Abstract
DNA sequence analysis dictates new interpretation of phylogenic trees. Taxa that were once thought to represent successive grades of complexity at the base of the metazoan tree are being displaced to much higher positions inside the tree. This leaves no evolutionary “intermediates” and forces us to rethink the genesis of bilaterian complexity.
A deep reorganization of the metazoan phylogenetic tree is presently taking place as a result of the input of molecular data. Far from being an exercise confined to a small circle of aficionados, the changing views on the pattern of animal interrelationships has profound consequences for understanding the underlying processes of animal diversification. As has repeatedly been stressed, we shall never be able to reason on the evolution of development and the way it has shaped animal diversity unless we have a reliable history of the path taken by this diversification. Here, we highlight the salient recent results based on genetic data, especially the displacement of taxa long thought to represent successive grades of complexity at the base of the metazoan tree, to much higher positions inside the tree. This leaves us with no evolutionary “intermediates” and forces us to rethink the genesis of bilaterian complexity. The reappraisal of animal evolution rests on several congruent approaches ranging from primary gene sequence analysis to qualitative molecular signatures within appropriate genes. Each of them, however, has its methodological difficulties; we shall, therefore, also try to briefly pinpoint the issues of contention and discuss the strength of the present view.
Preliminary Comments on the Reliability of Phylogenetic Trees.
To an outsider, the field of phylogenetic reconstruction may appear to be full of controversies and uncertainties. There have been acrimonious debates over the best methodology to use for reconstruction (phenetics vs. cladistics) and over the relative merits of morphological vs. molecular data. Worst of all, contradictory trees have kept pouring in, often with insufficient critical assessment. Obviously, we cannot review the whole field here, but we wish to emphasize three points to justify our reasoned optimism and confidence in the recent molecular phylogenies.
(i) Tree Reconstruction Has Improved.
Not only has the amount of molecular data increased exponentially, but we have become much more aware of the various difficulties and artifacts of phylogeny reconstruction (1, 2).
Among the artifacts plaguing molecular phylogenies, mutational saturation and unequal rates of evolution between homologous sequences are the most pervasive. Their combined effects are disastrous (3). This is the well studied long-branch attraction artifact. Its effects are all of the more pervasive than sequences are more saturated (i.e., they have accumulated multiple substitutions at certain nucleotide positions). Fortunately, there are now ways of identifying saturation as well as long branching taxa (3), which allow one to discard the most problematic data. In addition, a set of methods is emerging that aims at extracting the most meaningful information from sequences rather than pooling all substitutions (3).
Additionally, in recent years, next to primary genetic sequences, we have witnessed the emergence of a new type of genetic evidence, more qualitative in nature. These consist in genomic rearrangements, such as those in the mitochondrial genome (4) and transposition events of SINEs and LINEs in nuclear genomes (5). They are almost totally immune from homoplasy and therefore constitute very powerful “signatures” for kinship when they are found in the same arrangement in two taxa. Their main drawback is that, contrary to primary sequence, which can always be obtained, one has to rely on chance for these qualitative events to have occurred among the taxa under study!
It should be stressed that, although the set of difficulties just listed pertain to the quality of single datasets, phylogeneticists are often confronted with what seems to be difficulties related to the evolutionary process itself and that therefore can plague all datasets relative to a given problem. Of these, the most serious is “adaptive radiation.” Such a rapid splitting of lineages appears to have occurred repeatedly during evolution, and it renders reconstruction of the order of splits very difficult even with large amounts of sequence data. This emphasizes the notion of limit of resolution of a given data set. A molecule, such as rRNA, is not a “good” or “bad” phylogenetic indicator in itself. Its efficiency is to be evaluated against a particular historical set up. First, any gene may be unsuitable for very ancient events, if too variable, or for very recent events, if too conservative. A tradeoff must be found between the level of variability of the sequences used and the time interval one wishes to analyze. Second, even a gene that appears to be suitable for a given time interval may fail to resolve closely spaced speciation events within this interval. In these cases, contrary to intuition, simply increasing the amount of sequence will not allow to confidently resolve the order of emergence of taxa. as shown for ribosomal RNA (6, 7). In these cases, complementary information from either morphological or qualitative genomic modifications that have occurred in between these closely spaced nodes might prove to be the most useful.
