Background: The IGF-1 receptor is essential for cell growth and survival.
Results: A site for GSK3β phosphorylation in the C-terminal tail controls receptor kinase activity, trafficking, and signaling.
Conclusion: Serine phosphorylation restrains kinase activity and signaling.
Significance: Cell growth or survival may be selectively modulated by GSK-3β-mediated phosphorylation of IGF-1R.
Keywords: Cancer Biology, Cell Growth, Cell Signaling, Glycogen Synthase Kinase 3, Insulin-like Growth Factor (IGF)
Abstract
Insulin-like growth factor I receptor (IGF-1R) signaling is essential for cell, organ, and animal growth. The C-terminal tail of the IGF-1R exhibits regulatory function, but the mechanism is unknown. Here, we show that mutation of Ser-1248 (S1248A) enhances IGF-1R in vitro kinase activity, autophosphorylation, Akt/mammalian target of rapamycin activity, and cell growth. Ser-1248 phosphorylation is mediated by GSK-3β in a mechanism that involves a priming phosphorylation on Ser-1252. GSK-3β knock-out cells exhibit reduced IGF-1R cell surface expression, enhanced IGF-1R kinase activity, and signaling. Examination of crystallographic structures of the IGF-1R kinase domain revealed that the 1248SFYYS1252 motif adopts a conformation tightly packed against the kinase C-lobe when Ser-1248 is in the unphosphorylated state that favors kinase activity. S1248A mutation is predicted to lock the motif in this position. In contrast, phosphorylation of Ser-1248 will drive profound structural transition of the sequence, critically affecting connection of the C terminus as well as exposing potential protein docking sites. Decreased kinase activity of a phosphomimetic S1248E mutant and enhanced kinase activity in mutants of its predicted target residue Lys-1081 support this auto-inhibitory model. Thus, the SFYYS motif controls the organization of the IGF-1R C terminus relative to the kinase domain. Its phosphorylation by GSK-3β restrains kinase activity and regulates receptor trafficking and signaling.
Introduction
The IGF-1R3 is widely expressed in human tissues and, activated by either IGF-1 or -2, mediates a conserved signaling pathway necessary for cell and organismal growth (1). Reduced IGF-1R expression or IGF-1 signaling is associated with small stature, cell, and organ size (2). The IGF signaling pathway also promotes cell survival, proliferation, and migration (reviewed in Ref. 3); it harbors proteins encoded by several potent oncogenes (PI3K, Akt, mTOR); and it is regulated by key tumor suppressors (PTEN, TSC1, and p53) (4). Inhibitors of the IGF-1R and its signaling intermediates are in clinical testing in a range of cancers (5).
Although the IGF-1R shares structural similarity with the insulin receptor (IR), being most similar in the kinase domain (84% sequence identity) and juxtamembrane domains (61% sequence identity), the C-terminal tails are quite distinct (44% sequence identity) (6). These kinases exhibit a similar mechanism of activation. In the IGF-1R, sequential autophosphorylation of three tyrosines (Tyr-1135, Tyr-1131, and Tyr-1136) facilitates stabilization of the kinase activation loop (A-loop) in a position that promotes catalysis and subsequent phosphorylation of substrates (7).
The C-terminal tail of the IGF-1R has been proposed as a regulatory domain to potentially mediate differences in IGF-1R and IR signaling (8–10). Specific domains in the IGF-1R C-terminal tail, particularly the one encompassing Tyr-1250 and Tyr-1251, which are present in the IGF-1R and not in the IR, are essential for IGF-1R function (8, 11–15). Mutation of Tyr-1250/Tyr-1251 is sufficient to abrogate IGF-1-mediated cellular transformation, suppression of apoptosis, cell migration, MAPK activation, and association of the IGF-1R with the scaffolding protein RACK1 in cooperation with integrin signaling (8, 13, 15, 16). Deletion of the C-terminal tail enhances IGF-1R-mediated transformation and suppression of apoptosis (8, 17), and ectopic expression of a myristoylated peptide encompassing the entire C-terminal 108 amino acids (MyCF) induces apoptosis and inhibits clonogenic and tumorigenic growth in tumor cell and nude mouse models, respectively (18, 19).
However, the molecular mechanisms underlying regulatory functions of the C-terminal tail are not understood. There is no evidence for constitutive or IGF-1-mediated phosphorylation of Tyr-1250 or Tyr-1251. The tyrosines are flanked by serines Ser-1248 and Ser-1252 that are also present in the IR at positions Ser-1275 and Ser-1279. The function of these serines is unknown, but mutation of either Ser-1248 in the IGF-1R or Ser-1275 in the IR reduces RACK1 interaction with the IGF-1R and IR, respectively (20, 21). This observation and the requirement for Tyr-1250/Tyr-1251 in IGF-1R function suggest that the SFYYS motif is a site for regulatory phosphorylation events.
To address this, we investigated serine phosphorylation of the 1248SFYYS1252 motif in the IGF-1R and its effect on kinase activity and signaling. We also used the available crystal structures that, although truncated below amino acid 1256, encompass the SFYYS motif (7, 22–31) and enabled us to explore the structural basis for this motif in regulating IGF-1R activity. Our findings indicate that a direct regulatory interaction of the C terminus with the kinase domain can be controlled by phosphorylation of Ser-1248. Moreover, because phosphorylation of Ser-1248 is prevalent in the absence of IGF-1 and mediated by GSK-3β, we propose that phosphorylation of Ser-1248 restrains IGF-1R kinase activity.
EXPERIMENTAL PROCEDURES
Materials
Recombinant IGF-1 was from PeproTech (Rocky Hill, NJ). Anti-Akt, phospho-Akt, phospho-ERK1/2, phospho-p70 S6 kinase (Thr-389), p70 S6 kinase, phospho-4E-BP1 (Thr-37/46), and 4EB-P1 antibodies were from Cell Signaling (Beverly, Ma), and the anti-ERK, anti-β-catenin, and anti-GSK-3β monoclonal antibodies were from BD Transduction Laboratories (Heidelberg, Germany). Anti-IGF-1R polyclonal antibody was from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Anti-phosphotyrosine clone 4G10 and recombinant GSK-3β were from Millipore (Watford, UK).
Cell Culture, Transfection, and IGF-1-mediated Stimulation of Cells
MCF-7 cells, R− and R+ cells (mouse embryonic fibroblast (MEF) cell lines derived from IGF-1R knock-out mice (32)), and GSK-3β null MEFs, a kind gift of Dr. James Woodgett (33), were maintained in Dulbecco's modified Eagle's medium (BioWhittaker, Verviers, Belgium), supplemented with 10% (v/v) fetal bovine serum, 10 mm l-Glu, and 5 mg/ml penicillin/streptomycin. Where indicated, cells were transiently transfected with pcDNA3 plasmids encoding WT or mutant IGF-1R or empty pcDNA3 vector in 6-well plates using 1.5 μg of DNA and 10-cm plates for immunoprecipitations using 8 μg DNA.
Preparation of Cellular Protein Extracts and Immunoprecipitation
Cellular protein extracts were prepared by washing cells with phosphate-buffered saline and then scraping into lysis buffer consisting of 20 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1% Nonidet P-40 plus the tyrosine phosphatase inhibitor Na3VO4 (1 mm), and the protease inhibitors phenylmethylsulfonyl fluoride (1 mm), pepstatin (1 μm), and aprotinin (2 mm). For immunoprecipitation, lysates (700 μg of protein per sample) were incubated with 1 μg of polyclonal anti-IGF-1R antibody overnight at 4 °C, followed by addition of 20 μl of protein G-agarose beads for 3 h at 4 °C. Immunoprecipitates were washed three times with ice-cold lysis buffer, centrifuged at 3000 rpm for 3 min, heated in 15 μl of 2× concentrated Laemmli sample buffer, and separated by SDS-PAGE followed by Western blot analysis.
Western Blotting
Protein samples resolved by SDS-PAGE were transferred to nitrocellulose membranes, which were blocked for 1 h at room temperature in Tris-buffered saline containing 0.05% Tween 20 (TBS-T) and 5% milk (w/v). All primary antibody incubations were performed overnight at 4 °C, and secondary antibody incubations were for 1 h at room temperature. Alexa Fluor 680- and 800-coupled anti-rabbit and anti-mouse secondary antibodies (LI-COR Biosciences Cambridge, UK) were used for detection with the Odyssey infrared imaging system (LI-COR Biosciences, Cambridge, UK).
In-cell Western
Cells were seeded at 5 × 104 cells/well in 96-well tissue culture plates in complete medium for 24 h. Cells were immediately fixed with 100 μl of a freshly prepared fixing solution (3.7% formaldehyde/PBS) and incubated at room temperature for 20 min. The indicated samples were permeabilized (to control for total IGF-1R protein expression) with 50 μl of permeabilization solution (0.1% Triton X-100/PBS), washed once with PBS, and blocked in 50 μl of blocking buffer (5% goat serum/PBS) overnight at 4 °C. Primary antibody (anti-IGF-1R αIR3) was added for 2 h with gentle rocking followed by washing and incubation with secondary antibody (Alexa Fluor-800; 1:750 in blocking buffer). Syto60 (Invitrogen) was also included (1:10,000). Syto60 is a cell-permeable nucleic acid stain that fluoresces at 680 nm and is detected using the Odyssey infrared scanner. Cells were washed five times with PBS. Plates were dried, and IGF-1R (surface and total levels) intensity was quantified using the Odyssey software with normalization to Syto60 levels.
Two-dimensional Gel Electrophoresis
Cell samples to be resolved by two-dimensional electrophoresis were washed in 250 mm sucrose, 10 mm Tris, and lysed by scraping into 8 m urea, 4% CHAPS, 2% IPG Buffer, 40 mm DTT. 60 μg of each sample was loaded by rehydration onto 7-cm isoelectric focusing strips (IPG strips, GE Healthcare) with a pH range of 3–11 and focused to 55 kV. Strips were equilibrated with buffer composed of 50 mm Tris-HCl, pH 8.8, 6 m urea, 30% (v/v) glycerol, 2% (w/v) SDS, and 0.002% bromphenol blue, initially with 1% DTT (dithiothreitol), and then with 2.5% iodoacetamide. Isoelectric strips were resolved on 15% SDS-polyacrylamide gels and MyCF protein detected by Western blotting.
In Vitro Kinase Assays
R− cells were transiently transfected with pcDNA3 encoding WT or K1003R IGF-1R, and HEK 293T cells were transfected with MyCF WT and MyCF S1248A. At 48 h post-transfection for R− and 24 h for HEK 293T, cells were lysed and subjected to immunoprecipitation with anti-IGF-1R antibody. Immunocomplexes were washed in GSK-3β kinase buffer (5 mm MOPS, pH 7.2, 2.5 mm β-glycerophosphate, 1 mm EGTA, 400 μm EDTA, 4 mm MgCl2, 50 μm DTT, 0.4 mg/ml BSA) and incubated in the presence of active GSK-3β for 20 min at 30 °C in the presence of [γ-32P]ATP. Following incubation with GSK-3β and [γ-32P]ATP, immunocomplexes were washed three times in GSK-3β kinase buffer and resolved by SDS-PAGE.
