There is differential regulation of actin patch dynamics and endocytosis between the yeast and hyphal growth in Candida albicans. The mechanism involves phosphoregulation of the endocytic protein Sla1 by the CDK Cdc28–Cln3 and the actin-regulating kinase Prk1. The results establish the first molecular link between CDK and the endocytic machinery in fungi.
Abstract
A dynamic balance between targeted transport and endocytosis is critical for polarized cell growth. However, how actin-mediated endocytosis is regulated in different growth modes remains unclear. Here we report differential regulation of cortical actin patch dynamics between the yeast and hyphal growth in Candida albicans. The mechanism involves phosphoregulation of the endocytic protein Sla1 by the cyclin-dependent kinase (CDK) Cdc28–Cln3 and the actin-regulating kinase Prk1. Mutational studies of the CDK phosphorylation sites of Sla1 revealed that Cdc28–Cln3 phosphorylation of Sla1 enhances its further phosphorylation by Prk1, weakening Sla1 association with Pan1, an activator of the actin-nucleating Arp2/3 complex. Sla1 is rapidly dephosphorylated upon hyphal induction and remains so throughout hyphal growth. Consistently, cells expressing a phosphomimetic version of Sla1 exhibited markedly reduced actin patch dynamics, impaired endocytosis, and defective hyphal development, whereas a nonphosphorylatable Sla1 had the opposite effect. Taken together, our findings establish a molecular link between CDK and a key component of the endocytic machinery, revealing a novel mechanism by which endocytosis contributes to cell morphogenesis.
INTRODUCTION
Polarity is a fundamental property of cells, critical for the control of diverse cellular and developmental processes such as shape development in early plant and animal embryogenesis, intracellular movement of organelles and proteins, cell migration, and polarized growth of fungal cells (Slaughter et al., 2009; Tahirovic and Bradke, 2009; Dettmer and Friml, 2011; Hertzog and Chavrier, 2011; St Johnston and Sanson, 2011). A key feature of cell polarity is the asymmetric distribution of proteins and lipids in the plasma membrane (PM), which is mostly achieved through vesicular trafficking along cytoskeletal tracks (Orlando and Guo, 2009). Exocytosis deposits the content of secretory vesicles in the growth areas of the PM (Cai et al., 2007). It has become increasingly clear that endocytosis also plays an important role in establishing and maintaining cell polarity (Shivas et al., 2010; Lobert and Stenmark, 2011).
The molecules and pathways that determine cell polarity are highly conserved in eukaryotes and best understood in the budding yeast Saccharomyces cerevisiae (Slaughter et al., 2009). In G1 cells, the CDK Cdc28 in association with G1 cyclins triggers cell polarization by recruiting a set of polarity proteins, including the small GTPase Cdc42 and its regulators, to the presumptive bud site to initiate assembly of actin cables and delivery of secretory vesicles (Park and Bi, 2007; Slaughter et al., 2009). During the cell cycle, cells assume alternative states of polarized growth under the tight control of Cdc28: Cdc28–G1 cyclins drive apical extension of the bud, and later Cdc28–G2 cyclins promote isotropic growth. Near the end of mitosis, Cdc28 inactivation triggers repolarization of growth to the bud neck for cytokinesis and cell separation (Lew and Reed, 1993, 1995). In addition to actin cables, cortical actin patches also concentrate around the growth site, which is believed to play a crucial role in polarity maintenance through endocytic recycling of polarity proteins (Pruyne and Bretscher, 2000).
Polarized cell growth is a developmental mode in many organisms but is perhaps best exemplified in the hyphal growth of fungi, in which new materials are added exclusively and continuously to a small cell surface area, generating tubular cells (Momany, 2002). Polarization of the actin cytoskeleton is essential for filamentous fungi to achieve targeted secretion (Berepiki et al., 2011). Recently live-cell imaging experiments revealed that actin patches form in a subapical collar during the hyphal development of Aspergillus nidulans (Araujo-Bazan et al., 2008; Taheri-Talesh et al., 2008; Upadhyay and Shaw, 2008) and Neurospora crassa (Delgado-Alvarez et al., 2010; Berepiki et al., 2011). Because actin patches are the sites of endocytosis (Kaksonen et al., 2003), these findings led to the hypothesis that the actin collar performs endocytic recycling of polarity proteins to maintain the hyphal morphology (Shaw et al., 2011). During hyphal extension, exocytic vesicles carrying polarity proteins arrive at the growing apex and merge into the apical PM. As the cell growth advances, these proteins diffuse away from the growing tip and, if not removed in a timely manner, lead to growth in areas beyond the initially established growth zone, leading to loss of polarity (Marco et al., 2007).
The opportunistic human fungal pathogen Candida albicans is capable of switching growth mode between yeast and hyphae (Sudbery et al., 2004), rendering it an excellent model for the study of polarity control. Although the hyphal growth of C. albicans is regulated independently of cell cycle (Hazan et al., 2002), in recent years CDKs have been found to play crucial roles in the growth mode transition by directly targeting key components of the polarity machinery (Wang, 2009). C. albicans possesses three G1 cyclins—Ccn1, Cln3, and Hgc1—and their association with Cdc28 regulates different aspects of hyphal development (Loeb et al., 1999; Zheng et al., 2004; Bachewich and Whiteway, 2005; Chapa et al., 2005). The hyphal-specific Cdc28–Hgc1 kinase (Zheng et al., 2004) phosphorylates and inhibits Rga2, a GTPase-activating protein of Cdc42, leading to localized Cdc42 activation at the hyphal tip (Zheng et al., 2007). Cdc28–Hgc1 also targets the septins Cdc11 and Sep7 to maintain proper hyphal growth (Sinha et al., 2007; Gonzalez-Novo et al., 2008) and the transcriptional factor Efg1 to inhibit cell separation (Wang et al., 2009). More recently, Cdc28–Hgc1 was shown to phosphorylate Sec2, a guanine-nucleotide exchange factor of the Rab GTPase Sec4 that has a major role in exocytosis, to facilitate polarized secretion toward the hyphal tip (Bishop et al., 2010). Cdc28–Ccn1 also phosphorylates Cdc11 and Sec2, priming their further phosphorylation by Cdc28–Hgc1 (Sinha et al., 2007; Bishop et al., 2010). Cln3 is the only G1 cyclin essential for the viability of C. albicans, and its depletion initially results in filamentous growth (Bachewich and Whiteway, 2005; Chapa et al., 2005). In spite of all these findings, it remains unknown whether CDKs might regulate the dynamics of cortical actin patches and endocytosis in different growth modes. A previous global search of Cdc28 substrates in S. cerevisiae identified the endocytic protein Sla1 as a potential CDK target. However, their enzyme–substrate relationship has not been confirmed by independent means, and the biological significance of the phosphorylation remains undetermined.
In this study, by affinity purification and mass spectrometry (MS), we discover that Cdc28 in association with Cln3 phosphorylates Sla1 in C. albicans. Control of this phosphorylation plays an important role in regulating cortical actin patch dynamics and endocytosis when C. albicans switches between yeast and hyphal growth.
RESULTS
Sla1 physically interacts with the G1 cyclin Cln3
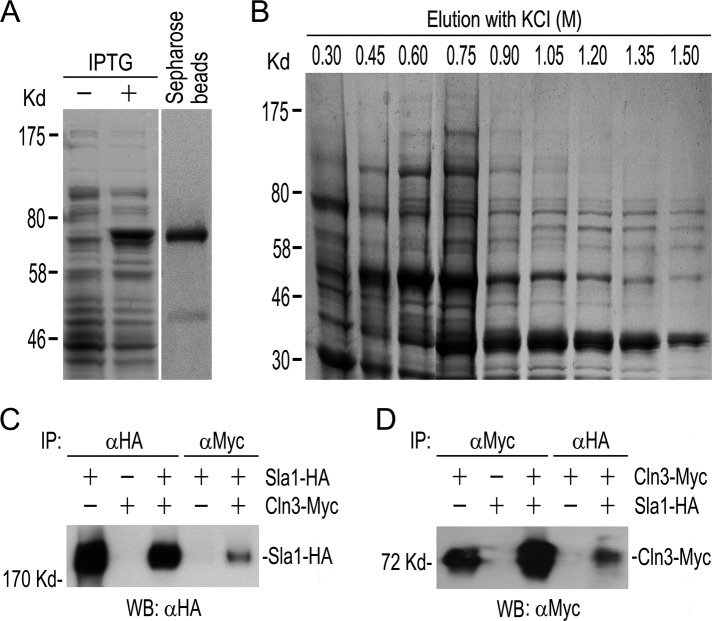
Depleting Cln3 causes filamentous growth in C. albicans (Bachewich and Whiteway, 2005; Chapa et al., 2005). To unravel the underlying mechanisms, we tried to first identify Cdc28–Cln3 substrates by affinity purification and MS identification of Cln3-binding proteins. To this end, we conjugated recombinant glutathione S-transferase (GST)–Cln3 to Sepharose beads (Figure 1A) to pull down proteins from C. albicans cell lysates. The bound proteins were eluted and separated by gel electrophoresis (Figure 1B). Protein bands were then excised for MS analysis. The results are given in Supplemental Table S3. Owing to the lack of a control experiment with the use of GST-bound beads, it was difficult to distinguish Cln3-associated proteins from GST-associated proteins. In fact, most of the GST-Cln3–associated proteins turned out to be chaperones, metabolic enzymes, and translation proteins, which were generally considered as nonspecific contaminants in affinity purification experiments (Archambault et al., 2004). However, the endocytic protein Sla1 attracted our special attention because S. cerevisiae Sla1 had been identified as a potential Cdc28 substrate (Holt et al., 2009). Therefore we performed coimmunoprecipitation (coIP) to test Cln3-Sla1 interaction using a strain coexpressing Myc-tagged Cln3 and HA-tagged Sla1. Western blot (WB) analysis showed that Myc antibody (αMyc) could pull down Sla1-HA together with Cln3-Myc (Figure 1C) and hemagglutinin (HA) antibody (αHA) could pull down Cln3-Myc together with Sla1-HA (Figure 1D), whereas such coIP was not detected in strains expressing only Sla1-HA or Cln3-Myc. The results demonstrate that Sla1 physically associates with Cln3 in vivo.
FIGURE 1:
Identification of Sla1 as a Cln3-interacting protein. (A) Expression and purification of GST-Cln3. E. coli cells expressing GST-Cln3 were induced with IPTG. Cell lysates were separated by gel electrophoresis and stained with CBB (left). The GST-Cln3 fusion was purified and bound to glutathione–Sepharose beads (right). (B) Affinity purification of GST-Cln3–associated proteins. The GST-Cln3–bound beads were incubated with C. albicans cell lysate, and the bound proteins were eluted with different concentrations of KCl, separated by gel electrophoresis, and stained with CBB. (C, D) CoIP between Sla1-HA and Cln3-Myc. Yeast extracts prepared from YPD cultures of GZY559 (CLN3-Myc), GZY584 (SLA1-HA), and GZY585 (CLN3-Myc SLA1-HA) were subjected to anti-Myc (αMyc) or αHA IP, followed by WB with αHA (C) or αMyc (D) antibodies.
