Abstract
Human immunodeficiency virus type 1 (HIV-1) is the etiological agent of AIDS. Chronic persistent infection is an important reason for the presence of “latent cell populations” even after Anti Retroviral Therapy (ART). We have analyzed the effect of ATP analogs in inhibiting cdk9/T1 complex in infected cells. A third generation drug named CR8#13 is an effective inhibitor of Tat activated transcription. Following drug treatment, we observed a decreased loading of cdk9 onto the HIV-1 DNA. We found multiple novel cdk9/T1 complexes present in infected and uninfected cells with one complex being unique to infected cells. This complex is sensitive to CR8#13 in kinase assays. Treatment of PBMC with CR8#13 does not kill infected cells as compared to Flavopiridol. Interestingly, there is a difference in sensitivity of various clades to these analogs. Collectively, these results point to targeting novel complexes for inhibition of cellular proteins that are unique to infected cells.
Keywords: HIV-1, ATP analog, Transcription, Tat, cdk9 and cyclin T1
Introduction
As of the end of 2008, there were an estimated 40–100 million people living with human immunodeficiency virus type 1 (HIV-1), globally. The development of an additional 18,000 new HIV-1 infections were calculated to take place daily, with 95% of these cases occurring in developing countries and 50% in women. Current projections suggest continued increase in the number of people infected with a possible resurgence of this pandemic in high-income countries, such as the United States. One possible reason for this resurgence is the problems associated with highly active anti-retroviral therapies (HAART) in HIV-1 treatment. HAART, a multi-drug therapy given to AIDS patients, has proven to be effective at lowering viral loads (<50 copies/mL in plasma) and improving an individual’s immune response to HIV-1 infection. However, limitations and concerns regarding cost, complexity of treatment, and long-term side effects have led to lower adherence to treatment regimens by some patients. Also, the development of drug-resistant mutants, either through a lack of protocol adherence or supervised/structured treatment interruptions (STIs), has increasingly become problematic (De Clercq, 2002; Simon et al., 2002).
Today, it is widely believed that the success of HIV-1 treatment will require targeting of other HIV-1 and/or host cellular proteins. To that end, we focused on inhibiting Tat activated HIV-1 transcription and post transcription events, as well as the interaction of Tat with the host cell cycle proteins. HIV-1 Tat interacts with many cellular factors, which may or may not be important for HIV-1 or cellular transcription. However, cdk9/cyclin T1 (P-TEFb) complex has emerged as the most important Tat binding partner that induces elongation of transcription from the HIV-1 LTR.
Expression of the HIV-1 proviral genome requires host cell transcription factors as well as a viral transactivator (Tat) protein (Greene and Peterlin, 2002; Karn, 1999; Karn and Stoltzfus, 2012; Mbonye and Karn, 2011). Tat stimulates formation of full-length transcripts from the HIV-1 promoter (Adams et al., 1994; Antoni et al., 1994) by promoting efficient transcript elongation (Greene and Peterlin, 2002; Karn, 1999). Tat associates with cdk9 (Yang et al., 1997; Zhu et al., 1997) through interaction with the cyclin partner, cyclin T1. Cyclin T1, in turn, interacts with the loop of TAR RNA (Wei et al., 1998). Also, the levels of cyclin T1 are dramatically up-regulated by two independent signaling pathways triggered by PMA and PHA in primary human PBLs, inducing these cells to enter and progress through the cell cycle (Garriga et al., 1998). Furthermore, cdk9 expression during the cell cycle is periodic and peaks at the G1/S border (Garriga et al., 2003; Kiernan et al., 2001), and cyclin T1 levels are dramatically reduced when cells are blocked at the G1/S phase of cell cycle (Kashanchi, unpublished data). Also, a growing body of evidence has indicated the role of yet another cdk/cyclin complex, namely cdk2/cyclin E, in Tat activated transcription. Cdk2/cyclin E is the major cdk/cyclin complex whose maximal activity is observed at the late G1/S boundary. In vivo studies on the involvement of the G1/S phase and cdk2 in HLM1 cells (HIV-1+/Tat−) have shown that Tat transfected cells produced far more infectious virus when blocked at G2/M as compared to G1/S (Ammosova et al., 2006; Ammosova et al., 2005; de la Fuente et al., 2003; Debebe et al., 2011; Kapasi and Spector, 2008; Nekhai et al., 2002).
Pharmacological inhibitors such as Flavopiridol represent the first-generation of selective cdk inhibitors, with 50% inhibitory concentrations (IC50) of 40 to 400 nM for all cdks tested (Dai and Grant, 2004). Interestingly, Flavopiridol tightly binds to cdk9 and inhibits Tat-dependent transcription in vitro as well as inhibiting HIV-1 replication in cultured cells (Chao et al., 2000). The second generation of cdk inhibitors such as R-roscovitine, blocks HIV-1 replication in chronically infected cells (Wang et al., 2001), possibly by directly blocking the activity of all three cdk/cyclin complexes. We recently used R-roscovitine (CYC202) and the acid metabolite M-CYC202 control and showed a potent transcriptional inhibition on the HIV-1 genome with low IC50 values. We also generated a 2nd and 3rd generation CYC202 inhibitor with potent activity against HIV-1. The 2nd generation drug, named CR8, was previously tested and found to effectively inhibit HIV-1 transcription (Guendel et al., 2010) and effectively kill infected cells. An analog of CR8, named CR8#13 (BJFP1154), was unique in that it displayed very little cytotoxicity toward infected cells, while causing the most significant decrease of HIV-1 transcription in HeLa cells (Carpio et al., 2010). Here we extend the effect and mechanism of CR8#13 in more depth in HIV-1 infected primary cells as well as multiple strains and clades of HIV-1. We also report on the presence of multiple novel cdk9 complexes, one of which is only present in HIV-1 infected cells. This complex can selectively be inhibited using ATP analogs.
Materials and Methods
Cell culture and reagents
CEM cells were obtained from ATCC (Manassas, VA). OM10.1 HIV-1 infected latent cells were obtained from the NIH AIDS Research and Reference Reagent Program. CEM cells were cultured and maintained in RPMI Medium 1640 containing 10% fetal bovine serum (FBS), 1% antibiotic solution (penicillin and streptomycin), and 1% glutamine (Invitrogen). The viability of cells was determined by trypan blue exclusion assay. To induce HIV-1 in OM10.1 cells, the cells were treated with TNF-α (10 ng/ml) for 2 hours, and then washed with phosphate-buffered saline (PBS). After virus stimulation, the cells were cultured in complete media and supernatants were collected and analyzed for presence of RT.
Phytohemagglutinin-activated PBMCs were kept in culture for 2 days prior to each infection. Isolation and treatment of PBMCs were performed by following the guidelines of the Centers for Disease Control. Approximately 2.5 × 106 PBMCs were infected with various HIV-1 strains (5 ng of p24 gag antigen). All viral isolates were obtained from the National Institutes of Health AIDS Research and Reference Reagent Program. After 8 hours of infection, cells were washed and fresh medium was added. Drug treatment was performed (only once) immediately after the addition of fresh medium. Samples were collected at various time points and stored at −20°C for RT assay. Histone H1 was purchased from Upstate Cell Signaling Solutions (Charlottesville, VA). Protein (G) and protein (A) agarose were purchased from Sigma (Atlanta, GA).
Transfections and CAT assays
HIV-1 LTR chloramphenicol acetyltransferase (CAT) and pcTat have previously been described (Agbottah et al., 2005; Agbottah et al., 2006; Berro et al., 2006; Kashanchi et al., 1994). HIV-LTR-CAT (5 µg) and CMV-Tat (1.5 µg) vectors were electroporated into CEM cells as described previously (Kashanchi et al., 1992). Extracts were prepared 18 hours later for CAT assay. Cells were harvested, washed once with phosphate-buffered saline (PBS) without Ca2+ and Mg2+, pelleted, and resuspended in 150 µl of 0.25 M Tris (pH 7.8). The cells were freeze-thawed three times, with vortexing after each thawing. The tubes were incubated for 5 minutes at 68°C and then centrifuged. The supernatants were transferred to 1.5-ml Eppendorf tubes. After one final spin, the supernatant was again transferred to 1.5-ml Eppendorf tubes and the protein concentration was determined. CAT assays were performed with 10 µg of protein.
Kinase assay
Cells were washed with PBS, and whole-cell lysates were prepared from the cells using whole cell lysis buffer (50 mM Tris–HCl, pH 7.5, 0.5 M NaCl, 1% NP-40, 0.1% SDS) supplemented with protease cocktail (Sigma). Protein concentration of lysates was measured by Bradford assay (Bio-Rad) and 1.0 mg of total protein was subjected to IP/Kinase reaction. Immunoprecipitations were carried out with the cell lysate and 5 µg of antibodies combined with 50 µl of 30% slurry of protein A + G agarose for 12 hours at 4°C in a TNN buffer containing 50 mM Tris–HCl, pH 7.5, 0.15 M NaCl, and 1% NP-40. The agarose beads were precipitated, washed with TNN buffer and used for the kinase assay. Care was taken to remove all detergents before the kinase assay. Kinase assay was performed at 37°C for 60 minutes in a kinase assay buffer (50 mM HEPES-KOH, pH 7.9, 10 mM MgCl2, 6 mM EGTA, 2.5 mM DTT) containing 1 µg of histone H1 as a substrate, 200 nM cold ATP, and 5 µCi of (γ-32P) ATP. CTD phosphorylation assay was performed at 32°C for 30 minutes in 20 µl of kinase assay buffer (50 mM HEPES-KOH, pH 7.9, 10 mM MgCl2, 6 mM EGTA, 2.5 mM DTT) containing 4 µg of CTD substrate, 250 nM cold ATP, and 5 µCi of (γ-32P)ATP. Reaction was stopped with SDS-loading buffer and resolved on 4–20% PAGE. Dried gel was exposed to phosphorimager screen.
Cell Viability Assay
PBMCs were seeded in 96 well plates at 50,000 cells per well and the appropriate inhibitors were added. Cell viability was measured using CellTiter-Glo Cell Luminescence Viability kit (Promega) as per manufacturer’s instructions. Briefly, an equal volume of CellTiter-Glo reagent (100 µl) was added to the cell suspension (100 µl). The plate was shaken for approximately 10 minutes on an orbital shaker at room temperature following which luminescence was detected using the GLOMAX multidetection system (Promega).
Cytoplasmic and nuclear extracts
The cytosolic extracts were prepared by resuspending the cells in 80 µl of Buffer A with 0.5% NP-40 for 10 minutes on ice. The nuclei were spun down at 5,000 g for 5 minutes and the supernatant was saved as the cytosolic extract (CE). The nuclei were washed once with 200 µl of Buffer A with 0.5% NP-40 and re-pelleted. The nuclei were resuspended in 80 µl of Buffer B (170) and incubated on ice for 10 minutes. The lysates were clarified by centrifugation at 20,000 g for 10 minutes. The sup was saved as the nuclear extract (NE). Western blotting was performed with 1/5 of the samples and the fraction of cdk9 in the cytosolic and nuclear extracts was determined by imaging the chemiluminescent signal on CCD camera (Bio-Rad).
Western blots
Cell extracts were resolved by SDS PAGE on a 4–20% tris-glycine gel (Invitrogen). Proteins were transferred to Immobilon membranes (Millipore) at 200 mA for 2 hours. Membranes were blocked with Dulbecco's PBS + 0.1% Tween-20 + 5% BSA. Primary antibody against specified antibodies was incubated with the membrane in PBS + 0.1% Tween-20 overnight at 4°C. Membranes were washed 3 times with PBS + 0.1% Tween-20 and incubated with HRP-conjugated secondary antibody for 1 hour. Presence of secondary antibody was detected by SuperSignal West Dura Extended Duration Substrate (Pierce).
