Mutations in the connexin-43 gap junction protein cause the developmental disease known as oculodentodigital dysplasia. Structure and function approaches are used to demonstrate that the nature of the missense mutation in the amino-terminal domain of connexin-43 governs the mechanism that leads to loss of connexin-43 function.
Abstract
Mutations in the gene encoding connexin-43 (Cx43) cause the human development disorder known as oculodentodigital dysplasia (ODDD). In this study, ODDD-linked Cx43 N-terminal mutants formed nonfunctional gap junction–like plaques and exhibited dominant-negative effects on the coupling conductance of coexpressed endogenous Cx43 in reference cell models. Nuclear magnetic resonance (NMR) protein structure determination of an N-terminal 23–amino acid polypeptide of wild-type Cx43 revealed that it folded in to a kinked α-helical structure. This finding predicted that W4 might be critically important in intramolecular and intermolecular interactions. Thus we engineered and characterized a W4A mutant and found that this mutant formed a regular, nonkinked α-helix but did not form functional gap junctions. Furthermore, a G2V variant peptide of Cx43 showed a kinked helix that now included V2 interactions with W4, resulting in the G2V mutant forming nonfunctional gap junctions. Also predicted from the NMR structures, a G2S mutant was found to relieve these interactions and allowed the protein to form functional gap junctions. Collectively, these studies suggest that the nature of the mutation conveys loss of Cx43 function by distinctly different mechanisms that are rooted in the structure of the N-terminal region.
INTRODUCTION
The gap junction family of 21 members selectively intermixes and oligomerizes to form connexons, which are frequently referred to as hemichannels (Sohl and Willecke, 2003). Connexons traffic to the cell surface, where they proceed to dock with connexons from a contacting cell to form an intercellular channel. These channels typically cluster and coalesce into tightly packed structures known as gap junctions (Laird, 2006). Gap junctional intercellular communication is a fundamental characteristic of almost all contacting cells in the human anatomy, establishing them as critical to human health (Laird, 2006). Each connexin consists of four α-helical transmembrane domains, two extracellular loops, and a cytoplasmic loop with the amino and carboxyl termini exposed to the cytoplasm (Laird and Revel, 1990). Each major domain of the connexin has been proposed to be important at regulating one or more functional aspects of the channel (Harris, 2001). In the case of the extracellular loops, they have been shown to be critical at establishing appropriate interconnexon docking domains through the formation of intramolecular disulfide bonds (Nakagawa et al., 2011; Rico et al., 2011). In connexin-43 (Cx43), the C-terminal domain is involved in binding numerous members of the gap junction proteome, as well as being the substrate for several protein kinases (Solan and Lampe, 2009; Laird, 2010). The cytoplasmic loop of Cx43 has been shown to interact with the C-terminal during pH-dependent channel closure (Hirst-Jensen et al., 2007). The present study focuses on the N-terminus of connexins, which has been reported to be important for transjunctional voltage-dependent gating of gap junction channels (Harris, 2001), regulating ion permeability (Harris, 2001; Oshima et al., 2011), and even regulating connexin oligomerization (Lagree et al., 2003). Crystal structure analysis of Cx26 revealed an α-helical structure within the first 10 amino acids of the molecule, followed by a critical bend at residues 11 and 12 that appeared to act like a hinge in allowing the N-terminus to fold into and plug the narrowest part of the pore in a voltage-dependent gating model (Maeda et al., 2009). In another study, the first 23 amino acids of the N-terminal of Cx37 were predominantly α -helical between glycine 5 and glutamate 16 (Kyle et al., 2009). This 11–amino acid domain was dispensable for Cx37 localization to the cell surface but was required for channel and hemichannel function (Kyle et al., 2008). Not surprisingly, mutations in the N-terminal domain of a number of connexins have been found to be causal of several human diseases, such as neurosensory hearing loss (e.g., Cx26, Cx30, and Cx31; Scott and Kelsell, 2011), Charcot–Marie–Tooth disease (e.g., Cx32; Latour et al., 1997), congenital cataracts (e.g., Cx46 and Cx50; Pfenniger et al., 2011), and oculodentodigital dysplasia (ODDD; e.g., Cx43; Paznekas et al., 2009).
Cx43 is by far the most widely found connexin in human anatomy, being found in more than 35 distinct cell and tissue types (Laird, 2006). Consequently Cx43 has also been the most rigorously studied member of the connexin family. Interest in examining Cx43 structure and function surged when primarily autosomal dominant mutations in the GJA1 gene were found to be casual of the developmental disorder ODDD (Paznekas et al., 2003). Not surprisingly, this disease manifests as a plethora of symptoms, but the most common affected tissues are localized to the hands, feet, eyes, facial bones, and teeth (Paznekas et al., 2003). Collectively there are >62 mutations associated with ODDD (Laird, 2008; Paznekas et al., 2009). The vast majority of these mutations are missense mutations, and 10 of these mutations occur within the N-terminal domain of Cx43 (Paznekas et al., 2009). Approximately 20 ODDD-linked mutants have been examined for their ability to form gap junction channels, and in nearly all cases the missense mutation renders Cx43 either completely or almost completely nonfunctional and also acts to dominantly inhibit the function of wild-type Cx43 (Flenniken et al., 2005; Gong et al., 2006, 2007; Dobrowolski et al., 2007; Manias et al., 2008; McLachlan et al., 2008; Musa et al., 2009; Paznekas et al., 2009; Churko et al., 2010; Toth et al., 2010). However, the vast majority of the mutants examined can traffic to the cell surface, suggesting that they pass the “quality control” mechanism used within the endoplasmic reticulum. We hypothesized that the nature and position of the mutation might inactivate Cx43 by employing different mechanisms and further provide insight into how specific motifs of Cx43 might regulate function.
To that end, we focused on the N-terminal domain, which encodes >15% of the reported ODDD-linked mutations (Paznekas et al., 2009). We synthesized a polypeptide encoding the first 23 amino acids of native Cx43, an ODDD-linked mutant peptide (G2V), and a mutant-containing peptide (W4A) that has not been reported to be linked to disease and used nuclear magnetic resonance (NMR) spectroscopy to resolve the structures. Through structure and functional analysis of these mutants and others we determined that the nature of the mutation within the N-terminal conveys loss of Cx43 function by distinctly different mechanisms that are rooted in the structure of the N-terminal domain.
RESULTS
Localization and functional status of ODDD-linked, N-terminal–specific Cx43 mutants
There are >62 Cx43 mutants linked to ODDD, with 10 of these missense mutations mapped to the N-terminal of Cx43 (Paznekas et al., 2009). Analysis of these N-terminal mutants revealed that some mutations would be considered relatively conservative (e.g., D3N or L7V), whereas others either changed the charge or hydrophobicity of the domain (e.g., G2V, Y17S; Table 1).
TABLE 1:
Peptides that mimic the N-terminal of Cx43.
| Cx43: MGDWSALGKLLDKVQAYSTAGGK |
| G2V: MVDWSALGKLLDKVQAYSTAGGK |
| D3N: MGNWSALGKLLDKVQAYSTAGGK |
| L7V: MGDWSAVGKLLDKVQAYSTAGGK |
| L11P: MGDWSALGKLPDKVQAYSTAGGK |
| Y17S: MGDWSALGKLLDKVQASSTAGGK |
| S18P: MGDWSALGKLLDKVQAYPTAGGK |
| W4A: MGDASALGKLLDKVQAYSTAGGK |
| G2S: MSDWSALGKLLDKVQAYSTAGGK |
Peptide amino acid substitutions that mimic Cx43-linked ODDD mutants or non-ODDD mutants are indicated in boldface.
