Abstract
Vertebrate innovations include neural crest cells and their derivatives, neurogenic placodes, an elaborate segmented brain, endoskeleton, and an increase in the number of genes in the genome. Comparative molecular and developmental data give new insights into the evolutionary origins of these characteristics and the complexity of the vertebrate body.
All chordates, at some stage in their life cycle, possess a hollow neural tube dorsal to a notochord, plus lateral muscle blocks. These characters unite tunicates (including ascidians), amphioxus, and vertebrates in the phylum Chordata. Comparative molecular and developmental analyses have refined this anatomical picture, suggesting that the neural tube of the common ancestor of chordates had three major subdivisions along the anterior-posterior axis (1), was patterned along the dorsoventral axis by hedgehog and Bmp signaling (2), and was probably segmented (3). These data suggest considerable complexity of the common ancestor of chordates. Vertebrates are more complex still, both morphologically and genetically, and are characterized by a considerable number of derived features (see Fig. 1). Here we review recent molecular and developmental data that give insight into the evolutionary origin of these vertebrate innovations, notably the neural crest, placodes, complex brain, skeleton, and additional genes.
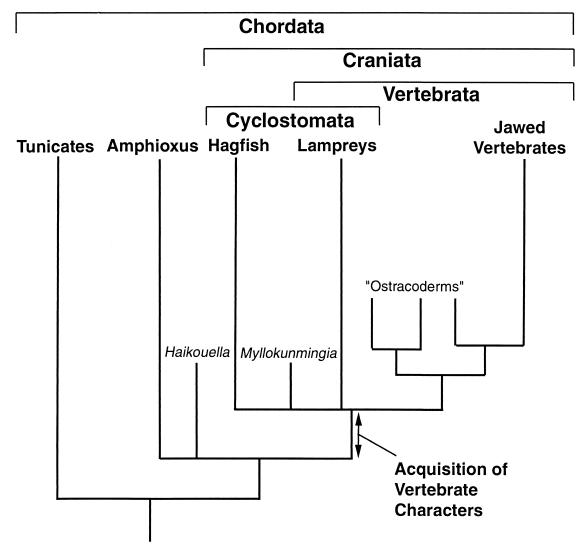
Figure 1.
What is a vertebrate? Phylogeny showing the relationship between living members of the Phylum Chordata (in bold, at top), plus the Cambrian fossils Myllokunmingia (12) (a craniate) and Haikouella (30) (a basal chordate). Some putative fossil chordates, including Pikaia and Cathaymyrus (possibly related to amphioxus) (31, 32) and the Euconodonts (25) (possible vertebrates) are omitted, as their precise relationships are less clear. Systematics reserves the taxon Vertebrata for those animals possessing vertebrae: that is, lampreys and jawed vertebrates, with hagfish excluded as a sister group. These three taxa are united by the term Craniata. Some molecular analyses, however, support monophyly of lampreys and hagfish (comprising the Cyclostomata) (33), which would make the Vertebrata paraphyletic. Here we depict the node at the base of hagfish, lampreys, and jawed vertebrates as unresolved to reflect this conflict between molecular and morphological data. We view the evolution of vertebrates as the acquisition of characters along the chordate stem lineage. These characters, established by comparison to outgroups (i.e., amphioxus and tunicates), include (i) neural crest cells and their derivatives; (ii) elaboration of placode-derived structures; (iii) elaboration of the brain, including rhombomeric segmentation; (iv) cartilage (and possibly mineralization); (v) the axial skeleton and the head skeleton; and (vi) a large increase in total number of genes in the genome. Other characters present only in jawed vertebrates, including paired appendages, hinged jaws, an adaptive immune system, and specialization of the axial skeleton along the anterior-posterior axis, were probably acquired later, on the jawed vertebrate stem lineage, and will not be considered here.
The Neural Crest.
Neural crest cells are a key vertebrate character. They form at the boundary between neural plate and surface ectoderm and migrate to contribute to many of the structures considered to be vertebrate novelties, including the cranium, branchial skeleton, and sensory ganglia. Experimentally, trunk neural crest cells can be induced from chick embryonic neural plate by application of Bmp-4 or -7 proteins (4), the genes encoding which are expressed by ectodermal cells. Neural crest cells themselves are marked by expression of a number of genes, including members of the Msx, slug/snail, Zic, Pax-3/7, and Distalless gene families. Lateral neural plate cells also express these genes in vertebrate embryos, suggesting some continuity of character between these two cell populations.