(ii) Molecular Kinship Cannot Be Caused by Chance.
Although the idea is perhaps not intuitive, establishing kinship in a phylogeny does not have the same status as denying it. The underlying principle is simple: When kinship, based on numerous nucleotide or amino acid similarities, is observed in a molecular phylogeny at terminal nodes of a tree, then the probability that such a conjunction occurs by chance is small and can only be interpreted as being caused by recent shared ancestry. In contrast, species can be pulled away from each other in a tree because of various artifacts, as indicated above. Thus, finding kinship is a positive, strong result whereas finding disjunction is at best indicative.
Indeed, this is a major characteristic of the new results: Discovering new and unexpected kinship relationships, between nematodes and arthropods, for example, is a stronger argument than merely claiming that nematodes should be placed at the base of protostomes and deuterostomes because they are different from them and less complex.
(iii) Congruence Is a Powerful Argument.
Phylogenetic reconstruction, as much as cosmology, is an exercise in which there cannot be direct experimental testing of hypotheses through reconstitution of evolutionary history in the laboratory. In such scientific disciplines, congruence between results obtained from independent data sets remains the most decisive argument. Such congruence has increasingly emerged in animal phylogeny, as will be seen below. We claim that the improvements just listed now allow one to reach quite a few solid conclusions, enabling an independent confrontation with the vast and precious amount of morphological data.
The New rRNA-Based Phylogeny in a Nutshell.
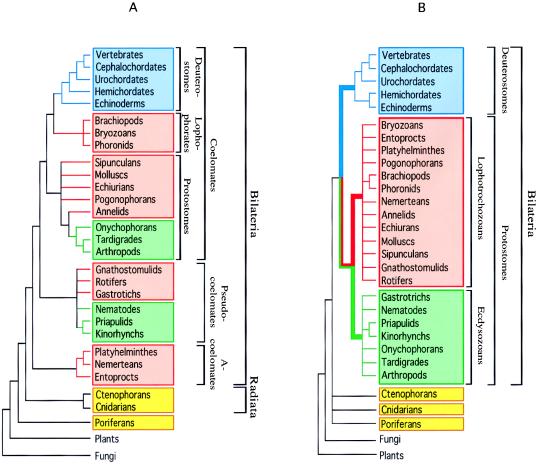
Small subunit ribosomal RNA remains the molecule for which the largest database is available for phylogenetic reconstruction. Starting with the work of Field et al. (8), it has progressively allowed revisions of metazoan phylogeny ranging from the shift of “superphyla” to details of intraphyla arrangement. Fig. 1 A and B summarize many of these modifications, based on the experimental work of several laboratories (9) with emphasis on those that appear to be robust when using the criteria discussed above. Fig. 1A, “traditional” animal phylogeny, is basically that reproduced in major precladistic zoology textbooks (10) following the work of Hyman (11). It is a good illustration of the long prevailing notion that animal evolution went from simple to complex through gradual steps, with extant animals actually representing grades of intermediate complexity supposed to have been those of their ancestors. Fig. 1B is exclusively based on rRNA. One can notice the depth of the reorganization by comparing the shift in colored rectangles between Fig. 1 A and B.
Figure 1.
Metazoan phylogenies. (A) The traditional phylogeny based on morphology and embryology, adapted from Hyman (11). (B) The new molecule-based phylogeny. A conservative approach was taken in B: i.e., some datasets provide resolution within some of the unresolved multifurcations displayed, but we have limited the extent of resolution displayed to that solidly provided by rRNA only.
The clear demarcation of Bilaterians.
All molecular phylogenies show the bilaterians as a monophyletic group clearly separated from sponges, cnidarians, and ctenophores. In the rRNA tree, bilaterians arise from a long stem, probably reflecting mutational acceleration in rRNA. Recent detailed studies based on rRNA involving sponges, cnidarians, and ctenophores (12–15) failed to fully resolve the phylogeny of the outgroups of Bilateria. They suggest a paraphyletic emergence of sponges at the base of the metazoan tree, followed by a monophyletic Ctenophora and a possibly paraphyletic Cnidaria. Use of protein coding genes such as EF1-α (16) or HSP 70 (17) yielded even less resolved trees.
The demise of Articulés and the birth of Lophotrochozoa.
