Abstract
Although the mechanisms of anterior-posterior axis formation are well understood in Drosophila, both embryological and molecular studies suggest significant variation in the mechanisms generating this axis within the Insecta class as a whole.
A great number of studies aimed at understanding the evolution of development have been carried out within insects. Without a doubt, this is largely because our detailed understanding of the genetic and molecular basis of pattern formation in the model insect, Drosophila melanogaster, provides an excellent starting point for a large number of comparative studies. In addition, insects are an evolutionarily diverse group of animals; almost one million species of insects have been described, and estimates of insect diversity place the total number of undescribed insect species at over twenty million. More importantly, there is an enormous range of morphological and developmental diversity found within this group of animals, extending from spectacularly colored butterflies, to stick insects, to horned beetles, to wingless silverfish, to miniscule parasitic wasps. Over the last few years, evolutionary studies within the insects have ranged from characterizing the genetic and molecular changes responsible for reproductive isolation between closely related species of Drosophila, to comparing gene expression patterns to understand the developmental basis for variation in appendage number among distantly related members of this group. A number of investigations have also focused on the evolution of the developmental process of segmentation. Finally, recent studies in a variety of insects have revealed interesting molecular changes in the process of axis formation.
Through a combination of, first, genetic followed by molecular and biochemical approaches over the last 20 years, we have come to a detailed understanding of how segmental pattern is established during Drosophila development. Through the sequential action of maternal coordinate, gap, pair-rule, and segment polarity genes, the Drosophila embryo is progressively subdivided into progressively smaller units, whose regional identities along the anterior-posterior axis are specified by the homeotic genes. Insect embryologists working primarily in the 1920s through the 1970s, however, discovered interesting differences in the mode of early pattern formation throughout insects (reviewed in ref. 1). Many of these early studies revealed the differences between so called long germ insects, such as Drosophila, in which the entire pattern of segments is established at the blastoderm stage, and short germ insects, such as the grasshopper and flour beetle, in which segments are added sequentially during an extended period of growth after the blastoderm stage. Once a basic outline of the mechanisms of segmentation was established for Drosophila, several researchers began to explore how the molecular tools developed in Drosophila might be used to shed light on the mechanisms of segmentation in other insects. These studies have revealed relatively well conserved expression patterns (and presumably functions) of genes of the segment polarity and homeotic classes. There are, however, some interesting differences in the expression patterns and regulation of pair-rule and gap genes in at least some groups of insects, suggesting that there have been evolutionary changes in the earlier steps of the segmentation hierarchy (2). Recently, particularly interesting insights have now been made into the evolution of the initial steps that establish anterior-posterior polarity in the developing egg.
Maternal Establishment of Polarity.
In Drosophila, two maternal systems, one acting from the anterior end and another acting from the posterior end, establish anterior-posterior positional information within the syncytial blastoderm shortly after fertilization. An anterior gradient of Bicoid protein acts to both activate zygotic transcription of the gap gene hunchback in the anterior end and repress the translation of uniformly distributed, maternally supplied caudal mRNA in the anterior. At the posterior end, a gradient of Nanos protein acts to repress the translation of uniformly distributed, maternally provided hunchback mRNA. To some extent, these systems reveal a level of redundancy. Both the bicoid and nanos systems generate an anterior domain of hunchback expression, but, whereas zygotic expression of hunchback via activation of transcription by bicoid is required, the role of nanos in axis formation can be made dispensable by simply eliminating maternally supplied hunchback mRNA (reviewed in ref. 3). At face value, this would seem to indicate that the anterior gradient system is of primary importance, and the posterior system, although clearly essential in Drosophila, may be thought of as somewhat secondary. By extension, we might then believe that the anterior gradient system would show greater evolutionary conservation that the posterior gradient system.