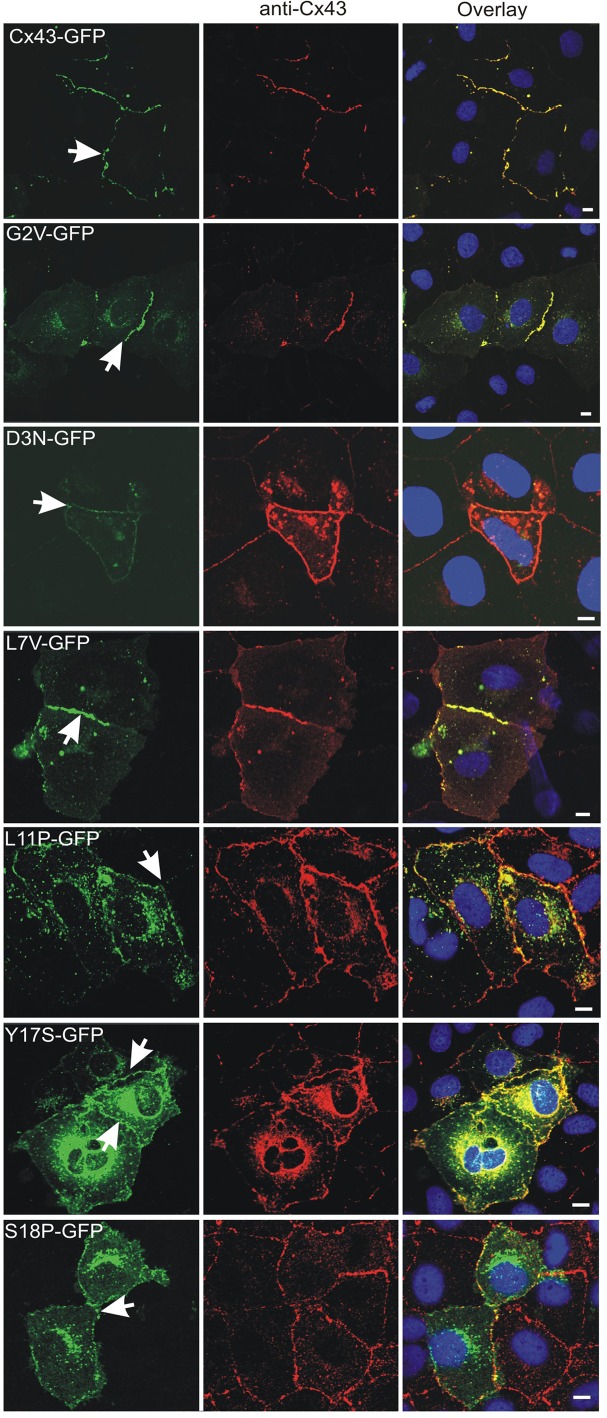
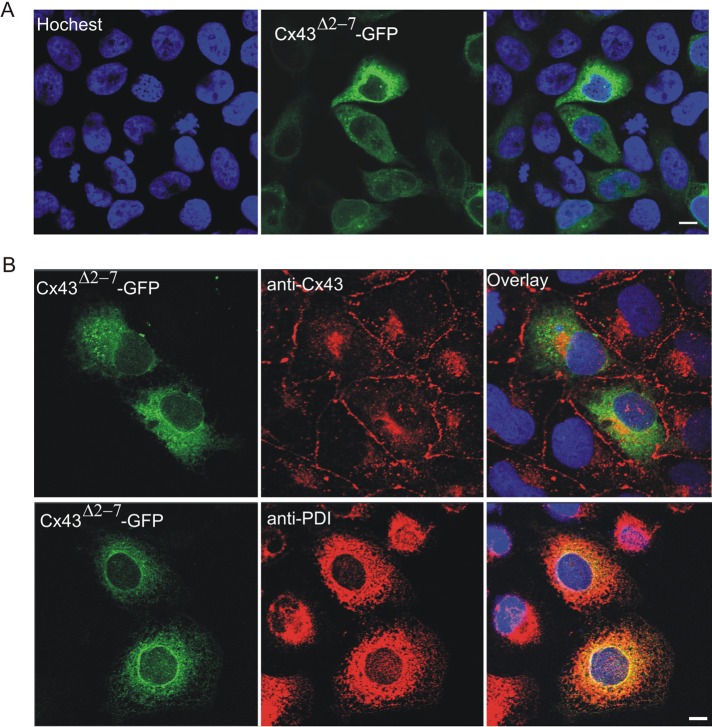
To begin to assess the role of N-terminal mutants, we expressed green fluorescent protein (GFP)–tagged G2V, D3N, L7V, L11P, Y17S, and S18P mutants in Cx43-positive normal rat kidney (NRK) cells to determine whether the mutant would be delivered to the cell surface and assembled into gap junction–like plaques (Figure 1). Similar to wild-type Cx43, the G2V, D3N, and L7V mutants all readily assembled into gap junction–like plaques. Although the L11P, Y17S, and S18P mutants did assemble into plaques, it appeared that the efficiency of this process was low, as large amounts of GFP-tagged mutants were retained within intracellular compartments. To further determine the importance of the N-terminus of Cx43 for gap junction plaque assembly and trafficking to the cell surface, we transfected HeLa (Figure 2A) and NRK (Figure 2B) cells with a construct encoding Cx43Δ2-7. Of interest, Cx43Δ2-7 was localized to an intracellular compartment consistent with the endoplasmic reticulum, as revealed by immunolabeling for protein disulfide isomerase (PDI; Figure 2B).
FIGURE 1:
Distribution of N-terminal Cx43 mutants in NRK cells. Cells were transfected with vectors encoding GFP-tagged Cx43 or the G2V, D3N, L7V, L11P, Y17S, or S18P mutant before immunolabeling with anti-Cx43 (red) antibodies. Hoechst 33342 was used to stain the nuclei Arrows indicate gap junction plaque-like structures at sites of cell–cell contact. Bars, 10 μm.
FIGURE 2:
Intracellular distribution of Cx43Δ2-7-GFP in HeLa and NRK cells. HeLa (A) or NRK (B) cells were transfected with a vector that encoded a GFP-tagged Cx43 mutant (Cx43Δ2-7-GFP) and immunolabeled with anti-Cx43 (red) or anti-PDI (red) antibodies. Hoechst 33342 was used to stain the nuclei. Bars, 10 μm.
To assess whether the N-terminal mutants possess the ability to form functional gap junction channels, we expressed full-length Cx43 or the mutants in intercellular gap junctional communication–deficient HeLa cells and assessed whether the cells were capable of passing microinjected Lucifer yellow to contacting cells (Figure 3A). Similar to wild-type cells, all mutant expressing cells could not form Lucifer yellow–permeable gap junction channels. Furthermore, dual whole-cell patch-clamp electrophysiology was used to show that all mutants were incapable of forming functional gap junction channels when expressed in gap junction communication–incompetent neuroblastoma (Neuro-2A [N2A]) cells (Figure 3C). We also examined whether the mutants would dominantly inhibit the gap junction channel function of coexpressed endogenous Cx43 by blocking Lucifer yellow dye transfer (Figure 3B). Again in all cases, the mutants significantly decreased the incidence of dye transfer, suggesting that they were dominant to the function of endogenous Cx43.
