Background: Dislocation of proteins from the endoplasmic reticulum to the cytosol is an essential cellular process.
Results: Dislocation of engineered proteins leads to reconstitution of split GFP fragments in living cells.
Conclusion: A GFP reporter has been developed for studying dislocation in living cells.
Significance: The strategy is broadly applicable to the study of transmembrane transport of proteins and likely also of viruses and toxins.
Keywords: ER Quality Control, ER-associated Degradation, Proteasome, Ubiquitin, Ubiquitin Ligase, Dislocation, Hrd1, Importin β, Retrotranslocation, p97/VCP
Abstract
Misfolded proteins in the endoplasmic reticulum (ER) are dislocated to the cytosol to be degraded by the proteasomes. Various plant and bacterial toxins and certain viruses hijack this dislocation pathway to exert their toxicity or to infect cells. In this study, we report a dislocation-dependent reconstituted GFP (drGFP) assay that allows, for the first time, imaging proteins dislocated from the ER lumen to the cytosol in living cells. Our results indicate that both luminal and membrane-spanning ER proteins can be fully dislocated from the ER to the cytosol. By combining the drGFP assay with RNAi or chemical inhibitors of proteins in the Hrd1 ubiquitin ligase complex, we demonstrate that the Sel1L, Hrd1, p97/VCP, and importin β proteins are required for the dislocation of misfolded luminal α-1 antitrypsin. The strategy described in this work is broadly applicable to the study of other types of transmembrane transport of proteins and likely also of viruses and toxins in living cells.
Introduction
Proteins destined for the secretory pathway are inserted into the membrane or lumen of the endoplasmic reticulum (ER),2 where they are processed and folded into their native conformation. Misfolded or unassembled proteins are retained in the ER and transported to the cytosol for proteasomal degradation, a process known as ER-associated protein degradation (ERAD) (1, 2). ERAD protects cells against the detrimental effects of ER stress and regulates many essential cellular functions, such as sterol homeostasis and calcium signaling (3–5).
The ER-to-cytosol transport, called dislocation or retrotranslocation, is one of the key steps in ERAD because the ubiquitin-proteasome system is localized in the cytosol (6–11). Various plant and bacterial toxins and certain viruses hijack the dislocation process to reach the cytosol, where they may exert their cytotoxicity or effectively infect cells (12–16). In addition, growing evidence suggests that certain ER-localized proteins, such as the EGF receptor, ErbB2, calreticulin, Nrf1, and OS9, dislocate first to the cytosol and then are imported into the nucleus when they function as transcriptional regulators (17–22). However, the mechanisms underlying dislocation remain poorly understood.
The development of an effective assay for dislocation should facilitate research on this important process. Dislocation is typically analyzed by biochemical approaches. One method is to detect the cytosolic localization of dislocated substrate proteins by subcellular fractionation and immunoblotting (6, 23). Another approach is to analyze dislocation indirectly by detecting substrate deglycosylation (for glycoproteins) or substrate ubiquitination (for luminal substrates), events that occur only when the substrates reach the cytosol (23–27). However, these methods are not always effective because the dislocation of certain substrates is tightly coupled with their ubiquitination and subsequent proteasomal degradation (28). The complexity of dislocation processes also requires more efficient assays. Nearly ninety proteins in mammalian cells are involved in ERAD or have high-confidence interactions with ERAD machineries (26, 29–32). The actual number of ERAD components could easily be over a hundred, as the actual components do not necessarily have to interact with the ERAD machinery with high affinity. These ERAD components are organized into various complexes that are centered on one of the many proven or potential ERAD E3 ubiquitin ligases, which dispose of misfolded proteins with various characteristics (33–35). Moreover, it is possible that a given E3 can organize different complexes that perform different roles in different cells (36, 37). It would be difficult and tedious to study the large number of potential regulators in many different dislocation complexes by the currently available biochemical methods. Therefore, the establishment of an efficient live cell assay for dislocation could significantly expedite the study of the molecular mechanisms of dislocation.
In this study, we report a novel split-GFP-based dislocation assay. In this assay, dislocation leads to the reconstitution of GFP fluorescence from its fragments. The reconstituted GFP serves as a reporter for the localization and quantity of dislocated substrates in living cells. By combining this approach with RNAi and chemical inhibitors of proteins in ERAD complexes, we demonstrate the feasibility of this assay for analyzing the mechanisms that underlie the dislocation of various types of substrates. Moreover, the strategy described herein should be widely applicable to the study of other types of transmembrane transport of proteins in living cells.
EXPERIMENTAL PROCEDURES
Plasmid Constructs
The plasmids pCMV-mGFP(1–10) and pCMV-mGFP(Cterm S11) were purchased from Theranostech, Inc. The lentiviral backbone plasmid pRRL-sinhCMV-ΔGFP and the two-helper plasmids, pVSVG and pDelta8.7, as well as the p53-GFP construct, have been described previously (32, 38). The plasmid pRRL-S1-10 was constructed by inserting the open reading frame (ORF) of mGFP S1-10 (NheI/BamH I fragment blunted with Klenow) from pCMV-mGFP(1-10) into the BamH I site (blunted) of pRRL-sinhCMV-ΔGFP. To construct plasmids expressing the SP-S11-tagged ATM or NHK, DNA fragments encoding an N-terminal signal peptide (the first 33 amino acids from ATM) followed by mGFP S11, a linker sequence (GDGGSGGGSAS), the ATM or NHK sequence without the N-terminal 25 amino acids and a C-terminal HA tag sequence were inserted into the EcoRI/NotI sites of pCMV-mGFP(Cterm S11) to replace the original sequence in the vector. A nuclear localization signal (NLS, PKKKRKV) was inserted by site-directed mutagenesis between the linker sequence and NHK sequence to generate a construct for expressing SP-S11-NLS-NHK-HA. These DNA fragments were also inserted after the FLAG tag sequence that was constructed in the pCIneo vector for expressing FLAG-SP-S11-ATM-HA, -NHK-HA, or -NLS-NHK-HA constructs. To make a construct for expressing S11-NHK-HA (no SP), a DNA fragment encoding S11, the linker sequence, the NHK sequence without the N-terminal 25 amino acids and a C-terminal HA tag sequence were inserted into the pCIneo vector. The FLAG-S11-NHK-HA and FLAG-S11-NLS-NHK-HA plasmids were constructed by inserting DNA fragments encoding S11-NHK-HA or S11-NLS-NHK-HA from the previous constructs into pFLAG-CMV-6a. The SP-S11-CD3δ-HA plasmid was constructed by replacing its SP sequence with the sequence encoding NHK SP, S11 and the linker sequence using pCIneo-CD3δ-HA as a template plasmid. Site-directed mutagenesis was used to replace the sequence encoding the C-terminal PRNKKSG sequence from the mouse CD3δ with the NLS to make the SP-S11-CD3δ-NLS-HA construct.
Antibodies
Monoclonal anti-gp78 (2G5) and rabbit polyclonal anti-Hrd1 antibodies have been described previously (39). Mouse anti-calnexin antibody was purchased from Affinity BioReagents. Goat anti-α1-antitrypsin antibody was purchased from Bethyl Laboratories. Mouse monoclonal anti-β-actin (AC-74), anti-HA (HA-7), anti-FLAG (M2), anti Lamin A/C, and rabbit anti-Sel1L antibodies were purchased from Sigma. Mouse monoclonal anti-GFP, rabbit anti-CHOP and HRP-conjugated anti-ubiquitin antibodies were purchased from Santa Cruz Biotechnology, Inc.
Chemicals and siRNAs
Importazole (IPZ) was a kind gift from Dr. Rebecca Heald (40). DBeQ was purchased from Sigma. Epoxomycin and MG132 were purchased from Calbiochem. All siRNAs were ordered from Ambion (Invitrogen), including Silencer® Negative Control #1, Sel1L (siRNA ID: 121516), Hrd1 (siRNA ID: 44536), gp78 (siRNA ID: 110866), and p97/VCP (siRNA ID: 214796) siRNAs.
