Abstract
The expression patterns of developmental genes provide new markers that address the homology of body parts and provide clues as to how body plans have evolved. Such markers support the idea that insect wings evolved from limbs but refute the idea that insect and crustacean jaws are fundamentally different in structure. They also confirm that arthropod tagmosis reflects underlying patterns of Hox gene regulation but they do not yet resolve to what extent Hox expression domains may serve to define segment homologies.
The goal of much evo-devo research is to understand how developmental mechanisms evolve to generate new body plans. A prerequisite is to understand how body plans themselves may best be compared. For macroevolutionary comparisions, this is no trivial task—a fact evident from century-old disputes as to the homology of parts. Molecular embryology provides new markers to address these old questions, markers that also provide clues to the molecular changes that underlie evolutionary transformations. Arthropods provide an excellent test case for this approach because their diversity is constrained by the literal straight jacket of a modular exoskeleton. I review the first fruits of such studies below. In some cases, these data clearly support or refute previous hypotheses, fulfulling much of their promise. In other cases, interpretation remains difficult. However, given the potential richness of this data set, there is good cause to be optimistic that further studies will resolve, rather than compound, ambiguities.
The Place of Arthropods in the Tree of Life.
The context for any study that seeks to understand the evolution of development must be a phylogeny, albeit uncertain. Arthropods are no longer considered to be the kin of annelids, but both molecular and morphological data support the traditional association between arthropods proper and the segmented lobopods, typified by Peripatus. This pan-arthropod grouping would now be placed by many within a larger assemblage of molting animals, the Ecdysozoa [refs. 1 and 2; see the article by Adoutte in this issue of PNAS (3)]. The basal radiation of the arthropods is not yet resolved, but both molecular and new morphological data support a close relationship between insects and crustaceans, to the exclusion of chelicerates (4–8). The position of the myriapods remains uncertain, although molecular analyses consistently place them outside an insect/crustacean clade.
Homology of Limbs and Segmentation.
Conserved details of engrailed gene expression support a common origin for segmentation within the arthropods. Engrailed protein marks the posterior parts of segments and, in all arthropods tested, limb buds arise at the boundary of engrailed expressing and non-expressing cells (9, 10). No studies have examined segmentation gene expression in other pan-arthropodan phyla.
The limbs of insects and myriapods have a single proximo-distal axis—they are uniramous. Crustacean limbs are frequently branched, with two (biramous) or more proximo-distal axes. These branched structures arise by the appearance of multiple growth foci at different dorso-ventral positions around the distal margin of the limb bud. However, all limb branches arise at the same interface between engrailed expressing and non-expressing cells: i.e., at the same A/P position (11, 12). This pattern provides no developmental support for a model of arthropod segment evolution that derives biramous limbs from the fusion of two primitive uniramous segments (13).
Current models propose that the proximo/distal axis of the Drosophila limb is specified by the overlap of decapentaplegic (dpp) and wingless signaling territories, with distal territories defined by expression of the distalless gene (14). Homologues of distalless and wingless are both involved in patterning the multiple branches of crustacean limbs, but the pattern of their expression in the most complex multiply branched limbs does not suggest a simple reiteration of the insect model in each limb branch (15, 16).
Molecular markers support the hypothesis that the wings of insects may derive from the dorsal branches of an ancestrally branched arthropod limb, and not from an extension of the notum, as has been proposed more recently. Two genes characteristically expressed in the developing wing of Drosophila, nubbin and apterous, are both expressed specifically in the dorsal lobe of the multiply branched limb of the branchiopod crustacean, Artemia (17).
The Mandible.