FIGURE 3:
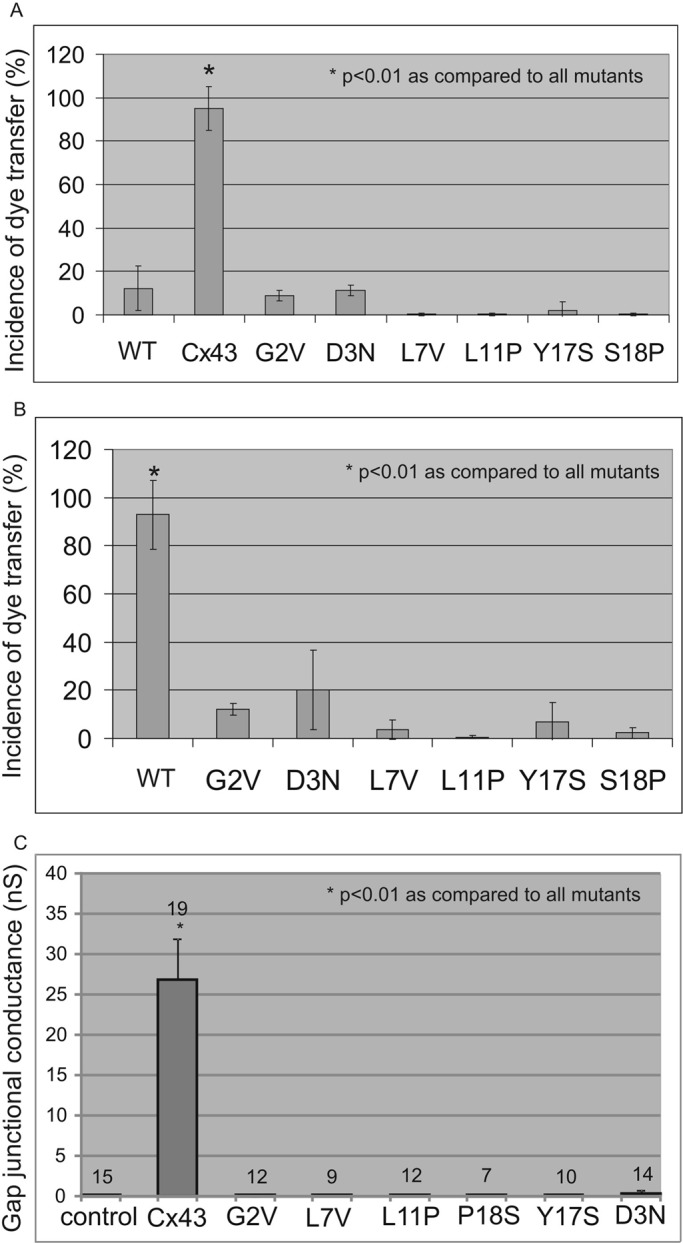
N-terminal–linked Cx43 mutants do not make functional intercellular channels and exhibit dominant-negative properties on coexpressed endogenous Cx43. HeLa (A) or NRK (B) cells expressing Cx43 or mutants were pressure microinjected with 5% Lucifer yellow, and the incidence of dye transfer to neighboring cells was assessed (n > 20 for each group). (C) N2A cells expressing wild-type or Cx43 mutants were patch-clamped to measure electrical coupling conductance. In all cases, the mutants failed to form functional gap junction channels (A, C) and exhibited dominant-negative properties on coexpressed Cx43 (B). WT, untransfected wild-type cells; control in C, untransfected N2A cells.
Structural analysis of N-terminal peptides mimicking wild-type and mutant Cx43
Because all the Cx43 mutants found throughout the N-terminal domain assembled into gap junction–like structures but failed to form functional channels, we reasoned that the mechanism behind the loss of function might be unique to the position of the mutation and the nature of the amino acid substitution. To begin to address this question, we first determined the structure of a 23-mer N-terminal peptide from wild-type Cx43 (Figure 4). In aqueous solution at neutral pH, NMR spectra of the N-terminal peptide from Cx43 displayed little regular secondary structure (unpublished data) but did show some chemical shifts that were consistent with α-helical character. Previous studies with a Cx26 N-terminal peptide showed that α-helical structure can be observed under similar conditions that can be stabilized using 30% trifluoroethanol (TFE) as a cosolvent (Arita et al., 2006). These observations are consistent with other studies, in which 30% TFE appears to be optimal for helix stability of N-terminal peptides from Cx32, Cx36, and Cx43 (Fort and Spray, 2009). The NMR assignment and three-dimensional structure of the wild-type Cx43 N-terminal peptide was determined in a mixture of water and a minimal amount of TFE (30%). The 10 lowest-energy structures were determined (Figure 4) using >600 distance restraints. The structures showed no distance violations of >0.5 Å and had acceptable bond and angle deviations from ideal geometry, with satisfactory Ramachandran statistics (Table 2). In all structures the wild-type Cx43 peptide displayed a short α-helix between residues W4 and L7, a turn centered at residues G8 and K9, and a longer α-helix between residues L10 and A20. The overall fold was very similar to that displayed by an N-terminal peptide of Cx32, which also folds back on itself (Kalmatsky et al., 2009). In Cx43, the turn was a result of interactions between W4 near the N-terminus of the peptide and L7 and L11 in the longer α-helix that was supported by numerous cross-peaks in nuclear Overhauser effect spectroscopy (NOESY) spectra (i.e., pairs of protons residing within 6 Å apart in the folded peptide) between the indole ring of W4 with the side chains from L7 and L11. The tryptophan indole ring (W4) in Cx43 packs against the side chains of both leucine residues, burying ∼100 Å of hydrophobic surface area. Similar interactions were observed between W3 and L6, L9, and L10 in the three-dimensional structure of the N-terminal peptide from Cx32 (Kalmatsky et al., 2009).
FIGURE 4:
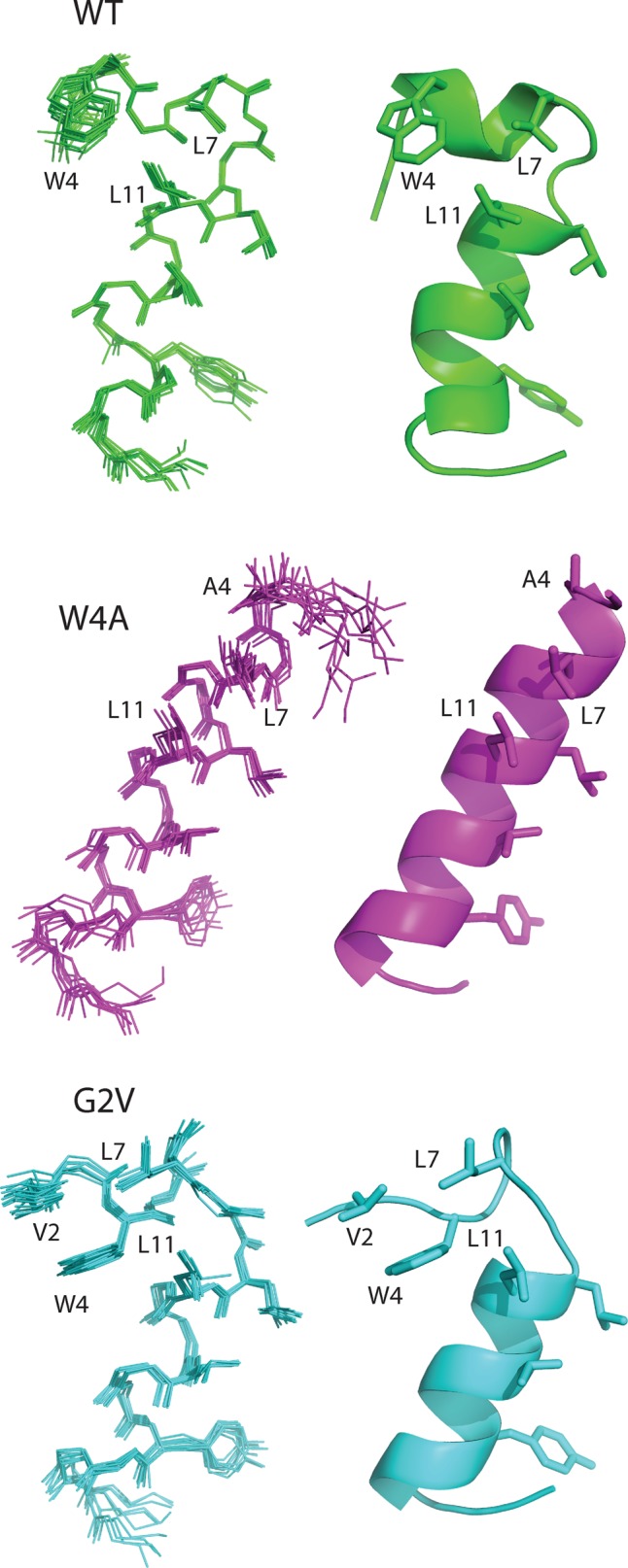
Ribbon representations of Cx43 N-terminal and mutant peptide structures as revealed by NMR analysis. The superpositions of the 10 best structures for the native 23–amino acid Cx43 N-terminal peptide (WT) and peptides containing single–amino acid substitutions (W4A, G2V) are shown (left), along with a ribbon structure of the most representative structure (right). In each case, the side chains of key residues (described in the text) are shown along with the one-letter amino acid name and sequence number. All peptides are shown in a similar orientation based on the superposition of residues D12–Y17.