Lentivirus Production
A previously reported protocol was used for the production of lentiviral particle expressing S1-10 (38). Briefly, HEK293T cells were co-transfected with pRRL-S1-10 and helper vectors pDelta8.7 and pVSVG in a ratio of 5:5:3 using Lipofectamine 2000 (Invitrogen). Forty-eight hours after transfection, virus-containing crude supernatant was collected and stored at −80 °C until use.
Establishment of a HeLa Cell Line Stably Expressing SP-S11-NHK-HA and S1-10
We first established a HeLa cell line stably expressing S1-10. HeLa cells were infected with the virus-containing crude supernatant. Forty-eight hours after infection, cells were re-seeded to a 96-well plate at a concentration of ∼1 cell/well. After an additional 2 weeks in culture, cells expressing S1-10 were confirmed by immunostaining and immunoblotting. Clones expressing S1-10 evenly in all cells were used for subsequent experiments.
HeLa cells stably expressing S1-10 were co-transfected with the SP-S11-NHK construct and pBABE-puro at a ratio of 20:1. Twenty-four hours after transfection, cells were selected with puromycin and cultured for an additional 2 weeks until single clones were visible. The single clones were separately transferred to 12-well plates for expansion. The expression levels of SP-S11-NHK-HA in these cells were analyzed by immunoblotting. Positive clones were treated with MG132 for 4 h to visualize drGFP fluorescence. Clones showing even fluorescence among cells after MG132 treatment were used for further experiments.
Immunofluorescence
Cells were fixed, either by methanol for 1 h at −20 °C or by 4% paraformaldehyde for 30 min at 4 °C, and blocked in 0.1% saponin plus 0.1% human serum albumin. Then, the cells were labeled with mouse monoclonal anti-HA or anti-GFP antibody, as indicated, for 1 h followed by labeling with Alexa® Fluor 594-conjugated goat anti-mouse IgG (H+L) for 1 h before fluorescence microscopy.
Plasma Membrane Permeabilization Assay
The plasma membrane permeabilization assay was performed according to a previous report (41). Briefly, cells cultured on chambered coverglass were treated as indicated. Then, the culture medium was replaced with KHM buffer (110 mm potassium acetate, 20 mm HEPES, 2 mm MgCl2). An equal volume of KHM buffer containing 20 μg/ml digitonin was added to the cells to permeabilize the plasma membranes. Images were acquired on a Zeiss Axiovert 200 m fluorescence microscope every 30 s immediately after the addition of digitonin for 3 min.
Preparation of Cytoplasmic and Nuclear Fractions
Cytoplasmic and nuclear fractions were prepared according to the previous report with minor modifications (42). Briefly, about 3 × 106 cells were suspended in 80 μl cold HB buffer (10 mm Tris pH 7.9, 1.5 mm MgCl2, 10 mm KCl, protease inhibitor mixture), and left on ice for 20 min. Then 20 μl of HB buffer containing 1% Triton X-100 was added to the cells. After vortexing for 5 s, the homogenate was spun at 1000 × g for 10 min. The supernatant represents the cytoplasmic fraction. The pellet was resuspended in HB buffer containing 0.2% Triton X-100, votexed for 5s and spun at 1000 × g for 10 min to remove the contaminated cytoplasmic fraction components. The pellet containing the nuclear fraction was then resuspended in buffer C (10 mm Tris pH 7.9, 1.5 mm MgCl2, 10 mm KCl, 400 mm NaCl, 0.4% Triton X-100, protease inhibitor mixture), vortexed at 4 °C for 30 min vigorously and spun at 20,000 × g for 15 min to prepare the nuclear extract. The cytoplasmic and nuclear fractions were further processed for immunoblotting as reported previously (32).
Live Cell Imaging and drGFP Measurement
Live cell images were acquired every 10 min under a 63x objective lens mounted on a Zeiss AxioObserver Z1 fluorescence microscope equipped with a high-sensitivity CCD camera (QuantEM 512SC; Photometrics, Tucson, AZ), environment control units and a Definitive Focus module. The relative intensities of drGFP were quantified either using ImageJ software or on a fluorescence microplate reader. HeLa cells stably expressing SP-S11-NHK-HA and S1-10 were seeded to black 96-well plate (Costar 3916). After overnight culture, the cells were treated with MG132 (10 μm) alone or MG132 (10 μm) together with IPZ (30 μm) and/or DBeQ (7.5 μm) for 4 h. Then the cells were washed once with PBS. The relative fluorescence units (RFU) of drGFP were measured with GloMax®-Multi+ Detection System using excitation = 488 nm and emission = 525 nm.
Proteinase K Protection Assay
HeLa cells stably expressing SP-S11-NHK-HA and S1-10 were transfected with siRNA targeting Sel1L or Hrd1 as indicated. Forty-eight hours after transfection, the cells were homogenized in fractionation buffer (50 mm Tris/HCl, pH 8.0, 1 mm 2-mercaptoethanol, 1 mm EDTA, 10 mm triethanolamine, and 0.32 m sucrose) by passing through a 27-gauge syringe 20 times. The homogenates were centrifuged at 1000 × g for 5 min to remove unbroken cells, cell debris and nuclei. The supernatant-containing the microsomes was treated with proteinase K (100 μg/ml) as indicated for 20 min on ice and lysed by adding 1% Triton X-100. The proteinase K-treated cells and untreated control cells were further processed for immunoblotting for indicated proteins, as we previously reported (32, 43).
In Vivo Ubiquitination
NHK ubiquitination was examined as previously reported (32). To study the effects of Sel1L and Hrd1 on NHK ubiquitination, HeLa cells stably expressing SP-S11-NHK and S1-10 were transfected with siRNA targeting Sel1L or Hrd1 as indicated. 48 h after transfection, cells were treated with MG132 (10 μm) for 4 h to inhibit proteasomal degradation. Then, the cells were harvested and lysed in 2% SDS. After boiling for 30 min, the lysates were diluted 20 times in cell lysis buffer, and the DNA was broken by sonication for 3 s. The cell debris and nuclei were removed by centrifugation. Two hundred micrograms of protein in the lysates was used for immunoprecipitation (IP) with an anti-HA Affinity Matrix (Roche). The precipitates were processed for immunoblotting. To study the effects of IPZ and DBeQ on NHK ubiquitination, HeLa cells stably expressing SP-S11-NHK and S1-10 were treated with MG132 alone or together with the chemicals, as indicated, for 4 h. Then, the cells were processed for IP as described (32).