Homologues of several putative neural crest cell inducers and markers have been cloned from amphioxus and/or tunicates, and their expression patterns have been compared with vertebrates. These studies have revealed that the single BMP-2/4 gene is expressed in non-neural ectoderm of both amphioxus (5) and ascidian (6) embryos, although the expression pattern differs in detail between taxa. Lateral neural plate cells in amphioxus embryos express members of the Msx (7), slug/snail (8), and Distalless (9) gene families whereas the latter gene is also expressed in ectoderm adjacent to (and overgrowing) the neural plate. In ascidians, Msx (10) and slug/snail (11) homologues are expressed in cells contributing to the lateral neural plate (and in some other tissues); the Pax-3/7 gene is expressed in the immediately adjacent ectoderm, after earlier expression in cells fated to contribute to neural plate (3). These studies demonstrate that the relative spatial expression patterns of many genes involved in neural crest induction were already present before the evolution of vertebrates. Evolutionary changes to the regulation of these genes, therefore, have probably not contributed directly to neural crest origins. The homologues of several other genes involved in neural crest induction have not yet been examined in amphioxus or tunicate embryos; it will be particularly informative if any of these show significant expression differences to vertebrates.
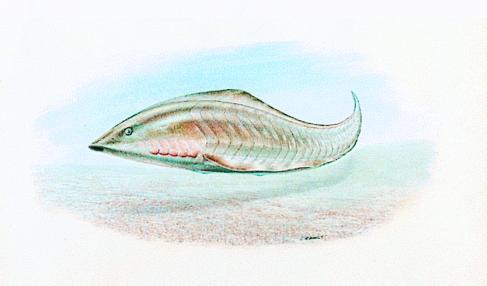
It is probably inappropriate to consider the evolution of neural crest cells as a single evolutionary step or a single vertebrate character. First, head and trunk neural crest have different developmental fates and potentials, and it is not yet clear whether they evolved as a single cell population or at different times in evolution. The existence of head neural crest in early vertebrate fossils such as Myllokunmingia (Fig. 2) is inferred by the presence of a complex branchial skeleton (12). Interestingly, no evidence of trunk neural crest derivatives is seen in these fossils (although this is not strong evidence for absence because preservation of such structures may be poor). Second, neural crest cells at one axial level may themselves be a diverse population, with cells that migrate at different times having different developmental properties and possibly distinct evolutionary origins. For example, the zebrafish mutant narrowminded specifically disrupts an early migrating cell population that contributes to spinal ganglia, without affecting later-migrating neural crest cells (13). Furthermore, this mutant also affects Rohon-Beard cells; these are not considered to be neural crest derivatives but are a dorsal neural population of cells involved in mechanosensory reception (and also present in amphioxus). This genetic link between early-migrating neural crest and Rohon-Beard cells suggests a possible common origin. An early step in the evolution of neural crest, therefore, may have been the origin of a specific dorsal neural cell population contributing to sensory processing; this would predate the divergence of the amphioxus and vertebrate lineages. The evolution of cell migration was probably of later evolutionary origin, as was the origin of neural crest cell lineages with primarily non-neural fates, such as melanocytes and connective tissue.
Figure 2.
Reconstruction of the early Cambrian craniate Myllokunmingia (12). [Reproduced with permission from John Sibbick (Copyright 1999 John Sibbick).]
Placodes.
Placodes are paired ectodermal thickenings that contribute to the formation of a number of specialized structures of the vertebrate head. They can be divided into the “sensory placodes,” which contribute to the eye, ear, lateral line, and olfactory organs, and the “neurogenic placodes,” which contribute sensory neurons to cranial ganglia. It is probable that the two types of placode have separate evolutionary origins. Sensory placodes form a variety of cell types, including neuroendocrine cells, sensory neurons, ciliated sensory receptors, and glia. They may also not be confined to vertebrates. Amphioxus has a putative homologue of the olfactory placode in the corpuscles of de Quatrefages, a specialized group of anterior ectoderm cells. These cells send axonal projections to the central nervous system along the most anterior nerve and are marked by expression of the homeobox gene AmphiMsx (7). This putative homology to olfactory placodes could be investigated further by functional studies on the amphioxus organs and by examination of olfactory receptor gene expression in amphioxus. Adult ascidians have sensory hair cells grouped into gelatinous cupular organs, located in the atrium of the adult; these cells have striking structural similarities to the sensory hair cells of the vertebrate acousticolateralis system (14). Furthermore, the ascidian atrium develops from a pair of ectodermal invaginations, topographically similar to sensory placodes; these atrial primordia, like vertebrate otic placodes, express members of the Pax-2/5/8 gene family (15). Thus, cellular organization, embryological origin, and gene expression all point to probable homology between the atrial primordia of ascidia and the otic placodes of vertebrates.