TABLE 2:
Structural statistics for N-terminal Cx43 peptides.
| WT | G2V | W4A | |
|---|---|---|---|
| rmsd from experimental distance restraints | |||
| Unambiguous NOEs (Å) | 0.0271 ± 0.0015 (570) | 0.0249 ± 0.0044 (678) | 0.0234 ± 0.0043 (544) |
| Number of violations > 0.5 Å | 0 | 0 | 0 |
| Ambiguous NOEs (Å) | 0.0259 ± 0.0024 (127) | 0.0413 ± 0.0062 (101) | 0.033 ± 0.004 (106) |
| Number of violations > 0.5 Å | 0 | 0 | 0 |
| Deviations from ideal geometry | |||
| Bonds (Å) | 0.0051 ± 0.0002 | 0.0051 ± 0.0003 | 0.0042 ± 0.0005 |
| Angles (deg) | 0.603 ± 0.011 | 0.611 ± 0.037 | 0.498 ± 0.033 |
| Impropers (deg) | 1.75 ± 0.08 | 1.35 ± 0.17 | 1.19 ± 0.12 |
| Ramachandran statistics | |||
| Most-favored regions (%) | 76.8 | 78.2 | 93.6 |
| Additional allowed regions (%) | 17.4 | 21.1 | 6.4 |
| Generously allowed regions (%) | 5.9 | 0.6 | 0.0 |
| Disallowed regions (%) | 0.0 | 0.0 | 0.0 |
| rmsd from mean structurea | |||
| Backbone atoms (Å) | 0.20 ± 0.04 | 0.31 ± 0.05 | 0.38 ± 0.18 |
| Heavy atoms (Å) | 0.52 ± 0.05 | 0.56 ± 0.04 | 0.67 ± 0.21 |
Statistics are listed for the 10 lowest-energy conformers of the wild-type (WT), G2V, and W4A substituted peptides. Number of experimental restraints is given in parentheses.
armsd is shown for the ordered portions of the structures as determined using protein structure validation software; WT (A3–A20), G2V(A3–A20), and W4A(S5-T19).
The bent-helical structure of the N-terminus of Cx43 (and Cx32) differs from the regular α-helix found for the N-terminus of Cx26 observed from its three-dimensional structure (Maeda et al., 2009). In the isolated peptide, W4 in Cx43 likely packs against L7 and L11 to minimize hydrophobic surface exposure in the absence of the remainder of the protein. This arrangement contrasts that of Cx26, in which W3 interacts with I33, M34, and V37 of transmembrane helix 1 (TM1) of an adjacent protomer in the hexamer. In the context of the intact Cx43 protein, W4 would probably be recruited by the analogous TM1 residues (I34, L35) and take on a similar regular α-helical structure as observed in Cx26 (Maeda et al., 2009). To test this idea, we substituted W4 with an alanine with the rationale that substitution to a smaller side-chain residue would relieve the interactions with L7 and L10, allowing the structure to assume a more elongated α-helix similar to that observed in Cx26. Using similar conditions as for the wild-type peptide, we determined the solution structure of the W4A Cx43 peptide by NMR spectroscopy (Figure 4). The W4A polypeptide formed a contiguous α-helix from residues S5 to T19, with excellent similarity among the 10 structures (root-mean-square deviation [rmsd], 0.38 Å). These findings indicated that the W4A substitution formed a regular, nonkinked α-helix. We believe that this structure will be more representative of the native N-terminus in Cx43.
The solution structure of an N-terminal peptide from Cx43 containing the G2V variant linked to ODDD was also determined. As with the wild-type and W4A peptides, the G2V peptide displayed excellent structural characteristics (rmsd, 0.31 ± 0.05 Å). Although the G2V peptide had a comparable helical structure to both wild-type and W4A peptides toward its C-terminus (D12–Y17; rmsd, 0.37 ± 0.19 Å), it showed a bent helix near its N-terminus similar to that shown by the wild-type structures. As described for the wild-type peptide, this is a result of W4 interactions with L7 and L11. However in G2V, further important hydrophobic interactions exist involving V2 that are not present in the wild-type peptide. Specifically, the glycine-substituted valine interacts with W4 by positioning its CH3 groups against the back side of the tryptophan indole, opposite from L7 and L11 (Figure 4). This has the effect of sandwiching the W4 indole ring between a series of hydrophobic residues. We propose that these additional interactions in the G2V mutant would inhibit the interactions of W4 with residues in TM1 and compromise gap junction function.
Predictive structure and function studies
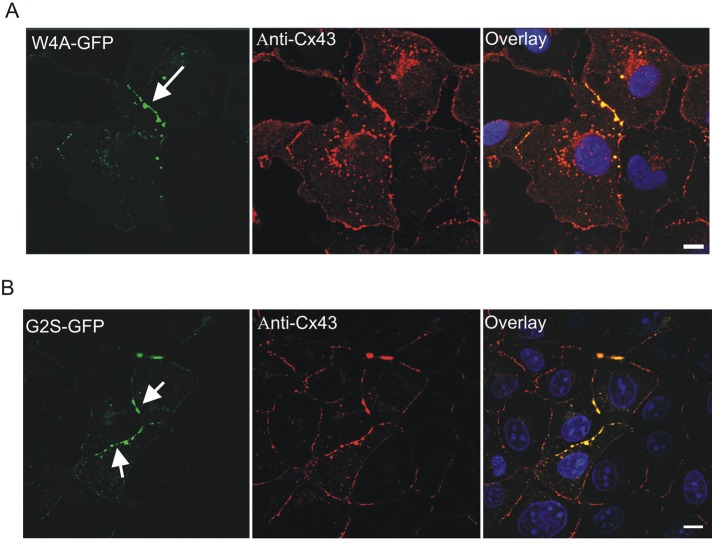
On the basis of the structures of the wild-type, W4A, and G2V Cx43 peptides, we designed experiments to test the Cx43 distribution and function of these and additional mutants. To assess how the W4A substitution would affect the distribution and function of Cx43, we engineered the Cx43 cDNA to encode this mutation and expressed the mutant in both gap junctional communication–deficient HeLa cells and Cx43-positive NRK cells. Of interest, the W4A mutant was found to form gap junction–like plaques (Figure 5A). However, like the G2V and all other N-terminal missense mutants, the W4A mutant did not form functional gap junctions and acted as a dominant negative on coexpressed Cx43 (Figure 6, A and B).
FIGURE 5:
Localization of the W4A and G2S Cx43 mutants in NRK cells. NRK cells were transfected with vectors encoding W4A-GFP (A) or G2S-GFP (B) and immunolabeled with anti-Cx43 antibodies (red) before Hoechst 33342 staining (blue). Arrows indicate that the mutants were localized to gap junction–like structures at cell–cell interfaces. Bars, 10 μm.
FIGURE 6:
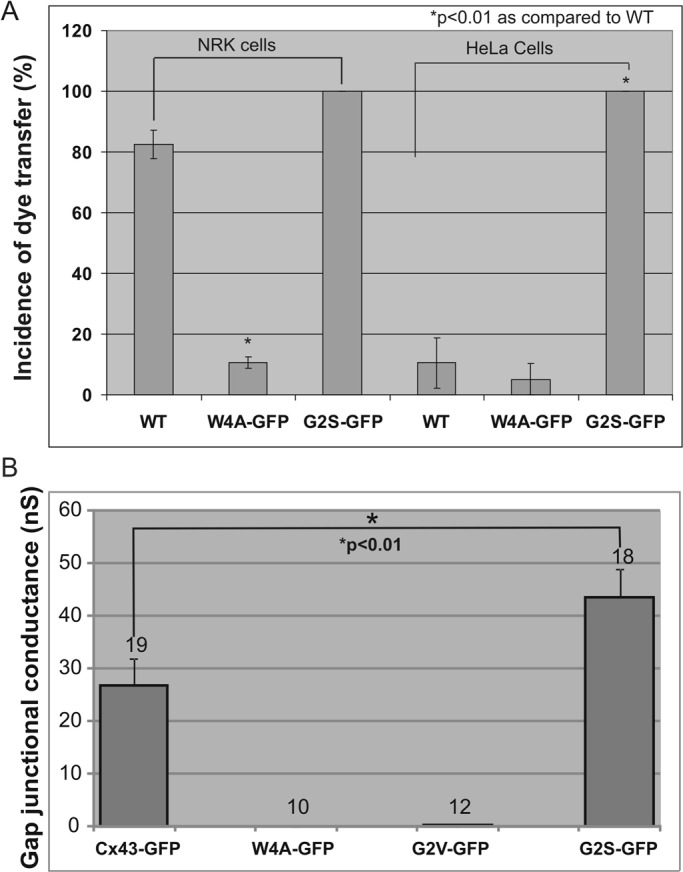
The G2S, but not the G2V or W4A, mutant forms functional gap junctions. (A) NRK and HeLa cells were untreated (WT) or engineered to express W4A-GFP or G2S-GFP before microinjection with 5% Lucifer yellow and assessment of dye transfer. (B) Gap junctional intercellular communication–deficient N2A cells were engineered to express Cx43-GFP, W4A-GFP, G2V-GFP, or G2S-GFP. Patch-clamp analysis was used to assess gap junction coupling conductance. Whereas the W4A and G2V mutants did not form functional gap junction channels, the G2S mutant exhibited functional conductance that even exceeded that of Cx43. The numbers above each column depict the number of cell pairs recorded.