Chromatin Immunoprecipitation Assay (ChIP)
Cells were seeded to early-mid log phase of growth and then processed for ChIP beginning with cross-linking proteins to DNA by 1.0% formaldehyde. Chromatin was sonicated five times for 20 seconds each, generating DNA fragments of about 500–100 base pairs. The sonicated supernatants containing the DNA were diluted with ChIP dilution buffer (0.01% SDS, 1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl pH 8.1, and 167 mM NaCl) to a total volume of 5.5 ml, and precleared by rotating for one hour at 4°C with ChIP prepared protein A/G beads (beads washed twice with 1 ml TNE50 + 0.1 % NP-40, resuspended in 650 µl; with addition of 40 µL of ssDNA (10 mg/ml) and 75 µl BSA (10 mg/ml)). No proteases or RNAses were used for the extraction. The extract was centrifuged at 3,000 rpm for 10 minutes at 4°C and the lysate was transferred to a fresh tube. Supernatant (500 µL) was reserved for input and then 10 µg of each antibodies indicated above were added to the reaction mixture. After overnight rotation at 4°C, the immune complexes were collected by addition of ChIP prepared protein A/G beads. After extensive washes, the immune complexes were eluted with 1% SDS/NaHCO3 solution for 30 minutes at room temperature. The eluted complexes were treated with NaCl solution and reverse cross-linked overnight. DNA was extracted using 1:1 phenol/chloroform (500 µL) followed by addition of 1 ml of absolute ethanol and 3M sodium acetate (50 µL) incubated at −20°C for at least 20–30 minutes. The solution was spun for 20 minutes at 14,000 rpm at 4°C followed by a 70% ethanol wash and 5 minute spin. DNA pellet was resuspended in 1X TE and stored at 4°C. Afterwards, DNA was purified by PCR purification Kit (BiONEER) and amplified by PCR. Primer pairs for quantitative-PCR analysis of chromatin immunoprecipitation amplified sequences flanked the HIV-1 LTR nuc-1 (+10 to +165) and Env (+8990 to +9120).
Reverse transcription (RT) assay for presence of virus
Viral supernatants (10 µl) were incubated in a 96-well plate with reverse transcriptase (RT) reaction mixture containing 1 × RT buffer (50 mM Tris-HCl, 1 mM DTT, 5 mM MgCl2, 20 mM KCl), 0.1% Triton, poly(A) (1U/ml), pd(T) (1U/ml), and [3H]TTP. The mixture was incubated overnight at 37°C, and 10 µl of the reaction mix was spotted on a DEAE Filtermat paper, washed 4 times with 5% Na2HPO4, 3 times with water, and then dried completely. RT activity was measured in a Betaplate counter (Wallac, Gaithersburg, MD). Cells were also processed for western blot analysis using anti-actin antibodies.
Size-exclusion chromatography
Early-mid log phaseHIV-1 infected J1-1 (which produce high titer RT positive virus present in the media) or uninfected Jurkat cells were pelleted for analysis. Cell pellets were washed twice with PBS without Ca2+ and Mg2+ and resuspended in lysis buffer [50 mM Tris–HCl (pH 7.5), 120 mM NaCl, 5 mM ethylenediaminetetraacetic acid, 0.5% NP-40, 50 mM NaF, 0.2 mM Na3VO4, 1 mM DTT, and one complete protease cocktail tablet/50 mL] and incubated on ice for 20 minutes, with gentle vortexing every 5 minutes. Lysates were then centrifuged at 4°C at 10,000 rpm for 10 minutes. Supernatants were transferred to a fresh tube and protein concentrations were determined using the Bradford protein assay (BioRad, Hercules, CA). Two milligrams of protein from each treatment was equilibrated and degassed in chromatography running buffer [0.2 M Tris–HCl (pH 7.5), 0.5 M NaCl, and 5% glycerol]. The lysates were run on a Superose 6 HR 10/30 size-exclusion chromatography column using the AKTA purifier system (GE Healthcare, Piscataway, NJ, USA). Flow-through was collected at 4°C at a flow rate of 0.3 mL/minute at 0.5 mL for approximately 70 fractions. Every 5th fraction was acetone precipitated using 4 volumes of ice-cold 100% acetone, incubating for 15 minutes on ice. Lysates were centrifuged at 4°C for 10 minutes at 12,000 rpm, supernatants were removed, and the pellets were allowed to dry for a few minutes at room temperature. The pellets were resuspended in Laemmli buffer and analyzed by immunoblotting for cyclin T1 (Santa Cruz Biotechnology Inc., H-245), cdk9 (Santa Cruz Biotechnology Inc., C-20), Hexim 1 (Santa Cruz Biotechnology Inc., H-66), ELL2 (Santa Cruz Biotechnology Inc., G-5), Brd4 (Santa Cruz Biotechnology Inc., H-250), p24 (NIH AIDS Research and Reference Reagent Program, 4121) and β-actin (Abcam., AB49900). Equal volume of sample fractions (# 14, 31, and 46) from both Jurkat and J1-1 were loaded on gels prior to western blots. The protein concentrations for these three fractions showed a typical bell shape curve where fraction #31 (peak of the bell) had the most proteins in either Jurkat or J1-1 preps.
Quantitative RT-PCR analysis
Aliquots of the sample fractions (50 µl) harvested after size-exclusion chromatographyof the lysates of J1.1 and Jurkat cells were incubated with DNaseI, RNase-Free (Roche) – 1U per sample for 1 hour at 37° C. DNase was then inactivated at 65° C for 15 minutes; total RNA was isolated using TRI Reagent-LS (MRC, Cincinnati, OH) according to the manufacturer's protocol. A 250 ng aliquot of total RNA was taken for RT reaction to generate cDNA with the GoScript Reverse Transcription System (Promega, Madison, WI) using 7SK-RTr reverse primer (5’-AAAAGAAAGGCAGACTGCC-3’). Subsequent quantitative real-time PCR analysis of 2 µl aliquots of the RT reaction mixes diluted to 10−1 and 10−2 was performed using iQ SYBR Green Supermix (BioRad, Hercules, CA) with the following pairs of primers: 7SK-F (5’-GACATCTGTCACCCCATTGA-3’) and 7SK-R (5’-GCGCAGCTACTCGTATACCC-3’) amplified 184 nt. fragment of the cDNA copy of 7SK snRNA. Real-time PCR reactions were carried out in duplicate using the PTC-200 Peltier Thermal Cycler with Chromo4 Continuous Fluorescence Detector (both from MJ Research) and Opticon Monitor 2.03 software.
Statistical analysis
All quantifications are based on data obtained from triplicate experiments. P-values were calculated by the student's t-test.
Results
3rd generation ATP analog inhibits HIV-1 activated transcription
We have previously shown that a set of drugs derived from 2,6,9-trisubstituted cdk inhibitory purines as well as 6-aminomethylenebiaryl analogs of CYC202 can down regulateHIV-1 activity (Carpio et al., 2010; Oumata et al., 2008). Among these inhibitors, CR8, an analog bearing a 2-pyridyl on position 4 of its phenyl ring, was created (Galons et al., 2010). Among the 18 CR8 derivatives tested in vitro, the compound CR8#13 (BJFP1154), exhibited the greatest transcriptional inhibition of the HIV-1 LTR in HeLa cells with no apparent cell toxicity. Here, we have extended the studies on the CR8#13 to better define the range of inhibition and other possible modes of action in HIV-1 infected cells.
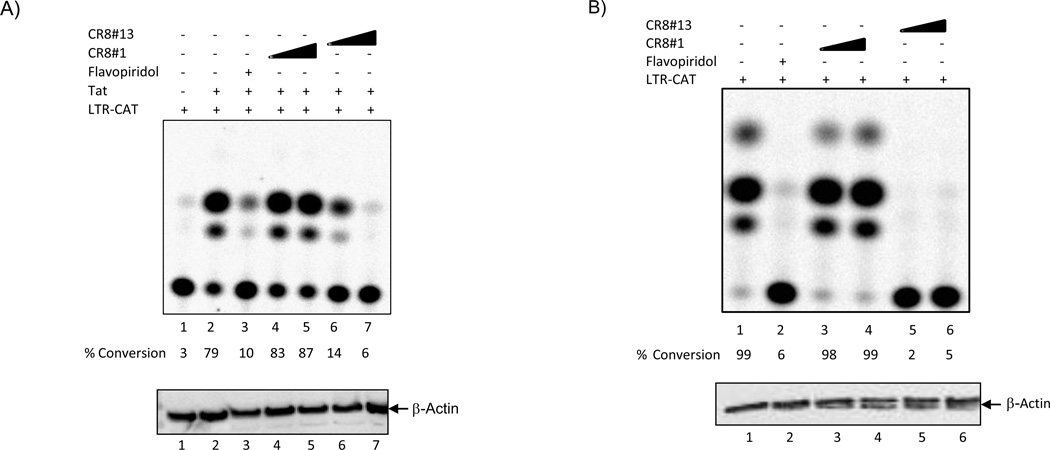
We initially tested the effect of two CR8 derivatives in cells transfected with a LTR-CAT reporter and asked whether Tat activated transcription could be inhibited by these ATP analogs. Data in Figure 1A show a titration of two drugs, CR8#1 (MRT033) and CR8#13, where low concentration of CR8#13 (50 nM, Lane 6) was effectively able to inhibit Tat activated transcription. The inhibition with CR8#13 was similar to a well known cdk9 inhibitor, Flavopiridol (50 nM, Lane 3). Cells treated with a low concentration of these drugs were not affected as measured by PARP western blot or changes in a few well known cdk9 regulated endogenous genes including CIITA, IL-8, CAD, MCL-1 and cyclin D (data not shown).
Figure 1. Effect of CR8#13 on the HIV-1 Promoter in Cells.
A) Early to mid log phase CEM cells were electroporated with either LTR-CAT alone (5µg) or with CMV-Tat (Pc Tat; 1.5µg). Cells were processed for CAT assay 48 hours later. Various concentrations of CR8#1 and CR8#13 (50 and 100 nM) were used immediately after plasmid transfection. Flavopiridol (50 nM) was used as a positive control for inhibition of cdk9/T1 in cells. Ten micrograms of total lysate was used in western blot for presence of β-actin. B) TNF-α treated OM10.1 cells were electroporated with LTR-CAT (5µg) and cells were processed for CAT assay 48 hours later. Similarly to Panel A, concentrations of drugs were used immediately after electroporation of cells. C) Same as Panel B, where OM10.1 cells were electroporated with LTR-CAT and processed 48 hours later for CAT assay. These cells were not treated with TNF-α and therefore no Tat should be available for activation of the LTR-CAT construct. D) Same as Panel A, where CEM cells were electroporated with 5 µg of either CMV-CAT, LTR-CAT (ΔNF-κB), or LTR-CAT (ΔTAR; TM26). Samples were treated with CR8#13 (100 nM) immediately after transfection and CAT assay performed 48 hours later. Ten microgram of total extract was used for β-actin western blot. E) Total cell extracts (TNF-α treated OM10.1; 2.5 mg) were used with 25 µg of α-cdk9 antibody for immunoprecipitation. Samples were incubated overnight at 4°C and protein A + G added next day for 2 hours. IPed material was washed twice with TNE150 + 0.1% NP-40 and twice kinase buffer. The IPed material was divided into 6 tubes with histone H1, kinase reaction mix, and drugs were added to each tube. Samples were incubated for 30 minutes/37°C followed by separation on 4–20% SDS/PAGE. Two concentrations of CR8#1 and CR8#13 (10 and 50 nM) were used for kinase reactions. Flavopiridol (10 nM) was used as a control. Counts are from Molecular Dynamics phosphorimager analysis. F) Similar to Panel E, where 10 µg of α-cdk2 and α-cdk4 antibodies were used for immunoprecipitation overnight followed by in vitro kinase assay next day using histone H1 as substrate. A final concentration of 50nM of CR8#13 was used for in vitro assay. The “K” units are in counts of 1000 from the phosphorimager (Molecular Dynamics software). "CE" and "NP" stand for cytosolic extract and nuclear pellet, respectively.
