Abstract
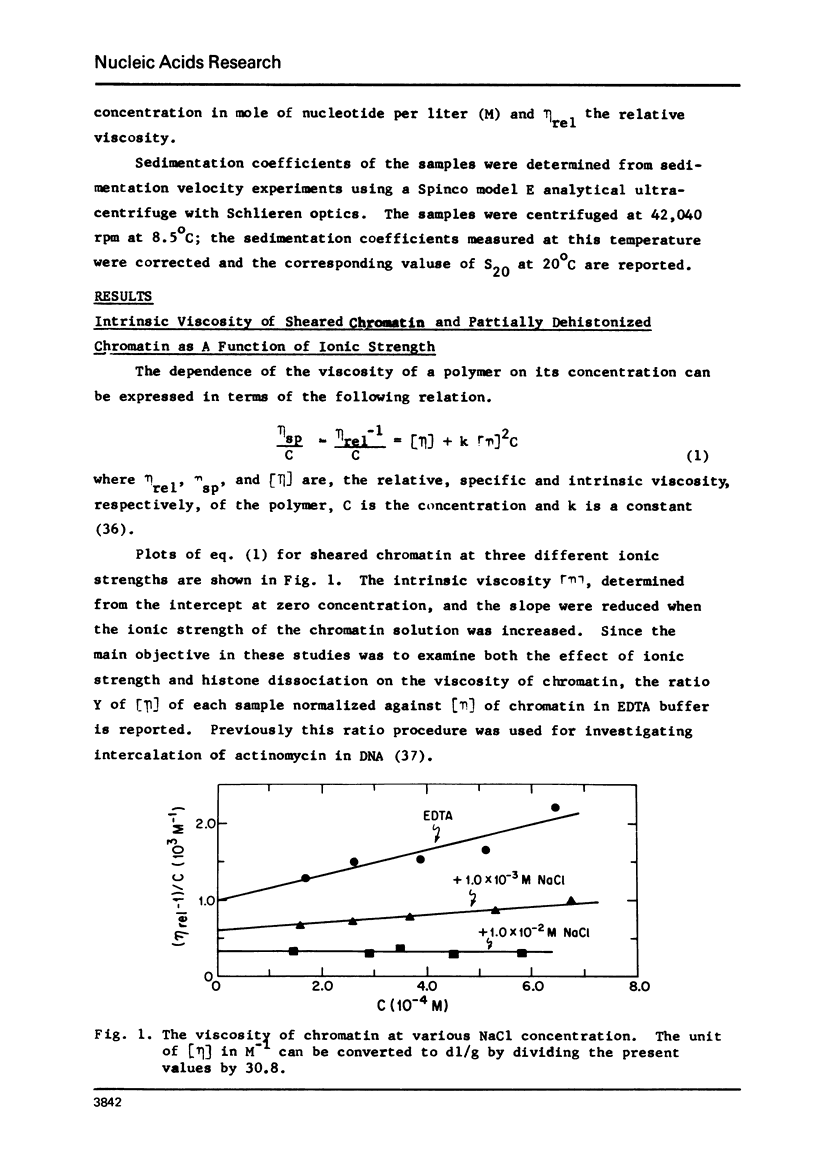
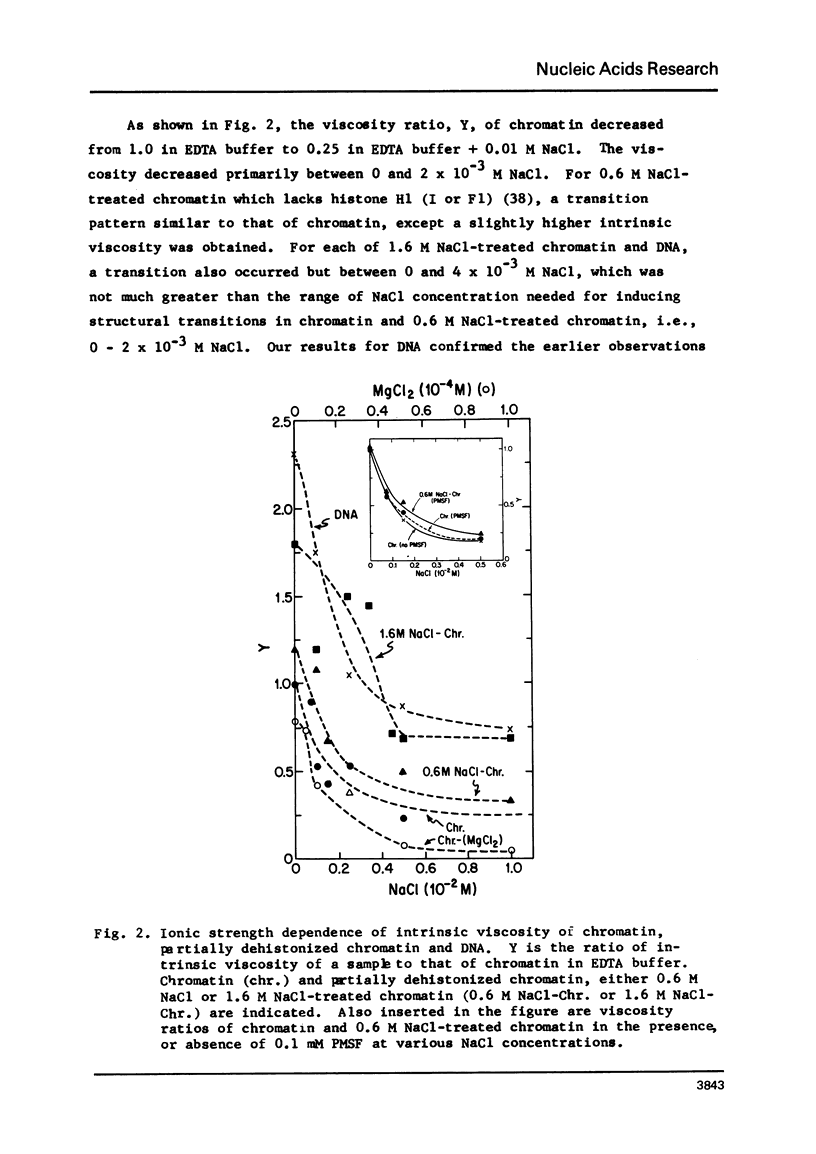
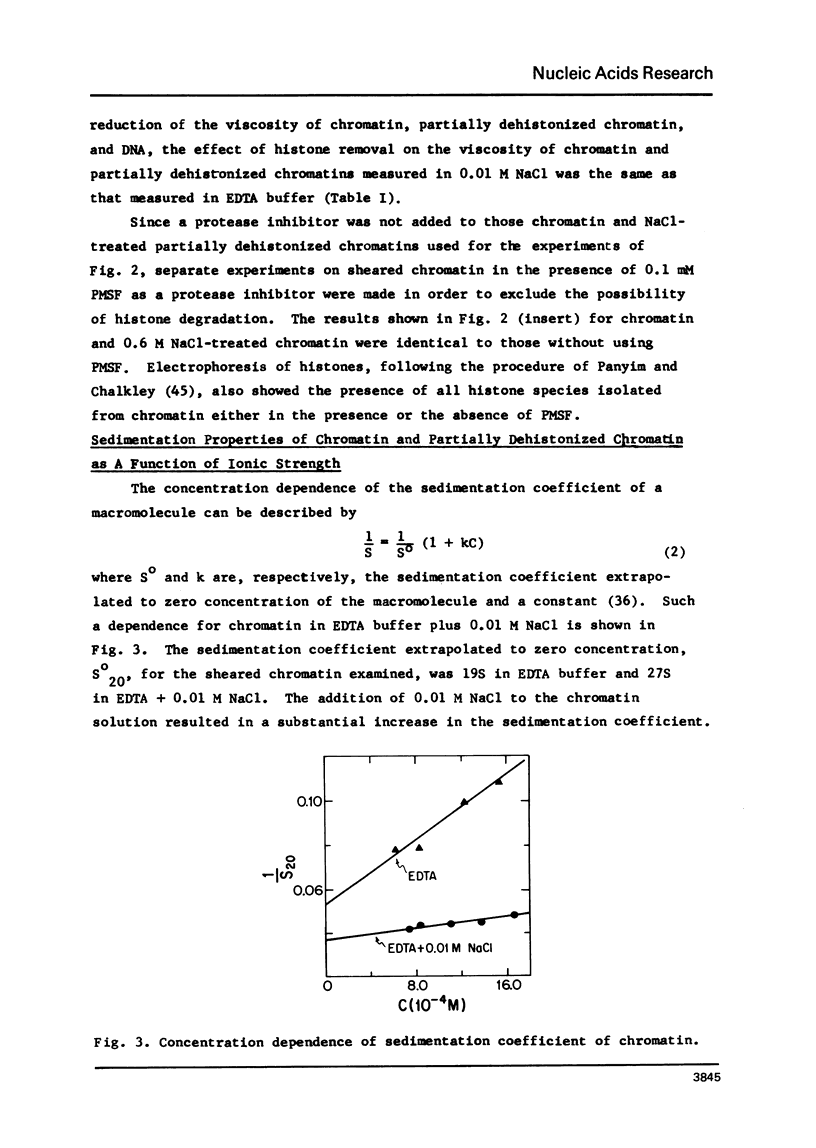
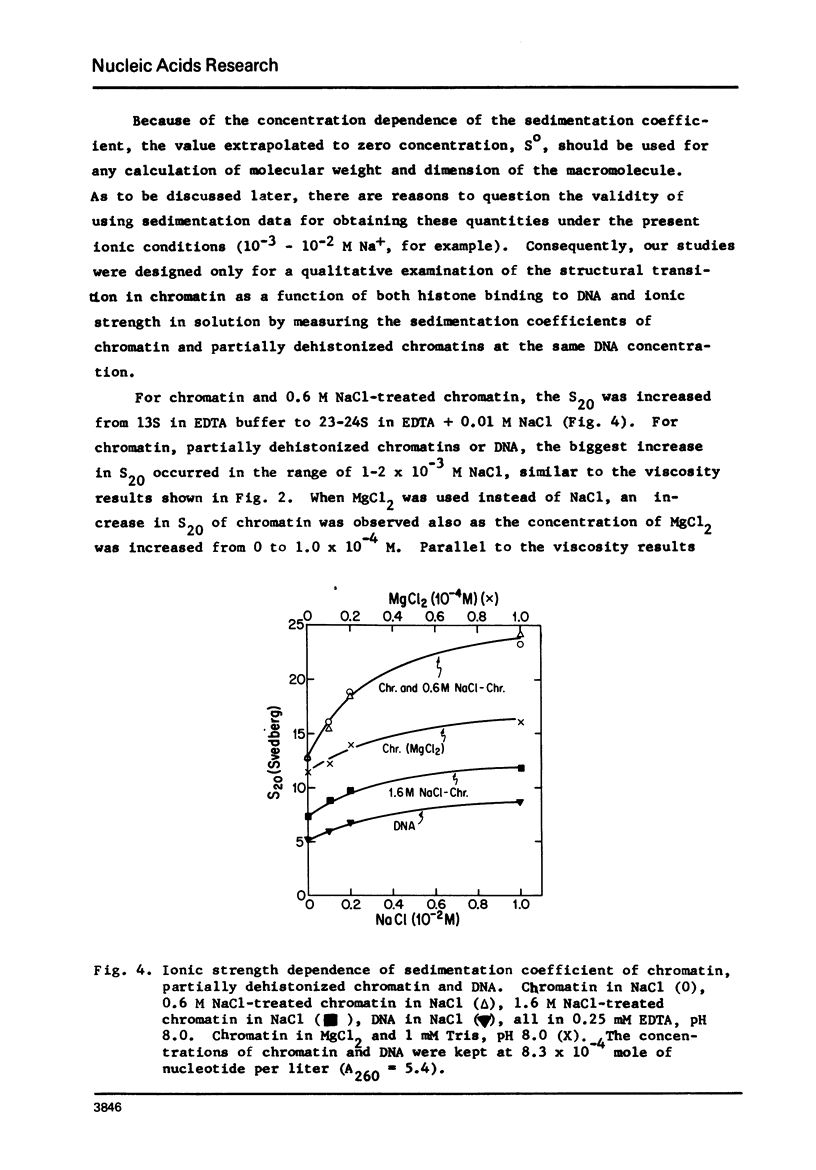
Structural transition in chromatin was measured as a function of counter ions in solution (NaCl or MgCl2) and of histones bound on the DNA. The addition of counter ions to aqueous solutions of chromatin, partially dehistonized chromatin, and DNA caused a drastic reduction in viscosity and a significant increase in sedimentation coefficient. Transitions occurred primarily at about 2 × 10−3 M NaCl and 1 × 10−5 M MgCl2 and are interpreted as a change in structure of chromatin induced by tight binding of cations (Na+ or Mg++) to DNA, either free or bound by histones, and is an intrinsic property of DNA rather than of the type of histone bound. At a given ionic condition, removal of histone H1 from chromatin had only a minor effect on the hydrodynamic properties of chromatin while removal of other histones caused a drastic change in these properties. An increase in the sedimentation coefficient of DNA was observed also for protamine. DNA complexes wherein the bound protein contains only unordered coil rather than the α-helices found in histones.
