Background: Activin receptor-like kinase 1 (ALK1) is an important regulator of normal blood vessel formation and pathological tumor angiogenesis.
Results: Crystal structure of ALK1-BMP9-ACTRIIB signaling complex and kinetic and thermodynamic properties of receptor-ligand interactions are described.
Conclusions: ALK1 achieves high specificity for BMP9/10 through unusual receptor positioning and unique receptor-ligand interface.
Significance: Structural and mechanistic insights into ALK1 signaling provide a framework for novel anti-angiogenic therapies.
Keywords: Angiogenesis, Bone Morphogenetic Protein (BMP), Crystal Structure, Surface Plasmon Resonance (SPR), Transforming Growth Factor β (TGFβ), ALK1, BMP10, BMP9
Abstract
Activin receptor-like kinase 1 (ALK1), an endothelial cell-specific type I receptor of the TGF-β superfamily, is an important regulator of normal blood vessel development as well as pathological tumor angiogenesis. As such, ALK1 is an important therapeutic target. Thus, several ALK1-directed agents are currently in clinical trials as anti-angiogenic cancer therapeutics. Given the biological and clinical importance of the ALK1 signaling pathway, we sought to elucidate the biophysical and structural basis underlying ALK1 signaling. The TGF-β family ligands BMP9 and BMP10 as well as the three type II TGF-β family receptors ActRIIA, ActRIIB, and BMPRII have been implicated in ALK1 signaling. Here, we provide a kinetic and thermodynamic analysis of BMP9 and BMP10 interactions with ALK1 and type II receptors. Our data show that BMP9 displays a significant discrimination in type II receptor binding, whereas BMP10 does not. We also report the crystal structure of a fully assembled ternary complex of BMP9 with the extracellular domains of ALK1 and ActRIIB. The structure reveals that the high specificity of ALK1 for BMP9/10 is determined by a novel orientation of ALK1 with respect to BMP9, which leads to a unique set of receptor-ligand interactions. In addition, the structure explains how BMP9 discriminates between low and high affinity type II receptors. Taken together, our findings provide structural and mechanistic insights into ALK1 signaling that could serve as a basis for novel anti-angiogenic therapies.
Introduction
TGF-β superfamily proteins are potent determinants of cell fate in metazoans. In mammals this family consists of more than 30 ligands, including bone morphogenetic proteins (BMP),5 growth and differentiation factors (GDF), activins, seven type I receptors (ALK1–7), and five type II receptors (ActRIIA, ActRIIB, BMPRII, TGFβRII, and AMHRII), as well as several co-receptors and accessory proteins (1). TGF-β family proteins commonly act by forming a ternary signaling complex that comprises dimeric ligand, two type I receptors, and two type II receptors. According to currently held views, signaling complexes can form in two ways: either the ligand binds first with high affinity to the type II receptor followed by recruitment of the type I receptor, as is generally observed for TGF-β and activin receptors (2, 3), or the ligand binds first with high affinity to the type I receptor, followed by type II receptor recruitment, as is typical of BMP receptors (4, 5). In either case, ligand binding leads to trans-phosphorylation of the type I receptor kinase domain by the apposed, constitutively active type II receptor kinase domain. Type I receptor phosphorylation then activates SMAD signal transduction pathways to regulate expression of target genes (1).
A remarkable feature of the TGF-β superfamily is the broad specificity possessed by most of its members, which results in a great combinatorial complexity of partnering between ligands, type I and type II receptors. For example, BMP2 has been shown to bind two type I receptors (ALK3, ALK6) with comparable affinities and three type II receptors (BMPRII, ActRIIA, and ActRIIB) also with comparable affinities (6, 7). Similarly, multiple ligands, including activin A and B, GDF8 and GDF11, bind to ActRIIB with comparable affinities (8). An interesting exception to this general pattern is the type I receptor ALK1, which seems to have a strikingly discriminate relationship with BMP9 and BMP10. Only ALK1, but not other type I receptors, binds these two ligands with high affinity in vitro (9). Conversely, BMP9 and BMP10, which exhibit 65% sequence identity at the protein level, appear to signal exclusively via ALK1 in endothelial cells (10–12), although published data suggest that they could utilize other low affinity type I receptors in other cell types where ALK1 is not expressed. In addition, it is unclear which type II receptors act in combination with ALK1/BMP9 and ALK1/BMP10. Based on chemical cross-linking and limited in vitro experiments BMPRII, ActRIIA, and ActRIIB have all been implicated in BMP9 signaling (10–13), but these interactions have not been analyzed systematically; even less is known about BMP10 type II receptor utilization.
ALK1 is predominantly expressed in vascular endothelial cells (14). Mutations in the ALK1 gene have been shown to cause hereditary hemorrhagic telangiectasia type 2 (HHT2), a vascular disorder characterized by abnormal capillaries, recurrent nosebleeds, mucocutaneous telangiectasias, and arteriovenous malformations in the brain, lungs, liver, and gastrointestinal tract (15). Multiple lines of genetic, pharmacological, and histopathological evidence support a critical role for ALK1 signaling in the regulation of blood vessel formation in tumors as well as normal tissues (16–19), explaining the growing interest in ALK1 as a therapeutic target to complement existing anti-angiogenesis treatments. Currently, a soluble ALK1 ligand trap (Acceleron Pharma) and an anti-ALK1 antibody (Pfizer) are in clinical trials for treatment of advanced solid tumors (20).
Given the biological and clinical importance of the ALK1 pathway, we sought to elucidate the molecular and biophysical basis of ALK1 complex formation. Here, we provide a kinetic and thermodynamic characterization of BMP9 and BMP10 binding to ALK1 and different type II receptors. Our data show that BMP9 displays a significant discrimination in type II receptor binding, whereas BMP10 does not. Importantly, we present the crystal structure of a fully assembled high-affinity complex between BMP9 and the extracellular domains of the type I receptor ALK1 and the type II receptor ActRIIB refined at 3.35 Å. The structure reveals the molecular determinants of ALK1 selectivity for BMP9 and BMP10, and the comparatively discriminate binding of BMP9 to its high affinity type II receptor. Last, we map extracellular HHT2 mutations to the ALK1 structure to explain the structural mechanism of HHT2 pathogenesis.
EXPERIMENTAL PROCEDURES
Construction of Expression Vectors
The expression constructs pAID4T ALK1.EK.Fc and pAID4T ActRIIB.EK.Fc contained the extracellular domain (ECD) of human ALK1 (residues 22–118) and ActRIIB (residues 19–134), respectively, followed by an enterokinase (EK) cleavage motif fused to the human IgG1 Fc domain. hBMP9 cDNA harboring the tissue plasminogen activator signal peptide was cloned into the pAID4 vector and transfected into a CHO DUKX B11 cell line engineered to express soluble PACE/Furin to facilitate propeptide cleavage.
Protein Expression and Purification
ALK1.EK.Fc and ActRIIB.EK.Fc were expressed by Gala Biotech using GnTI-deficient HEK293S cells (21). The conditioned media was purified over a MabSelect SuRe column (GE Healthcare). Purified ALK1 and ActRIIB ECDs were digested with enterokinase (prepared in house), treated with Endo Hf (New England Biolabs), and further purified on a Superdex S200 (GE Healthcare). A proprietary receptor-Fc fusion protein was used to prepare an affinity resin for BMP9 purification. BMP9-conditioned media was concentrated 10-fold before affinity purification. BMP9 was eluted from the affinity resin with 0.1 m glycine (pH 3.0) and further purified by reverse phase HPLC.
Ternary Complex Formation and Crystallization
To form the ternary complex, BMP9 (2 mg/ml) was combined with ALK1ECD (4.3 mg/ml) and ActRIIBECD (1.94 mg/ml) at a molar ratio of 1:3:3 in 50 mm HEPES, pH 7.5, 150 mm NaCl buffer containing 1% CHAPS. The complex was treated with carboxypeptidase B and Y (Worthington Biochemical). A 1:2:2 complex was isolated by SEC on a Superdex S75 (GE Healthcare) column. Trigonal crystals grew in 1–2 weeks at 20 °C in 20% PEG 3350, 0.2 m sodium malonate, and 0.1 m BisTris propane, pH 6.5.