For analysis of IGF-1R tyrosine kinase activity, R− cells were transiently transfected with pcDNA3 encoding WT, Tyr-1250/Tyr-1251, S1248A, S1252A, S1248E, K1081G, or K1081E IGF-1R. At 48 h post-transfection, cells (or 48 h in culture in the case of GSK-3β null MEFs) were lysed, and the lysates were sonicated for 10 min in the benchtop Elma TranSonic T310 waterbath sonicator. Dot blots of lysates were carried out with three dilutions of each, followed by immunoblotting with the anti-IGF-1R antibody, and the input of lysate per immunoprecipitation was adjusted accordingly. Protein G-agarose complexes obtained from immunoprecipitations were washed in kinase buffer (50 mm Hepes, pH 7.4, 10 mm MgCl2, 10 mm MnCl2) and then resuspended in 25 μl of a kinase reaction mixture containing ATP (0.03 mm final concentration), 2 μl of [γ-32P]ATP (5 μCi/μl), and 2 mg/ml of poly(Glu,Tyr) (Sigma). Following a 20-min incubation period, samples (5 μl) were removed to fresh tubes containing 9 μl of H2O and 35 μl of 20 mm EDTA, pH 7.4. Triplicate samples were then transferred to glass microfiber filters in 24-well plates and washed extensively with trichloroacetic acid (10%) containing 10 mm Na2HPO4. Following a final wash with 70% ethanol, the filters were dried, and 32P incorporation was measured in a scintillation counter (Beckman Instruments).
Foci Formation, Cell Attachment, and Size
R− cells stably transfected with pcDNA3 vector, IGF-1R, WT, or S1248A were plated on 6-well plates at a density of 400 cells per well in 6-well plates in complete media. After 12 days cells were washed in PBS and fixed in 96% ethanol and then stained with 0.05% crystal violet, 20% ethanol before being extensively washed in distilled H2O. Foci were quantified by eye and also by using the Odyssey infrared imaging system (LI-COR Biosciences, Cambridge, UK). Cells were measured for attachment to uncoated 24-well tissue culture plates by plating at a density of 1 × 105 cells/well. Cells were fixed and stained using the same methods as used with plating efficiency at the indicated time points, and representative images at 30 min were obtained using an inverted microscope and MetaMorph software. For size measurement cells were detached with trypsin/EDTA and analyzed using the Countess automated cell counter from Invitrogen.
Migration Assays
Transwell assays were performed using 6.5-mm transwell filter inserts with 5.0 m inserts (Costar, Cambridge, MA). Cells (at or near confluence) were trypsinized and cultured in fresh media 18 h prior to each assay. The cells were serum-starved for 4 h, harvested, washed twice, and then resuspended in serum-free DMEM. The lower wells of the Boyden chamber apparatus were loaded with DMEM containing 10% FBS, and 5 × 104 cells were added to each upper well. After 4 h at 37 °C, the cells on the upper surface of the membrane were removed by scraping so that only cells that had migrated through the membrane remained. The membrane was then fixed with methanol, stained with 0.1% crystal violet, and air-dried. Cell counts were obtained by counting all cells, and the averages of counts from five fields each from triplicate wells for each test condition were calculated.
IGF-1R Internalization Assays
Cells were seeded on tissue culture dishes at a density of 1 × 106 cells/10-cm plate for 24 h (∼80% confluent). Cells were washed with serum-free medium, starved for 4 h before stimulation with 10 ng/ml biotin-IGF-1, and incubated at 37 °C as indicated to induce endocytosis of the IGF-1R. Control cells were also incubated with biotin-IGF-1 but were incubated at 4 °C to prevent receptor internalization. Cells were then washed with acid (0.2 m acetic acid, 0.5 m NaCl) for 5 min at 4 °C to remove unbound biotin-IGF-1, washed with ice-cold PBS, and lysed in Nonidet P-40 lysis buffer. Following cell lysis, IGF-1R was immunoprecipitated using anti-IGF-1R, resolved by 15% SDS-PAGE, and transferred to nitrocellulose. IGF-1R was assessed for bound biotin-IGF-1 (visible at 8 kDa) using an anti-streptavidin secondary antibody (Alexa Fluor-800 bound) and detected using the Odyssey infrared scanner.
RESULTS
Mutation of Ser-1248 Increases IGF-1R Tyrosine Kinase Activity and Signaling Output
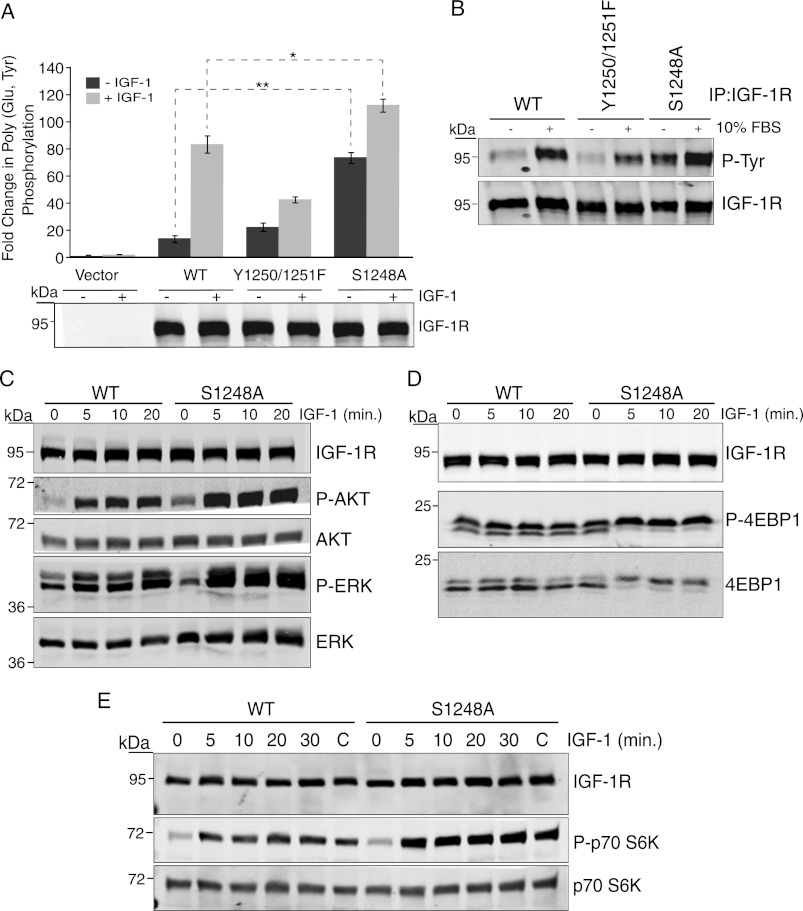
Previous work has demonstrated a requirement for Tyr-1250/Tyr-1251 and Ser-1248 in receptor function and association with the scaffolding protein RACK1 (8, 11, 13, 14, 16, 20). To determine whether Ser-1248 phosphorylation is important for function of this domain in the IGF-1R C terminus, we first compared the kinase activity of the S1248A mutant expressed in R− cells with that of the WT receptor and the Y1250F/Y1251F mutant in the presence and absence of IGF-1 stimulation. The S1248A mutant displayed 5.3-fold increased in vitro kinase activity toward the exogenous substrate poly(Glu,Tyr) compared with WT IGF-1R in the absence of IGF-1 stimulation and 1.35-fold increased kinase activity when stimulated with IGF-1 (Fig. 1A). In agreement with previous results (8), the Y1250F/Y1251F mutant exhibited 2-fold decreased poly(Glu,Tyr) phosphorylation compared with WT IGF-1R from cells stimulated with IGF-1 (Fig. 1A). Autophosphorylation of the IGF-1R was also increased in S1248A compared with WT and the Y1250F/Y1251F mutant, as indicated by increased phosphotyrosine content in Western blots (Fig. 1B). Interestingly, in agreement with the in vitro kinase activity (Fig. 1A), increased autophosphorylation of the S1248A mutant was evident when cells were starved from serum (Fig. 1B). Phosphorylation of Akt was also enhanced in serum-starved cells expressing the S1248A mutant compared with WT IGF-1R, both in the absence and presence of IGF-1 stimulation (Fig. 1C), whereas IGF-1-induced ERK phosphorylation was slightly increased. IGF-1-induced activation of the mTOR pathway was strongly increased as indicated by phosphorylation of S6K1 and 4E-BP1 (Fig. 1, D and E). Taken together, the data indicate that mutation of Ser-1248 increases IGF-1R kinase activity and signaling output and suggest that phosphorylation of Ser-1248 negatively regulates IGF-1R kinase activity.
FIGURE 1.
Mutation of Ser-1248 increases IGF-1R tyrosine kinase activity and signaling. A, clones of R− cells stably expressing pcDNA3 empty vector (Vector), pcDNA3 IGF-1R WT, or IGF-1R mutants Tyr-1250/Tyr-1251 and S1248A (R−/IGF-1R WT, R−/IGF-1R Y1250F/Y1251F, and R−/IGF-1R S1248A) were serum-starved for 4 h (−) or stimulated with IGF-1 for 15 min (+). Immunoprecipitated (IP) IGF-1R was assessed for in vitro kinase activity toward poly(Glu,Tyr) in the presence of [γ-32P]ATP. Data from three independent experiments are presented as the fold change in kinase activity of IGF-1R immunoprecipitates relative to control (immunoprecipitates from empty vector cells). Error bars reflect the standard deviation, **, p < 0.005, and *, p < 0.01, calculated by Student's t test. Levels of IGF-1R determined by immunoblotting with anti-IGF-1R antibody are indicated. B, R−/IGF-1R WT, R−/IGF-1R Y1250F/Y1251F, and R−/IGF-1R S1248A cells were serum-starved for 4 h or maintained in serum. IGF-1R was immunoprecipitated and then immunoblotted for anti-phosphotyrosine content. Blots were stripped and re-probed with anti-IGF-1R antibody. C, R−/IGF-1R WT, R−/IGF-1R Y1250F/Y1251F, and R−/IGF-1R S1248A cells were serum-starved for 4 h and then stimulated with IGF-1 for the indicated times. Cell lysates were immunoblotted with anti-IGF-1R, -phospho-Akt, -Akt, -phospho-ERK, and -ERK antibodies. D and E, Western blots as described in C were probed with anti-phospho-p70 S6 kinase, -p70 S6 kinase, -phospho-4E-BP1, and -4E-BP1 antibodies.