Cdc28–Cln3 phosphorylates Sla1
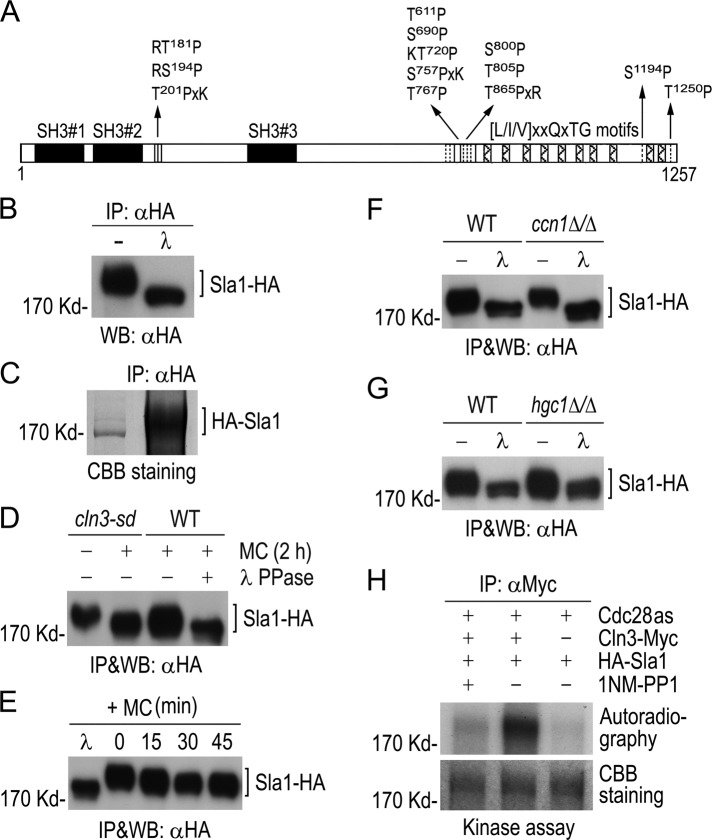
The Cln3 association with Sla1 suggests that Cdc28–Cln3 might phosphorylate Sla1. Consistently, Sla1 contains six perfect (S/TPxR/K or R/KS/TP) and seven minimal (S/TP) consensus CDK phosphorylation sites (Figure 2A). In addition, Sla1 also contains three SH3 domains in the N-terminal region and 10 putative recognition motifs of the Prk1 kinase in the C-terminal region (Figure 2A). Both N- and C-terminal regions of Sla1 are known to mediate interactions with actin-binding proteins and other endocytic proteins in S. cerevisiae (Tang et al., 2000; Zeng et al., 2001; Warren et al., 2002; Gourlay et al., 2003; Rodal et al., 2003; Di Pietro et al., 2010).
FIGURE 2:
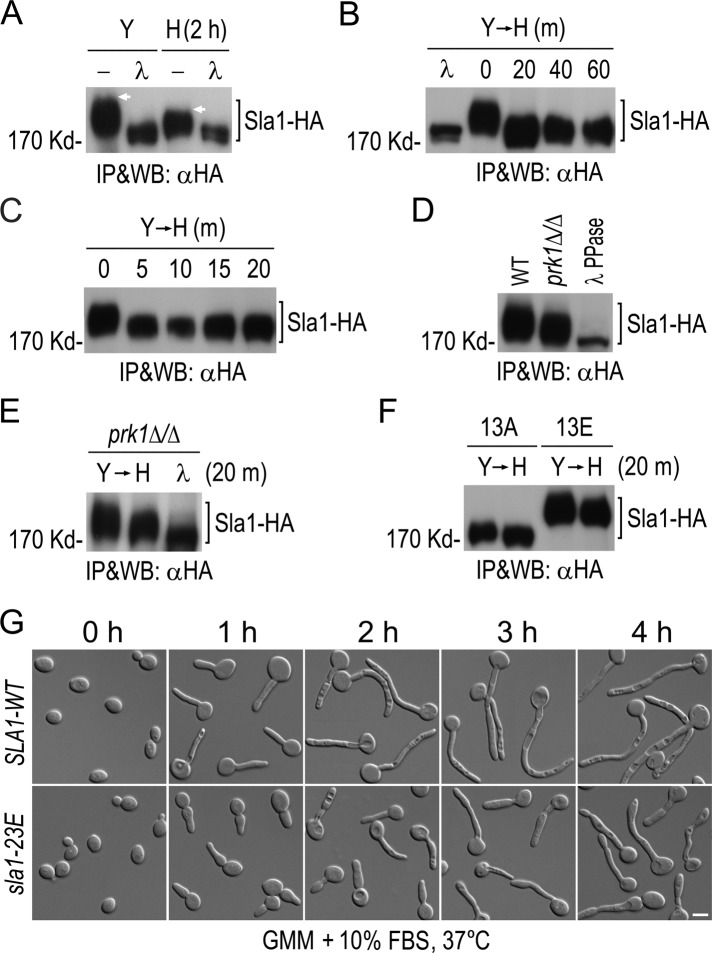
Phosphorylation of Sla1 by Cdc28−Cln3. (A) Schematic diagram of Sla1 showing CDK phosphorylation sites (vertical lines). Solid boxes indicate SH3 domains, and hatched boxes show putative Prk1 kinase recognition motifs. (B) Detection of phosphorylated Sla1 in vivo. Sla1-HA was immunoprecipitated from GZY584 (SLA1-HA) yeast cell lysate and subjected to λPP or mock treatment, followed by αHA WB analysis. (C) Purification of Sla1 for phospho-site mapping. HA-Sla1 was immunoprecipitated from YPG culture of GZY631 (PGAL1-HA-SLA1) yeast cells and electrophoresed on SDS–PAGE. After staining with CBB, the Sla1 band was excised for phospho-site mapping by MS. (D) Cln3-dependent phosphorylation of Sla1 in vivo. GMM culture of cln3-sd yeast cells expressing Sla1-HA (GZY622) was incubated with 0.5 mM Met and Cys (MC) at 30°C for 2 h to switch off CLN3 expression. Sla1-HA was immunoprecipitated from cell lysates for WB analysis. A WT strain (BWP17) expressing Sla1-HA (GZY584) was included as a control, and half of the immunoprecipitated Sla1-HA was treated with λPP. (E) Sla1 phosphorylation in Cln3-depleted cells. GZY622 cells were grown in GMM in the presence of 0.5 mM MC at 30°C, and samples were taken at 15-min intervals for IP and WB analysis of Sla1-HA using αHA. Part of Sla1-HA from the 0-min time point was treated with λPP for control. (F) Comparison of Sla1 phosphorylation between ccn1Δ/Δ and WT cells. Sla1-HA was immunoprecipitated from lysates of WT (GZY584) and ccn1Δ/Δ (GZY744) yeast cells and subjected to λPP or mock treatment for WB analysis. (G) Comparison of Sla1 phosphorylation between hgc1Δ/Δ and WT cells. WT (GZY584) and hgc1Δ/Δ (GZY624) yeast cells were induced with 20% FBS at 37°C for 2 h for hyphal growth. Sla1-HA was then immunoprecipitated and subjected to λPP or mock treatment for WB analysis. (H) In vitro phosphorylation of Sla1 by Cdc28−Cln3. Cln3-Myc was immunoprecipitated from GZY641 (cdc28as CLN3-Myc) yeast cells and incubated with immunopurified HA-Sla1 to perform in vitro kinase assay in the presence or absence of 25 μM 1NM-PP1. Immunoprecipitates from IS89 (cdc28as) cell lysates were used as a negative control. After electrophoresis, the gel was stained with CBB to visualize HA-Sla1 and dried onto a filter paper for autoradiography.
To determine whether Sla1 is a phosphoprotein in vivo, we pulled down Sla1-HA with αHA and then treated half of the precipitate with λ phosphatase (λPP). WB analysis showed that λPP-treated Sla1 migrated significantly faster in gel than the untreated protein (Figure 2B), indicating that Sla1 is phosphorylated in vivo. We next immunopurified Sla1-HA for phospho-site mapping by MS (Figure 2C). With 81% sequence coverage, 12 of the 13 consensus CDK sites were found to be phosphorylated on the Ser or Thr residue (Supplemental Figure S1).
To obtain evidence that Cln3 mediates Sla1 phosphorylation, we generated a cln3-sd mutant with one copy of CLN3 deleted and the other controlled by the MET3 promoter. As reported previously (Bachewich and Whiteway, 2005; Chapa et al., 2005), the cln3-sd cells switched from budding to filamentous growth (unpublished data) when CLN3 expression was repressed by methionine and cysteine (MC). Next we expressed Sla1-HA in the cln3-sd mutant and compared its phosphorylation level in the presence and absence of Cln3. As shown in Figure 2D, the level of Sla1 phosphorylation was significantly reduced at 2 h after switching off CLN3 expression. In fact, the Sla1 dephosphorylation was already detected at 15 min after the addition of MC (Figure 2E). However, in spite of prolonged growth in the presence of MC, the gel mobility of Sla1 from cln3-sd cells was still slightly slower than that of λPP-treated Sla1 (Figure 2, D and E), indicating possible involvement of other kinase(s) in Sla1 phosphorylation. We also determined whether Sla1 phosphorylation might depend on Ccn1 and Hgc1 and found that Sla1 phosphorylation was unaffected in ccn1∆/∆ (Figure 2F) and hgc1∆/∆ (Figure 2G) mutants compared with wild-type (WT) BWP17 cells.
Next we performed kinase assays to test whether Cdc28–Cln3 can phosphorylate Sla1 in vitro. We tagged Cln3 with Myc in a cdc28as mutant and immunopurified the Cdc28as–Cln3-Myc complex. Cdc28as is sensitive to the ATP analogue 1NM-PP1 (Bishop et al., 2000). We incubated the purified kinase complex with immunopurified HA-Sla1 in a kinase reaction containing γ-32P-ATP. Phosphorylated proteins were resolved on SDS–PAGE and visualized by autoradiography. Figure 2H shows that the Cdc28as–Cln3 complex phosphorylated HA-Sla1. In contrast, the αMyc precipitate from cells expressing only Cdc28as failed to do so. Furthermore, the in vitro phosphorylation was largely abolished by 1NM-PP1, indicating that Cdc28–Cln3 is responsible for Sla1 phosphorylation.
The sla1Δ/Δ mutant exhibits defects in hyphal growth, actin cytoskeleton organization, and endocytosis
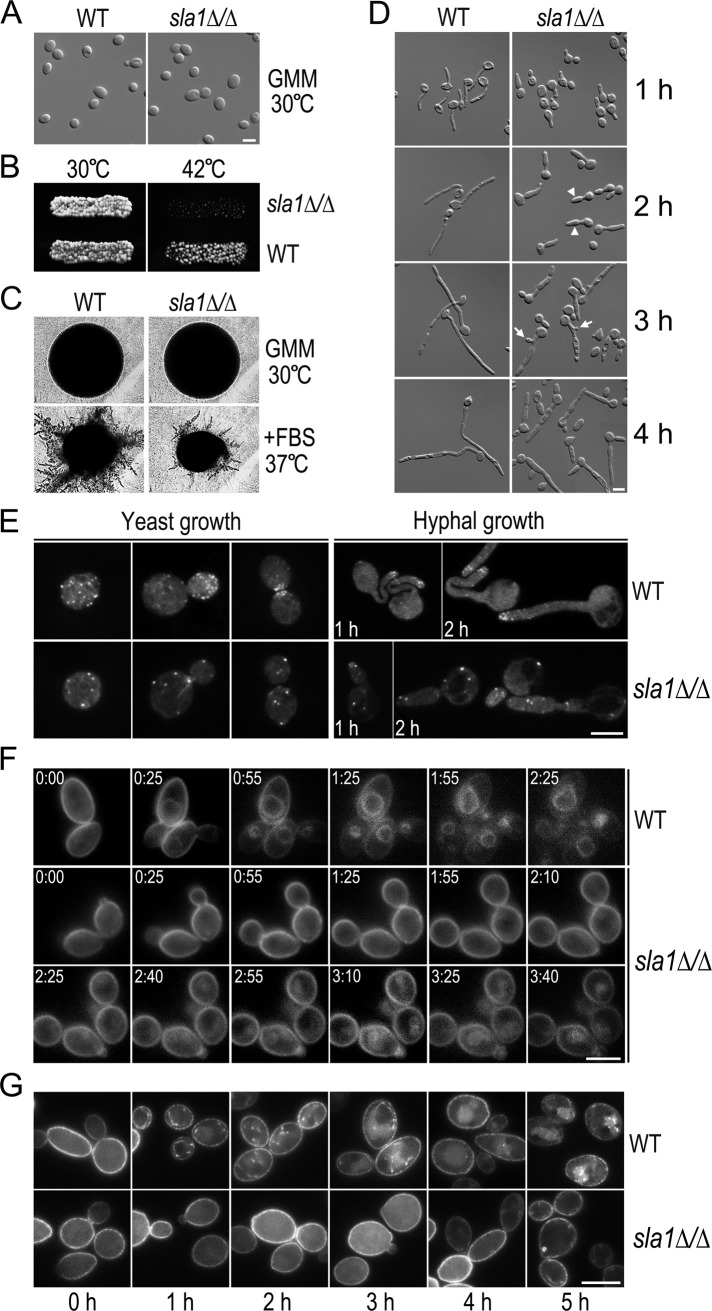
To start to investigate whether and how Sla1 phosphorylation regulates C. albicans morphogenesis, we first generated a sla1Δ/Δ mutant from the BWP17 (WT) strain. The sla1Δ/Δ mutant appeared to grow normally at 30°C in both liquid and solid medium, with morphologies indistinguishable from WT yeast cells (Figure 3, A and B). However, sla1Δ/Δ cells lost viability at 42°C (Figure 3B, right). Next we examined the morphology of sla1Δ/Δ cells under hyphal-inducing conditions. Compared with WT cells, which produced colonies with many long filaments on serum-containing solid medium at 37°C, sla1Δ/Δ cells formed colonies with much fewer and shorter filaments (Figure 3C). Consistent with a previous study (Reijnst et al., 2010), sla1Δ/Δ cells produced short filaments with significant swelling in parts apical to the first septum in liquid media (Figure 3D), in sharp contrast to the long and thin WT hyphae. Moreover, the sla1Δ/Δ mutant often formed a new bud in the middle of an elongated subapical cell. These data suggest that the sla1Δ/Δ mutant is defective in restricting growth to the hyphal tip and in the selection of new growth sites under hyphal growth conditions.