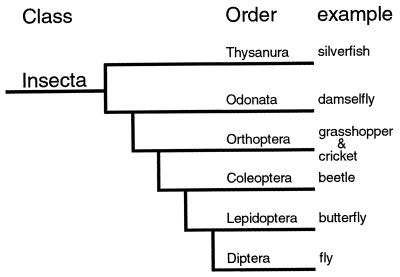
Both of the studies of classical insect embryologists (1), and recently published molecular analyses (4), however, argue against this view. Various manipulations of developing embryos of crickets and damselflies, phylogenetically primitive groups of insects (see Fig. 1 for a phylogeny of representative insect groups), reveal that it is the posterior pole of the egg that provides the signals specifying the axis of the embryo. In some way, the posterior pole of the embryos is able to establish the information generating the anterior pattern elements (head and to some extent thorax) that are specified during the blastoderm stage of short germ insects (1). Thus, these data support the notion that it is the posterior patterning system that might be evolutionarily ancient. In support of this notion, researchers have found nanos orthologs in a variety of animals, although in vertebrates such as Xenopus, their potential roles in axis formation and germ line differentiation have yet to be determined (5). Bicoid orthologs, however, have not yet been found outside of Diptera. This may merely reflect a technical problem; the homeodomain sequence of bicoid appears to be quickly evolving, and this would make orthologous genes, should they be present, difficult to both find and identify.
Figure 1.
Insect phylogeny. The phylogenetic position of several insect orders, and common names of a representative of each order, is shown.
Recently published data, however, provide an alternate explanation. When a bicoid ortholog was cloned from Megaselia abdita, a Dipteran relatively far removed phylogenetically from Drosophila, it was found that this gene was quite closely related to the zen gene from this same fly (4). Zen, the insect representative of the Hox3 family, is involved in specification of the extra-embryonic membranes of insect embryos (6). One implication of this work is that bicoid is a relatively new gene, having evolved somewhere within the higher insects via duplication from the zen gene. If this is true, how might axis formation work in insects without a bicoid gene? This question is best addressed by focusing on two other insects, the flour beetle Tribolium castaneum and the grasshopper Schistocerca americana.
Life Without Bicoid.
In Tribolium, several studies clearly indicate that some sort of early anterior gradient in this embryo is involved in setting up the anterior-posterior axis (7, 8). A Caudal protein gradient similar to that found in Drosophila is observed in this beetle (9). More significantly, the translation of Tribolium caudal mRNA expressed in Drosophila embryos is repressed by Drosophila Bicoid (7). This would seem to indicate that there must be a bicoid ortholog in Tribolium and that it presumably forms a gradient from the anterior end as in Drosophila, thereby establishing the initial gradient of Caudal protein. Should Tribolium prove to lack bicoid, however, could there be another explanation? In Diptera, zen is expressed in extra-embryonic membranes that are located in the dorsal part of the embryo. In Tribolium, however, this tissue is initially located at the anterior end of the egg, and, as in Drosophila, these cells express zen (6). It is possible that zen in Tribolium serves the function of both zen and bicoid in Drosophila. In this scenario, it would be the anterior expression of zen that sets up the initial Caudal gradient. In addition, hunchback is expressed in the anteriorly positioned extra-embryonic cells of the Tribolium embryo; this too could be under the control of zen. In support of this notion, Drosophila hunchback is also expressed in the extra-embryonic cells (dorsally located amnioserosa), possibly under the control of zen. Of course, Drosophila bicoid does more than just regulate the expression of hunchback, and any evolutionary scenario claiming bicoid as a derived feature of more phylogenetically derived insects must ultimately account for these functions as well (3).
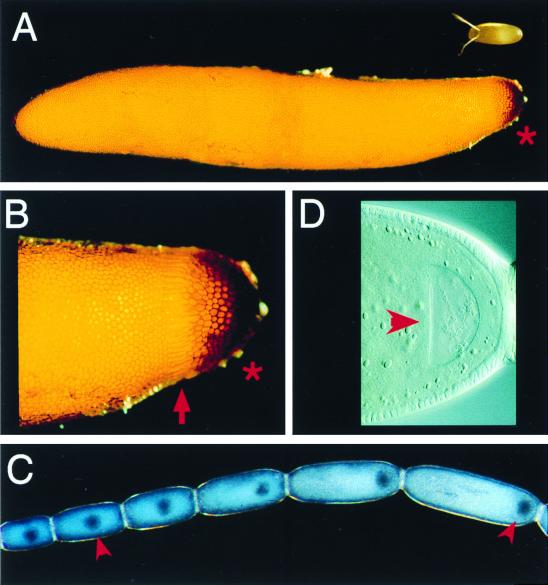
How is polarity established in even more distantly related insects, such as grasshoppers, in which it appears that a posterior gradient system establishes the axis of the embryo, with no clear evidence for an initial anterior gradient system? The egg of a grasshopper is approximately 0.6–0.7 cm long, about 10 times longer than the egg of Drosophila (Fig. 2A). The grasshopper embryo itself, however, forms from only the nuclei that arrive and condense to form a germ disk at the posterior end of the egg; the remaining nuclei form the extra-embryonic serosa. Although the initial germ disk is about the size of a Drosophila embryo, it is essentially the primordium of just the head, placing this embryo in the category of extreme short germ embryos (2). A small, posterior growth zone within the disk will later generate the cells of the entire thorax and abdomen. So how is the position of the germ disk within the egg determined, and how is its polarity established?