In insects, myriapods, and crustaceans, the first mouthpart segment is modified to form a biting jaw, the mandible. The structure of the mandible has been a key character supporting a phylogeny that groups the insects with the myriapods to the exclusion of the crustaceans. From the evidence of functional morphology, Sidnie Manton argued that the myriapod and insect mandibles were constructed from a whole limb whereas the crustacean mandible was derived from only the basal segment of the appendage (a so-called gnathobasic mandible). Developmental data do not agree with this interpretation. The insect mandible, uniquely among insect appendages, does not express distalless at any stage of its development, strongly suggesting that it does not correspond to a whole limb. However, distalless expression is also lost from the developing mandible of myriapods, and of those crustaceans that lack a mandibular palp (18, 19). By this criterion, the biting structures of all arthropod mandibles are gnathobasic in the adult. The nature of the mandible is therefore not useful for defining relationships between these three groups. It is, however, a character that unites myriapods, insects, and crustaceans (traditionally termed the mandibulate arthropods) to the exclusion of the chelicerates, where all of the limbs retain distal elements, and distalless expression.
Ancestral Patterns of Arthropod Segmentation.
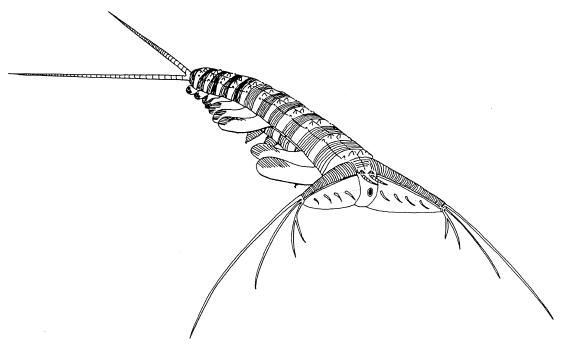
The ancestral arthropod has traditionally been envisaged as an animal with a large and somewhat ill defined number of similar trunk segments. However, current arthropod phylogenies suggest that we should look again at animals showing characteristics that may be interpreted as intermediate between those of arthropods and onychophorans. Several have been described from the Cambrian—most recently, Kerygmachela, from the Sirius Passat fauna of Greenland (20). This animal has a lobopod-like body, but spiny and/or segmented appendages at the anterior and posterior (Fig. 1). Perhaps the first jointed appendages of arthropods were the antennae and cerci, with trunk appendages derived by the transfer of developmental programs that first evolved to build these sensory structures.
Figure 1.
Proposed reconstruction of the Cambrian lobopod, Kerygmachela kierkegaardi, from ref. 20. This animal had typical onychophoran trunk appendages, but remarkably arthropod-like sensory appendages at front and back. [Reproduced with permission from the Royal Society of Edinburgh from Transactions of the Royal Society of Edinburgh: Earth Sciences, volume 89 (1999 for 1998), pp. 249–290.]
These putative intermediate Cambrian forms have relatively few trunk segments, often 11 (20). If these lie close to the arthropod stem lineage, then the ancestral arthropod may itself have had a relatively short trunk with a well defined segment number. This implies a mechanism generating a specific number of segments, not an indefinite budding process akin to that of annelids. The large and variable number of segments seen in many trilobites, some crustaceans, and some myriapods would then be a derived character, not the ancestral state. Indeed, among centipedes, the orders with large and variable segment numbers are derived, not basal (21).
The Ancestral Complement of Arthropod Hox Genes.
Comparison of the Hox gene complements of different phyla, and of different classes within the arthropods, suggests that the ancestral Hox cluster of the arthropods contained 10 linked genes, corresponding to the 8 canonical Hox genes of Drosophila and two more genes—one orthologous to the Hox3 gene of vertebrates, which in insects gave rise to the zen and bicoid genes, and one additional central gene that gave rise to the segmentation gene ftz of Drosophila and its relatives in other insects (refs. 2 and 22; C. Cook and M.A., unpublished work).
In chelicerate arthropods, the ftz and zen related genes, as well as all of the canonical Hox genes that have been analyzed, are expressed in restricted domains along the body axis (10, 23, 24). These expression patterns presumably reflect a conserved ancestral role for all of the Hox genes in the specification of axial position. It is not yet clear when in arthropod evolution the Hox3 and ftz-related genes acquired the new functions in embryonic patterning seen in higher insects, or lost their old functions.
Hox Genes, Tagmosis, and Segment Morphology.