RESULTS
Dislocation-dependent Reconstituted GFP (drGFP) in Living Cells
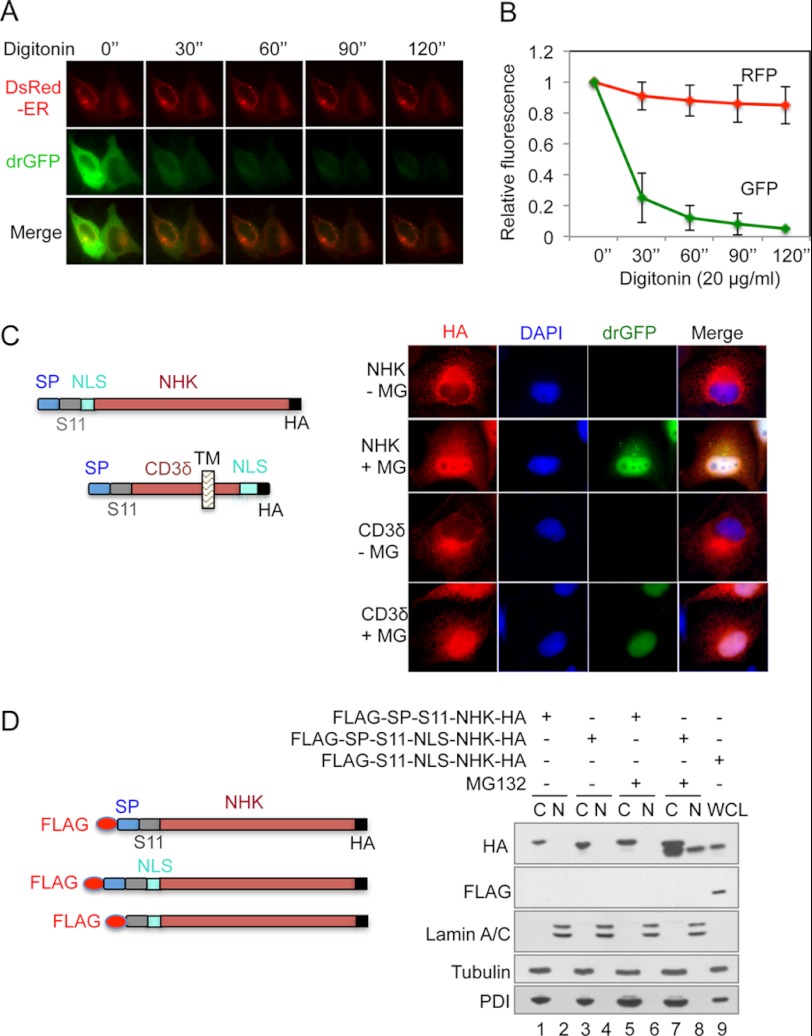
To study protein dislocation in living cells, we took advantage of the split-GFP system that had been developed from a well-folded variant of GFP (44). This GFP molecule can be split into two fragments: the C-terminal β-strand (S11), and the remaining 10 β-strands (S1-10). S11 can efficiently associate with S1-10 to reconstitute GFP when the two fragments meet. We hypothesized that if we expressed S11-tagged misfolded protein in the ER lumen and S1-10 in the cytosol, then when S11-tagged protein was dislocated from the ER lumen to the cytosol, the tagged S11 should have the opportunity to meet and reassemble with S1-10, resulting in reconstitution of the GFP (Fig. 1A). We first tested this hypothesis using the null Hong Kong variant of α-1-antitrypsin (NHK), a mutant protein that is retained in the ER lumen and then dislocated to the cytosol for degradation by the proteasomes (45, 46). A plasmid was constructed to express a modified version of NHK in which an S11 fragment was inserted after the signal peptide (SP) of NHK and an HA tag was added at the C terminus, resulting in the production of SP-S11-NHK-HA protein (Fig. 1A). Wild type α-1-antitrypsin (ATM) with the same tags (SP-S11-ATM-HA) was used as a negative control for dislocation, as ATM folds properly and therefore is not subject to dislocation and degradation (45, 46). We first determined whether the modified proteins could translocate into the ER as efficiently as their unmodified forms by examining their N-glycosylation, an event that occurs only when proteins are translocated into the ER (47). The modified proteins were expressed in HeLa cells. Extracts prepared from these cells were treated with Peptide: N-glycosidase F (PNGase F) to remove the N-glycans from ERAD substrates (48). Immunoblotting revealed that the sizes of both proteins were decreased after PNGase F treatment, indicating that N-glycans were removed from these proteins (Fig. 1B). SP cleavage is another indicator of ER translocation (49, 50). To determine whether the SP was cleaved, we further modified SP-S11-NHK-HA and SP-S11-ATM-HA by adding a FLAG tag at the N terminus of the SP. Thus, cleavage of SP would also remove the FLAG tag. We expressed these proteins in HeLa cells. Immunoblotting with anti-α-1-antitrypsin antibody confirmed their expression (Fig. 1C). Importantly, these proteins could not be detected by an anti-FLAG antibody even when proteasomal degradation was inhibited by epoxomycin (Fig. 1C, lanes 1, 2, 4, 5). A control, cytosolic version of NHK (FLAG-S11-NHK-HA) with the SP deleted was detected by both antibodies (Fig. 1C, lanes 3, 5). These results indicate that the modified NHK and ATM are subject to SP cleavage that results in the removal of their FLAG tags. Taken together, the evidence of efficient N-glycosylation and SP cleavage indicates that S11 tagging does not hinder the translocation of NHK or ATM into the ER.
FIGURE 1.
Detection of protein dislocation by dislocation-dependent reconstituted GFP (drGFP). A, schematic representation of the split-GFP-based method to monitor ER protein dislocation. S11-tagged NHK or CD3δ is expressed in the ER, and S1-10 is expressed in the cytosol. GFP can be re-assembled when S11-tagged proteins are dislocated to the cytosol. B, peptidyl-N-glycosidase (PNGase) F digestion. HeLa cells expressing SP-S11-tagged proteins as indicated were lysed and treated with PNGase F (20 units/μl) at 37 °C for 30 min. C, cleavage of signal peptides from S11-tagged ATM and NHK. HeLa cells expressing FLAG-SP-S11-ATM-HA, FLAG-SP-S11-NHK-HA, or FLAG-S11-NHK-HA were processed for IB. A group of cells was treated with epoxomycin (2 μm) for 4 h as indicated. D, HeLa cells stably expressing S1-10 were transfected with plasmids expressing FLAG-SP-S11-ATM-HA (ATM), FLAG-SP-S11-NHK-HA (NHK), or FLAG-S11-NHK-HA (NHK no SP). After overnight culture, the cells were treated with epoxomycin (2 μm) as indicated for 4 h, followed by immunofluorescence staining for HA (in red). E, as in B, except that SP-S11-CD3δ-HA was expressed in HeLa cells. F, as in D, except that SP-S11-CD3δ-HA was expressed in HeLa cells stably expressing S1-10, and the cells were treated with MG132 (10 μm) for 4 h. SP: signal peptide. Epox: epoxomycin. MG: MG132.
Next, we expressed SP-S11-NHK-HA or SP-S11-ATM-HA in HeLa cells that stably express S1-10 to determine whether we could detect GFP reconstitution by fluorescence microscopy. The stably expressed S1-10 was confirmed by immunofluorescence staining with an anti-GFP antibody (date now shown). Immunofluorescence staining for HA revealed the expression of SP-S11-NHK-HA and SP-S11-ATM-HA, but almost no GFP signal was observed in the cells (Fig. 1D, panels 1–3 and 7–9), likely as a result of the tight coupling of dislocation with proteasomal degradation. Indeed, treatment with the proteasome inhibitor epoxomycin (to block the degradation of dislocated proteins) led to significant increases in GFP fluorescence in cells expressing SP-S11-NHK-HA (Fig. 1D, panels 10–12), but not SP-S11-ATM-HA (Fig. 1D, panels 4–6). Time-lapse imaging revealed a time-dependent increase in GFP fluorescence in SP-S11-NHK-HA-expressing cells (supplemental Movie S1). These results are consistent with the fact that NHK, but not ATM, undergoes dislocation and degradation by the proteasome. When a cytosolic version (SP-deleted) of S11-NHK-HA was expressed in HeLa cells stably expressing S1-10, GFP fluorescence was readily observed in the cells even without epoxomycin treatment (Fig. 1D, panels 13–18). These results indicate that the GFP fluorescence in cells expressing SP-S11-NHK-HA and S1-10 resulted from the reassembly of the dislocated S11-NHK-HA with S1-10. This dislocation-dependent reconstituted GFP (drGFP) can, therefore, be used to monitor NHK dislocation in cells.
To determine whether the same strategy can be used to monitor the dislocation of membrane-spanning ERAD substrates, we chose to test CD3δ, a single transmembrane ERAD substrate with its N terminus in the lumen and its C terminus in the cytosol (51, 52). We generated a plasmid for expressing SP-S11-CD3δ-HA in which S11 coding sequence was inserted after the signal peptide so that the S11 region of the recombinant protein will locate in the lumen (Fig. 1A). PNGase F digestion results indicated that the modified CD3δ was efficiently translocated into the ER (Fig. 1E). Similar to the results observed for NHK, the co-expression of SP-S11-CD3δ-HA and S1-10 in HeLa cells did not directly result in GFP fluorescence (Fig. 1F). The generation of GFP fluorescence in these cells once again required proteasome inhibition by MG132 (Fig. 1F and supplemental Movie S2). These results indicate that the S11-tagging approach can also be used to monitor the dislocation of membrane-bound CD3δ in living cells.
