce the Fn/c of P53–174-GFP to a level equivalent to that of P53–139-GFP, indicating that the region P139–174 contains additional sequence(s) that contribute to P3 nuclear localization, potentially including basic residues forming part of the N-NLS sequence. To examine this, we analyzed the nucleocytoplasmic localization of P3-GFP carrying mutations to asparagine of the residues Lys139 and Arg144, which show high conservation between the P proteins of RabV strains and related lyssaviruses (data not shown). Live-cell CLSM analysis revealed no significant difference between the levels of nuclear accumulation of P3-(K139N)-GFP and WT P3-GFP (Fn/c of approximately 8 in both cases), with mutation of both residues resulting in only a moderate decrease (Fn/c of approximately 6; images not shown). This indicated that Arg144 makes a contribution to P3 nuclear accumulation, but that other residues are important, and residues, Lys54 and Arg55 are essential. Importantly, the fact that Lys54 and Arg55 are located within the N-NES indicates a physical overlap of sequences critical to the N-NES and N-NLS (Fig. 1).
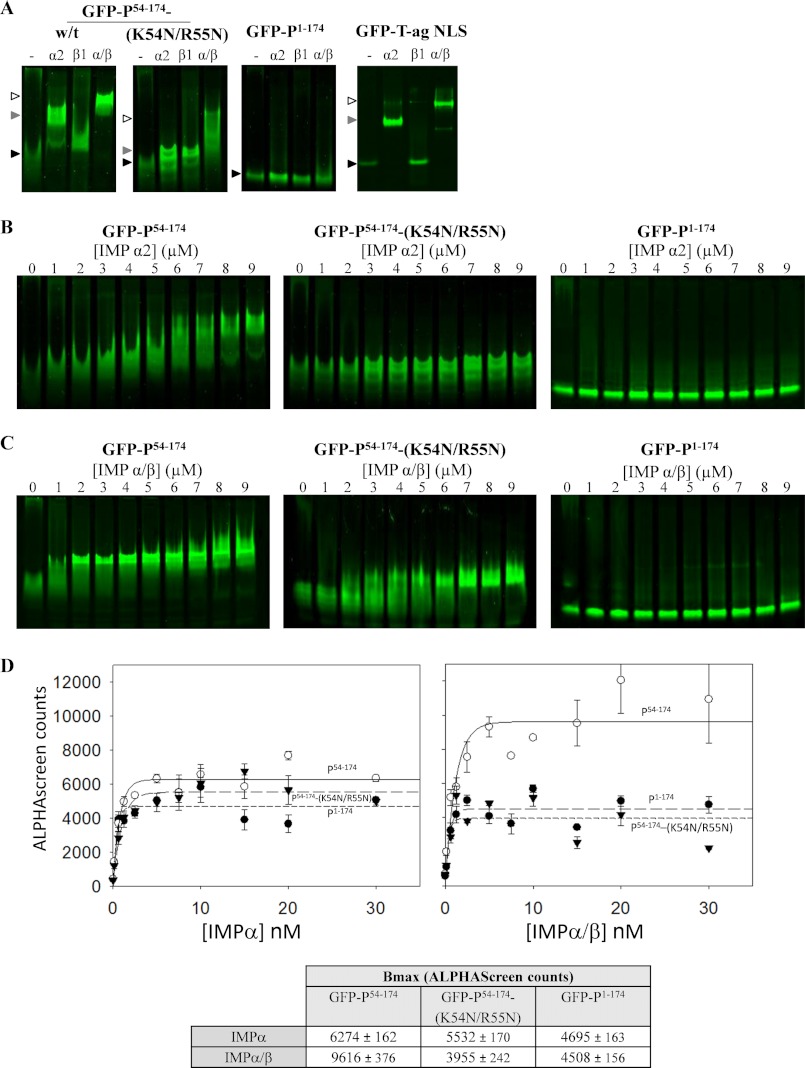
The P3 N-NLS Is Recognized by the IMPα/β Heterodimer with High Affinity
The above data indicate that P53–174 contains a previously unidentified NLS (N-NLS) that is responsible for the high nuclear accumulation of P3. To confirm that this region contains an IMP-recognized NLS (10), we tested the binding of purified recombinant GFP-fused proteins encoding the regions P54–174 and P54–174-(K54N/R55N) to GST-fused IMPα2 and IMPβ1 proteins (see supplemental Fig. S1), initially using native PAGE as described previously (50–52, 55–57) (Fig. 5).