Arthropod bodies are subdivided into distinct regions comprising arrays of functionally integrated and, to a greater or lesser extent, morphologically similar segments, termed tagmata (from the Greek regiment). Available data support the hypothesis that the abrupt and extensive changes in segment morphology that characterize the boundaries between tagmata reflect discontinuities in Hox gene expression (10, 25–32).
No Hox gene is known to be expressed in or anterior to the first appendage pair of any arthropod: i.e., the antennae of insects (corresponding to the first antenna of crustacea), or the eponymous cheliceral segment of chelicerates. Insect antennae require the absence of Hox gene expression for normal development, and it is to this state that appendage development defaults when Hox genes are deleted (33). Thus, we may surmise that the characteristic differences between the first appendage bearing segment of chelicerates and mandibulates are independent of Hox genes, and reflect other differences in the segment patterning machinery of these two arthropod groups.
In the prosoma of chelicerates, anterior Hox genes are expressed in extensively overlapping patterns (10, 30, 32), resembling more the patterns seen in vertebrates, and in the thorax and abdomen of insects, than the well resolved segment specific patterns observed for anterior Hox genes in the head of insects and crustaceans (29, 34). Comparison of Hox gene expression in the heads of several insect and crustacean species reveals considerable variation in the precise domains of Hox gene expression. Abzhanov and Kaufman (29) suggest that these restricted patterns have been derived independently from an ancestral pattern more similar to that seen in chelicerates, presumably as the morphology of anterior segments has become more diversified.
In comparisons between mandibulates and chelicerates, Hox gene expression is in general no guide to the form or function of trunk appendages. For example, “walking legs” express a quite different suite of Hox genes in the two groups. This contrasts markedly with the conserved relationship between the expression of some regulatory genes and the development of specific organs [e.g., the pax 6 gene and eyes (35)]. Perhaps this is because what distinguishes different appendages is not, in general, the possession of unique cell types, but more subtle aspects of tissue patterning and relative growth, the regulation of which may become linked to new transcription factors on relatively short evolutionary time scales. There is one possible exception to this rule: The most posterior Hox gene, Abdominal-B, is expressed in the genitalia in at least some insects, crustaceans, and chelicerates (25, 31).
Although Hox genes cannot in general be tied to particular morphologies, there are striking analogies in the way that Hox genes are used to pattern segments within the leg bearing tagmata of insects and spiders. In both cases, a regiment of fundamentally similar appendages are more or less subtly differentiated one from one another. In insects, all of the thoracic segments initially express Antennapedia, which in combination with other region-specific transcription factors (e.g., teashirt) appears to specify a thoracic ground state. The legs are differentiated one from one another by the locally modulated expression of other Hox genes [in this case, Sex combs reduced (Scr) and Ultrabithorax (Ubx) (36, 37)]. In the spiders, all of the leg buds express Deformed, and other anterior Hox genes, but in later development, the appendages are distinguished by distinct patterns of Scr expression, which is expressed only in the more posterior legs (30, 32). Perhaps we see here convergent evolution of the role of Hox genes, but using different members of the gene family in the two groups.
Segment Homologies Between Mandibulate and Chelicerate Arthropods.
The serial ordering of Hox gene expression along the body axis is largely conserved in arthropods, as it is in many other phyla. It is perhaps more remarkable that, if segments of insects and chelicerates are similarly numbered by counting from the first appendage-bearing segment backwards, then the anterior boundaries of expression for several of the Hox genes lie between the same pairs of segments—labial between segments 1 and 2, deformed between segments 2 and 3, etc. (10, 30, 32, 38) This pattern has led two groups to propose that the anterior limits of Hox gene expression are conserved ancestral characteristics that reflect segment homologies and, on this basis, to resolve between two long-standing models for segment organization in insect and chelicerate heads (10, 32). However, not all are convinced that Hox gene expression boundaries can be used as markers for segment homology (30). Data from the myriapods, and from other chelicerate and crustacean groups, are needed to resolve this question.