NHK and CD3δ Can Be Fully Dislocated from the ER to the Cytosol
Although there are reported examples of both membrane and luminal substrates being fully dislocated to the cytosol as revealed by subcellular fractionation assays, it remains controversial whether this full dislocation actually occurs in cells (28, 36). To assess the full dislocation of NHK, we examined the effects of plasma membrane permeabilization on NHK drGFP. If NHK was fully dislocated to the cytosol, drGFP would be released from the cell when the membrane was permeabilized, leading to decreases in fluorescence. We expressed SP-S11-NHK-HA in HeLa cells that stably express S1-10. An ER luminal DsRed molecule was co-expressed to monitor the integrity of the ER membrane. The cells were then treated with MG132 for 4 h, followed by plasma membrane permeabilization with digitonin to release the cytosol (41), and time-lapse imaging to determine the changes in drGFP and DsRed fluorescence intensities. Whereas the ER luminal DsRed signal remained almost constant, the drGFP signal was rapidly lost (Fig. 2, A and B), suggesting that the S11-NHK-HA molecule is fully dislocated to the cytosol and released from the cells upon plasma membrane permeabilization.
FIGURE 2.
Full dislocation of NHK and CD3δ in living cells. A, SP-S11-NHK-HA and an ER luminal DsRed were co-expressed in HeLa cells stably expressing and S1-10. The cells were treated with MG132 for 4 h followed by plasma membrane permeabilization with digitonin (20 μg/ml) to release the cytosol. Images were acquired every 30 s. B, measurement of drGFP and DsRed intensities in A. The data are presented as the means ± S.E. of 15 cells. The drGFP and DsRed intensities at time 0 were set as 1, respectively. C, left: schematic representation of SP-S11-NLS-NHK-HA and SP-S11-CD3δ-NLS-HA. NLS: nuclear localization signal. Right: HeLa cells expressing S1-10 and SP-S11-NLS-NHK-HA or SP-S11-CD3δ-NLS-HA were treated with MG132 (10 μm) for 4 h followed by immunofluorescence staining for HA (in red). The nucleus was labeled with DAPI. D, HeLa cells transfected with plasmids as indicated in the schematic representation were treated with MG132 (10 μm) for 4 h. The cells were then separated into cytoplasmic and nuclear fractions. C: cytoplasmic fraction. N: nuclear fraction. WCL: whole cell lysate.
Next, we wanted to demonstrate the full dislocation of proteins in intact living cells. To accomplish this goal, we included a nuclear localization signal (NLS) in SP-S11-NHK-HA and SP-S11-CD3δ-HA, resulting in the SP-S11-NLS-NHK-HA and SP-S11-CD3δ-NLS-HA proteins (Fig. 2C, left). We reasoned that if full dislocation occurred, the NLS would then direct the fully dislocated NHK and CD3δ to the nucleus. Because S11 and the HA tag are located in the N and C termini, respectively, co-localization of drGFP and HA immunofluorescence reflects dislocation of the whole molecule. HeLa cells co-expressing S1-10 and SP-S11-NLS-NHK-HA or SP-S11-CD3δ-NLS-HA were treated with MG132 and then stained for HA to show total S11-NLS-NHK-HA or S11-CD3δ-NLS-HA. As shown in Fig. 2C, NHK and CD3δ drGFP showed clear nuclear localization and, at the same time, a portion of HA staining (for total NHK or CD3δ) co-localized with the nucleus-localized NHK or CD3δ drGFP, suggesting that both NHK and CD3δ are fully dislocated from the ER to the cytosol and then imported into the nucleus. Time-lapse imaging revealed a continuous increase in NHK and CD3δ drGFP fluorescence in the nucleus following proteasome inhibition (supplemental Movies S3 and S4). Interestingly, CD3δ drGFP exhibited prominent nuclear localization after proteasome inhibition, even without the addition of an NLS (Fig. 1F and supplemental Movie S2), suggesting that full dislocation of this protein is not an artifact caused by the addition of an NLS. We further characterized the nucleus-localized NHK by biochemical approaches. HeLa cells expressing FLAG-SP-S11-NLS-NHK-HA were processed into nuclear and cytoplasmic fractions. Immunoblotting showed that a portion of the NHK was localized in the nuclear fraction when MG132 was used to prevent the degradation of the dislocated NHK (Fig. 2D, lanes 8 versus 7). The nuclear localization of NHK is dependent on the added NLS because NHK without the NLS cannot be detected in the nuclear fraction (Fig. 2D, lanes 8 versus 6). Importantly, the nucleus-localized NHK did not contain the FLAG tag, but the FLAG tag became detectable when the SP was deleted from FLAG-SP-S11-NLS-NHK-HA (Fig. 2D, lane 9). These results indicate that the SP is cleaved from the nuclear NHK. Taken together, these results suggest that NHK and CD3δ are fully dislocated from the ER to the cytosol during ERAD.
Knockdown of Sel1L, Hrd1, and p97/VCP but Not gp78 Inhibits NHK Dislocation and drGFP
To further validate the drGFP assay, we turned to the Hrd1 ubiquitin ligase complex (29, 53–56). The Hrd1 and Sel1L proteins in this complex have been shown to be critically required for NHK degradation (56), but it is not known whether they play a role in NHK dislocation. To answer this question, we established a HeLa cell line stably expressing SP-S11-NHK-HA and S1-10. We then knocked down the expression of Sel1L or Hrd1. Time-lapse imaging of drGFP was performed every 10 min for up to 4 h immediately after the addition of MG132. The results showed that MG132 treatment induced a fairly homogeneous increase in drGFP in all control knockdown cells (Fig. 3A, upper panel and supplemental Movie S5a), but the increase was markedly suppressed in cells with Sel1L or Hrd1 knockdown (Fig. 3A, middle and lower panels as well as supplemental Movie S5, b and c). Quantification using ImageJ, a free image-processing software program, revealed that the drGFP intensity gradually increased by ∼6.5-fold after a 4-h treatment with MG132 in control cells, whereas the increase was almost completely abolished by the knockdown of Sel1L or Hrd1 (Fig. 3, B and C). These results indicate that drGFP is capable of monitoring dynamic increases in NHK dislocation and that Hrd1 and Sel1L are essential for NHK drGFP.
FIGURE 3.
Knockdown of Sel1L or Hrd1 inhibits NHK dislocation. A, HeLa cells stably expressing SP-S11-NHK-HA and S1-10 were transfected with control siRNA or siRNA targeting Sel1L or Hrd1. Forty-eight hours after transfection, the cells were treated with MG132 (10 μm) for 4 h before being imaged with fluorescent microscope. B, cells were transfected as in A. Forty-eight hours after transfection, the cell culture medium was replaced with phenol red-free medium containing MG132 (10 μm). The drGFP images were acquired every 10 min for up to 4 h. C, measurement of drGFP intensity in B using ImageJ. The data are presented as the means ± S.E. of at least 30 cells. D, cells were transfected as in A. Forty-eight hours after transfection, the microsomes from these cells were prepared and treated with proteinase K (100 μg/ml) as indicated for 20 min on ice. Then, the microsomes were lysed and processed for IB. E, cells transfected as in A were processed for immunoprecipitation (IP) with an anti-HA antibody followed by IB with an anti-ubiquitin antibody to detect polyubiquitinated S11-NHK. The numbers in the polyubiquitin blot (PolyUb, lanes 4–6) show the relative density of polyubiquitinated S11-NHK-HA. WCL: whole cell lysate. F, HeLa cells stably expressing SP-S11-NHK-HA and S1-10 were subject to siRNA knockdown of Sel1L, Hrd1, gp78, or p97/VCP. NC: non-targeting control siRNA. Graph shows the relative drGFP fluorescence intensity in the knockdown cells measured on a fluorescence microplate reader. Blots show the efficiency of gp78 and p97/VCP knockdown and the levels of NHK.