FIGURE 5.
IMPα and α/β binding by P54–174 requires residues Lys54 and Arg55 and is inhibited by residues P1–53. A, the indicated recombinant His6-tagged GFP-RabV P protein derivatives or GFP-T-ag NLS (2 μm) were incubated without (−) or with 10 μm GST-IMPα2, GST-IMPβ1, or preformed heterodimers of GST-IMPα2/β1 for 15 min, before electrophoresis on a nondenaturing gel, and imaging. Interaction of GFP-RabV P protein derivatives or GFP-T-ag NLS with IMPs is indicated by altered mobility (“shift”) of the GFP band compared with the no IMP control (−); black arrows indicate non-IMP-associated protein; gray arrows, IMPα-associated protein; and white-filled arrows, IMPα/β-associated protein. B and C, GFP-RabV P protein derivatives (2 μm) were incubated with the indicated concentration of GST-IMPα2 (B) or GST-IMPα/β heterodimer (C) before electrophoresis. D, upper, the indicated His6-tagged GFP-P-protein derivatives were incubated with different concentrations of biotinylated IMPα2 or biotinylated IMPα/β heterodimer before conjugation to nickel chelate-coated donor beads and streptavidin-coated acceptor beads for AlphaScreen analysis (51). AlphaScreen assays were performed in triplicate and data (mean AlphaScreen count ± S.E. (error bars)) are from a single assay representative of three separate assays. The maximal binding (Bmax) values (± S.E.) from the curve fits for are shown in the table (D, lower).
GFP-P54–174 interacted with both IMPα2 and the IMPα/β dimer, as indicated by a change in the mobility in the native gel compared with in the absence of IMP addition, which was comparable with that observed for the well characterized IMPα/β-binding NLS of T-ag (50–52) (Fig. 5A). Consistent with the idea that Lys54 and Arg55 are critical to the N-NLS, mutation of these residues diminished the IMP interactions. Although there was some interaction of the proteins with GST-IMPβ1, this was to a markedly lower extent than with GST-IMPα or GST-IMPα/β, implying that P3 nuclear transport is likely to rely on the IMPα/β heterodimer. Native PAGE was also used to analyze binding of increasing amounts of GST-IMPα2 (Fig. 5B) or GST-IMPα/β heterodimer (Fig. 5C), results confirming markedly reduced binding to GFP-P54–174-(K54N/R55N) of IMPα and IMPα/β compared with GFP-P54–174. Notably, GFP-P54–174 showed a strong shift in mobility at concentrations of 1–2 μm IMPα/β, whereas a similar shift was only observed at 8–9 μm in the case of IMPα, consistent with the idea that P54–174 contains a classical IMPα/β-recognized NLS, where high affinity binding of IMPα is dependent on heterodimerization with IMPβ (see 22, 50, 52).
We also used the highly sensitive AlphaScreen assay to analyze binding (51), results indicating high affinity binding of IMPα/β to GFP-P54–174 (apparent dissociation constant, Kd, of 0.8 ± 0.15 nm) (Fig. 5D). Analysis also revealed that maximal binding (Bmax) of GFP-P54–174-(K54N/R55N) to IMPα/β was markedly lower than that of GFP-P54–174 (Fig. 5D). These data indicate that residues Lys54 and Arg55 are critical for IMP binding, representing the basis for the effects on nuclear transport efficiency observed in Fig. 4.
The N-terminal 52 Residues of P1 Inhibit N-NLS Activity
The above data indicate that P3 contains a functional N-NLS that is inactivated by the deletion of key residues from P4 and P5, accounting for the difference in nuclear localization of these isoforms. However, P1 contains the entire N-NLS sequence, but remains substantially less nuclear than P3 in cells treated with or without LMB (Fig. 2, A and C). This implies that the N-NLS is inhibited in the context of P1, through the presence of residues P1–52. To examine this, we performed native PAGE and AlphaScreen analyses to examine the binding of GFP-P1–174 to IMPα2 and the IMPα/β heterodimer as above (Fig. 5). Binding of both IMPα and IMPα/β to GFP-P1–174 was profoundly decreased compared with GFP-P54–174, such that little or no shift was evident in the native gels (Fig. 5, B and C). Quantitative analysis using AlphaScreen (Fig. 5D) confirmed this observation, with a Bmax for GFP-P1–174 binding to IMPα/β that was markedly decreased compared with that for GFP-P54–174. Clearly, the N-terminal 53 residues of P inhibit IMP binding to the N-NLS in vitro.