Full text
PDF










Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BAYLEY P. M., PRESTON B. N., PEACOCKE A. R. Thymus deoxyribonucleoprotein. II. Dissociation in sodium chloride solution. Biochim Biophys Acta. 1962 Jun 11;55:943–952. doi: 10.1016/0006-3002(62)90908-3. [DOI] [PubMed] [Google Scholar]
- Bartley J. A., Chalkley R. The viscosity of nucleohistone in urea. Biochim Biophys Acta. 1968 Jun 26;160(2):224–228. doi: 10.1016/0005-2795(68)90090-1. [DOI] [PubMed] [Google Scholar]
- Bartley J., Chalkley R. An approach to the structure of native nucleohistone. Biochemistry. 1973 Jan 30;12(3):468–474. doi: 10.1021/bi00727a017. [DOI] [PubMed] [Google Scholar]
- Bekhor I., Kung G. M., Bonner J. Sequence-specific interaction of DNA and chromosomal protein. J Mol Biol. 1969 Jan;39(2):351–364. doi: 10.1016/0022-2836(69)90322-2. [DOI] [PubMed] [Google Scholar]
- Carlson R. D., Olins A. L., Olins D. E. Urea denaturation of chromatin periodic structure. Biochemistry. 1975 Jul 15;14(14):3122–3125. doi: 10.1021/bi00685a013. [DOI] [PubMed] [Google Scholar]
- Chang C., Li H. J. Urea perturbation and the reversibility of nucleohistone conformation. Nucleic Acids Res. 1974 Aug;1(8):945–958. doi: 10.1093/nar/1.8.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark R. J., Felsenfeld G. Structure of chromatin. Nat New Biol. 1971 Jan 27;229(4):101–106. doi: 10.1038/newbio229101a0. [DOI] [PubMed] [Google Scholar]
- Crothers D. M. Calculation of binding isotherms for heterogenous polymers. Biopolymers. 1968 Apr;6(4):575–584. doi: 10.1002/bip.1968.360060411. [DOI] [PubMed] [Google Scholar]
- Finch J. T., Klug A. Solenoidal model for superstructure in chromatin. Proc Natl Acad Sci U S A. 1976 Jun;73(6):1897–1901. doi: 10.1073/pnas.73.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GIANNONI G., PEACOCKE A. R. Thymus deoxyribonucleoprotein. III. Sedimentation behaviour. Biochim Biophys Acta. 1963 Feb 26;68:157–166. doi: 10.1016/0006-3002(63)90131-8. [DOI] [PubMed] [Google Scholar]
- Griffith J. D. Chromatin structure: deduced from a minichromosome. Science. 1975 Mar 28;187(4182):1202–1203. doi: 10.1126/science.187.4182.1202. [DOI] [PubMed] [Google Scholar]
- Hanlon S., Johnson R. S., Chan A. Relationship between protein and DNA structure in calf thymus chromatin. I. Compositional aspects. Biochemistry. 1974 Sep 10;13(19):3963–3971. doi: 10.1021/bi00716a023. [DOI] [PubMed] [Google Scholar]
- Hewish D. R., Burgoyne L. A. Chromatin sub-structure. The digestion of chromatin DNA at regularly spaced sites by a nuclear deoxyribonuclease. Biochem Biophys Res Commun. 1973 May 15;52(2):504–510. doi: 10.1016/0006-291x(73)90740-7. [DOI] [PubMed] [Google Scholar]
- Hjelm R. P., Jr, Huang R. C. The role of histones in the conformation of DNA in chromatin as studied by circular dichroism. Biochemistry. 1974 Dec 17;13(26):5275–5283. doi: 10.1021/bi00723a004. [DOI] [PubMed] [Google Scholar]
- Kiryanov G. I., Manamshjan T. A., Polyakov V. Y., Fais D., Chentsov J. S. Levels of granular organization of chromatin fibres. FEBS Lett. 1976 Sep 1;67(3):323–327. doi: 10.1016/0014-5793(76)80557-1. [DOI] [PubMed] [Google Scholar]
- Lewis E. A., DeBuysere M. S., Rees A. W. Configuration of unsheared nucleohistone. Effects of ionic strength and of histone F1 removal. Biochemistry. 1976 Jan 13;15(1):186–192. doi: 10.1021/bi00646a029. [DOI] [PubMed] [Google Scholar]
