Abstract
Broadly cross neutralizing antibodies (NAbs) are generated in a group of HIV-1 infected individuals during the natural infection, but little is known about their prevalence in patients infected with viral subtypes from different geographical regions. We tested here the neutralizing efficiency of plasma antibodies from 80 HIV-1 infected antiretroviral drug naive patients against a panel of subtype-B and C tier 2 viruses. We detected cross-neutralizing antibodies in approximately 19–27% of the plasma, however the subtype-C specific neutralization efficiency predominated (p = 0.004). The neutralizing activity was shown to be exclusively mediated by the immunoglobulin G (IgG) fraction in the representative plasma samples. Epitope mapping of three, the most cross-neutralizing plasma (CNP) AIIMS206, AIIMS239 and AIIMS249 with consensus-C overlapping envelope peptides revealed ten different binding specificities with only V3 and IDR being common. The V3 and IDR were highly antigenic regions but no correlation between their reciprocal Max50 binding titers and neutralization was observed. In addition, the neutralizing activity of CNP was not substantially reduced by V3 and gp41 peptides except a modest contribution of MPER peptide. The MPER was rarely recognized by plasma antibodies though antibody depletion and competition experiments demonstrated MPER dependent neutralization in two out of three CNP. Interestingly, the binding specificity of one of the CNP (AIIMS206) overlapped with broadly neutralizing mAb 2F5 epitope. Overall, the data suggest that, despite the low immunogenicity of HIV-1 MPER, the antibodies directed to this region may serve as crucial reagents for HIV-1 vaccine design.
Introduction
The development of an immunogen capable of eliciting neutralizing antibody (NAb) response against HIV-1 remains elusive primarily due to enormous viral diversity [1]–[3]. The progress is hampered in part by inadequate information about the mechanisms associated with complex immune responses evoked during the natural course of HIV-1 infection [4]–[7]. Even the most promising antibody based vaccine candidates have shown effectiveness against limited number of HIV-1 strains [8]–[11]. However, results from a recent HIV-1 vaccine trial (RV144) in Thailand demonstrated for the first time, a vaccine induced protection in humans [12]. Although the study of immune correlates in the recipients of RV144 vaccine revealed that only high titer of plasma anti-V1/V2 antibodies correlated with lower risk of HIV infection, the induction of broadly cross-neutralizing antibodies still remains a main goal for the development of HIV vaccine [13]. Indeed, studies have shown that broad and potent NAb responses develop in the sera of a subset of HIV-1 infected individuals, and dissecting the nature of these responses may provide important clues for the design of new vaccine immunogens [14]–[17]. Analysis of the antibody response in HIV-1 infected individuals have revealed their specificities to all the HIV-1 proteins but antibodies directed mainly to the envelope glycoproteins (gp120 and gp41) are capable of mediating virus neutralization [16]–[18].
The non-covalently associated HIV-1 envelope glycoproteins, which mediate receptor binding and viral entry into the host cells [19], [20], remain the sole viral targets for neutralizing antibodies [16], [21], [22]. The CD4 binding site (CD4bs) and co-receptor binding region (third variable (V3) loop) of gp120 have been shown to serve as major vulnerable targets for HIV-1 neutralization [15], [16], [21], [23]–[27], although the role of other regions is also documented [28]–[31]. In comparison, the gp41 is highly conserved and is divided into three major regions, the extracellular region, the transmembrane (TM) domain and the cytoplasmic tail (CT) [32]. Antibody reactivity has been observed to be associated with different regions of the gp41 in HIV-1 infected donors, including the N and C-heptad regions [33], [34], immunodominant loop (ID) [35], [36], membrane proximal external region (MPER) [37]–[39] and C-terminal (CT) [40] of which the MPER constitutes the major target for broadly neutralizing antibodies on HIV-1 gp41 [22], [39].
Subtype-C remains the major HIV-1 infecting clade accounting for approximately 50% infections worldwide, with its primary centre being Africa followed by India [1]. Majority of the knowledge, as it relates to HIV-1 neutralizing antibody responses in subtype-C viruses, primarily come from African patients [41]–[44] with limited information on Indian subtype-C viruses despite the considerable differences between the viruses from these two geographical regions [45]–[48].
The study was aimed to examine the percentage of cross neutralizing plasma antibodies and identification of major linear antigenic regions in subtype-C HIV-1 infected Indian patients which has not yet been addressed in a great detail. Overall, our data suggest that, a good percentage of cross neutralizing antibodies circulate in these patients and the antibodies directed to the MPER region of HIV-1 gp41 envelope protein may serve as an important neutralization epitope.
Materials and Methods
Ethics Statement
The study was approved by the ethics committee of All India Institute of Medical Sciences (AIIMS) and the written informed consent was obtained from all the participants.
Plasma samples
Eighty HIV-1 seropositive patients enrolled in this study were recruited from the Regional STD Teaching Training & Research centre, Safdarjang hospital, and department of Medicine All India Institute of Medical Sciences (AIIMS), New Delhi, India. The patient samples were collected between the year 2008 and 2010. The HIV-1 seronegative samples were collected from the Blood bank, C.N. Centre, AIIMS, New Delhi. All the patients were tested and diagnosed hepatitis and tuberculosis negative. The whole blood samples of HIV-1 positive donors were collected in EDTA vacutainers, plasma was separated by centrifugation at 300 g and stored in aliquots at −80°C until use. The plasma samples were heat inactivated at 56°C for 1 h before using in the assays.
Monoclonal antibodies, peptides and recombinant proteins
The monoclonal antibodies (mAbs) used in this study were 447-52D, 1418 kindly provided by S. Zolla Pazner, and 2F5 and 4E10 (contributed by H. Katinger), were obtained through NIH AIDS Research and Reference Reagent Program (NIH, ARRRP). The 6 consensus subtype-B (con-B) MPER linear overlapping peptides and a complete set of 212 linear peptides (each 15 mer with an 11 amino acid overlap or a 4 amino acid walk) corresponding to the complete sequence of consensus subtype-C (con-C) gp120 and gp41 envelope proteins were obtained from the NIH, ARRRP. Three peptides corresponding to the con-C V3 (35 mer) (CTRPNNNTRKSIRIGPGQTFYATGDIIGDIRQAHC), 25 mer MPER (DLLALDSWKNLWNWFDITNWLWYIK) and 19 mer immunodominant region (IDR) (LGIWGCSGKLICTTAVPWN) were synthesized from Sigma Genosys, USA. The peptides had >95% purity. Three recombinant gp120s representing sequences of primary HIV-1 isolates from subtype-A, B and C (produced in 293 cells) were purchased from Immune Technology Corp. (New York, NY).
Determination of plasma viral load, CD4 count and total IgG levels
The plasma viral load was determined commercially (Lifeline Laboratories, New Delhi) using a real time PCR based method. The viral load was represented as RNA copies/ml of plasma. The CD4 counts were determined by flow cytometry. Total IgG content of the plasma was assessed by using a commercial ELISA kit from Ray Biotech, Inc USA. The clinical and demographic data are summarized in Table S1.
Purification of the polyclonal immunoglobulin G (IgG) fractions
Three plasma samples, AIIMS206, AIIMS239 and AIIMS249, identified as the most cross-neutralizing in this study, one plasma sample from HIV-1 seronegative healthy donor A1, were used for this experiment. Briefly, the polyclonal IgG was purified from the plasma samples by using protein A affinity chromatography (GE Healthcare) according to the manufacturer's instructions. IgG was eluted from the columns in 0.1 M citric acid, pH 3.0. Column fractions containing IgG were neutralized, pooled, and dialyzed against phosphate-buffered saline (PBS), pH 7.4. IgG purity was determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and the concentration was determined by measuring the relative absorbance at 280 nm and also by non-commercial total IgG quantitation ELISA.
Antibody binding assay (ELISA)
The test peptides (V3, MPER, IDR and con-C gp160 overlapping peptides) were immobilized on 96-well high binding ELISA plates (Corning) by an overnight incubation at 4°C of 100 µl of peptide per well diluted at 1 µg/ml concentration in 50 mM bicarbonate buffer (pH 9.6) except the MPER peptide which was adjusted equimolar to the V3 peptide concentration. Unbound peptides were removed by washing thrice with phosphate-buffered saline (PBS) containing 0.2% Tween 20 (Sigma-Aldrich) (PBST), and the plates were blocked with 200 µl of Roswell Park Memorial Institute media (RPMI) containing 15% fetal calf serum (FCS; Hyclone) and 2% bovine serum albumin (BSA; Sigma-Aldrich), incubated for 1 h at 37°C. Next, the plates were washed again three times and 100 µl of each plasma sample at different dilutions were added and incubated for 1.5 h at 37°C. Following 3 washings with PBST, the plates were further incubated for 1.5 h at 37°C with 100 µl (diluted 1/2000 in PBS containing 2% BSA) of alkaline-phosphatase conjugated anti-human IgG Fc (Southern Biotech). Finally the bound antibodies were detected by addition of alkaline phosphatase substrate (Sigma-Aldrich) in 10% diethanolamine buffer, and the colorimetric reaction was stopped by the addition of 6N NaOH. The optical density was read at 405 nm.
The 50% maximal binding of plasma antibodies (Max50) value was calculated for plasma that showed saturation for binding at low plasma dilutions. Initially all the plasma samples were tested at eight dilutions (dilution range: 1/30 to 1/100000) with V3, MPER and IDR but only six dilutions (1/300 to 1/100000) were considered for analysis as the first two dilutions (1/30 and 1/100) could not cross the cutoff for Max50 value which was three times the mean OD405 of the twenty healthy seronegative plasma samples at the lowest dilution plus three standard deviations. For epitope mapping with con-C gp160 overlapping peptides, a similar criterion was employed for Max50 determination with a different dilution range (1/100 to1/3000).
For binding of IgG fraction to con-C and B overlapping MPER peptides, the IgG was concentrated five times to that of original plasma volume that was purified, while for binding to recombinant gp120s the volume was adjusted to equal to that of the original volume of plasma used. For ELISA binding of IgG fractions, the coating concentration of the MPER overlapping peptides was also increased to 4 µg/ml. The binding of the plasma IgG fractions was performed by titrating the purified IgG fractions against MPER peptides and subtype-A, B and C recombinant gp120 proteins. The ELISA binding protocol was similar as above, except that the final reaction with substrate for gp120 ELISA was read in 20 minutes instead of 30 minutes. All the control mAbs were tested with peptides and proteins at a concentration ranging from 10 to 0.00003 µg/ml (12 dilutions). Experiments were carried out in triplicates and repeated at least two times for reproducibility.