FIGURE 3:
Defects of the sla1∆/∆ mutant. (A) Morphology of sla1Δ/Δ yeast cells. WT (BWP17) and sla1∆/∆ (GZY602) cells were grown in GMM at 30°C. Bar, 5 μm, in this and subsequent figures. (B) Temperature sensitivity of sla1Δ/Δ cells. WT and sla1Δ/Δ cells were grown on a GMM plate at 30°C and replicated to a new GMM plate for incubation at 42°C for 24 h. (C) Defective hyphal growth of sla1Δ/Δ cells on solid medium. WT and sla1∆/∆ cells were streaked on either a GMM plate at 30°C or a GMM plate containing 20% FBS at 37°C. After incubation for 24 h, single colonies were photographed. (D) Defective hyphal growth of sla1Δ/Δ cells in liquid medium. WT and sla1Δ/Δ yeast cells were induced for hyphal growth in YPD containing 20% of FBS at 37°C. Arrowheads and arrows indicate swelling hyphae and buds in subapical cells, respectively. (E) Actin cytoskeleton abnormality in sla1Δ/Δ cells. WT and sla1∆/∆ cells were either grown as yeast in YPD at 30°C or induced for hyphal formation in YPD with 20% of FBS at 37°C for 1–2 h. Cells were stained with rhodamine–phalloidin to visualize the actin cytoskeleton under a fluorescence microscope. (F) Defective FM4-64 uptake in sla1Δ/Δ cells. Fresh WT and sla1Δ/Δ yeast cells were incubated with 20 μM of FM4-64 at 30°C for 10 min before time-lapse microscopic analysis. Representative images taken at the indicated time points (h:min) are shown. (G) Delayed Arn1-GFP internalization in sla1Δ/Δ cells. WT (GZY746) and sla1∆/∆ cells (GZY747) expressing Arn1-GFP were grown in GMM at 30°C and then incubated with 200 μM FC to initiate Arn1-GFP internalization. Samples were taken at 1-h intervals for microscopy.
In S. cerevisiae, Sla1 was first characterized as an actin cytoskeletal protein essential for proper formation of cortical actin patches (Holtzman et al., 1993) and later found to be required for endocytosis (Warren et al., 2002). We found that the green fluorescent protein (GFP)–tagged C. albicans Sla1 localized as cortical spots partially overlapping with mCherry-tagged actin patches in both yeast and hyphae (Supplemental Figure S2), suggesting that Sla1 may regulate polarized growth via its role in actin cytoskeleton organization and endocytosis. To confirm this, we stained sla1Δ/Δ cells with rhodamine–phalloidin to visualize actin filaments. As shown in Figure 3E, sla1Δ/Δ cells had much fewer but slightly larger cortical actin patches than WT cells in both yeast and hyphal cells. Moreover, the actin patches in sla1Δ/Δ cells were apparently depolarized, unlike those in WT cells, which localized to regions of polarized growth such as small buds, bud necks, and hyphal tips (Figure 3E). We also labeled the actin filaments in sla1Δ/Δ cells with GFP-tagged Lifeact, a short peptide that specifically binds filamentous actin in vivo (Riedl et al., 2008), and obtained similar results (Supplemental Figure S3, A and B).
The aberrant actin cytoskeleton suggests that sla1Δ/Δ cells might also be defective in endocytosis. We first examined the fluid-phase endocytosis by Lucifer yellow (LY) uptake assay. Surprisingly, sla1Δ/Δ cells were able to accumulate LY in the vacuoles almost as efficiently as WT cells (Supplemental Figure S3C). Next we stained sla1Δ/Δ cells with FM4-64, a lipophilic dye commonly used to visualize PM uptake by endocytosis (Vida and Emr, 1995), and performed time-lapse microscopy. As shown in Figure 3F, the majority of WT cells (96.2%, n = 26) began to show FM4-64 staining of the vacuolar membrane at 25 min, and the vacuoles were clearly stained at 55 min. In stark contrast, it took ∼2.5 h in most sla1Δ/Δ cells (94.4%, n = 36) for the dye to reach the vacuoles, showing a significant retardation in endocytosis.
Next, to evaluate Sal1's role in receptor-mediated endocytosis, we examined the uptake of Arn1, a transporter of ferrichrome (FC)-type siderophores in C. albicans (Hu et al., 2002). Arn1 localizes to the PM and is internalized as intracellular spots when cells are exposed to FC (Hu et al., 2002). To visualize Arn1, we expressed Arn1-GFP from the MET3 promoter. Figure 3G shows that Arn1-GFP localized to the PM in both WT and sla1Δ/Δ cells in GMM without FC. At 1 h after the addition of FC, intracellular spots of Arn1-GFP were seen beneath the PM in most WT cells (94.9%, n = 39) and then moved away from the PM and appeared in vacuoles at later time points. In contrast, Arn1-GFP internalization in sla1Δ/Δ cells was dramatically delayed, as the intracellular spots could be detected in only 76.7% of mutant cells (n = 43) after 5 h of incubation with FC, indicating that Sla1 is critically required for receptor-mediated endocytosis. Taken together, our data revealed an important role for Sla1 in the polarized localization of cortical actin patches and endocytosis.
The phosphomimetic sla1-13E mutant has similar defects as the sla1Δ/Δ mutant
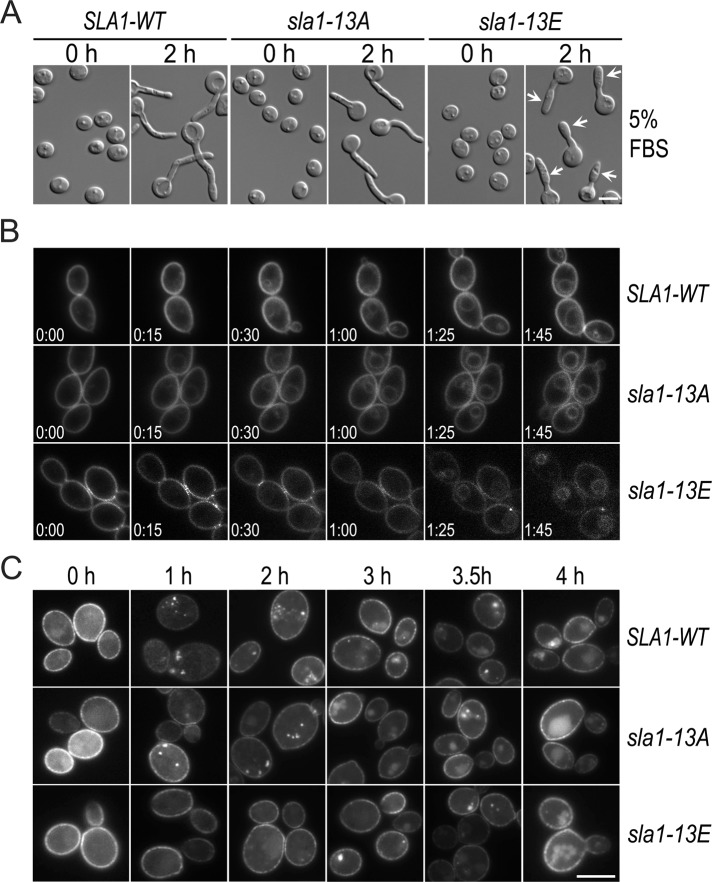
To reveal the physiological role of Cdc28−Cln3 phosphorylation of Sla1, we mutated the Ser/Thr residues in all 13 consensus CDK sites on Sla1 to either Ala (A) or Glu (E) to generate nonphosphorylatable (sla1-13A) and phosphomimetic (sla1-13E) versions of Sla1, respectively, and integrated the mutated genes with a C-terminal HA tag into sla1Δ/Δ cells. As a control, the WT SLA1 gene was also integrated into sla1Δ/Δ cells in the same way. The yeast cells of both sla1-13A and sla1-13E mutants exhibited no apparent morphological abnormality. However, abnormality was observed in the sla1-13E mutant during hyphal morphogenesis, which was particularly obvious at weaker hyphal-inducing conditions such as in media containing 5% fetal bovine serum (FBS): the filaments were significantly shorter, and cells apical to the first septum were wider than WT hyphae, reminiscent of the sla1Δ/Δ mutant (Figure 4A). In comparison, the sla1-13A mutant produced normal hyphae. The results suggest that Sla1 dephosphorylation has a role in maintaining polarized hyphal growth.
FIGURE 4:
Phenotypic characterization of the sla1-13A and sla1-13E mutants. (A) Hyphal morphology of the sla1-13A and sla1-13E cells. Elutriated G1 cells of SLA1-WT (GZY652), sla1-13A (GZY658), and sla1-13E (GZY670) were recovered in GMM at 30°C for 2 h before hyphal induction with 5% of FBS at 37°C. Arrows indicate swollen cells apical to the first septum. (B) FM4-64 uptake in sla1-13A and sla1-13E cells. Cells of GZY652 (SLA1-WT), GZY658 (sla1-13A), and GZY670 (sla1-13E) were grown in YPD overnight and refreshed at 30°C for 2 h, followed by incubation with 20 μM of FM4-64 at 30°C for 10 min before time-lapse microscopy. (C) Arn1-GFP internalization in sla1-13A and sla1-13E cells. SLA1-WT (GZY761), sla1-13A (GZY762), and sla1-13E (GZY763) cells expressing Arn1-GFP were grown in GMM at 30°C and then treated with 200 μM FC to initiate Arn1-GFP internalization.