It does not seem implausible that the anterior segments of the common arthropod ancestor already possessed unique molecular identities, defined by Hox genes, and that these may have become fixed, even if they were not reflected in overt specialization of appendages. They may have controlled patterns of cell specialization in the mesoderm or nervous system, and only subsequently acquired more extensive roles in the control of external segment morphology. The acquisition of such new roles has been well documented for subsequent evolutionary steps in the insect lineage [e.g., appendage suppression (39)]. However, I find it hard to maintain the argument (10) that segments can be homologized throughout the trunk by conserved patterns of Hox gene expression. It is clear that domains of Ubx/abdA Hox gene expression vary with respect to ordinal segment number, even in quite closely related crustacean groups (26).
Hox Genes and Segment Modification in Crustacea.
The crustacea in particular exhibit a wonderful diversity of segment specialization and tagmosis. This diversity has three aspects. One is the diversity of segments in the adult of a single species. This is at its most extreme in the Malacostraca, with as many as 14 clearly distinct segment types. We do not know in detail how this diversity is controlled, but all of the evidence suggests that it does not require the proliferation of Hox genes. It is likely that the required diversity of Hox codes is provided by increased complexity in the regulation of a constant set of Hox genes (40). One case in which it seems that the number of Hox genes may have changed is in the cirripedes—but this is a case of gene loss, not gain. Barnacles appear to have lost the Hox gene abdominal-A, concomitant with loss of abdominal segments (41).
A second aspect of segment diversity is that which has arisen between species. The diverse forms that any one segment exhibits in different species probably reflect, in large part, changes downstream of the Hox genes. However, when it is the organization of segment types along the body axis that varies between species, then it seems more likely that the Hox genes will be directly involved. Averof and Patel (26) have examined one such case of segment diversification—the recruitment of anterior thoracic segments to generate auxiliary feeding appendages called maxillipeds. This has occurred repeatedly in several crustacean lineages. In each case tested, this transformation has been accompanied by a shift in the limits of expression of Ubx/abd-A related Hox genes—from an inferred primitive boundary at the anterior of the first thoracic segment, to a more posterior segment.
A third aspect of segment diversity, all too easily forgotten by Drosophila geneticists, is the diversity of segment morphology during ontogeny. (The appendage morphology of maggots is not rich!) Indirect developing crustaceans are famous for their range of larval forms. In these larvae, the morphology of a single segment may change dramatically at specific stages in the life cycle, often associated with changes in locomotory or feeding behavior. Perhaps even more remarkably, tagmosis itself may be altered, with the pattern of segment similarity shifting between molts (42).
These striking changes may be achieved in two ways. The same Hox proteins may exert differential effects at different stages in development, perhaps because hormonal changes modify the combinatorial input that controls segment morphology. Alternatively, the axial extent of Hox gene expression may itself change at different stages of development. An exam-ple of this second mode has recently been demonstrated in Porcellio, an isopod crustacean.
In this pillbug (woodlouse) the series of larval forms has been compressed into a series of embryonic stages, but some of the morphological transitions characteristic of the indirect developing ancestor are still evident. For example, the first thoracic appendage develops as a walking appendage, identical to those of the more posterior segments until mid embryogenesis, whereupon it diverges from the pathway of its thoracic homologues, coming to form a maxilliped. Abzhanov and Kaufman (43) show that this transition is associated with a transition in the pattern of Hox gene expression—Scr protein is initially repressed in the first thoracic appendage, but later expressed. Intriguingly, and exceptionally for the Hox genes, the early regulation (repression) of Scr is at the level of translational control, not transcription.
Conclusions.
Evolutionary developmental studies are mapping the relationships between gene expression and the diversity of form within arthropods. We can begin to propose models for the underlying changes in developmental mechanisms. Techniques to manipulate gene expression in arthropods are developing fast, promising that the role of individual genes may soon be tested directly. However, we should beware of trying to explain too much, with too little. No one gene family—not even the Hox genes—will provide a sufficient tool to explain the whole of any major step in evolution.
