Background: TFIID/SAGA histone fold-containing subunits contain conserved additional domains of unknown function.
Results: The large TAF6 conserved C-terminal domain contains HEAT repeats and influences TAF6-TAF9 complex formation.
Conclusion: TAF6 C-terminal domain is important for TFIID assembly and may trigger signals from transcriptional effectors.
Significance: The noncanonical nature of TFIID/SAGA histone fold-containing subunits appears essential for assembly of these complexes.
Keywords: Crystal Structure, General Transcription Factors, Protein Complexes, Protein Structure, Protein-Protein Interactions, RNA Polymerase II, Structural Biology, SAGA, TAF6, TFIID
Abstract
The general transcription factor TFIID recognizes specifically the core promoter of genes transcribed by eukaryotic RNA polymerase II, nucleating the assembly of the preinitiation complex at the transcription start site. However, the understanding in molecular terms of TFIID assembly and function remains poorly understood. Histone fold motifs have been shown to be extremely important for the heterodimerization of many TFIID subunits. However, these subunits display several evolutionary conserved noncanonical features when compared with histones, including additional regions whose role is unknown. Here we show that the conserved additional C-terminal region of TFIID subunit TAF6 can be divided into two domains: a small middle domain (TAF6M) and a large C-terminal domain (TAF6C). Our crystal structure of the TAF6C domain from Antonospora locustae at 1.9 Å resolution reveals the presence of five conserved HEAT repeats. Based on these data, we designed several mutants that were introduced into full-length human TAF6. Surprisingly, the mutants affect the interaction between TAF6 and TAF9, suggesting that the formation of the complex between these two TFIID subunits do not only depend on their histone fold motifs. In addition, the same mutants affect even more strongly the interaction between TAF6 and TAF9 in the context of a TAF5-TAF6-TAF9 complex. Expression of these mutants in HeLa cells reveals that most of them are unstable, suggesting their poor incorporation within endogenous TFIID. Taken together, our results suggest that the conserved additional domains in histone fold-containing subunits of TFIID and of co-activator SAGA are important for the assembly of these complexes.
Introduction
Transcription initiation by eukaryotic RNA polymerase II requires the binding at the core promoters of several general transcription factors (TFIIA,2 -B, -D, -E, -F, -H) that correctly position RNA polymerase II at the transcription start site and subsequently initiate promoter clearance (1, 2). TFIID, a large multiprotein complex composed of 15 well conserved subunits (the TATA box-binding protein (TBP) and 14 TBP-associated factors, TAF1 to TAF14), binds specifically to core promoters and initiates the recruitment of the other general transcription factors and RNA polymerase II over the transcription start site (2–4).
Recruitment of TFIID is carried out by interaction of its subunits with transcriptional activators as well as its specific recognition of promoter elements such as the TATA box (TBP), the initiator (TAF1 and TAF2), as well as the downstream promoter element and the mutation ten element (TAF6 and TAF9) (4, 5). In metazoans, recognition of the doubly acetylated H4 histone tail by the tandem bromodomains of TAF1 and the trimethylated lysine 4 of histone H3 (H3K4me3) by TAF3 PHD (plant homeo domain) domain also contributes to TFIID recruitment at promoters (6, 7).
Recently, electron microscopy (EM) studies at 20 Å resolution have shed light on the TFIID mechanism (8). Upon recruitment by the Rap1 transcriptional activator, and in the presence of TFIIA, TFIID binds to a TATA box-containing promoter by inducing the formation of a DNA loop. This process appears to lock TFIID onto promoter DNA, suggesting the first steps of the assembly of the transcription preinitiation complex. However, despite these first results, the mode of action of TFIID at the molecular level still remains poorly understood. Notably, high resolution structural data on the whole complex are still crucially missing, such data being difficult to obtain due to the large size and the intrinsic flexibility of this complex.
Nevertheless, many biochemical and structural studies have provided information on the structure of submodules of TFIID as well as on TFIID assembly. Notably, the histone fold motif has been shown to play an essential role for the dimerization of several TAFs, and five different histone-like pairs (TAF4-TAF12, TAF6-TAF9, TAF10-TAF3, TAF10-TAF8, and TAF11-TAF13) have been characterized so far (9–17). It has been proposed that a histone octamer-like substructure (formed by the pairs TAF4-TAF12 and TAF6-TAF9) exists in TFIID, and such a complex has been reconstituted in vitro with the yeast proteins (18, 19). Moreover, immunolabeling experiments coupled to EM studies as well as biochemical studies have revealed that each histone-like pair in TFIID is present twice in the complex, each pair being found in two different lobes of TFIID (20, 21).
Histone fold-containing TAFs (HFTs) are not sufficient to form a stable subcomplex within TFIID, and the WD40 repeat-containing TAF5 subunit appears important for integrating HFTs into a single subcomplex (22, 23). In agreement, in yeast, TAF5 and four HFTs (TAF6, TAF9, TAF10, and TAF12) are shared between TFIID and the transcriptional co-activator SAGA, suggesting that they form the structural core of these complexes (24, 25). In metazoan SAGA, these HFTs are also shared, with the exception of TAF5 and TAF6, which are replaced by paralogues, namely TAF5L and TAF6L (25). In addition, the other HFTs specific to TFIID are replaced in SAGA by specific histone fold-containing subunits (Ada1, Spt3, and Spt7L), suggesting a way to form two different multiprotein transcriptional activators with the same structural core (4, 12).
Although the histone fold motifs of the HFTs have drawn most of the attention on these TAFs, those motifs have not kept the high positive charge of the canonical histones, suggesting that their primary role is not DNA binding but rather dimerization, and possibly multimerization. In addition, the HFTs often have additional regions whose role remains elusive. It is not clear whether these additional regions, the histone fold motifs, or both, are responsible for the assembly of higher order structures within TFIID.
TAF6 contains one of these additional regions. This region is located at its C terminus and has been evolutionary conserved. Surprisingly, despite this strong conservation, it has been proposed that this region is not essential for TFIID assembly (26). This result is, however, in contradiction with the fact that a human TFIID complex incorporating the TAF6 isoform TAF6δ, which lacks the central part of its histone fold domain, still retains all TAFs except TAF9 (27), suggesting that other regions of TAF6 are required for integration of this TAF within TFIID.
To address this issue, we have performed biochemical and structural studies on the conserved C-terminal region of TAF6. This C-terminal region appears to be formed by two domains: a small middle domain and a larger C-terminal domain. The crystal structure of the larger C-terminal domain of TAF6 from Antonospora locustae reveals that it is constituted of five HEAT repeats, a motif generally involved in protein/protein interactions. Surprisingly, mutations of conserved residues at the surface of this C-terminal domain in full-length human TAF6 cause the weakening of the interactions between TAF6 and TAF9. Moreover, introduction of these TAF6 mutants in the context of a TAF5-TAF6-TAF9 complex appears to weaken even further the TAF6-TAF9 complex, suggesting conformational changes in the TAF6-TAF9 complex upon TAF5 binding. Expression of these mutants in HeLa cells shows that many of them are less stable when compared with the wild type TAF6. These results suggest that these mutants are poorly incorporated within TFIID and submitted to degradation inside the cell. Taken together, our results suggest that (i) formation of histone fold-containing heterodimers within TFIID does not simply rely on the histone fold motif of each partner and (ii) TFIID assembly is an intricate process possibly requiring conformational changes that may be important for TFIID function.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification
The various constructs used were amplified by standard PCR procedures and inserted in the pnEA-tH (28) expression vector using NdeI and BamHI restriction sites. Expression of all proteins was made using Escherichia coli BL21(DE3) cells (Novagen) in 2× LB medium for native proteins or using B834(DE3) cells (Novagen) in M9 medium supplemented with selenomethionine (Sigma) for selenomethionylated proteins. Cells were grown at 37 °C to an absorbance of 0.3 at 600 nm, and the temperature was then switched to 25 °C. Growth was then carried on until cells reached an absorbance of 0.8–1.0 at 600 nm. Expression was induced by adding a final concentration of 1 mm isopropyl-β-d-thiogalactopyranoside (Euromedex), and cells were further grown overnight at 25 °C. Cells were collected by low speed centrifugation, resuspended in buffer A (10 mm Tris, pH 8.0; 400 mm NaCl), and lysed by sonication. The soluble fraction recovered by high speed centrifugation was mixed with 1 ml of Talon resin (Clontech). After a 1-h incubation, the supernatant was removed, and the resin was washed extensively with buffer A. The resin was then resuspended in 2 ml of buffer A, and bovine thrombin (Sigma) was added overnight at 4 °C for cleaving off the histidine tag. The supernatant was recovered and applied onto a gel filtration column HiLoad 16/60 Superdex 75 (Amersham Biosciences) equilibrated with buffer A. The purified proteins were concentrated on Microsep 10K Omega (Pall Filtron) to a final concentration of around 5 mg/ml as assayed with the Bio-Rad protein assay (Bio-Rad).