Enhanced Size, Growth, and Adherence of Cells Expressing S1248A Mutant
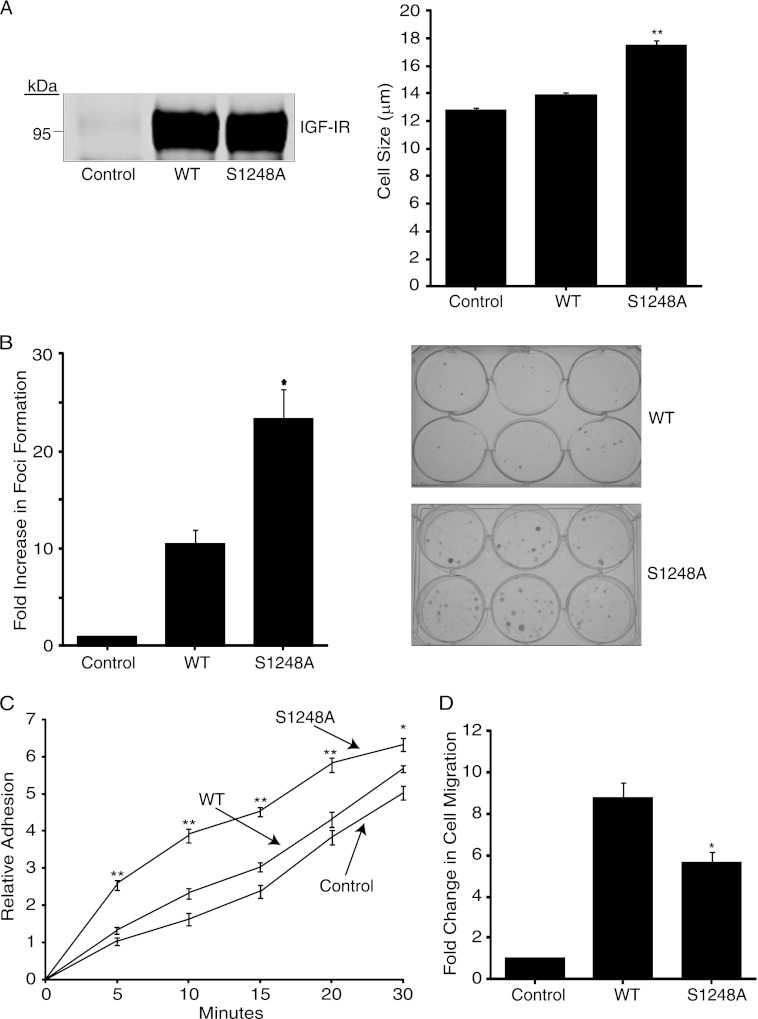
We next investigated the phenotype of R− cells expressing the S1248A mutant compared with WT IGF-1R. R− cells stably expressing the S1248A IGF-1R exhibited increased cell size compared with cells expressing WT IGF-1R or vector (Fig. 2A). No difference in rates of growth in monolayer cell cultures was observed (data not shown), but cells expressing S1248A cells exhibited an ∼2.5-fold increase in foci formation in low density cultures relative to cells expressing WT IGF-1R (Fig. 2B). Cells expressing S1248A also exhibited an increased rate of adherence to tissue culture plates (Fig. 2C). This was consistent with reduced migratory capacity in a Boyden chamber compared with those expressing WT IGF-1R (Fig. 2D). Overall, the data indicate that cells expressing the S1248A IGF-1R mutant exhibit enhanced growth potential, which is consistent with enhanced IGF-1R kinase activity and mTOR signaling.
FIGURE 2.
Cells expressing S1248A mutant exhibit enhanced size, increased adherence, and increased growth compared with WT IGF-1R. A, R− cells stably expressing WT or S1248A IGF-1R (R−/IGF-1R WT and R−/IGF-1R S1248A) were assessed for cell size using the Countess automated cell counter. Results are expressed in μm and are representative of three separate experiments. Error bars reflect the standard deviation, **, p < 0.005 was calculated by Student's t test. B, foci formation assay in which control, R−/IGF-1R WT, and R−/IGF-1R S1248A cells were seeded in multiple wells of a 6-well plate at a density of 400 cells/well for 12 days, after which colonies were stained with crystal violet and counted. Results represent three separate experiments and are presented as fold change in foci formation of IGF-1R-expressing cells compared with vector-transfected-R− cells (Control). *, p < 0.01 was calculated using Student's t test. Photographs of representative plates are shown in right panels. C, R−/IGF-1R WT and R−/IGF-1R S1248A were allowed to adhere to tissue culture plates in complete media for the indicated time at which point they were fixed and stained with crystal violet to assess relative cell number. Data are representative of three independent experiments; error bars represent the standard deviation between experiments. **, p < 0.005, and *, p < 0.01 were calculated using Student's t test. D, R− cells transiently transfected with empty vector, WT IGF-1R, or S1248A IGF-1R were seeded at 5 × 104 cells/chamber in Transwell chambers and allowed to migrate toward FBS for 24 h. Cells on the upper surface of the membrane were removed, and the membrane was stained with crystal violet. Five fields per membrane were counted. The data are representative of three separate experiments and are presented as the fold change in cell counts over vector-transfected-R− cells (Control). *, p < 0.01 was calculated using Student's t test.
Serine Phosphorylation of IGF-1R C Terminus
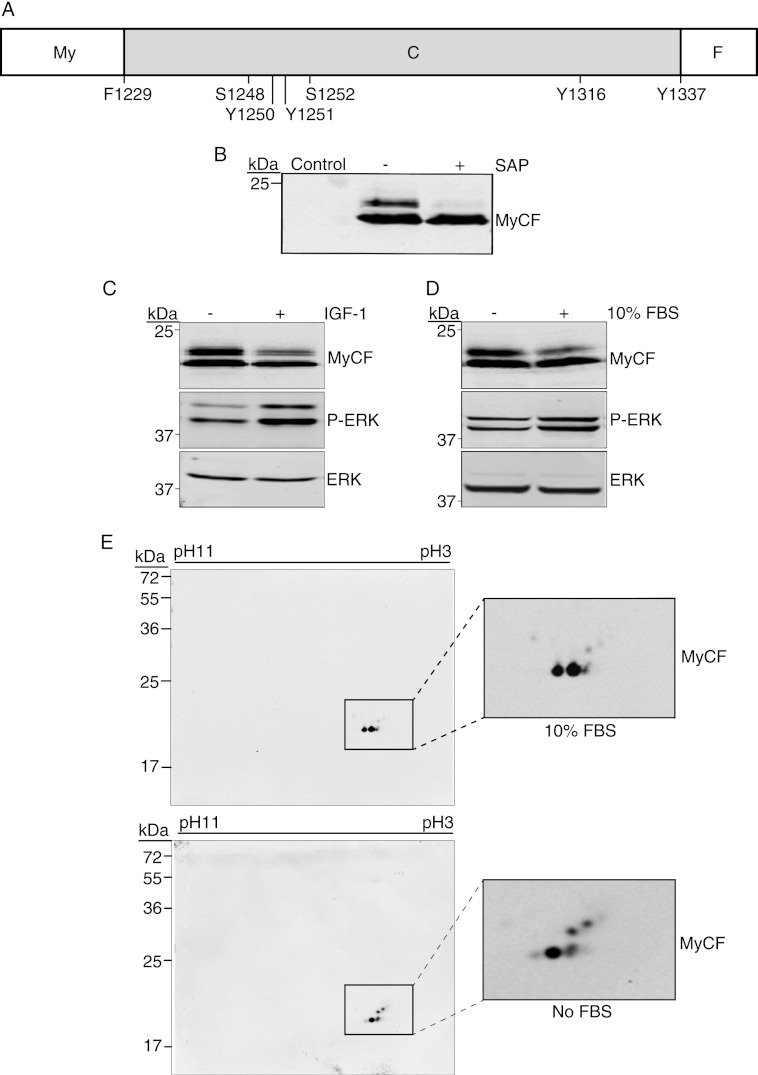
We next investigated whether Ser-1248 is phosphorylated under physiological conditions in cells cultures in the presence or absence of serum or IGF-1. Because the full-length IGF-1R has multiple phosphorylation sites throughout the cytoplasmic domain and, given the large mass of the IGF-1R β-chain (95 kDa), it is not possible to detect serine phosphorylation-induced mobility shifts on one- or two-dimensional SDS-PAGE. Therefore, we focused on those in the C terminus by using the MyCF expression construct, which encodes the entire C terminus (amino acids 1229–1337) plus a myristoylation sequence at the N terminus to promote membrane anchorage and a FLAG tag at the C terminus (Fig. 3A) (18, 19). The MyCF protein expressed in MCF-7 cells migrates as two bands in SDS-PAGE, one at the predicted mass of 20 kDa and a slower migrating band that we hypothesized represents a serine-phosphorylated form of the peptide (Fig. 3B). Exposure of cell lysates to shrimp alkaline phosphatase removed the slower migrating species, confirming that this is phosphorylated MyCF (Fig. 3B).
FIGURE 3.
Serine 1248 in IGF-1R C terminus is phosphorylated in cells. A, illustration depicting the MyCF peptide, which encodes the IGF-1R C terminus (C) (residues 1229–1337) with an N-terminal myristoylation (My), sequence, and C-terminal FLAG (F) tag. B, MCF-7 cells were transiently transfected with pcDNA3 (control) or pcDNA3 MyCF and cultured for 24 h in complete medium. Cell lysates, untreated or treated with shrimp alkaline phosphatase (SAP), were immunoblotted with anti-IGF-1R antibody to detect the MyCF peptide. C, MCF-7 cells transiently transfected with pcDNA3 MyCF WT or pcDNA3 MyCF S1248A were cultured in complete medium or serum-starved (−FBS) for 4 h prior to cell lysis. Lysates were immunoblotted with anti-IGF-1R, anti-phospho-ERK, or anti-ERK antibodies. D, MCF-7 cells transiently transfected with pcDNA3 MyCF were serum-starved for 4 h and stimulated with IGF-1 for 15 min (+ IGF-1). Cell lysates were immunoblotted as in C. E, two-dimensional PAGE analysis of total cell lysates prepared from MCF-7 cells transiently transfected with pcDNA3 MyCF WT cultured in the presence or absence of FBS.
We next compared the migratory patterns of the MyCF peptide extracted from cells that were serum-starved for 4 h (−) or stimulated with IGF-1 (Fig. 3C). The slower migrating (phosphorylated) MyCF band was evident in cells that were serum-starved for 4 h (Fig. 3C). The intensity of phosphorylated MyCF band was reduced when cells were stimulated with IGF-1. A similar reduction in intensity of this upper band was also evident in cells stimulated with FBS (Fig. 3D).
We also used two-dimensional gel electrophoresis to analyze migration of MyCF-expressing cell lysates derived from cells cultured in the presence or absence of serum. MyCF was detected by Western blotting at the pI of ∼4.6, as predicted by the on-line server Scansite 2.0 (36). As can be seen in Fig. 3E, WT MyCF is represented as a number of species with different pI values and mobility, and higher mobility species are present in cells cultured in the absence of serum. These species are not visible when these lysates are treated with shrimp alkaline phosphatase (supplemental Fig. 1). This is consistent with observations in one-dimensional SDS-PAGE (Fig. 3B) and the conclusion that these spots represent phosphorylated species. Overall, the data indicate that phosphorylation of the C terminus and potentially Ser-1248 is more prevalent in serum-starved conditions.
GSK-3β Phosphorylates Ser-1248 in Cells
We next investigated candidate kinases that may phosphorylate Ser-1248. GSK-3β is known to be active in serum-starved cells, is inactivated by IGF-1 stimulation, and is therefore a candidate negative regulatory kinase. The IGF-1R C-terminal region 1248SFYYS1252 matches the proposed consensus site for GSK-3β phosphorylation: (S/T)XXX(S/T), where the first Ser or Thr is the target residue; X is any amino acid, and the C-terminal Ser or Thr is the site of a priming phosphorylation, which may occur prior to GSK-3β-mediated phosphorylation of some substrates (37). Although not strictly required for every GSK-3β substrate, the priming phosphorylation increases the phosphorylation efficiency of GSK-3β by 100–1000-fold (38).