Acknowledgments
My thanks to Michalis Averof, Max Telford, and Chris Klingenberg for comments on the manuscript. Work in this laboratory has been supported principally by the Wellcome Trust and the Biotechnology and Biological Sciences Research Council of the United Kingdom.
References
- 1.Aguinaldo A M A, Turbeville J M, Linford L S, Rivera M C, Garey J R, Raff R A, Lake J A. Nature (London) 1997;387:489–493. doi: 10.1038/387489a0. [DOI] [PubMed] [Google Scholar]
- 2.De Rosa R, Grenier J, Andreeva T, Cook C, Adoutte A, Akam M, Carroll S, Balavoine G. Nature (London) 1999;399:772–775. doi: 10.1038/21631. [DOI] [PubMed] [Google Scholar]
- 3.Adoutte A, Balavoine G, Lartillot N, Lespinet O, Prud'homme B, de Rosa R. Proc Natl Acad Sci USA. 2000;97:4453–4456. doi: 10.1073/pnas.97.9.4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Freidrich M, Tautz D. Nature (London) 1995;376:165–167. doi: 10.1038/376165a0. [DOI] [PubMed] [Google Scholar]
- 5.Strausfeld N J. Brain Behav Evol. 1998;52:186–206. doi: 10.1159/000006563. [DOI] [PubMed] [Google Scholar]
- 6.Regier J C, Shultz J W. Mol Biol Evol. 1997;14:902–913. doi: 10.1093/oxfordjournals.molbev.a025833. [DOI] [PubMed] [Google Scholar]
- 7.Averof M, Akam M. Philos Trans R Soc London B. 1995;347:293–303. [Google Scholar]
- 8.Boore J L, Lavrov D V, Brown W M. Nature (London) 1998;392:667–668. doi: 10.1038/33577. [DOI] [PubMed] [Google Scholar]
- 9.Patel N H, Kornberg T B, Goodman C S. Development (Cambridge, UK) 1989;107:201–213. doi: 10.1242/dev.107.2.201. [DOI] [PubMed] [Google Scholar]
- 10.Damen W G M, Hausdorf M, Seyfarth E A, Tautz D. Proc Natl acad Sci USA. 1998;95:10665–10670. doi: 10.1073/pnas.95.18.10665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Panganiban G, Sebring A, Nagy L, Carroll S. Science. 1995;270:1363–1369. doi: 10.1126/science.270.5240.1363. [DOI] [PubMed] [Google Scholar]
- 12.Williams T A, Müller G B. Dev Genes Evol. 1996;206:161–168. doi: 10.1007/s004270050042. [DOI] [PubMed] [Google Scholar]
- 13.Emerson M J, Schram F R. Science. 1990;250:667–669. doi: 10.1126/science.250.4981.667. [DOI] [PubMed] [Google Scholar]
- 14.Lecuit T, Cohen S M. Nature (London) 1997;388:139–145. doi: 10.1038/40563. [DOI] [PubMed] [Google Scholar]
- 15.Williams T A. Dev Genes Evol. 1997;207:427–434. doi: 10.1007/s004270050133. [DOI] [PubMed] [Google Scholar]
- 16.Nulsen C, Nagy L. Dev Genes Evol. 1999;209:340–348. doi: 10.1007/s004270050262. [DOI] [PubMed] [Google Scholar]
- 17.Averof M, Cohen S M. Nature (London) 1997;385:627–630. doi: 10.1038/385627a0. [DOI] [PubMed] [Google Scholar]
- 18.Popadic A, Panganiban G, Rusch D, Shear W A, Kaufman T C. Dev Genes Evol. 1998;208:142–150. doi: 10.1007/s004270050165. [DOI] [PubMed] [Google Scholar]
- 19.Scholtz G, Mittmann B, Gerberding M. Int J Dev Biol. 1998;42:801–810. [PubMed] [Google Scholar]
- 20.Budd G. Trans R Soc Edinburgh Earth Sci. 1999 1998;89:249–290. [Google Scholar]
- 21.Arthur W. Evol Dev. 1999;1:62–69. doi: 10.1111/j.1525-142x.1999.t01-1-.x. [DOI] [PubMed] [Google Scholar]