Because the structure of the G2V peptide showed interactions with W4 that impede function, it was rationalized that a more conservative amino acid substitution at G2 could relieve this interaction and lead to the retention of Cx43 channel function or even result in gain of channel function. Thus we engineered a construct to encode a G2S substitution and expressed this Cx43 variant in HeLa and NRK cells. As predicted from the structural analysis, this mutant assembled into gap junction plaque structures (Figure 5B). Of importance, dye microinjection studies revealed that the G2S mutant was functional and failed to act as a dominant negative on coexpressed endogenous Cx43 (Figure 6A). To further validate that the G2S mutant formed functional gap junction channels, we performed dual whole-cell patch-clamp analysis on N2A cells engineered to express wild-type Cx43 or mutants. We found that cells expressing either the W4A or G2V mutant exhibited no significant coupling, whereas cells expressing the G2S mutant exhibited robust electrical coupling that exceeded that of wild-type Cx43, suggesting that the G2S mutant might in fact favor a more open channel state or that more channels are assembled (Figure 6B).
DISCUSSION
ODDD and N-terminal domain–specific Cx43 mutants
There is a plethora of Cx43 mutations that cause ODDD, with 15% of the mutations resulting in single–amino acid substitutions in the first 23 amino acids of Cx43 (Paznekas et al., 2009). These patients, in addition to having the classic ocular, dental, and digital developmental defects, also exhibited some uncommon features, such as cleft palate (25% of all N-terminal patients), hair problems (75%), hearing loss (50%), and/or neurological symptoms and urinary track defects (25%; Paznekas et al., 2009). For example, a patient harboring the G2V mutant was reported to have bilateral optociliary veins and umbilical hernia, which were also reported in a S18P mutant–harboring patient (de la Parra and Zenteno, 2007). In another study a D3N ODDD patient was reported to have a tethered tongue, heart murmur, kidney malfunction, and neurological symptoms (Churko et al., 2011). Thus it would seem that each ODDD patient who carries an N-terminal–specific mutation can exhibit symptoms that go beyond the common developmental disorders associated with ODDD, raising the question as to whether N-terminal–specific mutations might have variable mechanisms for inactivating Cx43 channels. Because the highly conserved N-terminal domain of connexins has been proposed to play multiple functional roles, ranging from initiating protein insertion and oligomerization to physically plugging the functional pore (Harris, 2001; Lagree et al., 2003; Oshima et al., 2011), it seemed plausible that a specific amino acid substitution might alter channel function by very different mechanisms. In the present study we combine NMR studies with mutant expression and functional studies to determine how the structural organization of the N-terminal dictates functional outcome, which might help to further explain the diversity of clinical symptoms manifested in ODDD patients.
Trafficking, localization, and function of N-terminal–specific mutants
In the present study all six naturally occurring ODDD-linked, N-terminal–specific mutants were capable of trafficking to cell surface and forming gap junction plaque-like structures, not unlike what was reported for the G21R and Y17S mutants (Roscoe et al., 2005; Lai et al., 2006; Gong et al., 2007). Of interest, the efficiency of plaque formation appeared to decrease when the amino acid substitution occurred more distal to the extreme amino terminus, as more of the L11P, Y17S, and S18P mutants were retained in intracellular compartments. Nevertheless, all point mutants were capable of membrane insertion and delivery to the cell surface, even in the absence of coexpressed wild-type Cx43. These findings raised the question as to how important the amino terminal is in general for Cx43 insertion, transport, and assembly into a plaque-like structure at the plasma membrane. To address this question, we expressed a Cx43Δ2-7 mutant and found that the loss of six amino acids from the N-terminal completely inhibited the ability of the mutant to exit the endoplasmic reticulum and localize to the plasma membrane. This was somewhat surprising, as recently it was reported that the substitution of amino acids 2–8 to alanines did not impair the ability of Cx37 to form plaque-like structures, and a deletion of nine amino acids or more was necessary to cause Cx37 to remain within an intracellular compartment (Kyle et al., 2008). Our studies suggest that smaller deletions of Cx43 N-terminal motifs are more potent at inhibiting the transport and assembly of Cx43, and both studies suggest that the N-terminal is critical for connexin assembly.
It is interesting that all six ODDD-linked, N-terminal mutants failed to form functional gap junctions, suggesting that the N-terminal domain is critical for proper channel assembly and function, as previously demonstrated for the G21R and D3N mutants (McLachlan et al., 2005; Roscoe et al., 2005; Gong et al., 2007; Churko et al., 2011). Of importance, these same mutants exhibited dominant-negative properties on coexpressed endogenous Cx43, suggesting that patients who harbor these mutants are likely surviving on far less than 50% of normal Cx43 function. Mechanistically it is likely that the mutants are co-oligomerizing with endogenous Cx43 and are able to transport to the cell surface and form plaque-like structures but that the resulting channel pores are incapable of passing small molecules. These studies raised the question as to how each mutant changes the structure of the N-terminus and whether the mutants cause similar or different structural changes.
N-terminal Cx43 mutations differentially alter structure
The structure of membrane protein motifs is always difficult to predict due to their anchoring in hydrophobic membrane bilayers. However, several lines of evidence suggest that NMR analysis of untethered polypeptide domains can prove useful in establishing structural information on connexin domains that are exposed to the cytoplasm, including intramolecular distances and α-helical formations (Duffy et al., 2002; Zhou et al., 2007, 2009; Chen et al., 2011). Cx43 belongs to the α-subfamily of connexins and contains 23 amino acids before the start of the first transmembrane domain according to most topological models (Sohl and Willecke, 2004). Polypeptides of the first 23 amino acids of native Cx43, amino acid substitutions that mimic ODDD mutants, and amino acid substitutions that do not mimic ODDD mutants were synthesized, and NMR spectra were obtained and analyzed. Wild-type Cx43 N-terminal peptide exhibited an amphipathic character, with an α-helix running between L10-A20 similar to that observed in the structure of an N-terminal peptide from Cx32 (Kalmatsky et al., 2009) and the crystal structure of Cx26 (Maeda et al., 2009). Surprisingly, a single turn centered at residues G8 and K9 resulted in the formation of a small hydrophobic cluster comprising W4, L7, and L11. A similar kinked-helix structure was observed for an N-terminal peptide from Cx32 (Kalmatsky et al., 2009). Within the hexameric structure of Cx43 it is unlikely that W4 retains this relationship with the remainder of the N-terminus. Instead, the N-terminus of Cx43 likely adopts an “unkinked” structure, elongating the helix and taking on interactions with residues in TM1 similar to those observed in Cx26. In line with this, substitution of W4 to a smaller side chain residue (alanine) relieved interactions with L7 and L11 and resulted in a rigid, contiguous α-helix that “unkinked” the N-terminus. This structure for the N-terminus would fit well with respect to the remaining transmembrane helices. In contrast, the G2V substitution resulted in a larger hydrophobic cluster, where W4 was now sandwiched between V2, L7, and L11. It is possible that the glycine-to-valine substitution might prevent W4 from interacting with I34 and L35 in TM1 of the adjacent Cx43 protein in the hexamer and compromise transjunctional exchange of molecules. The observation that a G2S substitution maintains Cx43 function supports this idea, suggesting that the glycine can be substituted without destroying Cx43 function.