We next examined the effect of both CR8#1 (control) and the CR8#13 (experimental) in HIV-1 latent cells. For this we utilized OM10.1 cells, where wild type virus is silent, and where addition of TNF-α activates the virus. The OM10.1 cells (promyelocytic) were first transfected with LTR-CAT, followed by treatment of transfected cells with TNF-α for 2 hours and then either CR8#1 or CR8#13 (50 and 100 nM) were added to the media for 48 hours. Here, the assumption is that as a result of TNF-α addition, the production of Tat mRNA and protein will activate the exogenously added LTR-CAT construct. As seen in Figure 1B, the construct was effectively activated in these cells (Lane #1). Treatment of cells with Flavopiridol or CR8#13 (50 nM) effectively wiped out transcription (Lanes 2 and 5) of LTR-CAT in these cells. A western blot performed using a β-actin antibody for both cell types (CEM and OM10.1) showed no change in the protein levels in these transient transfection experiments.
As another set of controls, we transfected LTR-CAT construct into OM10.1 cells without TNF-α activation. Here the rationale was to score only for basal transcription and not Tat activated transcription (from the endogenous virus). Results in Panel C show that there was basal transcription of LTR in these cells and an increasing concentration of CR8#13 (50 and 100 nM) caused only a modest reduction in basal transcription. We also performed another set of control experiments in CEM cells with constructs that were either a HIV-LTR NF-κB mutant, a TAR mutant (TM26; generous gift of Dr. A Kumar; (Zhou et al., 2004) or a CMV-CAT construct. Following transfection, cells were treated with CR8#13 and results in Panel D showed that, again, the level of inhibition on these constructs was less than 2 fold. This further argues that the CR8#13 may be unique to Tat activated transcription on wild type LTR.
We next asked whether these drugs could potentially inhibit cdk9 kinase activity in vitro. For that, we used cdk9 immunoprecipitated complexes from TNF-α treated OM10.1 cells (source of active kinase) and incubated them with histone H1 as a substrate. A number of substrates including histone H1 have been shown to be phosphorylated by cdk9 (O'Brien et al., 2010; O'Brien et al., 2012; Tian et al., 2012). Results in Panel E show that CR8#13 (10 and 50 nM; Lanes 4 and 5) effectively inhibited cdk9 activity to similar levels as Flavopiridol (10 nM; Lane6). Similar results were also observed when cdk9 was immunoprecipitated and analyzed by kinase assays with a CTD substrate (data not shown). Finally, we performed a similar experiment with active cdk2 and cdk4 enzymes immunoprecipitated from TNF-α treated OM10.1 cells followed by in vitro kinase assays. Results in Panel F show that CR8#13 selectively inhibits cdk9 but not cdk2 or cdk4 when isolated from infected cells. Collectively, these results reconfirm that CR8#13 is an effective inhibitor of Tat activated transcription in T-cells and other cell types.
Effect of CR8#13 on loading of cdk9 complex and its localization
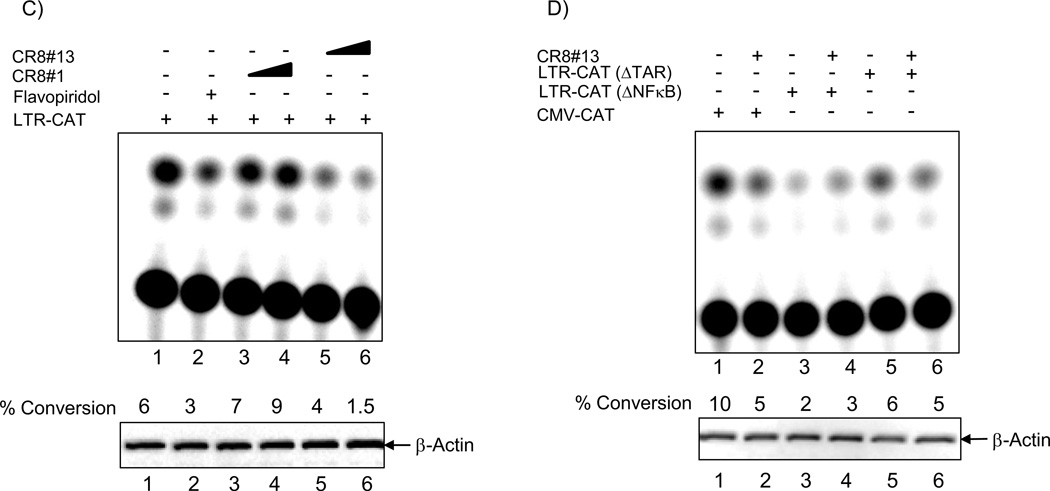
We have previously shown that the 2nd generation drug, CYC202, was able to inhibit HIV-1 transcription by interfering with loading of cdk9, but not cdk4, onto the HIV-1 genome (Agbottah et al., 2005). We specifically showed that siRNA interference followed by ChIP can be used to measure the binding of various cdk/cyclin complexes loaded onto the HIV-1 DNA in vivo (proof of principle experiments) and that CYC202 inhibited loading of cdk9 onto the LTR and Env regions. We therefore asked whether CR8#13 or the control CR8#1 could effectively down regulate levels of cdk9 protein on the LTR or the Env regions. Results in Figure 2, Panel A show that when cells are treated with CR8#13, there is an appreciable drop of cdk9 and Pol II on the LTR region (lanes 3 and 5). More importantly, these proteins were completely absent on the envelope region when treated with CR8#13. A dilution of PCR products (2 fold) was resolved on ethidium bromide containing agarose gels and relative signal intensities were measured using a phosphorimager. Measured signal intensities indicated a linear drop in the immunoprecipitated material for both LTR and Env regions. Finally, treatment with CR8#13 at these low concentrations did not push cells into apoptosis, as shown by the number of live cells in these 48 hour treatment experiment. Similar results were obtained when the relative numbers of live cells were measured using CellTiter-Glo assay from these drug treated cells (Panel C). Collectively, these results imply that CR8#13 is able to inhibit uploading of cdk9 and Pol II onto LTR and Env regions.
Figure 2. Effect of CR8#13 on cdk9 and Pol II occupancy on the HIV-1 genome.
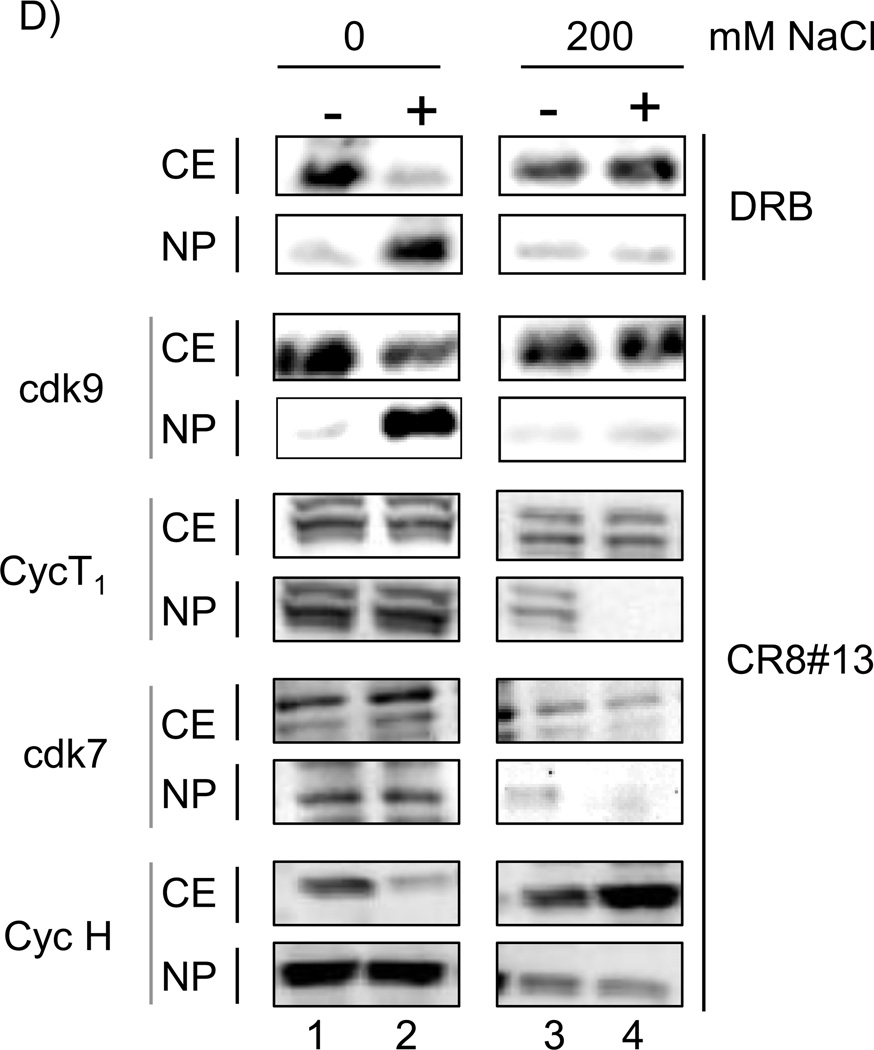
A) Inhibition of loading of cdk9 and Pol II onto the HIV-1 genome when using CR8#13 or CR8#1 (negative control). Following induction of OM10.1, cells were treated with 50 nM of either drug. Forty eight hours later samples were collected and treated for ChIP analysis and PCR using LTR and Env primers. Percent of live cells is included from treatment prior to ChIP assay using any of the two antibodies. Live cell numbers and percentages were calculated based on a trypan blue exclusion assay. B) ChIP samples were diluted 1:2 and run on an agarose gel (2%) at three concentrations. Specific band intensities were counted using a phosphorimager (Molecular Dynamics software) and plotted for both LTR and Env products. C) Induced OM10.1 samples were treated with CR8#1 or CR8#13 (similar to Panel A) and carried out to 48 hours. Samples were processed for viability using CellTiter-Glo assay. D) OM10.1 activated cells were either untreated or treated with DRB (100 µM) or CR8#13 (50 nM) concentrations for 48 hours (lanes 2 and 4). The cytosolic extracts were prepared and the nuclei were separated using centrifugation. Samples were run on a 4–20% gel and western blotting was performed with 1/5 of the samples and the fraction of cdk9 in the cytosolic extracts (CE) and nuclear pellets (NP) were determined. The top panel of DRB treated cells was used for western blot with α-cdk9 antibody. The CR8#13 treated cells were further processed for the presence of cdk9, cyclin T1, cdk7 and cyclin H.