Antibody depletion assay
In order to remove V3, MPER and IDR specific antibodies from the plasma, six passages on V3, MPER and IDR peptide-coated wells were carried out by using the ELISA binding protocol with some modifications described previously [49]. Briefly, we increased the binding capacity by coating the plates with 10 µg/ml peptide concentration. Second, to avoid the deleterious effects of detergent in subsequent neutralization cell cultures, the Tween 20 in the wash solution was replaced by 5% FCS. Third, sequentially, the plasma samples at 1∶30 dilution, were incubated for several successive passages (6 passages) on coated wells to remove the peptide specific antibodies. For this purpose, after 1 h of incubation at 37°C, the plasma were removed by pipetting and further incubated on other coated wells for an additional hour. For controls, the plasma were also mock depleted by serial passages on plates treated in parallel but not coated with peptides. The antibodies removed from the plasma and bound to the peptide-coated wells were then detected by incubation with alkaline phosphatase-conjugated anti-human IgG(Fc) and followed by adding substrate as described above. The diminution in OD405 throughout the successive passages could be monitored and correlated to the sequential depletion of peptide specific antibodies. The depletion percentage in the last passage was calculated as follows: percent depletion = 100−[100×(OD405 at the last passage/OD405 at the first passage)]. The plasma samples of two healthy HIV-1 seronegative individuals (A1 and A2) were used as ELISA controls for the depletion assay, and minimal upto 5% depletion was observed in the last passage. The depleted fractions of plasma were filtered through 0.45 mm-pore-size filters (Costar) before being tested for their neutralizing activity.
Primary isolates and pseudoviruses
The two newly generated primary isolates, AIIMS201 and AIIMS212 were isolated in our laboratory by previously described co-cultivation method [50]. Both the viruses belong to subtype-C, based on the partial envelope sequence of gp120 (C2–C5) (Andrabi et al, submitted). The envelope clones of subtype-C Du156.12, ZM109F.PB4, ZM53M.PB12, and subtype-B JRFL, RHPA4259.7, TRO.11 have been previously described [51], [52], and were obtained from the NIH, ARRRP. The pseudotyped viruses were produced by co-transfection of rev/env expression plasmid and an env-deficient HIV-1 backbone vector (pSG3ΔEnv) into exponentially dividing 293T cells in 6-well tissue culture plates (Corning Inc) using calcium phosphate method (Promega Inc). Pseudovirus-containing culture supernatants were harvested 48–53 hours post transfection, filtered (0.45 µm pore size) and stored at −80°C in 1 ml aliquots. The 50% tissue culture infectious dose (TCID50) was determined in TZM-bl cells.
Neutralization assay
Neutralization was measured as a reduction in luciferase gene expression after a single round of infection of JC53bl-13 cells, also known as TZM-bl cells (ARRRP; catalog no. 8129), with Env-pseudotyped viruses [53]. Briefly, 200 TCID50 of pseudovirus was incubated with heat inactivated diluted plasma samples (dilution range: 60 to 20000) in duplicates in a total volume of 100 µl for 1 hr at 37°C in 96-well flat-bottom culture plates. Freshly trypsinized cells (10,000 cells in 100 µl of growth medium containing 25 µg/ml DEAE Dextran and indianavir (1 µM) in case of primary isolates) were added to each well. One set of control wells received cells plus pseudovirus (virus control) and another set received cells only (background control). After 48 hours of incubation, luciferase activity was measured by using the Bright-Glo Luciferase Assay System (Promega Inc.). Fifty percent infective dose (ID50) values of the plasma were derived by determination of the plasma dilution that neutralized 50% of the infectious virus. Values were calculated through a dose-response curve fit with nonlinear function using GraphPad prism software (San Diego, CA). To compare the neutralization capacity of purified IgG fractions with the corresponding plasma, the IgG fractions for each sample were concentrated to a volume equal to that of the original volume of plasma that was purified.
In peptide interference neutralization assay, a test peptide was preincubated 30 minutes with plasma prior to the addition of virus. The final concentration of peptide in mixtures with plasma and virus was 20 µg/ml. To control for the possible direct effects of peptides on viral infection (i.e., in the absence plasma antibodies), assays were conducted in the presence of peptides (V3, MPER and IDR) at the same concentrations as those used in competition assays. In these experiments, the peptides had either little or no effect on virus infectivity. This internal control level of infection with each peptide was used as a baseline reference for peptide competition assays with plasmas. For competition assay validation, an anti-V3 antibody (447-52D) known to neutralize SF162 (a tier 1 sensitive subtype-B virus) was competed with two separate peptides corresponding to con-C and B V3 sequences (final peptide concentration 10 µg/ml) and tested for neutralization.
Statistical Analysis
Statistical analyses were performed using Graph Pad Prism 5 for Windows, Graph Pad Software, San Diego, California USA. A non-linear regression curve straight line was plotted using the method of least squares to determine the Max50 and ID50 values. Median reciprocal Max50 binding titers were compared using Wilcoxon matched pairs test or Mann-Whitney U test. Further the Spearman rank test was used to determine the correlation between the two variables. A p-value less than 0.05 was considered significant for this study.
Results
Characteristics of HIV-1 infected patients
The details of the 80 HIV-1 infected antiretroviral naïve patients recruited for this study is summarized in Table S1. The patients had been infected for different time periods, ranging from a few days up to seven years (based on time since 1st diagnosis). There were 30 females and 50 males within the age range of 20–57 years. The median viral load determined for 53 patients was 30800 (range = 156–2180000) RNA copies/ml plasma with a few patients having viral load below the detectable limits (<47 copies/ml). The median CD4 count was 337 (range = 14–966) cells/cubic millimeter (n = 80), while the mean plasma total IgG levels was 12.3 mg/ml (n = 65).
Neutralizing activity of the plasma antibodies from HIV-1 infected patients
In order to determine the neutralizing activity, we tested plasma antibodies against a panel of 5 subtype-C and 3 subtype-B tier 2 viruses (Table 1). Two of the new subtype-C isolates (AIIMS201 and AIIMS212) were tested and displayed resistance to neutralization by broadly neutralizing antibodies (Table S2), and were assigned as tier 2 viruses (Table 1). Overall the viruses were selected based on their subtype, geographical occurrence and resistance to neutralization. Presumably most of the patients were infected with subtype-C which is the major subtype in India [45], [46]. Indeed the subtyping of a few patients from this cohort showed a prominence of subtype-C infections (Andrabi et al, submitted) (Table 2).
Table 1. List of HIV-1 viruses used in this study.
| Virus | Tier | Subtype | Country of origin | 1Acc no. |
| AIIMS201 | 2 | C | India | JF300176 |
| AIIMS212 | 2 | C | India | JF300177 |
| ZM109F.PB4 | 2 | C | Zambia | AY424138 |
| Du156.12 | 2 | C | South Africa | DQ411852.1 |
| ZM53M.PB12 | 2 | C | Zambia | AY423984.2 |
| JRFL | 2 | B | USA | U63632 |
| TRO.11 | 2 | B | Italy | AY835445.1 |
| RHPA4259.7 | 2 | B | USA | AY835447 |
The viruses were selected based on resistance to neutralization, the clade they belongs to and the geographical origin.
Acc no.: Gene bank accession number.
Table 2. Neutralization potential of plasma antibodies from HIV-1 patients against subtype-B and C tier 2 viruses.
| AIIMS ID | Subtype | cAIIMS201 | cZM53M.PB12 | cDu156.12 | bRHPA4259.7 | bTRO.11 | cAIIMS212 | bJRFL | cZM109F.PB4 |
| AIIMS206 | ND | >20000 | 17766 | 3016 | 3313 | 4199 | 4022 | 3040 | 3243 |
| AIIMS213 | C | 1978 | 258 | 1940 | <60 | <60 | <60 | <60 | <60 |
| AIIMS239 | C | 783 | 1257 | 317 | 509 | 385 | 457 | 245 | 220 |
| AIIMS220 | ND | 106 | 480 | 2178 | 796 | 156 | <60 | 66 | 84 |
| AIIMS914 | ND | 2264 | 431 | <60 | 119 | 163 | <60 | 445 | 113 |
| AIIMS905 | ND | 2264 | <60 | <60 | 70 | 269 | <60 | 145 | <60 |
| AIIMS249 | ND | 457 | 369 | 189 | 120 | 734 | 520 | 291 | 189 |
| AIIMS210 | C | 1794 | <60 | 102 | <60 | <60 | <60 | 442 | <60 |