To examine the effect of the Sla1 mutants on cortical actin patches, we expressed Lifeact-GFP in SLA1-WT, sla1-13A, and sla1-13E cells. During yeast and hyphal growth, both mutants exhibited no significant defects in the number and polarized distribution of the actin patches (unpublished data). However, time-lapse microscopic examination of FM4-64 uptake revealed endocytic defects in sla1-13E cells (Figure 4B). Slightly earlier than SLA1-WT cells, sla1-13A cells began to show FM4-64 staining of the vacuolar membrane at 15 min (78.3%, n = 23), and the majority of the vacuoles (91.3%, n = 23) were clearly stained at 30 min. In contrast, most sla1-13E cells (92.9%, n = 28) took ∼85 min for the dye to reach the vacuoles. We further examined Arn1-GFP internalization upon the addition of FC (Figure 4C). Similar to SLA1-WT cells, sla1-13A cells (91.4%, n = 35) had Arn1-GFP spots beneath the PM at 1 h, which moved close to the cell center at 2 h and appeared in vacuoles at 3 h. In contrast, sla1-13E cells (92.3%, n = 39) began to have faint Arn1-GFP spots only at 2 h, which became brighter at 3 h and finally entered the vacuole at 4 h. These results demonstrated that the sla1-13E, but not the sla1-13A, mutant, is defective in endocytosis.
Taken together, our results show that the phosphomimetic modification of Sla1 at the CDK sites impairs hyphal development and endocytosis without obvious effects on the organization and localization of the actin cytoskeleton.
Sla1 is rapidly dephosphorylated upon hyphal induction
The hyphal defects in sla1-13E cells suggest requirement for Sla1 dephosphorylation during hyphal growth. To test this idea, we compared the gel mobility of Sla1-HA between yeast and hyphal cells. As shown in Figure 5A, Sla1 clearly exhibited a lower level of phosphorylation in hyphal cells induced for 2 h than in yeast cells. Next we examined the kinetics of Sla1 dephosphorylation. After initiating the hyphal induction, we collected aliquots of cells at 20-min intervals for WB analysis and detected Sla1 dephosphorylation at the first time point (Figure 5B). We then shortened the time intervals and found that Sla1 dephosphorylation occurred within the first 5 min of hyphal induction (Figure 5C). The data indicate the existence of a mechanism that rapidly dephosphorylates Sla1 during the yeast-to-hypha transition.
FIGURE 5:
Rapid dephosphorylation of Sla1 upon hyphal induction. (A) Sla1 phosphorylation in yeast and hyphae. WT cells expressing Sla1-HA (GZY584) were either grown as yeast in YPD at 30°C or induced for hyphal growth with 20% FBS at 37°C for 2 h. (B, C) Kinetics of Sla1 dephosphorylation after hyphal induction. GZY584 cells were grown in YPD at 30°C and then incubated with 20% FBS at 37°C. Samples were taken at 20- (B) or 5-min (C) intervals after the addition of FBS. (D) Comparison of Sla1 phosphorylation in WT and prk1Δ/Δ cells. Cells of GZY584 (SLA1-HA) and GZY750 (prk1∆/∆ SLA1-HA) were grown as yeast. (E) Comparison of Sla1 phosphorylation in the prk1∆/∆ yeast and hyphal cells. GZY750 cells were either grown as yeast or induced for hyphal growth for 20 min. (F) Comparison of Sla1-13A and Sla1-13E phosphorylation in yeast and hyphae. Cells of GZY658 (sla1-13A) and GZY670 (sla1-13E) were either grown as yeast or switched to hyphal growth for 20 min. In A–F, Sla1-HA was immunoprecipitated with αHA and subjected to λPP or mock treatment, followed by αHA WB analysis. (G) Defective hyphal growth of sla1-23E cells. Cells of GZY652 (SLA1-WT) and GZY727 (sla1-23E) were grown in GMM at 30°C and then induced for hyphal growth with 10% FBS at 37°C.
In S. cerevisiae, the Sla1 C-terminal region contains multiple L/I/V/MQxTG motifs, which are the recognition sites for the kinase Prk1 (Huang et al., 2003). Sla1 phosphorylation by Prk1 regulates the formation and function of the Pan1/Sla1/End3 complex (Zeng et al., 2001). Similarly, 10 L/I/V/MxxQxTG motifs also exist in the C-terminal region of C. albicans Sla1 (Figure 2A), some of which were found to be phosphorylated on Thr by phospho-site mapping (Supplemental Figure S1). Deletion of the C-terminal region of Sla1 containing all the Prk1 sites caused severe defects in hyphal formation (Supplemental Figure S4), suggesting that the C-terminal phosphorylation sites are present in a functionally important region of the protein. To test whether Sla1 undergoes Prk1-mediated phosphorylation, we immunoprecipitated Sla1-HA from a prk1Δ/Δ mutant and compared its gel mobility with that from WT cells. As shown in Figure 5D, there was a clear, albeit moderate, decrease in the phosphorylation level of Sla1 in the prk1Δ/Δ cells, suggesting that Prk1 also contributes to Sla1 phosphorylation.
To investigate whether Cdc28–Cln3- or Prk1-mediated phosphorylation is rapidly reduced during the yeast-to-hypha transition, we compared the phosphorylation level of Sla1-HA in prk1Δ/Δ cells grown under yeast and hyphal conditions. Figure 5E shows that Sla1-HA was still highly phosphorylated in prk1Δ/Δ cells during yeast growth, and the phosphorylation level exhibited a substantial decrease when cells were induced for hyphal growth, suggesting that the Cdc28–Cln3-mediated phosphorylation was down-regulated. We also observed a slight decrease in the phosphorylation level of Sla1-13A-HA and Sla1-13E-HA, in which all consensus CDK sites were mutated, upon the yeast-to-hypha transition (Figure 5F), suggesting that Sla1 phosphorylation by Prk1 and perhaps other kinases is down-regulated as well.
To reveal the biological importance of Sla1 dephosphorylation in hyphal development, we generated a sla1-23E mutant in which Ser and Thr residues within the 13 CDK sites and 10 Prk1 sites were all mutated into Glu to mimic constitutive phosphorylation and examined its hyphal morphology. The mutant grew like WT cells under yeast growing conditions (Figure 5G). However, when induced for hyphal growth (with 10% FBS), sla1-23E cells exhibited morphological defects, with many cells initially forming an elongated bud with a tapered tip that later grew into filaments with swollen tips and septal constrictions (Figure 5G). The results demonstrate the importance of Sla1 dephosphorylation for normal hyphal development.
Cdc28–Cln3 phosphorylation of Sla1 regulates Prk1 phosphorylation of Sla1 and Sla1 association with Pan1
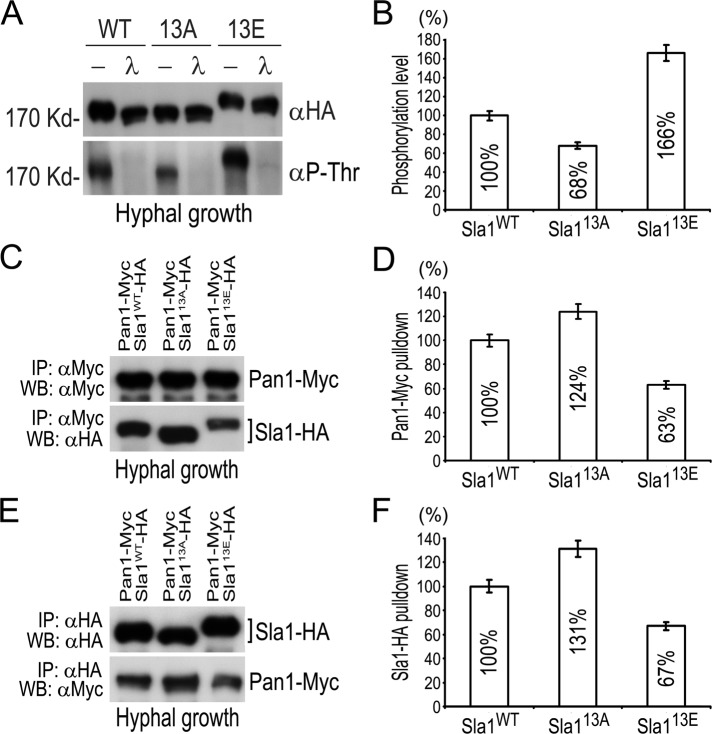
We showed that 12 of 13 consensus CDK sites in Sla1 were phosphorylated in vivo (Supplemental Figure S1). In contrast, although all the 10 Prk1 recognition motifs were covered by the mapping, only five were found to have phosphorylated Thr. To investigate whether the Cdc28–Cln3-mediated phosphorylation might affect Prk1 phosphorylation of Sla1, we immunoprecipitated the same amount of Sla1-13A and Sla1-13E proteins and probed them with an antibody (αP-Thr) specific for phospho-Thr. Because Prk1 only phosphorylates Thr residues (Huang et al., 2003), and all Thr residues in CDK sites had been mutated in Sla1-13A and Sla1-13E, the signal detected by αP-Thr would reflect the Prk1-mediated phosphorylation. Compared with that on Sla1-WT, the Prk1-mediated phosphorylation was decreased to 68% on Sla1-13A but increased to 166% on Sla1-13E during hyphal growth (Figure 6, A and B). Similar changes of Prk1-mediated phosphorylation on Sla1-13A and Sla1-13E were also observed during the yeast growth (unpublished data), suggesting that Cdc28–Cln3 phosphorylation of Sla1 enhances its phosphorylation by Prk1.
FIGURE 6:
Cdc28–Cln3 phosphorylation of Sla1 enhances its phosphorylation by Prk1 and reduces its association with Pan1. (A) Detection of Prk1-mediated phosphorylation of Sla1. Yeast cells of GZY652 (SLA1-WT), GZY658 (sla1-13A), and GZY670 (sla1-13E) were incubated for hyphal growth with 20% FBS at 37°C for 2 h. Sla1 (tagged with HA) was immunoprecipitated with αHA and subjected to λPP or mock treatment. For WB analysis, the membrane was first probed with αP-Thr and then stripped for reprobing with αHA. (B) Comparison of Prk1-mediated phosphorylation on Sla1-13A and Sla1-13E in A. The phosphorylation intensity of Sla1-WT-HA, Sla1-13A-HA, and Sla1-13E-HA (detected by αP-Thr) was measured by a densitometer and normalized against the amount of respective protein (detected by αHA). The results were then calculated as percentages against the normalized value of Sla1-WT-HA. (C) CoIP of Sla1-WT, Sla-13A, and Sla-13E with Pan1. Yeast cells of GZY718 (SLA1-WT-HA PAN1-Myc), GZY719 (sla1-13A-HA PAN1-Myc), and GZY720 (sla1-13E-HA PAN1-Myc) were incubated with 20% FBS at 37°C for 2 h. Cell extracts were prepared and subjected to αMyc IP, followed by immunoblotting with αMyc and αHA. (D) Comparison of the amounts of Sla1-13A and Sla1-13E coimmunoprecipitated by Pan1 in C. The amount of Sla1-WT-HA, Sla1-13A-HA, and Sla1-13E-HA was measured by a densitometer and normalized against the amount of Pan1-Myc used for coIP, respectively. The results were then calculated as percentages against the normalized value of Sla1-WT-HA. (E) CoIP of Pan1 with Sla1-WT, Sla1-13A, and Sla1-13E. Cells of GZY718, GZY719, and GZY720 were incubated with 20% FBS at 37°C for 2 h. Cell extracts were prepared and subjected to αHA IP, followed by WB with αHA and αMyc. (F) Comparison of the amounts of Pan1 coimmunoprecipitated with Sla1-13A-HA and Sla1-13E-HA in E. The amount of Pan1-Myc was measured by a densitometer and normalized against the amount of Sla1-WT-HA, Sla1-13A-HA, and Sla1-13E-HA used for coIP, respectively. The results were then calculated as percentages against the value for Sla1-WT-HA.