During ERAD, NHK must be dislocated before it can be ubiquitinated. A failure in dislocation should suppress NHK ubiquitination and lead to accumulation of NHK in the ER Lumen. To assess luminal accumulation of NHK, Sel1L and Hrd1 were knocked down in a HeLa cell line stably expressing SP-S11-NHK-HA and S1-10. The microsomal fraction was isolated and subjected to a proteinase K protection assay (43). As shown in Fig. 3, D and E (lanes 1–3), knockdown of either Sel1L or Hrd1 caused increases in NHK levels that were not sensitive to proteinase K digestion (Fig. 3D, lanes 4–6). As a positive control, proteinase K completely digested a cytosolic domain of calnexin in all cells, as reflected by the drop in its molecular weight (Fig. 3D, lanes 4–6). These results suggest that knockdown of either Sel1L or Hrd1 causes accumulation of NHK in the ER lumen. To determine whether NHK is ubiquitinated, Hrd1 or Sel1L knockdown cells were lysed in a denaturing buffer and processed for immunoprecipitation of NHK followed by immunoblotting for ubiquitin. As shown in Fig. 3E, knockdown of either Sel1L or Hrd1 caused an increase in total ubiquitination level (lanes 2 and 3 versus 1) but significantly inhibited NHK ubiquitination (lanes 5 and 6 versus 4). Thus, the decreases in NHK drGFP fluorescence correlate well with increases in luminal NHK and decreases in ubiquitinated NHK in Sel1L or Hrd1 knockdown cells, suggesting that Sel1L and Hrd1 are critically required for NHK dislocation.
gp78 is another E3 ubiquitin ligase for ERAD, but it is not involved in the degradation of NHK (57, 58). As predicted, the knockdown of gp78 did not cause accumulation of the NHK protein and had no inhibitory effect on NHK drGFP fluorescence (Fig. 3F) as measured with a fluorescence microplate reader after the treatment with MG132. On the other hand, knockdown of p97/VCP, a well-established dislocation factor (59, 60), significantly inhibited the formation of NHK drGFP accompanied by an increase in the steady level of NHK (Fig. 3F). These results are in agreement with the previously reported roles of gp78 and p97/VCP in the dislocation of NHK.
The drGFP Assay Reveals the Cooperation of p97/VCP and Importin β in NHK Dislocation
The RNAi results suggest that drGFP assay can be useful in the screening and functional analysis of proteins involved in certain substrate dislocation. We then asked if drGFP assay could also be used in the identification of chemical modulators of dislocation. We tested this idea by assessing the recently developed small molecule inhibitor of p97/VCP, N2, N4-dibenzylquinazoline-2, 4-diamine (DBeQ), and the inhibitor of importin β, importazole.
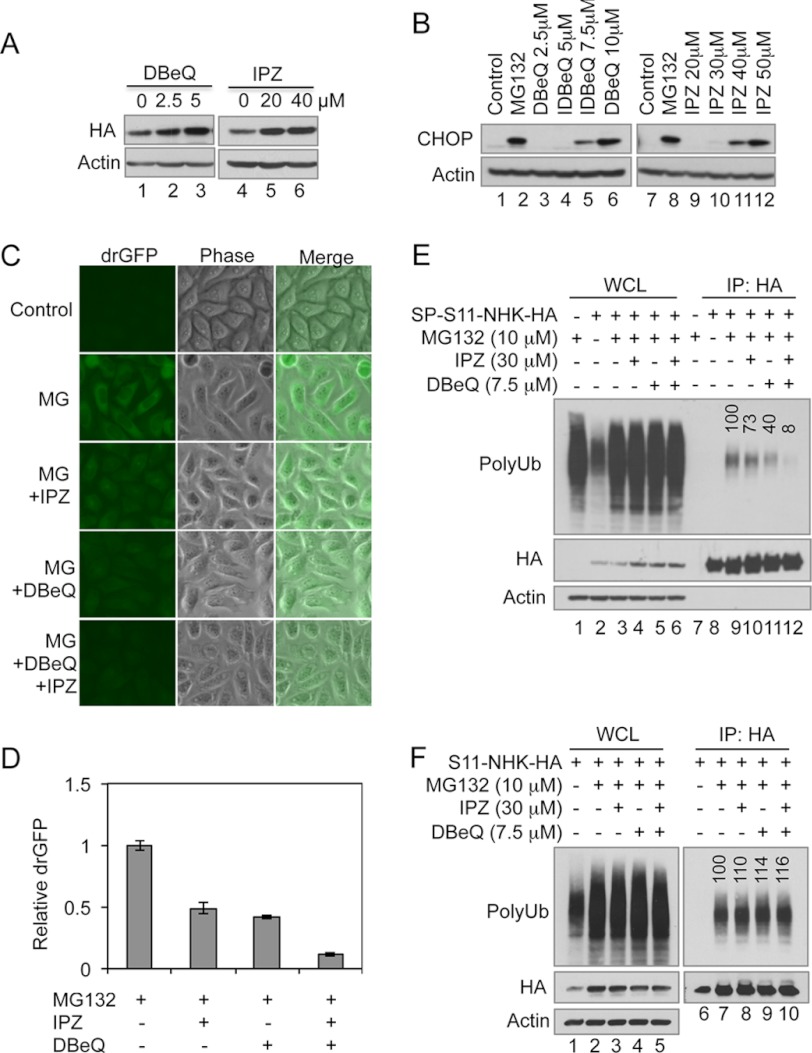
DBeQ inhibits the ATPase activity of p97/VCP (61). Our RNAi results have confirmed the role of p97/VCP in dislocation (Fig. 3F). However, the requirement for the ATPase activity of p97/VCP in dislocation has not been demonstrated in living cells. In initial characterization, we found that treatment with DBeQ stabilized SP-S11-NHK-HA proteins (Fig. 4A) and induced ER stress as indicated by up-regulation of CHOP (GADD153), a well-established marker of the unfolded protein response initiated by ER stress (Fig. 4B). This is consistent with previous reports (61).
FIGURE 4.
Synergistic inhibitory effects of inhibitors to p97/VCP and importin β on dislocation and ubiquitination of NHK. A, HeLa cells expressing SP-S11-NHK-HA were treated with importazole (IPZ) or DBeQ for 4 h as indicated. Treated cells were then processed for IB. B, HeLa cells were treated with MG132 (10 μm) or different amounts of IPZ or DBeQ as indicated for 4 h before processed for IB. C, HeLa cells stably expressing SP-S11-NHK-HA and S1-10 were treated with MG132 (10 μm) alone or MG132 (10 μm) together with IPZ (30 μm) and/or DBeQ (7.5 μm) for 4 h. The drGFP images were taken under the same parameters. D, HeLa cells stably expressing SP-S11-NHK-HA and S1-10 were treated as in C. The relative fluorescence units (RFU) of drGFP were measured with a fluorescence microplate reader. The data are presented as the means ± S.E. of 4 samples. The drGFP intensity increased after 4-h treatment with MG132 was set as 1. E, non-transfected HeLa cells or HeLa cells expressing SP-S11-NHK-HA were treated with inhibitors as indicated for 4 h. The IP and IB were performed as in Fig. 3. The numbers in PolyUb blot (lanes 9–12) indicate the relative density of polyubiquitinated S11-NHK-HA. F, as in E, except that HeLa cells expressing S11-NHK-HA (no SP) were used. WCL: whole cell lysate.
Importazole is a 2,4-diaminoquinazoline that specifically blocks importin-β-mediated nuclear import by interfering with the interaction between importin β and RanGTP (40). Importin β is a recently identified component of the ERAD machinery that regulates ERAD from the cytosolic side of the ER (32). It may cooperate with RanGDP in the degradation of NHK, likely by enhancing dislocation. We tested first whether importazole treatment affects the ERAD of NHK. Using the same experiments performed for DBeQ, we found that treatment with increasing amounts of importazole recapitulated the effects of DBeQ, including NHK stabilization and ER stress induction (Fig. 4, A and B). These results support our previous reports that the interaction between importin β and Ran plays an important role in NHK degradation (32).