The concept of Articulés, i.e., the clade uniting annelids and arthropods on the basis of shared segmentation of the body trunk, is as old as comparative anatomy (18) and has been maintained for nearly two centuries in all major textbooks and even in some recent cladistic treatments (19, 20) of metazoan phylogeny with two notable exceptions (21, 22). It has been known for about the same length of time, however, that annelids share with molluscs and several other unsegmented phyla a very typical mode of spiral cleavage of the egg, usually followed by the formation of a so-called trochophore larva. The split of Articulés into arthropods on one side and annelids on the other is now strongly supported by rRNA. The interesting consequence in terms of evolution of development is that it resurrects the question of the ancestry of segmentation in a new frame.
Not only did Articulés explode, but several groups were quite unexpectedly brought in the immediate proximity of annelids and molluscs. This is especially the case of the lophophorates, a group that comprised brachiopods, bryozoans, and phoronids, which all share a horseshoe-shaped feeding device made of ciliated tentacles. These were considered as basal deuterostomes in the majority of zoological textbooks and, especially in the case of brachiopods, even in the most recent cladistic analyses (23) because of the radial mode of cleavage of their egg and their trimeric coelom. rRNA, however, brought them robustly within protostomes and, more specifically, within the clade comprising annelids and molluscs (24). Hence, the name “Lophotrochozoa” for the whole clade uniting lophophore-bearing animals with those displaying a trochophore larva. Interestingly, removal of lophophorates from the deuterostomes induces a reevaluation of the scenario of early deuterostome evolution and genesis of the chordate body plan.
Acoelomates are Lophotrochozoans.
The other groups that were brought within lophotrochozoans are the flatworms (platyhelminths) and the nemerteans. These animals, lacking a coelome, segmentation, elaborate organs, and an anus (in flatworms) were considered, in a gradist perspective, to be the most primitive bilaterians. However, they were brought, through rRNA, inside the lophotrochozoans (25–28), with whom they share a spiral mode of cleavage and ciliated larval forms that may be considered as modified trochophores.
The burst of Pseudocoelomates and the birth of Ecdysozoa.
Perhaps the most striking result of the landmark study of Aguinaldo et al. (28) based on rRNA was the demonstration that nematodes had been misplaced outside of the bilaterians by a long-branch attraction artifact and that their true position was as a sister group of arthropods. This result came as the conclusion of a set of results, both molecular and morphological, showing that pseudocoelomates (or aschelminths) did not make up a monophyletic group. The various phyla grouped by Hyman (11) under this name comprised Rotifera, Gastrotricha, Kinorhyncha, Nematomorpha, and Nematoda. Priapulids are often attached to this group. They are all essentially small animals having some form of internal cavity but devoid of a true coelom. Both molecular and morphological analysis (23, 29–31) showed them to distribute between the two major protostome lines, but the nematodes resisted and remained very basal in molecular analyses because, previous to the analysis of Aguinaldo et al. (28), all of their representatives had extremely long branches. Displacement of the nematodes to a much higher position in the metazoan tree, if true, is a major result because it forces a complete reinterpretation of the data originating from their now fully sequenced genome as well as of all of their biological features: hence, the importance of confirming this placement with independent data, as described below.
In short, the subdivision of protostomes into two large group was confirmed and extended. As a sister group to lophotrochozoans, a vast group comprising nematodes, arthropods, and other phyla (kinorhynchs, priapulids, nematomorphs, etc.) emerged. These phyla all share the presence of a molting cuticle (albeit of very different composition, chitin in arthropods, collagen in nematodes), hence the name “Ecdysozoa” given to them (28).
The relative stability of deuterostomes.
In the face of the deep restructuring of protostome phylogeny, deuterostomes have remained stable as a monophyletic group but have undergone significant internal reorganization. Other than the removal of lophophorates, one major point is the joining of hemichordates with echinoderms (32). The important implication is that the existence of a sister group to chordates allows the reevaluation of competing scenarii of vertebrate origination.
The Lack of Resolution Within each of the Two Great Protostome Clades.