In S. cerevisiae, the C-terminal region of Sla1 interacts with Pan1 (Tang et al., 2000), an activator of the Arp2/3 complex (Duncan et al., 2001), and Prk1 phosphorylation of Sla1 negatively regulates their physical association (Zeng et al., 2001). To investigate whether such physical interaction is also regulated by Cdc28–Cln3 phosphorylation of Sla1, we performed coIP between Pan1-Myc and different versions of Sla1-HA. When equal amounts of Pan1-Myc were pulled down, WB analysis detected coIP of more Sla1-13A-HA and less Sla1-13E-HA than Sla1-WT-HA (Figure 6, C and D). Consistently, similar results were obtained in the reciprocal coIP experiment (Figure 6, E and F). Thus the results suggest that Cdc28–Cln3 phosphorylation of Sla1 negatively regulates the physical association between Sla1 and Pan1.
Cdc28–Cln3 phosphorylation of Sla1 regulates cortical actin patch dynamics
In S. cerevisiae, the early endocytic pathway requires coordinated spatiotemporal assembly of multiple factors in three phases: a nonmotile phase in which coat proteins, including Sla1, Sla2, Pan1, and Las17, are assembled into a complex at the PM; a slow-movement phase, in which Abp1 and Arp2/3 complex are recruited and actin polymerization is initiated to provide forces for membrane invagination and vesicle detachment; and a fast-movement phase, in which forces from actin polymerization propel the released endocytic vesicle into cytoplasm (Kaksonen et al., 2003). Because Sla1 is one of the early proteins to be recruited at the beginning of endocytosis to form an endocytic coat with other components, including two Arp2/3 complex activators, Pan1 and Las17 (Winter et al., 1999; Duncan et al., 2001), the absence or mutation of Sla1 would likely affect the coat assembly, disturb subsequent actin polymerization, and impair endocytosis. Thus we decided to investigate the dynamics of cortical actin patches in sla1 mutants during yeast and hyphal growth in C. albicans.
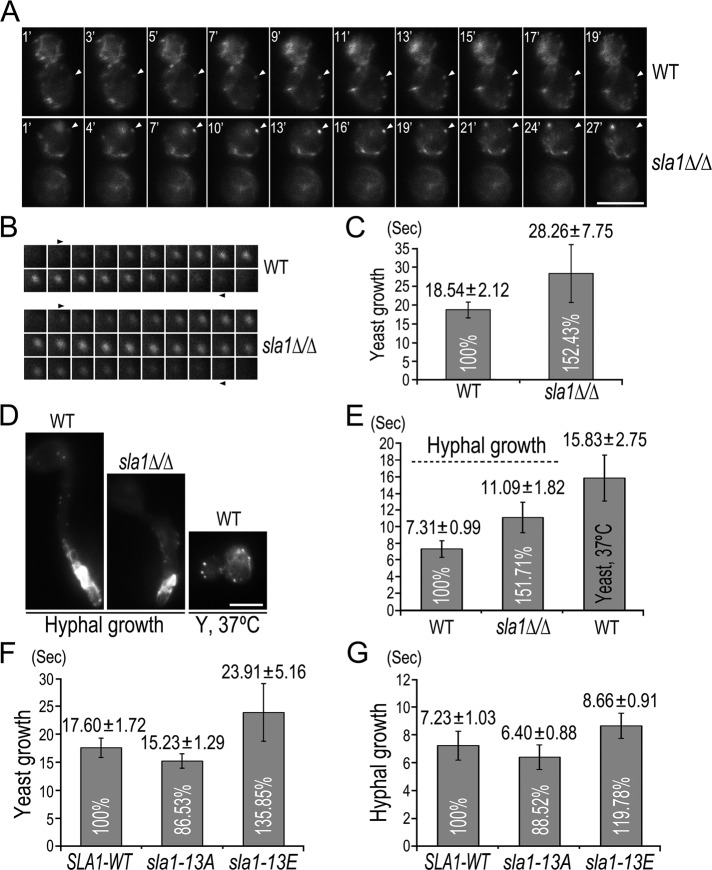
We first compared the actin patch dynamics between WT and the sla1∆/∆ mutant. Cells expressing Lifeact-GFP were subjected to time-lapse fluorescence microscopy. Movies were taken with a frame interval of 1 s for a duration of 2 min, and the time elapsed between the appearance and disappearance of individual patches was used to calculate the average lifespan of actin patches (Figure 7, A and B). In yeast growth, Lifeact-GFP patches showed dynamic movement in WT cells (Supplemental Movie S1), with a patch lifespan of ∼18 s (Figure 7C), whereas their movement in sla1Δ/Δ cells was much less dynamic (Supplemental Movie S2), with a prolonged lifespan of ∼28 s (Figure 7C). When induced for hyphal growth, Lifeact-GFP patches became highly dynamic in both WT and sla1Δ/Δ cells (Figure 7D and Supplemental Movies S3 and S4) and had much shorter lifespans (Figure 7E). Nevertheless, the patch lifespan was ∼50% longer in sla1Δ/Δ than in WT cells (Figure 7E). The data demonstrate that Sla1 is important for the proper dynamics of cortical actin patches during both yeast and hyphal growth.
FIGURE 7:
Regulation of actin patch dynamics by Cdc28–Cln3 phosphorylation of Sla1. (A) Live-cell imaging of Lifeact-GFP in yeast cells of WT (GZY730) and sla1∆/∆ (GZY733). Single frames from movies of GZY730 and GZY733 yeast cells were chosen to show the appearance to disappearance of an individual cortical actin patch (indicated by arrowheads). See Supplementary Movies S1 and S2. (B) Actin patch dynamics in WT and sla1∆/∆ yeast cells. Montages of a representative Lifeact-GFP patch were used to demonstrate the dynamics of cortical actin patches in WT and sla1∆/∆ yeast cells. (C) Lifespan of actin patches in WT and sla1∆/∆ yeast cells. Lifeact-GFP patches (n ≥ 35) in GZY730 and GZY733 yeast cells were visually examined for the time elapsed between patch appearance and disappearance, and the lifespan was calculated as the average time ± SD. (D, E) Live-cell imaging (D) and patch lifespan (E) of Lifeact-GFP in WT and sla1∆/∆ hyphal cells and in WT yeast cells grown at 37°C. Single frames from movies of GZY730 and GZY733 hyphal cells and GZY730 yeast cells grown at 37°C are shown in D. (F, G) Lifespan of actin patches in SLA1-WT, sla1-13A, and sla1-13E cells during yeast (F) and hyphal (G) growth. SLA1-WT (GZY739), sla1-13A (GZY740), and sla1-13E (GZY741) cells expressing Lifeact-GFP were grown as yeast or hyphae for live-cell imaging and patch lifespan calculation. The differences in patch lifetimes between each mutant and the wild type were statistically significant (p < 0.01).
We noted that cortical actin patches were generally more dynamic, with shorter lifespans in hyphal than in yeast cells. The patch lifespan of Lifeact-GFP was shortened from ∼18 to ∼7 s when WT cells switched from yeast to hyphal growth (Figure 7, C and E). To exclude the possibility that such change was caused solely by the increase of temperature, we grew WT cells at 37°C in the absence of FBS and found that the increase of temperature alone had a rather small effect on the dynamics of cortical actin patches, reducing the patch lifespan by only ∼3 s in comparison with that measured at 30°C (Figure 7E). The data suggest that hyphal growth may require accelerated actin patch turnover.
Next we examined the behaviors of Lifeact-GFP patches in SLA1-WT, sla1-13A, and sla1-13E cells. As expected, the SLA1-WT cells exhibited an almost identical pattern of patch dynamics and turnover as WT cells during both yeast and hyphal growth (Figure 7, F and G). However, there was a ∼13% decrease in the lifespan of Lifeact-GFP patches in both yeast and hyphal cells in the sla1-13A mutant (Figure 7, F and G). In contrast, the patch lifespan of Lifeact-GFP increased by ∼35% in sla1-13E yeast cells and ∼20% in sla1-13E hyphal cells (Figure 7, F and G). These data suggest that Cdc28–Cln3 phosphorylation of Sla1 negatively regulates actin patch dynamics.
DISCUSSION
The hyphal development of fungi is an extreme form of polarized growth that requires the polarity machinery to be maintained persistently at the growing tip. The CDK Cdc28 plays a central role in regulating C. albicans hyphal growth by phosphorylating several key components of the polarity machinery (Zheng et al., 2007; Bishop et al., 2010). In this study, we identified a new Cdc28 substrate, Sla1, which is a crucial part of the endocytic machine, establishing a new molecular link between Cdc28 and another fundamental cellular process critically involved in the maintenance of cell polarity.
Sla1 is a substrate of Cdc28–Cln3
In S. cerevisiae, Sla1 has been identified as a potential Cdc28 substrate in vivo (Holt et al., 2009). Our studies in C. albicans provide strong evidence demonstrating that Cdc28 phosphorylates Sla1. First, Sla1 physically associates with Cln3, as shown by in vitro affinity binding and coIP experiments. Second, Sla1 contains 13 consensus CDK sites, and 12 were found to be phosphorylated in vivo by MS. Third, Sla1 phosphorylation largely depends on Cln3, as Cln3 depletion greatly reduced Sla1 phosphorylation. Finally, purified Sla1 could be specifically phosphorylated by Cln3-associated Cdc28 in vitro. Taking these data together, we conclude that Cdc28–Cln3 phosphorylates Sla1. Several endocytic proteins have been reported as CDK substrates in other organisms. Amphiphysin 1 in rat brain participates in synaptic vesicle endocytosis and is phosphorylated by both Cdk5 and Cdc2 kinases (Floyd et al., 2001). The yeast amphiphysin homologue Rvs167 binds to the cyclin Pcl2 and is phosphorylated by the CDK Pho85 (Lee et al., 1998). Epsin, which has a role in clathrin-mediated endocytosis, is phosphorylated by the Cdc2 kinase (Chen et al., 1999). These data indicate that phosphoregulation of key components of the endocytic machine by CDKs is a common mechanism in the control of endocytosis.
The function of Sla1 in actin organization and endocytosis
Similar to its S. cerevisiae homologue, C. albicans Sla1 localizes as cortical spots that partially overlap with actin patches, consistent with a role for Sla1 in actin regulation. Abnormalities were indeed observed in sla1Δ/Δ cells, which contained much fewer and depolarized cortical actin patches compared with WT cells. Furthermore, the dynamics of cortical actin patches was significantly reduced and the patch lifespan increased in sla1∆/∆ cells. These data demonstrate that Sla1 is crucial for the proper polarization and dynamics of cortical actin patches.
Studies in S. cerevisiae have established cortical actin patches as the sites of endocytosis (Kaksonen et al., 2003), where Sla1 plays a key role through direct interaction with both actin-binding and other endocytic proteins (Tang et al., 2000; Warren et al., 2002; Gourlay et al., 2003; Rodal et al., 2003). Consistently, sla1 mutants also exhibit endocytosis defects in C. albicans. We demonstrated that Sla1 is required for efficient internalization of the membrane dye FM4-64 and the siderophore transporter Arn1. Of interest, the fluid-phase endocytosis in sla1Δ/Δ cells appeared normal, as shown by the LY uptake assay. This is in contrast to situations in S. cerevisiae in which defective fluid-phase endocytosis is always observed when other types of endocytosis are impaired. In line with our findings, it has been reported recently that the arp2Δ/Δ and arp2Δ/Δ arp3Δ/Δ mutants of C. albicans are defective in FM4-64 internalization but normal in LY uptake (Epp et al., 2010).