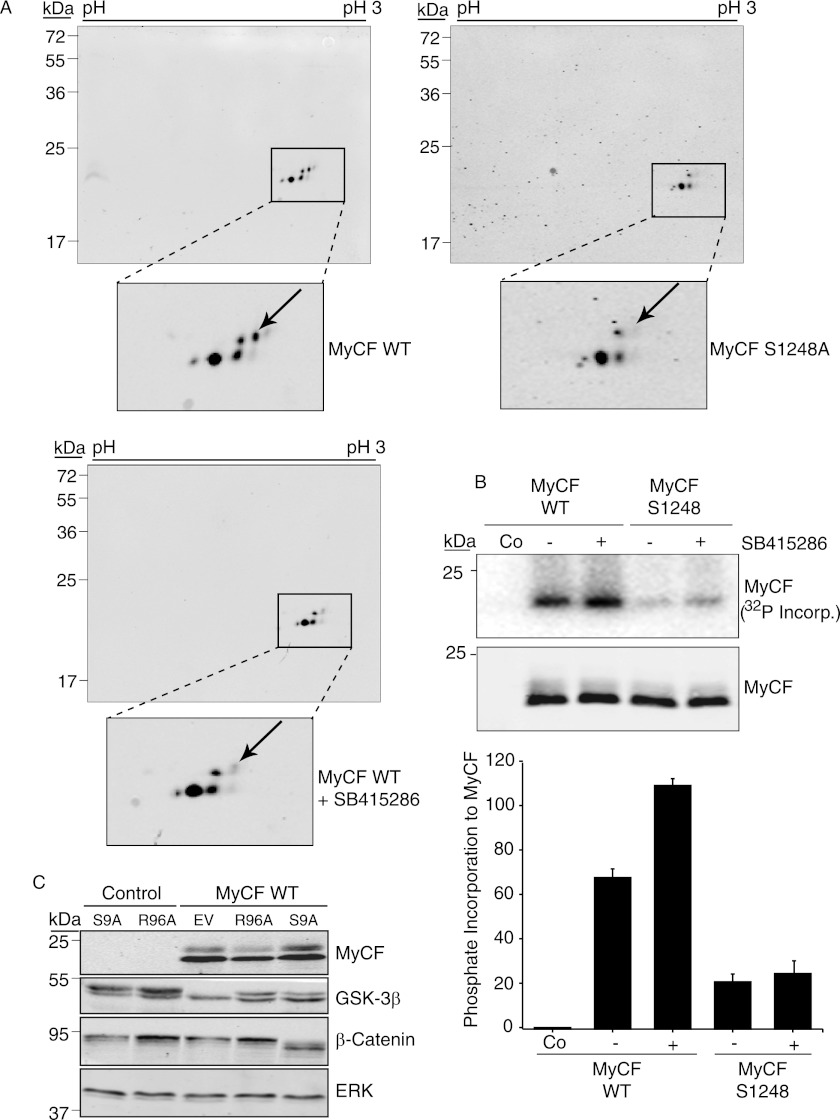
We first asked whether preincubation of cells with a GSK-3β inhibitor would affect the migration of phosphorylated MyCF species in two-dimensional gel electrophoresis. As can be seen in Fig. 4A, MyCF is detected as a number of species some of which represent phosphorylated species as determined in Fig. 3E. Interestingly, one of the species (Fig. 4A, indicated by arrow) is absent in cell lysates expressing the MyCF S1248A mutant. In cell lysates that were preincubated with the GSK-3β inhibitor SB415286, the same phosphorylated species was reduced compared with untreated WT MyCF cells. The observation that GSK-3β inhibition reduces abundance of the same MyCF phosphorylated species that is absent in resolution of the S1248A mutant indicates that GSK-3β phosphorylates the IGF-1R C terminus on Ser-1248.
FIGURE 4.
Phosphorylation of Ser-1248 by GSK-3β. A, two-dimensional PAGE analysis of total cell lysates prepared from serum-starved MCF-7 cells transiently transfected with pcDNA3 MyCF WT or pcDNA3 MyCF S1248A. Where indicated, cells expressing MyCF were pretreated with the GSK-3β inhibitor SB415286 for 1 h prior to lysis. MyCF was detected by immunoblotting with anti-IGF-1R antibody. B, HEK 293T cells were transiently transfected with pcDNA3 encoding MyCF or MyCF/S1248A and where indicated were incubated with GSK-3β inhibitor SB415286 for 1 h prior to cell lysis. IGF-1R immunoprecipitates were subsequently incubated in GSK-3β kinase buffer with active GSK-3β and [γ-32P]ATP for 20 min at 30 °C before analysis by autoradiography. MyCF expression levels were determined by immunoblotting with anti-IGF-1R antibody. Co, control. C, MCF-7 cells transfected with either pcDNA3 (control) or pcDNA3 MyCF were co-transfected with pcDNA3 vectors encoding Myc-tagged GSK-3β R96A or GSK-3β S9A or empty vector (EV) for 48 h. Cell lysates were immunoblotted with anti-IGF-1R (to detect MyCF), anti-GSK-3β, anti-β-catenin, and anti-ERK2 antibodies. Note: Myc-tagged GSK-3β proteins exhibit lower mobility than endogenous GSK-3β protein.
To further investigate whether Ser-1248 is a site for GSK-3β phosphorylation in cells, we carried out in vitro kinase assays with MyCF immunoprecipitates from HEK293T cells that had been pre-exposed to the GSK-3β inhibitor or not. As described previously for c-Abl and GSK-3β (39, 40), inhibition of the kinase in vivo may enhance availability of substrate sites for subsequent phosphorylation in vitro. As can be seen in Fig. 4B, immunoprecipitated WT MyCF can be phosphorylated by recombinant GSK-3β in vitro, and levels of [γ-32P]ATP incorporation were enhanced 1.6-fold in the presence of the GSK-3β inhibitor. In contrast there is minimal phosphorylation of the S1248A mutant, which includes several additional serines. This indicates that Ser-1248 is required for GSK-3β phosphorylation of MyCF. The data also indicate that serine 1248 is phosphorylated at least on a portion of MyCF in cells.
To investigate whether GSK-3β phosphorylates the IGF-1R C terminus in a mechanism that requires priming, we co-expressed GSK-3β mutants with the MyCF protein in MCF-7 cells and then assessed MyCF mobility in SDS-PAGE. GSK-3β S9A is a constitutively active mutant, which cannot be phosphorylated on Ser-9 and thereby inactivated. GSK-3β R96A is a dominant negative kinase that can only phosphorylate unprimed GSK-3β substrates such as Axin and Tau (38). GSK-3β R96A can thus be used to distinguish whether or not a GSK-3β substrate requires a priming phosphorylation. As can be seen in Fig. 4C, the control MyCF protein migrates as two species, and the upper one is indicative of phosphorylation (Fig. 3). However, when co-expressed with GSK-3β R96A, there is reduced abundance of the upper migrating species, whereas co-expression with S9A increases the abundance of the upper species. This suggests that GSK-3β R96A acts in a dominant negative fashion to reduce MyCF phosphorylation, which indicates that the IGF-1R SFYYS motif is a primed substrate for GSK-3β. Levels of β-catenin were assessed as a control for GSK-3β function. GSK-3β S9A promotes degradation of β-catenin, whereas overexpression of GSK-3β R96A promotes accumulation of β-catenin. Overall, we conclude that Ser-1248 within the SFYYS motif is a site for GSK-3β phosphorylation.
Phosphorylation of Ser-1248 by GSK-3β Suppresses IGF-1R Tyrosine Kinase Activity
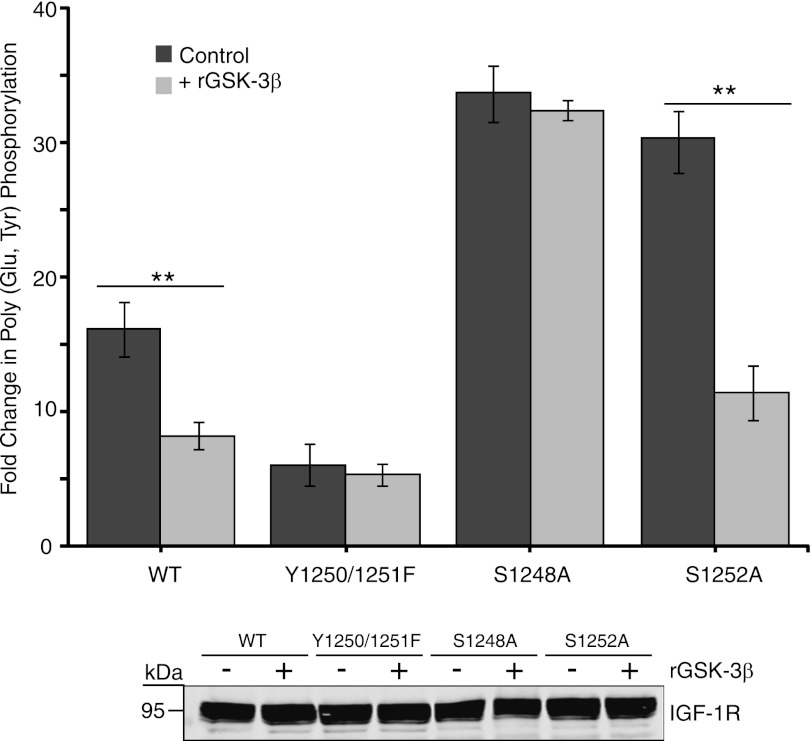
The IGF-1R S1248A mutant exhibits increased tyrosine kinase activity compared with WT IGF-1R (Fig. 1). To directly test whether the enhanced kinase activity of S1248A is due to loss of a GSK-3β phosphorylation site, we pre-phosphorylated IGF-1R immunoprecipitates in vitro with GSK-3β, and we then assessed tyrosine kinase activity toward poly(Glu,Tyr). Preincubation of WT IGF-1R immunoprecipitates with recombinant GSK-3β caused a 55% decrease in IGF-1R kinase activity toward poly(Glu,Tyr) (Fig. 5). The Y1250F/Y1251F mutant displayed inherently low kinase activity compared with wild type, which was unaffected by pre-phosphorylation with GSK-3β. IGF-1R S1248A kinase activity was also unaffected by pre-phosphorylation with GSK-3β, which is consistent with this serine being a site for GSK-3β phosphorylation. The tyrosine kinase activity of S1252A was comparable with that of the S1248A, which further indicates that Ser-1252 acts as the priming site for GSK-3β phosphorylation of Ser-1248. The requirement for priming can be overcome when GSK-3β is abundant or saturating, which would account for the observation that S1252A kinase activity was reduced by 65% following GSK-3β phosphorylation.
FIGURE 5.
GSK-3β phosphorylation of IGF-1R suppresses tyrosine kinase activity. R− cells expressing pcDNA3 empty vector, IGF-1R WT, or the Tyr-1250/Tyr-1251, S1248A, and S1252A mutants were assessed for in vitro kinase activity toward poly(Glu,Tyr) in the presence of [γ-32P]ATP. IGF-1R immunoprecipitates were preincubated, where indicated, with recombinant GSK-3β and 3 mm ATP (cold) for 30 min prior to addition of IGF-1R kinase reaction components. Data from three independent experiments are presented as the fold change in kinase activity of each IGF-1R immunoprecipitate over that of immunoprecipitates from cells expressing empty vector (set at 1). Mean and standard deviation are shown for kinase activity with no GSK-3β or preincubated with recombinant GSK-3β (**, p < 0.005 was calculated using Student's t test). IGF-1R input levels were confirmed by immunoblotting equivalent amounts of input fractions with anti-IGF-1R antibody (lower panel).
