The mitochondrial outer membrane protein Tom40 is the entry gate for imported proteins in essentially all eukaryotes. Trypanosomatids lack a conventional Tom40 and use instead a protein termed ATOM. pATOM36 is a novel essential component of the trypanosomal outer membrane protein import system that interacts with ATOM.
Abstract
The mitochondrial outer membrane protein Tom40 is the general entry gate for imported proteins in essentially all eukaryotes. Trypanosomatids lack Tom40, however, and use instead a protein termed the archaic translocase of the outer mitochondrial membrane (ATOM). Here we report the discovery of pATOM36, a novel essential component of the trypanosomal outer membrane protein import system that interacts with ATOM. pATOM36 is not related to known Tom proteins from other organisms and mediates the import of matrix proteins. However, there is a group of precursor proteins whose import is independent of pATOM36. Domain-swapping experiments indicate that the N-terminal presequence-containing domain of the substrate proteins at least in part determines the dependence on pATOM36. Secondary structure profiling suggests that pATOM36 is composed largely of α-helices and its assembly into the outer membrane is independent of the sorting and assembly machinery complex. Taken together, these results show that pATOM36 is a novel component associated with the ATOM complex that promotes the import of a subpopulation of proteins into the mitochondrial matrix.
INTRODUCTION
The acquisition of a bacterial endosymbiont by a primitive host cell and its subsequent conversion into the mitochondrion marks one of the most important transitions in biology—the advent of the eukaryotic cell (Embley and Martin, 2006). A defining aspect of mitochondria is their capability to import proteins from the cytosol, a process that is driven by a set of characteristic protein translocases (Neupert and Herrmann, 2007; Lithgow and Schneider, 2010; Schmidt et al., 2010; Endo et al., 2011).
In the outer membrane we find two such translocases. The most important one—the general entry gate for essentially all imported proteins—is the translocase of the outer mitochondrial membrane (TOM). It consists of the pore-forming core subunit Tom40 (Hill et al., 1998; Ahting et al., 2001), a β-barrel–structured protein, protein import receptors (such as Tom70, Tom20, and Tom22 in yeast), and of a number of smaller subunits. The other translocase in the outer membrane is the sorting and assembly machinery (SAM), whose function is the insertion of β-barrel proteins. Its core subunit is Sam50, which is itself a β-barrel protein (Chacinska et al., 2009).
Extensive studies have revealed that most components and much of the architecture of the TOM and the SAM are conserved between yeast and mammals (Hoogenraad et al., 2002; Dolezal et al., 2006; Schneider et al., 2008; Hewitt et al., 2011). However a more global analysis shows that only the core components of the mitochondrial protein translocases are conserved in all eukaryotes. Among these, Sam50 is the most highly conserved: it is found in all groups of eukaryotes and even has a bacterial orthologue, the Omp85-like protein BamA, that functions in inserting β-barrel proteins into the outer bacterial membrane (Bos et al., 2007). Tom40 is also conserved and found in virtually all eukaryotes; however, although the β-barrel structure of Tom40 would indicate a bacterial ancestry, no bacterial orthologue has been identified (Dolezal et al., 2006; Zeth, 2010).
The receptor subunits of the TOM complex are much less conserved. Thus, in plants Tom22 is severely truncated, and a conventional Tom20 is absent (Mac´asev et al., 2004). Instead we find a structural analogue of Tom20 that is coded in reverse, suggesting that the structural similarity between the mammalian and fungal Tom20 on one side and the plant Tom20 on the other side is due to convergent evolution and not to common descent (Perry et al., 2006). Tom70 appears to be missing in plants (Chan et al., 2006) but has recently been detected in some representatives of the Chromalveolata, which are unrelated to fungi, mammals, and plants, indicating that Tom70 might be more widespread than previously thought (Tsaousis et al., 2011).
Recent studies in a number of different protozoa revealed interesting but for the most part minor deviations to the canonical protein import systems described in yeast and mammals (Dolezal et al., 2005). It was therefore a surprise that trypanosomatids have a fundamentally different mitochondrial outer membrane translocation machinery. Trypanosomatids lack Tom40 (Pusnik et al., 2009) and instead employ an outer membrane protein translocation channel termed ATOM, for archaic translocase of the outer mitochondrial membrane (Pusnik et al., 2011). Unlike Tom40, ATOM has a bacterial orthologue. It shows similarities to a subgroup of the bacterial Omp85-like protein family that is distinct from the Sam50 orthologue BamA. This suggests that trypanosomatids have retained an archaic outer membrane protein import system that might have preceded the Tom40-based protein import channel.
The discovery of ATOM provided unexpected new insights into the evolution of the mitochondrial outer membrane protein import system and the evolution of eukaryotes in general. Moreover, it raised the question of what other factors might be required for protein translocation across the outer membrane of trypanosomal mitochondria. Using Trypanosoma brucei as an experimental model system, we discovered such a novel factor. This protein is an essential mitochondrial outer membrane protein that is required for import of a large subset but not all matrix proteins and therefore might have a receptor-like function.
RESULTS
A novel mitochondrial outer membrane protein of T. brucei
Mitochondrial protein import is known to be essential in all eukaryotes under all growth conditions (Baker and Schatz, 1991). Whereas oxidative phosphorylation is essential only in the insect stage of T. brucei, mitochondrial protein import is required throughout the life cycle. Outer mitochondrial membrane proteins that are essential for normal growth of both procyclic and bloodstream forms of the parasite are therefore prime candidates to be components of the mitochondrial protein import system. To find such candidates, we purified mitochondrial outer membranes from T. brucei and characterized their protein composition (unpublished data). Within the outer membrane proteome we found a relatively abundant protein encoded by the open reading frame (ORF) Tb927.7.5700, which in a recent global RNA interference (RNAi) analysis (Alsford et al., 2011) was found to be essential throughout the life cycle. The protein has a predicted molecular weight of 45.5 kDa, but global transcriptome analyses of T. brucei (Siegel et al., 2011), biochemical experiments, and phylogenetic profiling against other kinetoplastid genomes indicate that a gene-internal ATG (position 85 on the annotated amino acid sequence) is used as a start codon, resulting in a protein with a molecular weight of 35.7 kDa. For reasons outlined later, we termed the protein pATOM36 for peripheral subunit of the archaic protein translocase of the outer mitochondrial membrane of 36 kDa. pATOM36 has orthologues in all trypanosomatids, which on average are 49% identical to each other in protein sequence, and all are predicted to have an overall conserved secondary structure. We could find no evidence of related proteins outside the trypanosomatid lineage (TriTrypDB, http://tritrypdb.org), and the proteins have no obvious relationship to known protein domains as defined by the Conserved Domain Architecture Retrieval Tool (www.ncbi.nlm.nih.gov/Structure/lexington/lexington.cgi).
