Abstract
Porous silicon nanoparticles (PSiNPs) were synthesized by a silver-assisted electroless chemical etching of silicon nanowires generated on a silicon wafer. The rod-shaped particles (200-400 nm long and 100-200 nm in diameter) were derivatized with a cyclodextrin based nanovalve that is closed at physiological pH 7.4 but open at pH<6. Release profiles in water and tissue culture media show that no cargo leaks when the valves are closed and that release occurs immediately after acidification. In vitro studies using human pancreatic carcinoma PANC-1 cells proved that these PSiNPs are endocytosed and carry cargo molecules into the cells and release them in response to lysosomal acidity. These studies show that PSiNPs can serve as an autonomously functioning delivery platform in biological systems and open new possibilities for drug delivery.
Porous silicon-based materials are attracting much recent interest for bio-applications including sensing, imaging and drug delivery. 1-6 Particles of porous silicon with attractive properties of high surface area,7,8 intrinsic luminescence and bio-degradability 1a,2-4,11 have become a very promising scaffold for delivering drugs. Polymer-coated porous silicon nanoparticles (PSiNPs) have been shown to be an effective chemotherapeutic drug delivery system.1a The method for drug transportation in these studies was based on the electrostatic interaction and/or physical adsorption of specific drugs to the particles.1-4 A key challenge for drug delivery applications is to utilize and control the access to the pores of the PSiNPs in order to generate a stimulus-responsive system that can transport and release the payload on demand without premature release. Similar efforts on mesoporous silica nanoparticles (MSNPs) have led to the development of molecular nanovalves that control the pore openings.9 Because the surface of the PSiNPs is usually terminated with a native silica layer, it is reasonable to expect that nanovalve-controlled systems similar to those that have been developed on silica nanoparticles can also be adapted onto the PSiNP platform. In this communication, we demonstrate the first example of a mechanized PSiNPs system with access to the pore structure controlled by nanovalves chemically bonded on the surface of the PSiNPs. We further show that these PSiNPs can be used as an in vitro delivery system to the human pancreatic carcinoma PANC-1 cells where the cargo is confined in the particles at pH 7.4 and released only on exposure to the decreased pH in endocytic vesicles.
Traditionally, porous silicon is produced by applying a voltage bias to a silicon wafer in hydrofluoric acid containing solutions.10 In this study, the rod-like PSiNPs were prepared from breaking down porous silicon nanowires (PSiNWs), that were generated on a silicon wafer via a Ag-assisted electroless chemical etching method.8 Currently, several mechanisms have been proposed to explain the porosification process.8c,11,12 The mechanism we suggested her is based on our previous work8c and schematically illustrated in Figure 1A. Silver nanoparticles, which were pre-deposited on the silicon wafer (Figure 1A1), will etch pits down into the wafer. The nanowires were consequently formed as the result of the pit walls left between particles. In the process, the Ag nanoparticles at the bottom of these pits can re-dissolve in the etching solution, diffuse up and nucleate again (Figure 1A2-1A3) on the sidewalls of the as-formed silicon nanowires, serving as a new etching site and generating the pores (Figure 1A4). By using this method, PSiNWs can be generated without external electricity, and the nanowire lengths as well as their pore sizes can be easily tuned by controlling the reaction time and conditions.8c Figure 1B displays the cross sectional SEM image of an as-prepared PSiNW array on the substrate. The length of the nanowires is around 30 μm. The nanowires were separated from the substrate and broken into small fragments to form PSiNPs via sonication (Figure 1A5). PSiNPs with different sizes were then separated by using filtration membranes. The size chosen for this study was between 200-400 nm, which is suitable for conducting in vitro studies. Figure 1C shows the TEM image of PSiNPs after sonication, from which their average length is estimated to be about 300 nm, in agreement with the result obtained from dynamic light scattering measurements (Supporting Information Page S3). The pore structure can be clearly observed via HR-TEM (Figure 1C inset). N2 adsorbtion-desorption analysis gives a BET surface area of 293 m2/g and a pore volume of 0.98 cc/g, consistent with a previous report.8c These PSiNPs exhibit a relatively wide pore size distribution (Supporting Information Page S3) due to the variable sizes of the nucleated Ag nanoparticles on the sidewall of the silicon nanowires.