Increased IGF-1R Kinase Activity, Cell Size, and IGF-1 Signaling in GSK-3β−/− Cells
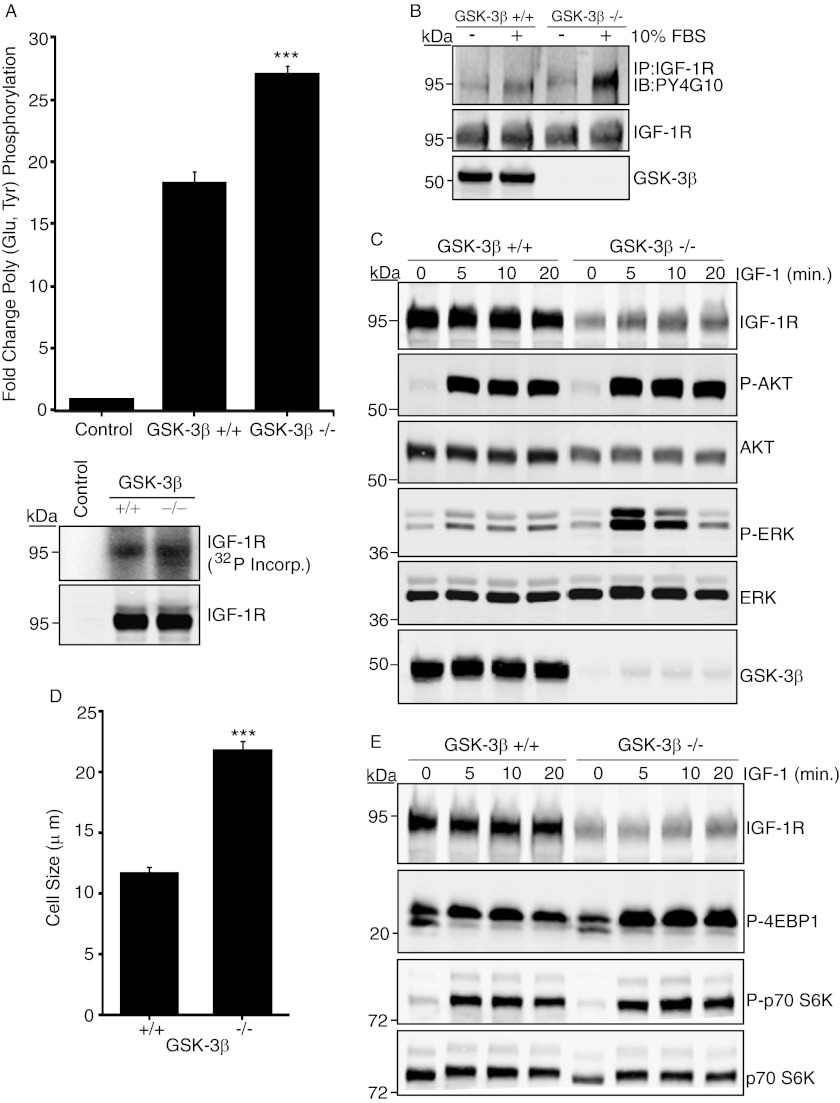
We next asked whether regulation of IGF-1R by GSK-3β has more general physiological consequences. To do this, we investigated IGF-1R activity in cells that are deficient in GSK-3β expression. GSK-3β−/− MEFs were first assessed for IGF-1R tyrosine kinase activity, autophosphorylation, and signaling responses. In vitro kinase assays with equivalent amounts of immunoprecipitated IGF-1R demonstrated significantly enhanced IGF-1R kinase activity in −/− cells compared with WT controls (Fig. 6A). Autophosphorylation of the IGF-1R was also enhanced (Fig. 6B). Although IGF-1R and AKT levels appeared to be significantly reduced in GSK-3β−/− cell extracts, IGF-1-induced phosphorylation of AKT and Erk was increased in −/− cells compared with controls (Fig. 6C). Activation of the mTOR pathway was also increased (Fig. 6E), and this correlated with increased cell size (Fig. 6D). Taken together, these data indicate that cells lacking GSK-3β exhibit increased activation of the IGF-1R and its signaling pathway. This supports the conclusion that GSK-3β regulation of the IGF-1R is a physiological event that has important consequences for restraining cell growth.
FIGURE 6.
GSK-3β−/− cells exhibit enhanced IGF-1R kinase activity and signaling. A, IGF-1R immunoprecipitates from WT or GSK-3β−/− cells were incubated with poly(Glu,Tyr) in the presence of [γ-32P]ATP. Data from three independent experiments are presented as the fold change in kinase activity of IGF-1R immunoprecipitates relative to negative control (protein G-agarose beads). ***, p < 0.001 using Student's t test. Lower panels show [γ-32P]ATP incorporation in the IGF-1R immunoprecipitates after the kinase assay and the levels of IGF-1R. B, WT or GSK-3β−/− cells were serum-starved for 4 h or maintained in serum as indicated. IGF-1R was immunoprecipitated (IP) from cell lysates that had been normalized for equal IGF-1R input followed by immunoblotting (IB) with anti-phosphotyrosine antibody. Blots were stripped and re-probed with the anti-IGF-1R antibody. Input samples were immunoblotted with anti-GSK-3β (lower panel). C, WT or GSK-3β−/− cells were serum-starved for 4 h and stimulated with IGF-1 for the indicated times, prior to immunoblotting with the indicated antibodies. D, WT or GSK-3β−/− cells were assessed for cell size using the Countess automated cell counter. Results are expressed in μm and are representative of three separate experiments. Error bars reflect the standard deviation; ***, p < 0.001 was calculated by Student's t test. E, lysates from WT or GSK-3β−/− cells were immunoblotted with anti-IGF-1R, phospho-4E-BP1 (Thr-37/46), phospho-p70 S6 kinase (Thr-389), and p70 S6 kinase antibodies.
Altered IGF-1R Expression and Internalization with S1248A Mutation and GSK-3β Deficiency
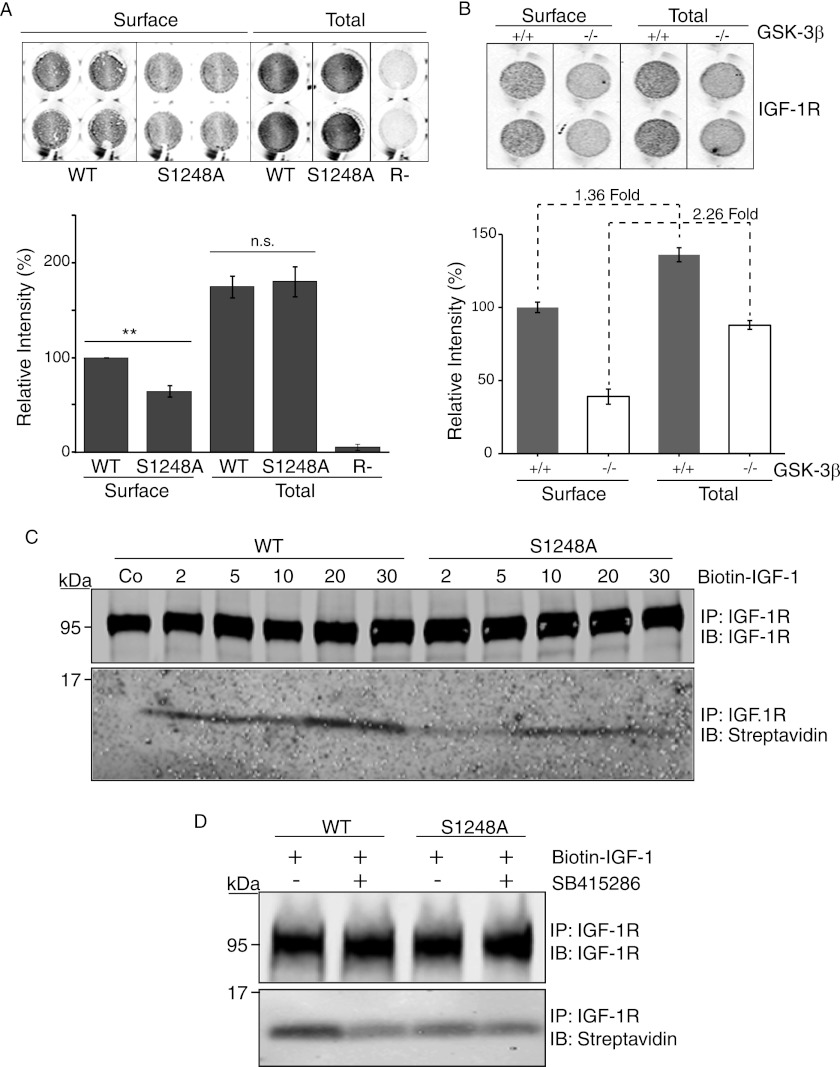
GSK-3β activity has previously been associated with receptor internalization (41, 42), and because GSK-3β null cells express lower IGF-1R levels, we investigated IGF-1R internalization using biotin-IGF-1 uptake. We first measured cell surface and total IGF-1R with the in cell Western method using a mAb to label IGF-1R in permeabilized (total) and nonpermeabilized (surface) cells. Although overall expression of WT and S1248A was equivalent, S1248A exhibited lower surface expression than WT IGF-1R (Fig. 7A). This suggests altered recycling of IGF-1R. This was also evident in GSK-3β null cells, which had overall lower IGF-1R expression than WT, but still exhibited even lower levels of IGF-1R expression at the cell surface than WT cells (44% compared with 73%) (Fig. 7B). It must be noted, however, that S1248A and GSK-3β null cells cannot be directly compared because clones of R− cells with equivalent levels of WT or S1248A were selected following transfection.
FIGURE 7.
IGF-1R trafficking is impaired in cells that express the S1248A mutant or are deficient in GSK-3β. A, R− cells expressing IGF-1R or IGF-1R/S2148A were assayed for cell surface IGF-1R expression by in cell Western blot. Cells were fixed and either permeabilized (Total) or not (Surface). Surface IGF-1R was detected with the αIR-3 antibody, and R− cells, which do not express IGF-1R, were used as a negative control. The lower panel depicts intensity of IGF-1R expression relative to syto60 nuclear stain. Error bars reflect standard deviation in IGF-1R staining, normalized to syto-60, from three separate experiments. **, p < 0.005 was calculated by Student's t test; n.s., not significant. B, GSK-3β WT (+/+) and GSK-3β-null (−/−) MEF cells were assayed for cell surface IGF-1R expression as in A. C, R−/IGF-1R WT- and R−/IGF-1R S1248A-expressing cells were stimulated with biotin-IGF-1 for the times indicated. The control sample was incubated at 4 °C. IGF-1R was immunoprecipitated from cell lysates and detected on Western blots using streptavidin. D, R−/IGF-1R WT- and R−/IGF-1R S1248A-expressing cells stimulated with biotin-IGF-1 for 20 min, and where indicated cells were pretreated with the GSK-3β inhibitor SB415286 for 1 h prior to cell lysis. Immunoprecipitated IGF-1R was assessed for bound IGF-1 using streptavidin.
IGF-1R internalization in S1248A cells was monitored using biotinylated IGF-1 (Fig. 7C). S1248A cells internalized a lower amount of IGF-1 than WT cells. Preincubation with the GSK-3β inhibitor reduced biotin-IGF-1 internalization in WT cells but did not affect S1248A cells (Fig. 7D). This, taken with the observation that S1248A internalizes biotin-IGF-1 less efficiently, suggests that phosphorylation of Ser-1248 by GSK-3β facilitates recycling of the IGF-1R between the cytosol and cell surface.
Model for Function of Ser-1248 in Phosphorylated and Unphosphorylated States Supported by Kinase Activity of S1248E and Kinase Domain Lys-1081 Mutants
We next explored a structural basis for the mechanism of IGF-1R kinase regulation by GSK-3β phosphorylation of the SFYYS motif in the C-terminal tail. To do this, we examined crystal structures of the IGF-1R kinase that, although C-terminally truncated after amino acid 1256, encompass the SFYYS motif. These include crystal structures with the kinase A-loop in various states of phosphorylation (from unphosphorylated to fully phosphorylated) and variously complexed with ATP mimetics, inhibitors, and phospho-acceptor peptide substrates (7, 22–31). In these structures, the unphosphorylated SFYYS motif universally adopts a conformation tightly packed against helices αI and αE of the kinase C-lobe (Fig. 8A, Position-1). The conformation is conserved in the cognate SFFHS region of the IR kinase (43).