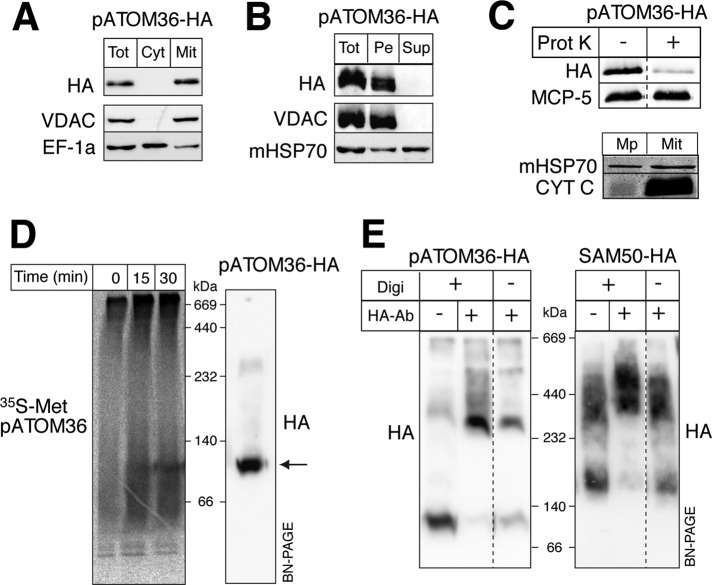
Digitonin fractionation of whole cells and carbonate extraction of gradient-purified mitochondria from cells expressing a C-terminally hemagglutinin (HA)-tagged version of pATOM36 (pATOM36-HA) showed that it is located in mitochondria and that it is an integral membrane protein (Figure 1, A and B).
FIGURE 1:
pATOM36 is an integral protein of the mitochondrial outer membrane. (A) Immunoblots of total (Tot), digitonin-extracted crude cytosolic (Cyt), and mitochondrial (Mit) fractions from a cell line expressing pATOM36 with an HA tag at its C-terminus (pATOM36-HA) probed with anti-HA antibodies (HA). VDAC and elongation factor 1a (EF-1a) serve as mitochondrial outer membrane and cytosolic marker, respectively. (B) Immunoblot of an alkaline carbonate extraction of pATOM36-HA–containing mitochondria. Pe, pellet; Sup, supernatant. The integral membrane protein VDAC and the soluble mHSP70 serve as markers. (C) Top, immunoblot of untreated and proteinase K-treated (50 μg/ml) pATOM36-HA–containing mitochondria. Mitochondrial carrier protein 5 (MCP-5) serves as a loading control. Bottom, immunoblot of isolated mitoplasts (Mp) having a disrupted outer membrane and the intact pATOM36-HA–containing mitochondria (Mit) shown at the top. CYT C and mHSP70 serve as markers for the intermembrane space and the matrix, respectively. (D) Left, import of in vitro–translated, 35S-Met–labeled pATOM36 into isolated mitochondria analyzed by BN-PAGE. Right, immunoblot of pATOM36-HA–containing mitochondrial extract separated on the same BN-PAGE. The main pATOM36 complex is indicated by the arrow. (E) Antibody shift experiments. Left, pATOM36-HA containing digitonin-lysed (+Digi) and intact mitochondria (−Digi) were incubated with polyclonal anti-HA antibodies (+HA, −antibody), separated by BN-PAGE, and analyzed by immunoblots using a monoclonal anti-HA antiserum (HA). Untreated pATOM36-HA (−HA, −antibody) complex is shown on the left as a control. Right, same as shown on the left, but SAM50-HA–expressing mitochondria were used.
To confirm that pATOM36 is localized in the outer mitochondrial membrane, we treated isolated mitochondria with proteinase K. Figure 1C shows that, as expected for an outer membrane protein, most of the tagged pATOM36 is digested under these conditions. The intactness of the mitochondria was assayed by comparing the ratios between the matrix protein mitochondrial heat shock protein 70 (mHSP70) and intermembrane space protein cytochrome C (CYT C). This ratio is expected to be high in mitoplasts, which are essentially devoid of CYT C, and low in intact mitochondria, where CYT C is retained in the intermembrane space (Schneider et al., 2007b).
Blue native gel electrophoresis (BN-PAGE) revealed that in vitro–translated 35S-Met–labeled pATOM36 is inserted into the outer membrane and forms a complex of ∼100 kDa when incubated with isolated pATOM36-HA–containing mitochondria (Figure 1D). Formation of this complex is time dependent, and comparisons with immunoblots from the same gels showed that it comigrates with the preexisting pATOM36-HA–containing complex.
Finally, to probe the topology of pATOM36, we performed antibody shift experiments. Intact and digitonin-solubilized pATOM36-HA–containing mitochondria were incubated with anti-HA antibodies. Subsequently the solubilized mitochondrial membranes were analyzed by BN-PAGE, and protein complexes were visualized on immunoblots using anti-HA antibodies. Figure 1E (left) shows that the pATOM36-HA–containing protein complex becomes shifted to a higher molecular weight independent of whether the antibody was added to intact or to digitonin-solubilized mitochondria. These results indicate that pATOM36 is a mitochondrial outer membrane protein whose C-terminus is facing the cytosol. As a control we did the same analysis using the previously characterized outer mitochondrial membrane protein SAM50 (Sharma et al., 2010). The BN-PAGE in Figure 1E (right) shows that when using mitochondria containing C-terminally HA-tagged SAM50 a shift of the SAM complex was only observed with digitonin-solubilized but not with intact mitochondria, indicating that the C-terminus of SAM50 is exposed to the intermembrane space, consistent with the known topology of other members of the Omp85-like family of proteins.
pATOM36 is not a β-barrel protein
The known receptors for mitochondrial protein import are anchored in the outer membrane via α-helices. The channel-building subunits of the outer membrane protein translocases, such as Tom40 and Sam50, on the other hand, are β-barrel–structured integral membrane proteins (Neupert and Herrmann, 2007; Chacinska et al., 2009).
Although pATOM36 sequences are predicted to be largely α-helical in nature, none of these α-helices has the hydrophobicity characteristics of “classic” transmembrane helices. It is worth noting that this is also true for other outer membrane proteins, including Tom6 and Tom7 (Allen et al., 2002). To experimentally investigate whether pATOM36 is a β-barrel protein, we used an RNAi cell line allowing inducible ablation of SAM50. The results in Figure 2 show that ablation of SAM50 causes a strong reduction of the levels of the voltage-dependent anion channel (VDAC), as expected, whereas the levels of pATOM36 are not affected. Thus insertion of pATOM36 into the outer membrane does not require SAM50.
FIGURE 2:
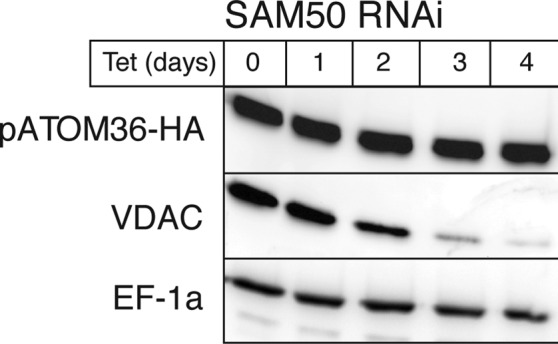
In vivo assembly of pATOM36 is independent of SAM50. Levels of pATOM36-HA in total cellular extracts are not affected, whereas the levels of the β-barrel protein VDAC declines during tet induction of SAM50 RNAi.
pATOM36 is a peripheral component of the ATOM complex
Does the essential outer membrane protein pATOM36 function in protein import?