Structure Determination
A 3.35-Å dataset was collected in-house (Rigaku FR-E+ SuperBrightTM/Cu anode) at 100 K on a single crystal equilibrated in mother liquor containing 15% ethylene glycol (for processing and refinement statistics, see supplemental Table S1). A MR solution, containing six ternary complexes in the asymmetric unit, was found with PHASER (22) using crystal structures of BMP9 (PDB 1ZKZ) (13), ActRIIB (PDB 2H62) (23), and an in-house structure of ALK16 as search models. The model was refined using PHENIX (22), applying NCS restraints to all equivalent residues. Manual building was performed in COOT (24). Electron density was continuous for all protomers, with the only disordered regions at the N and C terminus. No waters were picked in the final model, because of the resolution. The final structure has an Rfree of 26.1% and Rcryst of 21.9%. Ramachandran analysis by MolProbity (25) indicates 88% of residues reside in the most favorable regions. Contact maps and buried surface area values were calculated using the Protein Interfaces, Surfaces, and Assemblies (PISA) server (26). Structural figures were prepared using PyMOL (27). The atomic coordinates and structure factor amplitudes for the ALK1ECD/BMP9/ActRIIBECD structure have been deposited in the Protein Data Bank (PDB ID code 4FAO).
Surface Plasmon Resonance Analysis
Experiments were performed using a Biacore T100 biosensor (Biacore/GE Healthcare) essentially as described (28). Briefly, goat anti-human Fc-specific antibody was immobilized onto a Series S research grade CM5 sensor chip (Biacore/GE Healthcare) using standard amino-coupling chemistry. Receptor-Fc fusions were captured on the flow cell at a density of ∼100 Resonance units. For kinetic analysis at 25 and 37 °C, a concentrated series of BMP9 or BMP10 (RnD) were injected in duplicates or triplicates over the flow cells at a flow rate of 50 μl/ml. To obtain kinetic rate constants the corrected data were fit to a 1:1 interaction model using BiaEvaluation software (GE Healthcare). The equilibrium binding constant KD was determined by the ratio of binding rate constants kd/ka.
RESULTS
BMP9 Exhibits Strikingly Different Binding Kinetics and Affinities for Type II Receptors
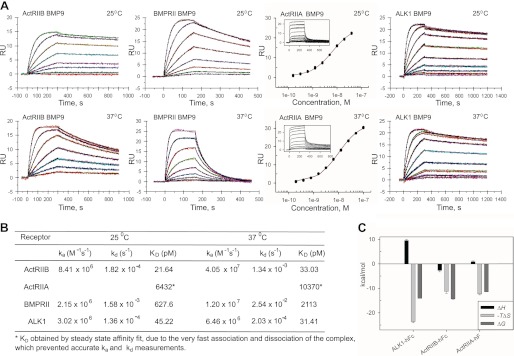
Previous studies have suggested that BMP9 signaling can occur via several type II receptors, namely BMPRII, ActRIIA, and ActRIIB (10–13); however, BMP9-receptor interactions have not been measured directly and thus the importance of a particular receptor interaction is difficult to ascertain. We therefore determined the relative affinities of BMP9 for its type II receptors using surface plasmon resonance (SPR). We captured ActRIIBECD-Fc, ActRIIAECD-Fc, and BMPRIIECD-Fc in separate trials on a CM5 chip with immobilized anti-hFc IgG to mimic the dimeric nature of the receptors on the cell surface and exposed each type II receptor to a concentration series of BMP9 at 25 °C. Our results reveal that BMP9 binds ActRIIB with ∼30-fold higher affinity (KD of 21.6 pm) than BMPRII (KD 627.6 pm) (Fig. 1). Interestingly, the BMP9 interaction with ActRIIA is ∼300-fold weaker (KD 6.43 nm) than with ActRIIB and is characterized by very fast association and dissociation of the complex. Other known type II receptors, TGFβRII and anti-Müllerian hormone type II receptor (AMHRII), did not show any binding to BMP9 at a concentration of 100 nm (data not shown). We repeated the experiment at 37 °C and confirmed the overall trend in receptor specificity at physiological temperature (Fig. 1). Notably, BMP9/ActRIIB binding was least affected (KD = 33.0 pm) by temperature, confirming the high specificity of this interaction. At 37 °C, the BMPRII/BMP9 interaction was ∼64-fold weaker (KD = 2.11 nm) compared with ActRIIB, mainly due to a significant decrease in receptor-ligand complex stability.
FIGURE 1.
Kinetic and thermodynamic analysis of BMP9 binding to ALK1, ActRIIB, ActRIIA, and BMPRII. A, SPR sensograms of ALK1ECD-Fc, ActRIIBECD-Fc, ActRIIAECD-Fc, and BMPRIIECD-Fc binding to BMP9 at 25 and 37 °C. Raw data (colored lines) were overlaid with a global fit to a 1:1 model with mass transport limitations (black lines). ActRIIAECD-Fc data were analyzed using affinity fit in SigmaPlot. B, kinetic parameters for receptor-ligand interactions. C, comparison of thermodynamic parameters of ALK1ECD-Fc, ActRIIBECD-Fc, and ActRIIAECD-Fc interactions with BMP9 at 25 °C.
BMP10 Exhibits Comparable Binding Kinetics and Affinities for Type II Receptors
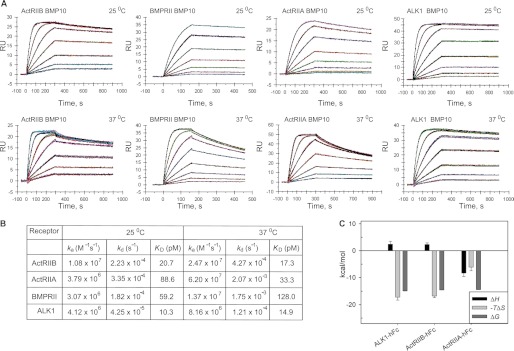
Because little is known about which type II receptors are involved in BMP10 signaling, we investigated the selectivity profile of BMP10 for type II receptors, in part to compare BMP10 selectivity to that of BMP9. We first screened known type II receptors (ActRIIA, ActRIIB, BMPRII, TGFβRII, and AMHRII) for binding to BMP10 using SPR. Whereas TGFβRII and AMHRII did not display any binding to BMP10 as expected (data not shown), all three receptors previously shown to bind BMP9 also bound BMP10. We therefore performed a detailed kinetic analysis of BMP10 interactions with ActRIIB, ActRIIA, and BMPRII, at 25 and 37 °C (Fig. 2). In contrast to BMP9, we found that BMP10 did not have a distinct preference for one of these three type II receptors at 25 °C, as these receptors bound BMP10 with very similar kinetics and high affinities (KD = 20.7, 88.6, and 59.2 pm, respectively). However, analysis of these interactions at 37 °C suggested that, at physiological temperature, BMP10 exhibits a slight preference for ActRIIB and ActRIIA (KD = 17.3 and 33.3 pm, respectively) over BMPRII. This is due mainly to the greater stability of the BMP10-ActRIIB complex and faster rate of BMP10-ActRIIA complex formation.
FIGURE 2.
Kinetic and thermodynamic analysis of BMP10 binding to ALK1, ActRIIB, ActRIIA, and BMPRII. A, SPR sensograms of ALK1ECD-Fc, ActRIIBECD-Fc, ActRIIAECD-Fc, and BMPRIIECD-Fc binding to BMP10 at 25 and 37 °C. B, kinetic parameters for receptor-ligand interactions. C, comparison of thermodynamic parameters of ALK1ECD-Fc, ActRIIBECD-Fc, and ActRIIAECD-Fc interactions with BMP10 at 25 °C.