Crystallization
The A. locustae TAF6C domain (residues 164–355) was crystallized by mixing 2 μl of protein solution with an equal volume of the reservoir solution containing 0.1 m CHES (Sigma), pH 9.5, and 10% (w/v) PEG3350 (Fluka). Crystals grew within a few days to reach an approximate size of 0.15 × 0.15 × 0.05 mm3. For data collection, crystals were briefly transferred in a cryoprotectant solution of 0.1 m CHES, pH 9.5, 10% (w/v) PEG3350, and 25% glycerol (Fluka) and quickly frozen in liquid ethane.
Data Collection and Structure Determination
Initial in-house data collection revealed that A. locustae TAF6C crystals belong to space groups P21. To solve the phase problem, crystals were grown with selenomethionylated protein. Due to the relatively rapid decay of the crystals upon x-ray exposure at synchrotron sources, only single wavelength anomalous diffraction experiments could be performed on these crystals at the selenium peak wavelength. Data collection on single native and selenomethionylated crystals was carried out on beamline ID14H4 at the European Synchrotron Radiation Facility. Data at 1.9 Å resolution for native and 2.4 Å resolution for selenomethionylated crystals were obtained (see Table 1). All data were processed and scaled using HKL2000 (29). Location of selenium atoms was done using Shake and Bake (30), their positions were refined within the phasing program SHARP (31), and the phases were further improved with the solvent flattening program SOLOMON (32).
TABLE 1.
Crystallographic statistics
| Native | Se-Met peak | |
|---|---|---|
| Data collection statistics | ||
| Wavelength (Å) | 0.9793 | 0.9793 |
| Space group | P21 | P21 |
| Cell constants | ||
| a (Å) | 33.79 | 33.76 |
| b (Å) | 50.69 | 50.67 |
| c (Å) | 67.29 | 67.42 |
| β (°) | 100.81 | 100.64 |
| Resolution (Å) | 50-1.90 | 50-2.38 |
| Reflections (measured/unique) | 113119/17911 | 83645/17423 |
| Redundancy (overall/last shell) | 3.7/3.3 | 1.9/1.6 |
| Completeness (%) (overall/last shell) | 99.5/99.6 | 98.5/89.1 |
| Rsym (%) (overall/last shell) | 4.6/34.1 | 3.4/7.4 |
| I/σ(I) (overall/last shell) | 30.6/3.8 | 22.4/10.1 |
| Refinement statistics | ||
| Resolution (Å) | 18.5-1.90 | |
| Number of protein atoms | 1577 | |
| Number of small molecule and water molecules atoms | 211 | |
| Number of reflections (F > 0) (all/test set) | 17887/915 | |
| R-factor (%) | 16.9 | |
| Rfree (%) | 21.5 | |
| Deviations from ideal geometry | ||
| Bonds (Å) | 0.01 | |
| Angles (°) | 0.95 | |
| Mean temperature factors (Å2) | 30.2 | |
Model Building and Refinement
Eight α-helices could be built in the initial single wavelength anomalous diffraction experimental electron density map at 2.4 Å resolution. Unfortunately, this information was not sufficient to improve the quality of the map. We therefore used this initial model and the native 1.9 Å resolution data for phase improvement within ARP/wARP (33). The resulting map was of sufficient quality to build the rest of the model. This model was further refined by cycles of manual building within TURBO-FRODO and Coot (34) and refinement with REFMAC (35) and BUSTER (36). The structure shows good deviations from ideal geometry (see Table 1), with no Ramachandran outliers.
The TAF6C domains from A. locustae, yeast, and human show almost identical values for pairwise identity (around 28%) and similarity (around 47%). Based on these numbers, we could confidently make models of yeast and human TAF6C domains by using MODELLER (37). As expected, major differences were located within the loop regions and in the last HEAT repeat, which shows lower sequence similarity. We used this information subsequently for the design of our mutants and chose residues for mutations that were located in regions where the confidence on model quality was high. The models are available upon request.
Purification of the Baculovirus-expressed Human TAF6-TAF9 and TAF5-TAF6-TAF9 Complexes
All hTAF6 mutants were constructed using nested PCR protocols, and the cDNAs were inserted in the pVL1392 vector to encode the full-length human TAF6 with a FLAG tag at the N-terminal end of all the mutant proteins. These vectors were used to produce baculoviruses using standard procedures.
Sf9 cells were simultaneously infected with the different baculoviruses that express His-hTAF5, FLAG-hTAF6, or FLAG-hTAF6 mutants, and hTAF9, respectively. All wild type constructs used to generate the baculoviruses were described previously (38).
Cells were lysed 48 h after infection in a buffer containing protease inhibitors, 400 mm KCl, 20 mm Tris-HCl (pH 7.9), 20% glycerol, and 2 mm DTT by three cycles of freeze-thawing. Cells were grown in a 75-cm2 Falcon flask, resuspended in 400 μl of buffer, lysed, and centrifuged, and the soluble proteins were used for immunoprecipitation.
Immunoprecipitation, Western Blot, and Coomassie Blue Staining Analyses of TAF6-TAF9 and TAF5-TAF6-TAF9 Complexes
First, soluble Sf9 cell extracts were diluted to 100 mm KCl with immunoprecipitation buffer (25 mm Tris-Cl, pH 7.9, 10% (v/v) glycerol, 0.1% Nonidet P-40, 0.5 mm dithiothreitol, 5 mm MgCl2). Next TAF6-containing complexes were isolated by immunoprecipitation with a mouse monoclonal M2 antibody (ANTI-FLAG® M2 affinity gel-purified immunoglobulin beads, Sigma-Aldrich, product number A2220). Immunoprecipitation was carried out by an overnight incubation of 250 μl of diluted protein extract with 100 μl of affinity-purified ANTI-FLAG® M2 antibody-coupled beads. Beads with M2 antibody-bound protein complexes were washed three times with immunoprecipitation buffer containing 500 mm KCl and twice with immunoprecipitation buffer containing 100 mm KCl. Immunoprecipitated proteins were eluted by an excess (2 mg/ml) of the corresponding FLAG epitope peptide and then boiled in sodium dodecyl sulfate (SDS) sample buffer and separated by SDS-polyacrylamide gel electrophoresis (PAGE).
Proteins were either visualized by staining the gels with Coomassie Blue or transferred to nitrocellulose membrane and probed with the indicated primary antibodies. Chemiluminescence detection was performed according to the manufacturer's instructions (Amersham Biosciences). The rabbit polyclonal antibody (pAb 2282) raised against hTAF9 and the monoclonal anti-hTAF6 (25TA 2G7) and anti-hTAF5 (1TA 1C2) antibodies were described previously (23, 39).
In Vivo Expression in HeLa Cells
For cell transfection assays, the wild type and mutant TAF6 proteins have been cloned into the pCDNA3.1 vector using the NheI/NotI restriction sites of this vector. The baculovirus vectors harboring these different proteins have been used as templates for the PCR reactions that were also used to FLAG tag the TAF6 proteins C-terminally. HeLa cells (5 × 106) were transfected in 90-mm dishes with 7 μg of the different expression vectors by using JetPEI (PolyPlus Transfection). Human HeLa cells were lysed 48 h after transfection in 100 μl of buffer/dish containing protease inhibitors, 400 mm KCl, 20 mm Tris-HCl (pH 7.9), 20% glycerol, and 2 mm DTT by three cycles of freeze-thawing. Extracts were then centrifuged, and the soluble proteins were used for immunoprecipitation (see above).
RESULTS
Domain Organization of TAF6
The homology of the TAF6 histone fold with histone H4 has rapidly retained most of the focus on this small N-terminal domain (17, 18, 40–42). However, the C-terminal region of TAF6 is also well conserved throughout evolution, with the exception of its very C-terminus (Fig. 1A). Specifically, the conserved C-terminal region of TAF6 is more than four times bigger than the histone fold motif and starts immediately after the histone fold, suggesting that these two regions may be functionally linked.
FIGURE 1.