To address this question, immunoprecipitations were performed to see whether pATOM36 interacts with ATOM or with SAM50, the only two components of the mitochondrial outer membrane known to be involved in mitochondrial protein import in T. brucei.
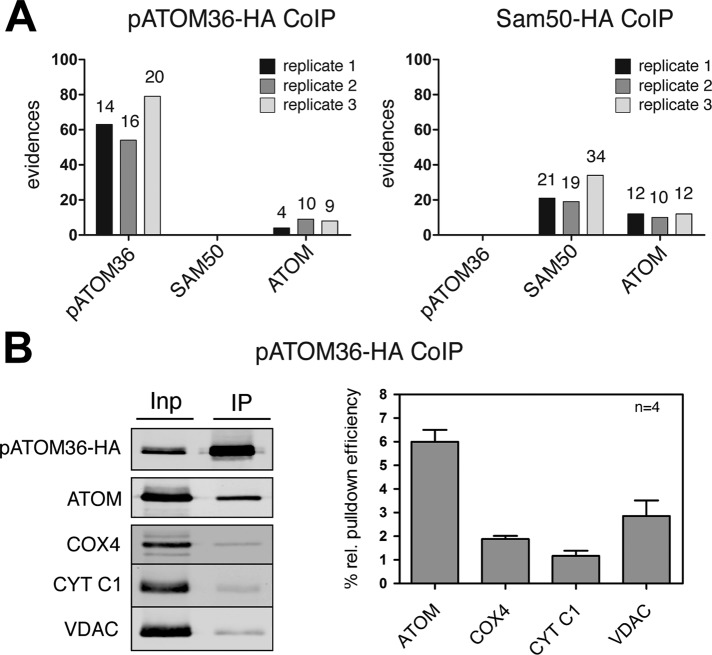
Gradient-purified mitochondria containing either HA-tagged pATOM36 or HA-tagged SAM50 were solubilized by digitonin and subjected to immunoprecipitation using anti-HA antibodies. The resulting eluates were analyzed for the presence of pATOM36, SAM50, and ATOM using mass spectrometry. Figure 3A shows that, as expected, the tagged proteins are efficiently recovered in the eluate. Moreover, a small amount of ATOM but not of SAM50 is reproducibly coprecipitated in the pATOM36-HA pull down. Consistent with these results, no pATOM36 is detected in the SAM50-HA pull down. Of interest, as in the case of pATOM36-HA, ATOM is also recovered. The experiment was performed in triplicate, and immunoprecipitations using wild-type mitochondria lacking any tagged proteins served as controls. To confirm these results, we repeated the pATOM36-HA immunoprecipitations and analyzed them by immunoblots. Figure 3B shows that a small but significant amount of ATOM is reproducibly recovered in the pellet fraction, whereas only background levels of the highly abundant membrane proteins cytochrome C1 (CYT C1) subunit 4 of cytochrome oxidase (COX4), and VDAC were detected. In summary, these experiments show that a fraction of pATOM36 is stably associated with ATOM but not with SAM50. The fact that HA-tagged ATOM is resolved as a 700-kDa complex on blue native gels (Pusnik et al., 2011) that is distinct from the pATOM36-HA complex (Figure 1D) suggests that pATOM36 is not a core component of the ATOM complex. A low-affinity, nonstoichiometric, and dynamic interaction with the ATOM complex might be expected if pATOM36 has a receptor function; in yeast, the canonical protein import receptors Tom20 and Tom70 are recovered in the TOM complex only at low-stringency conditions (Meisinger et al., 2001).
FIGURE 3:
pATOM36 is a peripheral component of the ATOM complex. (A) A 0.15% digitonin lysate of isolated, HA-tagged pATOM36 (left) and HA-tagged SAM50 containing mitochondria (right) were immunoprecipitated using anti-HA antibodies. The corresponding eluates were analyzed for the presence of pATOM36, SAM50, and ATOM using mass spectrometry. The graphs depict the number of evidences that were detected in the eluate of each experiment. The number of unique peptides identified for each protein is depicted on the top of each column. The experiment was performed in triplicate, and immunoprecipitations using wild-type mitochondria lacking any tagged proteins served as controls. pATOM36, SAM50, nor ATOM was recovered in the eluates of wild-type mitochondria. The only exception was ATOM that was detected by a single peptide/evidence in the first replicates of wild-type immunoprecipitates (data not shown). (B) A 0.15% digitonin lysate of isolated, HA-tagged pATOM36–containing mitochondria was immunoprecipitated using anti-HA antibodies and analyzed by immunoblots. Five percent of the total extract (Inp) and 95% of the bound fraction (IP) were analyzed by immunoblot using a monoclonal anti-HA antibody. The same samples were also analyzed with polyclonal ATOM, VDAC, COX4, and CYT C1 antisera. The experiment shown on the left was performed in quadruplicate and quantitated using the Odyssey infrared imaging system (Li-Cor Biosciences, Lincoln, NE). The means of the relative amounts of ATOM, VDAC, COX4, and CYT C1 that were recovered in the bound fraction were calculated. The pATOM36 that was recovered in the pellet was set to 100%. Standard errors are indicated.
pATOM36 is required for in vivo import of a subset of matrix proteins
To directly study whether pATOM36 functions in mitochondrial protein import, we prepared tetracycline (tet)-inducible RNAi cell lines. These cell lines show that ablation of the pATOM36 mRNA abolishes growth of both insect-stage and bloodstream forms of T. brucei (Figure 4).
FIGURE 4:

pATOM36 is essential for normal growth of procyclic and bloodstream forms of T. brucei. (A) Growth curve of an uninduced and tet-induced procyclic pATOM36 RNAi cell line. Right, Northern blot of total RNA isolated from uninduced and induced (2 d) RNAi cell lines probed for pATOM36 mRNA. The rRNA region of the ethidium bromide–stained gel (EtBr) was used as a loading control. (B) As in A, but results are for an uninduced and tet-induced pATOM36 RNAi cell line of the bloodstream form.
RNAi-mediated ablation of a protein import factor might be expected to cause an accumulation of precursor proteins in the cytosol and a concomitant decrease of the mitochondrial localized mature forms. However, previous experiments showed that accumulation of precursor is difficult to detect because mislocalized proteins that accumulate in the cytosol are often rapidly degraded. A decrease in the levels of newly synthesized imported proteins during induction of RNAi, on the other hand, was shown to be a highly sensitive proxy to quantify the inhibition of mitochondrial protein import (Pusnik et al., 2011).
In a first pATOM36 RNAi cell line, we tested import of newly synthesized mHSP70, for which, unlike for the other tested substrates shown later, we can see a time- and tetracycline-dependent accumulation of the precursor protein (Supplemental Figure S1). This indicates that pATOM36 is required for import of the mitochondrial matrix protein mHSP70.