We then determined the effects of DBeQ and importazole on dislocation using the drGFP assay. HeLa cells stably expressing SP-S11-NHK-HA and S1-10 were treated with MG132 or MG132 plus one or two of these inhibitors for 4 h. NHK drGFP fluorescence was examined by microscopy and measured with a fluorescence microplate reader after the treatments. It is worth noting that we chose to use concentrations of DBeQ and importazole that induce ER stress to a moderate extent (as shown in Fig. 4B) to determine whether they have a synergistic effect on NHK dislocation. In addition, the concentration of importazole (30 μm) used did not significantly block the nuclear import of GFP-tagged p53 (supplemental Fig. S1), which is consistent with its reported effects on the nuclear import of other proteins (40). The results show that the drGFP signal is significantly increased in cells treated with MG132 alone (Fig. 4, C and D). The increases were reduced by 51% when 30 μm importazole was present and by 58% when 7.5 μm DBeQ was used (Fig. 4, C and D), suggesting that both inhibitors inhibits the dislocation of NHK. Therefore, the drGFP assay results suggest that ATPase activity of p97/VCP and interaction between importin β and Ran are both required for NHK dislocation. Interestingly, the simultaneous use of DBeQ and importazole reduced the drGFP signal by 89% (Fig. 4, C and D), suggesting a synergistic inhibitory effect when these two inhibitors are combined. Next, we determined whether the synergistic effect correlated with the inhibition of NHK ubiquitination. Cells subjected to the same treatments described in Fig. 4, C and D were lysed in a denaturing buffer, and then NHK was immunoprecipitated and immunoblotted for ubiquitin. Whereas the total ubiquitination levels were increased to a similar level in the cells with different treatments (Fig. 4E, lanes 3–6 versus 2), NHK ubiquitination was inhibited in the cells treated with DBeQ, importazole, or both (Fig. 4E, lanes 10–12 versus 9). Importantly, DBeQ and importazole also synergistically inhibited NHK ubiquitination (Fig. 4E, lane 12) as they did in the drGFP assay (Fig. 4, C and D). Treatment with DBeQ and importazole did not inhibit, and, in fact, slightly increased the ubiquitination of SP-deleted NHK (Fig. 4F), a cytosolic protein that does not require dislocation for ubiquitination and proteasomal degradation. This result suggests that DBeQ and importazole inhibit NHK ubiquitination by inhibiting its dislocation but not by directly interfering in the ubiquitination process itself. Therefore, the results of the biochemical assays confirmed the drGFP assay results. The ATPase activity of p97/VCP and the importin β-Ran interaction are required for NHK dislocation, and it is likely that p97/VCP and importin β may cooperate in this dislocation process. However, it is surprising that DBeQ treatment inhibited NHK ubiquitination, as previous studies have shown that p97/VCP facilitates dislocation after the substrates have been ubiquitinated (24). We reason that in DBeQ-treated cells, the ATPase-inactivated p97/VCP may bind to the dislocation complex and block NHK dislocation, resulting in the inhibition of NHK ubiquitination.
DISCUSSION
Based on the reassembly property of the split-GFP system (44), we have established a dislocation-dependent reconstituted GFP (drGFP) assay. The usefulness of the drGFP assay for studying dislocation was validated by various biochemical approaches. More importantly, we have used this assay to make several important observations. First, we demonstrated that both the luminal substrate NHK and the membrane-spanning substrate CD3δ could be fully dislocated to the cytosol. Second, we found that importin β cooperates with p97/VCP to regulate NHK dislocation. Third, we showed that Hrd1 is not only functions as an E3 ubiquitin ligase but also plays an essential role in NHK dislocation. It may act as a dislocation channel, similar to its yeast counterpart (62). Fourth, we found that the Sel1L protein, the luminal substrate receptor, is also essential for NHK dislocation, suggesting that Sel1L may play a key role in the initiation of dislocation. Fifth, we showed that proteasome activity is not required for dislocation in living cells, which is consistent with previous reports using in vitro reconstitution assays (63). This finding is supported by the fact that the drGFP signal continuously increases when proteasome activity is inhibited. We can envision that this new assay will be appropriate for studying several novel aspects of dislocation that are otherwise difficult or not possible to study with the current biochemical methods. For example, drGFP can be used to rapidly monitor and quantify dislocation in living cells. It specifically enables the study of the fate, localization, and solubility (aggregation) of dislocated substrates under physiological and pathological conditions. More importantly, it can be combined with large-scale RNAi and compound screens for proteins and chemical modulators of dislocation of a given specific substrate protein. This assay has the potential to be adopted for studying the dislocation of all luminal substrates and all membrane substrates that have at least one terminus (N or C terminus) in the ER lumen. The luminal terminus of membrane-spanning substrates can serve as the site for S11 tagging. However, as with other biological assays, the drGFP assay has limitations. For example, a ubiquitination site too close to the S11 tag may interfere with the reassembly of S11 and S1-10. Thus, one should be cautious when an unexplainable negative result is obtained with the drGFP assay. Substrate folding state could be another factor that affects the drGFP assay. Misfolded substrates may embed S11 and block GFP reassembly. However, this may not be a concern; increasing evidence indicates that substrates dislocate in unfolded or partially unfolded states (26, 36, 64, 65). In addition, S11 and S1-10 reassembly is time- and concentration-dependent, which could decrease the sensitivity of the drGFP assay for dislocation. Nonetheless, the present study indicates that the drGFP assay is a simple and reliable assay and should significantly expedite the study of dislocation in ERAD.
The strategy described in this article should be widely applicable to the study of other transmembrane transport of proteins in living cells, such as protein transport across the membranes of mitochondria, Golgi, peroxisomes and lysosome, as well as plasma and nuclear membranes. Moreover, it is well known that certain toxins, such as ricin and cholera toxin, and viruses, such as Simian Virus 40 (SV40), enter cells through endocytosis followed by vesicular transport to the lumen of the ER. These toxins and viruses then co-opt the dislocation process to reach the cytosol, where they exert their cytotoxicity or infect cells (12–16). We envision that dislocation of these toxins or viruses can be readily monitored in living cells using S11 fused with the toxins or with a viral surface protein. In addition, many proteins, such as the EGF receptor, ErbB2, Calreticulin, OS9, and Nrf1, can translocate from the ER to the nucleus when they function as transcriptional regulators and regulate important cellular events, such as tumorigenesis and oxidative defense (17–22). It is generally thought that these proteins first utilize the ERAD dislocation machinery to enter the cytosol and are then imported into the nucleus. However, the underlying mechanisms of this pathway have not yet been established. Application of the drGFP assay should significantly facilitate research in this area. More importantly, the drGFP assays for toxins, viruses and oncogenes, once established, could serve as a simple platform for high-throughput screening of small molecule inhibitors, facilitating dislocation-targeted drug discovery for these specific targets.
Supplementary Material
Acknowledgments
We thank Dr. Rebecca Heald for the generous gift of importazole, Dr. Mariusz Karbowski for access to microscope for live cell imaging, and Dr. Mervyn J. Monteiro for helpful discussions.
This work was supported by NSF Grant 1120833 (to S. F.).

This article contains supplemental Movies S1–S5 and Fig. S1.
- ER
- endoplasmic reticulum
- drGFP
- dislocation-dependent reconstituted GFP
- ERAD
- ER-associated degradation
- NHK
- null Hong Kong variant of α-1-antitrypsin
- ATM
- wild type α-1-antitrypsin
- PNGase F
- peptide: N-glycosidase F
- DBeQ
- N2,N4-dibenzylquinazoline-2,4-diamine
- IPZ
- importazole
- RFU
- relative fluorescence unit.