The role of Cdc28 phosphorylation of Sla1 in polarized growth
A key finding of this study is Sla1 hypophosphorylation in hyphal growth, suggesting a role for Sla1 phosphorylation in controlling polarized growth. Indeed, the sla1-13E mutant that mimics constitutive phosphorylation at the CDK sites in Sla1 exhibited significant hyphal defects, whereas the nonphosphorylatable sla1-13A mutant grew normal hyphae. Like the sla1Δ/Δ mutant, neither one of the CDK-site mutants showed apparent morphological defects during the yeast growth. Strikingly, our live-cell imaging analysis revealed that actin patches were much more dynamic in hyphae than in yeast and that SLA1 deletion markedly reduced the patch dynamics, suggesting that Cdc28 phosphorylation of Sla1 might influence polarized growth by regulating the actin patch turnover. In support of this idea, we observed a significant decrease in the patch dynamics in sla1-13E cells, with the patch lifespan extended by 35% in hyphae and 20% in yeast in comparison with WT cells. In contrast, the patch dynamics was accelerated in the sla1-13A mutant, with the patch lifespan shortened by ∼13% in both yeast and hyphae. Consistent with the reduced patch dynamics, the sla1-13E yeast cells exhibited a severe retardation of endocytosis. In comparison, the sla1-13A yeast cells were normal in endocytosis. Given the fact that hyphal cells of sla1-13E exhibited a similar defect on patch behaviors as yeast cells, it is highly possible that the sla1-13E mutant also had impaired endocytosis under hyphal growth conditions. Taking these results together, we propose that Cdc28 phosphorylation of Sla1 regulates actin-mediated endocytosis in different growth modes of C. albicans in addition to other cellular processes discovered previously (Sinha et al., 2007; Zheng et al., 2007; Wang et al., 2009; Bishop et al., 2010).
How does Cdc28 negatively regulate endocytosis through Sla1 phosphorylation? The mechanism seems to intimately involve the actin-regulatory kinase Prk1 and its regulation of Sla1's interaction with proteins in the actin–endocytic complex (Cope et al., 1999; Zeng and Cai, 1999). In S. cerevisiae, Prk1 phosphorylates several endocytic coat proteins, including Sla1, Pan1, and Scd5, to facilitate disassembly of the coat complex (Zeng et al., 2007). Deletion of Prk1 causes abnormal assembly of the actin cytoskeleton and failure of endocytosis (Cope et al., 1999; Zeng and Cai, 1999). Furthermore, the coat proteins are proposed to be dephosphorylated by the type 1 phosphatase Glc7, and probably other phosphatases, before they can be reused for the next round of endocytosis (Zeng et al., 2007). Thus, timely and proper phosphorylation and dephosphorylation of some endocytic complex proteins is crucial for the efficiency and rate of endocytosis. C. albicans Sla1 also exhibits Prk1-dependent phosphorylation. We showed that deletion of the Prk1 phosphorylation region in Sla1 severely affected the hyphal morphology. Furthermore, Cdc28 phosphorylation of Sla1 enhanced Prk1 phosphorylation of Sla1 and, of importance, reduced Sla1's association with Pan1. However, it remains unclear how the CDK-enhanced Prk1 phosphorylation of Sla1 influences the endocytic process. Given that we observed correlations of Sla1 hyperphosphorylation with reduced actin patch dynamics and slower endocytosis, one possibility is that Sla1 phosphorylation might prevent its recruitment into the endocytic coat and thus delay the initiation of endocytosis.
Actin polymerization drives invagination of the PM and release/movement of endocytic vesicles (Kaksonen et al., 2003). Las17, the major activator of the actin-nucleating Arp2/3 complex (Winter et al., 1999), is recruited to endocytic sites as early as other coat proteins (Kaksonen et al., 2003). It was proposed that Sla1 interacts with Las17 (Rodal et al., 2003) and keeps it inactive so that actin assembly will not occur until the coat assembly is complete (Sun et al., 2006). Our observation of a prolonged lifespan of cortical actin patches in the sla1-13E mutant suggests that Cdc28 phosphorylation of Sla1 may affect actin assembly. It is possible that the association between Sla1 and Las17 is inhibited by Cdc28 and Prk1 phosphorylation of Sla1, resulting in earlier initiation of actin nucleation and thus an extended lifespan of actin patches. It is also possible that the Sla1 phosphorylation level affects the nature of the actin filaments assembled by the Arp2/3 complex, such as filaments with different dynamics or stability, which may determine actin patch dynamics and endocytosis rates (Ayscough, 2005).
Differential regulation of endocytosis in different growth modes of C. albicans
This study revealed differential regulation of the cortical actin patch dynamics between the yeast and hyphal growth in C. albicans. Of importance, we found significantly faster actin patch turnover in hyphae than in yeast cells, suggesting accelerated endocytosis during hyphal growth. Our data are consistent with a critical role for endocytosis in polarized growth proposed in recent reports (Steinberg, 2007a, b; Upadhyay and Shaw, 2008; Shaw et al., 2011). In polarized growth, such as hyphal tip growth and apical bud extension in yeasts, polarity molecules localize to a small area on the cell surface to direct localized growth. To maintain the polarity, the cell must have a mechanism that prevents the polarity molecules from diffusing away and accumulating to high concentrations in areas outside the established growth zone (Momany, 2002). Endocytosis is believed to play this role because actin patches, as the sites of endocytosis, are always found to concentrate around the growth sites in fungi and yeasts (Marco et al., 2007; Steinberg, 2007b; Upadhyay and Shaw, 2008). Recently high-resolution imaging analyses revealed actin patches forming a collar encircling the region closely behind the growing apex during the hyphal development of A. nidulans (Araujo-Bazan et al., 2008; Taheri-Talesh et al., 2008; Upadhyay and Shaw, 2008) and N. crassa (Delgado-Alvarez et al., 2010; Berepiki et al., 2011). This actin collar is believed to carry out endocytic removal of polarity molecules escaping the growth cap, thus restricting growth to the hyphal tip (Shaw et al., 2011). Here we propose that maintaining continuous hyphal growth requires not only polarizing actin patches to the subapical region but also accelerating endocytosis. This hypothesis is consistent with the increase in actin patch dynamics when C. albicans switches from yeast to hyphal growth and the retardation of actin patch turnover and endocytosis as a result of SLA1 deletion or phosphomimetic mutation of CDK sites of Sla1, which causes partial loss of polarized growth.
In summary, we have discovered a concrete molecular link between CDK and the endocytic machinery in the polymorphic fungus C. albicans and revealed a novel mechanism by which CDK regulates actin-mediated endocytosis in different growth modes via differential phosphorylation of Sla1.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions
Yeast strains and plasmids used in this study are listed in Supplemental Tables S1 and S2, respectively. Recombinant DNA manipulations were performed according to standard methods. Site-directed mutagenesis followed the manual of the QuikChange Multi Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA). Transformation of C. albicans was done following the protocol of the Fast Yeast Transformation Kit (Geno Technology, Maryland Heights, MO). Yeast cells were routinely grown at 30°C in 2% yeast extract, 1% peptone, and 2% glucose (YPD) or galactose (YPG) or glucose minimal medium (GMM; 6.79 g/l yeast nitrogen base without amino acids and 2% glucose) supplemented with appropriate amino acids (uridine 80 μg/ml, arginine 40 μg/ml, and histidine 40 μg/ml) for auxotrophic mutants. G1 cells were prepared by centrifugal elutriation using a J-26 XP elutriator (Beckman, Fullerton, CA). To shut down the MET3 promoter, both methionine and cysteine were added to the culture at a final concentration of 0.5 mM. For hyphal induction, yeast culture was 1:10 diluted into fresh medium containing 5–20% of FBS (HyClone Laboratories, Logan, UT) and incubated at 37°C.
Affinity purification of GST-Cln3 bound proteins
To express GST-Cln3 fusion protein, Escherichia coli BL21 was transformed with the plasmid pYGS837 and grown in 800 ml of LB medium containing 100 μg/ml ampicillin to OD600 of 0.5, followed by incubation with 1 mM isopropylthio-β-d-galactoside (IPTG) at 37°C for 4 h. Cells were collected by centrifugation and resuspended into 40 ml of cold phosphate-buffered saline (PBS). The suspension was sonicated on ice to break the cells, and the lysate was centrifuged at 15,000 rpm for 15 min. The supernatant was incubated at 4°C for 1 h with a 1-ml slurry of glutathione–Sepharose 4B beads (Amersham-Pharmacia Biotech, GE Healthcare Bio-Sciences, Piscataway, NJ) prewashed with cold PBS three times. The beads were then passed through a disposable column. The detained GST-Cln3–bound beads were washed with 25 ml of cold PBS three times and kept in 1 ml of PBS in the column at 4°C for further use. To perform affinity purification, 25 ml of cell lysate was prepared from 500 ml of YPD culture of BWP17 yeast cells grown at 30°C overnight and cleared of cell debris by centrifugation. The supernatant was precleaned by incubating with a 1-ml slurry of glutathione–Sepharose 4B beads at 4°C for 1 h and passing through a disposable column. The cleaned extract was collected and transferred to the column containing GST-Cln3–bound beads for further incubation at 4°C for 1 h. After washing with 25 ml of cold lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM KCl, 1% NP-40) five times, the GST-Cln3–bound beads were incubated with 1 ml KCl (0.15 M) at room temperature for 5 min to elute the bound proteins. The eluted proteins were allowed to flow through the column by gravity and collected. The elution was repeated with increasing concentrations of KCl (0.3, 0.45, 0.6, 0.75, 0.9, 1.05, 1.2, 1.35, and 1.5 M). Proteins from each elution were precipitated by incubating with 10% trichloroacetic acid for 10 min on ice, redissolved in 1× protein loading buffer (50 mM Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 1% β-mercaptoethanol, 12.5 mM EDTA, 0.02% bromophenol blue), electrophoresed on 12% PAGE gel, and visualized by Coomassie brilliant blue (CBB) staining.
Mass spectrometry analysis
MS analysis was performed by Center for Functional Genomics, University at Albany, State University of New York. The excised gel pieces containing protein bands were washed, reduced, alkylated, and in-gel tryptic digested. Proteolytic peptides were extracted from the gel. Peptide mixture was concentrated and reconstituted in 5% formic acid for liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis. Phosphopeptides were enriched using TiO2, and both fractions were analyzed by LC-MS/MS. PKL file was created using the MassLynx 3.5 software from Waters (Milford, MA) with a processing macro that smoothes, centroids, and assesses the quality of data. In-house MASCOT 2.2 from Matrix Science (London, United Kingdom) was used to assist in the interpretation of tandem mass spectra against other fungus subdatabases of the nonredundant databases (http://pubmlst.org/analysis). Phosphopeptide mapping was searched against targeted protein sequences.
Protein extraction, immunoprecipitation, and Western blotting
To prepare cell lysates, cells were harvested into 2-ml screw-cap microcentrifuge tubes by brief centrifugation to obtain pellets with a volume of ≤500 μl and resuspended in 500 μl of ice-cold lysis buffer (50 mM Tris-HCI, pH7.4, 150 mM KCI, 1% NP-40) containing protease inhibitor cocktail (Nacalai Tesque, Kyoto, Japan) and phosphatase inhibitor cocktail B (Santa Cruz Biotechnology, Santa Cruz, CA). After addition of an equal volume of acid-washed glass beads (Sigma-Aldrich, St. Louis, MO), cells were broken by six rounds of 60-s beating at 5000 rpm in a MicroSmash MS-100 beater (TOMY Medico, Tokyo, Japan) with 1 min of cooling on ice between rounds. The lysed cells were then centrifuged at 16,000 rpm for 15 min at 4°C, and supernatants were collected.
To perform IP, cell lysate was incubated with a 30-μl slurry of rabbit polyclonal HA or Myc beads (Santa Cruz Biotechnology) at 4°C for 1 h. After brief centrifugation at 8000 rpm, the beads were washed by resuspension in 800 μl of cold lysis buffer and subjected to another brief centrifugation. The washed beads were resuspended in 10 μl of 1× protein loading buffer and boiled for 6 min. For λPP treatment, the washed beads were incubated with 2 μl of 10× buffer, 2 μl 10 mM MnCI2, and 1 μl λPP (New England BioLabs, Hitchin, United Kingdom) at 30°C for 20 min before adding 1× protein loading buffer. Protein samples were separated by SDS–PAGE and subsequently transferred to a polyvinylidene fluoride membrane (Bio-Rad Laboratories, Hercules, CA).