Domain organization of TAF6 and sequence alignment of the TAF6C domain. A, schematic representation of the putative domain organization of TAF6 from E. cuniculi, A. locustae, S. cerevisiae, and Homo sapiens with their corresponding amino acid numbering. The colored boxes indicate the evolutionary conserved regions. HFD, histone fold domain; TAF6M, middle domain; TAF6C, C-terminal domain. B, multiple sequence alignment of the evolutionary conserved TAF6C domain from different species, as indicated. Human TAF6L was also included in the alignment. Different levels of gray shading indicate distinct levels of conservation. The numbering above and in the middle of the alignment corresponds to the TAF6 proteins from A. locustae and H. sapiens, respectively. The numbers at the end of each row correspond to the sequence displayed on the row. The red cylinders depict the α-helices observed in the structure of A. locustae TAF6C. + symbols above the sequences mark residues that are participating in the A. locustae TAF6C evolutionary conserved positive electrostatic patch. Symbols underneath the sequences indicate the residues mutated: m1 (*), m2 (YY), m3 (&), m4 (#), m5 (§), m6 (@), m7 (P), and m8 (A). Symbols that are circled indicate the residues that have been used to creates single point mutants m9 to m16 (see also Tables 2 and 3). The alignment was created with Aline (53). S. pombe, Schizosaccharomyces pombe.
To understand the functional role of the TAF6 C-terminal region, we have started its biochemical and structural characterization. The proteins from four organisms were used: human, Saccharomyces cerevisiae (thereafter termed yeast), and the two eukaryotic intracellular parasites Encephalitozoon cuniculi and A. locustae. We use these two latter organisms as model organisms for structural studies because their proteins are generally shorter than their higher eukaryotic orthologues but still retain the same domain organization (43–46). Specifically, the C-terminal region of the TAF6 proteins from these two organisms ends exactly where the C-terminal conservation of TAF6 finishes (Fig. 1A).
Despite numerous trials, initial crystallization attempts of the TAF6 conserved C-terminal region from the four organisms studied were unsuccessful. Sequence analysis suggested that this region can be divided into two domains. The first domain (TAF6 middle domain; TAF6M) encompasses about 70 residues immediately after the histone fold motif. TAF6M is separated from the second domain (TAF6 C-terminal domain; TAF6C) by a poorly conserved stretch of 20–50 residues. The TAF6C domain forms the larger part of the TAF6 conserved C-terminal region, spanning about 220 residues (Fig. 1, A and B).
Secondary structure prediction suggests that both TAF6M and TAF6C domains have different secondary structure content. Specifically, predictions do not clearly indicate the kind of secondary structure elements present in TAF6M, with propensities for α-helices, β-strands, and coil being relatively equal over the whole domain. In contrast, the TAF6C domain is clearly predicted as purely α-helical. We reasoned that the putatively poorly folded TAF6M domain could have prevented the crystallization of the full C-terminal region of TAF6. We therefore expressed, purified, and submitted to crystallization trials the TAF6C domains of the human, yeast, E. cuniculi, and A. locustae TAF6 proteins.
The Conserved TAF6C Domain Is Formed of Five HEAT Repeats
Plate-shaped crystals were obtained with the A. locustae construct. Despite their small thickness, some of the crystals showed diffraction better than 2 Å resolution at synchrotron sources, and a complete data set at 1.9 Å resolution could be collected from a single crystal with good data collection statistics (Table 1). Structure determination was carried out by collecting single wavelength anomalous diffraction data at 2.4 Å resolution on crystals grown with selenomethionine-substituted protein. After improvement of the initial single wavelength anomalous diffraction map by solvent flattening, a few helices could be located in the electron density but were not sufficient for improving the quality of the map. Therefore, these helices were positioned by rigid body in the 1.9 Å native map, and the electron density was improved using ARP/wARP (33).
The resulting map was of sufficient quality to build the rest of the protein. The final model includes all but the last C-terminal residue and 178 water molecules and has an R-factor and an Rfree of 16.9 and 22.3%, respectively, with good deviations from the ideal geometry (Table 1). Inspection of the electron density also revealed the presence of two CHES molecules at the interface of symmetry-related molecules, explaining the absolute requirement for CHES in the crystallization condition.
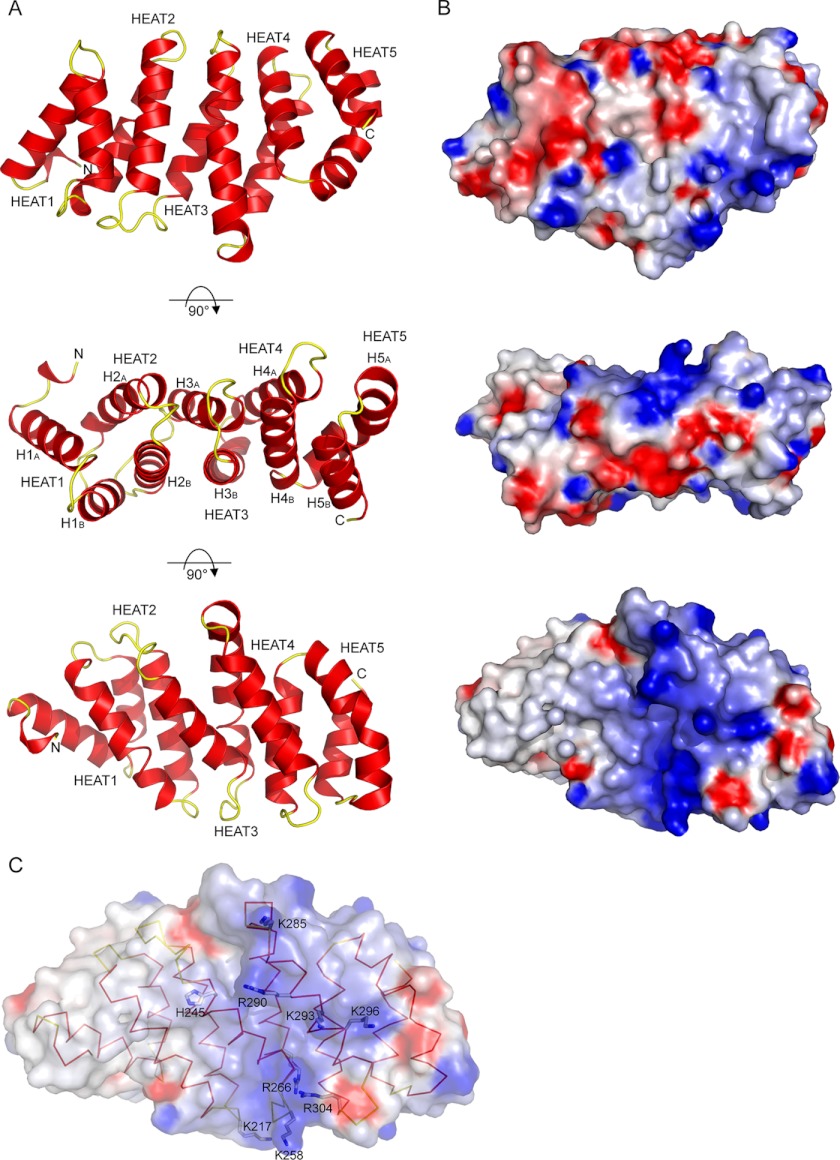
Analysis of the final model revealed the presence of five HEAT repeats tightly packed against each other, defining a single structural domain (Fig. 2A). HEAT repeats are formed of a single helical hairpin formed by two α-helices (αA and αB). HEAT repeats are known protein/protein interaction motifs that are found repeated several times in many proteins (47). Specifically, when found in large numbers in proteins, these repeats adopt a superhelix conformation. This is clearly not the case in the TAF6C domain due to the small number of repeats, which is not sufficient to adopt a helical conformation. In fact, this domain resembles more a paving stone with six faces (Fig. 2A).
FIGURE 2.
Structure of TAF6C domain. A, ribbon representations of the A. locustae TAF6C domain using three different views. α-Helices are colored red, and the loops connecting them are colored yellow. The five HEAT repeats composing the domain are labeled (HEAT1 to HEAT5). The different helices composing these HEAT repeats are labeled in the middle view according to the nomenclature used in Fig. 1B. B, electrostatic potential at the surface of the A. locustae TAF6C domain. The electrostatic potentials −7 and +7 kBT (kB, Boltzmann constant; T, temperature) are colored red and blue, respectively. The orientation of the protein in each view corresponds to the one showed in panel A. A clear positive electrostatic patch is observed in the surface represented in the lower view. C, detail of the residues participating in the positive electrostatic patch (A. locustae numbering). For clarity, the orientation of the structure is slightly tilted from the one used in the views of the lower panels in A and B. The figure was made by using GRASP (54) and PyMOL (55).