Neurogenic placodes contribute sensory neurons to cranial ganglia and may themselves be divided into two groups. The dorsolateral placodes (trigeminal and vestibular) develop from ectoderm lateral to the brain whereas the epibranchial placodes (geniculate, petrosal, and nodose) lie more ventrally, close to the pharyngeal pouches (16). Although epibranchial and dorsolateral have no identified homologues in amphioxus or tunicates, the generation of individual neurons from ectoderm is a character common to many animal taxa (17). It is not the neurogenic potential of ectoderm that is a vertebrate novelty, therefore, but the concentration of neurogenesis to discrete focal regions of ectoderm. Experiments show that the chick trigeminal placode is induced by signals from the neural plate (18) whereas pharyngeal endoderm is the source of the inductive signal (Bmp-7) for epibranchial placodes (16). In both cases, competence to respond to the inductive signal is widespread in cranial ectoderm, but absent from trunk ectoderm (16, 19). These data suggest that the focused neurogenesis of vertebrate epibranchial and dorsolateral placodes is achieved by a combination of localized inductive signals and restricted ectodermal competence.
In summary, the collective term “placodes” refers to some rather different structures, probably with different evolutionary origins. Some sensory placodes (at least the otic and olfactory) may have homologues in basal chordates. Even if this is so, it is apparent that they were elaborated considerably during early vertebrate evolution. Epibranchial and dorsolateral placodes appear to be new; we infer that their origin depended on the evolution of specific inductive signals.
Elaboration of the Vertebrate Brain.
A complex brain with specialized fore-, mid-, and hindbrain regions is characteristic of all vertebrates. Other chordates also possess a distinct structure at the rostral end of the neural tube, called the cerebral vesicle (amphioxus) or sensory vesicle (ascidian larvae). The extent of homology between these structures has long been controversial, but is central to discerning which aspects of brain organization are vertebrate innovations. Gene expression patterns, together with fine structural studies, have clarified the picture considerably. These studies reveal that embryos of vertebrates, amphioxus, and ascidia each have a distinct rostral domain of the neural tube, marked by Otx gene expression, separated from a more caudal region in which Hox genes are active. Between these two domains in vertebrates lies the isthmocerebellar region, including rhombomere 1 of the hindbrain and the midbrain-hindbrain boundary (MHB). This region is marked by expression of all three Pax 2/5/8 genes of vertebrates, both En genes, Wnt-1, and FGF-8. Strikingly, the single ascidian Pax 2/5/8 homologue is also expressed in cells between the Otx and Hox domains of Halocynthia (15). These comparisons led Wada et al. to propose a universal tripartite organization for chordate neural tubes, comprising Otx, Pax 2/5/8, and Hox domains; these correspond to the fore-/midbrain, isthmocerebellar/MHB, and hindbrain/spinal cord of vertebrates (15). The amphioxus Pax 2/5/8 gene is not expressed in the central domain, however, implying secondary modification in cephalochordates (or convergent evolution between tunicates and vertebrates) (20).
Despite the underlying homologies, the vertebrate brain is massively more complex. The fore-/midbrain region has been greatly expanded, for example, and shows evidence of compartmentalization not yet described in other chordates. The second subdivision includes the MHB in vertebrates: a structure with important patterning roles, probably mediated by FGF8 (21). It is unclear whether this role of the MHB is a vertebrate innovation. Two observations combine to suggest it might be: FGF8 inhibits vertebrate Hox gene expression in rhomobomere 1 of the hindbrain (21) whereas Hox-1 gene expression extends more rostrally in ascidia then in vertebrates (15). A more definitive statement on the origins of MHB signaling awaits cloning of FGF8 homologues from amphioxus and tunicates. The hindbrain region is also more complex in vertebrates than other chordates, notably through its stereotyped subdivision into rhombomeres with distinct developmental properties. Genes such as Krox-20 and kreisler play key roles in hindbrain segmentation, and in the control of rhombomere fate through modulation of Hox gene expression (22, 23). Current data suggest that this elaboration of segmentation is a vertebrate innovation because amphioxus and ascidian Hox genes do not show a similar pattern of modulation (15, 24), and amphioxus homologues of Krox-20 and kreisler are not expressed in appropriate patterns in the neural tube (R. D. Knight, W. Jackman, personal communication).