To confirm that the nuclear targeting function of the N-NLS is inhibited by residues P1–52 in vivo, we expressed P1–174-GFP and P53–174-GFP as well as P1-GFP, P3-GFP, and mutated versions thereof, in HeLa cells and imaged them live by CLSM (Fig. 6). Quantitative analysis indicated that P53–174-GFP showed almost 10-fold higher nuclear targeting ability than P1–174-GFP in cells treated either without or with LMB (Fig. 6, A and B). Importantly, although mutation of Lys54 and Arg55 significantly (p < 0.0001) diminished P3-GFP nuclear accumulation, it had negligible effect on nuclear localization of P1-GFP in cells treated either without or with LMB (Fig. 6, C and D), implying that the N-NLS plays no role in P1 nuclear import. Together, these data indicate that the N-NLS is inhibited in P1 by residues P1–52, and that, in a unique mechanism, truncation of these residues concomitantly inactivates the N-NES and activates the N-NLS to effect strong nuclear accumulation of P3.
FIGURE 6.
N-NLS activation requires the deletion of P1–52. A and C, HeLa cells were transfected to express the indicated proteins and treated with or without LMB. B and D, analysis of living cells was performed by CLSM to determine the Fn/c (mean ± S.E. (error bars), n ≥ 73 from three separate assays) as described in the legend to Fig. 2. Statistical analysis was according to the legend to Fig. 2.
DISCUSSION
Here we show definitively for the first time that RabV P protein contains a novel trafficking module, incorporating overlapping NES and NLS sequences, representing the basis of a unique regulatory mechanism. Specifically, residues P1–52 form part of the functional N-NES, but directly inhibit the function of the N-NLS sequence (identified for the first time in this study), to effect robust nuclear exclusion of the P1 isoform. Truncation of the module in P3 by deletion of P1–52 inactivates the N-NES and activates the N-NLS de novo, resulting in strong nuclear accumulation. Thus, this module enables the highly coordinated regulation of opposing nuclear trafficking sequences. This mechanism differs fundamentally from the previously accepted model of regulation of P protein isoform nuclear trafficking through the differential expression of distinct, functionally independent trafficking signals (30) and represents a novel and highly efficient strategy to control protein nucleocytoplasmic localization.
Because of the close proximity of P1–52 to the critical N-NLS residues Lys54 and Arg55, the inhibitory effect of this region is likely to be due to direct molecular masking, whereby P1–52 interacts with and/or occludes these key residues from recognition by IMPs, as implied by the direct binding studies where P1–174 shows low binding to IMPs in contrast to the clear, high affinity binding by P54–174 (Fig. 5D), but still enables interaction with CRM1. Alternatively, inhibition of the N-NLS may relate to local structural effects due to the presence or absence of P1–52. Indeed, the region P53–88 is predicted to be consistent with the flexible, “intrinsically disordered” regions of protein (58) that are believed to enable the formation of diverse molecular interactions (59–61), such that P1–52 could significantly impact on molecular interactions formed by this region.
Importantly, we found that the N-NLS is not a classical short monopartite or bipartite sequence, as P53–68 or P53–81, which are deleted from P4 and P5 and contain essential N-NLS residues, are not sufficient to recapitulate the nuclear import conferred by P3. In fact, N-NLS activity requires a large region (P53–174) suggestive of an extensive, conformation-dependent signal or a nonclassical bipartite NLS (9, 13) with an unconventionally long spacer region. Several basic residues/motifs in the C-terminal part of P53–174 could contribute to such a sequence. Thus, it is possible that regulation of the N-NLS involves broader effects of P1–52 on the structure of the P protein. Intriguingly, deletion of P1–52 from P3 also appears to be able to activate the microtubule-association sequence (MTAS) within the C-terminal domain (Fig. 1) (43), indicative of such effects on distal regions of P3 and suggesting that the presence or absence of P1–52 concomitantly regulates a number of distinct trafficking/regulatory sequences involved in P3-mediated IFN-antagonism (29, 30, 36–38, 42, 43). Importantly, the putative spacer of the N-NLS contains the RabV P dimerization domain (residues 91–131, Fig. 1), structural studies of which indicate the formation of two α-helices that form a hairpin (62) and may thus align distally located residues at the N and C termini of the N-NLS to form a bipartite signal. This is particularly intriguing as the function of the MTAS is dependent on dimerization (43), such that effects on the dimerization domain may contribute to the coordinated effect of the deletion of P1–52 on these sequences.