Kinetic and Thermodynamic Analysis of Receptor Discrimination
To permit a direct comparison of affinity measurements between the type I and II receptors, we also performed the SPR assay with captured ALK1ECD-Fc at 25 and 37 °C. Our results (Figs. 1 and 2) show that ALK1 binds BMP10 with ∼2–4-fold better affinity than BMP9 (KD = 10.3 pm for BMP10 and 45.2 pm for BMP9), which correlates well with previously published data (9). The temperature dependence of ligand binding to ALK1 was negligible, as shown by similar equilibrium binding constants at 37 °C (KD = 31.4 pm for BMP9 and 14.8 pm for BMP10) (Figs. 1B and 2B). In summary, our kinetic data reveal that BMP9 and BMP10 bind with picomolar affinities to both type I receptor ALK1 and type II receptor ActRIIB.
To investigate whether receptor discrimination is thermodynamically driven, we performed a thermodynamic characterization of BMP9 and BMP10 binding to ALK1 and ActRIIB by SPR on a Biacore T100 (see supplemental “Experimental Procedures” and Fig. S1) using the same receptor-Fc capture format described above. Thermodynamic parameters indicate that the free energies of binding (ΔG0) for BMP9 and BMP10 to their cognate type I and II receptors are almost identical within experimental error: −14.11 ± 0.02 kcal mol−1 for BMP9/ALK1 and −14.30 ± 0.30 kcal mol−1 for BMP9/ActRIIB; −14.88 ± 0.05 kcal mol−1 for BMP10/ALK1 and −14.52 ± 0.05 kcal mol−1 for BMP10/ActRIIB (Figs. 1C and 2C, and Table 1). Thus, there is no thermodynamic preference for the order in which either ligand (BMP9 or BMP10) binds type I receptor (ALK1) versus type II receptor (ActRIIB) in vitro.
TABLE 1.
Comparison of thermodynamic parameters (ΔH0, −TΔS0, ΔG0, and ΔCp) for receptor-ligand interactions at 25 °C
The confidence limits age given for a confidence probability of 0.95.
| ΔH0a | −TΔS0 | ΔG0a | ΔCpb | SFEc | |
|---|---|---|---|---|---|
| kcal mol−1 | kcal mol−1 | kcal mol−1 | kcal mol−1K−1 | ||
| ALK1-hFc/BMP9 | 9.5 ± 0.5 | −23.6 ± 0.5 | −14.11 ± 0.02 | 0.0 | 0.124 |
| ALK1-hFc/BMP10 | 2.4 ± 1.1 | −17.2 ± 1.1 | −14.88 ± 0.05 | 0.0 | 0.158 |
| ActRIIB-hFc/BMP9 | −2.7 ± 0.6 | −11.3 ± 0.8 | −14.30 ± 0.30 | −1.5 ± 0.1 | 0.102 |
| ActRIIB-hFc/BMP10 | 2.3 ± 0.6 | −16.8 ± 0.6 | −14.52 ± 0.03 | 0.0 | 0.087 |
| ActRIIA-hFc/BMP9 | 0.9 ± 0.3 | −12.3 ± 0.4 | −11.42 ± 0.02 | −0.47 ± 0.04 | 0.062 |
| ActRIIA-hFc/BMP10 | −8.3 ± 1.3 | −6.1 ± 1.4 | −14.40 ± 0.10 | −1.3 ± 0.2 | 0.103 |
a Calculated.
b Experimental.
c Standard fit error.
Structure of the Ternary Complex of ALK1ECD-BMP9-ActRIIBECD
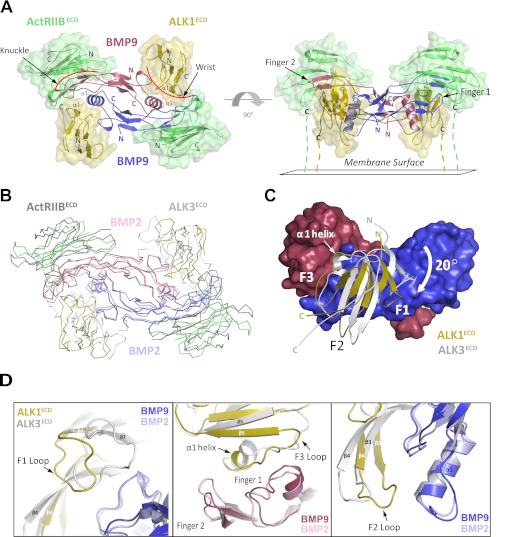
We determined the crystal structure of the high-affinity BMP9-ALK1ECD-ActRIIBECD complex at 3.35-Å resolution (see “Experimental Procedures” and supplemental Table S1). The choice of ActRIIB as the type II receptor in the complex was based on its high affinity for BMP9 in our SPR assays and not on the prevalence of specific receptor combinations in vivo. The structure is composed of one BMP9 dimer, two ALK1ECD molecules, and two ActRIIBECD molecules. The overall assembly of the complex resembles that of other reported BMP-receptor ternary complexes (6, 29), with the BMP9 homodimer located in the center of the complex, and two alternating molecules of ALK1 and ActRIIB bound on the outside (Fig. 3A). Specifically, one ActRIIB molecule binds on the outside convex face of each BMP9 protomer. Two antiparallel β-sheets on BMP9 (fingers 1 and 2, also known as the “knuckle” region) mediate binding to a concave hydrophobic patch on ActRIIB (6) (Fig. 3A). The ALK1 interface is formed through recognition of both BMP9 protomers on the “wrist” (6), with the majority of binding occurring through packing of the ALK1 α1 helix against the pre-helix loop and α1 helix of BMP9 (Fig. 3A). Consistent with other BMP ternary complexes, the receptors make no direct contact with one another.
FIGURE 3.
Structure of the ALK1ECD-BMP9-ActRIIBECD ternary complex. A, the ternary complex comprised of one BMP9 homodimer (red/blue), two ALK1ECD receptors (gold), and two ActRIIBECD receptors (green). The complex is shown as viewed from above (left) and from the side with the membrane below (right). B, superposition with the BMP2-ALK3ECD-ActRIIBECD ternary complex (shown in ribbon) via the ligands reveals differences in receptor positioning (BMP2 homodimer colored pink/light blue; ALK3ECD and ActRIIBECD receptors colored light gray and dark gray, respectively; ALK1ECD-BMP9-ActRIIBECD complex colored as in panel A). C, superposition of ALK1ECD and ALK3ECD at the BMP9 interface (shown in surface representation) reveals a 20° rotation of the ALK1 protomer pivoted around helix α1. D, close-up view of helix α1, F1, F2, and F3 loops from ALKECD and ALK3ECD (shown as schematic). Repositioning of these elements in ALK1 results in a novel set of receptor-ligand interactions.
Because there are multiple copies of the ternary complex in this crystal structure, we also observe a higher ordered assembly composed of a dimer of ternary complexes (supplemental Fig. S2). Dimerization occurs between ActRIIB molecules on adjacent ternary complexes, facilitating the formation of a large ring-shaped structure. Interaction at the dimer interface is mediated predominantly through Asn24, which is glycosylated and makes glycan-mediated contacts with the opposing ActRIIB protomer. Although not reported, the same dimerization is evident in the ternary structure of BMP2-ALK3ECD-ActRIIAECD upon analysis of crystal packing (PDB code 2G00) (29). Other published ternary structures within the family are not glycosylated, and thus do not exhibit this arrangement.