Figure 1.
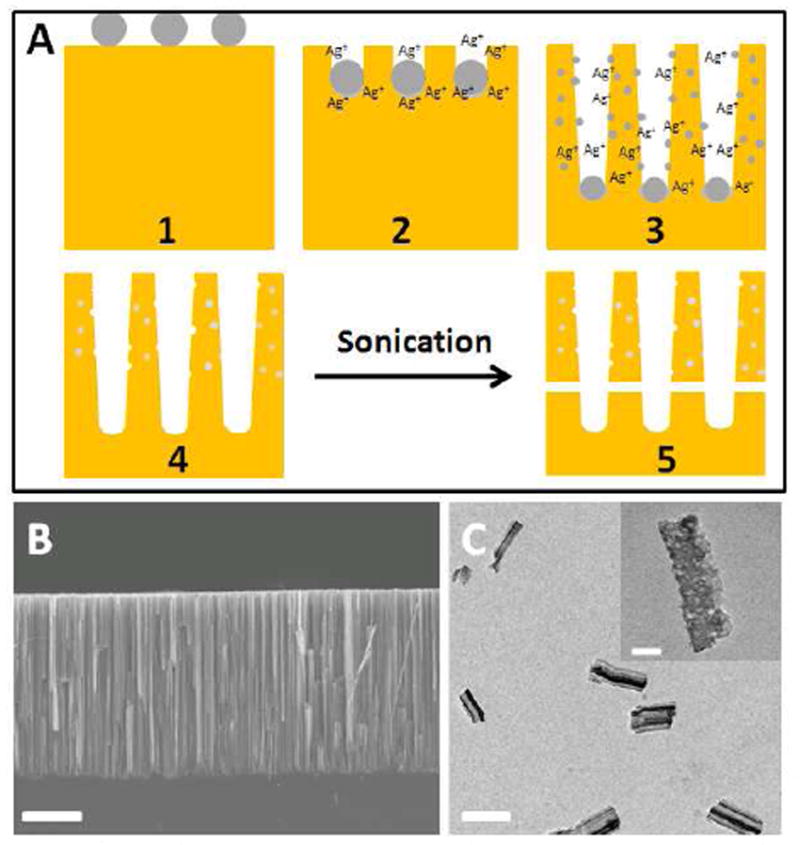
A) The generation of porous silicon nanowires by the etching process with silver nanoparticles. B) Cross sectional SEM image of a porous silicon nanowire array on the substrate. The scale bar is 10 μm. C) TEM image of the PSiNPs after sonication and filtration. The scale bar is 300 nm. A high resolution TEM image of a PSiNP is displayed as an inset with a scale bar of 50 nm.
In this study, a well-established pH-responsive nanovalve-system consisting of an aromatic amino group and a cyclodextrin cap9d,9f was chosen to be adapted to the PSiNPs. This nanovalve was shown to be tightly closed under physiological pH (pH 7.4) and to open autonomously under acidic conditions (pH<6) present in endosomal/lysomal vesicles. In order to attach the nanovalves, The PSiNPs were first derivatized with 3-iodopropyltrimethoxysilane and then coupled with a benzimidazole molecule under base catalysis (Figure 2). Solid state NMR spectrum proves that the modification was successful (Supporting Information Page S4). The modified PSiNPs exhibit a BET surface area of 282 m2/g and a pore volume of 0.95 cc/g, which is similar to those before modification.
Figure 2.