In 2007, a collaborative work between three labs (Maury, Price and Bensaude) showed that there are two forms of the cdk9/T1 complexes and they could be physically separated in HeLa cells when using any of the three cdk inhibitors including Flavopiridol (Alvocidib), DRB (5,6-dichloro-1-β-D-ribofuranosylbenzimidazole) or R-roscovitine (Seliciclib/CYC202)(Biglione et al., 2007). They also developed a rapid salt extraction assay that allowed them to determine the amount of large and free forms of cdk9/T1 present in cells. As controls, the differential salt extractability of the TFIIH subunits p62, cdk7 and cyclin H showed no change in nuclear vs. cytoplasmic fractions after drug treatment. Therefore, we decided to pursue similar experiments with CR8#13, and asked whether cdk9 localization was also altered in these drug treated cells. We treated cells with either DRB (100 µM, as positive control) or with CR8#13 (50 nM, experimental). We isolated cytoplasmic, as well as nuclear extracts and used differential salt extractions (0 and 200 mM), which were subsequently Western blotted for the presence of cdk9, cyclin T1, cdk7 and cyclin H in these extracts. Results in Figure 2, Panel D show that CR8#13 treated cells behaved very similarly to DRB treatment, and most of the cdk9 in CR8#13 treated cells became part of the large inactive cdk9 complex (lanes 3 and 4). We have consistently observed some 1–5% of cdk9 still remains in the nucleus when cells are either treated with DRB or CR8#13. We observed very little change in the cyclin T1 distribution in cytoplasmic vs. nuclear fractions in these HIV-1 infected OM10.1 cells. The only significant difference was at high salt (which extracts both large and free complexes) in drug treated cells, where the majority of cyclin T1 did not enter the nucleus (Lane 4). Interestingly, data for cdk7 and cyclin H were somewhat similar to HeLa data from Biglione et al., where salt extractability of these proteins was unaffected by the addition of CR8#13. Collectively, these results imply that the majority of the cdk9 is most likely unable to move to the nucleus when treated with these transcription inhibitors in infected cells and that may partially explain the loss of loading of cdk9 onto the HIV-1 DNA.
Presence of a novel small complex in HIV-1 infected cells
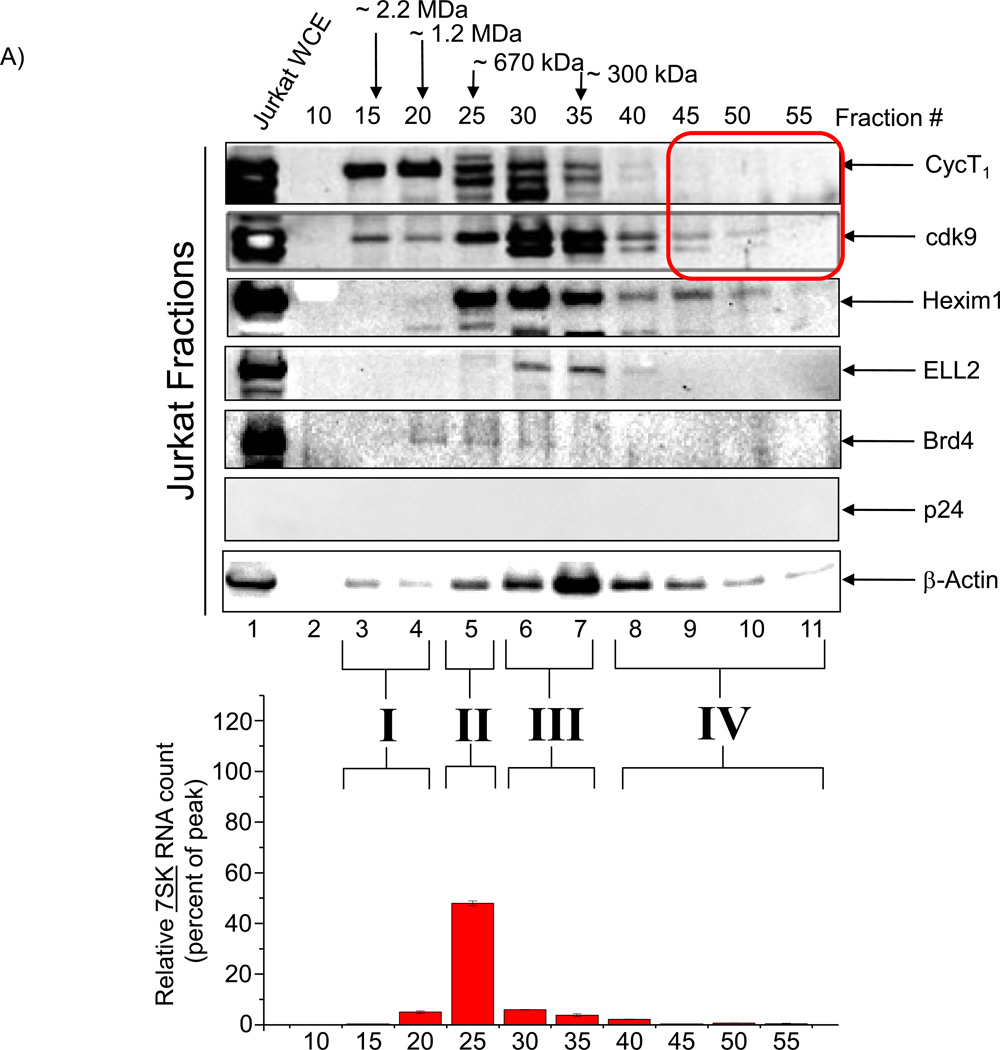
We next asked whether infected cells that express virus on a continuous basis would have a novel cdk9/T1 complex that may be differentially regulated by CR8#13. Here we used parental uninfected (Jurkat) and LAI infected Jurkat (J1-1) cells for separation of various cdk9 and cyclin T1 complexes. We used cells at early – mid log phase of growth and prepared total protein using Lysate buffer. The samples were then loaded onto a sizing column in the presence of high salt to minimize non-specific binding (500 mM NaCl). FPLC fractions were then precipitated and used for western blot analysis and kinase assays. Results of such an experiment are shown in Figure 3, where Jurkat uninfected fractions showed cyclin T1 complexes in fractions 15–35 and a similar profile was observed for cdk9 (Panel A). We performed Western blot analyses for Hexim 1 (inhibitor of P-TEFb), ELL2 (component of the super elongation complex) (He et al., 2010; Sobhian et al., 2010), Brd4 (cellular analog of HIV-1 Tat protein which activates cellular genes when complexed with cdk9/T1 complex), and RT/PCR for 7SK RNA (an abundant RNA molecule that, when bound to MePCE, LARP7, Hexim 1, and cdk9/T1 proteins, forms the large inactive complex called “7SK SNRNP Complex”). Results show that Hexim 1 eluted mostly at fractions 25–35; ELL2 eluted mostly at fractions 30–40; and Brd4 at fractions 20–30. It is interesting to note that both ELL2 and especially Brd4 did not elute from the WCE preps indicating that these proteins are mostly nuclear and bound to chromatin DNA (insoluble complexes). Interesting results were obtained with 7SK RT/PCR where the majority of the RNA molecule eluted at fraction 25. Based on these results we categorized the cdk9/T1 complexes as complex I (high molecular weight of ~2.2 MDa with minimal 7SK or Hexim 1), complex II (molecular weight of ~670 kDa with maximum 7SK and Hexim 1), complex III (molecular weight of ~450 kDa with minimal 7SK and highest β-actin) and complex IV (molecular weight of ~100 kDa with minimal Hexim 1 and low β-actin). It is important to note that the high amount of protein (peak of the bell shape curve) was present at fraction # ~30 (data not shown).
Figure 3. Presence of novel cdk9/cyclin T1 complexes in HIV-1 infected replicating cells.
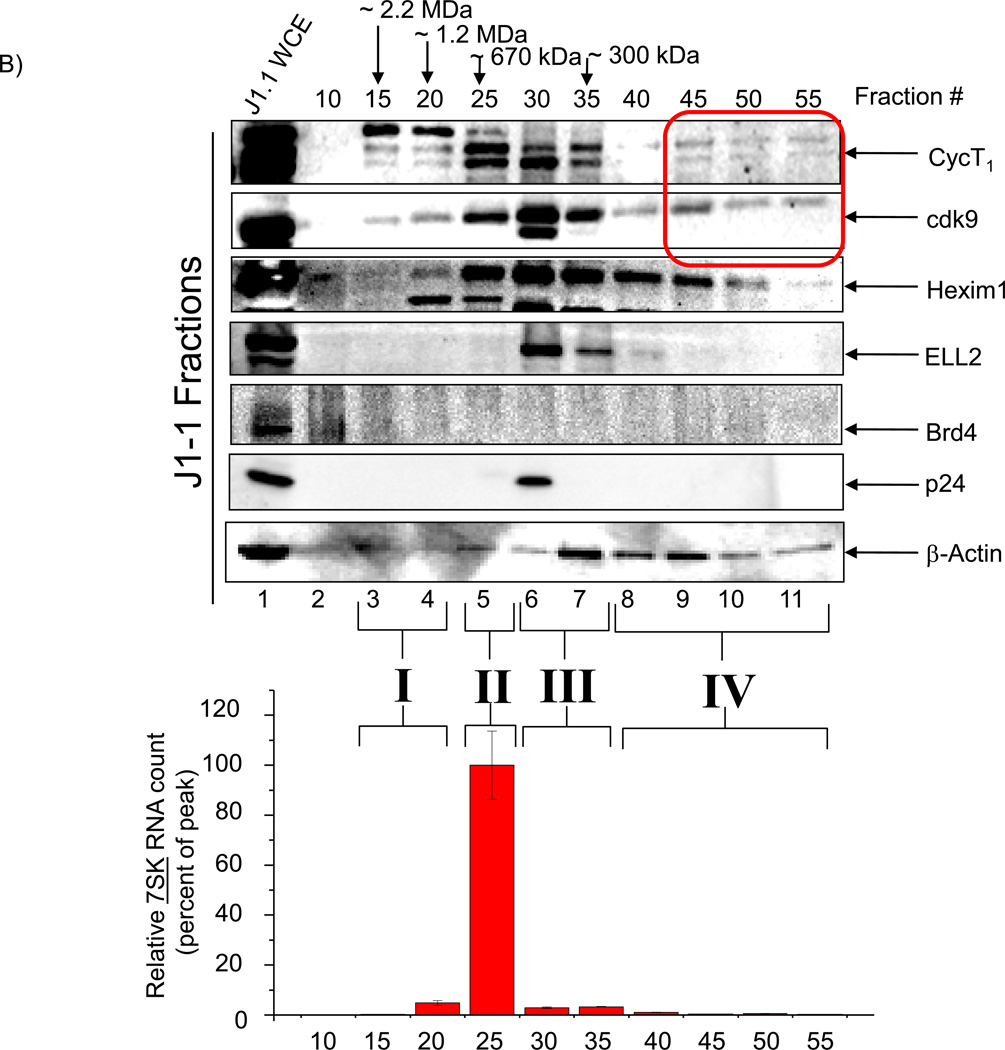
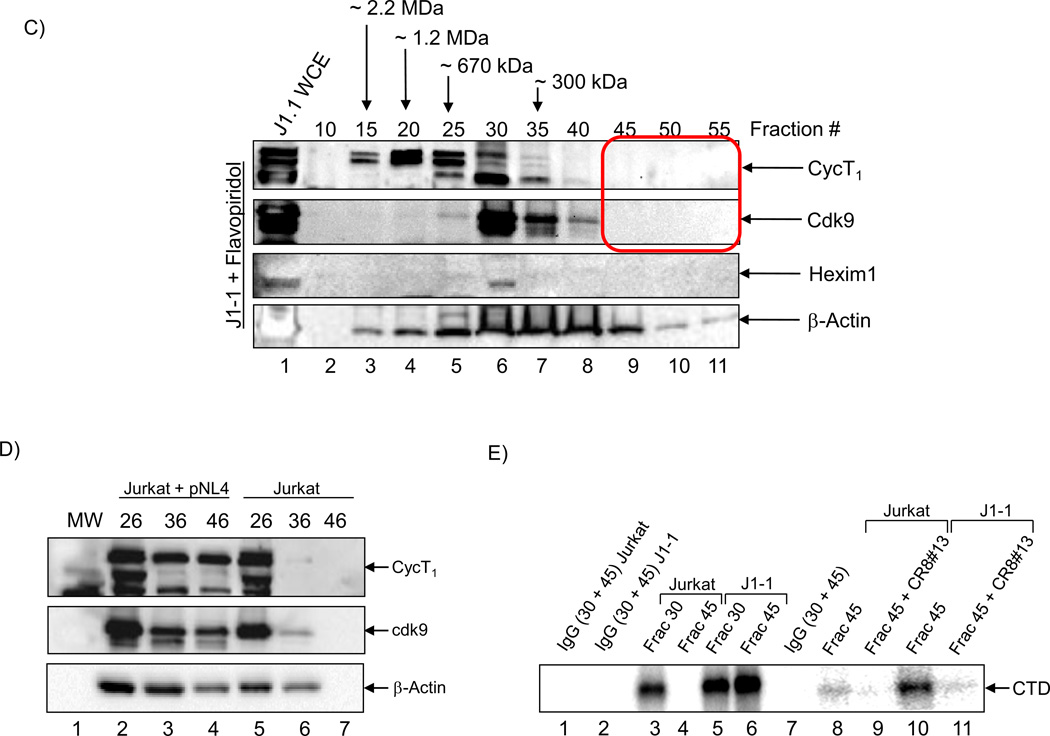
A) Total cell extracts from Jurkat were processed over a sizing column in presence of 500 mM salt. Every fifth fraction was used to precipitate proteins and western blot for presence of cyclin T1, cdk9, Hexim 1, ELL2, Brd4, p24 and β-actin. Total RNA isolated from equal aliquots of SEC fractions 10–55 of the lysates of Jurkat were quantitated by RT-SYBR Green real-time PCR with the primers specific for 7SK cellular snRNA sequence. Results are presented as a mean of two independent measurements and shown as a percent of max RNA count. Error bars show the standard error of mean values. Equal volumes of samples (half of each 500 µl fraction) were processed for western blots. Molecular weight markers of known positive controls were used to determine the range and size of the complexes. B) Similar to Panel A, however, J1-1 cell extracts were used for fractionations and then western blotted for presence of cyclin T1, cdk9, Hexim 1, ELL2, Brd4, p24 and β-actin. Similar RT/PCR was performed for 7SK RNA. A total of 4 complexes designated as complexes I, II, III and IV were observed in HIV-1 infected cells. C) HIV-1 infected J1-1 cells were treated with Flavopiridol (100 nM) and sample were processed for chromatography after 48 hours. Western blots were performed for cdk9, cyclin T1 and Hexim 1 and β-actin. D) Mid-log growing Jurkat cells were transfected with 20 µg of wild type pNL4-3 plasmid and samples were processed 5 days later for sizing column chromatography. Fractions representing complexes II (#26), III (#36), and IV (#46) were precipitated using acetone, dried and western blotted with antibodies against cyclin T1, cdk9, and β-actin. E) Kinase assay using cdk9 immunoprecipitates (J1-1 and Jurkat) followed by phosphorylation of GST-CTD. IgG immunoprecipitates of fractions 30 and 45 from both infected and uninfected cells were used as a control. CR8#13 at 10 nM concentration was used in lanes 9 and 11.