Because HEAT repeats are generally found repeated many times in proteins harboring this kind of motif, the question remains whether more repeats are present in TAF6. As already mentioned, the TAF6M domain is not predicted to have a high helical content and is separated from the TAF6C by a nonconserved region of variable length, thus suggesting that it adopts a different fold. Inspection of the conformation of the N-terminal residues in our TAF6C structure confirms that their packing is incompatible with the extension of the HEAT repeats at the N terminus of TAF6C. Although it cannot be excluded that the truncation of the protein leads to a different conformation of these N-terminal residues, the poor helical propensity and the poor conservation of the residues preceding the TAF6C domain argue against an extension of the HEAT repeats at the N terminus of the TAF6C.
The A. locustae TAF6 protein terminates exactly with the last HEAT repeat, which corresponds exactly to the position where the conservation of the TAF6 finishes (Figs. 1 and 2A). Based on the A. locustae structure and our multiple sequence alignment, we modeled the conserved yeast and human TAF6C domains, which revealed that both proteins can adopt the same fold without any steric clashes. Nonetheless, these latter two proteins are longer than the A. locustae and E. cuniculi proteins (about 50 and 250 additional residues for the yeast and human TAF6, respectively). Secondary structure predictions suggest the presence of helices in the remaining C-terminal region of yeast and human TAF6, notably in the regions immediately following the conserved TAF6C domain. Thus, it cannot be excluded that the yeast and human TAF6 proteins encode a small number of additional C-terminal HEAT repeats, but the length of the TAF6 protein precludes that a large number of these repeats are present in the TAF6 protein, as observed in other proteins.
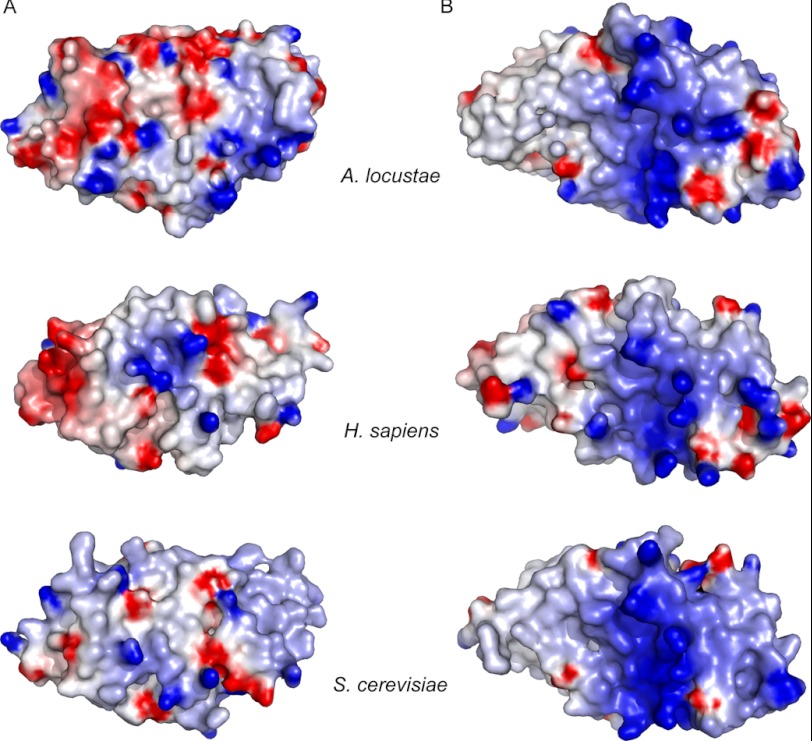
Analysis of the electrostatic potential at the surface of the TAF6C domain reveals a rather large positively charged patch on one face of the TAF6C domain that could be indicative of a specific electrostatic interaction surface (Fig. 2, B and C). Interestingly, analysis of the electrostatic potential at the surface of the modeled human and yeast TAF6C domains reveals that only this positive patch is conserved between these three domains (Fig. 3), suggesting that it is important for TFIID assembly and/or function.
FIGURE 3.
Electrostatic potential at the surface of A. locustae, H. sapiens, and S. cerevisiae TAF6C domains. The electrostatic potentials −7 and +7 kBT (kB, Boltzmann constant; T, temperature) are colored red and blue, respectively. The structures used for the calculations are those of A. locustae TAF6C crystallographic structure and the modeled structures of H. sapiens and S. cerevisiae TAF6C domains built by homology using the A. locustae structure. The two orientations showed correspond to the upper and lower orientations of A. locustae TAF6C displayed in Fig. 2B. A clear conserved positive patch is observed in the three proteins in panel B. In contrast, the other side of the protein (panel A) does not seem to have conserved electrostatic features.
The Conserved C-terminal Domain of Human TAF6 Is Important for Interactions with TAF9 and TAF5
We next sought to understand the role of the TAF6 HEAT repeats, considering their strong evolutionary conservation. From (i) the structure of the A. locustae TAF6C, (ii) the established models of the yeast and human TAF6C, and (iii) the multiple sequence alignment, we listed a certain number of exposed conserved or semiconserved residues that could play a role by acting in protein/protein interactions. Although many of these residues were located in the vicinity of the positive electrostatic patch, others were located on other faces of the TAF6C, suggesting that the TAF6C could mediate multiple interactions.
We decided to mutate these residues facewise (i.e. each face of the TAF6C paving stone being mutated independently from the others) in the human protein to investigate the role of each face of the TAF6C domain. Due to the presence of several conserved residues on all faces, multiple residues were initially mutated together. Another mutant was made according to a known mutant from the literature (48). Finally, two single mutants were made that involved half-buried and perfectly conserved residues. All mutants are listed in Table 2, and their locations are shown in Fig. 1B. To test the effect of these mutants, we used the TAF6-TAF9 complex, which is supposed to assemble through the histone fold motif of these two proteins, as well as the TAF5-TAF6-TAF9 complex, which appears to be sufficient to form the three-lobed structure of TFIID and is important for human TFIID assembly (22).
TABLE 2.
Facewise mutations in full-length human TAF6
| Mutant | Mutation(s) | Symbol (Fig. 1B) | Based on |
|---|---|---|---|
| m1 | H214A/E215A/S217A/E219A/Q220A/Q221A/L222A/Y223A/K225/E226A/E229A/D249A/Q254A/R258A | * | Conservation and surface exposure |
| m2 | Insertion of two tyrosines between Thr-228 and Glu-229 | YY | Phenotype in Caenorhabditis elegans |
| m3 | R284A/K287A/D291A/D331A/R335A/Q339A | & | Surface exposure |
| m4 | E298A/K299A/Y300A/H302A/E303A | # | Conservation and surface exposure |
| m5 | P306A/T310A/V313A/S314A/R315A | § | Conservation and surface exposure |
| m6 | R315A/Q316A/K357A/T358A/K361A | @ | Conservation and surface exposure |
| m7 | R330P | P | Conservation |
| m8 | Y374A | A | Conservation |
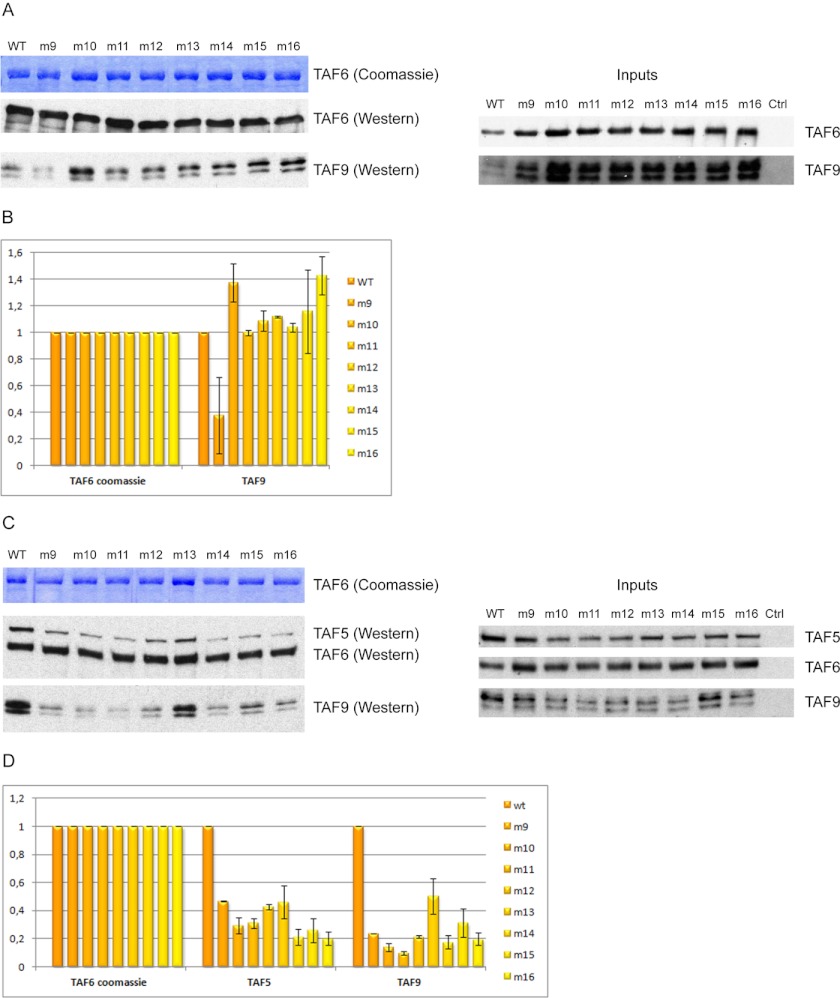
The human TAF6-TAF9 and TAF5-TAF6-TAF9 complexes, containing either wild type or mutated TAF6, were reconstituted by co-expressing all full-length proteins in insect cells using the baculovirus expression system. All TAF6 proteins (wild type and mutated) contained a FLAG tag for affinity purification. All the TAF6 mutants were expressed and soluble, showing that the mutations do not affect solubility of TAF6 (Fig. 4A). After affinity purification, the complexes were analyzed by SDS-PAGE and Coomassie Blue staining. From this initial analysis, the exact amounts of sample to be loaded on an SDS gel were estimated to keep the quantity of the different TAF6 proteins constant. This second gel was then used for Western blotting TAF9 and, if necessary, TAF5, thus enabling quantification of the relative abundance of each subunit in the complex.