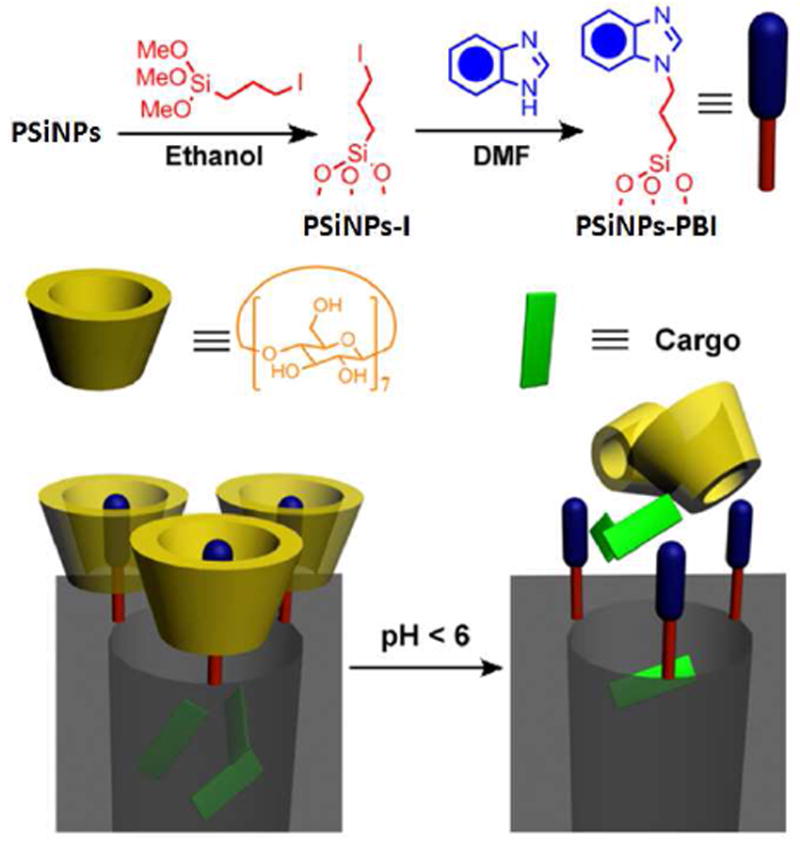
Schematic illustration of the synthesis and operation mechanism of the nanovalve. Top left: attachment of the stalk precursor to the nanoparticle. Top right: completion of the synthesis of the stalk. Bottom: binding of β-cyclodextrin cap to the neutral stalk (left) and release of the cap upon protonation of the stalk.
In order to demonstrate the operation of the nanovalves, the fluorescent biological staining dye Hoechst 33342 was chosen as a model cargo. The PSiNPs were loaded with the dye by soaking them in a concentrated Hoechst 33342 solution. β-cyclodextrin was then added into the solution to finish the full assembly of the nanovalves, followed by washing with water to remove dye molecules absorbed on the nanoparticles’ exteriors. Under neutral pH conditions, the benzimidazole stalk remains hydrophobic and therefore can bind to the cyclodextrin molecule via supramolecular interactions. By doing this, the bulky cyclic cyclodextrin molecules block the pore openings and function as gate keepers to prevent the cargo in the pore from leaking out. When the pH is lowered, the benzimidazole is protonated, and the binding constant between the stalk and the cyclodextrin drops dramatically, causing the cyclodextrin to dissociate. After the pores are unblocked, the cargo can diffuse out (Figure 2).
In order to verify the functional operation of the nanovalves, the cargo-loaded PSiNPs were placed in water and the pH was lowered to stimulate the opening of the nanovalves. The increased concentration of cargo dye released into the aqueous solution was monitored as a function of time by fluorescence spectroscopy to generate a release profile (Figure 3A). Similar to the behavior in the case of silica nanoparticles,9f these nanovalves on silicon nanoparticles remain tightly closed at pH 7.0, giving a flat baseline as illustrated in Figure 3A. Upon lowering the pH, the nanovalves opened and the cargo was released. Cargo release was completed in about 3 hr, which is much faster than that in the case of MCM-41. As a control experiment, PSiNPs with stalks but without capping components were used. In this case, all of the cargo should be removed from PSiNPs during washing. The flat profile for these PSiNPs observed upon acidification shows that no cargo was released as expected (Figure 3A).