When examining the HIV-1 infected J1-1 cells (Figure 3, Panel B), we observed a similar pattern for complexes I–III with some notable differences. For instance, the overall level of ELL2 in infected cells which was soluble was higher than uninfected cells, and the overall level of Brd4 was generally lower in infected cells (compare Lanes 1 and fractions between Jurkat and J1-1). There was also a dramatic increase in the 7SK levels in the infected cells in complex II in infected vs. uninfected cells. These results imply that when assaying for active P-TEFb complexes in HIV-1 infected cells, there needs to be a comprehensive examination of the components that control cdk9/T1 activity including the proteins tested here. Finally, complex IV was mostly abundant in infected cells and reduced (especially for cyclin T1) in uninfected cells.
The small complex IV was extremely sensitive to Flavopiridol as J1-1cells treated with this drug dramatically shifted complex IV to complex III for cdk9 and cyclin T1 (Figure 3, Panel C). We also observed movement of cdk9 larger complexes to complex III and an overall reduction in Hexim 1 and its concentration in complex III. Therefore, complexes IV and possibly complex I may be the true target of Flavopiridol in HIV-1 infected cells.
We next performed a transfection of wild type HIV-1 plasmid (pNL4-3) into Jurkat cells (for 5 days) and fractionated the whole cell extract over the sizing column. We were interested to see if we could detect the smaller complex IV in these cells. Results in Panel D show that both cyclin T1 and cdk9 were present in fractions 36 (complex III) and 46 (complex IV) from infected cells.
We then decided to further focus on the differences between complexes III and IV by immunoprecipitating fractions 30 and 45 from infected and uninfected cells and used them in an in vitro kinase assay using RNA Pol II CTD as a substrate. Results in Panel E show that both fractions # 30 from Jurkat and J1-1 were kinase active; however, only fraction #45 showed appreciable activity from J1-1 fractions (compare 4 and 6). Finally, we used CR8#13 in these in vitro kinase assays and were able to efficiently inhibit activity of cdk9 from fraction #45 (Lanes 10 and 11). Collectively, these results suggest that a novel cdk9/T1 complex may be present in HIV-1 infected cells that actively produce virus and that the CR8#13 at low concentrations could inhibit the kinase activity of this complex.
Decreased activity of cdk9 also regulates HIV-1 splicing
In recent years, cdk9 has been associated with not only transcription, but also splicing through CTD and other proteins (Bres et al., 2008; Lin et al., 2008; Mapendano et al., 2010; Pirngruber et al., 2009). RNA Pol II CTD functions both, as an assembly platform for and as a regulator of the transcription and pre-mRNA processing machineries. During transcription initiation, serine 5 of the CTD heptad repeat is phosphorylated to recruit capping enzymes. Serine 2 of the heptad repeat is then phosphorylated, leading to the recruitment of factors such as human SPT5 (hSPT5) and Tat-SF1 that are involved in subsequent steps needed for elongation and RNA splicing. Along these lines, we have previously shown that RNA Pol II CTD is phosphorylated at both serine 2 and 5 in the presence of Tat resulting in altered cdk9 substrate specificity (Zhou et al., 2003).
The Caputi lab has previously published convincing data on the effect of Tat and splicing (Jablonski et al., 2010) that is consistent with our previous results of how Tat could modulate not only transcription but also HIV-1 splicing through mechanisms, such as acetylation of lysine residues (Berro et al., 2008). The association with both elongation and splicing factors has led to the suggestion that factors such as Tat-SF1 can couple these two processes (Chen et al., 2009; Kim et al., 1999; Miller et al., 2009). Tat-SF1 has been shown to be associated with other transcription regulators such as Tat-CT1 and the transcription-splicing coupling factor, CA150 (Zhou et al., 1998). Tat-SF1 and hSPT5 are required for optimal Tat activation, as shown by immunodepletion with specific sera and complementation with recombinant proteins (Parada and Roeder, 1999). Expression of Tat-SF1 and hSPT5 specifically stimulates the transcriptional activity of Tat and modulates transcription elongation and splicing of HIV-1 RNA.
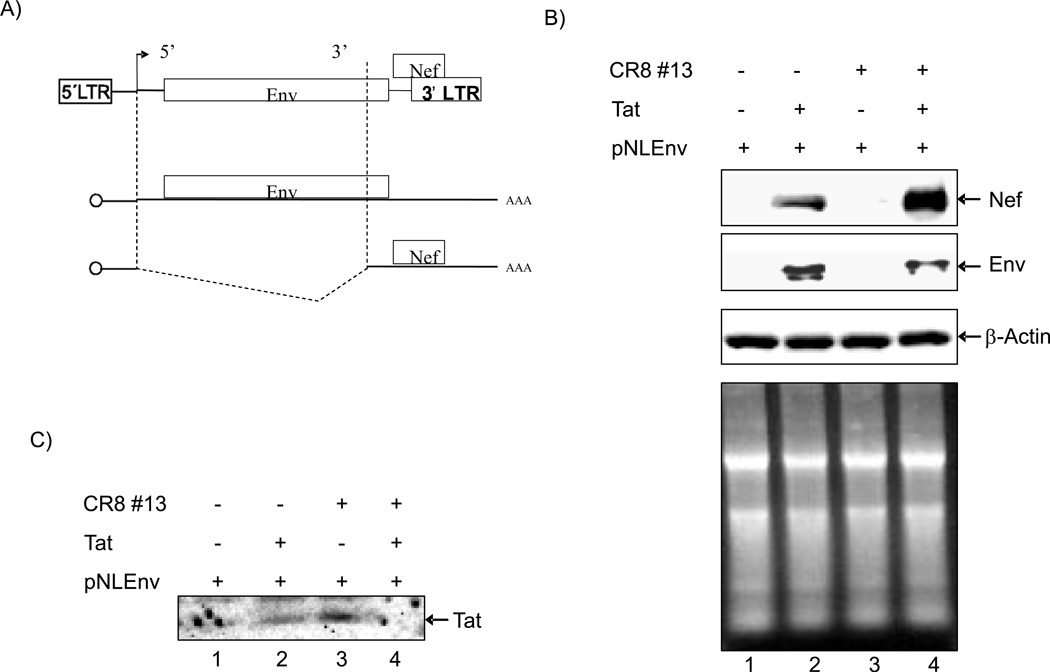
To show whether treatment with CR8#13 leads to not only regulation of transcription but also splicing, we performed an experiment using a HIV-1 vector with differential splicing patterns. In the plasmid pNLEnv, the Env gene is expressed from an unspliced mRNA, whereas Nef is made from a spliced mRNA (Figure 4, Panel A). We transfected 293T cells with pNLEnv DNA along with Tat alone or Tat plus CR8#13. The production of Nef and Env proteins were determined by Western blot analysis. Results in Figure 4, Panel B show that CR8#13 was able to increase doubly spliced Nef and lower Env proteins. This change in splicing pattern was Tat specific, since in the absence of Tat we observed virtually no rate of transcription or splicing. When testing for β-actin, we observed similar levels of proteins from both extracts. The total amount of RNA from these cells also did not change significantly after drug treatment. Finally, we performed a Tat western blot from transfected cells and were able to detect Tat (albeit at low levels) in these cells (Panel C). Collectively, these data indicate that factors that affect splicing may be regulated by cdk9 and in turn may explain, at least partially, the mechanisms of drug inhibition and downstream events leading to loss of viral production.
Figure 4. Effects of CR8#13 on HIV-1 splicing.
A) Schematic of the pNLEnv plasmid. Envelope gene is expressed from an unspliced mRNA, whereas Nef is made from a spliced mRNA. B) 293T cells were transfected with pc-Tat plasmid (1 µg) and/or CR8#13 (10 nM). Western blot analysis using antibodies against Nef, or Env were performed after 3 days. Actin was used as control. To detect RNA levels of Nef and Env after drug treatment, 100 ng of total RNA was used for cDNA synthesis and PCR for Nef and Env regions. Total RNA samples were stained with ethidium bromide and run on a 2% denaturing gel. C) pNLEnv vector (20 µg) was transfected with pc-Tat (10 µg) into 293T cells and treated with CR8#13 (10 nM) for 3 days. Soluble nuclear proteins were obtained and 50 µg of nuclear extract was run on 4–20% SDS/PAGE and western blotted with α-Tat rabbit polyclonal antibody (5 µg; generous gift from Dr. Bryan Cullen; (Zhou et al., 2004).