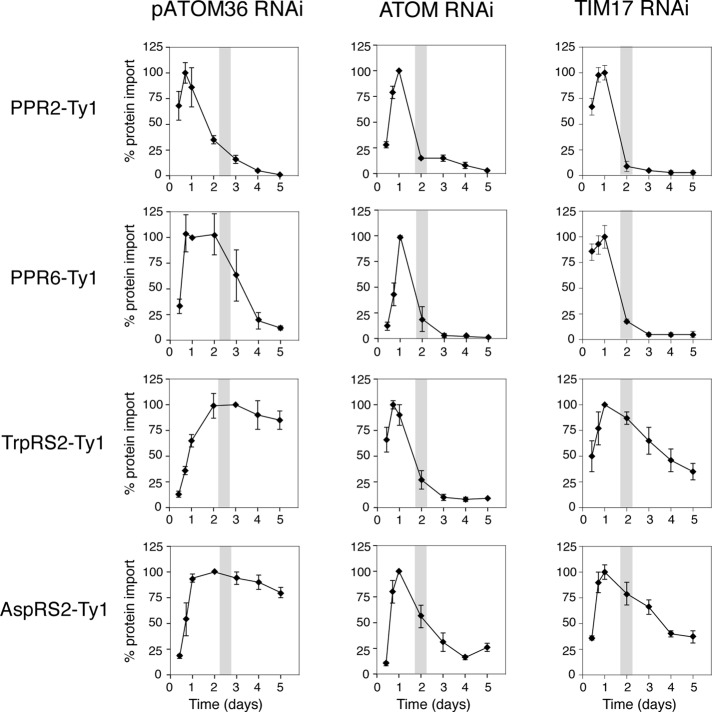
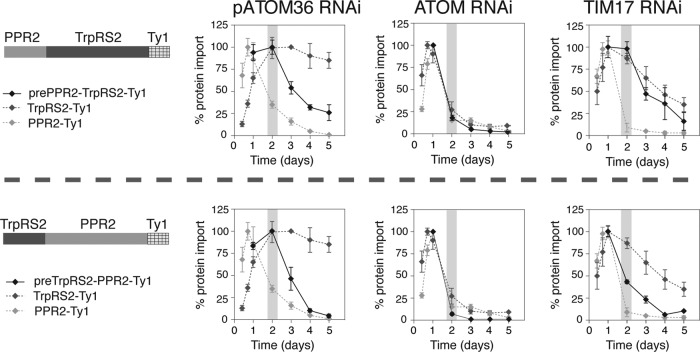
To determine whether pATOM36 is required for import of other matrix proteins, we produced cell lines in which inducible RNAi was combined with inducible expression of the tagged matrix proteins pentatricopeptide repeat protein 2 (PPR2) and PPR6, which belong to the same protein family but share very little sequence homology (Pusnik et al., 2007), and the mitochondrial tryptophanyl-tRNA (TrpRS2) (Charrière et al., 2006) and aspartyl-tRNA synthetases (AspRS2; Charrière et al., 2009). Each of these substrates was tested in the pATOM36 RNAi cell line and, as a control, also in cell lines ablated for the general outer membrane import channel ATOM (Pusnik et al., 2011) and the putative inner membrane translocation pore TIM17 (Gentle et al., 2007; Singha et al., 2008), both of which are expected to be required for import of all matrix proteins.
Figure 5 shows that ablation of either ATOM or TIM17 affects import of all tested substrates. Ablation of pATOM36, however, resulted in a selective reduction of newly synthesized PPR2 and PPR6, whereas the levels of TrpRS2 and AspRS2 were not affected. This suggests that pATOM36 is required for import of PPR2 and PPR6 but not for import of TrpRS2 and AspRS2. Moreover, the fact that in the pATOM36 RNAi cell lines, just as in the ATOM and TIM17 RNAi cell lines, the reduction of PPR2 and PPR6 precedes the growth arrest suggests that pATOM36 plays a direct role in import.
FIGURE 5:
pATOM36 is required for in vivo import of a subset of matrix proteins. Degradation of newly synthesized tagged matrix proteins during induction of RNAi of pATOM36 (first column), ATOM (second column), and TIM17 (third column) was used as a proxy for inhibition of mitochondrial protein import. The levels of indicated newly synthesized, C-terminally Ty1-tagged import substrates during pATOM36 RNAi were quantified by immunoblot analysis using antitag antibodies and normalized to the levels of cytosolic EF-1a. All experiments were replicated at least three times. The graphs depict the means and the standard errors of the normalized protein levels for each time point. Maximal expression was set to 100% for each tested substrate protein. Gray bar in graphs indicates the time of onset of growth arrest. The two control panels testing import of PPR2-Ty1 and TrpRS2-Ty1 in the ATOM RNAi cell line were published previously (Pusnik et al., 2011).
Moreover, Supplemental Figure S2 shows that RNAi-mediated ablation of pATOM36 for 48 h affects neither mitochondrial membrane potential nor organellar ATP production (Bochud-Allemann and Schneider, 2002), indicating that the observed inhibition of protein import is not due to a general mitochondrial dysfunction.
pATOM36 is required for in organello import of matrix proteins
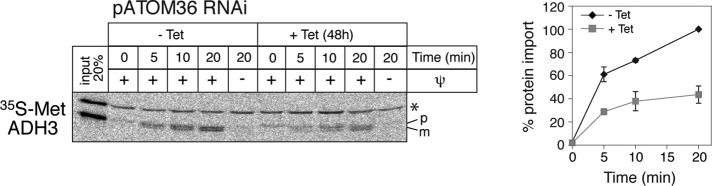
The requirement of pATOM36 for mitochondrial protein import was confirmed in vitro by comparing protein import into mitochondria isolated from noninduced and induced pATOM36 RNAi cell lines, respectively. Figure 6 shows that import of the in vitro–translated precursor of alcohol dehydrogenase III (Hauser et al., 1996) was greatly reduced in mitochondria isolated from pATOM36-ablated cells. Mitochondria used in this experiment were isolated 48 h after induction of RNAi, which is before the slow-growth phenotype becomes apparent (Figure 4), indicating that it is the impairment of protein import that causes the growth phenotype.