ALK1 Receptor Orientation Shapes a Novel Interaction Surface
Despite similarities in overall architecture, there are clear differences between the structure presented here and the structures of homologous BMP complexes determined previously. Superposition with the BMP2/ALK3ECD/ActRIIBECD ternary structure (PDB code 2H64) (23) via one ligand protomer (Fig. 3B, red protomer) shows that ALK1 is significantly displaced relative to ALK3 with Cα root mean squares differences of 3.74 and 4.31 Å for protomers C (61 atoms) and D (63 atoms), respectively (Fig. 3B). The receptors align closest along helix α1, although the helix is shifted by as much as 3.5 Å in ALK1 compared with equivalent residues on ALK3 (Fig. 3, C and D). This shift is offset partly by a parallel rigid body movement in BMP9, where the tips of fingers 1 and 2 shift by about 2.5 Å relative to BMP2 (Fig. 3D). Thus, ALK1 pivots around helix α1 by about 20° relative to ALK3. This rotation repositions several key structural elements (Fig. 3, C and D), notably the F2 loop (residues 57–63), which shifts by as much as 6 Å to lie within contacting distance of BMP9. This is of particular note, as the F2 loop does not participate in ligand recognition in other type I receptor complexes. The F1 loop (residues 36–44) also rearranges significantly, by as much as 9 Å, relative to ALK3 (Fig. 3, C and D), but in this case the effect is opposite; the loop moves away from the ligand interface and only loosely contacts BMP9. Alignment of this complex with the BMP2/ALK3ECD/ActRIIAECD ternary structure (PDB code 2G00) (29) and GDF5/ALK6ECD binary structure (PDB code 3EVS) (30) shows a similar trend in ALK1 orientation, validating expectations that the ALK1-ligand interface diverges from other type I receptor-BMP interfaces. The changes in ALK1 orientation therefore contribute to a unique set of receptor-ligand interactions.
ActRIIB also shifts relative to the BMP2/ALK3ECD/ActRIIBECD structure, but here a rigid body movement in fingers 1 and 2 of BMP9 mirrors the shift (Fig. 3B). Thus, ActRIIB is able to maintain canonical receptor-ligand contacts. Last, there are no significant conformational changes in BMP9 upon complex formation. Similar to BMP2, the BMP9 homodimer binds as a rigid unit, with a Cα root mean square difference of 1.01 Å compared with the apo structure (13). This is in contrast to activin, which is a highly flexible molecule that has been crystallized in multiple conformations (31, 32), and GDF5, which undergoes an induced conformational change upon receptor binding (30).
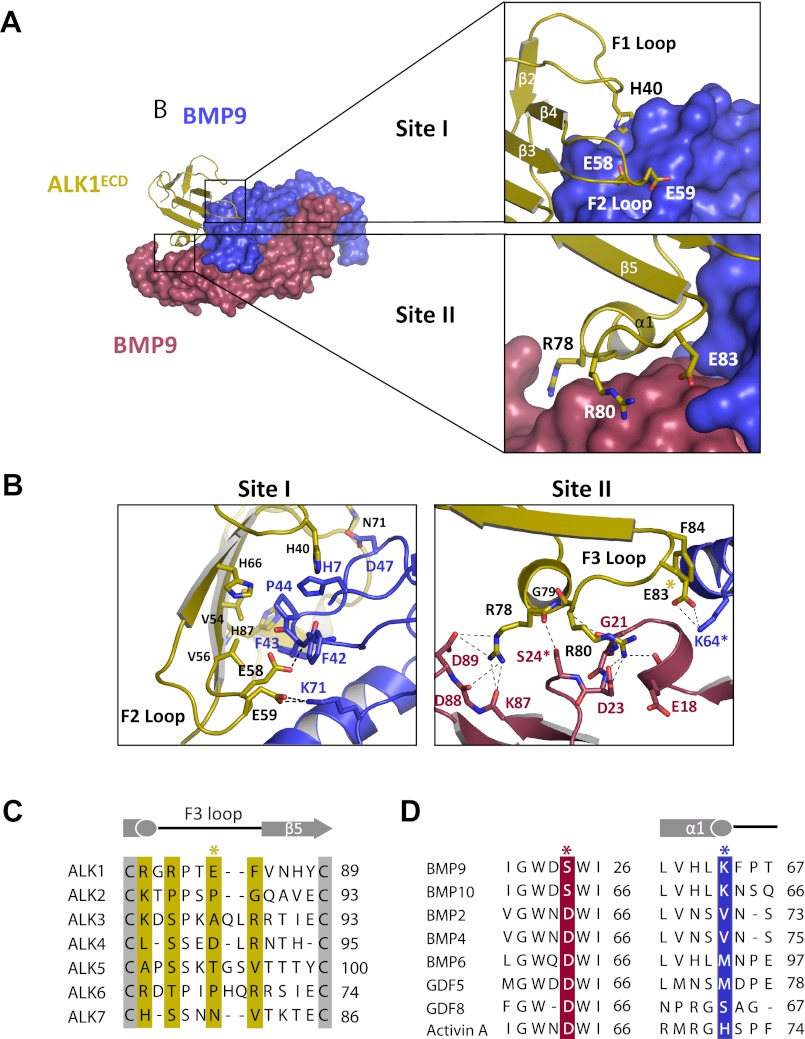
The BMP9-ALK1 Binding Interface
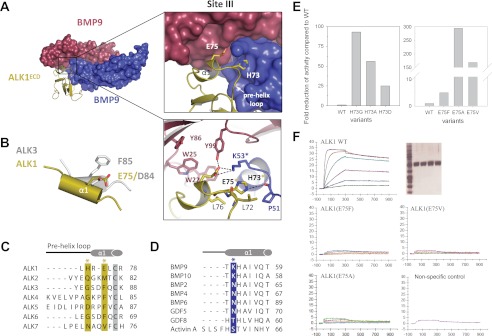
In the complex described here, one ALK1 molecule contacts two BMP9 protomers. The binding interface is broadly separated into three sites (sites I, II, and III) (supplemental Table S2). Sites I and III overlap with those of other type I receptor-ligand complexes, whereas site II is unique to ALK1.
The interface at site I comprises BMP9 residues that contribute to the pre-helix loop (Phe42, Phe43, and Pro44) and together with His7, His62, and Leu63 form a hydrophobic interface with ALK1 residues His40, Val54, Val56, Arg57, His66, and His87 (Fig. 4). Phe43 and Pro44 are conserved in BMP2 (Phe49, Pro50) and GDF5 (Phe54, Pro55) and form similar hydrophobic contacts with ALK3 and ALK6. Distinctively, residues in the F2 loop, including Glu58 and Glu59, directly contact the ligand (Fig. 4B), whereas the F2 loop does not participate in recognition in the BMP2-ALK3 or GDF5-ALK6 complexes, as Glu58 and Glu59 are replaced by the shorter amino acid aspartate in both ALK3 (Asp66 and Asp67) and ALK6 (Asp46 and Asp47) (supplemental Fig. S3). Thus, in ALK3 and ALK6, the F2 loop is too far away to mediate any ligand interactions, suggesting that the F2 loop may play a role in conferring specificity to the ALK1-BMP9 complex.
FIGURE 4.
Site I and II interactions at the ALK1/BMP9 interface. A, close-up of the interaction surfaces, with the color scheme retained from Fig. 3. ALK1 is shown in schematic, with key residues drawn as sticks; BMP9 is shown in surface representation. For clarity the overall complex is shown to the side. B, detailed close-up of the interfaces, showing specific polar contacts (dashed lines) between residues along the F2 and F3 loops of ALK1. Key residues are highlighted with asterisks. C, sequence alignment of type I receptors (ALK1–7), with contact residues highlighted in gold. D, sequence alignment of BMP9 and BMP10 with select TGF-β ligands, showing key contact residues from both BMP9 protomers (red and blue). Asterisks coincide with panel B.
Site II interactions are ostensibly unique to this complex and absent in the other known structures described to date, BMP2-ALK3 and GDF5-ALK6. Site II comprises a band of charged residues on ALK1 helix α1 and loop F3 that form a network of polar interactions with residues on fingers 1 and 2 of one BMP9 protomer and on helix α1 of the second BMP9 protomer, BMP9′ (Fig. 4). ALK1 Arg78 is particularly prominent in that it forms a network of salt bridges with BMP9 Asp89, along with hydrogen bonds to the backbone carbonyl of Lys87 and Asp88, and a main chain hydrogen bond to Ser24 (Fig. 4B). ALK1 Arg80 and Glu83 form a similarly prominent network of contacts with BMP9. Although Arg78 is conserved in ALK6 (Arg69) and conservatively replaced by a lysine in ALK3 (Lys88), there are no functional substitutes for ALK1 Arg80 (Ser90 in ALK3; Thr71 in ALK6) or Glu83 (Ala93 in ALK3; Pro74 in ALK6) (Fig. 4C). Thus, not surprisingly, the F3 loop is positioned too far away in the structures of ALK3 and ALK6 to make contact with the co-crystallized ligand. Also in contrast to those previous observations, ALK1 Glu83 and Phe84 contact the second BMP9 protomer, BMP9′ (Fig. 4B), where, conversely, several unique BMP9 residues interact with ALK1. Of these, Lys64 is invariant in BMP9 and BMP10, suggesting that the ALK1/Glu83-BMP9/Lys64 interaction is important for ALK1 ligand recognition (Fig. 4D). Ser24 is also invariant in BMP9 and BMP10 (Fig. 4D), whereas all other TGF-β ligands have an Asp in the corresponding position, potentially introducing an unfavorable charge interaction with the main chain carbonyl of Arg78.