Figure 3.
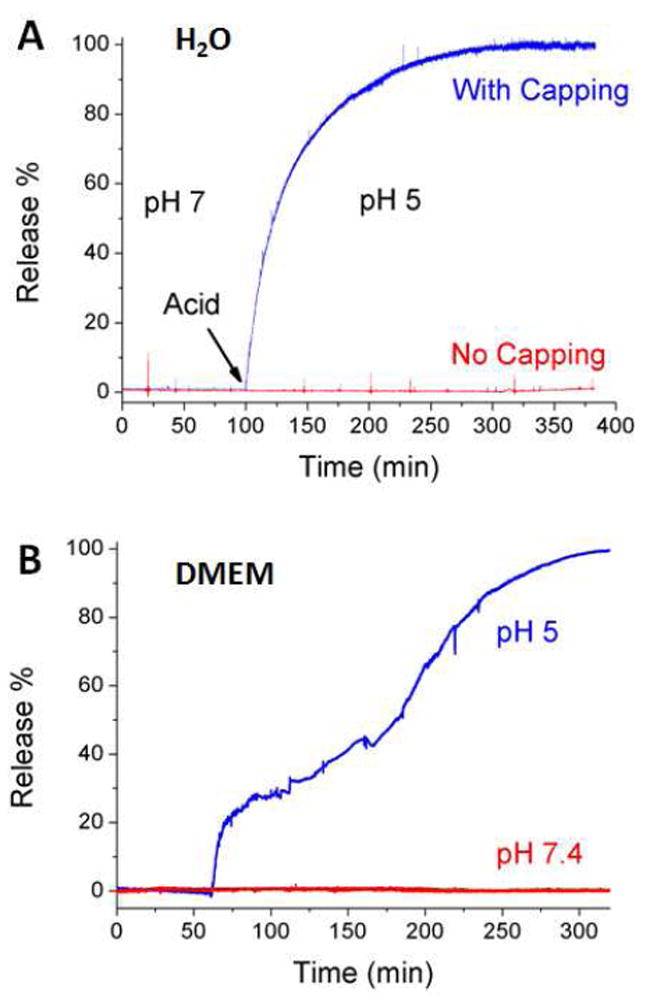
A) Plot of the release of Hoechst 33342 from PSiNPs in aqueous solution. Acid was added to adjust the solution pH. The red line shows the release profile from the control experiment where no cyclodextrin was used for capping. B) The release profile generated in tissue culture medium DMEM. The blue line shows the functioning of the nanovalve when acid was added at 60 min. The red line shows the absence of leakage when the pH is unchanged.
The loading capacity of these PSiNPs is dependent on the size of the cargo. In the case of Hoechst, a loading capacity of 1% w/w was determined using UV-vis absorbtion spectroscopy. This relatively low value is due to the small size of the Hoechst molecules. The nanovalve on the PSiNPs has a size of about 1.5 nm, thus is only capable of controlling the release of Hoechst from < 5.5 nm sized pores.13
Before conducting in vitro studies, it is important to verify that the nanovalve-modified PSiNPs remain functional in biological conditions where the large amount of buffering salt and proteins could potentially change the behavior of the nanovalves. In order to address this point, release profiles were generated in a DMEM cell culture medium with 10% FBS, using Hoechst-loaded PSiNPs (Figure 3B). The PSiNPs were stable in the cell culture medium for at least 5 hr, as no cargo leakage was observed. When the pH was lowered, the fluorescence intensity increased over time, indicating that the valves opened and released cargo. This result demonstrates that the nanovalve-modified PSiNPs are stable, functional and suitable for utilization as a drug delivery system.