FIGURE 4.
Effect of mutants with multiple amino acid changes in H. sapiens TAF6 on TAF6-TAF9 and TAF5-TAF6-TAF9 complex formation. A, Coomassie Blue staining of TAF6 and Western blotting of TAF6 and TAF9 from baculovirus-expressed and immunopurified FLAG-TAF6-TAF9 human complexes, isolated upon FLAG tag purification. The different lanes correspond to complexes incorporating full-length human TAF6, either wild type (WT) or with various mutations (m1 to m8). Each mutant harbors multiple amino acid changes (except for m7 and m8) described in Table 2. Inputs are displayed on the right side of the panel. B, graph displaying the amount of TAF9 (assessed upon quantification of the TAF9 Western blotting) bound to TAF6 upon complex formation in the presence of the WT or different m1–m8 mutants of TAF6. C, same as in panel A in the case of baculovirus-expressed and immunoprecipitated TAF5/FLAG-TAF6-TAF9 complexes isolated upon FLAG tag purification. Inputs are displayed on the right side of the panel. D, graph displaying the different amounts of TAF5 and TAF9 (assessed upon quantification of the TAF5 and TAF9 Western blots) bound to TAF6 upon complex formation in the presence of the WT or different m1–m8 mutants of TAF6. Error bars in panels B and D represent ± S.D.
None of the mutants showed a complete loss of interaction with TAF9 or TAF5-TAF9. Surprisingly, however, in the case of the TAF6-TAF9 complex, all mutants showed a weakening of the interaction because lower amounts of TAF9 were co-immunoprecipitated with the TAF6 mutants than with the wild type protein (Fig. 4, A and B). Specifically, the two mutants with single point mutations (mutants m7 and m8) showed the larger decrease of interaction with TAF9. The residues mutated in these two mutants are highly conserved residues in TAF6C and, contrary to most of the residues mutated in the other mutants, these residues are half-buried within the domain. It might therefore well be that these residues affect locally the structure of the TAF6C and cause a stronger effect on the complex than what would be expected by weakening direct interactions between the two partners. Nonetheless, altogether these results suggested that the TAF6C domain also participates in the overall interaction between TAF6 and TAF9.
We then analyzed the effect of the same mutants in the context of the TAF5-TAF6-TAF9 complex. Once again, none of the mutants caused a complete dissociation of the complex. However, all mutants were responsible for a weakening of the interaction because lower amounts of both TAF5 and TAF9 were co-immunoprecipitated with TAF6 mutants when compared with the wild type TAF6 (Fig. 4, C and D). Importantly, some of the mutants appeared to have different effects when introduced in the context of either the TAF6-TAF9 complex or the TAF5-TAF6-TAF9 complex. Firstly, mutant m1, which already reduced by more than 2-fold the quantity of TAF9 bound to TAF6 in the context of the dimeric complex, appeared to have an even stronger effect on TAF9 binding in the trimeric complex. Interestingly, this mutation had less effect on TAF5 binding, suggesting that TAF5 interaction with the TAF6-TAF9 complex may somehow weaken the interaction between these two HFTs, thus leading to a stronger loss of TAF9 (Fig. 4).
In contrast, the effect of the mutants m7 and m8, which were the most deleterious for the TAF6-TAF9 complex, was much milder for the ternary complex. As already mentioned, these two mutants could have destabilized, at least partially, the structure of the TAF6C, and thus have an indirect effect on TAF9 binding. In the context of the ternary complex, binding of TAF5 to TAF6 could stabilize TAF6C and thus restore binding of TAF9 to TAF6. In agreement, not only TAF9 binding, but also TAF5 binding to TAF6, was less affected.
The remaining mutants (m2 to m6) affected more strongly TAF5 binding, with a loss of about 60% of the bound protein (Fig. 4, C and D). Interestingly, these mutants can be split into two classes when considering their effect on TAF9 binding in the context of the TAF5-TAF6-TAF9 ternary complex. First, mutants m2 and m3 showed similar amounts of TAF9 binding as in the context of the dimeric complex. This suggests that the effect of these mutants on TAF5 and TAF9 binding is not coupled, i.e. that the mutants affect nonoverlapping binding surfaces for TAF5 and TAF9. Contrasting with mutants m2 and m3, mutants m4, m5, and m6 show a decrease of TAF9 binding when compared with their effect on the TAF6-TAF9 complex. This effect appears, however, not as strong as for mutant m1 but, once again, speaks in favor of a destabilization of the TAF6-TAF9 complex upon binding of TAF5, suggesting structural rearrangements occurring during TFIID assembly. Of note, mutants m4, m5, and m6 are located on the surface of TAF6C that has a strong positive charge.
Binding of Human TAF5 Modulates the Interaction between Human TAF6 and TAF9
The results obtained by these initial mutational studies apparently imply an unexpected role of TAF6C in TAF9 binding, which appears further modulated by TAF5 binding. Most of the mutants considered, however, were composed of a large number of mutations, suggesting that these strong changes could have, in fact, an indirect effect on dimeric and trimeric complex formation (e.g. due to a large change of the overall electrostatic charge of TAF6). To address this issue, we decided to create mutants with single point mutations that should have less of an effect on the physicochemical properties of the TAF6 protein. The residues to be mutated were chosen within the initial m1 and m5 mutants that showed strong, albeit different effects on TAF5 and TAF9 binding. The choice was also made considering the position of the residues to be mutated, looking for fully exposed residues, but also for residues that were not conserved between the TAF6 and TAF6L families. To increase the chances that these single mutations would still show an effect on complex formation, we decided to introduce larger changes than only alanines, either with bulkier side chains or, when applicable, residues observed in the human TAF6L protein (mutants are shown in Table 3, and their locations are shown in Fig. 1B).
TABLE 3.
Single point mutations in full-length human TAF6
| Mutant | Mutation | Based on |
|---|---|---|
| m9 | R315A | Loss of charge |
| m10 | S314E | TAF6 → TAF6L |
| m11 | T310K | Bulkier |
| m12 | T310Y | TAF6 → TAF6L |
| m13 | P306K | Bulkier |
| m14 | L222K | TAF6 → TAF6L |
| m15 | E219R | Inversion of charge |
| m16 | V313K | Bulkier |
In contrast to the results with the first set of mutants, analysis of the effect of these new single point mutants on the formation of the TAF6-TAF9 complex revealed that the mutations introduced hardly have any influence (Fig. 5, A and B). Mutant m9 (R315A) appears the only exception, causing a decrease of TAF9 binding, suggesting an important role for this residue in the formation of the TAF6-TAF9 complex. In strong contrast, introduction of these new single point mutants in the context of the TAF5-TAF6-TAF9 complex shows drastic effects on complex formation, affecting both TAF5 and TAF9 binding (Fig. 5, C and D). These results reinforce the idea that TAF5 binding to the TAF6-TAF9 complex causes destabilization of this latter complex and also suggest that exposed TAF6L residues could serve to discriminate between TAF5 and TAF5L in metazoans.