FIGURE 8.
IGF-1R kinase domain models and IGF-1R mutants illustrate potential impact of Ser-1248 phosphorylation. A, IGF-1R kinase domain (PDB code 1K3A) (7) is shown (gray ribbon) with the conformation of its unphosphorylated SFYYS motif indicated (Position-1, green ribbon). The unphosphorylated Ser-1248 and phosphorylated A-loop Tyr-1131 residues are marked (green sphere and orange stick, respectively). The trajectory for connection of the 81-residue C-terminal extension (absent in the crystallographic IGF-1R construct) lies proximal to Tyr(P)-1131. Charge opposition between Ser(P)-1248 and Asp-1091 will drive conformational reorganization of the C-terminal linker region following phosphorylation of Ser-1248 by GSK-3β. The linker may refold around the kinase domain as in position-2 (red ribbon), which is modeled on the corresponding region of the FGFR1 (PDB code 3GQL) (44), with Ser(P)-1248 (red sphere) favorably engaging Lys-1081 and targeting downstream acidic residues (1259EPEELD1264) to a basic surface near the binding sites for substrate (blue surface) and nucleotide (stick/orange sphere). Position-2 organization may confer an autoinhibitory function on the C terminus as established for the corresponding region of Tie2 (45, 46). Alternatively, phosphorylation of Ser-1248 may lift the pSFYYS linker region from the surface of the kinase domain as in position-3 (violet ribbon), where Ser(P)-1248 (violet sphere) and C-terminal residues may contribute to protein docking sites. The position-3 conformation is adopted by the C-terminal region of the FGFR1 kinase (PDB code 3GQI), where the cognate residue (Tyr-766) to IGF-1R Ser-1248 is phosphorylated and serves as a binding site for PLCγ Src homology 2 (44). Position-2 and -3 conformations are indicative of feasible structural reorganization arising from Ser-1248 phosphorylation rather than illustrative of definitively established post-phosphorylation conformations for IGF-1R. B, structural detail for the unphosphorylated SFYYS region highlighting Ser-1252 and its proximity to Glu-1254. Charge opposition between these residues following priming phosphorylation on Ser-1252 may destabilize the SFYYS conformation, assisting unfolding and subsequent phosphorylation of Ser-1248 by GSK-3β. C and D, immunoprecipitates of S1248E (C) and K1081G and K1081E mutants (D) mutants transiently expressed in R− cells were incubated with poly(Glu,Tyr) in the presence of [γ-32P]ATP and assessed for incorporated radioactivity. Data from three independent experiments are presented as the fold change in kinase activity of IGF-1R immunoprecipitates compared with immunoprecipitates from control (R−/vector) cells. Error bars reflect the standard deviation, ***, p < 0.001, and **, p < 0.005 were calculated using Student's t test. IGF-1R levels in each assay were determined by immunoblotting.
Presentation of the SFYYS motif is controlled substantially by the formation of two strong hydrogen bonds from Asp-1091 (αE) to the peptide backbone adjacent to Ser-1248 (Fig. 8B), and this results in a connection trajectory for the protein's 81-amino acid C-terminal extension that lies proximal to the position occupied by Tyr(P)-131 when the A-loop adopts the active conformation. Interestingly, Ser-1248 is not surface-exposed in the unphosphorylated state but is trapped against helix αE (Fig. 8B); its phosphorylation must therefore involve unfolding of the motif from the core kinase domain, and this may account in part for the required priming phosphorylation on Ser-1252. The latter is surface-exposed and lies proximal to the Glu-1254 side chain (Fig. 8B); charge opposition between Ser(P)-1252 and Glu-1254 will partially destabilize the crystallographically observed SFYYS conformation and may therefore assist unfolding of the sequence and subsequent phosphorylation on Ser-1248. Once phosphorylated, a second charge opposition between Ser(P)-1248 and Asp-1091 will block a return to the position-1 conformation (Fig. 8A, green) favored for the unphosphorylated motif. Phosphorylation of Ser-1248 will therefore result in a profound conformational change, and this suggests that the SFYYS motif is a key linker region that controls the presentation of the C terminus relative to the kinase domain.
At present the post-phosphorylation conformation of the SFYYS motif is unclear. An intriguing possibility is that Ser(P)-1248 may target the side chain of Lys-1081 and thereby promote wrapping of the C-terminal extension around the core kinase domain proximal to the kinase insert region (Fig. 8A, Position-2, red), potentially targeting downstream acidic residues (1259EPEELD1264) to a basic surface near to the substrate-binding site. The adoption of just such a trajectory is seen in the crystal structure (PDB code 3GQL) of an FGFR1 kinase construct, where a C-terminal aspartate docks to the cognate surface (44). Such folding of the C-terminal region in IGF-1R could feasibly exert a cis-autoinhibitory action by obstructing substrate docking, thus accounting for the reduced kinase activity associated with the phosphorylation of Ser-1248. Indeed, this type of autoinhibitory action by the C-terminal sequence is known to operate for Tie2, where a C-terminal trajectory similar to the FGFR1-like position-2 folding (Fig. 8A, red) is also observed in crystal structures (45, 46).
To test the concept that Ser(P)-1248 may associate with Lys-1081 and thereby facilitate an inhibitory interaction of the C-terminal tail with the kinase domain, we assessed the in vitro kinase activity of a phosphomimetic S1248E IGF-1R mutant and also of K1081G and K1081E IGF-1R mutants that would be predicted not to associate with a phosphorylated serine. As can be seen in Fig. 8C, the S1248E mutant exhibited decreased in vitro kinase activity and autophosphorylation. In contrast, both the K1081G and K1081E mutants exhibited increased in vitro kinase activity and autophosphorylation (Fig. 8D). These results indicate that Ser(P)-1248 and Lys-1081 contribute to regulation of IGF-1R kinase activity and support the hypothesis that this may be mediated by folding of the C-terminal tail around the substrate site as outlined above.
Alternative mechanisms by which the C-terminal region might exert an autoinhibitory action are also conceivable, however, and in principle these could include mechanisms where the C-terminal presentation regulated by phosphorylation of Ser-1248 allows a trans-inhibitory action across protomers within a multimeric receptor assembly. Thus, rather than wrapping tightly around the kinase core (Fig. 8A, Position-2), the pSFYYS motif may lift from the surface as illustrated in position-3 (Fig. 8A, violet), an arrangement that could also result in exposure of the motif and/or downstream residues as a protein-docking site (e.g. for RACK1, see above). Significantly, the corresponding region of the FGFR1 kinase responds in such a way upon phosphorylation of the cognate residue (Tyr-766) to Ser-1248, with the exposed phosphotyrosine and flanking residues serving as a binding site for the PLCγ Src homology 2 domain in this instance (PDB code 3GQI) (44).
Although the post-phosphorylation conformation of the pSFYYS motif has yet to be established, it is clear from analysis of the available crystal structures that phosphorylation of Ser-1248 must drive a profound structural transition in this key linker region. In contrast, introduction of the S1248A mutation is unlikely to significantly alter the structure from that observed for the unphosphorylated SFYYS linker because the overall package of interactions responsible for maintenance of its conformation, and especially the two hydrogen bonds from Asp-1091, will be largely retained. The primary impact of S1248A mutation is thus likely to be the loss of a phosphorylation site, which acts as a switch for movement of the C-terminal linker from the crystallographic Position-1 (Fig. 8A, green). S1248A mutation is therefore likely to “lock” the linker region in Position-1, blocking its conformational reorganization relative to WT enzyme under conditions where phosphorylation is possible. This is consistent with our finding of enhanced kinase activity with the S1248A mutant (Fig. 1A) in serum-starved conditions and the effects of GSK-3β inhibition or deletion in enhancing the kinase activity of WT IGF-1R.
DISCUSSION
Here, we demonstrate that serine phosphorylation of the SFYYS motif in the IGF-1R C terminus regulates IGF-1R kinase activity. The overall findings are summarized as follows. In the presence of IGF-1 stimulation, Ser-1248 is unphosphorylated, and the C terminus is organized in a manner that favors IGF-1R kinase activity. GSK-3β-mediated phosphorylation of Ser-1248 in the absence of IGF-1 stimulation suppresses IGF-1R activity by promoting conformational reorganization of the C terminus. This results in direct auto-inhibitory action of the C-terminal region on the kinase or potentially indirect regulation facilitated by protein docking. This regulatory mechanism could prevent latent activation of the receptor in the absence of ligand or while the receptor is trafficked within the cell.
These findings offer important new insights into regulation of IGF-1R activation, shed light on many previous studies that proposed a regulatory role for the IGF-1R C-terminal tail (13, 17–19, 47, 48), and show a role for serine phosphorylation in this regulation (10). Our study is the first to demonstrate phosphorylation of the SFYYS motif in cells and to demonstrate the consequences of this for kinase activity and signaling via the PI3K-mTOR pathway for cell growth. The data also suggest a mechanism by which naturally occurring mutations of the IGF-1R C terminus and kinase domain could lead to enhanced kinase activity with consequences for cell/organ growth as well as for cancer.
Unlike most protein kinases, GSK-3β displays significant activity in unstimulated cells, and many GSK-3β substrates (for example glycogen synthase, IRS-1, β-catenin, and eIF2B) are phosphorylated under these conditions (49, 50). Upon agonist stimulation, several GSK-3β substrates become dephosphorylated (51, 52), and our findings suggest a similar mechanism for the IGF-1R. Our observation of increased IGF-1R kinase activity and signaling output in GSK-3β−/− cells strongly supports this conclusion. The phenotypes of GSK-3β−/− knock-out mice have previously been linked to several de-regulated receptor tyrosine kinase signaling pathways and transcription factors (33, 53–55). However, the phenotypes are also consistent with lack of IGF-1 receptor regulation, possibly by reduced phosphorylation of Ser-1248. GSK-3β−/− knock-out mice die late in embryonic development. This perinatal death has been attributed to heart failure caused by de-regulated proliferation of cardiomyocyte progenitors leading to greatly thickened ventricular walls (54). A critical role for IGF-1 signaling in heart development and repair is now well established, and interestingly, the over-growth phenotype of the Igf2R null mouse is also linked to defects in heart development (56). Thus, we propose that GSK-3β regulation of IGF-1R signaling is critical during embryonic development.
Although the S1248A mutant may enhance IGF-1R kinase activity and signaling due to a decreased threshold for IGF-1-induced activation because this mutant has its C-terminal region locked in the “active” position, it could also affect receptor trafficking. Indeed, we observed apparently lower expression levels of the IGF-1R in GSK-3β−/− cells, lower cell surface expression on both cells, and reduced efficiency of IGF-1 uptake in S1248A cells. GSK-3β has been shown to regulate internalization of the α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid receptor (41) and the N-methyl-d-aspartic acid receptor (42), which interestingly contains a candidate GSK-3β phosphorylation motif (SDRYS) in the NR2B subunit.
The crystallographic structures indicate that phosphorylation of Ser-1248 would lead to a profound change in conformation of the SFYYS motif and alter the organization of the C terminus with respect to the kinase domain. The decreased activity of the phosphomimetic S1248E mutant and increased activity of Lys-1081 (the residue that Ser(P)-1248 is predicted to target) mutants support this model. However, without crystal structures that include the entire C-terminal tail, we can only model this interaction.