FIGURE 6:
pATOM36 is required for in vitro import of matrix proteins. Inhibition of in vitro import into isolated mitochondria from induced pATOM36 RNAi cells. In vitro–translated, 35S-Met–labeled alcohol dehydrogenase 3 (ADH3) precursor was imported for the indicated times into mitochondria isolated from uninduced (–Tet) and induced (+Tet) pATOM36 RNAi cell lines and analyzed by SDS–PAGE. The Coomassie-stained gel is shown as a loading control. All import reaction were proteinase treated. Input, 20% of added substrate; ψ, membrane potential. The band indicated by the asterisk is a labeled protein that is template independently synthesized in this batch of reticulocyte lysate. The positions of the precursor (p) and mature (m) forms of the protein are indicated.
pATOM36 function depends on the N-terminal presequence-containing domain of substrate proteins
The fact that some but not all matrix proteins require pATOM36 to be imported in vivo is surprising. To investigate which determinants in the substrate proteins are responsible for the pATOM36 dependence of protein import, we produced two variants of PPR2 and TrpRS2. In these variants the N-terminal 45 amino acids, which include the presequence, were swapped between the two proteins, resulting in two variants termed prePPR2-TrpRS2 and preTrpRS2-PPR2, respectively. Figure 7 shows that import of TrpRS2 indeed becomes dependent on pATOM36 when its 45 N-terminal amino acids are replaced by the equivalent region of PPR2. However, the converse experiment shows that the 45 N-terminal amino acids of TrpRS2 are not sufficient to make import of PPR2 independent of pATOM36. The two columns on the right of Figure 7 show that both fusion proteins, as expected, still required ATOM and TIM17 for import.
FIGURE 7:
pATOM36 function depends on a N-terminal presequence-containing domain of substrate proteins. Degradation of newly synthesized tagged substrate proteins during induction of pATOM36 (first column), ATOM (second column), and TIM17 (third column) RNAi was measured by immunoblots and used as a proxy for inhibition of mitochondrial protein import. Top, Ty1-tagged prePPR2-TrpRS2 consisting of the N-terminal 45 amino acids of PPR2 followed by TrpRS2 lacking the N-terminal 45 amino acids was used as a substrate. Bottom, Ty1-tagged preTrpRS2-PPR2 consisting of the N-terminal 45 amino acids of TrpRS2 followed by PPR2 lacking the N-terminal 45 amino acids was used as a substrate. All experiments were replicated at least three times. The graphs depict the means and the standard errors of the protein levels normalized to EF-1a for each time point. Maximal expression set to 100% for each tested substrate protein. Gray bar in graphs indicates the time of onset of growth arrest. The results for the tagged parent proteins TrpRS-Ty1 and PPR-Ty1 as shown in Figure 3 are shown in gray for comparison.
These results show that the interaction of precursor proteins with pATOM36 is of complex nature. Apparently pATOM36 recognizes a signal in the N-terminal 45 amino acids of PPR2, a region that includes the presequence. However, the fact that the N-terminal 45 amino acids of TrpRS2 are not sufficient to abolish the pATOM36 dependence of import indicates that other regions of the protein are also involved in the recognition process. A comparison of the N-terminal regions of TrpRS2 and AspRS2—two substrates that are independent of pATOM36—with the corresponding region of all other tested substrates did not reveal obvious differences.
DISCUSSION
The capability to import proteins from the cytosol is one of the key features that distinguish mitochondria from the bacterial endosymbiont from which they derive. It allowed the organelle to use proteins whose genes had been transferred to the nucleus, and current models for mitochondrial evolution envisage that the mitochondrial protein import system was established at a very early stage during mitochondrial evolution (Szklarczyk and Huynen, 2010). Consistent with this notion, the core components of the four hetero-oligomeric protein complexes that mediate protein import have been conserved in essentially all eukaryotes (Dolezal et al., 2006; Lithgow and Schneider, 2010; Hewitt et al., 2011). Given that trypanosomatids have a fundamentally different outer membrane preprotein translocase, termed ATOM (Schneider et al., 2008; Pusnik et al., 2011), we anticipated that 1) other outer membrane proteins would serve as partners to assist ATOM function and 2) these partner proteins would not necessarily be homologous to the TOM subunits of other eukaryotes.
We have now characterized pATOM36, another novel integral outer membrane protein that is required for mitochondrial protein import in T. brucei. Like ATOM, pATOM36 is essential for growth and viability and appears to be restricted to the trypanosomatids.
To allow detection of pATOM36 on immunoblots, most of our biochemical experiments shown in Figures 1–3 relied on a cell line in which one allele of pATOM36 was marked with an HA tag. Because it is possible that the tag alters the properties of pATOM36 we produced a cell line in which also the second allele is HA tagged (Supplemental Figure S3). The resulting cells grew as well as wild-type cells, indicating that C-terminally tagged pATOM36 is functional.
The component parts of the TOM complex
Besides Tom40, the canonical TOM consists of a number of functionally specialized subunits. The small α-helical proteins Tom5, Tom6, and Tom7 are tightly associated with Tom40 and help to stabilize the import pore. In addition, Tom20 and Tom70 function as import receptors, and both of these proteins contain α-helical domains characterized by tetratricopeptide repeat motifs (Endo et al., 2011). In yeast, neither Tom20 nor Tom70 is essential for growth, indicating that they have partly redundant functions: deletion of both Tom20 and Tom70 is lethal (Lithgow et al., 1994). Tom22 functions as both a protein import receptor that cooperates with Tom20 and as a general organizer of the TOM complex. Lack of Tom22 therefore severely interferes with normal growth (van Wilpe et al., 1999).
The import receptor Tom20 is a relatively recent addition to the TOM complex, with the protein found in yeast having homologues in all fungi and animals but not in other organisms (Likic´ et al., 2005; Dolezal et al., 2006). A quite distinct protein, also called Tom20, is found in the TOM complex in all plants and green algae (Perry et al., 2006). These proteins share no common ancestor, and yet each is composed of multiple α-helices: seven including the transmembrane domain for the Tom20 from animals and eight including the transmembrane domain for the Tom20 from plants (Abe et al., 2000; Perry et al., 2006). Both the animal Tom20 and the plant Tom20 are responsible for binding the mitochondrial-targeting sequence of substrate mitochondrial precursor proteins. In an evolutionary sense, this “prefilter” that binds mitochondrial-targeting sequences before the core TOM complex was a late addition, evolving after the divergence of the plant and animal lineages (Macasev et al., 2004; Perry et al., 2006). Independently, an equivalent, nonhomologous α-helical architecture evolved in these protein import receptors.
pATOM36 promotes the import of a subset of mitochondrial proteins
Although there is no sequence similarity between pATOM36 and the canonical import receptors Tom20 and Tom70, there are a number of shared features. A fraction of pATOM36 is associated with the ATOM complex (Figure 3)—the outer membrane protein translocase of trypanosomes—just as Tom20 and Tom70 are peripheral components of the canonical TOM complex (Meisinger et al., 2001). Antibody shift experiments (Figure 1E) indicate that pATOM36, similar to Tom20 and Tom70 (Neupert and Herrmann, 2007; Endo et al., 2011), appears to be anchored with its C-terminal part exposed to the cytosol. Finally, and most important, functional analyses show that each of the three proteins is required for import of a subset of but not all proteins into mitochondria.
Whereas the Tom20 import receptor binds simply to the mitochondrial targeting sequence, the Tom70 import receptor binds to multiple regions of its substrate proteins, whether or not they have mitochondrial targeting sequences (Brix et al., 2000; Chan et al., 2006; Yamamoto et al., 2009). This division of labor between the receptors provides a sophisticated means of recognizing the broad diversity of sequence characteristics in the ∼1000 proteins imported into mitochondria. What features in the substrate proteins determine the pATOM36 dependence of import is unknown, but the in vivo results shown in Figure 7 suggest that the N-terminal 45 amino acids contribute to it.