| AIIMS232 | C | 210 | 440 | 510 | 214 | <60 | 316 | 244 | <60 |
| AIIMS211 | ND | 68 | <60 | 222 | 1396 | <60 | 124 | <60 | <60 |
| AIIMS225 | C | 208 | 90 | 1128 | 96 | <60 | 62 | <60 | <60 |
| AIIMS901 | ND | 208 | 183 | 291 | <60 | 472 | 177 | 308 | <60 |
| AIIMS287 | C | <60 | 838 | 302 | <60 | <60 | <60 | 264 | <60 |
| AIIMS288 | ND | <60 | 752 | 108 | <60 | <60 | <60 | 76 | <60 |
| AIIMS904 | ND | 618 | 235 | <60 | <60 | <60 | <60 | 76 | <60 |
| AIIMS254 | C | 70 | <60 | 206 | 446 | 162 | <60 | <60 | 104 |
| AIIMS289 | ND | <60 | 138 | 352 | 210 | <60 | <60 | 124 | 88 |
| AIIMS212 | C | <60 | <60 | <60 | 656 | <60 | <60 | <60 | <60 |
| AIIMS248 | ND | 226 | 88 | 298 | <60 | 108 | 88 | <60 | <60 |
| AIIMS284 | ND | <60 | 334 | 250 | <60 | <60 | <60 | <60 | 78 |
| AIIMS906 | ND | 418 | 70 | 62 | <60 | 142 | <60 | <60 | <60 |
| AIIMS911 | ND | <60 | 72 | <60 | <60 | <60 | <60 | 463 | <60 |
| AIIMS909 | ND | 405 | <60 | <60 | <60 | 89 | 85 | 75 | <60 |
| AIIMS250 | ND | 110 | 160 | 230 | <60 | <60 | 84 | <60 | <60 |
| AIIMS291 | ND | <60 | 292 | 84 | <60 | <60 | <60 | <60 | 116 |
| AIIMS219 | ND | <60 | <60 | <60 | 366 | <60 | <60 | <60 | <60 |
| AIIMS299 | ND | 108 | 74 | <60 | 78 | <60 | <60 | 66 | 264 |
| AIIMS234 | C | <60 | <60 | 190 | <60 | <60 | 182 | <60 | <60 |
| AIIMS255 | C | 146 | 90 | <60 | <60 | <60 | 168 | <60 | 64 |
| AIIMS913 | ND | <60 | 281 | <60 | <60 | <60 | <60 | <60 | <60 |
| AIIMS300 | ND | 158 | <60 | <60 | 112 | <60 | <60 | <60 | 98 |
| AIIMS281 | A | <60 | <60 | <60 | <60 | <60 | 98 | 204 | <60 |
| AIIMS283 | ND | <60 | 228 | <60 | <60 | <60 | <60 | <60 | <60 |
| AIIMS264 | C | 78 | <60 | 204 | <60 | <60 | <60 | <60 | <60 |
| AIIMS910 | ND | 68 | 209 | <60 | <60 | <60 | <60 | <60 | <60 |
| AIIMS205 | ND | <60 | 202 | 66 | <60 | <60 | <60 | <60 | <60 |
| AIIMS242 | ND | <60 | 170 | 64 | <60 | 64 | 82 | <60 | <60 |
| AIIMS274 | ND | <60 | 74 | 126 | <60 | 100 | <60 | <60 | 78 |
| AIIMS277 | ND | <60 | <60 | 80 | 152 | <60 | <60 | 86 | <60 |
| AIIMS290 | C | <60 | 184 | <60 | <60 | <60 | <60 | <60 | 72 |
| AIIMS276 | ND | <60 | 148 | 82 | <60 | <60 | <60 | 62 | <60 |
| AIIMS224 | C | <60 | <60 | 160 | <60 | <60 | <60 | <60 | <60 |
| AIIMS208 | C | <60 | <60 | 156 | <60 | <60 | <60 | <60 | <60 |
| AIIMS241 | C | 72 | <60 | 136 | <60 | <60 | 64 | <60 | <60 |
| AIIMS253 | C | 120 | <60 | <60 | <60 | <60 | 88 | <60 | <60 |
| AIIMS297 | ND | 80 | 66 | <60 | <60 | <60 | 110 | <60 | <60 |
| AIIMS275 | C | <60 | <60 | 82 | <60 | <60 | <60 | <60 | 100 |
| AIIMS230 | ND | <60 | <60 | 120 | <60 | <60 | <60 | <60 | <60 |
| AIIMS286 | ND | <60 | <60 | <60 | <60 | <60 | 62 | 110 | <60 |
| AIIMS273 | ND | <60 | 76 | <60 | 92 | <60 | <60 | <60 | <60 |
| AIIMS235 | A | 82 | <60 | <60 | <60 | <60 | 84 | <60 | <60 |
| AIIMS294 | C | <60 | <60 | <60 | 70 | <60 | <60 | 94 | <60 |
| AIIMS203 | ND | <60 | <60 | 94 | <60 | <60 | <60 | <60 | <60 |
| AIIMS908 | ND | <60 | <60 | <60 | <60 | 94 | <60 | <60 | <60 |
| AIIMS293 | C | <60 | <60 | <60 | <60 | <60 | <60 | <60 | 86 |
| AIIMS231 | ND | <60 | <60 | <60 | <60 | 74 | 68 | <60 | <60 |
| AIIMS226 | C | <60 | <60 | <60 | <60 | <60 | 78 | <60 | <60 |
| AIIMS272 | ND | <60 | 76 | <60 | <60 | <60 | <60 | <60 | <60 |
| AIIMS216 | ND | <60 | <60 | 70 | <60 | <60 | <60 | <60 | <60 |
| AIIMS285 | ND | <60 | 70 | <60 | <60 | <60 | <60 | <60 | <60 |
| AIIMS271 | ND | <60 | <60 | <60 | <60 | <60 | <60 | 68 | <60 |
| AIIMS298 | C | 66 | 62 | <60 | <60 | <60 | <60 | <60 | <60 |
| AIIMS233 | ND | <60 | <60 | 66 | <60 | <60 | <60 | <60 | <60 |
| AIIMS915 | ND | 65 | 61 | <60 | <60 | <60 | <60 | <60 | <60 |
| AIIMS207 | ND | <60 | <60 | <60 | <60 | <60 | <60 | <60 | <60 |
| AIIMS218 | ND | <60 | <60 | <60 | <60 | <60 | <60 | <60 | <60 |
| AIIMS223 | ND | <60 | <60 | <60 | <60 | <60 | <60 | <60 | <60 |
| AIIMS237 | ND | <60 | <60 | <60 | <60 | <60 | <60 | <60 | <60 |
| AIIMS244 | ND | <60 | <60 | <60 | <60 | <60 | <60 | <60 | <60 |
| AIIMS270 | ND | <60 | <60 | <60 | <60 | <60 | <60 | <60 | <60 |
| AIIMS279 | A | <60 | <60 | <60 | <60 | <60 | <60 | <60 | <60 |
| AIIMS280 | ND | <60 | <60 | <60 | <60 | <60 | <60 | <60 | <60 |
| AIIMS282 | ND | <60 | <60 | <60 | <60 | <60 | <60 | <60 | <60 |
| AIIMS292 | ND | <60 | <60 | <60 | <60 | <60 | <60 | <60 | <60 |
| AIIMS295 | C | <60 | <60 | <60 | <60 | <60 | <60 | <60 | <60 |
| AIIMS296 | C | <60 | <60 | <60 | <60 | <60 | <60 | <60 | <60 |
| AIIMS902 | ND | <60 | <60 | <60 | <60 | <60 | <60 | <60 | <60 |
| AIIMS903 | ND | <60 | <60 | <60 | <60 | <60 | <60 | <60 | <60 |
| AIIMS907 | ND | <60 | <60 | <60 | <60 | <60 | <60 | <60 | <60 |
| AIIMS912 | ND | <60 | <60 | <60 | <60 | <60 | <60 | <60 | <60 |
The cross neutralizing activities of plasma antibodies from 80 HIV-1 infected drug naïve individuals from north India against the tier 2 subtype-B and C viruses indicated on the top of the table. Each virus is designated with its subtype (subtype-C and subtype-B; superscript alphabet on left). The AIIMS ID of the patient samples is given on the left of the table followed by the subtype of viruses they were infected with (C- subtype-C, A- subtype-A and ND- not determined). The numerical values in boxes are the 50% neutralization titers (ID50) defined as the dilution of plasma which neutralized 50% of viral infection in the assay. For clarity, this information is coded: ID50>1000 (Bold), ID50 = 61–1000 (Italic) and <60, where ID50 was not reached. Each experiment was performed at least two independent times.
Of the 80 plasma samples, 64(80%) were able to neutralize at least one virus while 16(20%) did not show any neutralization. Nevertheless 20(25%) plasma samples were found to neutralize ≥50% viruses tested (Table 2). Only three plasma samples AIIMS206, AIIMS239 and AIIMS249 were able to neutralize all the eight viruses tested, AIIMS206 being the most potent. These three plasma samples are represented as most cross-neutralizing plasma (CNP) of tier 2 viruses tested here. In terms of overall neutralization frequency, 190 of 640 virus/plasma combinations showed neutralizing activity (approximately 29.6%) (Table 2). Among all the viruses, ZM53M.PB12, Du156.12 (subtype-C) were most sensitive while TRO.11 (subtype-B) and ZM109F.PB4 (subtype-C) were most resistant, neutralized by 35(43.7%), 34(42.5%) and 15(18.7%), 16(20%) plasma samples respectively. Although the cross-clade neutralizing activity was observed in most of the neutralizing plasma, however the subtype-specific neutralization predominated, plasma antibodies being more effective against subtype-C than subtype-B viruses (p = 0.004).
Plasma immunoglobulin-G (IgG) fractions mediate the neutralizing activity
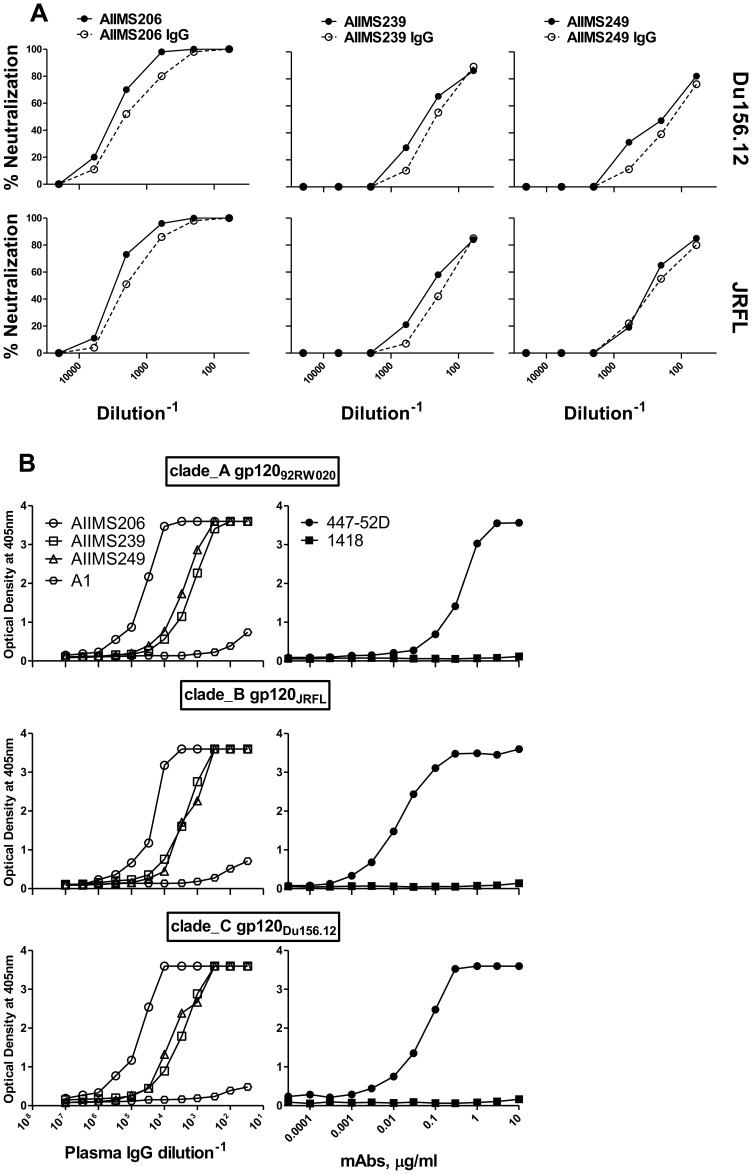
To determine the component of the HIV-1-infected donor plasma responsible for the neutralizing activity, we purified polyclonal IgG from three CNP, AIIMS206, AIIMS239, AIIMS249 and a healthy seronegative plasma sample A1 on protein A-Sepharose columns. The neutralization capacity of each column fraction was compared to the degree of neutralization in the original plasma. We tested IgG fractions of these plasma with two env-pseudotyped viruses from subtype-C (Du156.12) and B (JRFL) in a neutralization assay. In each case, we observed comparable neutralizing activity in the original plasma and purified IgG (Figure 1A). The IgG fractions were also tested for ELISA binding with three recombinant gp120 proteins representing subtype-A (92RW020), B (JRFL) and C (Du156.12) viruses. Consistent with neutralization results, we observed that the IgG fractions from HIV-1 plasma retained cross reactive binding to envelope gp120s (Figure 1B). In addition, we observed little or no binding or neutralization activity in the IgG-depleted fraction (data not shown).
Figure 1. Neutralization and binding of IgG fractions from cross-neutralizing (CNP) HIV-1 plasma.
(A) Neutralization curves of IgG fractions purified from CNP on protein-A. The neutralization capacity of purified polyclonal IgG (open circle) is compared with those of the original plasma (solid circle). To allow the comparison, the purified IgG were concentrated to volumes equal to that of the original volume of plasma run over the column. Neutralization of CNP, AIIMS206, AIIMS239 and AIIMS249 column fractions is compared for the subtype C (Du156.12) and subtype B (JRFL) isolates. (B) The binding pattern of IgG fractions of CNP along with IgG from healthy control plasma A1. The ELISA binding was carried out with envelope recombinant gp120 proteins from subtype-A (92RW020), subtype-B (JRFL) and subtype-C (Du156.12) isolates. The anti-V3 (447-52D) and anti-B19 (1418) monoclonal antibodies were used as assay controls.