For WB analysis, the membrane was first incubated with blocking solution (5% bovine serum albumin) at room temperature for 1 h or at 4°C overnight. After a brief rinse with PBS containing 0.1% (vol/vol) Tween-20 (PBST), the membrane was incubated with PBST containing a 1:1000 diluted primary antibody (mouse monoclonal HA or Myc antibody; Santa Cruz Biotechnology) at room temperature for 1 h, followed by three rounds of 5-min wash with PBST. The membrane was then incubated with PBST containing a 1:2000 diluted secondary antibody (horseradish peroxidase–linked anti–mouse immunoglobulin G from sheep; GE Healthcare UK, Chalfont St. Giles, United Kingdom). After three rounds of 5-min wash with PBST, the membrane was immersed in Pierce ECL WB substrate solution (Thermo Scientific, Rockford, IL) and exposed to X-film (Fujifilm, Tokyo, Japan).
Purification of HA-Sla1 and in vitro kinase assay
To purify HA-Sla1, 50 ml of cell lysate was prepared from 1 l of YPG culture of GZY631 yeast cells (grown at 30°C overnight) and centrifuged at 15,000 rpm for 15 min. The supernatant was collected and incubated with a 50-μl slurry of EZview red anti-HA affinity gel (Sigma-Aldrich) at 4°C overnight. The HA-Sla1–bound gel was washed five times with 1 ml of lysis buffer by resuspension in the buffer, followed by brief centrifugation at 8000 rpm. For phospho-site mapping, the washed gel was boiled in 50 μl of 1× protein loading buffer for 6 min. The denatured proteins were separated with 6% SDS–PAGE. After staining with CBB, the HA-Sla1 band was excised for MS analysis. For in vitro kinase assay, the washed gel was incubated with 200 μl (1 mg/ml in PBS) of freshly prepared HA peptide solution (Sigma-Aldrich) for 10 min on ice. After brief centrifugation at 8000 rpm, the supernatant was recovered, and the concentration of eluted HA-Sla1 was determined by a NanoDrop 1000 Spectrophotometer (Thermo Scientific).
To perform the in vitro kinase assay, aliquots of cell lysate prepared from YPD culture of GZY641 cells were incubated with rabbit polyclonal Myc beads (Santa Cruz Biotechnology) to precipitate the Cln3-Myc−Cdc28as kinase. The cell lysate prepared from IS89 cells was also used as a negative control. After washing twice with the lysis buffer, the beads were resuspended in 10 μl of 3× kinase buffer (150 mM Tris-HCl, pH 7.5, 30 mM MgCl2, 3 mM dithiothreitol, 3 mM ethylene glycol tetraacetic acid, 0.3% Tween-20, and 3 mM glycerophosphate) with or without 25 mM 1MN-PP1. Subsequently, 2 μl of 1 mM ATP, 2 μl of γ-32P-ATP (6000 Ci/mmol; PerkinElmer Life Sciences, Waltham, MA), 5 μg of the eluted HA-Sla1, and H2O were added to a final volume of 30 μl. The reaction was performed at 37°C for 30 min and terminated by adding 6 μl of 6× protein loading buffer and boiling for 6 min. After electrophoresis, the gel was stained with CBB to visualize the protein band of HA-Sla1 and then dried onto a filter paper for autoradiography.
Visualization of actin cytoskeleton and endocytosis assays
To stain actin filaments with rhodamine–phalloidin (200 U/ml; Molecular Probes, Invitrogen, Eugene, OR), yeast and hyphal cells were collected, suspended in PK buffer (100 mM KH2PO4 and 100 mM K2HPO4, pH 6.4), and fixed with 3.7% formaldehyde at room temperature for 1 h. After one wash with PK buffer, cells were incubated in the same buffer containing 0.1% Triton X-100 at room temperature for 30 min. Cells were then washed twice with PBS and incubated with rhodamine–phalloidin (1:50 dilution in PBS) overnight at 4°C. Cells were finally washed with PBS five times and resuspended in PBS for visualization with a DMRXA2 microscope (Leica, Wetzlar, Germany) equipped with a CoolSnap HQ2 digital camera (Roper Scientific, Tucson, AZ). All images were acquired using MetaMorph 7.5 software (MDS Analytical Technologies, Downington, PA). To directly visualize actin cytoskeleton, we harvested yeast or hyphal cells expressing Lifeact-GFP and resuspended them in Stopmix (1 mM NaN3, 50 mM NaF, 10 mM EDTA, 0.9% NaCl) for fixation before microscopy.
For LY uptake assay, cells grown in GMM at 30°C were harvested, resuspended in GMM containing 5 mg/ml LY CH dilithium (Sigma-Aldrich), and incubated at 30°C for 20, 40, and 60 min, respectively. Cells were then washed with PBS five times and resuspended in PBS for microscopy. To follow the internalization of Arn1-GFP, we grew cells in 1 ml GMM at 30°C to log phase. Then we added 2 μl of 100 mM ferrichrome (FC; Sigma-Aldrich) to initiate the internalization of Arn1-GFP. Samples were taken at 1-h intervals, and cells were collected and resuspended in Stopmix for microscopy.
FM4-64 uptake and live-cell imaging
To monitor the uptake of FM4-64 (Molecular Probes), overnight YPD cultures of yeast cells were inoculated (1:10) into fresh YPD medium and grown at 30°C for 2 h. Cells were harvested, resuspended in GMM, and incubated with 20 μM of FM4-64 for 10 min at 30°C. Cells were then adhered to the surface of a glass slide precoated with 2% agarose, covered with a coverslip, and sealed with Vaseline. The slide was kept inside a chamber (temperature constant at 30°C) during time-lapse imaging under an Axiovert 200M microscope (Zeiss, Jena, German) equipped with a CoolSnap HQ2 digital camera and controlled by MetaMorph 7.0 software. Images were acquired at 1 frame/5 min for 4 h, with an exposure time of 300 ms for each frame.
For live-cell imaging, yeast cells expressing Lifeact-GFP were grown in GMM at 30°C overnight. Cells were harvested, resuspended in fresh GMM or GMM containing 20% FBS, and adhered to glass slides precoated with 2% agarose. Each slide was covered with a coverslip and sealed with Vaseline. The slides were either kept at 30°C to maintain yeast growth or incubated at 37°C for 2 h for hyphal development and subjected to fluorescence imaging using the Zeiss Axiovert 200M microscope. Images were acquired continuously for 2 min at 1 frame/s with an exposure time of 100 ms for each frame and made into a movie. To determine the patch lifetime of Lifeact-GFP, we visually analyzed 35 patches from different cells of each strain for the time elapsed between patch appearance and disappearance and calculated the patch lifetime as the average time ± SD.
Supplementary Material
Acknowledgments
We thank Judith Berman for providing the mCherry plasmid and Peter Sudbury for the Lifeact-GFP plasmid. We also thank members of the Wang lab for critical reading of the manuscript. This work was funded by the Agency for Sciences, Technology and Research of Singapore.
Abbreviations used:
- CBB
Coomassie brilliant blue
- CDK
cyclin-dependent kinase
- coIP
coimmunoprecipitation
- FBS
fetal bovine serum
- FC
ferrichrome
- GFP
green fluorescent protein
- GMM
glucose minimal medium
- GST
glutathione S-transferase
- HA
hemagglutinin
- IP
immunoprecipitation
- IPTG
isopropylthio-β-d-galactoside
- LY
lucifer yellow
- MC
methionine and cysteine
- MS
mass spectrometry
- PBS
phosphate-buffered saline
- PM
plasma membrane
- λPP
λ phosphatase
- SD
standard deviation
- WB
Western blot
- WT
wild type
- YPD
yeast extract, peptone, and dextrose
- YPG
yeast extract, peptone, and galactose
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-03-0231) on July 11, 2012.
REFERENCES
- Araujo-Bazan L, Penalva MA, Espeso EA. Preferential localization of the endocytic internalization machinery to hyphal tips underlies polarization of the actin cytoskeleton in Aspergillus nidulans. Mol Microbiol. 2008;67:891–905. doi: 10.1111/j.1365-2958.2007.06102.x. [DOI] [PubMed] [Google Scholar]
- Archambault V, Chang EJ, Drapkin BJ, Cross FR, Chait BT, Rout MP. Targeted proteomic study of the cyclin-Cdk module. Mol Cell. 2004;14:699–711. doi: 10.1016/j.molcel.2004.05.025. [DOI] [PubMed] [Google Scholar]
- Ayscough KR. Defining protein modules for endocytosis. Cell. 2005;123:188–190. doi: 10.1016/j.cell.2005.10.008. [DOI] [PubMed] [Google Scholar]
- Bachewich C, Whiteway M. Cyclin Cln3p links G1 progression to hyphal and pseudohyphal development in Candida albicans. Eukaryot Cell. 2005;4:95–102. doi: 10.1128/EC.4.1.95-102.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berepiki A, Lichius A, Read ND. Actin organization and dynamics in filamentous fungi. Nat Rev Microbiol. 2011;9:876–887. doi: 10.1038/nrmicro2666. [DOI] [PubMed] [Google Scholar]
- Bishop A, Lane R, Beniston R, Chapa-y-Lazo B, Smythe C, Sudbery P. Hyphal growth in Candida albicans requires the phosphorylation of Sec2 by the Cdc28-Ccn1/Hgc1 kinase. EMBO J. 2010;29:2930–2942. doi: 10.1038/emboj.2010.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop AC, et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- Cai H, Reinisch K, Ferro-Novick S. Coats, tethers, Rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Dev Cell. 2007;12:671–682. doi: 10.1016/j.devcel.2007.04.005. [DOI] [PubMed] [Google Scholar]
- Chapa B, Bates S, Sudbery P. The G1 cyclin Cln3 regulates morphogenesis in Candida albicans. Eukaryot Cell. 2005;4:90–94. doi: 10.1128/EC.4.1.90-94.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Slepnev VI, Di Fiore PP, De Camilli P. The interaction of epsin and Eps15 with the clathrin adaptor AP-2 is inhibited by mitotic phosphorylation and enhanced by stimulation-dependent dephosphorylation in nerve terminals. J Biol Chem. 1999;274:3257–3260. doi: 10.1074/jbc.274.6.3257. [DOI] [PubMed] [Google Scholar]
- Cope MJ, Yang S, Shang C, Drubin DG. Novel protein kinases Ark1p and Prk1p associate with and regulate the cortical actin cytoskeleton in budding yeast. J Cell Biol. 1999;144:1203–1218. doi: 10.1083/jcb.144.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado-Alvarez DL, Callejas-Negrete OA, Gomez N, Freitag M, Roberson RW, Smith LG, Mourino-Perez RR. Visualization of F-actin localization and dynamics with live cell markers in Neurospora crassa. Fungal Genet Biol. 2010;47:573–586. doi: 10.1016/j.fgb.2010.03.004. [DOI] [PubMed] [Google Scholar]
- Dettmer J, Friml J. Cell polarity in plants: when two do the same, it is not the same. Curr Opin Cell Biol. 2011;23:686–696. doi: 10.1016/j.ceb.2011.09.006. [DOI] [PubMed] [Google Scholar]
- Di Pietro SM, Cascio D, Feliciano D, Bowie JU, Payne GS. Regulation of clathrin adaptor function in endocytosis: novel role for the SAM domain. EMBO J. 2010;29:1033–1044. doi: 10.1038/emboj.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan MC, Cope MJ, Goode BL, Wendland B, Drubin DG. Yeast Eps15-like endocytic protein, Pan1p, activates the Arp2/3 complex. Nat Cell Biol. 2001;3:687–690. doi: 10.1038/35083087. [DOI] [PubMed] [Google Scholar]