In addition to the autoinhibitory mechanisms discussed under “Results” (Fig. 8), another likely consequence of the conformational change is the exposure of protein docking site(s) on the C terminus. The scaffolding protein RACK1 is an obvious candidate for docking upon phosphorylation of Ser-1248 because the S1248A mutant has reduced interaction with RACK1 (20). Several signaling proteins associated with IGF-1 signaling that can constitutively associate with RACK1 (IRS-2 and Shp-2) are recruited to RACK1 in response to IGF-1 (IRS-1 and Shc) or are released from RACK1 in response to IGF-1 (PP2A and Src) (16, 20, 34, 57). Thus, RACK1 could participate in regulating IGF-1R kinase activity and also facilitate PKC recruitment to the IGF-1R (35). However, because RACK1 association with the IGF-1R is dependent on integrin ligation, we conclude that this regulatory interaction occurs in the context of integrin signaling.
Tyr-1250 and Tyr-1251 have been suggested to mediate signaling responses that distinguish the IGF-1R from the IR, where the cognate residues are Phe-1277 and His-1278. Ser-1275 and Ser-1279 in the IR SFFHS motif occupy the same relative positions as Ser-1248 and Ser-1252 in the IGF-1R SFYYS motif. Substitution of Phe-1277/His-1278 in the IR with YY has been shown to endow the IR with enhanced anti-apoptotic activity in 32D cells (9). However, substitution of both tyrosines was required for this effect, suggesting a conformational component. The crystallographic structures indicate that Tyr-1250/Tyr-1251 are exposed (Fig. 8B), suggesting they may potentially be phosphorylated when Ser-1248 is unphosphorylated and the kinase is in an active configuration. The observation that pre-phosphorylation of the Y1250F/Y1251F mutant with GSK-3β did not decrease its in vitro kinase activity suggests that these tyrosines already contribute to the GSK-3β-mediated effects on kinase regulation mediated by Ser-1248 phosphorylation. Although there is no evidence for phosphorylation of Tyr-1250/Tyr-1251, we cannot rule out the possibility that phosphorylation of these residues could modulate the effects of Ser-1248 phosphorylation on IGF-1R function.
In summary, we have provided a rational model for regulation of IGF-1R kinase activation by phosphorylation of the C-terminal tail and modulation of its phosphorylation on Ser-1248 by GSK-3β. It will be of significant interest to determine how this mechanism is controlled in different cells and how it contributes to IGF-1R and potentially IR signaling.
Supplementary Material
Acknowledgments
We are grateful to Kurt Tidmore for preparing illustrations and to colleagues in the Cell Biology Laboratory for helpful discussions.
Note Added in Proof
An in-frame deletion of S1248 in the IGF-IR has been identified as a somatic mutation in renal cell carcinoma (58), and recapitulation of this mutation results in enhanced basal MAPK and Akt signaling (59).
This work was supported by Science Foundation Ireland, The Health Research Board, and Cancer Research Ireland.

This article contains supplemental Fig. 1.
- IGF-1R
- insulin-like growth factor I receptor
- mTOR
- mammalian target of rapamycin
- MEF
- mouse embryonic fibroblast
- PDB
- Protein Data Bank
- IR
- insulin receptor.
REFERENCES
- 1. Baserga R. (2007) Is cell size important? Cell Cycle 6, 814–816 [DOI] [PubMed] [Google Scholar]
- 2. Liu J. P., Baker J., Perkins A. S., Robertson E. J., Efstratiadis A. (1993) Mice carrying null mutations of the genes encoding insulin-like growth factor I (Igf-1) and type 1 IGF receptor (Igf1r). Cell 75, 59–72 [PubMed] [Google Scholar]
- 3. O'Connor R. (2003) Regulation of IGF-I receptor signaling in tumor cells. Horm. Metab. Res. 35, 771–777 [DOI] [PubMed] [Google Scholar]
- 4. Feng Z., Hu W., de Stanchina E., Teresky A. K., Jin S., Lowe S., Levine A. J. (2007) The regulation of AMPKβ1, TSC2, and PTEN expression by p53. Stress, cell, and tissue specificity, and the role of these gene products in modulating the IGF-1-AKT-mTOR pathways. Cancer Res. 67, 3043–3053 [DOI] [PubMed] [Google Scholar]
- 5. Gualberto A., Pollak M. (2009) Emerging role of insulin-like growth factor receptor inhibitors in oncology: early clinical trial results and future directions. Oncogene 28, 3009–3021 [DOI] [PubMed] [Google Scholar]
- 6. Ullrich A., Gray A., Tam A. W., Yang-Feng T., Tsubokawa M., Collins C., Henzel W., Le Bon T., Kathuria S., Chen E. (1986) Insulin-like growth factor I receptor primary structure. Comparison with insulin receptor suggests structural determinants that define functional specificity. EMBO J. 5, 2503–2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Favelyukis S., Till J. H., Hubbard S. R., Miller W. T. (2001) Structure and autoregulation of the insulin-like growth factor 1 receptor kinase. Nat. Struct. Biol. 8, 1058–1063 [DOI] [PubMed] [Google Scholar]
- 8. O'Connor R., Kauffmann-Zeh A., Liu Y., Lehar S., Evan G. I., Baserga R., Blättler W. A. (1997) Identification of domains of the insulin-like growth factor I receptor that are required for protection from apoptosis. Mol. Cell. Biol. 17, 427–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen H., Yan G. C., Gishizky M. L. (1998) Identification of structural characteristics that contribute to a difference in antiapoptotic function between human insulin and insulin-like growth factor 1 receptors. Cell Growth Differ. 9, 939–947 [PubMed] [Google Scholar]
- 10. Li S., Termini J., Hayward A., Siddle K., Zick Y., Koval A., LeRoith D., Fujita-Yamaguchi Y. (1998) The carboxyl-terminal domain of insulin-like growth factor-I receptor interacts with the insulin receptor and activates its protein-tyrosine kinase. FEBS Lett. 421, 45–49 [DOI] [PubMed] [Google Scholar]
- 11. Hongo A., D'Ambrosio C., Miura M., Morrione A., Baserga R. (1996) Mutational analysis of the mitogenic and transforming activities of the insulin-like growth factor I receptor. Oncogene 12, 1231–1238 [PubMed] [Google Scholar]
- 12. Li S., Resnicoff M., Baserga R. (1996) Effect of mutations at serines 1280–1283 on the mitogenic and transforming activities of the insulin-like growth factor I receptor. J. Biol. Chem. 271, 12254–12260 [DOI] [PubMed] [Google Scholar]
- 13. Esposito D. L., Blakesley V. A., Koval A. P., Scrimgeour A. G., LeRoith D. (1997) Tyrosine residues in the C-terminal domain of the insulin-like growth factor-I receptor mediate mitogenic and tumorigenic signals. Endocrinology 138, 2979–2988 [DOI] [PubMed] [Google Scholar]
- 14. Blakesley V. A., Koval A. P., Stannard B. S., Scrimgeour A., LeRoith D. (1998) Replacement of tyrosine 1251 in the carboxyl terminus of the insulin-like growth factor-I receptor disrupts the actin cytoskeleton and inhibits proliferation and anchorage-independent growth. J. Biol. Chem. 273, 18411–18422 [DOI] [PubMed] [Google Scholar]
- 15. Brodt P., Fallavollita L., Khatib A. M., Samani A. A., Zhang D. (2001) Cooperative regulation of the invasive and metastatic phenotypes by different domains of the type I insulin-like growth factor receptor β subunit. J. Biol. Chem. 276, 33608–33615 [DOI] [PubMed] [Google Scholar]
- 16. Kiely P. A., Leahy M., O'Gorman D., O'Connor R. (2005) RACK1-mediated integration of adhesion and insulin-like growth factor I (IGF-I) signaling and cell migration are defective in cells expressing an IGF-I receptor mutated at tyrosines 1250 and 1251. J. Biol. Chem. 280, 7624–7633 [DOI] [PubMed] [Google Scholar]
- 17. Surmacz E., Sell C., Swantek J., Kato H., Roberts C. T., Jr., LeRoith D., Baserga R. (1995) Dissociation of mitogenesis and transforming activity by C-terminal truncation of the insulin-like growth factor-I receptor. Exp. Cell Res. 218, 370–380 [DOI] [PubMed] [Google Scholar]
- 18. Hongo A., Yumet G., Resnicoff M., Romano G., O'Connor R., Baserga R. (1998) Inhibition of tumorigenesis and induction of apoptosis in human tumor cells by the stable expression of a myristoylated COOH terminus of the insulin-like growth factor I receptor. Cancer Res. 58, 2477–2484 [PubMed] [Google Scholar]
- 19. Liu Y., Lehar S., Corvi C., Payne G., O'Connor R. (1998) Expression of the insulin-like growth factor I receptor C terminus as a myristoylated protein leads to induction of apoptosis in tumor cells. Cancer Res. 58, 570–576 [PubMed] [Google Scholar]
- 20. Kiely P. A., Sant A., O'Connor R. (2002) RACK1 is an insulin-like growth factor 1 (IGF-1) receptor-interacting protein that can regulate IGF-1-mediated Akt activation and protection from cell death. J. Biol. Chem. 277, 22581–22589 [DOI] [PubMed] [Google Scholar]
- 21. Zhang W., Zong C. S., Hermanto U., Lopez-Bergami P., Ronai Z., Wang L. H. (2006) RACK1 recruits STAT3 specifically to insulin and insulin-like growth factor 1 receptors for activation, which is important for regulating anchorage-independent growth. Mol. Cell. Biol. 26, 413–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pautsch A., Zoephel A., Ahorn H., Spevak W., Hauptmann R., Nar H. (2001) Crystal structure of bisphosphorylated IGF-1 receptor kinase. Insight into domain movements upon kinase activation. Structure 9, 955–965 [DOI] [PubMed] [Google Scholar]
- 23. Munshi S., Kornienko M., Hall D. L., Reid J. C., Waxman L., Stirdivant S. M., Darke P. L., Kuo L. C. (2002) Crystal structure of the Apo, unactivated insulin-like growth factor-1 receptor kinase. Implication for inhibitor specificity. J. Biol. Chem. 277, 38797–38802 [DOI] [PubMed] [Google Scholar]
- 24. Munshi S., Hall D. L., Kornienko M., Darke P. L., Kuo L. C. (2003) Structure of apo, unactivated insulin-like growth factor-1 receptor kinase at 1.5 Å resolution. Acta Crystallogr. D Biol. Crystallogr. 59, 1725–1730 [DOI] [PubMed] [Google Scholar]
- 25. Velaparthi U., Wittman M., Liu P., Stoffan K., Zimmermann K., Sang X., Carboni J., Li A., Attar R., Gottardis M., Greer A., Chang C. Y., Jacobsen B. L., Sack J. S., Sun Y., Langley D. R., Balasubramanian B., Vyas D. (2007) Discovery and initial SAR of 3-(1H-benzo[d[rsqb]imidazol-2-yl)pyridin-2(1H)-ones as inhibitors of insulin-like growth factor 1-receptor (IGF-1R). Bioorg. Med. Chem. Lett. 17, 2317–2321 [DOI] [PubMed] [Google Scholar]