FIGURE 5.
Effect of mutants with single amino acid changes in H. sapiens TAF6 on TAF6-TAF9 and TAF5-TAF6-TAF9 complex formation. A, Coomassie Blue staining of TAF6 and Western blotting of TAF6 and TAF9 from baculovirus-expressed and immunopurified human TAF6-TAF9 human complexes. The different lanes correspond to complexes incorporating full-length human TAF6 either with WT or with single mutations (m9 to m16). Each mutant is described in Table 3. Inputs are displayed on the right side of the panel. B, graph displaying the amount of TAF9 (assessed upon quantification of the TAF9 Western blotting) bound to TAF6 upon complex formation in the presence of the WT or different m9–m16 mutants of TAF6. C, same as in panel A in the case of baculovirus-expressed TAF5-TAF6-TAF9 complexes isolated upon FLAG tag purification of TAF6. Inputs are displayed on the right side of the panel. D, graph displaying different amounts of TAF5 and TAF9 (assessed upon quantification of the TAF5 and TAF9 Western blots) bound to TAF6 upon complex formation in the presence of the WT or different m9–m16 mutants of TAF6. Error bars in panels B and D represent ± S.D.
In Vivo Importance of the TAF6C Domain
We next investigated whether the TAF6 mutants would incorporate in human TFIID complex. For this purpose, vectors expressing FLAG-tagged human TAF6, either wild type or mutant forms, were transfected in HeLa cells. 48 h after transfection, whole cell extracts (WCEs) were prepared, and anti-FLAG purifications were carried out to test how the exogenously expressed TAF6 proteins incorporate in TFIID complexes. Only mutant m1, which harbored many mutations, was not considered for this study.
Surprisingly, when analyzing the overexpression levels of the transfected FLAG-tagged mutants in WCEs in comparison with endogenous TBP levels, we reproducibly observed lower amounts of several mutants when compared with the overexpressed wild type TAF6 (Fig. 6, A and B). This suggests that the mutations might cause partial misincorporation of TAF6 in endogenous TFIID and would thus lead to its partial degradation.
FIGURE 6.
Multiple and single amino acid mutations in the H. sapiens TAF6C domain affect TAF6 stability and endogenous TFIID assembly in HeLa cells. A, eukaryotic expression vectors expressing the indicated FLAG-tagged TAF6 wild type and different mutant proteins (see also Tables 2 and 3) were transfected into HeLa cells. 48 h following transfection, cells were collected, and WCEs were made. WCEs were normalized by Western blot analysis for equal TBP amounts (lower panel), and then the expression of wild type and mutant FLAG-tagged TAF6 proteins was analyzed by Western blot using an anti-FLAG antibody. Ctrl, control. B, several representative blots were scanned, and the quantities of FLAG-tagged wild type and mutant TAF6 proteins are represented in the graphs. The y axis shows arbitrary units. Error bars represent ± S.D. C, FLAG-TAF6-containing TFIID complexes were purified from HeLa WCEs by an anti-FLAG immunoprecipitation and analyzed by Western blotting for the presence of various subunits (as indicated). Wild type FLAG-TAF6 and the tested mutants are able to incorporate in TFIID complexes. D, model summarizing the influence of the TAF6C domain and the TAF5 protein on TAF6-TAF9 assembly. The model has been established with the structure of the Drosophila TAF6-TAF9 histone-like pair (Protein Data Bank (PDB) 1TAF) and the A. locustae TAF6C structure (this study; PDB 4ATG). HFD, histone fold domain.
Nevertheless, we further analyzed some mutants by carrying out FLAG purification to test their capability to incorporate in endogenous TFIID complexes. Three mutants were chosen according to their increasing stability in HeLa cells: m3, m6, and m9. Analysis by Western blotting for the presence of other TFIID subunits in these anti-FLAG immunoprecipitations did not reveal a total loss or strong diminution of these subunits, with a possible small effect on TAF5 for mutants m6 and m9 (Fig. 6C), suggesting that stabilization of the TAF6 mutants occurs upon complex formation. Taken together, these observations suggest that the observed diminution of the quantities of each mutant is most likely due to the difficulties encountered by the mutants to be incorporated into endogenous TFIID, leading to their partial degradation, a phenomenon already observed upon knockdown of different TFIID subunits in Drosophila (26).
Due to the complexity of TFIID assembly, it is difficult to directly compare these in vivo results with the ones obtained on the reconstitution of the TAF6-TAF9 and TAF5-TAF6-TAF9 subcomplexes by overexpression in insect cells. However, one important aspect concerns the results obtained with TAF6 single point mutants. We showed that these mutants had little effect on TAF6-TAF9 complex formation but, unexpectedly, had strong effects on TAF5-TAF6-TAF9 complex formation (Figs. 4 and 5). In the in vivo context, these mutants also appear to show an effect, albeit less pronounced than the mutants with several mutations. Thus, results obtained on the TAF6-TAF9 and TAF5-TAF6-TAF9 subcomplexes appear to be confirmed in vivo (Fig. 6D).
DISCUSSION
Numerous biochemical and structural studies have shown that the histone fold motif plays a major role in TFIID and SAGA assembly, enabling the formation of five distinct histone-like pairs (9, 11–17, 20). Interestingly, almost all HFTs harbor various additional regions that most likely convey structural and/or functional roles. For instance, the metazoan-specific TAF3 PHD domain and TAF4 homology domain (TAFH) have been shown to recognize epigenetic marks and activators (6, 7, 49–51). Other additional regions of HFTs have been conserved throughout eukaryotic evolution and are more likely to be involved in broader structural and functional roles of TFIID. These roles, however, remain to be determined.
We provide here results on the structure and the role of one of these conserved additional regions in the C-terminal region of TAF6. This region, which immediately follows the histone fold motif of TAF6, can be divided in two domains: a small middle domain (TAF6M), which is separated from a larger C-terminal domain (TAF6C) by a loop varying in sequence and in length. The structure of the TAF6C domain from A. locustae reveals that this domain is composed of five HEAT repeats that appear conserved from yeast to human. HEAT repeats (and the related ARM repeat) are known protein/protein interaction motifs that are repeated many times in a wide variety of proteins, forming superhelices that interact extensively with their target proteins (47). These motifs are also used by other transcriptional effectors to bind their targets, as shown recently for the Iws1 and Mot1 proteins (44, 52).
Importantly, our mutational analyses suggest that the TAF6C domain modulates the TAF6/TAF9 interaction. This modulation provides a more dynamic view of HFT assembly. Clearly, the TAF6C domain is not the driving force of the interaction between TAF6 and TAF9. Indeed, the human TAF6δ isoform is not able to interact with TAF9, although it still harbors the TAF6C domain (27). In addition, our mutations are only weakening the interaction between these two TAFs. The molecular basis for this modulation is not clear. One possibility could be that the TAF6C domain interacts directly with the histone-like pair. Another possibility could be that TAF6C/TAF6C interactions stabilize the TAF6-TAF9 complex. It cannot be excluded that a combination of these two possibilities is at work for modulating the TAF6/TAF9 interaction.
Moreover, the addition of TAF5 to the TAF6-TAF9 complex appears to enhance the modulator effect of TAF6C. Indeed, single point mutants of TAF6C exert an effect on the formation of the TAF6-TAF9 complex only when TAF5 is present. This result highlights once again the importance of TAF5 for TFIID assembly. Specifically, TAF5 is essential for the formation of a larger TFIID subcomplex containing most HFTs and contributes to the three-lobed architecture of TFIID (22). In addition, TAF5 is the only non-HFT subunit of TFIID shared with yeast SAGA. Interestingly, single point mutants, where residues of human TAF6 (TFIID-specific) are changed into residues present in human TAF6L (SAGA-specific), also show strong effects upon TAF5 binding (mutants m10, m12, and m14). Considering that in humans, TFIID-specific TAF5 is replaced by SAGA-specific TAF5L, our results suggest that specific residues of TAF5L most likely accommodate the changes observed between TAF6 and TAF6L. Thus, human SAGA, in contrast to yeast SAGA, seems to have evolved specific TAFs for dedicated functions.
Importantly, overexpression of the various mutants, but not of wild type TAF6, in HeLa cells appears to affect TAF6 stability, suggesting that these mutants are poorly incorporated into TFIID and partially degraded, as already observed upon knockdown of specific TAFs in Drosophila (26). Interestingly, even the single point mutants of TAF6 are affected, thus paralleling and corroborating the results obtained with these mutants upon reconstitution of the TAF5-TAF6-TAF9 complex.