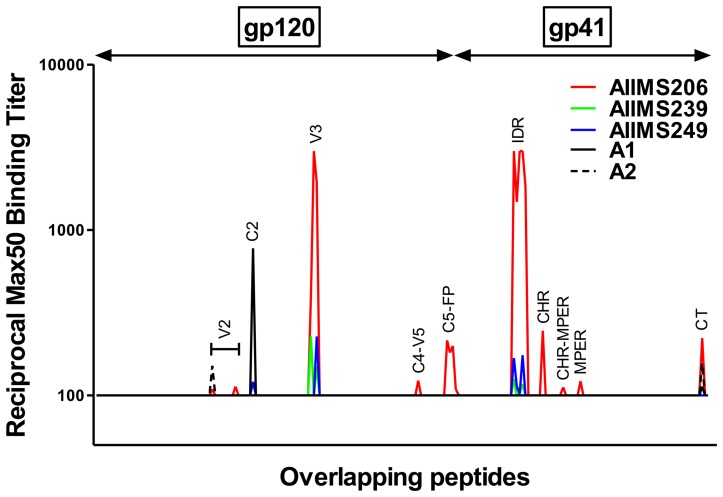
Epitope mapping of CNP with consensus-C gp160 overlapping peptides
To identify the role of linear antigenic epitopes on HIV-1 clade C envelope protein in virus neutralization, we tested the reactivity of three CNP samples (AIIMS206, AIIMS239 and AIIMS249) along with two control plasma from healthy seronegative donors with 212 con-C gp160 overlapping peptides in an ELISA binding assay (Table S3a; S3b, Figure 2). We were particularly interested in mapping the antigenic regions common for binding to these CNP. Based on the Max50 ELISA binding titers, the immunoreactivity of CNP mapped to the second variable (V2), second constant (C2), third variable (V3), fourth constant-fifth variable (C4-V5), fifth constant (C5) regions of gp120 and fusion protein (FP), immunodominant region (IDR), C-heptad region (CHR), membrane proximal external region (MPER) and C-terminal (CT) of gp41 protein. Among CNP, AIIMS206 showed highest number of binding specificities (nine), while only two binding sites were shared by all the three CNP (V3 and IDR) (Figure 2).
Figure 2. Epitope mapping of polyclonal antibodies from three cross-neutralizing plasma with consensus-C HIV-1 gp160 overlapping peptides.
ELISA binding of polyclonal plasma antibodies from three most cross-neutralizing plasma (CNP) AIIMS206, AIIMS239 and AIIMS249 and two healthy seronegative individuals (A1 and A2) to 212 consensus-C HIV-1 envelope glycoprotein gp160 (gp120 and gp41) overlapping linear peptides (15 mer each with 11 amino acid overlap or 4 amino acid walk). The plasma were reacted at 4 dilutions (dilution range: 100 to 3000) and reciprocal Max50 binding titers were calculated using least square regression method. The plasma showed reactivity to second variable (V2), second constant (C2), third variable (V3), fourth constant-fifth variable (C4-V5), fifth constant (C5) regions of gp120 and fusion protein (FP), immunodominant region (IDR), C-heptad region (CHR), membrane proximal external region (MPER) and C-terminal (CT) of gp41 protein.
For further analysis we took MPER in addition to V3 and IDR peptides (common binding specificities for CNP) based on the binding of AIIMS206 and considering that MPER is recognized by broadly neutralizing antibodies [22], [39]. We tested 80 plasma samples for ELISA binding with the con-C V3 (35 mer), IDR (19 mer) and MPER (24 mer) peptides and observed a high percentage of samples reaching Max50 binding titers with V3 (99%), IDR (95%) as compared to MPER (56%). The median Max50 binding titers were also higher with V3 (8706) and IDR (2382) in comparison to MPER (451) (Figure S1). To determine the possible role of anti-V3, anti-IDR and anti-MPER antibodies in neutralization, we compared the Max50 antibody binding titers specific to these three regions with mean ID50 neutralization titers of each plasma sample with all the viruses tested. We did not find any correlation between the neutralization potential and anti-V3 (p = 0.52), anti-IDR (p = 0.59) and anti-MPER (p = 0.97) antibody binding titers (Figure S2). We also did not find any subtype-B or C specific correlation of neutralization with binding. Nonetheless, we observed a modest positive correlation of MPER specific Max50 binding titers with Du156.12 (a subtype-C virus) (p = 0.04) and a negative correlation with JRFL (a subtype-B virus) (p = 0.02) neutralization titers (Figure S2).
Depletion and competition of V3, IDR and MPER directed plasma antibodies
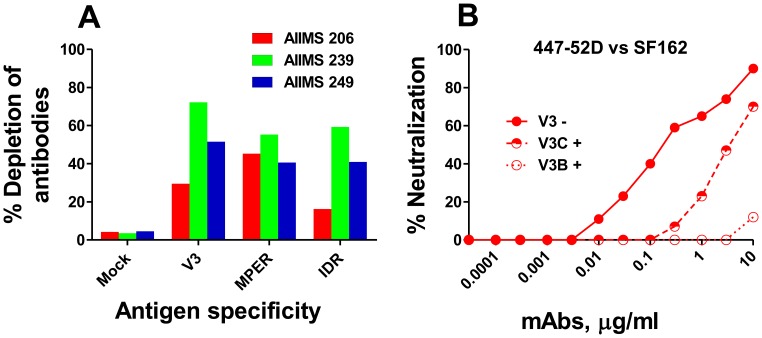
In order to evaluate the participation of anti-V3, IDR and MPER specific antibodies in viral neutralization, we depleted and competed the plasma antibodies from three CNP mentioned above, with peptides specific to these regions. For antibody depletion, we passed the CNP over antigen coated ELISA plates for six passages and observed a substantial removal (16% to 72%) of antigen specific antibodies (Figure 3A). The mock depleted plasma showed minimal antibody loss. For competition, plasma samples were pre-incubated with peptides at a final peptide concentration of 20 µg/ml. Both depleted ‘D’ and competed ‘+’ plasma samples were subsequently tested for neutralization with all eight viruses (Table 3).
Figure 3. Depletion and competition of plasma antibodies from cross-neutralizing plasma.
The depletion and competition of antibodies from three cross-neutralizing plasma (CNP) (AIIMS206, AIIMS239 and AIIMS249) was carried out with V3 (35 mer), MPER (25 mer) and IDR (19 mer) specific peptides. The antibodies from CNP were depleted by passing plasma samples (at 1∶30 dilution) over antigen coated ELISA plates (peptide coating concentration, 10 µg/ml) six times and the percent depletion of antibodies to each region was calculated as: percent depletion = 100−[100×(OD405 at the last passage/OD405 at the first passage)]. The percentage antibody depletion for CNP against each peptide is shown (A). As control, mock depletion of CNP was carried out on the uncoated plates which showed minimal effect on antibody depletion (A). For competition, CNP were preincubated 30 minutes with the same peptides at a final peptide concentration of 20 µg/ml. Both depleted (designated as ‘D’) and competed (designated as ‘+’) plasma were subsequently tested for neutralization with all eight viruses (data shown in Table 3). To validate the competition assay, we tested 447-52D an anti-V3 antibody known to neutralize SF162 (a tier 1 sensitive subtype-B virus) along with two peptides corresponding to consensus-C and B V3 sequences (at a final concentration, 10 µg/ml) (B).
Table 3. Effect of V3, MPER and IDR peptides on ID50 neutralization titers of the cross-neutralizing plasma in depletion and competition experiments.
| aID50 | bID50 fold decrease | ||||||||||||||
| Plasma ID | Virus | Untreated plasma | Mock D | V3 D | V3+ | MPER D | MPER+ | IDR D | ID+ | V3 D | V3+ | MPER D | MPER+ | IDR D | ID+ |
| AIIMS206 | AIIMS201 | >20000 | 10597 | 13143 | >20000 | 8362 | 1370 | 17776 | 10891 | 0.806 | 1 | 1.2 | 14.5 | 0.59 | 1.83 |
| AIIMS212 | 4022 | 3684 | 3965 | 3565 | 4345 | 862 | 3807 | 3123 | 0.929 | 1.12 | 0.84 | 4.66 | 0.96 | 1.28 | |
| Du156.12 | 3106 | 2398 | 2479 | 2172 | 2648 | 1903 | 2172 | 2479 | 0.967 | 1.43 | 0.9 | 1.63 | 1.1 | 1.25 | |
| ZM109F.PB4 | 3243 | 2737 | 2737 | 2924 | 2829 | 3123 | 2280 | 2924 | 1 | 1.1 | 0.96 | 1.03 | 1.2 | 1.1 | |
| ZM53M.PB12 | 17766 | 14259 | 12495 | 11697 | 8982 | 4641 | 7129 | 8670 | 1.14 | 1.51 | 1.58 | 3.82 | 2 | 2.04 | |
| JRFL | 3040 | 2648 | 2829 | 3123 | 3123 | 3448 | 3456 | 3564 | 0.936 | 0.97 | 0.84 | 0.88 | 0.76 | 0.85 | |
| RHPA4259.7 | 3313 | 2829 | 1967 | 1724 | 2320 | 1967 | 2398 | 2400 | 1.43 | 1.92 | 1.21 | 1.68 | 1.17 | 1.38 | |
| TRO.11 | 4199 | 3564 | 2737 | 3228 | 2479 | 1281 | 2737 | 2102 | 1.3 | 1.3 | 1.43 | 3.27 | 1.3 | 1.99 | |
| AIIMS239 | AIIMS201 | 783 | 543 | 508 | 476 | 390 | 476 | 640 | 731 | 1.06 | 1.64 | 1.39 | 1.64 | 0.84 | 1.07 |
| AIIMS212 | 457 | 331 | 353 | 445 | 281 | 150 | 342 | 431 | 0.93 | 1.02 | 1.17 | 3.04 | 0.96 | 1.06 | |
| Du156.12 | 317 | 254 | 230 | 281 | 223 | 101 | 263 | 238 | 1.1 | 1.12 | 1.13 | 3.1 | 0.96 | 1.33 | |
| ZM109F.PB4 | 220 | 230 | 215 | 230 | 108 | 123 | 195 | 246 | 0.92 | 0.95 | 2.12 | 1.78 | 1.17 | 0.89 | |
| ZM53M.PB12 | 1257 | 1324 | 525 | 620 | 119 | 120 | 921 | 755 | 2.52 | 2.02 | 11.12 | 10.47 | 1.43 | 1.66 | |
| JRFL | 245 | 290 | 281 | 223 | 140 | 78 | 290 | 223 | 1.03 | 1.09 | 2.07 | 3.14 | 1 | 1.09 | |
| RHPA4259.7 | 509 | 445 | 320 | 310 | 171 | 111 | 310 | 246 | 1.39 | 1.64 | 2.6 | 4.58 | 1.43 | 2.06 | |
| TRO.11 | 385 | 310 | 286 | 290 | 127 | 104 | 320 | 310 | 1.08 | 1.32 | 2.44 | 3.7 | 0.96 | 1.24 | |
| AIIMS249 | AIIMS201 | 457 | 390 | 476 | 662 | 417 | 525 | 419 | 476 | 0.819 | 0.69 | 0.93 | 0.87 | 0.93 | 0.96 |
| AIIMS212 | 520 | 525 | 640 | 492 | 290 | 230 | 640 | 862 | 0.82 | 1.05 | 1.81 | 2.26 | 0.82 | 0.6 | |
| Du156.12 | 189 | 170 | 166 | 159 | 156 | 106 | 149 | 140 | 1.02 | 1.18 | 1.08 | 1.78 | 1.14 | 1.35 | |
| ZM109F.PB4 | 189 | 223 | 177 | 230 | 171 | 119 | 140 | 160 | 1.25 | 0.82 | 1.3 | 1.58 | 1.59 | 1.18 | |
| ZM53M.PB12 | 369 | 342 | 342 | 340 | 353 | 202 | 353 | 365 | 1 | 1.08 | 0.96 | 1.82 | 0.96 | 1.01 | |
| JRFL | 291 | 281 | 300 | 246 | 246 | 215 | 310 | 271 | 0.936 | 1.18 | 1.14 | 1.35 | 0.906 | 1.07 | |
| RHPA4259.7 | 120 | 111 | 115 | 115 | 123 | 119 | 111 | 108 | 0.965 | 1.04 | 0.9 | 1 | 1 | 1.11 | |
| TRO.11 | 734 | 755 | 707 | 662 | 755 | 599 | 731 | 720 | 1.06 | 1.1 | 1 | 1.22 | 1.03 | 1.01 | |
The table describes the change in ID50 neutralization titers of the three cross-neutralizing plasma CNP (AIIMS206, AIIMS239 and AIIMS249) against 5 subtype-C and 3 subtype-B viruses upon depletion and competition with V3, MPER and IDR specific peptides.