Site III presumably holds a key ALK1 specificity determinant. It is present in all type I receptor complex structures and consists of residues along the ALK1 pre-helix loop and helix α1, as well as residues from the α1 helix of one BMP9 protomer and the concave face of the second protomer, BMP9′ (Fig. 5A). This site is largely hydrophobic in the BMP2/ALK3 and GDF5/ALK6 structures, characterized by a conserved phenylalanine on helix α1 (Phe85 in ALK3; Phe66 in ALK6). This phenylalanine makes a “knob into hole” packing interaction into a hydrophobic pocket on the ligand. ALK1 is the only type I receptor that does not have a phenylalanine or equivalent hydrophobic residue in this position, but instead contains a glutamate (Glu75), thus raising the question of how the charged residue in this position can fulfill a similar role? The structure described here shows that Glu75 does not point into the conserved hydrophobic pocket as presumed, but, due to the unique receptor positioning, aligns structurally with Asp84 (ALK3) and Asp65 (ALK6) at the n-1 position on helix α1 (Fig. 5B). Thus, ALK1 Glu75 fulfills a similar role as the aspartate residues in ALK3 and ALK6, forming a hydrogen bond with Tyr99 on the periphery of the hydrophobic pocket. Importantly, Glu75 forms additional salt bridges with Lys53 on helix α1 of the second BMP9 protomer, BMP9′. This lysine is “locked” into the binding site by His73 on the pre-helix loop of ALK1. Significantly, sequence and structural alignments indicate that the interactions of His73 and Glu75 with Lys53 are found exclusively at the BMP9 and BMP10/ALK1 interface, therefore suggesting that these three residues are key specificity determinants (Fig. 5, C and D). Consistent with this view, substitution of Glu75 with phenylalanine as in ALK3-ALK6 or valine as in ALK7 almost completely abolishes BMP9 binding in an SPR assay (Fig. 5F) and reduces cell signaling activity by ∼5-and ∼170-fold, respectively, compared with wild-type ALK1 (Fig. 5E). Additionally, replacement of ALK1 His73 by glycine (ALK3, -4, and -6) significantly reduces BMP9 binding as determined by SPR (data not shown) and impairs cell signaling activity ∼93-fold, whereas substitution by aspartate (ALK5) reduces BMP9 signaling ∼25-fold.
FIGURE 5.
Site III interactions at the ALK1/BMP9 interface. A, close-up of the interaction surface, with color scheme retained from Fig. 3. ALK1 specificity residues His73 and Glu75 and Lys53 from BMP9 are highlighted, with polar contacts drawn as dashed lines. B, overlay of the α1 helix from ALK1 and ALK3 showing the positioning of Glu75 (ALK1) with respect to Asp84 (ALK3) and Phe85 (ALK3). C, sequence alignment of type I receptors (ALK1–7), with ALK1 residues His73 and Glu75 highlighted in gold. D, sequence alignment of BMP9 and BMP10 with select TGF-β ligands, showing the invariant lysine residue Lys53 (blue). Asterisks coincide with panel A. E, cell based, and F, SPR analysis of ALK1 mutants showing effects of His73 and Glu75 substitutions on BMP9 binding and signaling. Proteins were expressed in COS-1 cells, purified by affinity chromatography and showed similar purity (SDS-PAGE given in the upper right panels shows: MW markers (first lane), ALK1-WT (second lane), ALK1(E75F) (third lane), ALK1(E75V) (fourth lane), and ALK1(E75A (fifth lane). SPR analysis was performed with Biacore T100 at 37 °C. Proteins were captured on ani-hFC IgG chip at similar levels. Positive control (WT ALK1) and negative control (unrelated protein that does no bind to BMP9) are given for comparison.
A comparison of the interfacial residues from BMP9 with the sequence of BMP10 confirms that the two ligands share functional similarity, as 16 of the 24 interacting residues are fully conserved (supplemental Fig. S4). Five of the remaining 8 residues are conservatively substituted, and these substitutions may account for the small difference in binding of the two ligands to ALK1. Last, Glu18 and Lys87 in BMP9 (Lys17 and Leu86 in BMP10) interact with ALK1 through main chain atoms. Asp89 of BMP9, which is involved in Site II interactions, is absent in BMP10.
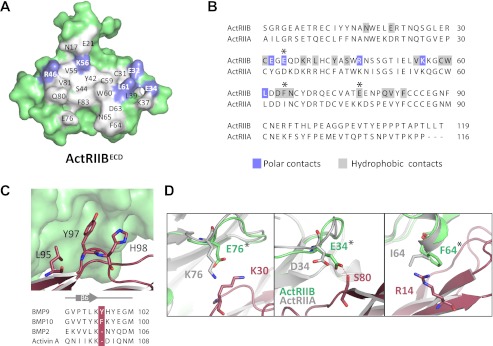
The BMP9/ActRIIB Binding Interface
Unlike the BMP9/ALK1 interface, the BMP9/ActRIIB interface closely resembles that of other type II receptor-ligand complexes. The majority of the ActRIIB residues that interact with BMP9 are also involved in interactions with Activin A (32). A total surface of ∼860 Å2 per BMP9 protomer and ∼880 Å2 per ActRIIBECD are buried at the interface, with 23 residues from BMP9 and 22 residues from ActRIIB participating in the interaction (Fig. 6 and supplemental Table S3). The majority of the interactions are hydrophobic with relatively few polar contacts. The conserved ActRIIB residues Tyr42, Trp60, and Phe83 make up the core of the hydrophobic ligand-binding site and BMP9 residues Ala28, Leu85, and Leu95 bind this site as expected (supplemental Fig. S5). These are the same residues that have been previously identified as important for ActRIIB binding to BMP2 and Activin A (29). However, uniquely observed here is an insertion of BMP9 Tyr97 (or BMP10 Phe95) that creates a small bulge, which changes the structure of an otherwise conserved β strand (Fig. 6C). Tyr97 buries into a hydrophobic pocket formed by the conserved ActRIIB Cys31-Cys59 disulfide on one side and Leu61 on the other side, whereas forming a hydrogen bond with the backbone carbonyl of ActRIIB Glu32 (supplemental Fig. S5). Also of note is a partial helix formed by ActRIIB Asp63, Phe64, and Asn65. ActRIIB Phe64 is located so that it can form a cation-π interaction with BMP9 Arg14 (supplemental Fig. S5). Similar interactions, although not as well defined, are also found in co-structures of BMP2 with ActRIIB and activin A with ActRIIB (23, 32).
FIGURE 6.
ActRIIB/BMP9 interface and basis for ActRIIB selectivity. A, peeled-away surface of ActRIIB showing residues involved in BMP9 binding. Polar (blue) and hydrophobic (gray) contacts are highlighted. B, structure-based sequence alignment of ActRIIB with ActRIIA. ActRIIB contacts are colored as in panel A. C, insertion of Tyr97 in BMP9 causes a loop out in the backbone structure. D, interaction of ActRIIB residues Arg14, Lys30, and Ser80 with BMP9 (residues shown at sticks). Alignment of the corresponding residues from ActRIIA (colored gray) suggests a mechanism for receptor discrimination. Asterisks coincide with panel B.