- Li H. J. A model for chromatin structure. Nucleic Acids Res. 1975 Aug;2(8):1275–1289. doi: 10.1093/nar/2.8.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. J., Bonner J. Interaction of histone half-molecules with deoxyribonucleic acid. Biochemistry. 1971 Apr 13;10(8):1461–1470. doi: 10.1021/bi00784a030. [DOI] [PubMed] [Google Scholar]
- Li H. J., Chang C., Evagelinou Z., Weiskopf M. Circular dichroism of histone-bound regions in chromatin. Biopolymers. 1975 Jan;14(1):211–226. doi: 10.1002/bip.1975.360140115. [DOI] [PubMed] [Google Scholar]
- Li H. J., Chang C., Weiskopf M. Helix-coil transition in nucleoprotein-chromatin structure. Biochemistry. 1973 Apr 24;12(9):1763–1772. doi: 10.1021/bi00733a016. [DOI] [PubMed] [Google Scholar]
- Li H. J. Thermal denaturation of nucleohistones--effects of formaldehyde reaction. Biopolymers. 1972;11(4):835–847. doi: 10.1002/bip.1972.360110408. [DOI] [PubMed] [Google Scholar]
- Li H. J., Wickett R., Craig A. M., Isenberg I. Conformational changes in histone IV. Biopolymers. 1972 Feb;11(2):375–397. doi: 10.1002/bip.1972.360110206. [DOI] [PubMed] [Google Scholar]
- Manning G. S. On the application of polyelectrolyte "limiting laws" to the helix-coil transition of DNA. I. Excess univalent cations. Biopolymers. 1972;11(5):937–949. doi: 10.1002/bip.1972.360110502. [DOI] [PubMed] [Google Scholar]
- Miller P., Kendall F., Nicolini C. Thermal denaturation of sheared and unsheared chromatin by absorption and circular dichroism measurements. Nucleic Acids Res. 1976 Aug;3(8):1875–1881. doi: 10.1093/nar/3.8.1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller W., Crothers D. M. Studies of the binding of actinomycin and related compounds to DNA. J Mol Biol. 1968 Jul 28;35(2):251–290. doi: 10.1016/s0022-2836(68)80024-5. [DOI] [PubMed] [Google Scholar]
- Nicolini C., Baserga R., Kendall F. DNA structure in sheared and unsheared chromatin. Science. 1976 May 21;192(4241):796–798. doi: 10.1126/science.1265482. [DOI] [PubMed] [Google Scholar]
- Noll M. Subunit structure of chromatin. Nature. 1974 Sep 20;251(5472):249–251. doi: 10.1038/251249a0. [DOI] [PubMed] [Google Scholar]
- Ohlenbusch H. H., Olivera B. M., Tuan D., Davidson N. Selective dissociation of histones from calf thymus nucleoprotein. J Mol Biol. 1967 Apr 28;25(2):299–315. doi: 10.1016/0022-2836(67)90143-x. [DOI] [PubMed] [Google Scholar]
- Olins D. E., Bryan P. N., Harrington R. E., Hill W. E., Olins A. L. Conformational states of chromatin nu bodies induced by urea. Nucleic Acids Res. 1977 Jun;4(6):1911–1931. doi: 10.1093/nar/4.6.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panyim S., Chalkley R. The heterogeneity of histones. I. A quantitative analysis of calf histones in very long polyacrylamide gels. Biochemistry. 1969 Oct;8(10):3972–3979. doi: 10.1021/bi00838a013. [DOI] [PubMed] [Google Scholar]
- Paoletti R. A., Huang R. C. Characterization of sea urchin sperm chromatin and its basic proteins. Biochemistry. 1969 Apr;8(4):1615–1625. doi: 10.1021/bi00832a043. [DOI] [PubMed] [Google Scholar]
- Polacow I., Cabasso L., Li H. J. Histone redistribution and conformational effect on chromatin induced by formaldehyde. Biochemistry. 1976 Oct 19;15(21):4559–4565. doi: 10.1021/bi00666a002. [DOI] [PubMed] [Google Scholar]
- Rill R., Van Holde K. E. Properties of nuclease-resistant fragments of calf thymus chromatin. J Biol Chem. 1973 Feb 10;248(3):1080–1083. [PubMed] [Google Scholar]