Figure 2.
Grasshopper eggs and oocytes. Shown are photos of grasshopper eggs (A and B) and freshly dissected oocytes (C and D). Posterior is to the right in all panels. (A) In the top right corner is a Drosophila egg for comparison. The grasshopper egg shows clear polarity in the maternally produced chorion, with a specialized region of chorion at the posterior end (asterisk). Higher magnification view in B shows that there is a ring of micropyle openings near the posterior as well (arrow). The initial germ disk forms from nuclei that reach this most posterior region, and the disk itself condenses into the region under the darkened chorion (asterisk). A region of a developing grasshopper ovariole is shown in C. Grasshoppers have panoistic ovaries in which there are no nurse cells. As an oocyte matures and enlarges, the oocyte nucleus (arrowheads) moves toward the posterior end of the oocyte. At late stages of oogenesis (D), the oocyte nucleus (arrowhead) is positioned at the extreme posterior end of the egg.
Life Without an Anterior Gradient.
To answer the first half of this question, it is important to note that, in grasshoppers, and in other insects such as crickets and damselflies, the position at which the initial germ disk will form is correlated with the earlier position of the oocyte nucleus during the late stages of oogenesis. For example, during the later stages of cricket oogenesis, the oocyte nucleus is located in a characteristic position about one-third of the way from the posterior end of the embryo. It is in this same position that the embryonic disk will form after fertilization. In the grasshopper S. americana, the oocyte nucleus migrates to the extreme posterior end of the egg during oogenesis (Fig. 2 C and D). Again, this correlates with the position where the germ disk will later form (Fig. 2B). Even more importantly, UV irradiation experiments in cricket embryos suggest that there is indeed a special property intrinsic to the area of the egg where the initial embryonic disk forms (reviewed in ref. 1). If a UV beam is used to destroy the nuclei that arrive in the area where the embryonic disk normally forms, other nuclei, which otherwise would have formed extra-embryonic cells, simply shift into the vacancy and go on to form a normal embryonic disk capable of forming a complete embryo. It is possible that, during the later stages of oogenesis in crickets and grasshoppers, the oocyte nucleus releases some particular protein or mRNA into the adjacent egg cytoplasm that ultimately acts directly on the nuclei that later arrive in this region after fertilization. Alternatively, the oocyte nucleus might signal to the overlying follicle cells, which then signal back to the egg to establish a unique positional identity, possibly in a manner similar to Drosophila in which localized gurken mRNA from the oocyte nucleus acts to control dorsal-ventral pattern (10).
As this initial disk is established, axis formation might then proceed in a manner that requires only a posterior gradient. Because the initial germ disk is “just a head,” it is possible that all of the cells of the disk and extra-embryonic tissue initially express genes associated with the anterior end of Tribolium embryos (possibly hunchback and zen). Indeed, the expression of zen in the grasshopper is consistent with this model (6). A posterior gradient of factors, such as Caudal and Nanos, might then be responsible for establishing the posterior growth zone, thereby generating an anterior-posterior axis without the need for an anterior gradient system. Clearly, this is a simplistic model, and one thing that certainly remains unexplained, both for the grasshopper and Tribolium, is how pattern is set up within the growing abdomen. Any type of gradient patterning the abdomen would have to form, be maintained, and function in a cellular context, and not in a syncytial environment as in Drosophila.