Blue native gel electrophoresis shows that the ATOM complex is ∼700 kDa (Pusnik et al., 2011), suggesting a hetero-oligomeric structure. Because there is a group of matrix proteins whose import does not depend on pATOM36, we hypothesize that additional protein import receptors are likely to exist on the mitochondrial surface in trypanosomes. Identification and functional analysis of further peripheral and core factors of the ATOM complex promise more insights into the fundamental features of mitochondrial protein import and the selective pressures that drove its evolution.
MATERIALS AND METHODS
Transgenic cell lines
Procyclic transgenic cell lines are based on T. brucei brucei 29-13 grown at 27°C in SDM-79 supplemented with 15% fetal calf serum (FCS) and the required antibiotics. The bloodstream T. brucei NYSM line was cultivated at 37°C in HMI-9 medium containing 10% FCS. Transformation, cloning, and selection of transgenic cell lines were done as described (McCulloch et al., 2004). Procyclic RNAi cell lines for ATOM, TIM17, and SAM50 have been described (Gentle et al., 2007; Pusnik et al., 2011; Tschopp et al., 2011). RNAi of pATOM36 was done by using a pLew-100–derived stem-loop construct (Bochud-Allemann and Schneider, 2002). As an insert, we used a 451–base pair fragment (nucleotides 724–1174) of the pATOM36 ORF (Tb927.7.5700).
To detect pATOM36, we in situ–tagged one of its alleles with a single HA tag at its C-terminus according to Shen et al. (2001; Supplemental Figure S3). In the case of SAM50, one allele carries a single C-terminal HA tag and the other allele was knocked out. Tet-inducible expression of C-terminally Ty1-tagged substrate proteins (Bastin et al., 1996; Figures 5 and 7) was achieved using pLew-100–derived constructs carrying various antibiotic resistance markers. The Ty1 tag was detected by the monoclonal BB2 antibody (Diagenode, Liège, Belgium), and the HA tag was visualized by the monoclonal antibody HA11 (Covance Research Products, Princeton, NJ).
Antibody shift assays
Mitochondria containing either HA-tagged pATOM36 or SAM50, respectively, were isolated under isotonic conditions (Schneider et al., 2007b). For each type of mitochondria two types of experiments were performed. In the first one intact mitochondria were resuspended in import buffer (Hauser et al., 1996) containing 5 mg/ml fatty acid–free bovine serum albumin and 1 μl of anti-HA polyclonal antibody (LabForce, Nunningen, Switzerland) was added. The reaction was incubated for 15 min on ice, and the mitochondria were reisolated. The supernatant was discarded, and the pellet was resuspended in 50 μl of 1.5% digitonin (Sigma-Aldrich, St. Louis, MO; Pusnik et al., 2011) and resolved by 6–16.5% BN-PAGE. In the second case the mitochondrial samples were first solubilized in import buffer containing 1.5% digitonin before it was incubated with 1 μl of the anti-HA polyclonal antibody for 15 min on ice and resolved on the same BN gel. The gel was then transferred to polyvinylidene fluoride membrane (Millipore, Billerica, MA) and analyzed by immunoblotting using monoclonal anti-HA antiserum (Santa Cruz Biotechnology, Santa Cruz, CA).
Immunoprecipitations
Isotonically isolated mitochondria (240 μg; Hauser et al., 1996) containing C-terminally HA-tagged pATOM36 were solubilized for 15 min on ice in 300 μl of lysis buffer: 20 mM Tris-HCl, pH 7.4, 0.1 mM EDTA, 200 mM KCl containing 0.15% (wt/vol) digitonin, and a protease inhibitor cocktail (Roche Applied Science, Indianapolis, IN). The lysate was cleared by centrifugation (16,000 × g, 4°C), and the supernatant was incubated for 2.5 h at 4°C with 75 μl of a 1:1 slurry of anti-HA agarose (Roche Applied Science). Subsequently, the beads where washed three times in 750 μl of lysis buffer containing 0.1% (wt/vol) digitonin before elution with 60 μl of SDS sample buffer. For mass spectrometric analysis (Figure 3A) the samples were precipitated using acetone and tryptically digested in 60% (vol/vol) methanol and 20 mM NH4HCO3. The resulting peptides were analyzed by nano high-performance liquid chromatography–electrospray tandem mass spectrometry as described (Gebert et al., 2011). For peptide and protein identification, data sets were correlated with the TriTryp Database 3.1 using MaxQuant, version 1.0.13.13 (Cox and Mann, 2008). A false discovery rate of <1% was applied, and the number of evidences given by MaxQuant was used a quantitative measure.
For immunoblot analysis cell lysates corresponding to 4 μg and eluate corresponding to 76 μg of mitochondria were separated on SDS polyacrylamide gels, transferred to nitrocellulose, and analyzed with the indicated antisera (Figure 3B).
In vitro import and assembly of precursor proteins
The 35S-labeled precursor proteins were produced using the in vitro translation kit TNT T7 Quick for PCR DNA from Promega (Madison, WI). Import of precursor proteins into isotonically isolated mitochondria of T. brucei was done as described (Hauser et al., 1996), except that the import buffer contained 5 mg/ml fatty acid–free bovine serum albumin. In vitro assembly of 35S-labeled pATOM36 into complexes and subsequent analysis on BN-PAGE were done as described for yeast, except that 1.5% (wt/vol) of digitonin was used for solubilization (Stojanovski et al., 2007).
Miscellaneous
Digitonin extractions to produce a crude mitochondrial fraction and ATP production assays were done as described (Schneider et al., 2007a).
Supplementary Material
Acknowledgments
We thank E. Horn and B. Schönfisch for technical assistance and F. Voncken of the University of Hull (Hull, United Kingdom) for the gift of the MCP-5 antiserum. The study was funded by grants from the Swiss National Foundation (31003A_121937 to A.S.), the Bundesministerium für Bildung und Forschung (Dynamo; to C.M.), the Deutsche Forschungsgemeinschaft (to C.M. and B.W.), the Excellence Initiative of the German Federal and State Governments (EXC 294 BIOSS; to C.M and B.W.), and the Peter and Traudl Engelhorn Foundation (to M.N.). T.L. is a Federation Fellow of the Australian Research Council.
Abbreviations used:
- AspRS2
mitochondrial aspartyl-tRNA synthetase
- ATOM
translocase of the outer mitochondrial membrane
- BN-PAGE
blue native gel electrophoresis
- COX4
cytochrome oxidase subunit 4
- CYT C
cytochrome C
- CYT C1
cytochrome C1
- FCS
fetal calf serum.
- HA
hemagglutinin
- mHSP70
mitochondrial heat shock protein 70
- ORF
open reading frame
- pATOM36
peripheral subunit of ATOM of 35 kDa
- PPR2
pentatricopeptide protein 2
- RNAi
RNA interference
- SAM
sorting and assembly machinery
- TOM
translocase of the outer mitochondrial membrane
- VDAC
voltage-dependent anion channel
- TrpRS2
mitochondrial tryptophanyl-tRNA synthetase
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-02-0107) on July 11, 2012.