ID50; The numerical values are the ID50 neutralization titers of CNP after depletion (‘D’) (passing plasma over antigen coated ELISA plates @10 µg/ml) and competition ‘+’ (incubating CNP with specific peptides @20 µg/ml) along with untreated and mock (passed CNP on uncoated ELISA plates) controls.
ID50 fold decrease; Plasma ID50 fold decrease for depletion assay was calculated as, ID50 with mock depleted plasma/ID50 with V3, MPER and IDR depleted plasma, and for competition assay as ID50 with untreated plasma/ID50 with V3, MPER and IDR treated plasma. The more than threefold change in ID50 neutralization titer was arbitrarily taken as positive. The CNP AIIMS206 and AIIMS239 showed dependence on MPER (Bold) directed antibodies especially with peptide competition while V3 and IDR showed minimal effect on neutralization as compared to untreated and mock plasma controls.
Based on the fact that multiple epitope specificities contribute to overall neutralization of HIV-1, we possibly may not anticipate a complete removal of neutralizing activity by knocking out antibodies directed to a single antibody specificity, however it may slightly alter the potency. Assuming the above for the analysis, we arbitrarily took a threefold change in the ID50 neutralization titer as positive in the depletion and competition experiments. Based on this criteria, the CNP AIIMS206 and AIIMS239 showed dependence on the MPER directed antibodies with four and six viruses respectively, effect being more prominent with peptide competition (Table 3). The V3 and IDR peptides showed minimal effect on the viral neutralization as compared to untreated and mock depleted plasma controls (Table 3). A partial and complete inhibitory effect on neutralization of SF162 by 447-52D was observed with V3-C and V3-B peptides respectively (Figure 3B).
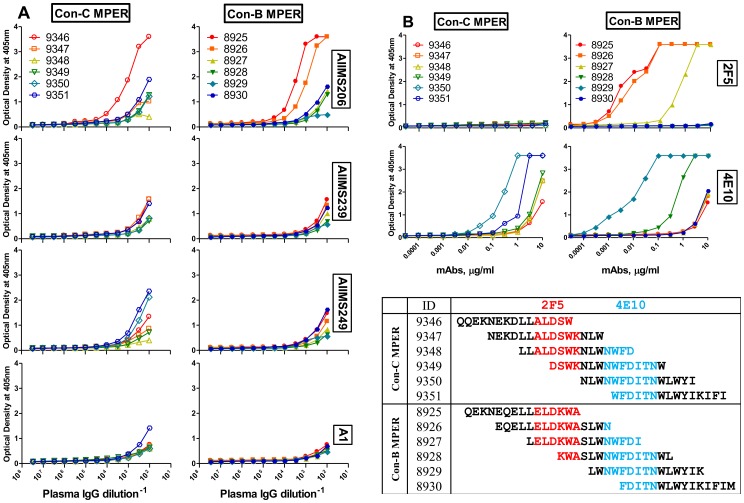
Epitope mapping of IgG fractions from the CNP with overlapping MPER peptides
The functional assessment of two out of three CNP, AIIMS206 and AIIMS239 showed MPER antibody directed neutralization dependence with a few viruses, however the precise target epitope within MPER remained to be identified. In this context, we titrated the IgG fractions of the CNP from a higher starting concentration (five times higher than the original plasma), against a set of six each overlapping peptides corresponding to the con-C and B MPER sequences in an ELISA binding assay. The binding curves of CNP show that AIIMS206 exhibited strong reactivity with peptides of both subtype-C and B, while AIIMS239 and AIIMS249 showed no or weak reactivity (Figure 4A). The binding assay was carried out in presence of two MPER specific mAbs, 2F5 and 4E10, known to be broadly neutralizing. The control antibodies 2F5 and 4E10 whose epitopes within MPER have been previously defined [22], showed specific binding to their respective core epitopes (Figure 4B). Interestingly, the reactivity pattern of the CNP, AIIMS206 defined an epitope within MPER, which was overlapping with the mAb 2F5 epitope. However the binding specificity was slightly different as it reacted with subtype-C peptides while 2F5 did not (Figure 4A–B).
Figure 4. Mapping of IgG fractions with overlapping MPER peptides.
(A) ELISA binding curves of IgG fractions from three cross-neutralizing plasma samples, AIIMS206, AIIMS239, AIIMS249 and a healthy seronegative control plasma A1, with linear consensus-C and consensus-B overlapping MPER peptides. The MPER overlapping peptides were coated at 4 µg/ml and the purified IgG fractions were concentrated five times to the volume that of the original plasma (used for IgG purification) and then reacted with peptides at 12 dilutions. (B) Binding pattern of two MPER specific broadly neutralizing mAbs 2F5 and 4E10 with same MPER peptides, which show a specific binding to their respective core epitopes. The 4B (lower) shows a schematic arrangement of consensus-C and B MPER overlapping peptides and the core epitope previously defined for 2F5 (red) and 4E10 (blue) are indicated. Among the CNP, AIIMS206 exhibited strong reactivity with peptides of both subtype-C and B and its binding region was overlapping with 2F5 epitope.
Discussion
Dissecting the specificities of the anti-HIV-1 neutralizing antibodies will assist in identifying the targets for an HIV-1 subunit vaccine. Broadly cross-neutralizing antibodies against HIV-1 are detected in 5–25% of plasma samples and their specificities are different and still analyzed [54]–[56]. We employed here a systematic approach of binding and neutralization to identify the antigenic targets recognized by HIV-1 specific envelope antibodies from the subtype-C infected individuals from India. We found high antigenicity of V3 and IDR as compared to MPER. However, we did not observe any correlation of neutralization with their reciprocal Max50 binding titers, though functional assessment showed a modest MPER dependent neutralization in two out of three cross neutralizing plasma.
The neutralizing activity of the plasma samples was evaluated against a panel of 3 subtype-B and 5 subtype-C tier 2 viruses. We found that 19–44% plasma samples contained neutralizing antibodies depending upon the virus. The neutralizing activity of three representative plasma samples was shown to be mediated by the IgG fractions, consistent with previous reports [57], [58]. Overall the plasma samples exhibited higher neutralizing capability of subtype-C compared to subtype-B viruses, though a substantial proportion of the plasma samples showed cross-clade neutralization potential. A potential reason for this subtype-C specific activity may be due to the limited envelope diversity in infecting viruses which would help greater sharing of the neutralization determinants [59]. Similar results were previously observed in studies conducted on HIV-1 individuals from South Africa [60], [61], Thailand [62], [63], and France [64]. The property of plasma samples showing a better neutralizing capacity against region specific viruses may in part be attributed to the involvement of the plasma antibodies effective against the lineage specific envelope domains in neutralization [47], [65]. Together these data suggest that neutralizing antibody-based vaccine immunogens might face less problems at the regional level to overcome epitope diversity.
This study was also aimed to identify and characterize the major antigenic targets and their relevance with viral neutralization in subtype-C Indian patients. Based on the immunoreactivity of three CNP (AIIMS206, AIIMS239 and AIIMS249) to linear overlapping envelope peptides, the binding specificities mapped to ten different regions on HIV-1 envelope glycoprotein (five each on gp120 and gp41). These regions have also been previously identified in subtype-B and C viruses as potentially important targets for neutralizing antibodies and serotyping [16], [21], [22]. We focussed here on three regions only (V3, IDR, MPER), however the other regions that were less immunodominant could also possibly function as neutralization epitopes and need to be studied in detail. The MPER is highly conserved target of broadly neutralizing mAbs 2F5, 4E10 and Z13, while V3 is a semiconserved target of cross-neutralizing mAb 447-52D [22], [24] and the IDR is not known as a target for bNAbs though highly antigenic [35], [36].
We observed a high immunodominance of V3 and IDR over MPER based on the relative binding titers. The Max50 binding titers for IDR and V3 were approximately 5 to 15 times respectively higher than MPER. Interestingly, a good proportion (56%) of the patient plasma reached Max50 binding value to MPER peptide which is unexpected due to low immunogenicity of this region [66], [67]. However, this high percentage of plasma binding to MPER should not be confused with the presence of 2F5 and 4E10 like bNAbs which are rare in HIV-1 infected individuals [22], [66], [68]. Since the binding was carried out with whole MPER (25mer) peptide in this study, it is possible that we would have scored the non-neutralizing binding antibodies whose epitopes overlap with the epitopes of bNAbs [69], [70].
We were unable to find any statistical correlation of antibody binding titers directed to V3 and IDR with the neutralizing activity, though high antibody titers were recorded in almost all the infected plasma. The functional assessment of these antibodies (V3 and IDR specific), from three CNP showed a little or no effect on neutralization, suggesting that these antibodies possibly do not contribute to neutralization. The findings are in consensus with studies showing that IDR directed antibodies have little role in HIV-1 neutralization [22]. With respect to the V3, a number previous studies carried out in HIV-1 subtype-A, B and C infected individuals have shown a similar effect, wherein despite the binding capability of plasma antibodies to autologous and heterologous V3 peptides, they fail to display any neutralization [29], [57], [60], [71]. The ubiquitous presence of anti-V3 antibodies could be attributed to recognition of “decoy” V3 epitopes exposed on defective monomeric envelope forms, with masking of V3 on the native envelope trimers possibly affecting the accessibility of these antibodies [72]–[75], thereby preventing neutralizing activity.