The experiments with the Drosophila melanogaster TAFs also suggested that the TAF6 C-terminal region is not required for TFIID assembly (26). In these experiments, the Drosophila TAF6 was divided into an N-terminal part and a C-terminal part. The N-terminal part, which was shown to be necessary for TFIID assembly, is in fact composed of the histone fold motif, the TAF6M domain, the following loop, and the first HEAT repeat of the TAF6C domain. Therefore, it is difficult to delineate a clear picture from these experiments. Notably, it remains unclear which part of TAF6 is really required for TFIID assembly. Specifically, our mutants located in the first HEAT repeat (m1, m2, m14, and m15) show strong effects on the stability of the TAF5-TAF6-TAF9 complex. Because the human TAF6δ isoform is incorporated within TFIID, but not TAF9 (27), this suggests in fact that the TAF6M domain and the first HEAT repeat of the TAF6C domain are the most important regions of TAF6 for its incorporation within TFIID. In agreement, knockdown of Drosophila TAF9 does not lead to any degradation of TAF6, whereas knockdown of TAF1, TAF2, TAF4, or TAF5 causes almost complete disappearance of Drosophila TAF6 in WCEs (26).
Although the remaining HEAT repeats of TAF6C appear, from the Drosophila experiments, to be nonessential for TAF6 incorporation in TFIID, it cannot be excluded that these repeats also participate, albeit less strongly, in this incorporation. Our mutational data suggest such a role. On the other hand, because HEAT repeats are protein/protein interaction motifs, they may also be involved in other kinds of interactions. Specifically, these repeats could, in the context of the full TFIID, interact with TFIID protein partners and/or DNA. Because the middle HEAT repeats of TAF6C define the surface having a strongly conserved positive electrostatic charge, it is likely that this surface is engaged in such interactions. Because our mutational data suggest structural rearrangements occurring at the TAF6/TAF9 interface upon TAF5 binding, it is tempting to speculate that binding of other TAFs, transcriptional effectors, and/or DNA to TAF6C might also trigger conformational rearrangements (Fig. 6D).
In agreement, it is interesting to note that binding of TFIID to DNA upon recruitment by Rap1 and in the presence of TFIIA induces conformational changes within TFIID (8). Thus, conformational rearrangements between subunits, as suggested by our data in the case of the formation of the TAF5-TAF6-TAF9 subcomplex, could be essential. The possibility to change from one conformation to another upon recognition of a biological target could facilitate major rearrangements and be of paramount importance for TFIID and SAGA function.
Acknowledgments
We thank members of the European Synchrotron Radiation Facility-European Molecular Biology Laboratory (ESRF-EMBL) joint structural biology groups for the use of their beamline facilities and for help during data collection.
This work was supported by institutional funds from the CNRS, the INSERM, and the Université de Strasbourg. Further funding was provided by the European Commission SPINE2-Complexes project (Contract LSHG-CT-2006-031220) (to C. R. and D. M.) and by grants from the Agence Nationale de la Recherche (ANR) (ANR-09-BLAN-0052) and the European Union (EU) (EUTRACC, LSHG-CT-2007-037445) (to L. T.).
The atomic coordinates and structure factors (code 4ATG) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- TFII
- general transcription factor II
- TBP
- TATA box-binding protein
- TAF
- TBP-associated factor
- HFT
- histone fold-containing TAF
- CHES
- N-cyclohexyl-2-aminoethane-sulfonic acid
- WCE
- whole cell extract
- SAGA
- Spt-Ada-Gcn5 acetyltransferase.
REFERENCES
- 1. Roeder R. G. (1996) The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci. 21, 327–335 [PubMed] [Google Scholar]
- 2. Thomas M. C., Chiang C. M. (2006) The general transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol. 41, 105–178 [DOI] [PubMed] [Google Scholar]
- 3. Burley S. K., Roeder R. G. (1996) Biochemistry and structural biology of transcription factor IID (TFIID). Annu. Rev. Biochem. 65, 769–799 [DOI] [PubMed] [Google Scholar]
- 4. Cler E., Papai G., Schultz P., Davidson I. (2009) Recent advances in understanding the structure and function of general transcription factor TFIID. Cell. Mol. Life Sci. 66, 2123–2134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Juven-Gershon T., Hsu J. Y., Theisen J. W., Kadonaga J. T. (2008) The RNA polymerase II core promoter: the gateway to transcription. Curr. Opin. Cell Biol. 20, 253–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jacobson R. H., Ladurner A. G., King D. S., Tjian R. (2000) Structure and function of a human TAFII250 double bromodomain module. Science 288, 1422–1425 [DOI] [PubMed] [Google Scholar]
- 7. Vermeulen M., Mulder K. W., Denissov S., Pijnappel W. W., van Schaik F. M., Varier R. A., Baltissen M. P., Stunnenberg H. G., Mann M., Timmers H. T. (2007) Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 131, 58–69 [DOI] [PubMed] [Google Scholar]
- 8. Papai G., Tripathi M. K., Ruhlmann C., Layer J. H., Weil P. A., Schultz P. (2010) TFIIA and the transactivator Rap1 cooperate to commit TFIID for transcription initiation. Nature 465, 956–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Birck C., Poch O., Romier C., Ruff M., Mengus G., Lavigne A. C., Davidson I., Moras D. (1998) Human TAFII28 and TAFII18 interact through a histone fold encoded by atypical evolutionary conserved motifs also found in the SPT3 family. Cell 94, 239–249 [DOI] [PubMed] [Google Scholar]
- 10. Lavigne A. C., Gangloff Y. G., Carré L., Mengus G., Birck C., Poch O., Romier C., Moras D., Davidson I. (1999) Synergistic transcriptional activation by TATA-binding protein and hTAFII28 requires specific amino acids of the hTAFII28 histone fold. Mol. Cell. Biol. 19, 5050–5060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gangloff Y. G., Pointud J. C., Thuault S., Carré L., Romier C., Muratoglu S., Brand M., Tora L., Couderc J. L., Davidson I. (2001) The TFIID components human TAFII140 and Drosophila BIP2 (TAFII155) are novel metazoan homologues of yeast TAFII47 containing a histone fold and a PHD finger. Mol. Cell. Biol. 21, 5109–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gangloff Y. G., Romier C., Thuault S., Werten S., Davidson I. (2001) The histone fold is a key structural motif of transcription factor TFIID. Trends Biochem. Sci. 26, 250–257 [DOI] [PubMed] [Google Scholar]
- 13. Gangloff Y. G., Sanders S. L., Romier C., Kirschner D., Weil P. A., Tora L., Davidson I. (2001) Histone folds mediate selective heterodimerization of yeast TAFII25 with TFIID components yTAFII47 and yTAFII65 and with SAGA component ySPT7. Mol. Cell. Biol. 21, 1841–1853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gangloff Y. G., Werten S., Romier C., Carré L., Poch O., Moras D., Davidson I. (2000) The human TFIID components TAFII135 and TAFII20 and the yeast SAGA components ADA1 and TAFII68 heterodimerize to form histone-like pairs. Mol. Cell. Biol. 20, 340–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Werten S., Mitschler A., Romier C., Gangloff Y. G., Thuault S., Davidson I., Moras D. (2002) Crystal structure of a subcomplex of human transcription factor TFIID formed by TATA binding protein-associated factors hTAF4 (hTAFII135) and hTAF12 (hTAFII20). J. Biol. Chem. 277, 45502–45509 [DOI] [PubMed] [Google Scholar]
- 16. Thuault S., Gangloff Y. G., Kirchner J., Sanders S., Werten S., Romier C., Weil P. A., Davidson I. (2002) Functional analysis of the TFIID-specific yeast TAF4 (yTAFII48) reveals an unexpected organization of its histone-fold domain. J. Biol. Chem. 277, 45510–45517 [DOI] [PubMed] [Google Scholar]
- 17. Xie X., Kokubo T., Cohen S. L., Mirza U. A., Hoffmann A., Chait B. T., Roeder R. G., Nakatani Y., Burley S. K. (1996) Structural similarity between TAFs and the heterotetrameric core of the histone octamer. Nature 380, 316–322 [DOI] [PubMed] [Google Scholar]
- 18. Hoffmann A., Chiang C. M., Oelgeschläger T., Xie X., Burley S. K., Nakatani Y., Roeder R. G. (1996) A histone octamer-like structure within TFIID. Nature 380, 356–359 [DOI] [PubMed] [Google Scholar]