Molecular Basis of BMP9 Selectivity for ActRIIB over ActRIIA
Our SPR results show that BMP9 binds ActRIIB with ∼300-fold greater affinity than ActRIIA. In contrast, the closely related ligand BMP10 has a more relaxed receptor binding profile and is able to bind ActRIIB and ActRIIA with similar, high affinities. Alignment of the BMP9 and BMP10 sequences reveals that nearly half of the residues (11 of 23) at the BMP9/ActRIIB interface differ between the two ligands (supplemental Fig. S4). Also, only 11 of 22 interfacial residues are conserved between ActRIIA and ActRIIB (Fig. 6B). Thus, it is not surprising that BMP9 and BMP10 show differences in type II receptor binding. Notably, BMP9 residues Arg14, Lys30, and Ser80 are likely to hinder binding to ActRIIA (Fig. 6D). Lys30 is within van der Waals bonding distance of ActRIIB Glu76, but in ActRIIA this glutamate is replaced by a lysine (Lys76), which introduces a potential steric clash (Fig. 6D) and the prospect of charge repulsion. BMP10 has a proline (Pro29) in place of Lys30, suggesting a mechanism for recognition tolerance at this position. BMP9 Ser80 is also likely involved in receptor discrimination as it forms a hydrogen bond with ActRIIB Glu34; replacement of Glu34 with aspartate in ActRIIA (Asp34) likely abolishes this stabilizing interaction. Last, ActRIIB Phe64, which forms a cation-π interaction with BMP9 Arg14, is replaced by isoleucine in ActRIIA (Ile64). Loss of this stabilizing interaction may be partially compensated for in BMP10 with the aromatic tyrosine (Tyr13) in place of BMP9 Arg14.
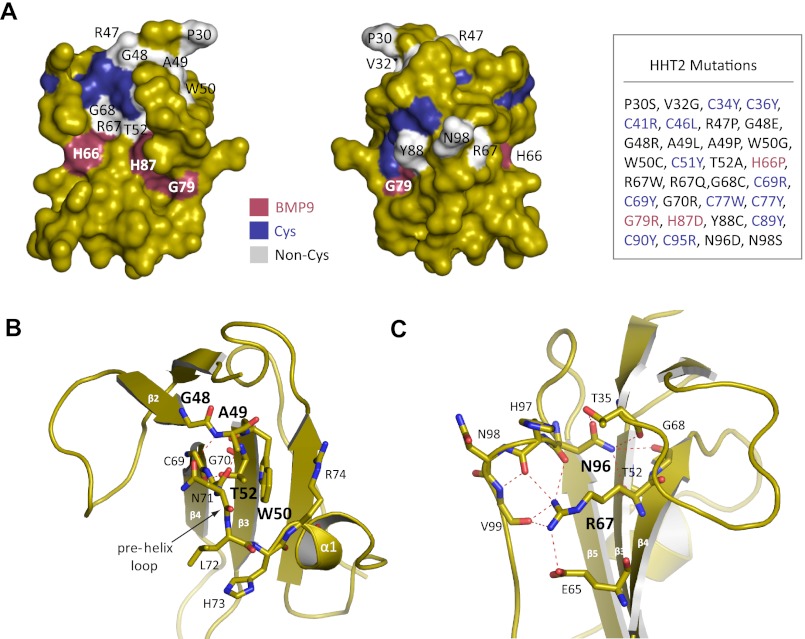
Mapping HHT2 Mutations to the ALK1 Structure
Mutations in the ALK1 gene are associated with HHT2, an autosomal dominant vascular dysplasia (15). We mapped 32 HHT2 mutations (www.hhtmutation.org) located in the ALK1 ECD to the ALK1 structure as determined here and were able to confirm which mutations play a role in destabilizing core structural elements and which ones disrupt interactions with BMP9 (Fig. 7A). Consistent with previous predictions, mutations that map to the highly conserved cysteine residues disrupt the core intramolecular disulfide bonds (33, 34). We found that only three mutations (H66P, G79R, and H87D) directly affect residues at the BMP9 interface. His66 and His87 form part of the conserved hydrophobic pocket in site I (Fig. 4B); mutation of His66 (H66P) is expected to deform strand β4, whereas mutation of His87 (H87D) introduces an unfavorable negative charge directly into the binding site. ALK1 Gly79 is on the F3 recognition loop (Fig. 4B); mutation to a bulky charged residue (G79R) would be predicted to cause clashing at the BMP9 interface and prevent binding. The remaining mutations are distal to the BMP9 binding site and are predicted to have an effect on protein folding and stability. Notably, Gly48, Ala49, and Trp50 connect β2-β3 strands and, along with Arg47 on β2 and Thr52 on β3, stabilize the conformation of the pre-helix loop and helix α1, which harbor BMP9 recognition residues Asn71, Leu72, His73, and Glu75 (Fig. 7B). Mutations of Arg47, Gly48, and Ala49 to bulkier (G48R, G48E, A49L) or conformation-restrained (R47P, A49P) residues would deform the backbone and ablate these stabilizing interactions. Indeed, introduction of the corresponding mutations in ALK1 ECD by site-directed mutagenesis resulted in protein misfolding and aggregation, which led to abrogated binding to BMP9.7 Mutation of Thr52 (T52A) would also lose stabilizing contacts with Asn71. Similarly, Trp50 mutations (W50G, W50C) eliminate stacking interactions with residues on helix α1 (notably Arg74) and introduce an unpaired cysteine. Other residues of note include Arg67 and Asn96, which form a large network of charged interactions between residues on strand β4 and residues at the C terminus (Fig. 7C). ALK1 Arg67 forms hydrogen bonds with residues Asn96, His97, and Val99, as well as a salt bridge to a neighboring Glu65. Similarly, ALK1 Asn96, which is invariant in the type I receptors, forms a salt bridge and backbone contact to Thr35, as well as a hydrogen bond to Thr52 and Gly68. Adding negative charge (N96D and R67Q) or removing charge (R67W) would be expected to disrupt this network of interactions.
FIGURE 7.
Mapping HHT2 mutations to the ALK1 structure. A, surface mapping of HHT2 mutations (Cys residues shown in blue; non-Cys residues in gray; BMP9 contact residues in red). A corresponding list of HHT2 mutations is shown to the right. B, close-up view of stabilizing interactions for ALK1 residues Gly48, Ala49, Trp50, and Thr52 and C, Arg67 and Asn96.
DISCUSSION
ALK1 has long been described as an “orphan” receptor as its signaling partners remained obscure. In an early attempt to identify its signaling partners, chemical cross-linking experiments suggested that TGF-β can interact with ALK1 in endothelial cells (16); however, direct binding of TGF-β to ALK1 could not be confirmed with SPR (9). More recently, BMP9 and BMP10 were identified as ALK1 ligands (9–11, 13, 35). In a thorough kinetic and thermodynamic analysis, we conclusively demonstrate here that ALK1 binds BMP9 and BMP10 with high affinity and specificity. Interestingly, whereas the affinities, kinetics, and Gibbs free energies of binding (ΔG0) for ALK1/BMP9 and ALK1/BMP10 are very similar, the thermodynamic signatures of interactions are different. The BMP9-ALK1 complex is driven by strongly favored entropy (−TΔS0 = −23.6 ± 0.5 kcal mol−1), which is significantly offset by positive enthalpy (ΔH0 = 9.5 ± 0.5 kcal mol−1), whereas the BMP10-ALK1 complex is characterized by a favorable input of entropy (−TΔS0 = −17.2 ± 1.1 kcal mol−1) and only mild offset of positive enthalpy (ΔH0 = 2.4 ± 1.1 kcal mol−1).