Cartilage, Bones, and Teeth.
Vertebrate cartilage and mineralized tissues are used for protection, predation, and endoskeletal support. There are no similar tissues in amphioxus or tunicates, where endoskeletal support is provided by a vacuolated notochord. The two earliest craniate fossils, Myllokunmingia (Fig. 2) and Haikouichthys from the early Cambrian, have a cartilaginous branchial skeleton but show no evidence of biomineralization (12). Late Cambrian fossils, the Euconodonts, show extensive biomineralization (in the tooth-like conodont elements), a probable cartilaginous head skeleton, but still no evidence of an axial skeleton (25). These observations suggest that cartilage predates biomineralization, and that a cranial skeleton evolved before the axial skeleton.
There are few insights into the molecular changes underlying the origins of cartilage or biomineralization. The origins of the axial skeleton seem more tractable because comparisons can be made to mesoderm development in outgroup taxa. In amphioxus, the entire mesoderm is segmented. In lampreys and jawed vertebrates, only the paraxial and intermediate mesoderm is fully segmented, although the lateral plate mesoderm of lamprey embryos shows traces of shallow segmental grooves (M. J. Cohn, personal communication). This suggests a progressive dorsal restriction of mesodermal segmentation in vertebrate evolution, which may be linked to increasing complexity of mesodermal patterning. Within the somite, it is the ventromedial region (the sclerotome) that produces the axial skeleton of vertebrates. The somites of amphioxus are also subdivided into distinct regions, although there is no histological evidence for sclerotome (Fig. 3). Furthermore, AmphiPax-1/9, a gene orthologous to the ancestor of the vertebrate sclerotome markers Pax-1 and Pax-9, is not expressed in amphioxus somites (26). Vertebrate sclerotome is induced by hedgehog (and possibly chordin) signaling from the notochord competing with Bmp signaling from more lateral cells. Because at least two of these signals are present in amphioxus (2, 5), the evolution of sclerotome probably involved changes downstream of these signals. The co-option of Pax gene expression into the somite is likely to be one of several such changes.
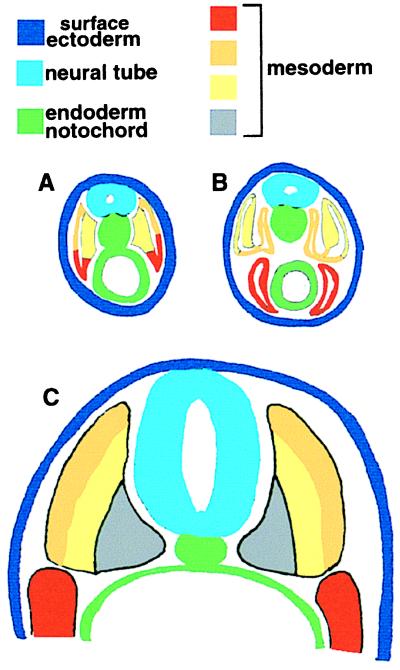
Figure 3.
Mesodermal subdivision in amphioxus (A and B) compared with a schematic vertebrate (C). A and B show progressive stages in the development of amphioxus mesoderm. A ventrolateral zone of amphioxus mesoderm (red) grows down to surround the gut. Homology of this zone to the lateral plate mesoderm of vertebrates (red in C) is supported by site of origin and fate. Medial amphioxus somite cells form myotome (yellow) whereas dorsolateral cells (orange) eventually move to surround the notochord. On the basis of position of origin, these may be homologous to the myotome and dermatome of vertebrates (yellow and orange, respectively, in C), although these cells do not contribute to a dermis in amphioxus. The vertebrate sclerotome (gray in C) has no equivalent in amphioxus and is a novelty linked with the evolution of the axial skeleton. A and B are adapted from ref. 34.
Conclusions: Toward a Causal Understanding of Vertebrate Origins.