One of the salient finding of this study was contribution of MPER specific antibodies in neutralization by two CNP, AIIMS206 and AIIMS239. Although we did not find any correlation of neutralization with MPER specific antibody binding titers except a modest positive and negative correlation with Du156.12 and JRFL respectively. However, the ID50 neutralization titers in the functional assays dropped considerably (up to fourteen fold) in two out of three CNP with a few viruses. Incidentally, the effect on viral neutralization was more prominent with competition than antibody depletion though AIIMS239 showed a uniform trend with ZM53M.PB12 (subtype-C) virus. This biased effect could not be explained, however a possible explanation is that inhibition neutralization assays were performed in solution, and therefore, it is likely that all the specific antibodies will be competed better by MPER peptides in solution compared to those adsorbed on the coated plates (in depletion) which might present the MPER peptide in a slightly different conformation. Interestingly, studies have shown that, in solution, linear peptides can efficiently inhibit binding of V3 mAbs to native gp120 proteins [76], [77].
Remarkably, the neutralization dependence on the MPER region in these two plasma samples did not show a clade specific inhibitory effect, rather viruses from both the subtype-B and C got affected. This finding is interesting because the plasma antibodies were competed by the same MPER peptide, which implicates the conservation of cross clade binding targets within the MPER, as have been previously appreciated [22], [39], [78], [79]. It is also noteworthy that three of the viruses affected by two competed plasma were common, while others were different, suggesting that the specificities of MPER directed antibodies in these two plasma samples would be possibly similar with some differences. However, the mapping of the IgG fraction from CNP with con-C and B MPER overlapping peptides did not show any reactivity for AIIMS239, possibly due to its very low antibody titers. Nonetheless, the CNP AIIMS206 showed binding to peptides containing 2F5 epitope ELDKWA in con-B MPER and also reacted to its con-C counterpart to which mAb 2F5 failed to bind, since it lacks a complete 2F5 epitope. Taken together, we observed MPER reactive antibodies in the CNP AIIMS206 whose epitopes overlap with mAb 2F5, however displays slightly different specificity. Indeed, a number of groups have recently shown that anti-MPER antibodies are found in samples with neutralization breadth that in some cases were identified as 2F5-like [79], or 4E10-like, Z13-like [7], [80]. Moreover, we were unable to completely remove the neutralizing activity from any of the three CNP sample, which suggests that MPER may not solely contribute towards the viral neutralization, instead the possibility that antibodies specific for oligomeric forms of envelope (quaternary epitopes) may define this activity [81]–[85].
Overall, our data reveals that broadly cross-neutralizing Abs can be detected in approximately 19–27% of the plasma samples derived from subtype-C HIV-1 infected Indian patients. The neutralization profiles of the plasma will allow the identification of HIV-1 donors for the production of neutralizing mAbs and also will help to guide the design of new vaccine immunogens. We explore here for the first time the antigencity of major immunogenic sites on HIV-1 infecting Indian patients and our results suggest the importance of MPER directed antibodies for HIV-1 neutralization.
Supporting Information
Relative reactivity of plasma antibodies form HIV-1 infected individuals against V3, MPER and IDR peptides. Relative anti-V3, anti-MPER and anti-IDR antibody titers in 80 HIV-1 infected drug naive patients. The plasma were reacted with V3 loop (35 mer), MPER (24 mer) and IDR (19 mer) peptides at six dilutions (dilution range: 300 to 100000) in an ELISA binding assay. The colour bars represent the reciprocal 50% binding (Max50) titers against V3 (red), MPER (green) and IDR (blue) regions. The Max50 binding titers were calculated by least square regression method using Graphpad Prism 5.
(TIF)
Association of Neutralization by plasma antibodies with Max50 ELISA binding titers to major antigenic regions on HIV-1 envelope. The reciprocal mean ID50 neutralization titers of all the tested viruses (black), subtype-C (gold) and subtype-B (aqua) viruses and were compared by spearman rank correlation with Max50 ELISA binding titers to third variable region (V3: red), membrane proximal external region (MPER: green) and immunodominant region (IDR: blue) of envelope glycoprotein gp160. Also the same statistical test was used to compare the Max50 binding values and mean neutralization titers of individual viral isolates. The analysis was done with 80 HIV-1 plasma samples and the p-values are given for each category.
(TIF)
Demographic and clinical data of 80 HIV-1 infected drug naive patients recruited for the study.
(DOC)
The neutralizing activity of broadly neutralizing antibodies against viruses tested in this study. The neutralizing activity of broadly neutralizing monoclonal antibodies bNAbs (indicated on the top) was assessed against the six reference subtype_B and C and two new subtype_C viruses (left), using the TZM-bl cell assay. The mAb 1418 specific to parvovirus B19 protein was used as negative control in neutralization assay. The numerical values below the mAbs represent IC50 neutralization titers (which is the amount of mAbs (µg/ml) needed for 50% neutralization) against each virus. The IC50 values which are shown in each cell are in coded: IC50<1 µg/ml (Bold); IC50, 1–30 µg/ml (Italic); IC50>30 indicates that IC50 was not achieved. The two new isolates (AIIMS201 and AIIMS212) showed resistance to neutralization by bNAbs and were assigned as tier 2 viruses in this study.
(DOC)
A. Epitope mapping of polyclonal antibodies from cross-neutralizing plasma (CNP) with overlapping linear peptides corresponding to HIV-1 consensus-C gp120. The polyclonal antibodies from three CNPs (AIIMS206, AIIMS239 and AIIMS249) and two seronegative healthy donors (A1 and A2) were reacted with 15 mer linear overlapping peptides (11 amino acid overlap) corresponding to the HIV-1 consensus-C gp120 amino acid sequence (sequences for each peptide are provided on the left of table), at four dilutions (dilution range: 100 to 3000) in an ELISA binding assay. The numerical values in boxes are the reciprocal Max50 binding titers, calculated by Graphpad Prism 5 using least square regression method. The information is coded: (Bold) Max50>1000, (Italic) Max50 = 101–1000 and unfilled Max50<100 indicates the Max50 was not achieved. B. Epitope mapping of polyclonal antibodies from cross-neutralizing plasma (CNP) with overlapping linear peptides corresponding to HIV-1 consensus-C gp41. The polyclonal antibodies from three CNPs (AIIMS206, AIIMS239 and AIIMS249) and two seronegative healthy donors (A1 and A2) were reacted with 15 mer linear overlapping peptides (11 amino acid overlap) corresponding to the HIV-1 consensus-C gp41 amino acid sequence (sequences for each peptide are provided on the left of table), at four dilutions (dilution range: 100 to 3000) in an ELISA binding assay. The numerical values in boxes are the reciprocal Max50 binding titers, calculated by Graphpad Prism 5 using least square regression method. The information is coded: (Bold) Max50>1000, (Italic) Max50 = 101–1000 and unfilled Max50<100 indicates the Max50 was not achieved.
(DOC)
Acknowledgments
We profoundly thank all the study participants. We acknowledge Prof. Miroslaw K. Gorny for critically reviewing the manuscript and Prof. Susan Zolla Pazner for constant technical advice and support. The Fogarty AIDS International Training & Research Program (AITRP) fellowship (USA), RF/SRF fellowship provided by Indian Council of Medical Research (ICMR) to Raiees Andrabi is highly acknowledged.
Funding Statement
This work was funded by Department of Biotechnology India (DBT) (BT/PR 10511/MED/29/66/2008) and Indian Council of Medical Research (ICMR) (61/7/2008-BMS). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Hemelaar J, Gouws E, Ghys PD, Osmanov S (2006) Global and regional distribution of HIV-1 genetic subtypes and recombinants in 2004. AIDS 20: W13–23 doi:10.1097/01.aids.0000247564.73009.bc. [DOI] [PubMed] [Google Scholar]
- 2. Taylor BS, Sobieszczyk ME, McCutchan FE, Hammer SM (2008) The challenge of HIV-1 subtype diversity. N Engl J Med 358: 1590–1602 doi:10.1056/NEJMra0706737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lynch RM, Shen T, Gnanakaran S, Derdeyn CA (2009) Appreciating HIV type 1 diversity: subtype differences in Env. AIDS Res Hum Retroviruses 25: 237–248 doi:10.1089/aid.2008.0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liu P, Overman RG, Yates NL, Alam SM, Vandergrift N, et al. (2011) Dynamic antibody specificities and virion concentrations in circulating immune complexes in acute to chronic HIV-1 infection. J Virol 85: 11196–11207 doi:10.1128/JVI.05601-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Haynes BF, Montefiori DC (2006) Aiming to induce broadly reactive neutralizing antibody responses with HIV-1 vaccine candidates. Expert Rev Vaccines 5: 347–363 doi:10.1586/14760584.5.3.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Parren PW, Gauduin MC, Koup RA, Poignard P, Fisicaro P, et al. (1997) Relevance of the antibody response against human immunodeficiency virus type 1 envelope to vaccine design. Immunol Lett 57: 105–112. [DOI] [PubMed] [Google Scholar]
- 7. Sather DN, Armann J, Ching LK, Mavrantoni A, Sellhorn G, et al. (2009) Factors associated with the development of cross-reactive neutralizing antibodies during human immunodeficiency virus type 1 infection. J Virol 83: 757–769 doi:10.1128/JVI.02036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Burton DR, Desrosiers RC, Doms RW, Koff WC, Kwong PD, et al. (2004) HIV vaccine design and the neutralizing antibody problem. Nat Immunol 5: 233–236 doi:10.1038/ni0304-233. [DOI] [PubMed] [Google Scholar]
- 9. McMichael AJ, Hanke T (2003) HIV vaccines 1983–2003. Nat Med 9: 874–880 doi:10.1038/nm0703-874. [DOI] [PubMed] [Google Scholar]
- 10. Koff WC (2011) HIV vaccine development: Challenges and opportunities towards solving the HIV vaccine-neutralizing antibody problem. Vaccine Available: http://www.ncbi.nlm.nih.gov/pubmed/22100891. Accessed 2012 January 28. [DOI] [PubMed] [Google Scholar]
- 11. Kim JH, Rerks-Ngarm S, Excler J-L, Michael NL (2010) HIV vaccines: lessons learned and the way forward. Curr Opin HIV AIDS 5: 428–434 doi:10.1097/COH.0b013e32833d17ac. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, et al. (2009) Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med 361: 2209–2220 doi:10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 13. Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, et al. (2012) Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med 366: 1275–1286 doi:10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Corti D, Langedijk JPM, Hinz A, Seaman MS, Vanzetta F, et al. (2010) Analysis of memory B cell responses and isolation of novel monoclonal antibodies with neutralizing breadth from HIV-1-infected individuals. PLoS ONE 5: e8805 doi:10.1371/journal.pone.0008805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lynch RM, Tran L, Louder MK, Schmidt SD, Cohen M, et al. (2012) The Development of CD4 Binding Site Antibodies During HIV-1 Infection. Journal of virology Available: http://www.ncbi.nlm.nih.gov/pubmed/22573869. Accessed 2012 May 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mascola JR, Montefiori DC (2010) The role of antibodies in HIV vaccines. Annu Rev Immunol 28: 413–444 doi:10.1146/annurev-immunol-030409-101256. [DOI] [PubMed] [Google Scholar]