*Present address: Division of Cell Biology, Medical University Innsbruck, 6020 Innsbruck, Austria.
REFERENCES
- Abe Y, Shodai T, Muto T, Mihara K, Torii H, Nishikawa S-I, Endo T, Kohda D. Structural basis of presequence recognition by the mitochondrial protein import receptor Tom20. Cell. 2000;100:551–560. doi: 10.1016/s0092-8674(00)80691-1. [DOI] [PubMed] [Google Scholar]
- Ahting U, Thieffry M, Engelhardt H, Hegerl R, Neupert W, Nussberger S. Tom40, the pore-forming component of the protein-conducting TOM channel in the outer membrane of mitochondria. J Cell Biol. 2001;153:1151–1160. doi: 10.1083/jcb.153.6.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen R, Egan B, Gabriel K, Beilharz T, Lithgow T. A conserved proline residue is present in the transmembrane-spanning domain of Tom7 and other tail-anchored protein subunits of the TOM translocase. FEBS Lett. 2002;514:347–350. doi: 10.1016/s0014-5793(02)02433-x. [DOI] [PubMed] [Google Scholar]
- Alsford S, Turner DJ, Obado SO, Sanchez-Flores A, Glover L, Berriman M, Hertz-Fowler C, Horn D. High-throughput phenotyping using parallel sequencing of RNA interference targets in the African trypanosome. Genome Res. 2011;21:915–924. doi: 10.1101/gr.115089.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker KP, Schatz G. Mitochondrial proteins essential for viability mediate protein import into yeast mitochondria. Nature. 1991;349:205–208. doi: 10.1038/349205a0. [DOI] [PubMed] [Google Scholar]
- Bastin P, Bagherzadeh A, Matthews KR, Gull K. A novel epitope tag system to study protein targeting and organelle biogenesis in Trypanosoma brucei. Mol Biochem Parasitol. 1996;77:235–239. doi: 10.1016/0166-6851(96)02598-4. [DOI] [PubMed] [Google Scholar]
- Bochud-Allemann N, Schneider A. Mitochondrial substrate level phosphorylation is essential for growth of procyclic Trypanosoma brucei. J Biol Chem. 2002;277:32849–32854. doi: 10.1074/jbc.M205776200. [DOI] [PubMed] [Google Scholar]
- Bos MP, Robert V, Tommassen J. Biogenesis of the Gram-negative bacterial outer membrane. Annu Rev Microbiol. 2007;61:191–214. doi: 10.1146/annurev.micro.61.080706.093245. [DOI] [PubMed] [Google Scholar]
- Brix J, Ziegler GA, Dietmeier K, Schneider-Mergener J, Schulz GE, Pfanner N. Identification of a 25 kDa core domain with a specific binding site for preproteins. J Mol Biol. 2000;303:479–488. doi: 10.1006/jmbi.2000.4120. [DOI] [PubMed] [Google Scholar]
- Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138:628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan NC, Likic´ VA, Waller RF, Mulhern TD, Lithgow T. The C-terminal TPR domain of Tom70 defines a family of mitochondrial protein import receptors found only in animals and fungi. J Mol Biol. 2006;358:1010–1022. doi: 10.1016/j.jmb.2006.02.062. [DOI] [PubMed] [Google Scholar]
- Charrière F, Helgadóttir S, Horn EK, Söll D, Schneider A. Dual targeting of a single tRNATrp requires two different tryptophanyl-tRNA synthetases in Trypanosoma brucei. Proc Natl Acad Sci USA. 2006;103:6847–6852. doi: 10.1073/pnas.0602362103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrière F, O'Donoghue P, Helgadóttir S, Maréchal-Drouard L, Cristodero M, Horn EK, Söll D, Schneider A. Dual targeting of a tRNAAsp requires two different aspartyl-tRNA synthetases in Trypanosoma brucei. J Biol Chem. 2009;284:16210–16217. doi: 10.1074/jbc.M109.005348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Dolezal P, Likic V, Tachezy J, Lithgow T. Evolution of the molecular machines for protein import into mitochondria. Science. 2006;313:314–318. doi: 10.1126/science.1127895. [DOI] [PubMed] [Google Scholar]
- Dolezal P, Smíd O, Rada P, Zubácová Z, Bursac´ D, Suták R, Nebesárová J, Lithgow T, Tachezy J. Giardia mitosomes and trichomonad hydrogenosomes share a common mode of protein targeting. Proc Natl Acad Sci USA. 2005;102:10924–10929. doi: 10.1073/pnas.0500349102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Embley TM, Martin W. Eukaryotic evolution, changes and challenges. Nature. 2006;440:623–630. doi: 10.1038/nature04546. [DOI] [PubMed] [Google Scholar]
- Endo T, Yamano K, Kawano S. Structural insight into the mitochondrial protein import system. Biochim Biophys Acta. 2011;1808:955–970. doi: 10.1016/j.bbamem.2010.07.018. [DOI] [PubMed] [Google Scholar]
- Gebert N, et al. Dual function of Sdh3 in the respiratory chain and TIM22 protein translocase of the mitochondrial inner membrane. Mol Cell. 2011;44:811–818. doi: 10.1016/j.molcel.2011.09.025. [DOI] [PubMed] [Google Scholar]
- Gentle IE, et al. Conserved motifs reveal details of ancestry and structure in the small TIM chaperones of the mitochondrial intermembrane space. Mol Biol Evol. 2007;24:1149–1160. doi: 10.1093/molbev/msm031. [DOI] [PubMed] [Google Scholar]
- Hauser R, Pypaert M, Häusler T, Horn EK, Schneider A. In vitro import of proteins into mitochondria of Trypanosoma brucei and Leishmania tarentolae. J Cell Sci. 1996;109:517–523. doi: 10.1242/jcs.109.2.517. [DOI] [PubMed] [Google Scholar]
- Hewitt V, Alcock F, Lithgow T. Minor modifications and major adaptations: the evolution of molecular machines driving mitochondrial protein import. Biochim Biophys Acta. 2011;1808:947–954. doi: 10.1016/j.bbamem.2010.07.019. [DOI] [PubMed] [Google Scholar]
- Hill K, Model K, Ryan MT, Dietmeier K, Martin F, Wagner R, Pfanner N. Tom40 forms the hydrophilic channel of the mitochondrial import pore for preproteins. Nature. 1998;395:516–521. doi: 10.1038/26780. [DOI] [PubMed] [Google Scholar]
- Hoogenraad NJ, Ward LA, Ryan MT. Import and assembly of proteins into mitochondria of mammalian cells. Biochim Biophys Acta. 2002;1592:97–105. doi: 10.1016/s0167-4889(02)00268-9. [DOI] [PubMed] [Google Scholar]
- Likic´ VA, et al. Patterns that define the four domains conserved in known and novel isoforms of the protein import receptor Tom20. J Mol Biol. 2005;347:81–93. doi: 10.1016/j.jmb.2004.12.057. [DOI] [PubMed] [Google Scholar]