An observation repeatedly made when using rRNA data is of the extreme difficulty in resolving the branching order of phyla within the lophotrochozoans and the ecdysozoans. This is so much the case that even groups that are strongly believed to be monophyletic on the basis of morphological data, such as molluscs, emerge as polyphyletic in these trees (33). We have argued elsewhere that, within both branches, the phyla have emerged in a relatively rapid historical succession, thus leading to a case in which rRNA reaches its limits of resolution (34). We would like to stress that, if this view is correct, it leads to a profound reappraisal of the Cambrian explosion: Instead of corresponding to the rapid diversification of all of the bilaterian phyla, the explosion would have occurred simultaneously in three already well separated and poorly diversified lineages (the lophotrochozoan stem line, the ecdysozoan one, and the deuterostome one), implying that such an explosion would have been caused not by a single “internal” genetic innovation but, more likely, by an “external” (i.e., ecological) set of events.
The Gist of Supporting Evidence from Hox Genes.
The 60-amino acid homeodomain of the Hox genes and its flanking sequences contain specific phylogenetic information in the form of signature amino acids or short characteristic peptides (35). Together with the laboratories of Carroll and Akam, we recently sequenced genes from the Hox complex of a number of key taxa to look for signatures that might allow to test the new rRNA phylogeny (36). No less than five Hox genes show signatures supporting the close affinity of brachiopods and annelids. Two of these signatures are also recognized in flatworms. In parallel, one strongly conserved “posterior” gene supports the close relationship between nematodes, priapulids, and arthropodes. The existence of each of the two protostome clades is thus greatly strengthened by the Hox data.
Are Acoels the Most Primitive Bilaterians?
It has recently been claimed by Ruiz-Trillo et al. (37) on the basis of rRNA data that acoel worms did not belong to the Platyhelminthes but instead formed a separate lineage emerging at the base of the Bilateria. This would be an important result, invalidating all of the previous conclusions and resurrecting the idea of an extant evolutionary intermediate of “simple” design. However, despite the precautions that were taken to ensure the reliability of this conclusion, there are reasons to be doubtful. First, the data of Ruiz-Trillo et al. (37) show that a substantial amount of saturation already affects the rRNA genes of the acoels they used, raising the risk of a long-branch attraction artifact, pulling acoels to the bottom of the tree. Second, there are morphological characters and even molecular studies [summarized by Peterson et al. (38)] linking acoels to nemertodermatids, which themselves clearly belong to lophotrochozoans, along with all other platyhelminths. Third, we have isolated from the acoel Childia a number of Hox sequences that bear clear lophotrochozoan signatures (N.L., B.P., and A.A., unpublished work). Finally, Berney et al. (39) have very recently shown, by using EF1-α sequences, that acoels are closely related to platyhelminths within the lophotrochozoans, both on the basis of full sequence phylogenetic analysis and through a shared sequence signature with three triclad platyhelminths.
A New Perspective on Animal Evolution.
The new molecular based phylogeny has several important implications. Foremost among them is the disappearance of “intermediate” taxa between sponges, cnidarians, ctenophores, and the last common ancestor of bilaterians or “Urbilateria.“ Several lineages previously thought to branch at the base of the bilaterian tree, most notably acoelomates (such as platyhelminths) and pseudocoelomates (such as nematodes) are now embedded within or next to groups that display elaborate morphologies and complex genomic arrays. The implication is that these groups are secondarily simplified, in their morphology as well as at the molecular level. A corollary is that we have a major gap in the stem leading to the Urbilataria. We have lost the hope, so common in older evolutionary reasoning, of reconstructing the morphology of the “coelomate ancestor” through a scenario involving successive grades of increasing complexity based on the anatomy of extant “primitive” lineages. For example, the traditional view of an acoel-like ancestor, progressively acquiring a coelome, differentiated internal organs, segments, and so on must be abandoned. In this respect, the situation is not unlike the new perspective emerging on the phylogeny of eukaryotes as a whole (40), in which most of the formerly intermediate taxa have been pulled upwards.