- 17. Stamatatos L, Morris L, Burton DR, Mascola JR (2009) Neutralizing antibodies generated during natural HIV-1 infection: good news for an HIV-1 vaccine? Nat Med 15: 866–870 doi:10.1038/nm.1949. [DOI] [PubMed] [Google Scholar]
- 18. Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, et al. (2011) Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477: 466–470 doi:10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dalgleish AG, Beverley PC, Clapham PR, Crawford DH, Greaves MF, et al. (1984) The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature 312: 763–767. [DOI] [PubMed] [Google Scholar]
- 20. Wyatt R, Sodroski J (1998) The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280: 1884–1888. [DOI] [PubMed] [Google Scholar]
- 21. Pantophlet R, Burton DR (2006) GP120: target for neutralizing HIV-1 antibodies. Annu Rev Immunol 24: 739–769 doi:10.1146/annurev.immunol.24.021605.090557. [DOI] [PubMed] [Google Scholar]
- 22. Montero M, van Houten NE, Wang X, Scott JK (2008) The membrane-proximal external region of the human immunodeficiency virus type 1 envelope: dominant site of antibody neutralization and target for vaccine design. Microbiol Mol Biol Rev 72: 54–84 table of contents. doi:10.1128/MMBR.00020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li Y, Migueles SA, Welcher B, Svehla K, Phogat A, et al. (2007) Broad HIV-1 neutralization mediated by CD4-binding site antibodies. Nat Med 13: 1032–1034 doi:10.1038/nm1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zolla-Pazner S, Cohen SS, Krachmarov C, Wang S, Pinter A, et al. (2008) Focusing the immune response on the V3 loop, a neutralizing epitope of the HIV-1 gp120 envelope. Virology 372: 233–246 doi:10.1016/j.virol.2007.09.024. [DOI] [PubMed] [Google Scholar]
- 25. Hioe CE, Wrin T, Seaman MS, Yu X, Wood B, et al. (2010) Anti-V3 monoclonal antibodies display broad neutralizing activities against multiple HIV-1 subtypes. PLoS ONE 5: e10254 doi:10.1371/journal.pone.0010254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wu X, Yang Z-Y, Li Y, Hogerkorp C-M, Schief WR, et al. (2010) Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329: 856–861 doi:10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Balla-Jhagjhoorsingh SS, Willems B, Heyndrickx L, Heyndrickx L, Vereecken K, et al. (2011) Characterization of neutralizing profiles in HIV-1 infected patients from whom the HJ16, HGN194 and HK20 mAbs were obtained. PLoS ONE 6: e25488 doi:10.1371/journal.pone.0025488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, et al. (1996) Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J Virol 70: 1100–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Moore PL, Gray ES, Choge IA, Ranchobe N, Mlisana K, et al. (2008) The c3-v4 region is a major target of autologous neutralizing antibodies in human immunodeficiency virus type 1 subtype C infection. J Virol 82: 1860–1869 doi:10.1128/JVI.02187-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Walker LM, Phogat SK, Chan-Hui P-Y, Wagner D, Phung P, et al. (2009) Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326: 285–289 doi:10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. McLellan JS, Pancera M, Carrico C, Gorman J, Julien J-P, et al. (2011) Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature 480: 336–343 doi:10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gabuzda DH, Lever A, Terwilliger E, Sodroski J (1992) Effects of deletions in the cytoplasmic domain on biological functions of human immunodeficiency virus type 1 envelope glycoproteins. J Virol 66: 3306–3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xu JY, Gorny MK, Palker T, Karwowska S, Zolla-Pazner S (1991) Epitope mapping of two immunodominant domains of gp41, the transmembrane protein of human immunodeficiency virus type 1, using ten human monoclonal antibodies. J Virol 65: 4832–4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Poumbourios P, McPhee DA, Kemp BE (1992) Antibody epitopes sensitive to the state of human immunodeficiency virus type 1 gp41 oligomerization map to a putative alpha-helical region. AIDS Res Hum Retroviruses 8: 2055–2062. [DOI] [PubMed] [Google Scholar]
- 35. Chiodi F, von Gegerfeldt A, Albert J, Fenyö EM, Gaines H, et al. (1987) Site-directed ELISA with synthetic peptides representing the HIV transmembrane glycoprotein. J Med Virol 23: 1–9. [DOI] [PubMed] [Google Scholar]
- 36. Gnann JW Jr, Nelson JA, Oldstone MB (1987) Fine mapping of an immunodominant domain in the transmembrane glycoprotein of human immunodeficiency virus. J Virol 61: 2639–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Muster T, Steindl F, Purtscher M, Trkola A, Klima A, et al. (1993) A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J Virol 67: 6642–6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Buchacher A, Predl R, Strutzenberger K, Steinfellner W, Trkola A, et al. (1994) Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res Hum Retroviruses 10: 359–369. [DOI] [PubMed] [Google Scholar]
- 39. Zwick MB, Labrijn AF, Wang M, Spenlehauer C, Saphire EO, et al. (2001) Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J Virol 75: 10892–10905 doi:10.1128/JVI.75.22.10892-10905.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cleveland SM, Buratti E, Jones TD, North P, Baralle F, et al. (2000) Immunogenic and antigenic dominance of a nonneutralizing epitope over a highly conserved neutralizing epitope in the gp41 envelope glycoprotein of human immunodeficiency virus type 1: its deletion leads to a strong neutralizing response. Virology 266: 66–78 doi:10.1006/viro.1999.0041. [DOI] [PubMed] [Google Scholar]
- 41. Gray ES, Moore PL, Choge IA, Decker JM, Bibollet-Ruche F, et al. (2007) Neutralizing antibody responses in acute human immunodeficiency virus type 1 subtype C infection. J Virol 81: 6187–6196 doi:10.1128/JVI.00239-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gray ES, Taylor N, Wycuff D, Moore PL, Tomaras GD, et al. (2009) Antibody specificities associated with neutralization breadth in plasma from human immunodeficiency virus type 1 subtype C-infected blood donors. J Virol 83: 8925–8937 doi:10.1128/JVI.00758-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li B, Decker JM, Johnson RW, Bibollet-Ruche F, Wei X, et al. (2006) Evidence for potent autologous neutralizing antibody titers and compact envelopes in early infection with subtype C human immunodeficiency virus type 1. J Virol 80: 5211–5218 doi:10.1128/JVI.00201-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rong R, Li B, Lynch RM, Haaland RE, Murphy MK, et al. (2009) Escape from autologous neutralizing antibodies in acute/early subtype C HIV-1 infection requires multiple pathways. PLoS Pathog 5: e1000594 doi:10.1371/journal.ppat.1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shankarappa R, Chatterjee R, Learn GH, Neogi D, Ding M, et al. (2001) Human immunodeficiency virus type 1 env sequences from Calcutta in eastern India: identification of features that distinguish subtype C sequences in India from other subtype C sequences. J Virol 75: 10479–10487 doi:10.1128/JVI.75.21.10479-10487.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jere A, Tripathy S, Agnihotri K, Jadhav S, Paranjape R (2004) Genetic analysis of Indian HIV-1 nef: subtyping, variability and implications. Microbes Infect 6: 279–289 doi:10.1016/j.micinf.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 47. Kulkarni SS, Lapedes A, Tang H, Gnanakaran S, Daniels MG, et al. (2009) Highly complex neutralization determinants on a monophyletic lineage of newly transmitted subtype C HIV-1 Env clones from India. Virology 385: 505–520 doi:10.1016/j.virol.2008.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lakhashe S, Thakar M, Godbole S, Tripathy S, Paranjape R (2008) HIV infection in India: epidemiology, molecular epidemiology and pathogenesis. J Biosci 33: 515–525. [DOI] [PubMed] [Google Scholar]
- 49. Spenlehauer C, Saragosti S, Fleury HJ, Kirn A, Aubertin AM, et al. (1998) Study of the V3 loop as a target epitope for antibodies involved in the neutralization of primary isolates versus T-cell-line-adapted strains of human immunodeficiency virus type 1. J Virol 72: 9855–9864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mascola JR, Louder MK, Winter C, Prabhakara R, De Rosa SC, et al. (2002) Human immunodeficiency virus type 1 neutralization measured by flow cytometric quantitation of single-round infection of primary human T cells. J Virol 76: 4810–4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li M, Gao F, Mascola John R, Stamatatos L, Polonis VR, et al. (2005) Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol 79: 10108–10125 doi:10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Seaman MS, Janes H, Hawkins N, Grandpre LE, Devoy C, et al. (2010) Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J Virol 84: 1439–1452 doi:10.1128/JVI.02108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Montefiori DC (2005) Evaluating neutralizing antibodies against HIV, SIV, and SHIV in luciferase reporter gene assays. Curr Protoc Immunol Chapter 12: Unit 12.11 doi:10.1002/0471142735.im1211s64. [DOI] [PubMed] [Google Scholar]
- 54. Walker LM, Simek MD, Priddy F, Gach JS, Wagner D, et al. (2010) A limited number of antibody specificities mediate broad and potent serum neutralization in selected HIV-1 infected individuals. PLoS Pathog 6: e1001028 doi:10.1371/journal.ppat.1001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mikell I, Sather DN, Kalams SA, Altfeld M, Alter G, et al. (2011) Characteristics of the earliest cross-neutralizing antibody response to HIV-1. PLoS Pathog 7: e1001251 doi:10.1371/journal.ppat.1001251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sather DN, Stamatatos L (2010) Epitope specificities of broadly neutralizing plasmas from HIV-1 infected subjects. Vaccine 28 Suppl 2: B8–12 doi:10.1016/j.vaccine.2009.07.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dhillon AK, Donners H, Pantophlet R, Johnson WE, Decker JM, et al. (2007) Dissecting the neutralizing antibody specificities of broadly neutralizing sera from human immunodeficiency virus type 1-infected donors. J Virol 81: 6548–6562 doi:10.1128/JVI.02749-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Li Y, Svehla K, Louder MK, Wycuff D, Phogat S, et al. (2009) Analysis of neutralization specificities in polyclonal sera derived from human immunodeficiency virus type 1-infected individuals. J Virol 83: 1045–1059 doi:10.1128/JVI.01992-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lakhashe SK, Kulkarni SS, Thakar MR, Ghate MV, Paranjape RS (2007) Extensive cross-reactive neutralizing antibody response in Indian patients with limited genetic diversity of HIV-1. Virology 359: 295–301 doi:10.1016/j.virol.2006.09.019. [DOI] [PubMed] [Google Scholar]