Effect of CR8#13 in a HIV-1 infected cell line and infected PBMCs
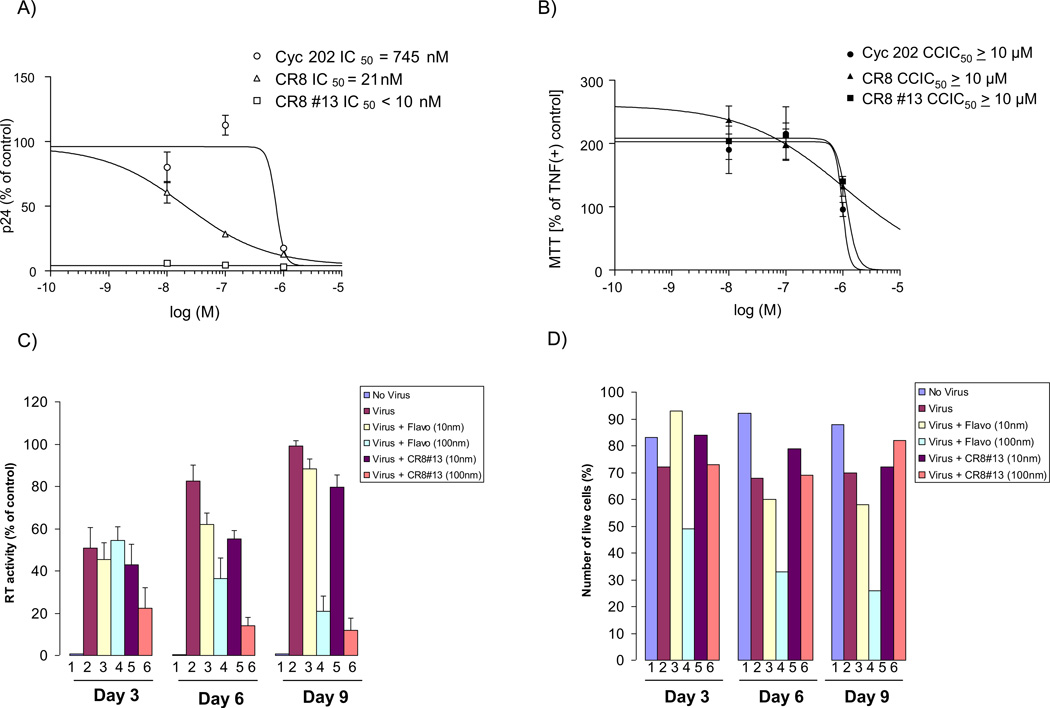
We next designed experiments to determine IC50 and MTT for cells treated with a titration of CR8#13 and other inhibitors. TNF-α activated OM10.1 cells (to activate the latent virus) were cultured for five days in the presence of the drug (one time treatment only) and supernatants were collected and examined for HIV-1 replication using p24 ELISA. The IC50 for CR8#13 was less than 10 nM in these infected cells (Figure 5, Panel A). These results indicate that there is a ~3 log difference between the inhibition on HIV-1 and cellular toxicity (Figure 5, Panel B). We next performed similar experiments in PBMCs infected with HIV-1 Thailand strain and followed the infected PBMCs up to days 3, 6 and 9 for presence of RT in the sups and also counted the number of live cells at each time point. We used Flavopiridol (10 and 100 nM) as positive control for inhibition of HIV-1 in these cells. Results in Panel C show that Flavopiridol at concentration of 100 nM (lanes 4) showed clear inhibition of virus by days 6 and 9. Similar results were obtained for CR8#13 treated cells where 100 nM concentrations effectively inhibited more than 90% of the RT activity in the supernatant (day 9). However, a more interesting story emerged when we counted the number of live cells after infection.
Figure 5. Effect of CR8#13 in latently infected cells and its effect in PBMCs.
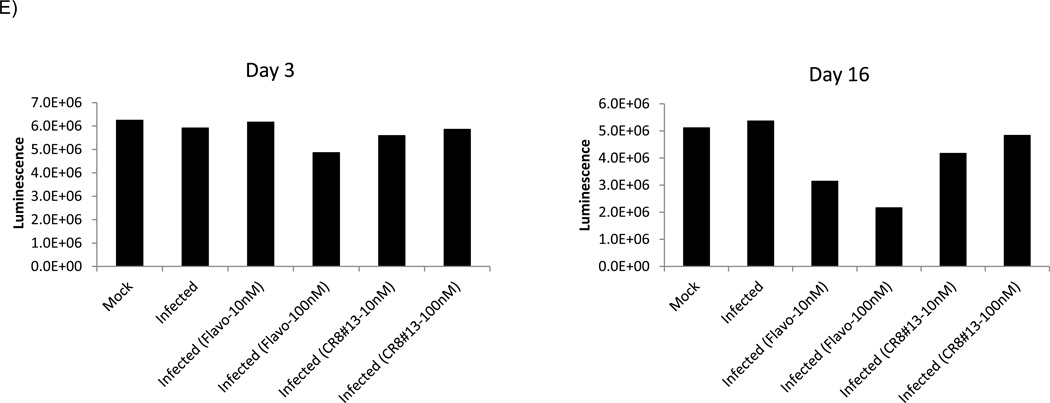
A) CYC202, CR8 and CR8#13 were used to treat activated OM10.1 cells. Cells were grown to mid-log phase of growth and treated with TNF-α for 2 hours to induce HIV-1 expression. TNF-α was then removed and cells were subsequently treated with drugs at 0.001, 0.01, 0.1, 1, 10 and 100 µM in complete media. Five days later, sups were collected and processed for p24 HIV-1 Gag ELISA. B) At day five cells were processed for MTT assay. Analysis was carried out with Prizm software using non-liner regression curve-fit of sigmoidal dose-response with variable slope. C & D) PHA-activated PBMC (5 × 106) were infected with THA/92/00 strain (MOI: 0.1). After 8 hours, unadsorbed virus was washed away and cells were treated with 10 or 100 nM of either Flavopiridol or CR8#13. Supernatants were collected every 3 days for RT analysis. Number of live cells was also plotted from each date and treatment (trypan blue exclusion assay). E &F) Similar to panel C, where PHA-activated PBMC cells (Lonza) were infected with the THA/92/00 strain and treated with various concentrations of Flavopiridol or CR8#13 cells. Samples were assayed using CellTiter-Glo after either 3 or 16 days.
Results in panel D showed that Flavopiridol at 100 nM was beginning to show increased toxicity at Days 6 and 9 (above what HIV-1 infection in PBMCs would normally show). This is consistent with the effect of Flavopiridol in long term cultures (Biglione et al., 2007). However, CR8#13 treated cells did not show appreciable cell death in Days 6 or 9 in these infected cells. Finally, we performed a similar experiment with another set of activated PBMC cells and treated these infected cells with Flavopiridol and CR8#13 at varying concentrations. To utilize an alternate assay to measure viable cells, we used CellTiter-Glo viability analysis for both early (day 3) and late (day 16) samples (Panel E). Again we observed a better rate of survivability in cells that were treated with CR8#13 as compared to Flavopiridol. Collectively, these results show that when using a low concentration of CR8#13, the level of HIV-1 expression is decreased, but cell death is not the reason for this apparent inhibition.
Effect of drug treatment on HIV-1 variants
Globally circulating HIV-1 sequences have been classified as major group M (subdivided in to 11 distinct subtypes named A–K) and two highly divergent groups named O and N. After the first description of the HIV-1 recombinant isolates, a substantial proportion of viral strains isolated from different geographical regions have been determined to comprise inter-subtype recombinants, and currently available full-length sequences of isolates classified as subtypes E, G, and I have been found to embody different regions of distinct HIV-1 clades (Koulinska et al., 2001). We next asked whether CR8#13 could differentially inhibit various HIV-1 clades due to nucleotide variations in the promoter or Tat open reading frame of these viruses. Subtype clade variations are apparent within the promoter region and Tat of main subtypes B, C, and E, which have been shown to include alterations in the TATA box, the NF-κB enhancer, and the TAR element, as well as other modulatory elements such as Sp1, USF, and NF-AT binding sites (Jeeninga et al., 2000; Montano et al., 1998). Also, the Tat activator from subtype E, compared to Tat B or C, possesses the highest transactivation capacity (Roof et al., 2002).
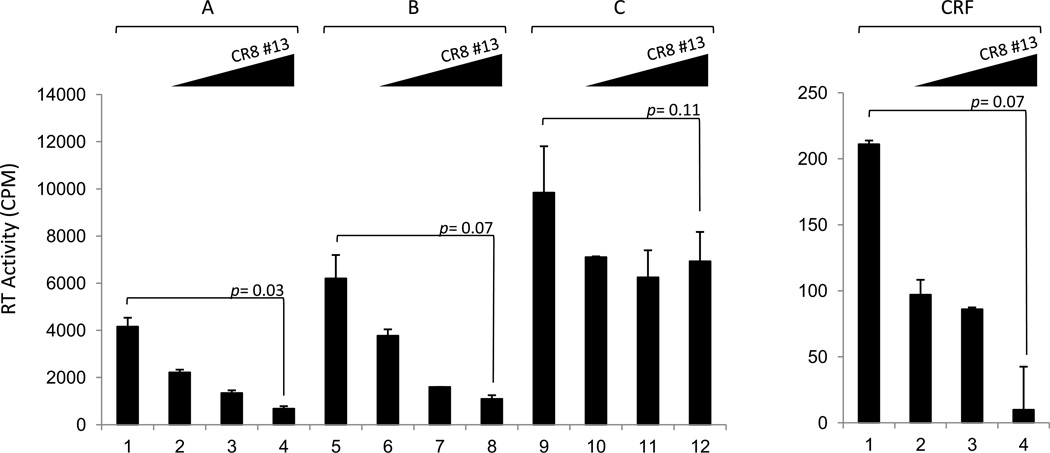
Along these lines, we performed infections using 4 different HIV-1 isolates. Activated PBMCs were infected with subtypes A, B, C and CRF AB. Two hours after adsorption/infection, cells were treated with a titration of CR8#13. Samples were collected after 7 days for RT assay. Results in Figure 6 show that all four strains of virus had different kinetics of replication in these cells. For instance, clade C replicated best and CRF replicated the least in our assays. Interestingly, a titration of CR8#13 showed effective inhibition for clades A, B and CRF but not so much for clade C (lanes 10–12). These results are consistent with previous results where differences with Tat half life and activity of the LTRs are observed in these clades. (Berro et al., 2006; Campbell et al., 2010; Desfosses et al., 2005; Gandhi et al., 2009; Li et al., 2008; Rao et al., 2008; Roof et al., 2002; Samikkannu et al., 2009; Wong et al., 2010). Collectively, these data indicate that transcription inhibitors may be clade specific and the activity of both LTR and Tat may dictate the efficacy of these agents.
Figure 6. Varying response of viral clades to CR8#13 treatment.
Approximately 5 × 106 PBMCs were infected with Subtype A: 92UG037.1; Subtype B: HxB2; Subtype C: 96BW05.02; and CRF AB: KAL153 (5 ng of p24 gag antigen/virus). After 2 hours of infection, cells were washed and fresh medium was added. Drug treatment (CR8#13; 0.1, 1, and 10 nM) was performed (only once) immediately after the addition of fresh medium. Samples were collected at day 7 for RT assay.