In addition to regulating the activity of the N-NES, N-NLS, and MTAS, residues P1–52 also contain sequence critical for binding to the viral polymerase L (P1–19), essential to P1 function as a co-factor in cytoplasmic genome replication/transcription (29). The deletion of this site from isoforms P2–P5 is believed to enable accessory functions, such as antagonism of IFN signaling, which depend on differential subcellular localization (29, 63). Consistent with this hypothesis, recent data indicate that P2, which contains the N-NES and is cytoplasmic, plays a more important role than P1 in inhibiting IFN signaling by arresting STAT1 in the cytoplasm (37, 63, 64). Importantly, our data (this report and see Ref. 43) indicate that the deletion of residues P1–52 in P3 does not simply produce a truncated version of P1/P2, but in fact generates a very different protein species, suggesting that this isoform has adopted specific unique properties which enable distinct intranuclear/microtubule-dependent roles in IFN antagonism (29, 35, 37–39, 43). In addition, our finding that nuclear localization of the region P53–174 is increased upon treatment with LMB (Fig. 3, B and D) suggests that this region contains additional NES activity which, together with the DLC-AS (Fig. 4, C and D), can modulate nuclear localization through the N-NLS. Thus, subcellular trafficking of RabV P isoforms involves complicated and intricate mechanisms, indicating an exquisite requirement for highly regulated nuclear trafficking in RabV infection, presumably to enable efficient shutdown of host immune responses.
In conclusion, we have identified a novel nuclear trafficking module for efficient co-ordinated regulation of the nucleocytoplasmic localization of protein isoforms. Future research will evaluate the role of the N-NLS/N-NES module in the context of infectious virus and pathogenicity in vivo, to evaluate its potential as a target for novel antiviral therapeutics/vaccines against RabV, the cause of more than 55,000 human fatalities annually (29, 65). This work should also assist in the delineation of the regulation of trafficking of other cellular and viral protein isoforms to provide important insights into the regulation of proteins involved in diverse cellular pathways and pathological processes.
Supplementary Material
Acknowledgments
We thank Monash Micro Imaging for providing microscopy instrumentation and technical support and Cassandra David for tissue culture.
This work was supported by National Health and Medical Research Council, Australia, Project Grant 1003244 (to G. W. M.), and Senior Principal Research Fellowship 1002486 (to D. A. J.), and by Australian Research Council Discovery Project DP110101749 (to G. W. M. and D. A. J.).

This article contains supplemental Fig. S1.
- NLS
- nuclear localization signal
- C-NLS
- NLS in the C-terminal domain
- CLSM
- confocal laser scanning microscopy
- DLC-AS
- dynein light chain-association sequence
- IMP
- importin
- LMB
- leptomycin-B
- MTAS
- microtubule-association sequence
- NES
- nuclear export signal
- N-NES
- NES close to the N terminus
- N-NLS
- NLS in the N-terminal region
- PML
- promyelocytic leukemia
- RabV
- rabies virus
- T-ag
- T-antigen.