Human pancreatic carcinoma PANC-1 cells were studied in order to prove that the nanovalve-modified PSiNPs can be taken up by cells and deliver their cargo inside them. The cells were incubated at 37 °C with the fluorescein-labeled PSiNPs that were loaded with Hoechst 33342. Figure 4A shows that after incubation for three hours, the Hoechst dye was released and stained the cell nucleus. This process is relatively fast and is consistent with the abiotic release profile. In order to further prove that it is the endocytosed PSiNPs that opened and released the dye, several control experiments were conducted. First, the cells were incubated at 4 °C, where the energy-dependant endocytosis pathway is inactive, with the same amount of PSiNPs. In principle, if it is the nanovalves that control the release of the dye, then non-endocytosed PSiNPs should not stain the cells because they do not experience a pH change. As shown in Figure 4B, no staining was observed after 3 hrs’ incubation at low temperature, proving that the dye is not released. Furthermore, contact between PSiNPs and the cell membrane does not cause the nanovalves to open. This result also suggests that the endocytosis process is necessary for opening the nanovalves. Secondly, in order to further investigate the functioning of the mechanized PSiNPs, a competition experiment was conducted. The cells were treated with the same amount of Hoechst-loaded FITC-labeled PSiNPs and increasing amounts of plain PSiNPs with no payload or FITC-labeling. If the particles are indeed taken up primarily via a specific energy dependant uptake process, the amount of FITC-labeled particle uptake (and nuclear staining) should decrease as the concentration of unlabelled particles increases. Figure 4C and 4D prove this assumption by showing much less FITC signal along with less nuclear staining. This result demonstrates that the nanovalves on the PSiNPs do not open in cell culture medium at 37 °C, and also proves that the endocytosis process is necessary to enable the particles to reach lysosomes and trigger the release of the cargo.
Figure 4.
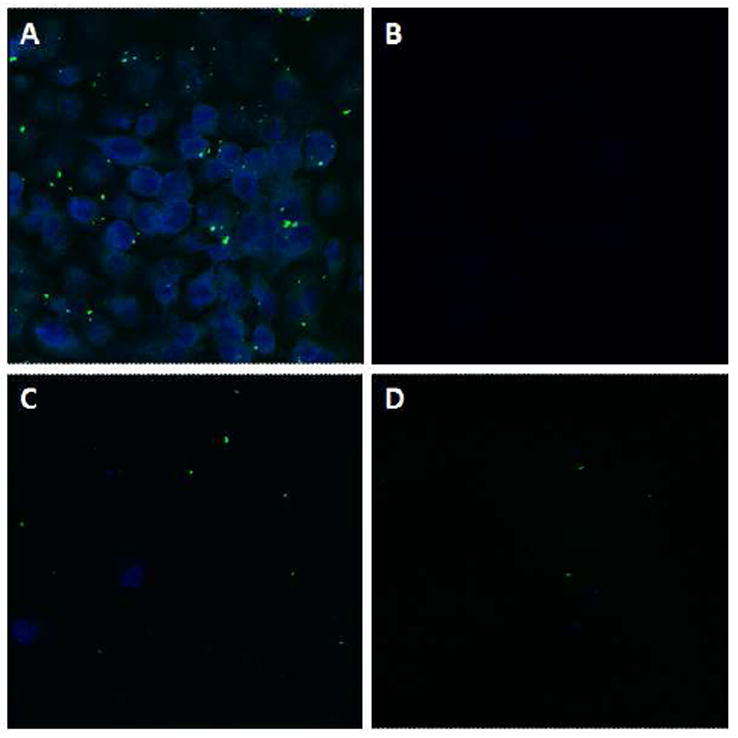
Confocal images of PANC-1 cells incubated with PSiNPs. A) Cells treated with 20 μg/ml of FITC-PSiNPs loaded with Hoechst 33342 at 37 °C. B) Same experiment conducted at 4 °C. No staining was observed after incubation at low temperature. C) and D) Competition tests: cells were treated with 20 μg/ml of FITC-PSiNPs loaded with Hoechst 33342 and C) 60μg/mL or D) 200μg/ml of plain PSiNPs at 37 °C.