In the above sections, we have highlighted developmental differences that underlie some of the key anatomical distinctions between vertebrates and their closest relatives. Because all evolutionary change must be based on genetic change, we have asked whether it is possible to identify candidate molecular changes that cause each developmental difference. In some cases (e.g., neural crest origins), the candidate molecular changes are not evident, despite extensive comparative work. In other cases, evidence points to changes in the regulation of specific genes (e.g., Hox, Krox, FGF-8, Pax-1/9). If we wish to probe deeper into vertebrate origins, therefore, it is important to consider how gene regulation evolves, and how genes can acquire new developmental roles.
In the context of vertebrate evolution, it is now very clear that numerous gene families expanded by gene duplication on the vertebrate stem lineage (27). These include gene families encoding transcription factors (Hox, ParaHox, En, Otx, Msx, Pax, Dlx, HNF3, bHLH), signaling molecules (hh, IGF, BMP), and others (dystrophin, cholinesterase, actin, keratin). In each case, vertebrates possess more copies than did the common ancestor of chordates, as inferred by comparison with amphioxus and/or ascidia. The mechanism by which this duplication occurred is controversial (polyploidy, multiple gene duplications, or both), but its prevalence can hardly be disputed. Indeed, increased genetic complexity can be validly viewed as a vertebrate innovation.
One long-standing hypothesis is that gene duplications promote the evolution of new functions because duplicate genes are freed from the constraining effects of natural selection by redundancy (28). This, however, has proven hard to test because there are many examples of genes evolving multiple roles without gene duplication, and a fundamentally different route of increasing gene number by duplication is if these roles become partitioned between daughter genes (e.g., by differential silencing of enhancers) (29). Nevertheless, the widespread duplication of developmental genes in early vertebrate evolution could conceivably be one of the key events underlying the evolution of vertebrate innovations, linking an increase in morphological complexity with an increase in genetic complexity via the intermediary of developmental control.
Acknowledgments
We thank A. Graham and I. Mason for helpful discussions, and John and Bridgette Sibbick for permission to reproduce the reconstruction of Myllokunmingia. The authors' work is funded by the Medical Research Council and the Biotechnology and Biological Sciences Research Council.
References
- 1.Williams N A, Holland P W H. Brain Behav Evol. 1998;52:177–185. doi: 10.1159/000006562. [DOI] [PubMed] [Google Scholar]
- 2.Shimeld S M. Am Zool. 1999;3:641–649. [Google Scholar]
- 3.Wada H, Holland P W H, Satoh N. Nature (London) 1996;384:123. doi: 10.1038/384123a0. [DOI] [PubMed] [Google Scholar]
- 4.Liem K F, Tremml G, Roelink H, Jessell T M. Cell. 1995;82:969–979. doi: 10.1016/0092-8674(95)90276-7. [DOI] [PubMed] [Google Scholar]
- 5.Panapoulou G D, Clark M D, Holland L Z, Lehrach H, Holland N D. Dev Dyn. 1998;213:130–139. doi: 10.1002/(SICI)1097-0177(199809)213:1<130::AID-AJA13>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 6.Miya T, Morita K, Suzuki A, Ueno N, Satoh N. Development (Cambridge, UK) 1997;124:5149–5159. doi: 10.1242/dev.124.24.5149. [DOI] [PubMed] [Google Scholar]
- 7.Sharman A C, Shimeld S M, Holland P W H. Dev Genes Evol. 1999;209:260–263. doi: 10.1007/s004270050251. [DOI] [PubMed] [Google Scholar]
- 8.Langeland J A, Tomsa J M, Jackman W R, Kimmel C B. Dev Genes Evol. 1998;208:569–577. doi: 10.1007/s004270050216. [DOI] [PubMed] [Google Scholar]