Predicted structure determination of gap junction channels
Connexins are considered to share a common architecture for gap junction channel formation (Sohl and Willecke, 2004). For example, the N-terminal is 45% conserved between Cx43 and Cx26. According to the published Cx26 structure and homology comparisons with human Cx43 (Maeda et al., 2009), any amino acid substitution within the amino termini is predicted to change the structure of the N-terminus and the subsequent interactions between the side chains. As for Cx43, the G2V substitution results in additional hydrophobic interactions with W4. This likely has two consequences. First, the altered structure might disrupt key hydrogen bonds that have been proposed to play an important role in forming a circular hydrogen bond network to stabilize the pore funnel observed in Cx26. In the Cx26 protein structure, D2 in each protomer forms a hydrogen bond with T5 of the neighboring protomer that stabilize the funnel structure. In Cx43, the interaction of V2 in the G2V mutant might prevent W4 from interactions with residues in TM1, which might ultimately regulate channel opening. In support of this interpretation, both dye coupling and electrical conductance analysis revealed that the G2V mutants formed nonfunctional gap junction channels and exhibited dominant-negative properties on coexpressed Cx43.
On the basis of on our NMR studies, we found that W4A peptide forms a contiguous α-helix from residues S5 to T19 with excellent similarity among the 10 structures analyzed (rmsd, 0.38 Å). Although this structure aligns well with that for Cx26, the W4A mutant lost the ability to form functional channels. In Cx26, W3 makes important contacts with I33, M34, and V37 in TM1 of an adjacent protomer that is believed to maintain the channel in an “open” conformation. Substitution of M34 causes channel defects in Cx26. It would be expected that W4 in Cx43 has similar contacts with TM1 (I34, L35), and its substitution to alanine disrupts similar contacts, presumable closing the channel. Not surprisingly, the W4A mutant was indeed found to be nonfunctional. To further assess the possible recovery of the channel structure, we found that G2S mutant not only was able to form functional channels but also uniquely exhibited gain-of-macroscopic coupling levels exceeding those of wild-type Cx43. Given that serine (CH2OH) has a small side chain similar to glycine as found in many other α connexins, this might be important in regulating the open state of the channel. Thus it appears that maintaining a small amino acid residue in the second amino acid position of Cx43 is essential for channel function.
In summary, we found that all ODDD-linked, N-terminal Cx43 mutants can traffic to the cell surface and assemble into nonfunctional gap junction–like structures. Furthermore, all ODDD-linked mutants exhibit dominant-negative properties on coexpressed Cx43, suggesting that ODDD patients are functioning on far less than 50% of normal Cx43 gap junction channel function. NMR structure analysis showed the importance of positions G2 and W4 in channel formation, regulation, and function. Our results further suggest that an elongated α-helical structure at the N-terminus of Cx43 is likely essential for Cx43 channel formation and function. Furthermore, the integrity of the N-terminus is necessary for Cx43 trafficking to cell surface, and conserved strategic amino acid substitutions can occur in the N-terminus that can even enhance channel function. Finally, the nature of the ODDD-linked mutations within the N-terminal conveys loss of Cx43 function by distinctly different mechanisms that are rooted in the structure of the N-terminal domain.
MATERIALS AND METHODS
Cells and reagents
NRK, mouse N2A, and human cervical tumor (HeLa) cells obtained from the American Type Culture Collection (Manassas, VA) were grown in high-glucose (4500 mg glucose/l) DMEM (Invitrogen, Burlington, Canada) supplemented with 10% fetal bovine serum, 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM l-glutamine (except N2A cells). Cells were cultured within a humidified environment that maintained 5% CO2 and a temperature of 37°C.
Mutant DNA constructs
Human Cx43 mutants (G2V, D3N, L7V, L11P, Y17S, S18P, W4A, and G2S) untagged and/or GFP tagged were constructed or special purchased from Norclone Biotech Industries (London, Canada). Briefly, mutations were generated that changed the code of the amino acid Gly (G) to Val (V) or Ser (S) at the 2nd position, Asp (D) to Asn (N) at the 3rd position, Leu (L) to Val (V) at the 7th position, Leu (L) to Pro (P) at the 11th position, Try (Y) to Ser (S) at 17th position, Ser (S) to Pro (P) at the 18th position, and Trp (W) to Ala (A) at the 4th position. All cDNA constructs were cloned into the pEGFP-N1 vector (BD Biosciences, Clontech, La Jolla, CA), resulting in all mutants being tagged with GFP. The cDNA construct encoding wild-type Cx43-GFP was previously engineered. The fidelity of the constructs was confirmed by DNA sequencing at the Robarts Research Institute DNA Sequencing Facility (London, Canada) using an Applied Biosystems (Foster City, CA) 3730 analyzer.
Engineering of GFP-tagged Cx43Δ2-7 construct
To engineer GFP-tagged Cx43Δ2-7 (where residues 2–7 were deleted from the N-terminal), we designed a forward primer by using 5′-TCC AAT GGT ACC ATG GGC AAA CTC CTT-3′, which added a KpnI restriction enzyme site, and a reverse primer 5′-CGC GGA TCC TTG ATC TCC AGG TCA TCA G-3′, which added a BamHI restriction enzyme site. PCR products were cloned into the pEGFP-N1 vector. DNA sequencing confirmed the deletion of the amino acids 2–7 from the N-terminal of human Cx43, with the addition of a seven–amino acid linker connecting, in-frame, the Cx43 mutant to GFP.
Transfections and immunofluorescent labeling
For transient transfections, NRK, HeLa, and N2A cells were grown to ∼70% confluency in 60-mm culture dishes containing glass coverslips. To prepare the transfection mixtures, we used 250 μl of OptiMEM-I medium (Life Technologies, Burlington, Canada). Between 2 and 5 μg of cDNA was added to one tube, and 3 μl of Lipofectamine 2000 (Invitrogen) was added to a second tube. The tubes were combined within 5 min, gently mixed, and allowed to sit at room temperature for 15 min. The transfection mixtures were then added to the cells. After 4 h, the transfection mixture was removed and replaced with regular media for 48 h.
For immunolabeling studies, a mixture of 80% methanol/20% acetone was added to cultured cells for 20 min at 4°C and then replaced with phosphate-buffered saline (PBS). Cells were treated with 2% bovine serum albumin in PBS for 30 min to block nonspecific binding sites. A rabbit anti-Cx43 polyclonal antibody (1:500 dilution; Sigma-Aldrich, Oakville, Canada) and an anti-PDI (1:500 dilution; Stressgen, Enzo Life Sciences, San Diego, CA) antibody (an endoplasmic reticulum marker) were used. Coverslips were exposed to primary antibodies and fluorescent-tagged secondary antibodies (rabbit Alexa 555 [Invitrogen]) for 1 h at room temperature. Cell nuclei were stained for 5 min with Hoechst 33342 (10 μg/ml) and then rinsed in doubly distilled (dd) H2O before mounting.
Dye transfer
Cells expressing Cx43 or Cx43 mutants were randomly pressure injected with 5% Lucifer yellow dye dissolved in ddH2O (Molecular Probes, Eugene, OR) using an Eppendorf Femtojet automated pressure microinjector until the cell brightly fluoresced. Images were collected 1–3 min after injection using OpenLab software (Quorum Technologies, Guelph, Canada). The instances of dye transfer were quantified.
Patch-clamp electrophysiology
Functional gap junction coupling between paired N2A cells expressing fluorescent protein–tagged mutant and/or wild-type Cx43 was assessed using the dual whole-cell patch-clamp technique as previously described (Gong et al., 2006, 2007). Isolated cell pairs with green fluorescent plaques (representing GFP-tagged Cx43 or mutant Cx43) at cell–cell contacts were chosen for patch-clamp recording. The cells were constantly perfused in an external solution containing 140 mM NaCl, 5 mM KCl, 2 mM CsCl2, 2 mM CaCl2, 1 mM MgCl2, 5 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 5 mM d-glucose, 2 mM pyruvate, and 1 mM BaCl2, pH 7.4. Junctional current was measured between cell pairs by using the dual whole-cell voltage-clamp technique with Axopatch 200B patch-clamp amplifiers (Axon Instruments, Union City, CA) at room temperature (22–25°C). Recording patch pipettes were prepared with a puller (PP-83; Narishige Scientific Instrument, Tokyo, Japan). The patch pipette had a resistance of 2.5–5.5 MΩ when filled with an internal solution containing 130 mM CsCl2, 10 mM ethylene glycol tetraacetic acid, 0.5 mM CaCl2, 3 mM MgATP, 2 mM Na2ATP, and 10 mM HEPES, pH titrated to 7.2 with CsOH. Data were low-pass filtered at 2 kHz and were acquired using pClamp9 software (Axon Instruments) and digitized at a 5- to 10-kHz sampling rate. Each cell of a pair was initially held at a common holding potential of 0 mV. To evaluate junctional coupling, we applied 7-s hyperpolarizing pulses from the holding potential of 0 to −20 mV to one cell to establish a transjunctional voltage gradient, and we measured the junctional current in the second cell. Macroscopic junctional conductance (gj) was calculated from gj = Ij/Vj, where Ij is the measured junctional current and Vj is transjunctional voltage. The experimenter was blind to the connexin subtypes when performing the patch-clamp experiments. Student's t test was performed with Excel (Microsoft, Redmond, WA).