Because ALK1-specific ligands have not been recognized until recently, it has not been possible to determine which type II receptors partner with ALK1 to form a functional signaling complex. Three different type II receptors, BMPRII, ActRIIA, and ActRIIB, had been suggested for BMP9 signaling, and thus indirectly implicated in ALK1 signaling (11–13); less is known about type II receptors involved in BMP10 signaling. Here we identify the type II receptors that interact with BMP9 and BMP10, and consequently form part of the ALK1 signaling pathway. Among receptor candidates suggested previously, we find that BMP9 is more selective for the type II receptor ActRIIB. Binding to the putative cognate type II receptor BMPRII is significantly diminished, whereas binding to ActRIIA appears to be transient in vitro, due to the extremely fast dissociation of the BMP9-ActRIIA complex. This is especially true at physiological temperatures, where BMP9 barely binds ActRIIA, whereas its affinity for ActRIIB remains unchanged. BMP10, however, exhibits a less discriminating type II receptor binding profile. We show that BMP10 can bind ActRIIB, ActRIIA, and BMPRII with similar affinities; although its binding profile is slightly more stringent at physiological temperatures. Nonetheless, BMP10 likely has the ability to form signaling complexes with ALK1 and all three type II receptors ActRIIA, ActRIIB, and BMPRII in vivo. Thus despite apparent sequence homology and possibly similar functions BMP9 and BMP10 could act through different type II receptors.
BMP9 and BMP10 are also noteworthy as potentially the only TGF-β ligands that can bind to both type I and II receptors with high affinity. As shown here, both ligands bind their cognate type I and II receptors with almost indistinguishable kinetic and thermodynamic profiles. Moreover, we have not observed any significant cooperativity between ECDs of ALK1 and ActRIIB receptors, as the affinity of ActRIIB for BMP9 in complex with ALK1 is similar to the affinity of ActRIIB for the uncomplexed ligand (supplemental Fig. S6). These data are corroborated by the absence of any conformational changes in the structure of ALK1, ActRIIB, or BMP9 upon complex formation, as well as the absence of any direct contacts between the receptor ECDs. This situation contrasts with other known TGF-β signaling complexes, which follow a two-state binding model where the ligand first binds with high affinity to either the type I or II receptor (as the case may be), followed by recruitment of the lower affinity receptor. Thus, our results suggest a new, nondiscriminative mechanism of ALK1-BMP9-ActRIIB complex assembly that is different from other TGF-β family signaling complexes. The significance of this finding in vivo remains to be determined. Critical analysis of literature data suggests that the type I/type II receptor combination in the BMP9 signaling might be cell type specific. It is possible that high affinity of BMP9 for ActRIIB is essential for signaling in cells where ALK1 is not expressed; whereas other lower affinity type II receptors might be recruited to a preformed ALK1-BMP9 complex in endothelial cells.
To better understand the structural basis for the strict ligand-receptor specificity in ALK1 signaling, we elucidated the structure of the fully assembled ALK1-BMP9-ActRIIB signaling complex. The structure shows that ALK1 achieves its particular specificity through a unique receptor orientation and a distinct set of interfacial contacts, as could not be predicted from previously available structures. ALK1 does not make the canonical knobs into hole packing interaction conserved among all other type I receptors, but instead a distinctive conformation of its helix α1 positions ALK1 Glu75 as would be ALK3 Asp84 and ALK6 Asp65 and not, as expected, in position of ALK3 Phe85 and ALK6 Phe66. Indeed, replacement of ALK1 Glu75 with a hydrophobic residue to mimic the canonical phenylalanine in this position abrogates the ability of ALK1 to bind BMP9. A consequence of this ALK1-specific conformation is that helix α1 and loop F3 bulge outward to within contacting distance of the second BMP9 protomer. ALK1 loop F2 also contacts the ligand, whereas it does not participate in recognition in the BMP2-ALK3 or GDF5-ALK6 complexes. Thus, given the unique set of receptor-ligand interactions as well as the offset in receptor positioning, it is not surprising that ALK1 has such a discriminating ligand binding profile.
The structure presented here also clarifies the structural consequences of specific pathogenic mutations associated with HHT2. HHT2 is a rare disease characterized by vascular malformations in the lung, liver, and brain that is often referred to as a conformational disease, as the mutant protein is rendered nonfunctional through misfolding or aggregation (15, 36). In the absence of a structure, however, the molecular mechanism of pathogenesis cannot be confirmed. Here we show that the majority of HHT2 mutations affect residues that are integral to the core structure, whereas only three residues are predicted to have a direct effect in BMP9 binding. These results may be useful for building a more comprehensive understanding of the contribution of particular mutations to the clinical pathology and severity of the disease in HHT2 patients.
Over the last decade, our understanding of the molecular mechanisms of angiogenesis has improved dramatically. This has led to the approval of several anti-angiogenic therapies targeting the VEGF pathway for treatment of various cancers. Although highly effective, these therapies do give rise to tumors that resist anti-VEGF therapies. Notably, it has been suggested that ALK1 is responsible for compensatory signaling and vascularization in tumors that are refractory to VEGF inhibition. Thus it is not surprising that application of ALK1ECD-Fc resulted in blockade of several VEGF but also non-VEGF angiogenic pathways, suppression of tumor growth in MCF7 mammary adenocarcinoma (9), and antitumor activity in patients with advanced solid tumors in Phase I clinical trials,8 emphasizing that development of ALK1 directed therapies could provide a mechanism to block the escape pathways of tumor progression, especially of VEGF refractory tumors. Taken together our results provide the first structural and mechanistic insight into ALK1 angiogenic signaling response, and as such establish a firm structural basis for a novel class of angiogenesis inhibitors.
Supplementary Material
Acknowledgments
We are very grateful to John Quisel and John Knopf for support and scientific discussion and Mark Alexander for editorial assistance with the manuscript. We thank Kathleen Tomkinson and Danielle Larocque for creating stable cell lines and Dianne Mitchell and Roselyne Castonguay for preparing HHT2 mutants.
S. A. T., E. M.-H., P. L., D. S., J. L., J. U., K. L., K. W. U., J. S., R. K., and A. V. G. are full time employees of Acceleron Pharma and/or have ownership interest in the company.

This article contains supplemental Tables S1–S3, Figs. S1–S6, and “Experimental Procedures”.
The atomic coordinates and structure factors (code 4FAO) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
S. A. Townson and E. Martinez-Hackert, unpublished data.
R. Castonguay and A. V. Grinberg, unpublished data.
Bendell, J. C., Gordon, M. S., Hurwitz, H., Condon, C. H., Wilson D., Yang, Y., Kumar, R., Pearsall, R. S., Solban, N., and Attie, K. M. (2011) A phase 1 dose escalating study of ACE-041, a novel inhibitor of ALK1-mediated angiogenesis, in patients with advanced solid tumors, ASCO poster.
- BMP
- bone morphogenetic protein
- GDF
- growth and differentiation factor
- HHT2
- hereditary hemorrhagic telangiectasia type 2
- ECD
- extracellular domain
- EK
- enterokinase
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- Endo Hf
- recombinant protein fusion of endoglycosidase H and maltose binding protein
- SPR
- surface plasmon resonance
- AMHRII
- anti-Müllerian hormone type II receptor.