- Epp E, Walther A, Lepine G, Leon Z, Mullick A, Raymond M, Wendland J, Whiteway M. Forward genetics in Candida albicans that reveals the Arp2/3 complex is required for hyphal formation, but not endocytosis. Mol Microbiol. 2010;75:1182–1198. doi: 10.1111/j.1365-2958.2009.07038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd SR, Porro EB, Slepnev VI, Ochoa GC, Tsai LH, De Camilli P. Amphiphysin 1 binds the cyclin-dependent kinase (cdk) 5 regulatory subunit p35 and is phosphorylated by cdk5 and cdc2. J Biol Chem. 2001;276:8104–8110. doi: 10.1074/jbc.M008932200. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Novo A, Correa-Bordes J, Labrador L, Sanchez M, Vazquez de Aldana CR, Jimenez J. Sep7 is essential to modify septin ring dynamics and inhibit cell separation during Candida albicans hyphal growth. Mol Biol Cell. 2008;19:1509–1518. doi: 10.1091/mbc.E07-09-0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourlay CW, Dewar H, Warren DT, Costa R, Satish N, Ayscough KR. An interaction between Sla1p and Sla2p plays a role in regulating actin dynamics and endocytosis in budding yeast. J Cell Sci. 2003;116:2551–2564. doi: 10.1242/jcs.00454. [DOI] [PubMed] [Google Scholar]
- Hazan I, Sepulveda-Becerra M, Liu H. Hyphal elongation is regulated independently of cell cycle in Candida albicans. Mol Biol Cell. 2002;13:134–145. doi: 10.1091/mbc.01-03-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzog M, Chavrier P. Cell polarity during motile processes: keeping on track with the exocyst complex. Biochem J. 2011;433:403–409. doi: 10.1042/BJ20101214. [DOI] [PubMed] [Google Scholar]
- Holt LJ, Tuch BB, Villen J, Johnson AD, Gygi SP, Morgan DO. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science. 2009;325:1682–1686. doi: 10.1126/science.1172867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzman DA, Yang S, Drubin DG. Synthetic-lethal interactions identify two novel genes, SLA1 and SLA2, that control membrane cytoskeleton assembly in Saccharomyces cerevisiae. J Cell Biol. 1993;122:635–644. doi: 10.1083/jcb.122.3.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu CJ, Bai C, Zheng XD, Wang YM, Wang Y. Characterization and functional analysis of the siderophore-iron transporter CaArn1p in Candida albicans. J Biol Chem. 2002;277:30598–30605. doi: 10.1074/jbc.M204545200. [DOI] [PubMed] [Google Scholar]
- Huang B, Zeng G, Ng AY, Cai M. Identification of novel recognition motifs and regulatory targets for the yeast actin-regulating kinase Prk1p. Mol Biol Cell. 2003;14:4871–4884. doi: 10.1091/mbc.E03-06-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaksonen M, Sun Y, Drubin DG. A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell. 2003;115:475–487. doi: 10.1016/s0092-8674(03)00883-3. [DOI] [PubMed] [Google Scholar]
- Lee J, Colwill K, Aneliunas V, Tennyson C, Moore L, Ho Y, Andrews B. Interaction of yeast Rvs167 and Pho85 cyclin-dependent kinase complexes may link the cell cycle to the actin cytoskeleton. Curr Biol. 1998;8:1310–1321. doi: 10.1016/s0960-9822(07)00561-1. [DOI] [PubMed] [Google Scholar]
- Lew DJ, Reed SI. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J Cell Biol. 1993;120:1305–1320. doi: 10.1083/jcb.120.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew DJ, Reed SI. Cell cycle control of morphogenesis in budding yeast. Curr Opin Genet Dev. 1995;5:17–23. doi: 10.1016/s0959-437x(95)90048-9. [DOI] [PubMed] [Google Scholar]
- Lobert VH, Stenmark H. Cell polarity and migration: emerging role for the endosomal sorting machinery. Physiology (Bethesda) 2011;26:171–180. doi: 10.1152/physiol.00054.2010. [DOI] [PubMed] [Google Scholar]
- Loeb JD, Sepulveda-Becerra M, Hazan I, Liu H. A G1 cyclin is necessary for maintenance of filamentous growth in Candida albicans. Mol Cell Biol. 1999;19:4019–4027. doi: 10.1128/mcb.19.6.4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco E, Wedlich-Soldner R, Li R, Altschuler SJ, Wu LF. Endocytosis optimizes the dynamic localization of membrane proteins that regulate cortical polarity. Cell. 2007;129:411–422. doi: 10.1016/j.cell.2007.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momany M. Polarity in filamentous fungi: establishment, maintenance and new axes. Curr Opin Microbiol. 2002;5:580–585. doi: 10.1016/s1369-5274(02)00368-5. [DOI] [PubMed] [Google Scholar]
- Orlando K, Guo W. Membrane organization and dynamics in cell polarity. Cold Spring Harb Perspect Biol. 2009;1:a001321. doi: 10.1101/cshperspect.a001321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HO, Bi E. Central roles of small GTPases in the development of cell polarity in yeast and beyond. Microbiol Mol Biol Rev. 2007;71:48–96. doi: 10.1128/MMBR.00028-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruyne D, Bretscher A. Polarization of cell growth in yeast. J Cell Sci. 2000;113:571–585. doi: 10.1242/jcs.113.4.571. [DOI] [PubMed] [Google Scholar]
- Reijnst P, Jorde S, Wendland J. Candida albicans SH3-domain proteins involved in hyphal growth, cytokinesis, and vacuolar morphology. Curr Genet. 2010;56:309–319. doi: 10.1007/s00294-010-0301-7. [DOI] [PubMed] [Google Scholar]
- Riedl J, et al. Lifeact: a versatile marker to visualize F-actin. Nat Methods. 2008;5:605–607. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodal AA, Manning AL, Goode BL, Drubin DG. Negative regulation of yeast WASp by two SH3 domain-containing proteins. Curr Biol. 2003;13:1000–1008. doi: 10.1016/s0960-9822(03)00383-x. [DOI] [PubMed] [Google Scholar]
- Shaw BD, Chung DW, Wang CL, Quintanilla LA, Upadhyay S. A role for endocytic recycling in hyphal growth. Fungal Biol. 2011;115:541–546. doi: 10.1016/j.funbio.2011.02.010. [DOI] [PubMed] [Google Scholar]
- Shivas JM, Morrison HA, Bilder D, Skop AR. Polarity and endocytosis: reciprocal regulation. Trends Cell Biol. 2010;20:445–452. doi: 10.1016/j.tcb.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha I, Wang YM, Philp R, Li CR, Yap WH, Wang Y. Cyclin-dependent kinases control septin phosphorylation in Candida albicans hyphal development. Dev Cell. 2007;13:421–432. doi: 10.1016/j.devcel.2007.06.011. [DOI] [PubMed] [Google Scholar]
- Slaughter BD, Smith SE, Li R. Symmetry breaking in the life cycle of the budding yeast. Cold Spring Harb Perspect Biol. 2009;1:a003384. doi: 10.1101/cshperspect.a003384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Johnston D, Sanson B. Epithelial polarity and morphogenesis. Curr Opin Cell Biol. 2011;23:540–546. doi: 10.1016/j.ceb.2011.07.005. [DOI] [PubMed] [Google Scholar]
- Steinberg G. Hyphal growth: a tale of motors, lipids, and the Spitzenkorper. Eukaryot Cell. 2007a;6:351–360. doi: 10.1128/EC.00381-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg G. On the move: endosomes in fungal growth and pathogenicity. Nat Rev Microbiol. 2007b;5:309–316. doi: 10.1038/nrmicro1618. [DOI] [PubMed] [Google Scholar]
- Sudbery P, Gow N, Berman J. The distinct morphogenic states of Candida albicans. Trends Microbiol. 2004;12:317–324. doi: 10.1016/j.tim.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Sun Y, Martin AC, Drubin DG. Endocytic internalization in budding yeast requires coordinated actin nucleation and myosin motor activity. Dev Cell. 2006;11:33–46. doi: 10.1016/j.devcel.2006.05.008. [DOI] [PubMed] [Google Scholar]
- Taheri-Talesh N, Horio T, Araujo-Bazan L, Dou X, Espeso EA, Penalva MA, Osmani SA, Oakley BR. The tip growth apparatus of Aspergillus nidulans. Mol Biol Cell. 2008;19:1439–1449. doi: 10.1091/mbc.E07-05-0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahirovic S, Bradke F. Neuronal polarity. Cold Spring Harb Perspect Biol. 2009;1:a001644. doi: 10.1101/cshperspect.a001644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang HY, Xu J, Cai M. Pan1p, End3p, and S1a1p, three yeast proteins required for normal cortical actin cytoskeleton organization, associate with each other and play essential roles in cell wall morphogenesis. Mol Cell Biol. 2000;20:12–25. doi: 10.1128/mcb.20.1.12-25.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay S, Shaw BD. The role of actin, fimbrin and endocytosis in growth of hyphae in Aspergillus nidulans. Mol Microbiol. 2008;68:690–705. doi: 10.1111/j.1365-2958.2008.06178.x. [DOI] [PubMed] [Google Scholar]
- Vida TA, Emr SD. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A, Raniga PP, Lane S, Lu Y, Liu H. Hyphal chain formation in Candida albicans: Cdc28-Hgc1 phosphorylation of Efg1 represses cell separation genes. Mol Cell Biol. 2009;29:4406–4416. doi: 10.1128/MCB.01502-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. CDKs and the yeast-hyphal decision. Curr Opin Microbiol. 2009;12:644–649. doi: 10.1016/j.mib.2009.09.002. [DOI] [PubMed] [Google Scholar]
- Warren DT, Andrews PD, Gourlay CW, Ayscough KR. Sla1p couples the yeast endocytic machinery to proteins regulating actin dynamics. J Cell Sci. 2002;115:1703–1715. doi: 10.1242/jcs.115.8.1703. [DOI] [PubMed] [Google Scholar]
- Winter D, Lechler T, Li R. Activation of the yeast Arp2/3 complex by Bee1p, a WASP-family protein. Curr Biol. 1999;9:501–504. doi: 10.1016/s0960-9822(99)80218-8. [DOI] [PubMed] [Google Scholar]
- Zeng G, Cai M. Regulation of the actin cytoskeleton organization in yeast by a novel serine/threonine kinase Prk1p. J Cell Biol. 1999;144:71–82. doi: 10.1083/jcb.144.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng G, Huang B, Neo SP, Wang J, Cai M. Scd5p mediates phosphoregulation of actin and endocytosis by the type 1 phosphatase Glc7p in yeast. Mol Biol Cell. 2007;18:4885–4898. doi: 10.1091/mbc.E07-06-0607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng G, Yu X, Cai M. Regulation of yeast actin cytoskeleton-regulatory complex Pan1p/Sla1p/End3p by serine/threonine kinase Prk1p. Mol Biol Cell. 2001;12:3759–3772. doi: 10.1091/mbc.12.12.3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Wang YM, Wang Y. Hgc1, a novel hypha-specific G1 cyclin-related protein regulates Candida albicans hyphal morphogenesis. EMBO J. 2004;23:1845–1856. doi: 10.1038/sj.emboj.7600195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng XD, Lee RT, Wang YM, Lin QS, Wang Y. Phosphorylation of Rga2, a Cdc42 GAP, by CDK/Hgc1 is crucial for Candida albicans hyphal growth. EMBO J. 2007;26:3760–3769. doi: 10.1038/sj.emboj.7601814. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.