- 9.Holland N D, Panganiban G, Henyey E L, Holland L Z. Development (Cambridge, UK) 1996;122:2911–2920. doi: 10.1242/dev.122.9.2911. [DOI] [PubMed] [Google Scholar]
- 10.Ma L, Swalla B J, Zhou J, Dobias S L, Bell J R, Chen J, Maxson R E, Jeffery W R. Dev Dyn. 1996;205:308–318. doi: 10.1002/(SICI)1097-0177(199603)205:3<308::AID-AJA10>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 11.Erives A, Corbo J C, Levine M. Dev Biol. 1998;194:213–225. doi: 10.1006/dbio.1997.8810. [DOI] [PubMed] [Google Scholar]
- 12.Shu D-G, Luo H L, Conway Morris S, Zhang X-L, Hu S-X, Chen L, Han J, Zhu M, Li Y, Chen L-Z. Nature (London) 1999;402:42–46. [Google Scholar]
- 13.Artinger K B, Chitnis A B, Mercola M, Driever W. Development (Cambridge, UK) 1999;126:3969–3979. doi: 10.1242/dev.126.18.3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bone Q, Ryan K P. J Zool. 1978;186:417–429. [Google Scholar]
- 15.Wada H, Saiga H, Satoh N, Holland P W H. Development (Cambridge, UK) 1998;125:1113–1122. doi: 10.1242/dev.125.6.1113. [DOI] [PubMed] [Google Scholar]
- 16.Begbie J, Brunet J-F, Rubenstein J L R, Graham A. Development (Cambridge, UK) 1999;126:895–902. doi: 10.1242/dev.126.5.895. [DOI] [PubMed] [Google Scholar]
- 17.Baker C V H, Bronner-Fraser M. Mech Dev. 1997;69:13–29. doi: 10.1016/s0925-4773(97)00129-9. [DOI] [PubMed] [Google Scholar]
- 18.Stark M R, Sechrist J, Bronner-Fraser M, Marcelle C. Development (Cambridge, UK) 1997;124:4287–4295. doi: 10.1242/dev.124.21.4287. [DOI] [PubMed] [Google Scholar]
- 19.Baker C V H, Stark M R, Marcelle C, Bronner-Fraser M. Development (Cambridge, UK) 1999;126:147–156. doi: 10.1242/dev.126.1.147. [DOI] [PubMed] [Google Scholar]
- 20.Kozmik Z, Holland N D, Kalousova A, Paces J, Schubert M, Holland L Z. Development (Cambridge, UK) 1999;126:1295–1304. doi: 10.1242/dev.126.6.1295. [DOI] [PubMed] [Google Scholar]
- 21.Irving C, Mason I. Development (Cambridge, UK) 2000;127:177–186. doi: 10.1242/dev.127.1.177. [DOI] [PubMed] [Google Scholar]
- 22.Nonchev S, Vesque C, Maconochie M, Seitanidou T, Ariza-McNaughton L, Frain M, Marshall H, Sham M H, Krumlauf R, Charnay P. Development (Cambridge, UK) 1996;122:543–554. doi: 10.1242/dev.122.2.543. [DOI] [PubMed] [Google Scholar]
- 23.Manzanares M, Cordes S, Kwan C T, Sham M H, Barsh G S, Krumlauf R. Nature (London) 1997;387:191–195. doi: 10.1038/387191a0. [DOI] [PubMed] [Google Scholar]
- 24.Wada H, Garcia-Fernàndez J, Holland P W H. Dev Biol. 1999;213:131–141. doi: 10.1006/dbio.1999.9369. [DOI] [PubMed] [Google Scholar]
- 25.Donoghue P C J, Purnell M A, Aldridge R J. Lethaia. 1998;31:211–219. [Google Scholar]
- 26.Holland N D, Holland L Z, Kozmik Z. Mol Mar Biol Biotech. 1995;4:206–214. [PubMed] [Google Scholar]
- 27.Holland P W H. Semin Cell Dev Biol. 1999;10:541–547. doi: 10.1006/scdb.1999.0335. [DOI] [PubMed] [Google Scholar]
- 28.Ohno S. Evolution by Gene Duplication. Heidelberg: Springer; 1970. [Google Scholar]
- 29.Force A, Lynch M, Pickett F B, Amores A, Yan Y L, Postlethwait J. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen J-Y, Huang D-Y, Li C-W. Nature (London) 1999;402:518–522. [Google Scholar]
- 31.Briggs D E G, Erwin D H, Collier F J. The Fossils of the Burgess Shale. Washington, DC: Smithsonian; 1994. [Google Scholar]
- 32.Shu D-G, Conway Morris S, Zhang X-L. Nature (London) 1996;384:156–157. [Google Scholar]
- 33.Mallatt J, Sullivan J. Mol Biol Evol. 1998;15:1706–1718. doi: 10.1093/oxfordjournals.molbev.a025897. [DOI] [PubMed] [Google Scholar]
- 34.Holland L Z, Pace D A, Blink M L, Kene M, Holland N D. Dev Biol. 1995;171:665–676. doi: 10.1006/dbio.1995.1313. [DOI] [PubMed] [Google Scholar]