REFERENCES
- 1. Massagué J. (1998) TGF-β signal transduction. Annu. Rev. Biochem. 67, 753–791 [DOI] [PubMed] [Google Scholar]
- 2. Attisano L., Wrana J. L., Montalvo E., Massagué J. (1996) Activation of signalling by the activin receptor complex. Mol. Cell. Biol. 16, 1066–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wrana J.L., Attisano L., Wieser R., Ventura F., Massagué J. (1994) Mechanism of activation of the TGF-β receptor. Nature. 370, 341–347 [DOI] [PubMed] [Google Scholar]
- 4. Koenig B. B., Cook J. S., Wolsing D. H., Ting J., Tiesman J. P., Correa P. E., Olson C.A., Pecquet A. L., Ventura F., Grant R. A. (1994) Characterization and cloning of a receptor for BMP-2 and BMP-4 from NIH 3T3 cells. Mol. Cell. Biol. 14, 5961–5974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kirsch T., Sebald W., Dreyer M. K. (2000) Crystal structure of the BMP-2-BRIA ectodomain complex. Nat. Struct. Biol. 7, 492–496 [DOI] [PubMed] [Google Scholar]
- 6. Sebald W., Nickel J., Zhang J. L., Mueller T. D. (2004) Molecular recognition in bone morphogenetic protein (BMP)/receptor interaction. Biol. Chem. 385, 697–710 [DOI] [PubMed] [Google Scholar]
- 7. Chen D., Zhao M., Mundy G. R. (2004) Bone morphogenetic proteins. Growth Factors 22, 233–241 [DOI] [PubMed] [Google Scholar]
- 8. Sako D., Grinberg A. V., Liu J., Davies M. V., Castonguay R., Maniatis S., Andreucci A. J., Pobre E. G., Tomkinson K. N., Monnell T. E. (2010) Characterization of the ligand binding functionality of the extracellular domain of activin receptor type IIb. J. Biol. Chem. 285, 21037–21048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mitchell D., Pobre E.G., Mulivor A. W., Grinberg A. V., Castonguay R., Monnell T. E., Solban N., Ucran J. A., Pearsall R. S., Underwood K. W. (2010) ALK1-Fc inhibits multiple mediators of angiogenesis and suppresses tumor growth. Mol. Cancer Ther. 9, 379–388 [DOI] [PubMed] [Google Scholar]
- 10. David L., Mallet C., Mazerbourg S., Feige J. J., Bailly S. (2007) Identification of BMP9 and BMP10 as functional activators of the orphan activin receptor-like kinase 1 (ALK1) in endothelial cells. Blood 109, 1953–1961 [DOI] [PubMed] [Google Scholar]
- 11. Scharpfenecker M., van Dinther M., Liu Z., van Bezooijen R. L., Zhao Q., Pukac L., Löwik C. W., ten Dijke P. (2007) BMP-9 signals via ALK1 and inhibits bFGF-induced endothelial cell proliferation and VEGF-stimulated angiogenesis. J. Cell Sci. 120, 964–972 [DOI] [PubMed] [Google Scholar]
- 12. Upton P. D., Davies R. J., Trembath R. C., Morrell N. W. (2009) Bone morphogenetic protein (BMP) and activin type II receptors balance BMP9 signals mediated by activin receptor-like kinase-1 in human pulmonary artery endothelial cells. J. Biol. Chem. 284, 15794–15804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brown M. A, Zhao Q., Baker K. A., Naik C., Chen C., Pukac L., Singh M., Tsareva T., Parice Y., Mahoney A. (2005) Crystal structure of BMP-9 and functional interactions with pro-region and receptors. J. Biol. Chem. 280, 25111–25118 [DOI] [PubMed] [Google Scholar]
- 14. Seki T., Yun J., Oh S. P. (2003) Arterial endothelium-specific activin receptor-like kinase 1 expression suggests its role in arterialization and vascular remodeling. Circ. Res. 93, 682–689 [DOI] [PubMed] [Google Scholar]
- 15. Abdalla S. A., Letarte M. (2006) Hereditary haemorrhagic telangiectasia. Current views on genetics and mechanisms of disease. J. Med. Genet. 43, 97–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Oh S. P., Seki T., Goss K. A., Imamura T., Yi Y., Donahoe P. K., Li L., Miyazono K., ten Dijke P., Kim S., Li E. (2000) Activin receptor-like kinase 1 modulates transforming growth factor-β 1 signaling in the regulation of angiogenesis. Proc. Natl. Acad. Sci. U.S.A. 97, 2626–2631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Urness L. D., Sorensen L. K., Li D. Y. (2000) Arteriovenous malformations in mice lacking activin receptor-like kinase-1. Nat. Genet. 26, 328–331 [DOI] [PubMed] [Google Scholar]
- 18. Goumans M. J., Valdimarsdottir G., Itoh S., Rosendahl A., Sideras P., ten Dijke P. (2002) Balancing the activation state of the endothelium via two distinct TGF-β type I receptors. EMBO J. 21, 1743–1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hu-Lowe D. D., Chen E., Zhang L., Watson K. D., Mancuso P., Lappin P., Wickman G., Chen J. H., Wang J., Jiang X., Amundson K., Simon R., Erbersdobler A., Bergqvist S., Feng Z., Swanson T. A., Simmons B. H., Lippincott J., Casperson G. F., Levin W. J., Stampino C. G., Shalinsky D. R., Ferrara K. W., Fiedler W., Bertolini F. (2011) Targeting activin receptor-like kinase 1 inhibits angiogenesis and tumorigenesis through a mechanism of action complementary to anti-VEGF therapies. Cancer Res. 71, 1362–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cunha S. I., Pietras K. (2011) ALK1 as an emerging target for antiangiogenic therapy. Blood 117, 6999–7006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Reeves P. J., Callewaert N., Contreras R., Khorana H. G. (2002) Structure and function in rhodopsin. High-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc. Natl. Acad. Sci. U.S.A. 99, 13419–13424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Weber D., Kotzsch A., Nickel J., Harth S., Seher A., Mueller U., Sebald W., Mueller T. D. (2007) A silent H-bond can be mutationally activated for high-affinity interaction of BMP-2 and activin type IIB receptor. BMC Struct. Biol. 7, 6–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Emsley P., Cowtan K. (2004) COOT. Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 25. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) MolProbity, all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Krissinel E., Henrick K. (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D 60, 2256–2268 [DOI] [PubMed] [Google Scholar]
- 27. DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific, San Carlos, CA [Google Scholar]
- 28. Castonguay R., Werner E. D., Matthews R. G., Presman E., Mulivor A. W., Solban N., Sako D., Pearsall R. S., Underwood K. W., Seehra J., Kumar R., Grinberg A. V. (2011) Soluble endoglin specifically binds bone morphogenetic proteins 9 and 10 via its orphan domain, inhibits blood vessel formation, and suppresses tumor growth. J. Biol. Chem. 286, 30034–30046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Allendorph G. P., Vale W. W., Choe S. (2006) Structure of the ternary signaling complex of a TGF-β superfamily member. Proc. Natl. Acad. Sci. U.S.A. 103, 7643–7648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kotzsch A., Nickel J., Seher A., Sebald W., Müller T. D. (2009) Crystal structure analysis reveals a spring-loaded latch as molecular mechanism for GDF5-type I receptor specificity. EMBO J. 28, 937–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Greenwald J., Vega M. E., Allendorph G. P., Fischer W. H., Vale W., Choe S. (2004) A flexible activin explains the membrane-dependent cooperative assembly of TGF-β family receptors. Mol. Cell 15, 485–489 [DOI] [PubMed] [Google Scholar]
- 32. Thompson T. B., Woodruff T. K., Jardetzky T. S. (2003) Structures of an ActRIIB-activin A complex reveal a novel binding mode for TGF-β ligand-receptor interactions. EMBO J. 22, 1555–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ricard N., Bidart M., Mallet C., Lesca G., Giraud S., Prudent R., Feige J. J., Bailly S. (2010) Functional analysis of the BMP9 response of ALK1 mutants from HHT2 patients. A diagnostic tool for novel ACVRL1 mutations. Blood 116, 1604–1612 [DOI] [PubMed] [Google Scholar]
- 34. Scotti C., Olivieri C., Boeri L., Canzonieri C., Ornati F., Buscarini E., Pagella F., Danesino C. (2011) Bioinformatic analysis of pathogenic missense mutations of activin receptor like kinase 1 ectodomain. PLoS One 6, e26431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cunha S.I., Pardali E., Thorikay M., Anderberg C., Hawinkels L., Goumans M.J., Seehra J., Heldin C.H., ten Dijke P., Pietras K. (2010) Genetic and pharmacological targeting of activin receptor-like kinase 1 impairs tumor growth and angiogenesis. J. Exp. Med. 207, 85–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fernandez-L. A., Fernandez-Lopez A., Garrido-Martin E. M., Sanz-Rodriguez F., Pericacho M., Rodriguez-Barbero A., Eleno N., Lopez-Novoa J. M., Düwell A., Vega M. A., Bernabeu C., Botella L. M. (2007) Gene expression fingerprinting for human hereditary hemorrhagic telangiectasia. Hum. Mol. Genet. 16, 1515–1533 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.