REFERENCES
- 1. Buchberger A., Bukau B., Sommer T. (2010) Protein quality control in the cytosol and the endoplasmic reticulum: brothers in arms. Mol. Cell 40, 238–252 [DOI] [PubMed] [Google Scholar]
- 2. Vembar S. S., Brodsky J. L. (2008) One step at a time: endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell Biol. 9, 944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hampton R. Y., Garza R. M. (2009) Protein quality control as a strategy for cellular regulation: lessons from ubiquitin-mediated regulation of the sterol pathway. Chem. Rev. 109, 1561–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Goldstein J. L., DeBose-Boyd R. A., Brown M. S. (2006) Protein sensors for membrane sterols. Cell 124, 35–46 [DOI] [PubMed] [Google Scholar]
- 5. Lu J. P., Wang Y., Sliter D. A., Pearce M. M., Wojcikiewicz R. J. (2011) RNF170 protein, an endoplasmic reticulum membrane ubiquitin ligase, mediates inositol 1,4,5-trisphosphate receptor ubiquitination and degradation. J. Biol. Chem. 286, 24426–24433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wiertz E. J., Jones T. R., Sun L., Bogyo M., Geuze H. J., Ploegh H. L. (1996) The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell 84, 769–779 [DOI] [PubMed] [Google Scholar]
- 7. Sommer T., Jentsch S. (1993) A protein translocation defect linked to ubiquitin conjugation at the endoplasmic reticulum. Nature 365, 176–179 [DOI] [PubMed] [Google Scholar]
- 8. Ward C. L., Omura S., Kopito R. R. (1995) Degradation of CFTR by the ubiquitin-proteasome pathway. Cell 83, 121–127 [DOI] [PubMed] [Google Scholar]
- 9. Jensen T. J., Loo M. A., Pind S., Williams D. B., Goldberg A. L., Riordan J. R. (1995) Multiple proteolytic systems, including the proteasome, contribute to CFTR processing. Cell 83, 129–135 [DOI] [PubMed] [Google Scholar]
- 10. McCracken A. A., Brodsky J. L. (1996) Assembly of ER-associated protein degradation in vitro: dependence on cytosol, calnexin, and ATP. J. Cell Biol. 132, 291–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hiller M. M., Finger A., Schweiger M., Wolf D. H. (1996) ER degradation of a misfolded luminal protein by the cytosolic ubiquitin-proteasome pathway. Science 273, 1725–1728 [DOI] [PubMed] [Google Scholar]
- 12. Tsai B., Rapoport T. A. (2002) Unfolded cholera toxin is transferred to the ER membrane and released from protein disulfide isomerase upon oxidation by Ero1. J. Cell Biol. 159, 207–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Spooner R. A., Watson P. D., Marsden C. J., Smith D. C., Moore K. A., Cook J. P., Lord J. M., Roberts L. M. (2004) Protein disulfide-isomerase reduces ricin to its A and B chains in the endoplasmic reticulum. Biochem. J. 383, 285–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yu M., Haslam D. B. (2005) Shiga toxin is transported from the endoplasmic reticulum following interaction with the luminal chaperone HEDJ/ERdj3. Infect. Immun. 73, 2524–2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kothe M., Ye Y., Wagner J. S., De Luca H. E., Kern E., Rapoport T. A., Lencer W. I. (2005) Role of p97 AAA-ATPase in the retrotranslocation of the cholera toxin A1 chain, a non-ubiquitinated substrate. J. Biol. Chem. 280, 28127–28132 [DOI] [PubMed] [Google Scholar]
- 16. Schelhaas M., Malmström J., Pelkmans L., Haugstetter J., Ellgaard L., Grünewald K., Helenius A. (2007) Simian Virus 40 depends on ER protein folding and quality control factors for entry into host cells. Cell 131, 516–529 [DOI] [PubMed] [Google Scholar]
- 17. Lin S. Y., Makino K., Xia W., Matin A., Wen Y., Kwong K. Y., Bourguignon L., Hung M. C. (2001) Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat. Cell Biol. 3, 802–808 [DOI] [PubMed] [Google Scholar]
- 18. Liao H. J., Carpenter G. (2007) Role of the Sec61 translocon in EGF receptor trafficking to the nucleus and gene expression. Mol. Biol. Cell 18, 1064–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang Y. N., Yamaguchi H., Huo L., Du Y., Lee H. J., Lee H. H., Wang H., Hsu J. M., Hung M. C. (2010) The translocon Sec61β localized in the inner nuclear membrane transports membrane-embedded EGF receptor to the nucleus. J. Biol. Chem. 285, 38720–38729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Afshar N., Black B. E., Paschal B. M. (2005) Retrotranslocation of the chaperone calreticulin from the endoplasmic reticulum lumen to the cytosol. Mol. Cell Biol. 25, 8844–8853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Baek J. H., Mahon P. C., Oh J., Kelly B., Krishnamachary B., Pearson M., Chan D. A., Giaccia A. J., Semenza G. L. (2005) OS-9 interacts with hypoxia-inducible factor 1alpha and prolyl hydroxylases to promote oxygen-dependent degradation of HIF-1α. Mol. Cell 17, 503–512 [DOI] [PubMed] [Google Scholar]
- 22. Steffen J., Seeger M., Koch A., Krüger E. (2010) Proteasomal degradation is transcriptionally controlled by TCF11 via an ERAD-dependent feedback loop. Mol. Cell 40, 147–158 [DOI] [PubMed] [Google Scholar]
- 23. Tiwari S., Weissman A. M. (2001) Endoplasmic reticulum (ER)-associated degradation of T cell receptor subunits. Involvement of ER-associated ubiquitin-conjugating enzymes (E2s). J. Biol. Chem. 276, 16193–16200 [DOI] [PubMed] [Google Scholar]
- 24. Nakatsukasa K., Huyer G., Michaelis S., Brodsky J. L. (2008) Dissecting the ER-associated degradation of a misfolded polytopic membrane protein. Cell 132, 101–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ernst R., Mueller B., Ploegh H. L., Schlieker C. (2009) The otubain YOD1 is a deubiquitinating enzyme that associates with p97 to facilitate protein dislocation from the ER. Mol. Cell 36, 28–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang Q., Liu Y., Soetandyo N., Baek K., Hegde R., Ye Y. (2011) A ubiquitin ligase-associated chaperone holdase maintains polypeptides in soluble states for proteasome degradation. Mol. Cell 42, 758–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Greenblatt E. J., Olzmann J. A., Kopito R. R. (2011) Derlin-1 is a rhomboid pseudoprotease required for the dislocation of mutant α-1 antitrypsin from the endoplasmic reticulum. Nat. Struct. Mol. Biol. 18, 1147–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nakatsukasa K., Brodsky J. L. (2008) The recognition and retrotranslocation of misfolded proteins from the endoplasmic reticulum. Traffic 9, 861–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mueller B., Klemm E. J., Spooner E., Claessen J. H., Ploegh H. L. (2008) SEL1L nucleates a protein complex required for dislocation of misfolded glycoproteins. Proc. Natl. Acad. Sci. U.S.A. 105, 12325–12330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jo Y., Sguigna P. V., DeBose-Boyd R. A. (2011) Membrane-associated ubiquitin ligase complex containing gp78 mediates sterol-accelerated degradation of 3-hydroxy-3-methylglutaryl-coenzyme A reductase. J. Biol. Chem. 286, 15022–15031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Christianson J. C., Olzmann J. A., Shaler T. A., Sowa M. E., Bennett E. J., Richter C. M., Tyler R. E., Greenblatt E. J., Wade Harper J., Kopito R. R. (2011) Defining human ERAD networks through an integrative mapping strategy. Nat. Cell Biol. 14, 93–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhong Y., Wang Y., Yang H., Ballar P., Lee J. G., Ye Y., Monteiro M. J., Fang S. (2011) Importin β interacts with the endoplasmic reticulum-associated degradation machinery and promotes ubiquitination and degradation of mutant α1-antitrypsin. J. Biol. Chem. 286, 33921–33930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Carvalho P., Goder V., Rapoport T. A. (2006) Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell 126, 361–373 [DOI] [PubMed] [Google Scholar]