Peptide synthesis and sample preparation
Peptides corresponding to the N-terminal first 23 residues (MGDWSALGKLLDKVQAYSTAGGK) or Cx43 peptides with the G2V or W4A amino acid substitutions were high-performance liquid chromatography purified, and purity was confirmed by electron ionization spectrometry (Genemed Synthesis, San Antonio, TX). NMR-ready samples were prepared by dissolving each peptide (∼1 mM) in buffer containing 50 mM sodium phosphate, pH 5.2, 100 mM NaCl, and 30% H2O/10% 2.2.2-trifluoroethanol-D3 D2O/30% TFE-d3 (vol/vol; Cambridge Isotope Laboratories, Andover, MA).
NMR spectroscopy
All experiments were acquired on an Inova 600-MHz spectrometer (Varian, Palo Alto, CA) equipped with a 13C-enhanced triple-resonance cold probe at 12ºC. Two-dimensional homonuclear 1H–1H total correlation spectroscopy (TOCSY; mixing time τm = 80 ms; Bax and Davis, 1985) and 1H–1H NOESY (τm = 200 ms; States et al., 1982) spectra were collected using spectral widths of 7000 Hz in both dimensions with 1104 and 552 complex points in t2 and t1 dimensions, respectively. Water suppression was achieved with the WATERGATE pulse sequence (Piotto et al., 1992). All spectra were processed with NMRPipe (Delaglio et al., 1995) and analyzed with NMRView (Johnson and Blevins, 1994). A 60° shifted sine bell window function was applied to each dimension and then zero filled to 2048 points before Fourier transformation. Sequence-specific 1H resonance assignments were obtained using TOCSY and NOESY spectra following standard procedures.
Structure calculations
The standard protocol of ARIA2.2 was used for iterated automatic NOE assignment and structure calculations (Rieping et al., 2007). Distance information was obtained from the volumes of cross-peaks in NOESY spectra and used as input along with chemical shift information for each peptide. Initial manual NOE assignments were used only when unambiguous assignment could be obtained. Each structure calculation was initialized from 20 randomly generated structures followed by eight iterations using the input data. The 10 lowest-energy structures in the final iteration were refined using the water refinement routine in ARIA2.2, submitted to the Protein Structure Validation Suite (http://psvs-1_4-dev.nesg.org) for structure quality evaluation, and analyzed and displayed using PyMOL (www.pymol.org).
Acknowledgments
This work was supported by the Canadian Institutes of Health Research (D.W.L., D.B., and G.S.S.) and the Canada Research Chair Program (D.W.L. and G.S.S).
Abbreviations used:
- Cx
connexin
- Cx43
connexin-43
- NMR
nuclear magnetic resonance
- ODDD
oculodentodigital dysplasia
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-02-0128) on July 18, 2012.
*These authors contributed equally to this study.
REFERENCES
- Arita K, Akiyama M, Aizawa T, Umetsu Y, Segawa I, Goto M, Sawamura D, Demura M, Kawano K, Shimizu H. A novel N14Y mutation in connexin26 in keratitis-ichthyosis-deafness syndrome: analyses of altered gap junctional communication and molecular structure of N terminus of mutated connexin26. Am J Pathol. 2006;169:416–423. doi: 10.2353/ajpath.2006.051242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bax A, Davis DG. MLEV-17 based two-dimensional homonuclear magetization transfer spectroscopy. J Magn Reson. 1985;65:355–360. [Google Scholar]
- Chen Y, et al. Molecular interaction and functional regulation of connexin50 gap junctions by calmodulin. Biochem J. 2011;435:711–722. doi: 10.1042/BJ20101726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churko JM, Langlois S, Pan X, Shao Q, Laird DW. The potency of the fs260 connexin43 mutant to impair keratinocyte differentiation is distinct from other disease-linked connexin43 mutants. Biochem J. 2010;429:473–483. doi: 10.1042/BJ20100155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churko JM, Shao Q, Gong XQ, Swoboda KJ, Bai D, Sampson J, Laird DW. Human dermal fibroblasts derived from oculodentodigital dysplasia patients suggest that patients may have wound-healing defects. Hum Mut. 2011;32:456–466. doi: 10.1002/humu.21472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Parra DR, Zenteno JC. A new GJA1 (connexin 43) mutation causing oculodentodigital dysplasia associated to uncommon features. Ophthalmic Genet. 2007;28:198–202. doi: 10.1080/13816810701538620. [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Dobrowolski R, Sommershof A, Willecke K. Some oculodentodigital dysplasia-associated Cx43 mutations cause increased hemichannel activity in addition to deficient gap junction channels. J Membr Biol. 2007;219:9–17. doi: 10.1007/s00232-007-9055-7. [DOI] [PubMed] [Google Scholar]
- Duffy HS, Sorgen PL, Girvin ME, O'Donnell P, Coombs W, Taffet SM, Delmar M, Spray DC. pH-dependent intramolecular binding and structure involving Cx43 cytoplasmic domains. J Biol Chem. 2002;277:36706–36714. doi: 10.1074/jbc.M207016200. [DOI] [PubMed] [Google Scholar]
- Flenniken AM, et al. A Gja1 missense mutation in a mouse model of oculodentodigital dysplasia. Development. 2005;132:4375–4386. doi: 10.1242/dev.02011. [DOI] [PubMed] [Google Scholar]
- Fort AG, Spray DC. Trifluoroethanol reveals helical propensity at analogous positions in cytoplasmic domains of three connexins. Biopolymers. 2009;92:173–182. doi: 10.1002/bip.21166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong XQ, Shao Q, Langlois S, Bai D, Laird DW. Differential potency of dominant negative connexin43 mutants in oculodentodigital dysplasia. J Biol Chem. 2007;282:19190–19202. doi: 10.1074/jbc.M609653200. [DOI] [PubMed] [Google Scholar]
- Gong XQ, Shao Q, Lounsbury CS, Bai D, Laird DW. Functional characterization of a GJA1 frameshift mutation causing oculodentodigital dysplasia and palmoplantar keratoderma. J Biol Chem. 2006;281:31801–31811. doi: 10.1074/jbc.M605961200. [DOI] [PubMed] [Google Scholar]
- Harris AL. Emerging issues of connexin channels: biophysics fills the gap. Q Rev Biophys. 2001;34:325–472. doi: 10.1017/s0033583501003705. [DOI] [PubMed] [Google Scholar]
- Hirst-Jensen BJ, Sahoo P, Kieken F, Delmar M, Sorgen PL. Characterization of the pH-dependent interaction between the gap junction protein connexin43 carboxyl terminus and cytoplasmic loop domains. J Biol Chem. 2007;282:5801–5813. doi: 10.1074/jbc.M605233200. [DOI] [PubMed] [Google Scholar]
- Johnson BA, Blevins RA. NMRView: A computer program for the visualization and analysis of NMR data. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- Kalmatsky BD, Bhagan S, Tang Q, Bargiello TA, Dowd TL. Structural studies of the N-terminus of connexin 32 using 1H NMR spectroscopy. Arch Biochem Biophys. 2009;490:9–16. doi: 10.1016/j.abb.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyle JW, Berthoud VM, Kurutz J, Minogue PJ, Greenspan M, Hanck DA, Beyer EC. The N terminus of connexin37 contains an alpha-helix that is required for channel function. J Biol Chem. 2009;284:20418–20427. doi: 10.1074/jbc.M109.016907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyle JW, Minogue PJ, Thomas BC, Domowicz DA, Berthoud VM, Hanck DA, Beyer EC. An intact connexin N-terminus is required for function but not gap junction formation. J Cell Sci. 2008;121:2744–2750. doi: 10.1242/jcs.032482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagree V, Brunschwig K, Lopez P, Gilula NB, Richard G, Falk MM. Specific amino-acid residues in the N-terminus and TM3 implicated in channel function and oligomerization compatibility of connexin43. J Cell Sci. 2003;116:3189–3201. doi: 10.1242/jcs.00604. [DOI] [PubMed] [Google Scholar]