- Lithgow T, Junne T, Wachter C, Schatz G. Yeast mitochondria lacking the two import receptors Mas20p and Mas70p can efficiently and specifically import precursor proteins. J Biol Chem. 1994;269:15325–15330. [PubMed] [Google Scholar]
- Lithgow T, Schneider A. Evolution of macromolecular import pathways in mitochondria, hydrogenosomes and mitosomes. Philos Trans R Soc Lond B Biol Sci. 2010;365:799–817. doi: 10.1098/rstb.2009.0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mac´asev D, Whelan J, Newbigin E, Silva-Filho MC, Mulhern TD, Lithgow T. Tom22’, an 8-kDa trans-site receptor in plants and protozoans, is a conserved feature of the TOM complex that appeared early in the evolution of eukaryotes. Mol Biol Evol. 2004;21:1557–1564. doi: 10.1093/molbev/msh166. [DOI] [PubMed] [Google Scholar]
- McCulloch R, Vassella E, Burton P, Boshart M, Barry JD. Transformation of monomorphic and pleomorphic Trypanosoma brucei. Methods Mol Biol. 2004;262:53–86. doi: 10.1385/1-59259-761-0:053. [DOI] [PubMed] [Google Scholar]
- Meisinger C, Ryan MT, Hill K, Model K, Lim JH, Sickmann A, Müller H, Meyer HE, Wagner R, Pfanner N. Protein import channel of the outer mitochondrial membrane: a highly stable Tom40-Tom22 core structure differentially interacts with preproteins, small Tom proteins, and import receptors. Mol Cell Biol. 2001;21:2337–2348. doi: 10.1128/MCB.21.7.2337-2348.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupert W, Herrmann JM. Translocation of proteins into mitochondria. Annu Rev Biochem. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- Perry AJ, Hulett JM, Likic´ VA, Lithgow T, Gooley PR. Convergent evolution of receptors for protein import into mitochondria. Curr Biol. 2006;16:221–229. doi: 10.1016/j.cub.2005.12.034. [DOI] [PubMed] [Google Scholar]
- Pusnik M, Charrière F, Mäser P, Waller RF, Dagley MJ, Lithgow T, Schneider A. The single mitochondrial porin of Trypanosoma brucei is the main metabolite transporter in the outer mitochondrial membrane. Mol Biol Evol. 2009;26:671–680. doi: 10.1093/molbev/msn288. [DOI] [PubMed] [Google Scholar]
- Pusnik M, Schmidt O, Perry AJ, Oeljeklaus S, Niemann M, Warscheid B, Lithgow T, Meisinger C, Schneider A. Mitochondrial preprotein translocase of trypanosomatids has a bacterial origin. Curr Biol. 2011;21:1738–1743. doi: 10.1016/j.cub.2011.08.060. [DOI] [PubMed] [Google Scholar]
- Pusnik M, Small I, Read LK, Fabbro T, Schneider A. Pentatricopeptide repeat proteins in Trypanosoma brucei function in mitochondrial ribosomes. Mol Cell Biol. 2007;27:6876–6888. doi: 10.1128/MCB.00708-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt O, Pfanner N, Meisinger C. Mitochondrial protein import: from proteomics to functional mechanisms. Nat Rev Mol Cell Biol. 2010;11:655–667. doi: 10.1038/nrm2959. [DOI] [PubMed] [Google Scholar]
- Schneider A, Bouzaidi-Tiali N, Chanez A-L, Bulliard L. ATP production in isolated mitochondria of procyclic Trypanosoma brucei. Methods Mol Biol. 2007a;372:379–387. doi: 10.1007/978-1-59745-365-3_27. [DOI] [PubMed] [Google Scholar]
- Schneider A, Bursac´ D, Lithgow T. The direct route: a simplified pathway for protein import into the mitochondrion of trypanosomes. Trends Cell Biol. 2008;18:12–18. doi: 10.1016/j.tcb.2007.09.009. [DOI] [PubMed] [Google Scholar]
- Schneider A, Charrière F, Pusnik M, Horn EK. Isolation of mitochondria from procyclic Trypanosoma brucei. Methods Mol Biol. 2007b;372:67–80. doi: 10.1007/978-1-59745-365-3_5. [DOI] [PubMed] [Google Scholar]
- Sharma S, Singha UK, Chaudhuri M. Role of Tob55 on mitochondrial protein biogenesis in Trypanosoma brucei. Mol Biochem Parasitol. 2010;174:89–100. doi: 10.1016/j.molbiopara.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Arhin GK, Ullu E, Tschudi C. In vivo epitope tagging of Trypanosoma brucei genes using a one step PCR-based strategy. Mol Bioch Parasitol. 2001;113:171–173. doi: 10.1016/s0166-6851(00)00383-2. [DOI] [PubMed] [Google Scholar]
- Siegel TN, Gunasekera K, Cross GA, Ochsenreiter T. Gene expression in Trypanosoma brucei: lessons from high-throughput RNA sequencing. Trends Parasitol. 2011;27:434–441. doi: 10.1016/j.pt.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singha UK, Peprah E, Williams S, Walker R, Saha L, Chaudhuri M. Characterization of the mitochondrial inner membrane protein translocator Tim17 from Trypanosoma brucei. Mol Biochem Parasitol. 2008;159:30–43. doi: 10.1016/j.molbiopara.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojanovski D, Pfanner N, Wiedemann N. Import of proteins into mitochondria. Methods Cell Biol. 2007;80:783–806. doi: 10.1016/S0091-679X(06)80036-1. [DOI] [PubMed] [Google Scholar]
- Szklarczyk R, Huynen MA. Mosaic origin of the mitochondrial proteome. Proteomics. 2010;22:4012–4024. doi: 10.1002/pmic.201000329. [DOI] [PubMed] [Google Scholar]
- Tsaousis AD, Gaston D, Stechmann A, Walker PB, Lithgow T, Roger AJ. A functional Tom70 in the human parasite Blastocystis sp.: implications for the evolution of the mitochondrial import apparatus. Mol Biol Evol. 2011;28:781–791. doi: 10.1093/molbev/msq252. [DOI] [PubMed] [Google Scholar]
- Tschopp F, Charrière F, Schneider A. In vivo study in Trypanosoma brucei links mitochondrial transfer RNA import to mitochondrial protein import. EMBO Rep. 2011;12:825–832. doi: 10.1038/embor.2011.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wilpe S, et al. Tom22 is a multifunctional organizer of the mitochondrial preprotein translocase. Nature. 1999;401:485–489. doi: 10.1038/46802. [DOI] [PubMed] [Google Scholar]
- Yamamoto H, et al. Roles of Tom70 in import of presequence-containing mitochondrial proteins. J Biol Chem. 2009;284:31635–31646. doi: 10.1074/jbc.M109.041756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeth K. Structure and evolution of mitochondrial outer membrane proteins of beta-barrel topology. Biochim Biophys Acta. 2010;1797:1292–1299. doi: 10.1016/j.bbabio.2010.04.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.