REFERENCES
- 1. Chumakov S. P., Prasolov V. S. (2010) Organization and regulation of nucleocytoplasmic transport. Mol. Biol. 44, 186–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cook A., Bono F., Jinek M., Conti E. (2007) Structural biology of nucleocytoplasmic transport. Annu. Rev. Biochem. 76, 647–671 [DOI] [PubMed] [Google Scholar]
- 3. Fulcher A. J., Jans D. A. (2011) Regulation of nucleocytoplasmic trafficking of viral proteins: an integral role in pathogenesis? Biochim. Biophys. Acta 1813, 2176–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Stewart M. (2007) Molecular mechanism of the nuclear protein import cycle. Nat. Rev. Mol. Cell Biol. 8, 195–208 [DOI] [PubMed] [Google Scholar]
- 5. Poon I. K., Jans D. A. (2005) Regulation of nuclear transport: central role in development and transformation? Traffic 6, 173–186 [DOI] [PubMed] [Google Scholar]
- 6. Deonarain R., Chan D. C., Platanias L. C., Fish E. N. (2002) Interferon-α/β-receptor interactions: a complex story unfolding. Curr. Pharm. Des. 8, 2131–2137 [DOI] [PubMed] [Google Scholar]
- 7. la Cour T., Kiemer L., Mølgaard A., Gupta R., Skriver K., Brunak S. (2004) Analysis and prediction of leucine-rich nuclear export signals. Protein Eng. Des. Sel. 17, 527–536 [DOI] [PubMed] [Google Scholar]
- 8. Ström A. C., Weis K. (2001) Importin-β-like nuclear transport receptors. Genome Biol. 2, REVIEWS3008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lange A., McLane L. M., Mills R. E., Devine S. E., Corbett A. H. (2010) Expanding the definition of the classical bipartite nuclear localization signal. Traffic 11, 311–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lange A., Mills R. E., Lange C. J., Stewart M., Devine S. E., Corbett A. H. (2007) Classical nuclear localization signals: definition, function, and interaction with importin α. J. Biol. Chem. 282, 5101–5105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Reich N. C., Liu L. (2006) Tracking STAT nuclear traffic. Nat. Rev. Immunol. 6, 602–612 [DOI] [PubMed] [Google Scholar]
- 12. Conti E., Uy M., Leighton L., Blobel G., Kuriyan J. (1998) Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin? Cell 94, 193–204 [DOI] [PubMed] [Google Scholar]
- 13. Giesecke A., Stewart M. (2010) Novel binding of the mitotic regulator TPX2 (target protein for Xenopus kinesin-like protein 2) to importin-α. J. Biol. Chem. 285, 17628–17635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hodel M. R., Corbett A. H., Hodel A. E. (2001) Dissection of a nuclear localization signal. J. Biol. Chem. 276, 1317–1325 [DOI] [PubMed] [Google Scholar]
- 15. Kalderon D., Richardson W. D., Markham A. F., Smith A. E. (1984) Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature 311, 33–38 [DOI] [PubMed] [Google Scholar]
- 16. Claudiani P., Riano E., Errico A., Andolfi G., Rugarli E. I. (2005) Spastin subcellular localization is regulated through usage of different translation start sites and active export from the nucleus. Exp. Cell Res. 309, 358–369 [DOI] [PubMed] [Google Scholar]
- 17. Fagioli M., Alcalay M., Pandolfi P. P., Venturini L., Mencarelli A., Simeone A., Acampora D., Grignani F., Pelicci P. G. (1992) Alternative splicing of PML transcripts predicts coexpression of several carboxy-terminally different protein isoforms. Oncogene 7, 1083–1091 [PubMed] [Google Scholar]
- 18. Rodríguez E., Martignetti J. A. (2009) The Krüppel traffic report: cooperative signals direct KLF8 nuclear transport. Cell Res. 19, 1041–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chenik M., Chebli K., Blondel D. (1995) Translation initiation at alternate in-frame AUG codons in the rabies virus phosphoprotein mRNA is mediated by a ribosomal leaky scanning mechanism. J. Virol. 69, 707–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Harcourt B. H., Tamin A., Ksiazek T. G., Rollin P. E., Anderson L. J., Bellini W. J., Rota P. A. (2000) Molecular characterization of Nipah virus, a newly emergent paramyxovirus. Virology 271, 334–349 [DOI] [PubMed] [Google Scholar]