- 19. Selleck W., Howley R., Fang Q., Podolny V., Fried M. G., Buratowski S., Tan S. (2001) A histone fold TAF octamer within the yeast TFIID transcriptional coactivator. Nat. Struct. Biol. 8, 695–700 [DOI] [PubMed] [Google Scholar]
- 20. Leurent C., Sanders S., Ruhlmann C., Mallouh V., Weil P. A., Kirschner D. B., Tora L., Schultz P. (2002) Mapping histone fold TAFs within yeast TFIID. EMBO J. 21, 3424–3433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sanders S. L., Garbett K. A., Weil P. A. (2002) Molecular characterization of Saccharomyces cerevisiae TFIID. Mol. Cell. Biol. 22, 6000–6013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Leurent C., Sanders S. L., Demény M. A., Garbett K. A., Ruhlmann C., Weil P. A., Tora L., Schultz P. (2004) Mapping key functional sites within yeast TFIID. EMBO J. 23, 719–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Demény M. A., Soutoglou E., Nagy Z., Scheer E., Jànoshàzi A., Richardot M., Argentini M., Kessler P., Tora L. (2007) Identification of a small TAF complex and its role in the assembly of TAF-containing complexes. PLoS One 2, e316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Grant P. A., Schieltz D., Pray-Grant M. G., Steger D. J., Reese J. C., Yates J. R., 3rd, Workman J. L. (1998) A subset of TAFIIs are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell 94, 45–53 [DOI] [PubMed] [Google Scholar]
- 25. Nagy Z., Tora L. (2007) Distinct GCN5/PCAF-containing complexes function as co-activators and are involved in transcription factor and global histone acetylation. Oncogene 26, 5341–5357 [DOI] [PubMed] [Google Scholar]
- 26. Wright K. J., Marr M. T., 2nd, Tjian R. (2006) TAF4 nucleates a core subcomplex of TFIID and mediates activated transcription from a TATA-less promoter. Proc. Natl. Acad. Sci. U.S.A. 103, 12347–12352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bell B., Scheer E., Tora L. (2001) Identification of hTAFII80δ links apoptotic signaling pathways to transcription factor TFIID function. Mol. Cell 8, 591–600 [DOI] [PubMed] [Google Scholar]
- 28. Diebold M. L., Fribourg S., Koch M., Metzger T., Romier C. (2011) Deciphering correct strategies for multiprotein complex assembly by co-expression: application to complexes as large as the histone octamer. J. Struct. Biol. 175, 178–188 [DOI] [PubMed] [Google Scholar]
- 29. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Method Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 30. Weeks C. M., Miller R. (1999) The design and implementation of SnB v2.0. J. Appl. Crystallogr. 32, 120–124 [Google Scholar]
- 31. Vonrhein C., Blanc E., Roversi P., Bricogne G. (2007) Automated structure solution with autoSHARP. Methods Mol. Biol. 364, 215–230 [DOI] [PubMed] [Google Scholar]
- 32. Abrahams J. P., Leslie A. G. (1996) Methods used in the structure determination of bovine mitochondrial F1-ATPase. Acta Crystallogr. D Biol. Crystallogr. 52, 30–42 [DOI] [PubMed] [Google Scholar]
- 33. Langer G., Cohen S. X., Lamzin V. S., Perrakis A. (2008) Automated macromolecular model building for x-ray crystallography using ARP/wARP version 7. Nat. Protoc. 3, 1171–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 35. Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 36. Blanc E., Roversi P., Vonrhein C., Flensburg C., Lea S. M., Bricogne G. (2004) Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr. D Biol. Crystallogr. 60, 2210–2221 [DOI] [PubMed] [Google Scholar]
- 37. Sali A., Potterton L., Yuan F., van Vlijmen H., Karplus M. (1995) Evaluation of comparative protein modeling by MODELLER. Proteins 23, 318–326 [DOI] [PubMed] [Google Scholar]
- 38. Dubrovskaya V., Lavigne A. C., Davidson I., Acker J., Staub A., Tora L. (1996) Distinct domains of hTAFII100 are required for functional interaction with transcription factor TFIIF β (RAP30) and incorporation into the TFIID complex. EMBO J. 15, 3702–3712 [PMC free article] [PubMed] [Google Scholar]
- 39. Frontini M., Soutoglou E., Argentini M., Bole-Feysot C., Jost B., Scheer E., Tora L. (2005) TAF9b (formerly TAF9L) is a bona fide TAF that has unique and overlapping roles with TAF9. Mol. Cell. Biol. 25, 4638–4649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hisatake K., Ohta T., Takada R., Guermah M., Horikoshi M., Nakatani Y., Roeder R. G. (1995) Evolutionary conservation of human TATA-binding-polypeptide-associated factors TAFII31 and TAFII80 and interactions of TAFII80 with other TAFs and with general transcription factors. Proc. Natl. Acad. Sci. U.S.A. 92, 8195–8199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kokubo T., Gong D. W., Wootton J. C., Horikoshi M., Roeder R. G., Nakatani Y. (1994) Molecular cloning of Drosophila TFIID subunits. Nature 367, 484–487 [DOI] [PubMed] [Google Scholar]
- 42. Weinzierl R. O., Ruppert S., Dynlacht B. D., Tanese N., Tjian R. (1993) Cloning and expression of Drosophila TAFII60 and human TAFII70 reveal conserved interactions with other subunits of TFIID. EMBO J. 12, 5303–5309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Katinka M. D., Duprat S., Cornillot E., Méténier G., Thomarat F., Prensier G., Barbe V., Peyretaillade E., Brottier P., Wincker P., Delbac F., El Alaoui H., Peyret P., Saurin W., Gouy M., Weissenbach J., Vivarès C. P. (2001) Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature 414, 450–453 [DOI] [PubMed] [Google Scholar]
- 44. Diebold M. L., Koch M., Loeliger E., Cura V., Winston F., Cavarelli J., Romier C. (2010) The structure of an Iws1/Spt6 complex reveals an interaction domain conserved in TFIIS, Elongin A, and Med26. EMBO J. 29, 3979–3991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Diebold M. L., Loeliger E., Koch M., Winston F., Cavarelli J., Romier C. (2010) Noncanonical tandem SH2 enables interaction of elongation factor Spt6 with RNA polymerase II. J. Biol. Chem. 285, 38389–38398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Romier C., James N., Birck C., Cavarelli J., Vivarès C., Collart M. A., Moras D. (2007) Crystal structure, biochemical, and genetic characterization of yeast and E. cuniculi TAFII5 N-terminal domain: implications for TFIID assembly. J. Mol. Biol. 368, 1292–1306 [DOI] [PubMed] [Google Scholar]
- 47. Andrade M. A., Petosa C., O'Donoghue S. I., Müller C. W., Bork P. (2001) Comparison of ARM and HEAT protein repeats. J. Mol. Biol. 309, 1–18 [DOI] [PubMed] [Google Scholar]
- 48. Zhou J., Zwicker J., Szymanski P., Levine M., Tjian R. (1998) TAFII mutations disrupt Dorsal activation in the Drosophila embryo. Proc. Natl. Acad. Sci. U.S.A. 95, 13483–13488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. van Ingen H., van Schaik F. M., Wienk H., Ballering J., Rehmann H., Dechesne A. C., Kruijzer J. A., Liskamp R. M., Timmers H. T., Boelens R. (2008) Structural insight into the recognition of the H3K4me3 mark by the TFIID subunit TAF3. Structure 16, 1245–1256 [DOI] [PubMed] [Google Scholar]
- 50. Wang X., Truckses D. M., Takada S., Matsumura T., Tanese N., Jacobson R. H. (2007) Conserved region I of human coactivator TAF4 binds to a short hydrophobic motif present in transcriptional regulators. Proc. Natl. Acad. Sci. U.S.A. 104, 7839–7844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wright K. J., Tjian R. (2009) Wnt signaling targets ETO coactivation domain of TAF4/TFIID in vivo. Proc. Natl. Acad. Sci. U.S.A. 106, 55–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wollmann P., Cui S., Viswanathan R., Berninghausen O., Wells M. N., Moldt M., Witte G., Butryn A., Wendler P., Beckmann R., Auble D. T., Hopfner K. P. (2011) Structure and mechanism of the Swi2/Snf2 remodeller Mot1 in complex with its substrate TBP. Nature 475, 403–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bond C. S., Schüttelkopf A. W. (2009) ALINE: a WYSIWYG protein-sequence alignment editor for publication-quality alignments. Acta Crystallogr. D Biol. Crystallogr. 65, 510–512 [DOI] [PubMed] [Google Scholar]
- 54. Nicholls A., Sharp K. A., Honig B. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11, 281–296 [DOI] [PubMed] [Google Scholar]
- 55. DeLano W. L. (2010) The PyMOL Molecular Graphics System, version 1.3r1, Schrödinger, LLC, New York [Google Scholar]