- 34. Denic V., Quan E. M., Weissman J. S. (2006) A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell 126, 349–359 [DOI] [PubMed] [Google Scholar]
- 35. Bernasconi R., Galli C., Calanca V., Nakajima T., Molinari M. (2010) Stringent requirement for HRD1, SEL1L, and OS-9/XTP3-B for disposal of ERAD-LS substrates. J. Cell Biol. 188, 223–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bagola K., Mehnert M., Jarosch E., Sommer T. (2011) Protein dislocation from the ER. Biochim. Biophys. Acta 1808, 925–936 [DOI] [PubMed] [Google Scholar]
- 37. Eura Y., Yanamoto H., Arai Y., Okuda T., Miyata T., Kokame K. (2012) Derlin-1 deficiency is embryonic lethal, Derlin-3 deficiency appears normal, and Herp deficiency is intolerant to glucose load and ischemia in mice. PLoS One 7, e34298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Apostolou A., Shen Y., Liang Y., Luo J., Fang S. (2008) Armet, a UPR-upregulated protein, inhibits cell proliferation and ER stress-induced cell death. Exp. Cell Res. 314, 2454–2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ballar P., Shen Y., Yang H., Fang S. (2006) The role of a novel p97/valosin-containing protein-interacting motif of gp78 in endoplasmic reticulum-associated degradation. J. Biol. Chem. 281, 35359–35368 [DOI] [PubMed] [Google Scholar]
- 40. Soderholm J. F., Bird S. L., Kalab P., Sampathkumar Y., Hasegawa K., Uehara-Bingen M., Weis K., Heald R. (2011) Importazole, a small molecule inhibitor of the transport receptor importin-β. ACS Chem. Biol. 6, 700–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lorenz H., Hailey D. W., Wunder C., Lippincott-Schwartz J. (2006) The fluorescence protease protection (FPP) assay to determine protein localization and membrane topology. Nat. Protoc. 1, 276–279 [DOI] [PubMed] [Google Scholar]
- 42. Li M., Brooks C. L., Wu-Baer F., Chen D., Baer R., Gu W. (2003) Mono- versus polyubiquitination: differential control of p53 fate by Mdm2. Science 302, 1972–1975 [DOI] [PubMed] [Google Scholar]
- 43. Ballar P., Zhong Y., Nagahama M., Tagaya M., Shen Y., Fang S. (2007) Identification of SVIP as an endogenous inhibitor of endoplasmic reticulum-associated degradation. J. Biol. Chem. 282, 33908–33914 [DOI] [PubMed] [Google Scholar]
- 44. Cabantous S., Terwilliger T. C., Waldo G. S. (2005) Protein tagging and detection with engineered self-assembling fragments of green fluorescent protein. Nat. Biotechnol. 23, 102–107 [DOI] [PubMed] [Google Scholar]
- 45. Hosokawa N., Tremblay L. O., You Z., Herscovics A., Wada I., Nagata K. (2003) Enhancement of endoplasmic reticulum (ER) degradation of misfolded Null Hong Kong alpha1-antitrypsin by human ER mannosidase I. J. Biol. Chem. 278, 26287–26294 [DOI] [PubMed] [Google Scholar]
- 46. Wu Y., Swulius M. T., Moremen K. W., Sifers R. N. (2003) Elucidation of the molecular logic by which misfolded α1-antitrypsin is preferentially selected for degradation. Proc. Natl. Acad. Sci. U.S.A. 100, 8229–8234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hebert D. N., Bernasconi R., Molinari M. (2010) ERAD substrates: which way out? Semin. Cell Dev. Biol. 21, 526–532 [DOI] [PubMed] [Google Scholar]
- 48. Suzuki T., Park H., Lennarz W. J. (2002) Cytoplasmic peptide:N-glycanase (PNGase) in eukaryotic cells: occurrence, primary structure, and potential functions. FASEB J. 16, 635–641 [DOI] [PubMed] [Google Scholar]
- 49. Blobel G. (1980) Intracellular protein topogenesis. Proc. Natl. Acad. Sci. U.S.A. 77, 1496–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Brodsky J. L., Hamamoto S., Feldheim D., Schekman R. (1993) Reconstitution of protein translocation from solubilized yeast membranes reveals topologically distinct roles for BiP and cytosolic Hsc70. J. Cell Biol. 120, 95–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sun Z. Y., Kim S. T., Kim I. C., Fahmy A., Reinherz E. L., Wagner G. (2004) Solution structure of the CD3ϵδ ectodomain and comparison with CD3ϵγ as a basis for modeling T cell receptor topology and signaling. Proc. Natl. Acad. Sci. U.S.A. 101, 16867–16872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhong X., Shen Y., Ballar P., Apostolou A., Agami R., Fang S. (2004) AAA ATPase p97/valosin-containing protein interacts with gp78, a ubiquitin ligase for endoplasmic reticulum-associated degradation. J. Biol. Chem. 279, 45676–45684 [DOI] [PubMed] [Google Scholar]
- 53. Kikkert M., Doolman R., Dai M., Avner R., Hassink G., van Voorden S., Thanedar S., Roitelman J., Chau V., Wiertz E. (2004) Human HRD1 is an E3 ubiquitin ligase involved in degradation of proteins from the endoplasmic reticulum. J. Biol. Chem. 279, 3525–3534 [DOI] [PubMed] [Google Scholar]
- 54. Ye Y., Shibata Y., Kikkert M., van Voorden S., Wiertz E., Rapoport T. A. (2005) Recruitment of the p97 ATPase and ubiquitin ligases to the site of retrotranslocation at the endoplasmic reticulum membrane. Proc. Natl. Acad. Sci. U.S.A. 102, 14132–14138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Schulze A., Standera S., Buerger E., Kikkert M., van Voorden S., Wiertz E., Koning F., Kloetzel P. M., Seeger M. (2005) The ubiquitin-domain protein HERP forms a complex with components of the endoplasmic reticulum associated degradation pathway. J. Mol. Biol. 354, 1021–1027 [DOI] [PubMed] [Google Scholar]
- 56. Christianson J. C., Shaler T. A., Tyler R. E., Kopito R. R. (2008) OS-9 and GRP94 deliver mutant α1-antitrypsin to the Hrd1-SEL1L ubiquitin ligase complex for ERAD. Nat. Cell Biol. 10, 272–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Fang S., Ferrone M., Yang C., Jensen J. P., Tiwari S., Weissman A. M. (2001) The tumor autocrine motility factor receptor, gp78, is a ubiquitin protein ligase implicated in degradation from the endoplasmic reticulum. Proc. Natl. Acad. Sci. U.S.A. 98, 14422–14427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hosokawa N., Wada I., Nagasawa K., Moriyama T., Okawa K., Nagata K. (2008) Human XTP3-B forms an endoplasmic reticulum quality control scaffold with the HRD1-SEL1L ubiquitin ligase complex and BiP. J. Biol. Chem. 283, 20914–20924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bays N. W., Hampton R. Y. (2002) Cdc48-Ufd1-Npl4: stuck in the middle with Ub. Curr. Biol. 12, R366–71 [DOI] [PubMed] [Google Scholar]
- 60. Tsai B., Ye Y., Rapoport T. A. (2002) Retro-translocation of proteins from the endoplasmic reticulum into the cytosol. Nat. Rev. Mol. Cell Biol. 3, 246–255 [DOI] [PubMed] [Google Scholar]
- 61. Chou T. F., Brown S. J., Minond D., Nordin B. E., Li K., Jones A. C., Chase P., Porubsky P. R., Stoltz B. M., Schoenen F. J., Patricelli M. P., Hodder P., Rosen H., Deshaies R. J. (2011) Reversible inhibitor of p97, DBeQ, impairs both ubiquitin-dependent and autophagic protein clearance pathways. Proc. Natl. Acad. Sci. U.S.A. 108, 4834–4839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Carvalho P., Stanley A. M., Rapoport T. A. (2010) Retrotranslocation of a misfolded luminal ER protein by the ubiquitin-ligase Hrd1p. Cell 143, 579–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lee R. J., Liu C. W., Harty C., McCracken A. A., Latterich M., Römisch K., DeMartino G. N., Thomas P. J., Brodsky J. L. (2004) Uncoupling retro-translocation and degradation in the ER-associated degradation of a soluble protein. EMBO J. 23, 2206–2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Nishikawa S. I., Fewell S. W., Kato Y., Brodsky J. L., Endo T. (2001) Molecular chaperones in the yeast endoplasmic reticulum maintain the solubility of proteins for retrotranslocation and degradation. J. Cell Biol. 153, 1061–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Claessen J. H., Ploegh H. L. (2011) BAT3 guides misfolded glycoproteins out of the endoplasmic reticulum. PLoS One 6, e28542. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.