- Rinehart F. P., Hearst J. E. The ionic strength dependence of s o 20 w of phage DNA in NH 4 AC. Arch Biochem Biophys. 1972 Oct;152(2):712–722. doi: 10.1016/0003-9861(72)90267-6. [DOI] [PubMed] [Google Scholar]
- Sahasrabuddhe C. G., Van Holde K. E. The effect of trypsin on nuclease-resistant chromatin fragments. J Biol Chem. 1974 Jan 10;249(1):152–156. [PubMed] [Google Scholar]
- Sander C., Ts'o P. O. Interaction of nucleic acids. 8. Binding of magnesium ions by nucleic acids. J Mol Biol. 1971 Jan 14;55(1):1–21. doi: 10.1016/0022-2836(71)90276-2. [DOI] [PubMed] [Google Scholar]
- Seligy V., Miyagi M. Studies of template activity of chromatin isolated from metabolically active and inactive cells. Exp Cell Res. 1969 Nov;58(1):27–34. doi: 10.1016/0014-4827(69)90113-x. [DOI] [PubMed] [Google Scholar]
- Shaw B. R., Herman T. M., Kovacic R. T., Beaudreau G. S., Van Holde K. E. Analysis of subunit organization in chicken erythrocyte chromatin. Proc Natl Acad Sci U S A. 1976 Feb;73(2):505–509. doi: 10.1073/pnas.73.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih T. Y., Lake R. S. Studies on the structure of metaphase and interphase chromatin of Chinese hamster cells by circular dichroism and thermal denaturation. Biochemistry. 1972 Dec 5;11(25):4811–4817. doi: 10.1021/bi00775a026. [DOI] [PubMed] [Google Scholar]
- Smart J. E., Bonner J. Selective dissociation of histones from chromatin by sodium deoxycholate. J Mol Biol. 1971 Jun 28;58(3):651–659. doi: 10.1016/0022-2836(71)90030-1. [DOI] [PubMed] [Google Scholar]
- Smart J. E., Bonner J. Studies on the role of histones in the structure of chromatin. J Mol Biol. 1971 Jun 28;58(3):661–674. doi: 10.1016/0022-2836(71)90031-3. [DOI] [PubMed] [Google Scholar]
- Tsai Y. H., Ansevin A. T., Hnilica L. S. Association of tissue-specific histones with deoxyribonucleic acid. Thermal denaturation of native, partially dehistonized, and reconstituted chromatins. Biochemistry. 1975 Mar 25;14(6):1257–1265. doi: 10.1021/bi00677a026. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. J., Bakayev V. V., Georgiev G. P. Heterogeneity of chromatin subunits in vitro and location of histone H1. Nucleic Acids Res. 1976 Feb;3(2):477–492. doi: 10.1093/nar/3.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. J., Ilyin Y. V., Georgiev G. P. Very long stretches of free DNA in chromatin. Nature. 1974 Aug 16;250(467):602–606. doi: 10.1038/250602a0. [DOI] [PubMed] [Google Scholar]
- Whitlock J. P., Jr, Simpson R. T. Removal of histone H1 exposes a fifty base pair DNA segment between nucleosomes. Biochemistry. 1976 Jul 27;15(15):3307–3314. doi: 10.1021/bi00660a022. [DOI] [PubMed] [Google Scholar]
- Wickett R. R., Li H. J., Isenberg I. Salt effects on histone IV conformation. Biochemistry. 1972 Aug 1;11(16):2952–2957. doi: 10.1021/bi00766a005. [DOI] [PubMed] [Google Scholar]
- Wilhelm F. X., de Murcia G. M., Champagne M. H., Daune M. P. Conformational changes of histones and DNA during the thermal denaturation of nucleoprotein. Eur J Biochem. 1974 Jun 15;45(2):431–443. doi: 10.1111/j.1432-1033.1974.tb03567.x. [DOI] [PubMed] [Google Scholar]
- Yu S. S., Li H. J. Helix-coil transition and conformational studies of protamine-DNA complexes. Biopolymers. 1973 Dec;12(12):2777–2788. doi: 10.1002/bip.1973.360121211. [DOI] [PubMed] [Google Scholar]
- Zimm B. H., Crothers D. M. SIMPLIFIED ROTATING CYLINDER VISCOMETER FOR DNA. Proc Natl Acad Sci U S A. 1962 Jun;48(6):905–911. doi: 10.1073/pnas.48.6.905. [DOI] [PMC free article] [PubMed] [Google Scholar]