Future Prospects.
Just as with so many questions in insect evolution, comparative studies of gene expression in a range of species should continue to provide important insights into the evolution of insect axis formation. However, the emergence of several new techniques should greatly accelerate the rate and depth of future analyses. First, the continuing development of Tribolium as a genetic model system should surely provide important insights, especially in identifying genes involved in pattern formation in a cellular environment that would not have been found in genetic screens of Drosophila (11). Second, the development of viral systems for gene misexpression (12, 13), double-stranded RNA interference techniques (14), and a hopefully broadly applicable system for creating transgenic insects (15) should allow us to design experiments to functionally test the specific roles of various genes in the evolution of pattern formation. It is also particularly important that researchers continue to take advantage of as many different groups of insects as possible; this is the only way we can adequately address the evolutionary questions facing us. The last few years have seen rapid growth in the field of evolution and development; hopefully, its future results will justify our current enthusiasm.
Acknowledgments
I thank Greg Davis and Sabbi Lall for helpful comments on this manuscript.
References
- 1.Sander K. In: Advances in Insect Physiology. Treherne J E, Berridge M J, Wiggelsworth V B, editors. New York: Academic; 1976. pp. 125–238. [Google Scholar]
- 2.Patel N H. Science. 1994;266:581–590. doi: 10.1126/science.7939712. [DOI] [PubMed] [Google Scholar]
- 3.Deardon P, Akam M. Curr Biol. 1999;9:R591–R594. doi: 10.1016/s0960-9822(99)80381-9. [DOI] [PubMed] [Google Scholar]
- 4.Stauber M, Jäckle H, Schmidt-Ott U. Proc Natl Acad Sci USA. 1999;96:3786–3789. doi: 10.1073/pnas.96.7.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mosquera L, Forristall C, Zhou Y, King M L. Development (Cambridge, UK) 1993;117:377–386. doi: 10.1242/dev.117.1.377. [DOI] [PubMed] [Google Scholar]
- 6.Falciani F, Hausdorf B, Schröder R, Akam M, Tautz D. Proc Natl Acad Sci USA. 1996;93:8479–8484. doi: 10.1073/pnas.93.16.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wolff C, Schulz R, Tautz D, Klingler M. Development (Cambridge, UK) 1998;125:3645–3654. doi: 10.1242/dev.125.18.3645. [DOI] [PubMed] [Google Scholar]
- 8.Wolf C, Sommer R, Scröder R, Glaser G, Tautz D. Development (Cambridge, UK) 1995;121:4227–4236. doi: 10.1242/dev.121.12.4227. [DOI] [PubMed] [Google Scholar]
- 9.Schulz C, Schröder R, Hausdorf B, Wolff C, Tautz D. Dev Genes Evol. 1998;208:283–289. doi: 10.1007/s004270050183. [DOI] [PubMed] [Google Scholar]
- 10.Neuman-Silberberg F S, Schüpbach T. Cell. 1993;75:165–174. [PubMed] [Google Scholar]
- 11.Maderspacher F, Bucher G, Klingler M. Dev Genes Evol. 1998;208:558–568. doi: 10.1007/s004270050215. [DOI] [PubMed] [Google Scholar]
- 12.Oppenheimer D, MacNicol A, Patel N H. Curr Biol. 1999;9:1288–1296. doi: 10.1016/s0960-9822(00)80050-0. [DOI] [PubMed] [Google Scholar]
- 13.Lewis D L, DeCamillis M A, Brunetti C R, Halder G, Kassner V, Selegue J, Higgs S, Carroll S B. Curr Biol. 1999;9:1279–1287. doi: 10.1016/s0960-9822(00)80049-4. [DOI] [PubMed] [Google Scholar]
- 14.Brown S J, Mahaffey J P, Lorenzen M D, Denell R E, Mahaffey J W. Evol Dev. 1999;1:11–15. doi: 10.1046/j.1525-142x.1999.99013.x. [DOI] [PubMed] [Google Scholar]
- 15.Berghammer A J, Klingler M, Wimmer E A. Nature (London) 1999;402:370–371. doi: 10.1038/46463. [DOI] [PubMed] [Google Scholar]