How then can we attempt to reconstruct the path to the Urbilateria? Four research programs can be advocated (also see ref. 41). First, through comparison of extant terminal taxa belonging to the two big branches, protostomes and deuterostomes, and careful evaluation of all of their possible homologous characters, we should be able to reconstruct an image of the urbilaterian. That approach is certainly feasible in terms of genetic homologies because establishment of homology at the sequence level is fairly straightforward, and, once several genome programs of metazoans are completed, we should be able to identify the minimal gene content that was present in the urbilaterian. It will prove more tricky to establish that these genes, especially the ones involved in development, were carrying out the same function in the ancestor. More elaborate strategies will be needed on that point because recruitment of gene networks to different functions may have been widespread during evolution. Interestingly, the new phylogeny draws attention to a badly understudied group of animals, the lophotrochozoans, which holds great promise for this reconstruction. None of the major model organisms presently belongs to this large animal branch. The new phylogeny thus provides a plea for a major effort toward the developmental and genomic study of a lophotrochozoan model.
The second approach will consist of better characterizing the outgroups to bilaterians, sponges, cnidarians, and ctenophores. A good start has already been made (42–45). Following the same reasoning as above, comparison of diploblasts vs. bilateria gene content should enable us to characterize the basal metazoan genome and hence the one from which the urbilaterian was progressively constructed.
Indeed, the demise of the gradist interpretation of early bilaterian evolution does not mean, of course, that the last common ancestor has not itself been the result of a progressive construction, possibly through an extended period. It means that we do not have extant representatives of these stages. This emphasizes the importance of the third approach, that of paleaontological investigations on the pre-Cambrian and early Cambrian, which may well yield the crucial missing information (41, 46).
A fourth approach to reconstruct the steps toward the last common ancestor was recently introduced (47). Through comparative analysis of early development in all metazoan phyla, these authors were led to conclude that the ancestral mode of development was one in which a small animal of the size and design of extant larvae was first constructed on the basis of short-range cell-to-cell interactions. The later appearance in these micrometazoans of undifferentiated set aside cells with much greater multiplication potential, together with the recruitment of genetically based large scale patterning mechanisms (such as those using the Hox complex) would have enabled the evolution of the adult body plans of the major animal phyla. This stimulating hypothesis is now opened to some forms of experimental testing because it makes specific predictions as to the gene networks involved at each stage. At any rate, it offers the first “post gradist” scenario accounting for the genesis of the urbilaterian.
The new phylogeny nicely accounts for the innumerable homologies that have been disclosed between genes belonging to taxa of the three branches. Because all of the bilaterians are now seen to descend from the same ancestor, they have shared the stock of genes possessed by this ancestor. Now, the fact that these homologies are so numerous implies that the genome of this ancestor was a quite elaborate one. In fact, some very crude evaluations can now be made: The number of genes is now known precisely or carefully estimated in several invertebrate phyla belonging both to protostomes (Caenorhabditis elegans, 20,000; Drosophila melanogaster, over 12,000) or to deuterostomes (Ciona intestinalis, 16,000). One may therefore wander to suggest that the basal metazoan genome was made up of 15–20,000 genes. The ≈70,000 genes of Vertebrates would then reflect the two massive duplication events that are thought to have occurred early in their history. If such is the case, then, except for vertebrates, morphological innovation within bilaterians would not have relied so much on generation of new genes as on tinkering with an already existing array. The new phylogeny thus reemphasizes the importance of evolution of developmental regulatory networks (48, 49).
Acknowledgments
Work in our laboratory has been supported by Centre National de la Recherche Scientifique, Université Paris-Sud, and “Programme Génome” from Centre National de la Recherche Scientifique. We thank Claus Nielsen and Eric Davidson for comments on the manuscript.