In summary, we have demonstrated that pH-sensitive nanovalves can be successfully grafted onto the surface of PSiNPs and function as valves to control the pore openings. These mechanized PSiNPs are endocytosed and release their cargo molecules inside the cells. Further studies regarding controlling the larger sized pores, optimizing the system, and understanding the biological process are in progress.
Supplementary Material
Acknowledgments
This study was funded by the US Public Health Service Grants, RO1 CA133697 by US DOD DTRA 1-08-1-0041 and by the National Science Foundation CHE 0809384. X.D. acknowledges the support by the NIH Director’s New Innovator Award Program, part of the NIH Roadmap for Medical Research, through Grant 1DP2OD004342-01.
Footnotes
Supporting Information Available: Materials and methods used in the experiments, high-resolution TEM image of PSiNPs showing pore structures, N2 adsorption-desorption isotherms, dynamic light scattering, 13C CPMS-NMR spectra of the PSiNPs, fluorescence stability of Hoechst 33342 dye and pH-dependant release studies. These can be found free of charge via Internet at http://www.acs.org.
References
- 1.(a) Park J-H, Gu L, von Maltzahn G, Ruoslahti E, Bhatia SN, Sailor MJ. Nature Materials. 2009;8:331–336. doi: 10.1038/nmat2398. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Sailor MJ, Wu EC. Adv Funct Mater. 2009;19:3195–3208. [Google Scholar]; (c) Cheng L, Anglin E, Cunin F, Kim D, Sailor MJ, Falkenstein I, Tammewar A, Freeman WR. Br J Ophthalmol. 2008;92:705–711. doi: 10.1136/bjo.2007.133587. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Anglin EJ, Cheng L, Freeman WR, Sailor MJ. Adv Drug Deliv Rev. 2008;60:1266–1277. doi: 10.1016/j.addr.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Wu EC, Park J-H, Park J, Segal E, Cunin F, Sailor MJ. ACS Nano. 2008;2:2401–2409. doi: 10.1021/nn800592q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.(a) Bimbo LM, Sarparanta M, Santos HA, Airaksinen AJ, Makila E, Laaksonen T, Peltonen L, Lehto V-P, Hirvonen J, Salonen J. ACS Nano. 2010;4:3023–3032. doi: 10.1021/nn901657w. [DOI] [PubMed] [Google Scholar]; (b) Salonen J, Laitinen L, Kaukonen AM, Tuura J, Björkqvist M, Heikkilä T, Vähä-Heikkilä K, Hirvonen J, Lehto V-P. J Ctrl Rel. 2005;108:362–374. doi: 10.1016/j.jconrel.2005.08.017. [DOI] [PubMed] [Google Scholar]
- 3.De Angelis F, Pujia A, Falcone C, Iaccino E, Palmieri C, Liberale C, Mecarini F, Candeloro P, Luberto L, de Laurentiis A, Das G, Scala G, Di Fabrizio E. Nanoscale. 2010;2:2230–2236. doi: 10.1039/c0nr00161a. [DOI] [PubMed] [Google Scholar]
- 4.(a) Ferrati S, Mack A, Chiappini C, Liu X, Bean AJ, Ferrari M, Serda RE. Nanoscale. 2010;2:1512–1520. doi: 10.1039/c0nr00227e. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Tasciotti E, Liu X, Bhavane R, Plant K, Leonard AD, Price BK, Cheng MM, Decuzzi P, Tour JM, Robertson F, Ferrari M. Nature Nano-technology. 2008;3:151–157. doi: 10.1038/nnano.2008.34. [DOI] [PubMed] [Google Scholar]; (c) Serda RE, Ferrati S, Godin B, Tasciotti E, Liu XW, Ferrari M. Nanoscale. 2009;1:250–259. doi: 10.1039/b9nr00138g. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Serda RE, Mack A, Pulikkathara M, Zaske AM, Chiappini C, Fakhoury JR, Webb D, Godin B, Conyers JL, Liu XW, Bankson JA, Ferrari M. Small. 2010;6:1329–1340. doi: 10.1002/smll.201000126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosso-Vasic M, Spruijt E, Popovic Z, Overgaag K, van Lagen B, Grandidier B, Vanmaekelbergh D, Dominguez-Gutierrez D, De Cola L, Zuilhof H. J Mater Chem. 2009;19:5926–5933. [Google Scholar]