- 21. Jayakar H. R., Whitt M. A. (2002) Identification of two additional translation products from the matrix (M) gene that contribute to vesicular stomatitis virus cytopathology. J. Virol. 76, 8011–8018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Forwood J. K., Brooks A., Briggs L. J., Xiao C. Y., Jans D. A., Vasudevan S. G. (1999) The 37-amino acid interdomain of Dengue virus NS5 protein contains a functional NLS and inhibitory CK2 Site. Biochem. Biophys. Res. Commun. 257, 731–737 [DOI] [PubMed] [Google Scholar]
- 23. Ghildyal R., Ho A., Wagstaff K. M., Dias M. M., Barton C. L., Jans P., Bardin P., Jans D. A. (2005) Nuclear import of the respiratory syncytial virus matrix protein is mediated by importin β1 independent of importin α. Biochemistry 44, 12887–12895 [DOI] [PubMed] [Google Scholar]
- 24. Rodriguez J. J., Horvath C. M. (2004) Host evasion by emerging paramyxoviruses: Hendra virus and Nipah virus V proteins inhibit interferon signaling. Viral Immunol. 17, 210–219 [DOI] [PubMed] [Google Scholar]
- 25. Rodriguez J. J., Parisien J. P., Horvath C. M. (2002) Nipah virus V protein evades αand γ interferons by preventing STAT1 and STAT2 activation and nuclear accumulation. J. Virol. 76, 11476–11483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rodriguez J. J., Wang L. F., Horvath C. M. (2003) Hendra virus V protein inhibits interferon signalling by inhibiting STAT-1 and STAT-2 nuclear accumulation. J. Virol. 77, 11842–11845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shaw M. L., Cardenas W. B., Zamarin D., Palese P., Basler C. F. (2005) Nuclear localization of the Nipah virus W protein allows for inhibition of both virus- and Toll-like receptor 3-triggered signaling pathways. J. Virol. 79, 6078–6088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shaw M. L., García-Sastre A., Palese P., Basler C. F. (2004) Nipah virus V and W proteins have a common STAT1-binding domain yet inhibit STAT1 activation from the cytoplasmic and nuclear compartments, respectively. J. Virol. 78, 5633–5641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Oksayan S., Ito N., Moseley G., Blondel D. (2012) Subcellular trafficking in rhabdovirus infection and immune evasion: a Novel target for therapeutics. Infect. Disord. Drug Targets 12, 38–58 [DOI] [PubMed] [Google Scholar]
- 30. Pasdeloup D., Poisson N., Raux H., Gaudin Y., Ruigrok R. W., Blondel D. (2005) Nucleocytoplasmic shuttling of the rabies virus P protein requires a nuclear localization signal and a CRM1-dependent nuclear export signal. Virology 334, 284–293 [DOI] [PubMed] [Google Scholar]
- 31. Bouttier M., Goncalvès C., Journo C., Letienne J., Piña M., Vitour D. (2008) Viruses and interferon: mechanisms of interferon induction and strategies to escape interferon response. Virologie 12, 159–173 [DOI] [PubMed] [Google Scholar]
- 32. Davey N. E., Travé G., Gibson T. J. (2011) How viruses hijack cell regulation. Trends Biochem. Sci. 36, 159–169 [DOI] [PubMed] [Google Scholar]
- 33. Gale M., Jr., Sen G. C. (2009) Viral evasion of the interferon system. J. Interferon Cytokine Res. 29, 475–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ahmed M., McKenzie M. O., Puckett S., Hojnacki M., Poliquin L., Lyles D. S. (2003) Ability of the matrix protein of vesicular stomatitis virus to suppress β interferon gene expression is genetically correlated with the inhibition of host RNA and protein synthesis. J. Virol. 77, 4646–4657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chelbi-Alix M. K., Vidy A., El Bougrini J., Blondel D. (2006) Rabies viral mechanisms to escape the IFN system: the viral protein P interferes with IRF-3, Stat1, and PML nuclear bodies. J. Interferon Cytokine Res. 26, 271–280 [DOI] [PubMed] [Google Scholar]