- 60. Bures R, Morris L, Williamson C, Ramjee G, Deers M, et al. (2002) Regional clustering of shared neutralization determinants on primary isolates of clade C human immunodeficiency virus type 1 from South Africa. J Virol 76: 2233–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rademeyer C, Moore PL, Taylor N, Martin DP, Choge IA, et al. (2007) Genetic characteristics of HIV-1 subtype C envelopes inducing cross-neutralizing antibodies. Virology 368: 172–181 doi:10.1016/j.virol.2007.06.013. [DOI] [PubMed] [Google Scholar]
- 62. Mascola JR, Louder MK, Surman SR, Vancott TC, Yu XF, et al. (1996) Human immunodeficiency virus type 1 neutralizing antibody serotyping using serum pools and an infectivity reduction assay. AIDS Res Hum Retroviruses 12: 1319–1328. [DOI] [PubMed] [Google Scholar]
- 63. Louisirirotchanakul S, Beddows S, Cheingsong-Popov R, Shaffer N, Mastro TD, et al. (1998) Characterization of sera from subjects infected with HIV-1 subtypes B and E in Thailand by antibody binding and neutralization. J Acquir Immune Defic Syndr Hum Retrovirol 19: 315–320. [DOI] [PubMed] [Google Scholar]
- 64. Braibant M, Brunet S, Costagliola D, Rouzioux C, Agut H, et al. (2006) Antibodies to conserved epitopes of the HIV-1 envelope in sera from long-term non-progressors: prevalence and association with neutralizing activity. AIDS 20: 1923–1930 doi:10.1097/01.aids.0000247113.43714.5e. [DOI] [PubMed] [Google Scholar]
- 65. Binley JM, Wrin T, Korber B, Zwick MB, Wang M, et al. (2004) Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J Virol 78: 13232–13252 doi:10.1128/JVI.78.23.13232-13252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Haynes BF, Fleming J, St Clair EW, Katinger H, Stiegler G, et al. (2005) Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science 308: 1906–1908 doi:10.1126/science.1111781. [DOI] [PubMed] [Google Scholar]
- 67. Andrabi R, Choudhary AK, Bala M, Kalra R, Prakash SS, et al. (2011) Relative reactivity of HIV-1 polyclonal plasma antibodies directed to V3 and MPER regions suggests immunodominance of V3 over MPER and dependence of high anti-V3 antibody titers on virus persistence. Arch Virol 156: 1787–1794 doi:10.1007/s00705-011-1053-5. [DOI] [PubMed] [Google Scholar]
- 68. Alam SM, Scearce RM, Parks RJ, Plonk K, Plonk SG, et al. (2008) Human immunodeficiency virus type 1 gp41 antibodies that mask membrane proximal region epitopes: antibody binding kinetics, induction, and potential for regulation in acute infection. J Virol 82: 115–125 doi:10.1128/JVI.00927-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Frey G, Chen J, Rits-Volloch S, Freeman MM, Zolla-Pazner S, et al. (2010) Distinct conformational states of HIV-1 gp41 are recognized by neutralizing and non-neutralizing antibodies. Nat Struct Mol Biol 17: 1486–1491 doi:10.1038/nsmb.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Nicely NI, Dennison SM, Spicer L, Scearce RM, Kelsoe G, et al. (2010) Crystal structure of a non-neutralizing antibody to the HIV-1 gp41 membrane-proximal external region. Nat Struct Mol Biol 17: 1492–1494 doi:10.1038/nsmb.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Choudhary AK, Andrabi R, Prakash SS, Kumar R, Choudhury SD, et al. (2012) Neutralization potential of the plasma of HIV-1 infected Indian patients in the context of anti-V3 antibody content and antiretroviral theraphy. J Microbiol 50: 149–154 doi:10.1007/s12275-012-1246-y. [DOI] [PubMed] [Google Scholar]
- 72. Pinter A, Honnen WJ, He Y, Gorny MK, Zolla-Pazner S, et al. (2004) The V1/V2 domain of gp120 is a global regulator of the sensitivity of primary human immunodeficiency virus type 1 isolates to neutralization by antibodies commonly induced upon infection. J Virol 78: 5205–5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Krachmarov C, Pinter A, Honnen WJ, Gorny MK, Nyambi PN, et al. (2005) Antibodies that are cross-reactive for human immunodeficiency virus type 1 clade a and clade B v3 domains are common in patient sera from Cameroon, but their neutralization activity is usually restricted by epitope masking. J Virol 79: 780–790 doi:10.1128/JVI.79.2.780-790.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Moore PL, Crooks ET, Porter L, Zhu P, Cayanan CS, et al. (2006) Nature of nonfunctional envelope proteins on the surface of human immunodeficiency virus type 1. J Virol 80: 2515–2528 doi:10.1128/JVI.80.5.2515-2528.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Davis KL, Gray ES, Moore PL, Decker JM, Salomon A, et al. (2009) High titer HIV-1 V3-specific antibodies with broad reactivity but low neutralizing potency in acute infection and following vaccination. Virology 387: 414–426 doi:10.1016/j.virol.2009.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Moore JP, Ho DD (1993) Antibodies to discontinuous or conformationally sensitive epitopes on the gp120 glycoprotein of human immunodeficiency virus type 1 are highly prevalent in sera of infected humans. J Virol 67: 863–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Moore JP, Cao Y, Ho DD, Koup RA (1994) Development of the anti-gp120 antibody response during seroconversion to human immunodeficiency virus type 1. J Virol 68: 5142–5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Zwick MB, Jensen R, Church S, Wang M, Stiegler G, et al. (2005) Anti-human immunodeficiency virus type 1 (HIV-1) antibodies 2F5 and 4E10 require surprisingly few crucial residues in the membrane-proximal external region of glycoprotein gp41 to neutralize HIV-1. J Virol 79: 1252–1261 doi:10.1128/JVI.79.2.1252-1261.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Shen X, Parks RJ, Montefiori DC, Kirchherr JL, Keele BF, et al. (2009) In vivo gp41 antibodies targeting the 2F5 monoclonal antibody epitope mediate human immunodeficiency virus type 1 neutralization breadth. J Virol 83: 3617–3625 doi:10.1128/JVI.02631-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Binley JM, Lybarger EA, Crooks ET, Seaman MS, Gray E, et al. (2008) Profiling the specificity of neutralizing antibodies in a large panel of plasmas from patients chronically infected with human immunodeficiency virus type 1 subtypes B and C. J Virol 82: 11651–11668 doi:10.1128/JVI.01762-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Pancera M, McLellan JS, Wu X, Zhu J, Changela A, et al. (2010) Crystal structure of PG16 and chimeric dissection with somatically related PG9: structure-function analysis of two quaternary-specific antibodies that effectively neutralize HIV-1. J Virol 84: 8098–8110 doi:10.1128/JVI.00966-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Changela A, Wu X, Yang Y, Zhang B, Zhu J, et al. (2011) Crystal structure of human antibody 2909 reveals conserved features of quaternary structure-specific antibodies that potently neutralize HIV-1. J Virol 85: 2524–2535 doi:10.1128/JVI.02335-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Pejchal R, Doores KJ, Walker LM, Khayat R, Huang P-S, et al. (2011) A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science 334: 1097–1103 doi:10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Spurrier B, Sampson JM, Totrov M, Li H, O'Neal T, et al. (2011) Structural analysis of human and macaque mAbs 2909 and 2.5B: implications for the configuration of the quaternary neutralizing epitope of HIV-1 gp120. Structure 19: 691–699 doi:10.1016/j.str.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Wu X, Changela A, O'Dell S, Schmidt SD, Pancera M, et al. (2011) Immunotypes of a quaternary site of HIV-1 vulnerability and their recognition by antibodies. J Virol 85: 4578–4585 doi:10.1128/JVI.02585-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Relative reactivity of plasma antibodies form HIV-1 infected individuals against V3, MPER and IDR peptides. Relative anti-V3, anti-MPER and anti-IDR antibody titers in 80 HIV-1 infected drug naive patients. The plasma were reacted with V3 loop (35 mer), MPER (24 mer) and IDR (19 mer) peptides at six dilutions (dilution range: 300 to 100000) in an ELISA binding assay. The colour bars represent the reciprocal 50% binding (Max50) titers against V3 (red), MPER (green) and IDR (blue) regions. The Max50 binding titers were calculated by least square regression method using Graphpad Prism 5.
(TIF)
Association of Neutralization by plasma antibodies with Max50 ELISA binding titers to major antigenic regions on HIV-1 envelope. The reciprocal mean ID50 neutralization titers of all the tested viruses (black), subtype-C (gold) and subtype-B (aqua) viruses and were compared by spearman rank correlation with Max50 ELISA binding titers to third variable region (V3: red), membrane proximal external region (MPER: green) and immunodominant region (IDR: blue) of envelope glycoprotein gp160. Also the same statistical test was used to compare the Max50 binding values and mean neutralization titers of individual viral isolates. The analysis was done with 80 HIV-1 plasma samples and the p-values are given for each category.
(TIF)
Demographic and clinical data of 80 HIV-1 infected drug naive patients recruited for the study.
(DOC)
The neutralizing activity of broadly neutralizing antibodies against viruses tested in this study. The neutralizing activity of broadly neutralizing monoclonal antibodies bNAbs (indicated on the top) was assessed against the six reference subtype_B and C and two new subtype_C viruses (left), using the TZM-bl cell assay. The mAb 1418 specific to parvovirus B19 protein was used as negative control in neutralization assay. The numerical values below the mAbs represent IC50 neutralization titers (which is the amount of mAbs (µg/ml) needed for 50% neutralization) against each virus. The IC50 values which are shown in each cell are in coded: IC50<1 µg/ml (Bold); IC50, 1–30 µg/ml (Italic); IC50>30 indicates that IC50 was not achieved. The two new isolates (AIIMS201 and AIIMS212) showed resistance to neutralization by bNAbs and were assigned as tier 2 viruses in this study.
(DOC)
A. Epitope mapping of polyclonal antibodies from cross-neutralizing plasma (CNP) with overlapping linear peptides corresponding to HIV-1 consensus-C gp120. The polyclonal antibodies from three CNPs (AIIMS206, AIIMS239 and AIIMS249) and two seronegative healthy donors (A1 and A2) were reacted with 15 mer linear overlapping peptides (11 amino acid overlap) corresponding to the HIV-1 consensus-C gp120 amino acid sequence (sequences for each peptide are provided on the left of table), at four dilutions (dilution range: 100 to 3000) in an ELISA binding assay. The numerical values in boxes are the reciprocal Max50 binding titers, calculated by Graphpad Prism 5 using least square regression method. The information is coded: (Bold) Max50>1000, (Italic) Max50 = 101–1000 and unfilled Max50<100 indicates the Max50 was not achieved. B. Epitope mapping of polyclonal antibodies from cross-neutralizing plasma (CNP) with overlapping linear peptides corresponding to HIV-1 consensus-C gp41. The polyclonal antibodies from three CNPs (AIIMS206, AIIMS239 and AIIMS249) and two seronegative healthy donors (A1 and A2) were reacted with 15 mer linear overlapping peptides (11 amino acid overlap) corresponding to the HIV-1 consensus-C gp41 amino acid sequence (sequences for each peptide are provided on the left of table), at four dilutions (dilution range: 100 to 3000) in an ELISA binding assay. The numerical values in boxes are the reciprocal Max50 binding titers, calculated by Graphpad Prism 5 using least square regression method. The information is coded: (Bold) Max50>1000, (Italic) Max50 = 101–1000 and unfilled Max50<100 indicates the Max50 was not achieved.
(DOC)