- Lai A, Le DN, Paznekas WA, Gifford WD, Jabs EW, Charles AC. Oculodentodigital dysplasia connexin43 mutations result in non-functional connexin hemichannels and gap junctions in C6 glioma cells. J Cell Sci. 2006;119:532–541. doi: 10.1242/jcs.02770. [DOI] [PubMed] [Google Scholar]
- Laird DW. Life cycle of connexins in health and disease. Biochem J. 2006;394:527–543. doi: 10.1042/BJ20051922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird DW. Closing the gap on autosomal dominant connexin-26 and connexin-43 mutants linked to human disease. J Biol Chem. 2008;283:2997–3001. doi: 10.1074/jbc.R700041200. [DOI] [PubMed] [Google Scholar]
- Laird DW. The gap junction proteome and its relationship to disease. Trends Cell Biol. 2010;20:92–101. doi: 10.1016/j.tcb.2009.11.001. [DOI] [PubMed] [Google Scholar]
- Laird DW, Revel JP. Biochemical and immunochemical analysis of the arrangement of connexin43 in rat heart gap junction membranes. J Cell Sci. 1990;97:109–117. doi: 10.1242/jcs.97.1.109. [DOI] [PubMed] [Google Scholar]
- Latour P, et al. New mutations in the X-linked form of Charcot-Marie-Tooth disease. Eur Neurol. 1997;37:38–42. doi: 10.1159/000117403. [DOI] [PubMed] [Google Scholar]
- Maeda S, Nakagawa S, Suga M, Yamashita E, Oshima A, Fujiyoshi Y, Tsukihara T. Structure of the connexin 26 gap junction channel at 3.5 A resolution. Nature. 2009;458:597–602. doi: 10.1038/nature07869. [DOI] [PubMed] [Google Scholar]
- Manias JL, Plante I, Gong XQ, Shao Q, Churko J, Bai D, Laird DW. Fate of connexin43 in cardiac tissue harbouring a disease-linked connexin43 mutant. Cardiovasc Res. 2008;80:385–395. doi: 10.1093/cvr/cvn203. [DOI] [PubMed] [Google Scholar]
- McLachlan E, Manias JL, Gong XQ, Lounsbury CS, Shao Q, Bernier SM, Bai D, Laird DW. Functional characterization of oculodentodigital dysplasia-associated Cx43 mutants. Cell Commun Adhes. 2005;12:279–292. doi: 10.1080/15419060500514143. [DOI] [PubMed] [Google Scholar]
- McLachlan E, Plante I, Shao Q, Tong D, Kidder GM, Bernier SM, Laird DW. ODDD-linked Cx43 mutants reduce endogenous Cx43 expression and function in osteoblasts and inhibit late stage differentiation. J Bone Miner Res. 2008;23:928–938. doi: 10.1359/jbmr.080217. [DOI] [PubMed] [Google Scholar]
- Musa FU, Ratajczak P, Sahu J, Pentlicky S, Fryer A, Richard G, Willoughby CE. Ocular manifestations in oculodentodigital dysplasia resulting from a heterozygous missense mutation (L113P) in GJA1 (connexin 43) Eye. 2009;23:549–555. doi: 10.1038/eye.2008.77. [DOI] [PubMed] [Google Scholar]
- Nakagawa S, Gong XQ, Maeda S, Dong Y, Misumi Y, Tsukihara T, Bai D. Asparagine 175 of connexin32 is a critical residue for docking and forming functional heterotypic gap junction channels with connexin26. J Biol Chem. 2011;286:19672–19681. doi: 10.1074/jbc.M110.204958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima A, Tani K, Toloue MM, Hiroaki Y, Smock A, Inukai S, Cone A, Nicholson BJ, Sosinsky GE, Fujiyoshi Y. Asymmetric configurations and N-terminal rearrangements in connexin26 gap junction channels. J Mol Biol. 2011;405:724–735. doi: 10.1016/j.jmb.2010.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paznekas WA, et al. Connexin 43 (GJA1) mutations cause the pleiotropic phenotype of oculodentodigital dysplasia. Am J Hum Genet. 2003;72:408–418. doi: 10.1086/346090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paznekas WA, et al. GJA1 mutations, variants, and connexin 43 dysfunction as it relates to the oculodentodigital dysplasia phenotype. Hum Mutat. 2009;30:724–733. doi: 10.1002/humu.20958. [DOI] [PubMed] [Google Scholar]
- Pfenniger A, Wohlwend A, Kwak BR. Mutations in connexin genes and disease. Eur J Clin Invest. 2011;41:103–116. doi: 10.1111/j.1365-2362.2010.02378.x. [DOI] [PubMed] [Google Scholar]
- Piotto M, Saudek V, Sklenar V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J Biomol NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- Rico F, Oshima A, Hinterdorfer P, Fujiyoshi Y, Scheuring S. Two-dimensional kinetics of inter-connexin interactions from single-molecule force spectroscopy. J Mol Biol. 2011;412:72–79. doi: 10.1016/j.jmb.2011.07.013. [DOI] [PubMed] [Google Scholar]
- Rieping W, Habeck M, Bardiaux B, Bernard A, Malliavin TE, Nilges M. ARIA2: automated NOE assignment and data integration in NMR structure calculation. Bioinformatics. 2007;23:381–382. doi: 10.1093/bioinformatics/btl589. [DOI] [PubMed] [Google Scholar]
- Roscoe W, Veitch GI, Gong XQ, Pellegrino E, Bai D, McLachlan E, Shao Q, Kidder GM, Laird DW. Oculodentodigital dysplasia-causing connexin43 mutants are non-functional and exhibit dominant effects on wild-type connexin43. J Biol Chem. 2005;280:11458–11466. doi: 10.1074/jbc.M409564200. [DOI] [PubMed] [Google Scholar]
- Scott CA, Kelsell DP. Key functions for gap junctions in skin and hearing. Biochem J. 2011;438:245–254. doi: 10.1042/BJ20110278. [DOI] [PubMed] [Google Scholar]
- Sohl G, Willecke K. An update on connexin genes and their nomenclature in mouse and man. Cell Commun Adhes. 2003;10:173–180. doi: 10.1080/cac.10.4-6.173.180. [DOI] [PubMed] [Google Scholar]
- Sohl G, Willecke K. Gap junctions and the connexin protein family. Cardiovasc Res. 2004;62:228–232. doi: 10.1016/j.cardiores.2003.11.013. [DOI] [PubMed] [Google Scholar]
- Solan JL, Lampe PD. Connexin43 phosphorylation: structural changes and biological effects. Biochem J. 2009;419:261–272. doi: 10.1042/BJ20082319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- States DJ, Haberkorn RA, Ruben DJ. A two dimensional nuclear Overhauser experiment with pure absorption phase in four quadrants. J Magn Reson. 1982;48:236–292. [Google Scholar]
- Toth K, Shao Q, Lorentz R, Laird DW. Decreased levels of Cx43 gap junctions result in ameloblast dysregulation and enamel hypoplasia in Gja1Jrt/+ mice. J Cell Physiol. 2010;223:601–609. doi: 10.1002/jcp.22046. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Yang W, Lurtz MM, Chen Y, Jiang J, Huang Y, Louis CF, Yang JJ. Calmodulin mediates the Ca2+-dependent regulation of Cx44 gap junctions. Biophys J. 2009;96:2832–2848. doi: 10.1016/j.bpj.2008.12.3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Yang W, Lurtz MM, Ye Y, Huang Y, Lee HW, Chen Y, Louis CF, Yang JJ. Identification of the calmodulin binding domain of connexin 43. J Biol Chem. 2007;282:35005–35017. doi: 10.1074/jbc.M707728200. [DOI] [PubMed] [Google Scholar]