- 36. Ito N., Moseley G. W., Blondel D., Shimizu K., Rowe C. L., Ito Y., Masatani T., Nakagawa K., Jans D. A., Sugiyama M. (2010) Role of interferon antagonist activity of rabies virus phosphoprotein in viral pathogenicity. J. Virol. 84, 6699–6710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vidy A., Chelbi-Alix M., Blondel D. (2005) Rabies virus P protein interacts with STAT1 and inhibits interferon signal transduction pathways. J. Virol. 79, 14411–14420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Vidy A., El Bougrini J., Chelbi-Alix M. K., Blondel D. (2007) The nucleocytoplasmic rabies virus P protein counteracts interferon signaling by inhibiting both nuclear accumulation and DNA binding of STAT1. J. Virol. 81, 4255–4263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Blondel D., Regad T., Poisson N., Pavie B., Harper F., Pandolfi P. P., De Thé H., Chelbi-Alix M. K. (2002) Rabies virus P and small P products interact directly with PML and reorganize PML nuclear bodies. Oncogene 21, 7957–7970 [DOI] [PubMed] [Google Scholar]
- 40. Shimizu K., Ito N., Mita T., Yamada K., Hosokawa-Muto J., Sugiyama M., Minamoto N. (2007) Involvement of nucleoprotein, phosphoprotein, and matrix protein genes of rabies virus in virulence for adult mice. Virus Res. 123, 154–160 [DOI] [PubMed] [Google Scholar]
- 41. Faria P. A., Chakraborty P., Levay A., Barber G. N., Ezelle H. J., Enninga J., Arana C., van Deursen J., Fontoura B. M. (2005) VSV disrupts the Rae1/mrnp41 mRNA nuclear export pathway. Mol. Cell 17, 93–102 [DOI] [PubMed] [Google Scholar]
- 42. Moseley G. W., Filmer R. P., DeJesus M. A., Jans D. A. (2007) Nucleocytoplasmic distribution of rabies virus P-protein is regulated by phosphorylation adjacent to C-terminal nuclear import and export signals. Biochemistry 46, 12053–12061 [DOI] [PubMed] [Google Scholar]
- 43. Moseley G. W., Lahaye X., Roth D. M., Oksayan S., Filmer R. P., Rowe C. L., Blondel D., Jans D. A. (2009) Dual modes of rabies P-protein association with microtubules: a novel strategy to suppress the antiviral response. J. Cell Sci. 122, 3652–3662 [DOI] [PubMed] [Google Scholar]
- 44. Moseley G. W., Roth D. M., DeJesus M. A., Leyton D. L., Filmer R. P., Pouton C. W., Jans D. A. (2007) Dynein light chain association sequences can facilitate nuclear protein import. Mol. Biol. Cell 18, 3204–3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Baliga B. C., Colussi P. A., Read S. H., Dias M. M., Jans D. A., Kumar S. (2003) Role of prodomain in importin-mediated nuclear localization and activation of caspase-2. J. Biol. Chem. 278, 4899–4905 [DOI] [PubMed] [Google Scholar]
- 46. Lahaye X., Vidy A., Pomier C., Obiang L., Harper F., Gaudin Y., Blondel D. (2009) Functional characterization of negri bodies (NBs) in rabies virus-infected cells: evidence that NBs are sites of viral transcription and replication. J. Virol. 83, 7948–7958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Roth D. M., Moseley G. W., Glover D., Pouton C. W., Jans D. A. (2007) A microtubule-facilitated nuclear import pathway for cancer regulatory proteins. Traffic 8, 673–686 [DOI] [PubMed] [Google Scholar]
- 48. Roth D. M., Moseley G. W., Pouton C. W., Jans D. A. (2011) Mechanism of microtubule-facilitated “fast track” nuclear import. J. Biol. Chem. 286, 14335–14351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Moseley G. W., Leyton D. L., Glover D. J., Filmer R. P., Jans D. A. (2010) Enhancement of protein transduction-mediated nuclear delivery by interaction with dynein/microtubules. J. Biotechnol. 145, 222–225 [DOI] [PubMed] [Google Scholar]
- 50. Forwood J. K., Harley V., Jans D. A. (2001) The C-terminal nuclear localization signal of the sex-determining region Y (SRY) high mobility group domain mediates nuclear import through importin β1. J. Biol. Chem. 276, 46575–46582 [DOI] [PubMed] [Google Scholar]
- 51. Wagstaff K. M., Jans D. A. (2006) Intramolecular masking of nuclear localization signals: analysis of importin binding using a novel AlphaScreen-based method. Anal. Biochem. 348, 49–56 [DOI] [PubMed] [Google Scholar]