- 6.Vaccari L, Canton D, Zaffaroni N, Villa R, Tormen M, di Fabrizio E. Microelectronic Engineering. 2006;83:1598–1601. [Google Scholar]
- 7.Hochbaum AI, Gargas D, Hwang YJ, Yang PD. Nano Lett. 2009;9:3550–3554. doi: 10.1021/nl9017594. [DOI] [PubMed] [Google Scholar]
- 8.(a) Qu YQ, Zhong X, Li YJ, Liao L, Huang Y, Duan XF. J Mater Chem. 2010;20:3590–3594. doi: 10.1039/c0jm00493f. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Qu YQ, Liao L, Li YJ, Zhang H, Huang Y, Duan XF. Nano Lett. 2009;9:4539–4543. doi: 10.1021/nl903030h. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Zhong X, Qu YQ, Lin YC, Liao L, Duan XF. ACS Appl Mater Inter. 2011;3:261–270. doi: 10.1021/am1009056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.(a) Patel K, Angelos S, Dichtel WR, Coskun A, Yang YW, Zink JI, Stoddart JF. J Am Chem Soc. 2008;130:2382–2383. doi: 10.1021/ja0772086. [DOI] [PubMed] [Google Scholar]; (b) Nguyen TD, Leung KCF, Liong M, Pentecost CD, Stoddart JF, Zink JI. Org Lett. 2006;8:3363–3366. doi: 10.1021/ol0612509. [DOI] [PubMed] [Google Scholar]; (c) Angelos S, Yang YW, Patel K, Stoddart JF, Zink JI. Angew Chem Int Ed. 2008;47:2222–2226. doi: 10.1002/anie.200705211. [DOI] [PubMed] [Google Scholar]; (d) Du L, Liao S, Khatib HA, Stoddart JF, Zink JI. J Am Chem Soc. 2009;131:15136–15142. doi: 10.1021/ja904982j. [DOI] [PubMed] [Google Scholar]; (e) Angelos S, Khashab NM, Yang YW, Trabolsi A, Khatib HA, Stoddart JF, Zink JI. J Am Chem Soc. 2009;131:12912–12914. doi: 10.1021/ja9010157. [DOI] [PubMed] [Google Scholar]; (f) Meng H, Xue M, Xia T, Zhao Y-L, Tamanoi F, Stoddart JF, Zink JI, Nel AE. J Am Chem Soc. 2010;132:12690–12697. doi: 10.1021/ja104501a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cullis AG, Canham LT, Calcott PD. J Appl Phys. 1997;82:909–965. [Google Scholar]
- 11.Chiappini C, Liu X, Fakhoury JR, Ferrari M. Adv Funct Mater. 2010;20:2231–2239. doi: 10.1002/adfm.201000360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang Z, Geyer N, Werner P, de Boor J, Gösele U. Adv Mater. 2011;23:285–308. doi: 10.1002/adma.201001784. [DOI] [PubMed] [Google Scholar]
- 13.The size of Hoechst molecule is about 2.5 nm by 0.75 nm. At the pore opening, the nanovalve occupies part of the space and blocks the pore. The size of nanovalve in this work is about 1.5 nm. Assuming that there are 2 nanovalves per pore opening; about 3 nm of the pore diameter will then be blocked. As a result, only pores with sizes smaller than 5.5 nm will be utilized to trap Hoechst molecules. Any Hoechst stored in larger pores will have enough space to move in and out freely, and they will be removed from the PSiNPs during the washing process. When a larger molecule (Doxorubicin) was used as cargo, a loading capacity of 3% was determined under the same conditions.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