References
- 1.Swofford D L, Olsen G J, Waddell P J, Hillis D M. Molecular Systematics. Sunderland, MA: Sinauer; 1996. pp. 407–514. [Google Scholar]
- 2.Felsenstein J. Annu Rev Genet. 1988;22:521–565. doi: 10.1146/annurev.ge.22.120188.002513. [DOI] [PubMed] [Google Scholar]
- 3.Philippe H, Laurent J. Curr Opin Genet Dev. 1998;8:616–623. doi: 10.1016/s0959-437x(98)80028-2. [DOI] [PubMed] [Google Scholar]
- 4.Boore J L, Brown W M. Curr Opin Genet Dev. 1998;8:668–674. doi: 10.1016/s0959-437x(98)80035-x. [DOI] [PubMed] [Google Scholar]
- 5.Nikaido M, Rooney A P, Okada N. Proc Natl Acad Sci USA. 1999;96:10261–10266. doi: 10.1073/pnas.96.18.10261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Philippe H, Chenuil A, Adoutte A. Development (Cambridge, UK) 1994;1994,Suppl.:15–25. [Google Scholar]
- 7.Abouheif E, Zardoya R, Meyer A. J Mol Evol. 1998;47:394–405. doi: 10.1007/pl00006397. [DOI] [PubMed] [Google Scholar]
- 8.Field K G, Olsen G J, Lane D J, Giovannoni S J, Ghiselin M T, Raff E C, Pace N R, Raff R A. Science. 1988;239:748–753. doi: 10.1126/science.3277277. [DOI] [PubMed] [Google Scholar]
- 9.Adoutte A, Balavoine G, Lartillot N, de Rosa R. Trends Genet. 1999;15:104–108. doi: 10.1016/s0168-9525(98)01671-0. [DOI] [PubMed] [Google Scholar]
- 10.Barnes R D. In: Invertebrate Zoology. Barnes R D, editor. Philadelphia: Saunders College; 1985. pp. 58–70. [Google Scholar]
- 11.Hyman L H. The Invertebrates. New York: McGraw–Hill; 1940. pp. 22–43. [Google Scholar]
- 12.Wainright P O, Hinkle G, Sogin M L, Stickel S K. Science. 1993;260:340–342. doi: 10.1126/science.8469985. [DOI] [PubMed] [Google Scholar]
- 13.Cavalier-Smith T, Allsopp M T E P, Chao E E, Boury-Esnault N, Vacelet J. Can J Zool. 1996;74:2031–2045. [Google Scholar]
- 14.Collins A G. Proc Natl Acad Sci USA. 1998;95:15458–15463. doi: 10.1073/pnas.95.26.15458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim J, Kim W, Cunningham C W. Mol Biol Evol. 1999;16:423–427. doi: 10.1093/oxfordjournals.molbev.a026124. [DOI] [PubMed] [Google Scholar]
- 16.Kobayashi M, Wada H, Satoh N. Mol Phylogenet Evol. 1996;5:414–422. doi: 10.1006/mpev.1996.0036. [DOI] [PubMed] [Google Scholar]
- 17.Borchiellini C, Boury-Esnault N, Vacelet J, Le Parco Y. Mol Biol Evol. 1998;15:647–655. doi: 10.1093/oxfordjournals.molbev.a025968. [DOI] [PubMed] [Google Scholar]
- 18.Cuvier G. Le Règne Animal Distribué Selon son Organisation. Paris: Deterville; 1817. [Google Scholar]
- 19.Nielsen C. Animal Evolution: Interrelationships of the Living Phyla. New York: Oxford Univ. Press; 1995. [Google Scholar]
- 20.Brusca R C, Brusca G J. Invertebrates. Sunderland, MA: Sinauer; 1990. pp. 879–889. [Google Scholar]
- 21.Eernisse D J, Albert J S, Anderson F E. Syst Biol. 1992;41:305–330. [Google Scholar]
- 22.Zrzavy J, Mihluka S, Kepka P, Bezdek A, Tietz D. Cladistics. 1998;14:249–285. doi: 10.1111/j.1096-0031.1998.tb00338.x. [DOI] [PubMed] [Google Scholar]
- 23.Nielsen C, Scharff N, Eibye-Jacobsen D. Biol J Linn Soc. 1996;57:385–410. [Google Scholar]
- 24.Halanych K M, Bacheller J D, Aguinaldo A M A, Liva S M, Hillis D M, Lake J A. Science. 1995;267:1641–1643. doi: 10.1126/science.7886451. [DOI] [PubMed] [Google Scholar]
- 25.Caranza S, Baguna J, Riutort M. Mol Biol Evol. 1997;14:485–497. doi: 10.1093/oxfordjournals.molbev.a025785. [DOI] [PubMed] [Google Scholar]