- 52. Wagstaff K. M., Dias M. M., Alvisi G., Jans D. A. (2005) Quantitative analysis of protein-protein interactions by native PAGE/fluorimaging. J. Fluoresc. 15, 469–473 [DOI] [PubMed] [Google Scholar]
- 53. Kosugi S., Hasebe M., Tomita M., Yanagawa H. (2009) Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc. Natl. Acad. Sci. U.S.A. 106, 10171–10176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Poisson N., Real E., Gaudin Y., Vaney M. C., King S., Jacob Y., Tordo N., Blondel D. (2001) Molecular basis for the interaction between rabies virus phosphoprotein P and the dynein light chain LC8: dissociation of dynein-binding properties and transcriptional functionality of P. J. Gen. Virol. 82, 2691–2696 [DOI] [PubMed] [Google Scholar]
- 55. Fulcher A. J., Dias M. M., Jans D. A. (2010) Binding of p110 retinoblastoma protein inhibits nuclear import of simian virus SV40 large tumor antigen. J. Biol. Chem. 285, 17744–17753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kaur G., Delluc-Clavieres A., Poon I. K., Forwood J. K., Glover D. J., Jans D. A. (2010) Calmodulin-dependent nuclear import of HMG-box family nuclear factors: importance of the role of SRY in sex reversal. Biochem. J. 430, 39–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kaur G., Jans D. A. (2011) Dual nuclear import mechanisms of sex determining factor SRY: intracellular Ca2+ as a switch. FASEB J. 25, 665–675 [DOI] [PubMed] [Google Scholar]
- 58. Gerard F. C., Ribeiro Ede A., Jr., Leyrat C., Ivanov I., Blondel D., Longhi S., Ruigrok R. W., Jamin M. (2009) Modular organization of rabies virus phosphoprotein. J. Mol. Biol. 388, 978–996 [DOI] [PubMed] [Google Scholar]
- 59. Uversky V. N. (2011) Intrinsically disordered proteins from A to Z. Int. J. Biochem. Cell Biol. 43, 1090–1103 [DOI] [PubMed] [Google Scholar]
- 60. Uversky V. N., Shah S. P., Gritsyna Y., Hitchcock-DeGregori S. E., Kostyukova A. S. (2011) Systematic analysis of tropomodulin/tropomyosin interactions uncovers fine-tuned binding specificity of intrinsically disordered proteins. J. Mol. Recognit. 24, 647–655 [DOI] [PubMed] [Google Scholar]
- 61. Xue B., Williams R. W., Oldfield C. J., Goh G. K., Dunker A. K., Uversky V. N. (2010) Viral disorder or disordered viruses: do viral proteins possess unique features? Protein Pept. Lett. 17, 932–951 [DOI] [PubMed] [Google Scholar]
- 62. Ivanov I., Crépin T., Jamin M., Ruigrok R. W. (2010) Structure of the dimerization domain of the rabies virus phosphoprotein. J. Virol. 84, 3707–3710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Marschalek A., Drechsel L., Conzelmann K. K. (2012) The importance of being short: the role of rabies virus phosphoprotein isoforms assessed by differential IRES translation initiation. Eur. J. Cell Biol. 91, 17–23 [DOI] [PubMed] [Google Scholar]
- 64. Brzózka K., Finke S., Conzelmann K. K. (2006) Inhibition of interferon signaling by rabies virus phosphoprotein P: activation-dependent binding of STAT1 and STAT2. J. Virol. 80, 2675–2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Schnell M. J., McGettigan J. P., Wirblich C., Papaneri A. (2010) The cell biology of rabies virus: using stealth to reach the brain. Nat. Rev. Microbiol. 8, 51–61 [DOI] [PubMed] [Google Scholar]
- 66. Raux H., Flamand A., Blondel D. (2000) Interaction of the rabies virus P protein with the LC8 dynein light chain. J. Virol. 74, 10212–10216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Chenik M., Chebli K., Gaudin Y., Blondel D. (1994) In vivo interaction of rabies virus phosphoprotein (P) and nucleoprotein (N): existence of two N-binding sites on P protein. J. Gen. Virol. 75, 2889–2896 [DOI] [PubMed] [Google Scholar]
- 68. Chenik M., Schnell M., Conzelmann K. K., Blondel D. (1998) Mapping the interacting domains between the rabies virus polymerase and phosphoprotein. J. Virol. 72, 1925–1930 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.