- 22.Stauber M, Jäckle H, Schmidt-Ott U. Proc Natl Acad Sci USA. 1999;96:3786–3789. doi: 10.1073/pnas.96.7.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Telford M, Thomas R H. Dev Genes Evol. 1998;208:591–594. doi: 10.1007/s004270050219. [DOI] [PubMed] [Google Scholar]
- 24.Telford M. Curr Biol. 2000;10:349–352. doi: 10.1016/s0960-9822(00)00387-0. [DOI] [PubMed] [Google Scholar]
- 25.Averof M, Akam M. Nature (London) 1995;376:420–423. doi: 10.1038/376420a0. [DOI] [PubMed] [Google Scholar]
- 26.Averof M, Patel N H. Nature (London) 1997;388:682–686. doi: 10.1038/41786. [DOI] [PubMed] [Google Scholar]
- 27.Akam M, Averof M, Castelli-Gair J, Dawes R, Falciani F, Ferrier D. In: The Evolving Role of Hox Genes in Arthropods. Akam M, Holland P, Ingham P, Wray G, editors. Cambridge, U.K.: Company of Biologists; 1994. pp. 209–215. [PubMed] [Google Scholar]
- 28.Abbott M K, Kaufman T. Genetics. 1986;114:919–942. doi: 10.1093/genetics/114.3.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abzhanov A, Kaufman T C. Proc Natl Acad Sci USA. 1999;96:10224–10229. doi: 10.1073/pnas.96.18.10224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abzhanov A, Popodic A, Kaufman T C. Evol Dev. 1999;1:77–89. doi: 10.1046/j.1525-142x.1999.99014.x. [DOI] [PubMed] [Google Scholar]
- 31.Damen W G M, Tautz D. J Exp Zool. 1999;285:85–91. doi: 10.1002/(sici)1097-010x(19990415)285:1<85::aid-jez10>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 32.Telford M J, Thomas R H. Proc Natl Acad Sci USA. 1998;95:10671–10675. doi: 10.1073/pnas.95.18.10671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beeman R W, Stuart J J, Brown S J, Denell R E. BioEssays. 1993;15:439–444. doi: 10.1002/bies.950150702. [DOI] [PubMed] [Google Scholar]
- 34.Rogers B T, Kaufman T C. Int Rev Cytol. 1997;174:1–84. doi: 10.1016/s0074-7696(08)62115-4. [DOI] [PubMed] [Google Scholar]
- 35.Halder G, Callaerts P, Gehring W J. Curr Opin Genes Dev. 1995;5:602–609. doi: 10.1016/0959-437x(95)80029-8. [DOI] [PubMed] [Google Scholar]
- 36.Rogers B T, Peterson M D, Kaufman T C. Development (Cambridge, UK) 1997;124:149–157. doi: 10.1242/dev.124.1.149. [DOI] [PubMed] [Google Scholar]
- 37.Stern D. Nature (London) 1998;396:463–466. doi: 10.1038/24863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Damen W G M, Tautz D. Invertebr Reprod Dev. 1999;36:203–209. [Google Scholar]
- 39.Palopoli M F, Patel N H. Curr Biol. 1998;8:587–590. doi: 10.1016/s0960-9822(98)70228-3. [DOI] [PubMed] [Google Scholar]
- 40.Akam M. Int J Dev Biol. 1998;42:445–451. [PubMed] [Google Scholar]
- 41.Mouchel-Vielh E, Rogolot C, Gibert J-M, Deutsch J. Mol Phylogenet Evol. 1998;9:382–389. doi: 10.1006/mpev.1998.0498. [DOI] [PubMed] [Google Scholar]
- 42.Gurney R. The larvae of Decapod Crustaceans. London: The Ray Society; 1942. [Google Scholar]
- 43.Abzhanov A, Kaufman T. Development (Cambridge, UK) 1999;126:1121–1126. doi: 10.1242/dev.126.6.1121. [DOI] [PubMed] [Google Scholar]