- 26.Campos A, Cummings M P, Peyes J L, Laclette J P. Mol Phylogenet Evol. 1998;10:1–10. doi: 10.1006/mpev.1997.0483. [DOI] [PubMed] [Google Scholar]
- 27.Littlewood D T, Telford M J, Clough K A, Rohde K. Mol Phylogenet Evol. 1998;9:72–79. doi: 10.1006/mpev.1997.0448. [DOI] [PubMed] [Google Scholar]
- 28.Aguinaldo A M A, Turbeville J M, Linford L S, Rivera M C, Garey J R, Raff R A, Lake J A. Nature (London) 1997;387:489–493. doi: 10.1038/387489a0. [DOI] [PubMed] [Google Scholar]
- 29.Mackey L Y, Winnepenninckx B, De Wachter R, Backeljau T, Emschermann P, Garey J R. J Mol Evol. 1996;42:552–559. doi: 10.1007/BF02352285. [DOI] [PubMed] [Google Scholar]
- 30.Winnepenninckx B, Backeljau T, Mackey L Y, Brooks J M, De Wachter R, Sudhir K, Garey J R. Mol Biol Evol. 1995;12:1132–1137. doi: 10.1093/oxfordjournals.molbev.a040287. [DOI] [PubMed] [Google Scholar]
- 31.Wallace R L, Ricci C, Melone G. Invertebr Biol. 1996;115:104–112. [Google Scholar]
- 32.Wada H, Satoh N. Proc Natl Acad Sci USA. 1994;91:1801–1804. doi: 10.1073/pnas.91.5.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Winnepennickx B, Backeljau T, De Wachter R. Mol Biol Evol. 1996;13:1306–1317. doi: 10.1093/oxfordjournals.molbev.a025577. [DOI] [PubMed] [Google Scholar]
- 34.Balavoine G, Adoutte A. Science. 1998;280:397–398. [Google Scholar]
- 35.Balavoine G. Am Zool. 1998;38:843–858. [Google Scholar]
- 36.de Rosa R, Grenier J K, Andreeva T, Cook C E, Adoutte A, Akam M, Carroll S B, Balavoine G. Nature (London) 1999;399:772–776. doi: 10.1038/21631. [DOI] [PubMed] [Google Scholar]
- 37.Ruiz-Trillo I, Riutort M, Littlewood D T, Herniou E A, Baguna J. Science. 1999;283:1919–1923. doi: 10.1126/science.283.5409.1919. [DOI] [PubMed] [Google Scholar]
- 38.Peterson K J, Cameron R A, Davidson E H. Dev Biol. 2000;219:1–17. doi: 10.1006/dbio.1999.9475. [DOI] [PubMed] [Google Scholar]
- 39.Berney, C., Pawlowski, J. & Zaninetti, L. (2000) Mol. Biol. Evol., in press. [DOI] [PubMed]
- 40.Philippe H, Adoutte A. In: Evolutionary Relationships Among Protozoa. Combs G H, Vickerman K, Sleigh M A, Warren A, editors. London: Chapman & Hall; 1998. pp. 25–56. [Google Scholar]
- 41.Knoll A H, Carroll S B. Science. 1999;284:2129–2137. doi: 10.1126/science.284.5423.2129. [DOI] [PubMed] [Google Scholar]
- 42.Finnerty J R, Martindale M Q. Evol Dev. 1999;1:16–23. doi: 10.1046/j.1525-142x.1999.99010.x. [DOI] [PubMed] [Google Scholar]
- 43.Galliot B, de Vargas C, Miller D. Dev Genes Evol. 1999;209:186–197. doi: 10.1007/s004270050243. [DOI] [PubMed] [Google Scholar]
- 44.Martinez D E, Dirksen M L, Bode P M, Jamrich M, Steele R E, Bode H R. Dev Biol. 1997;192:523–536. doi: 10.1006/dbio.1997.8715. [DOI] [PubMed] [Google Scholar]
- 45.Muller W E, Kruse M, Blumbach B, Skorokhod A, Muller M. Gene. 1999;238:179–193. doi: 10.1016/s0378-1119(99)00226-7. [DOI] [PubMed] [Google Scholar]
- 46.Conway Morris S. Development (Cambridge, UK) 1994;1994,Suppl.:1–13. [Google Scholar]
- 47.Peterson K J, Cameron R A, Davidson E H. BioEssays. 1997;19:623–631. doi: 10.1002/bies.950190713. [DOI] [PubMed] [Google Scholar]
- 48.Jacob F. Science. 1977;196:1161–1166. doi: 10.1126/science.860134. [DOI] [PubMed] [Google Scholar]
- 49.Carroll S B. Nature (London) 1995;376:479–485. doi: 10.1038/376479a0. [DOI] [PubMed] [Google Scholar]