Discussion
HIV-1 virology and pathogenetic mechanisms of infection are continuously being investigated. A detailed understanding of HIV-1 structure and how it establishes infection and causes AIDS are crucial not only to identify and develop new effective drugs and vaccines, but also to define strategies for the diagnosis of HIV infection. Further, studies on the different viral subtypes and recombinant forms have shown that marked differences in the infection cycle may occur based on the phylogenetic and geographic origin of HIV-1 isolates. This is key for the design of new preventive and therapeutic approaches aimed at counteracting molecules essential for virus cycle. These acquisitions have also enabled the development of novel drugs aimed at an earlier and more accurate inhibition of virus antigens and virus specific replication steps in biological samples.
In the current manuscript we show that 3rd generation derivatives of R-roscovitine, such as CR8#13, are effective in inhibiting HIV-1 replication in cell culture systems. The main mechanism of action is at the level of LTR activation by Tat and this inhibition is directly linked to inhibition of cdk9/T1 kinase activity from infected cells (Figure 1). One of the unique features of our study is the isolation of active cdk9/T1 complex from infected cells which may have a different set of properties/substrate specificity compared to uninfected cells.
Our data regarding factor occupancy also show that both cdk9 and RNA Pol II uploading onto the LTR and Env region is severely decreased in the presence of CR8#13. This may be partially because cdk9 trafficking is altered in these drug treated cells (Figure 2B), which further reinforces the significance of localization for Tat activated transcription to occur. Current studies in our lab and a number of other labs are pursuing this line of research and asking whether Tat or modified Tat can control movement of proteins from cytoplasm to nucleus.
When looking for substrates for CR8#13 in uninfected cells, we were surprised to find that a new cdk9/T1 complex may exist in HIV-1 chronically infected cells. Data in Figure 3 indicate that when cells produce active virus there are multiple novel cdk9/T1 complexes including a small complex that is unique to infected cells, kinase active, and sensitive to CR8#13 treatment. This is a unique window of opportunity to tackle cellular proteins such as cdk9/T1 that are altered as a result of infection. To our knowledge no other lab has examined cdk9/T1 complexes that are formed in cells that actively produce virus, making this observation fairly unique. Western blot analysis of these fractions have shown Hexim 1 (a negative regulator of cdk9/T1 complex) to be present in complex III making the small complex IV more attractive as a target of inhibition. Current proteomic experiments are in progress to define protein partners of the cdk9 complex in complexes I–IV.
Our analysis of splicing has also shown that Tat is involved in the regulation of mRNA (and possibly pri-microRNA such as an extended TAR) regulation in infected cells. It is intriguing to speculate that CR8#13 regulation of cdk9 not only controls transcriptional elongation but also regulates splicing of HIV-1 mRNA in infected cells. Finally, we have utilized CR8#13 in various infectious settings using 3 different clades and one recombinant form of virus. We were surprised to see that CR8#13 inhibits some clades better than others (Figure 6). Our current understanding of the LTR and Tat from these clades indicates the presence of minor nucleotide changes in LTR and increased lysine residues in some, but not all, Tats (i.e. clade C). Therefore, regulation of LTR in these clades may require complexes that are not always dependent on cdk9/T1 activity. This would point to an examination of various clades in both T-cells and monocyte derived macrophages MDM for their requirements in using cdk9/T1 complexes. Alternately, a different cdk9/T1 complex (i.e., complex I) may be the dominant complex in binding to clades such as clade C, which would have a different rate of transcription, splicing events and varying sensitivity to ATP analogs. Future experiments will determine the complexes present on various LTRs from these clades and how they may be sensitive to transcription inhibitors.
Conclusions
A third generation derivative of R-roscovitine named CR8#13 is an effective inhibitor of Tat activated transcription.
CR8#13 interferes with the loading of cdk9 onto the HIV-1 promoter and this may be a critical component that contributes to the anti-viral efficacy of CR8#13.
We have found that a novel cdk9/T1 complex that is present only in cells that actively express and shed virus. It was striking to observe that this novel cdk9/T1 complex is sensitive to CR8#13 as determined by in vitro kinase assays.
Treatment of PBMC with CR8#13 does not cause mortality of infected cells in long term cultures as compared to other cdk9 inhibitors such as Flavopiridol.
Finally, we show that various clades of HIV-1 may be differentially sensitive to these ATP analogs.
Acknowledgements
We would like to thank the members of the Kashanchi lab for experiments and assistance with the manuscript. This research was also funded by the “Conseil Régional de Bretagne”, ‘Fonds de Maturation’, « Développement de molécules inhibitrices de kinases à activité anti-SIDA. Optimisation du mode d'administration par voie orale/intrapéritonéale et validation sur modèle animal » to (LM). Further support came from grants from the George Mason University funds to FK and NIH grants AI043894, AI078859, and AI074410-01.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams M, Sharmeen L, Kimpton J, Romeo JM, Garcia JV, Peterlin BM, Groudine M, Emerman M. Cellular latency in human immunodeficiency virus-infected individuals with high CD4 levels can be detected by the presence of promoter-proximal transcripts. Proc Natl Acad Sci U S A. 1994;91:3862–3866. doi: 10.1073/pnas.91.9.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agbottah E, de La Fuente C, Nekhai S, Barnett A, Gianella-Borradori A, Pumfery A, Kashanchi F. Antiviral activity of CYC202 in HIV-1-infected cells. J Biol Chem. 2005;280:3029–3042. doi: 10.1074/jbc.M406435200. [DOI] [PubMed] [Google Scholar]
- Agbottah E, Zhang N, Dadgar S, Pumfery A, Wade JD, Zeng C, Kashanchi F. Inhibition of HIV-1 virus replication using small soluble Tat peptides. Virology. 2006;345:373–389. doi: 10.1016/j.virol.2005.09.062. [DOI] [PubMed] [Google Scholar]
- Ammosova T, Berro R, Jerebtsova M, Jackson A, Charles S, Klase Z, Southerland W, Gordeuk VR, Kashanchi F, Nekhai S. Phosphorylation of HIV-1 Tat by CDK2 in HIV-1 transcription. Retrovirology. 2006;3:78. doi: 10.1186/1742-4690-3-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammosova T, Berro R, Kashanchi F, Nekhai S. RNA interference directed to CDK2 inhibits HIV-1 transcription. Virology. 2005;341:171–178. doi: 10.1016/j.virol.2005.06.041. [DOI] [PubMed] [Google Scholar]
- Antoni BA, Stein SB, Rabson AB. Regulation of human immunodeficiency virus infection: implications for pathogenesis. Adv Virus Res. 1994;43:53–145. doi: 10.1016/s0065-3527(08)60047-0. [DOI] [PubMed] [Google Scholar]
- Berro R, Kehn K, de la Fuente C, Pumfery A, Adair R, Wade J, Colberg-Poley AM, Hiscott J, Kashanchi F. Acetylated Tat regulates human immunodeficiency virus type 1 splicing through its interaction with the splicing regulator p32. J Virol. 2006;80:3189–3204. doi: 10.1128/JVI.80.7.3189-3204.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berro R, Pedati C, Kehn-Hall K, Wu W, Klase Z, Even Y, Geneviere AM, Ammosova T, Nekhai S, Kashanchi F. CDK13, a new potential human immunodeficiency virus type 1 inhibitory factor regulating viral mRNA splicing. J Virol. 2008;82:7155–7166. doi: 10.1128/JVI.02543-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biglione S, Byers SA, Price JP, Nguyen VT, Bensaude O, Price DH, Maury W. Inhibition of HIV-1 replication by P-TEFb inhibitors DRB, seliciclib and flavopiridol correlates with release of free P-TEFb from the large, inactive form of the complex. Retrovirology. 2007;4:47. doi: 10.1186/1742-4690-4-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bres V, Yoh SM, Jones KA. The multi-tasking P-TEFb complex. Curr Opin Cell Biol. 2008;20:334–340. doi: 10.1016/j.ceb.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell GR, Loret EP, Spector SA. HIV-1 clade B Tat, but not clade C Tat, increases X4 HIV-1 entry into resting but not activated CD4+ T cells. J Biol Chem. 2010;285:1681–1691. doi: 10.1074/jbc.M109.049957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpio L, Klase Z, Coley W, Guendel I, Choi S, Van Duyne R, Narayanan A, Kehn-Hall K, Meijer L, Kashanchi F. microRNA machinery is an integral component of drug-induced transcription inhibition in HIV-1 infection. J RNAi Gene Silencing. 2010;6:386–400. [PMC free article] [PubMed] [Google Scholar]
- Chao SH, Fujinaga K, Marion JE, Taube R, Sausville EA, Senderowicz AM, Peterlin BM, Price DH. Flavopiridol inhibits P-TEFb and blocks HIV-1 replication. J Biol Chem. 2000;275:28345–28348. doi: 10.1074/jbc.C000446200. [DOI] [PubMed] [Google Scholar]
- Chen Y, Yamaguchi Y, Tsugeno Y, Yamamoto J, Yamada T, Nakamura M, Hisatake K, Handa H. DSIF, the Paf1 complex, and Tat-SF1 have nonredundant, cooperative roles in RNA polymerase II elongation. Genes Dev. 2009;23:2765–2777. doi: 10.1101/gad.1834709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Y, Grant S. Small molecule inhibitors targeting cyclin-dependent kinases as anticancer agents. Curr Oncol Rep. 2004;6:123–130. doi: 10.1007/s11912-004-0024-3. [DOI] [PubMed] [Google Scholar]
- De Clercq E. New developments in anti-HIV chemotherapy. Biochim Biophys Acta. 2002;1587:258–275. doi: 10.1016/s0925-4439(02)00089-3. [DOI] [PubMed] [Google Scholar]
- de la Fuente C, Maddukuri A, Kehn K, Baylor SY, Deng L, Pumfery A, Kashanchi F. Pharmacological cyclin-dependent kinase inhibitors as HIV-1 antiviral therapeutics. Curr HIV Res. 2003;1:131–152. doi: 10.2174/1570162033485339. [DOI] [PubMed] [Google Scholar]
- Debebe Z, Ammosova T, Breuer D, Lovejoy DB, Kalinowski DS, Kumar K, Jerebtsova M, Ray P, Kashanchi F, Gordeuk VR, Richardson DR, Nekhai S. Iron chelators of the di-2-pyridylketone thiosemicarbazone and 2-benzoylpyridine thiosemicarbazone series inhibit HIV-1 transcription: identification of novel cellular targets--iron, cyclin-dependent kinase (CDK) 2, and CDK9. Mol Pharmacol. 2011;79:185–196. doi: 10.1124/mol.110.069062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desfosses Y, Solis M, Sun Q, Grandvaux N, Van Lint C, Burny A, Gatignol A, Wainberg MA, Lin R, Hiscott J. Regulation of human immunodeficiency virus type 1 gene expression by clade-specific Tat proteins. J Virol. 2005;79:9180–9191. doi: 10.1128/JVI.79.14.9180-9191.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galons H, Oumata N, Meijer L. Cyclin-dependent kinase inhibitors: a survey of recent patent literature. Expert Opin Ther Pat. 2010;20:377–404. doi: 10.1517/13543770903524284. [DOI] [PubMed] [Google Scholar]