- 26. Mayer S. C., Banker A. L., Boschelli F., Di L., Johnson M., Kenny C. H., Krishnamurthy G., Kutterer K., Moy F., Petusky S., Ravi M., Tkach D., Tsou H. R., Xu W. (2008) Lead identification to generate isoquinolinedione inhibitors of insulin-like growth factor receptor (IGF-1R) for potential use in cancer treatment. Bioorg. Med. Chem. Lett. 18, 3641–3645 [DOI] [PubMed] [Google Scholar]
- 27. Wu J., Li W., Craddock B. P., Foreman K. W., Mulvihill M. J., Ji Q. S., Miller W. T., Hubbard S. R. (2008) Small molecule inhibition and activation-loop trans-phosphorylation of the IGF1 receptor. EMBO J. 27, 1985–1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Miller L. M., Mayer S. C., Berger D. M., Boschelli D. H., Boschelli F., Di L., Du X., Dutia M., Floyd M. B., Johnson M., Kenny C. H., Krishnamurthy G., Moy F., Petusky S., Tkach D., Torres N., Wu B., Xu W. (2009) Lead identification to generate 3-cyanoquinoline inhibitors of insulin-like growth factor receptor (IGF-1R) for potential use in cancer treatment. Bioorg. Med. Chem. Lett. 19, 62–66 [DOI] [PubMed] [Google Scholar]
- 29. Wittman M. D., Carboni J. M., Yang Z., Lee F. Y., Antman M., Attar R., Balimane P., Chang C., Chen C., Discenza L., Frennesson D., Gottardis M. M., Greer A., Hurlburt W., Johnson W., Langley D. R., Li A., Li J., Liu P., Mastalerz H., Mathur A., Menard K., Patel K., Sack J., Sang X., Saulnier M., Smith D., Stefanski K., Trainor G., Velaparthi U., Zhang G., Zimmermann K., Vyas D. M. (2009) Discovery of a 2,4-disubstituted pyrrolo[1,2-f][1,2,4]triazine inhibitor (BMS-754807) of insulin-like growth factor receptor (IGF-1R) kinase in clinical development. J. Med. Chem. 52, 7360–7363 [DOI] [PubMed] [Google Scholar]
- 30. Nemecek C., Metz W. A., Wentzler S., Ding F. X., Venot C., Souaille C., Dagallier A., Maignan S., Guilloteau J. P., Bernard F., Henry A., Grapinet S., Lesuisse D. (2010) Design of potent IGF1-R inhibitors related to bis-aza-indoles. Chem. Biol. Drug Des. 76, 100–106 [DOI] [PubMed] [Google Scholar]
- 31. Sampognaro A. J., Wittman M. D., Carboni J. M., Chang C., Greer A. F., Hurlburt W. W., Sack J. S., Vyas D. M. (2010) Proline isosteres in a series of 2,4-disubstituted pyrrolo[1,2-f][1,2,4]triazine inhibitors of IGF-1R kinase and IR kinase. Bioorg. Med. Chem. Lett. 20, 5027–5030 [DOI] [PubMed] [Google Scholar]
- 32. Sell C., Dumenil G., Deveaud C., Miura M., Coppola D., DeAngelis T., Rubin R., Efstratiadis A., Baserga R. (1994) Effect of a null mutation of the insulin-like growth factor I receptor gene on growth and transformation of mouse embryo fibroblasts. Mol. Cell. Biol. 14, 3604–3612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hoeflich K. P., Luo J., Rubie E. A., Tsao M. S., Jin O., Woodgett J. R. (2000) Requirement for glycogen synthase kinase-3β in cell survival and NF-κB activation. Nature 406, 86–90 [DOI] [PubMed] [Google Scholar]
- 34. Adams D. R., Ron D., Kiely P. A. (2011) RACK1, a multifaceted scaffolding protein. Structure and function. Cell Commun. Signal. 9, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hermanto U., Zong C. S., Li W., Wang L. H. (2002) RACK1, an insulin-like growth factor I (IGF-I) receptor-interacting protein, modulates IGF-I-dependent integrin signaling and promotes cell spreading and contact with extracellular matrix. Mol. Cell. Biol. 22, 2345–2365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Obenauer J. C., Cantley L. C., Yaffe M. B. (2003) Scansite 2.0. Proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 31, 3635–3641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dajani R., Fraser E., Roe S. M., Young N., Good V., Dale T. C., Pearl L. H. (2001) Crystal structure of glycogen synthase kinase 3β. Structural basis for phosphate-primed substrate specificity and autoinhibition. Cell 105, 721–732 [DOI] [PubMed] [Google Scholar]
- 38. Frame S., Cohen P., Biondi R. M. (2001) A common phosphate-binding site explains the unique substrate specificity of GSK3 and its inactivation by phosphorylation. Mol. Cell 7, 1321–1327 [DOI] [PubMed] [Google Scholar]
- 39. Kiely P. A., Baillie G. S., Barrett R., Buckley D. A., Adams D. R., Houslay M. D., O'Connor R. (2009) Phosphorylation of RACK1 on tyrosine 52 by c-Abl is required for insulin-like growth factor I-mediated regulation of focal adhesion kinase. J. Biol. Chem. 284, 20263–20274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Farghaian H., Turnley A. M., Sutherland C., Cole A. R. (2011) Bioinformatic prediction and confirmation of β-adducin as a novel substrate of glycogen synthase kinase 3. J. Biol. Chem. 286, 25274–25283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Du J., Wei Y., Liu L., Wang Y., Khairova R., Blumenthal R., Tragon T., Hunsberger J. G., Machado-Vieira R., Drevets W., Wang Y. T., Manji H. K. (2010) A kinesin signaling complex mediates the ability of GSK-3β to affect mood-associated behaviors. Proc. Natl. Acad. Sci. U.S.A. 107, 11573–11578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen P., Gu Z., Liu W., Yan Z. (2007) Glycogen synthase kinase 3 regulates N-methyl-d-aspartate receptor channel trafficking and function in cortical neurons. Mol. Pharmacol. 72, 40–51 [DOI] [PubMed] [Google Scholar]
- 43. Hubbard S. R., Wei L., Ellis L., Hendrickson W. A. (1994) Crystal structure of the tyrosine kinase domain of the human insulin receptor. Nature 372, 746–754 [DOI] [PubMed] [Google Scholar]
- 44. Bae J. H., Lew E. D., Yuzawa S., Tomé F., Lax I., Schlessinger J. (2009) The selectivity of receptor tyrosine kinase signaling is controlled by a secondary SH2 domain binding site. Cell 138, 514–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shewchuk L. M., Hassell A. M., Ellis B., Holmes W. D., Davis R., Horne E. L., Kadwell S. H., McKee D. D., Moore J. T. (2000) Structure of the Tie2 RTK domain. Self-inhibition by the nucleotide binding loop, activation loop, and C-terminal tail. Structure 8, 1105–1113 [DOI] [PubMed] [Google Scholar]
- 46. Niu X. L., Peters K. G., Kontos C. D. (2002) Deletion of the carboxyl terminus of Tie2 enhances kinase activity, signaling, and function. Evidence for an autoinhibitory mechanism. J. Biol. Chem. 277, 31768–31773 [DOI] [PubMed] [Google Scholar]
- 47. Blakesley V. A., Kalebic T., Helman L. J., Stannard B., Faria T. N., Roberts C. T., Jr., LeRoith D. (1996) Tumorigenic and mitogenic capacities are reduced in transfected fibroblasts expressing mutant insulin-like growth factor (IGF)-I receptors. The role of tyrosine residues 1250, 1251, and 1316 in the carboxyl terminus of the IGF-I receptor. Endocrinology 137, 410–417 [DOI] [PubMed] [Google Scholar]
- 48. Leahy M., Lyons A., Krause D., O'Connor R. (2004) Impaired Shc, Ras, and MAPK activation but normal Akt activation in FL5.12 cells expressing an insulin-like growth factor I receptor mutated at tyrosines 1250 and 1251. J. Biol. Chem. 279, 18306–18313 [DOI] [PubMed] [Google Scholar]
- 49. Rubinfeld B., Albert I., Porfiri E., Fiol C., Munemitsu S., Polakis P. (1996) Binding of GSK3β to the APC-β-catenin complex and regulation of complex assembly. Science 272, 1023–1026 [DOI] [PubMed] [Google Scholar]
- 50. Frame S., Cohen P. (2001) GSK3 takes centre stage more than 20 years after its discovery. Biochem. J. 359, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Welsh G. I., Miller C. M., Loughlin A. J., Price N. T., Proud C. G. (1998) Regulation of eukaryotic initiation factor eIF2B. Glycogen synthase kinase-3 phosphorylates a conserved serine that undergoes dephosphorylation in response to insulin. FEBS Lett. 421, 125–130 [DOI] [PubMed] [Google Scholar]
- 52. Suzuki Y., Lanner C., Kim J. H., Vilardo P. G., Zhang H., Yang J., Cooper L. D., Steele M., Kennedy A., Bock C. B., Scrimgeour A., Lawrence J. C., Jr., DePaoli-Roach A. A. (2001) Insulin control of glycogen metabolism in knockout mice lacking the muscle-specific protein phosphatase PP1G/RGL. Mol. Cell. Biol. 21, 2683–2694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Force T., Woodgett J. R. (2009) Unique and overlapping functions of GSK-3 isoforms in cell differentiation and proliferation and cardiovascular development. J. Biol. Chem. 284, 9643–9647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kerkela R., Kockeritz L., Macaulay K., Zhou J., Doble B. W., Beahm C., Greytak S., Woulfe K., Trivedi C. M., Woodgett J. R., Epstein J. A., Force T., Huggins G. S. (2008) Deletion of GSK-3β in mice leads to hypertrophic cardiomyopathy secondary to cardiomyoblast hyperproliferation. J. Clin. Invest. 118, 3609–3618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Voskas D., Ling L. S., Woodgett J. R. (2010) Does GSK-3 provide a shortcut for PI3K activation of Wnt signaling? Biol. Rep. 2, 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ludwig T., Eggenschwiler J., Fisher P., D'Ercole A. J., Davenport M. L., Efstratiadis A. (1996) Dev. Biol. 177, 517–535 [DOI] [PubMed] [Google Scholar]
- 57. Kiely P. A., O'Gorman D., Luong K., Ron D., O'Connor R. (2006) Insulin-like growth factor I controls a mutually exclusive association of RACK1 with protein phosphatase 2A and β1 integrin to promote cell migration. Mol. Cell. Biol. 26, 4041–4051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Greenman C., Stephens P., Smith R., Dalgliesh G. L., Hunter C., Bignell G., Davies H., Teague J., Butler A., Stevens C., Edkins S., O'Meara S., Vastrik I., Schmidt E. E., Avis T., Barthorpe S., Bhamra G., Buck G., Choudhury B., Clements J., Cole J., Dicks E., Forbes S., Gray K., Halliday K., Harrison R., Hills K., Hinton J., Jenkinson A., Jones D., Menzies A., Mironenko T., Perry J., Raine K., Richardson D., Shepherd R., Small A., Tofts C., Varian J., Webb T., West S., Widaa S., Yates A., Cahill D. P., Louis D. N., Goldstraw P., Nicholson A. G., Brasseur F., Looijenga L., Weber B. L., Chiew Y. E., DeFazio A., Greaves M. F., Green A. R., Campbell P., Birney E., Easton D. F., Chenevix-Trench G., Tan M. H., Khoo S. K., Teh B. T., Yuen S. Y., Leung S. Y., Wooster R., Futreal P. A., Stratton M. R. (2007) Patterns of somatic mutation in human cancer genomes. Nature 446, 153–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Craddock B. P., Miller W. T. (2012) Effects of somatic mutations in the C terminus of insulin-like growth factor I receptor on activity and signaling. J. Signal. Trans. 20121–20127 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.