- Gandhi N, Saiyed Z, Thangavel S, Rodriguez J, Rao KV, Nair MP. Differential effects of HIV type 1 clade B and clade C Tat protein on expression of proinflammatory and antiinflammatory cytokines by primary monocytes. AIDS Res Hum Retroviruses. 2009;25:691–699. doi: 10.1089/aid.2008.0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garriga J, Bhattacharya S, Calbo J, Marshall RM, Truongcao M, Haines DS, Grana X. CDK9 is constitutively expressed throughout the cell cycle, and its steady-state expression is independent of SKP2. Mol Cell Biol. 2003;23:5165–5173. doi: 10.1128/MCB.23.15.5165-5173.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garriga J, Peng J, Parreno M, Price DH, Henderson EE, Grana X. Upregulation of cyclin T1/CDK9 complexes during T cell activation. Oncogene. 1998;17:3093–3102. doi: 10.1038/sj.onc.1202548. [DOI] [PubMed] [Google Scholar]
- Greene WC, Peterlin BM. Charting HIV's remarkable voyage through the cell: Basic science as a passport to future therapy. Nat Med. 2002;8:673–680. doi: 10.1038/nm0702-673. [DOI] [PubMed] [Google Scholar]
- Guendel I, Agbottah ET, Kehn-Hall K, Kashanchi F. Inhibition of human immunodeficiency virus type-1 by cdk inhibitors. AIDS Res Ther. 2010;7:7. doi: 10.1186/1742-6405-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He N, Liu M, Hsu J, Xue Y, Chou S, Burlingame A, Krogan NJ, Alber T, Zhou Q. HIV-1 Tat and host AFF4 recruit two transcription elongation factors into a bifunctional complex for coordinated activation of HIV-1 transcription. Mol Cell. 2010;38:428–438. doi: 10.1016/j.molcel.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonski JA, Amelio AL, Giacca M, Caputi M. The transcriptional transactivator Tat selectively regulates viral splicing. Nucleic Acids Res. 2010;38:1249–1260. doi: 10.1093/nar/gkp1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeeninga RE, Hoogenkamp M, Armand-Ugon M, de Baar M, Verhoef K, Berkhout B. Functional differences between the long terminal repeat transcriptional promoters of human immunodeficiency virus type 1 subtypes A through G. J Virol. 2000;74:3740–3751. doi: 10.1128/jvi.74.8.3740-3751.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapasi AJ, Spector DH. Inhibition of the cyclin-dependent kinases at the beginning of human cytomegalovirus infection specifically alters the levels and localization of the RNA polymerase II carboxyl-terminal domain kinases cdk9 and cdk7 at the viral transcriptosome. J Virol. 2008;82:394–407. doi: 10.1128/JVI.01681-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karn J. Tackling Tat. J Mol Biol. 1999;293:235–254. doi: 10.1006/jmbi.1999.3060. [DOI] [PubMed] [Google Scholar]
- Karn J, Stoltzfus CM. Transcriptional and Posttranscriptional Regulation of HIV-1 Gene Expression. Cold Spring Harb Perspect Med. 2012;2 doi: 10.1101/cshperspect.a006916. a006916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashanchi F, Duvall JF, Brady JN. Electroporation of viral transactivator proteins into lymphocyte suspension cells. Nucleic Acids Res. 1992;20:4673–4674. doi: 10.1093/nar/20.17.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashanchi F, Shibata R, Ross EK, Brady JN, Martin MA. Second-site long terminal repeat (LTR) revertants of replication-defective human immunodeficiency virus: effects of revertant TATA box motifs on virus infectivity, LTR-directed expression, in vitro RNA synthesis, and binding of basal transcription factors TFIID and TFIIA. J Virol. 1994;68:3298–3307. doi: 10.1128/jvi.68.5.3298-3307.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiernan RE, Emiliani S, Nakayama K, Castro A, Labbe JC, Lorca T, Nakayama Ki K, Benkirane M. Interaction between cyclin T1 and SCF(SKP2) targets CDK9 for ubiquitination and degradation by the proteasome. Mol Cell Biol. 2001;21:7956–7970. doi: 10.1128/MCB.21.23.7956-7970.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JB, Yamaguchi Y, Wada T, Handa H, Sharp PA. Tat-SF1 protein associates with RAP30 and human SPT5 proteins. Mol Cell Biol. 1999;19:5960–5968. doi: 10.1128/mcb.19.9.5960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koulinska IN, Ndung'u T, Mwakagile D, Msamanga G, Kagoma C, Fawzi W, Essex M, Renjifo B. A new human immunodeficiency virus type 1 circulating recombinant form from Tanzania. AIDS Res Hum Retroviruses. 2001;17:423–431. doi: 10.1089/088922201750102508. [DOI] [PubMed] [Google Scholar]
- Li W, Huang Y, Reid R, Steiner J, Malpica-Llanos T, Darden TA, Shankar SK, Mahadevan A, Satishchandra P, Nath A. NMDA receptor activation by HIV-Tat protein is clade dependent. J Neurosci. 2008;28:12190–12198. doi: 10.1523/JNEUROSCI.3019-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Coutinho-Mansfield G, Wang D, Pandit S, Fu XD. The splicing factor SC35 has an active role in transcriptional elongation. Nat Struct Mol Biol. 2008;15:819–826. doi: 10.1038/nsmb.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapendano CK, Lykke-Andersen S, Kjems J, Bertrand E, Jensen TH. Crosstalk between mRNA 3' end processing and transcription initiation. Mol Cell. 2010;40:410–422. doi: 10.1016/j.molcel.2010.10.012. [DOI] [PubMed] [Google Scholar]
- Mbonye U, Karn J. Control of HIV latency by epigenetic and non-epigenetic mechanisms. Curr HIV Res. 2011;9:554–567. doi: 10.2174/157016211798998736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller HB, Saunders KO, Tomaras GD, Garcia-Blanco MA. Tat-SF1 is not required for Tat transactivation but does regulate the relative levels of unspliced and spliced HIV-1 RNAs. PLoS One. 2009;4:e5710. doi: 10.1371/journal.pone.0005710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montano MA, Nixon CP, Essex M. Dysregulation through the NF-kappaB enhancer and TATA box of the human immunodeficiency virus type 1 subtype E promoter. J Virol. 1998;72:8446–8452. doi: 10.1128/jvi.72.10.8446-8452.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekhai S, Zhou M, Fernandez A, Lane WS, Lamb NJ, Brady J, Kumar A. HIV-1 Tat-associated RNA polymerase C-terminal domain kinase, CDK2, phosphorylates CDK7 and stimulates Tat-mediated transcription. Biochem J. 2002;364:649–657. doi: 10.1042/BJ20011191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien SK, Cao H, Nathans R, Ali A, Rana TM. P-TEFb kinase complex phosphorylates histone H1 to regulate expression of cellular and HIV-1 genes. J Biol Chem. 2010;285:29713–29720. doi: 10.1074/jbc.M110.125997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien SK, Knight KL, Rana TM. Phosphorylation of histone H1 by P-TEFb is a necessary step in skeletal muscle differentiation. J Cell Physiol. 2012;227:383–389. doi: 10.1002/jcp.22797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oumata N, Bettayeb K, Ferandin Y, Demange L, Lopez-Giral A, Goddard ML, Myrianthopoulos V, Mikros E, Flajolet M, Greengard P, Meijer L, Galons H. Roscovitine-derived, dual-specificity inhibitors of cyclin-dependent kinases and casein kinases 1. J Med Chem. 2008;51:5229–5242. doi: 10.1021/jm800109e. [DOI] [PubMed] [Google Scholar]
- Parada CA, Roeder RG. A novel RNA polymerase II-containing complex potentiates Tat-enhanced HIV-1 transcription. EMBO J. 1999;18:3688–3701. doi: 10.1093/emboj/18.13.3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirngruber J, Shchebet A, Johnsen SA. Insights into the function of the human P-TEFb component CDK9 in the regulation of chromatin modifications and co-transcriptional mRNA processing. Cell Cycle. 2009;8:3636–3642. doi: 10.4161/cc.8.22.9890. [DOI] [PubMed] [Google Scholar]
- Rao VR, Sas AR, Eugenin EA, Siddappa NB, Bimonte-Nelson H, Berman JW, Ranga U, Tyor WR, Prasad VR. HIV-1 clade-specific differences in the induction of neuropathogenesis. J Neurosci. 2008;28:10010–10016. doi: 10.1523/JNEUROSCI.2955-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roof P, Ricci M, Genin P, Montano MA, Essex M, Wainberg MA, Gatignol A, Hiscott J. Differential regulation of HIV-1 clade-specific B, C, and E long terminal repeats by NF-kappaB and the Tat transactivator. Virology. 2002;296:77–83. doi: 10.1006/viro.2001.1397. [DOI] [PubMed] [Google Scholar]
- Samikkannu T, Saiyed ZM, Rao KV, Babu DK, Rodriguez JW, Papuashvili MN, Nair MP. Differential regulation of indoleamine-2,3-dioxygenase (IDO) by HIV type 1 clade B and C Tat protein. AIDS Res Hum Retroviruses. 2009;25:329–335. doi: 10.1089/aid.2008.0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon V, Vanderhoeven J, Hurley A, Ramratnam B, Louie M, Dawson K, Parkin N, Boden D, Markowitz M. Evolving patterns of HIV-1 resistance to antiretroviral agents in newly infected individuals. AIDS. 2002;16:1511–1519. doi: 10.1097/00002030-200207260-00008. [DOI] [PubMed] [Google Scholar]
- Sobhian B, Laguette N, Yatim A, Nakamura M, Levy Y, Kiernan R, Benkirane M. HIV-1 Tat assembles a multifunctional transcription elongation complex and stably associates with the 7SK snRNP. Mol Cell. 2010;38:439–451. doi: 10.1016/j.molcel.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Yang J, Brasier AR. Two-step cross-linking for analysis of protein-chromatin interactions. Methods Mol Biol. 2012;809:105–120. doi: 10.1007/978-1-61779-376-9_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, de la Fuente C, Deng L, Wang L, Zilberman I, Eadie C, Healey M, Stein D, Denny T, Harrison LE, Meijer L, Kashanchi F. Inhibition of human immunodeficiency virus type 1 transcription by chemical cyclin-dependent kinase inhibitors. J Virol. 2001;75:7266–7279. doi: 10.1128/JVI.75.16.7266-7279.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei P, Garber ME, Fang SM, Fischer WH, Jones KA. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- Wong JK, Campbell GR, Spector SA. Differential induction of interleukin-10 in monocytes by HIV-1 clade B and clade C Tat proteins. J Biol Chem. 2010;285:18319–18325. doi: 10.1074/jbc.M110.120840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Gold MO, Tang DN, Lewis DE, Aguilar-Cordova E, Rice AP, Herrmann CH. TAK, an HIV Tat-associated kinase, is a member of the cyclin-dependent family of protein kinases and is induced by activation of peripheral blood lymphocytes and differentiation of promonocytic cell lines. Proc Natl Acad Sci U S A. 1997;94:12331–12336. doi: 10.1073/pnas.94.23.12331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Deng L, Kashanchi F, Brady JN, Shatkin AJ, Kumar A. The Tat/TAR-dependent phosphorylation of RNA polymerase II C-terminal domain stimulates cotranscriptional capping of HIV-1 mRNA. Proc Natl Acad Sci U S A. 2003;100:12666–12671. doi: 10.1073/pnas.1835726100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Deng L, Lacoste V, Park HU, Pumfery A, Kashanchi F, Brady JN, Kumar A. Coordination of transcription factor phosphorylation and histone methylation by the P-TEFb kinase during human immunodeficiency virus type 1 transcription. J Virol. 2004;78:13522–13533. doi: 10.1128/JVI.78.24.13522-13533.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Chen D, Pierstorff E, Luo K. Transcription elongation factor P-TEFb mediates Tat activation of HIV-1 transcription at multiple stages. EMBO J. 1998;17:3681–3691. doi: 10.1093/emboj/17.13.3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Pe'ery T, Peng J, Ramanathan Y, Marshall N, Marshall T, Amendt B, Mathews MB, Price DH. Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes Dev. 1997;11:2622–2632. doi: 10.1101/gad.11.20.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]