Abstract
DNA is replicated in a defined temporal order that is developmentally regulated and constitutes a unique and stable fingerprint of a given cell type. Recently, we developed a robust assay to profile replication timing genome wide that can be applied to essentially any proliferating cell population. Asynchronously cycling cells are pulse labeled with the nucleotide analog 5-bromo-2-deoxyuridine (BrdU). The cells are sorted into S-phase fractions on the basis of DNA content using flow cytometry. BrdU-labeled DNA from each fraction is immunoprecipitated (BrdU IP), amplified, differentially labeled and co-hybridized to a whole-genome comparative genomic hybridization microarray (or sequenced). Since the basic steps of this protocol have been detailed elsewhere, here we focus on problems encountered when adapting this protocol to different cell types or tissue sources and modifications that have been successfully applied to troubleshoot these problems. There is an increasing demand for such studies to address how replication is regulated during development, its relationship to chromatin architecture and other chromosome functions, and the relevance of cell culture models to regulation in the native organismal niche.
Keywords: Replication Timing, Cell Cycle, Chromosome structure, BrdU immunoprecipitation, Flow Cytometry, Genomics
1. Introduction
All eukaryotic organisms replicate their DNA in a defined temporal order that is evolutionarily conserved, but the mechanisms regulating this “replication timing program” and its biological significance remain fundamental mysteries [1–3]. Recently, analyses of replication timing (RT) genome wide have provided a means to test many longstanding hypotheses as to how this program is executed and regulated during development. Comparison of many cell types and analyses during directed differentiation of stem cells has revealed that at least half the genome experiences changes in RT during development, generally coordinated with transcriptional regulation, creating cell type specific RT patterns [4–8]. Most of these changes occur coordinately across 400–800 kb segments of chromosomes, supporting the concept of replication domains [5,8,9]. Genome-wide RT profiles can be aligned to other chromosome properties. By far the closest alignment is to long-range chromatin interaction maps [9], defined by chromatin conformation capture [10], suggesting that replication domains represent structural and functional units of genome organization and a simplified readout for Hi-C. Several protocols have been developed to measure RT genome wide [11]. The protocol most commonly used in our laboratory is optimized for the rapid assessment of the global RT program in many different types of cells or experimental conditions and has been described in detail [12], including bioinformatic scripts to perform basic analyses with the resulting data.
There is an increasing need for comparative genomic studies of different cell types, diseased vs. normal tissue, differentiation intermediates and studies in the context of a whole animal, in order to understand normal developmental regulation and its alterations in disease. Moreover, replication timing analysis is now sufficiently robust and simplified that many different experimental conditions (drug treatments, gene knockouts or knockdowns) can be queried for their effects on replication timing genome wide. However, some preparations are not amenable to certain steps of the standard protocol. Here, we focus on how to identify and troubleshoot these problems, and alternative protocols that can be implemented to solve them.
2. Outline of protocol
The study of cell cycle regulation requires cell synchronization, which can be performed prospectively or retroactively (prior to or after cell collection). If the goal is to directly compare many different cell types, prospective cell synchrony is impractical because it requires developing a different synchronization regime for each cell type [3,13]. Many years ago, we developed a retroactive synchronization method that can be applied to any proliferating cell type that can be dissociated into a single cell suspension [14,15]. This protocol has been optimized for genome wide analysis and has been described in detail, so this report focuses on variables to modify for different cell types [12]. A brief synopsis of the method is as follows: Newly synthesized DNA is first pulse-labeled with 5’-bromo-2’-deoxyuridine (BrdU) while the cells are growing unperturbed in the native conditions to be queried, including intact animals and tissues (unpublished results), provided that the labeling times are first optimized using methods such as BrdU-immunofluorescence [16]. Cells are then dissociated into a single-cell suspension. DNA is subsequently stained with one of several fluorescent dyes, and cells are retroactively synchronized into early and late S phase fractions based on their increasing DNA content during S phase. Cells are fixed in Ethanol and the DNA that was synthesized either early or late during S phase is then purified, sheared by sonication, and the BrdU-substituted nascent DNA isolated by immunoprecipitation (BrdU-IP) [12]. A series of DNA sites that are known to replicate at specific times are then analyzed by PCR to verify the quality of the nascent strands, which can then be subjected to whole-genome amplification and microarray hybridization or next generation sequencing library preparation [4,7,12]. Once the cells are sorted into early and late fractions, the source of the starting material does not influence subsequent steps, which are all performed as described [12]. It should be noted that although labeling of non-replicative DNA synthesis such as DNA repair is a theoretical concern, the amount of signal from this is negligible compared to the signal from DNA replication as evidenced by the extremely low levels of BrdU-substituted DNA when G1-phase cells are sorted [14,15] and the absence of localized spikes in BrdU-IP signals that would be indicative of patches of DNA repair.
3. Considerations
3.1 Cellular material
The choice of starting cellular material will influence how you BrdU label, collect and stain for fluorescence activated cell sorting (FACS). The only limitation is that the cells should be proliferating. We have analyzed dozens of cell lines, differentiation intermediates derived from stem cells, and tissue explants from patients with different diseases [5,8,17,18], unpublished results). The primary variables are the number of starting cells, the method of cellular dissociation and the method of DNA staining. It is also important that your cells have a reasonably uniform karyotype, since synchronization is based upon DNA content. In our experience, only a significant mixture of diploid and tetraploid cells can preclude success [19], and we have profiled many aneuploid cancers (unpublished results). It is recommended that you run a preliminary flow cytometry cell cycle analysis with your material before attempting the full protocol. We will suggest ways to modify each step of the protocol as it is discussed.
3.2 Cell number
The number of starting cells you will need is directly dependent upon the percentage of cells in S phase. The central goal is to acquire 10,000 cells in early and late S phase. We continue to optimize this aspect of the protocol, but presently we have concluded that somewhere between 5,000 and 8,000 cells is the limit to permit an effective BrdU IP. If you cannot obtain this number of cells with the standard protocol, you can consider longer BrdU labeling times, higher concentrations of BrdU, or larger sorting windows (discussed below). When cell number is unlimited (e.g. immortalized cell lines) or when it is not possible to perform a preliminary flow cytometry cell cycle analysis (e.g. primary human patient samples) we recommend 5 to 10 million cells, which is sufficient for several experiments even when there are fewer than 5% of cells in S phase. With precious samples, and using the standard protocol, we have generated genome-wide profiles with as few as 200,000 starting cells with 25–30 percent in S phase.
3.3 BrdU labeling time
In almost all cases, you will want to label your cells in their native unperturbed environment first, and then dissociate them. In some cases (such as tissue from patients) it is impossible to do so and cells must be isolated first and then labeled. In either case, we label all mammalian cultures of any type for 2 hours with 50 µM BrdU. With few exceptions, S phase in mammalian cells is 8–12 hours, and a 2 hour labeling time ensures sufficient BrdU to provide an effective BrdU-IP while still being less than half the duration of the sorting window. Recently, we discovered that, for some cell types, increasing the BrdU concentration resulted in good quality BrdU IP even with less than 10,000 cells. This may be due to different nucleotide pools in different cell types. Therefore, if the standard concentration does not give robust results, the BrdU concentration can be empirically adjusted. With mouse ES cells, we have found that 90 minutes is sufficient, but 60 minute labeling periods can give erratic results, which we attribute to increased noise in the BrdU-IP due to the inevitable presence of contaminating unsubstituted DNA. In principle, a double-IP could eliminate most contamination, but we have found the yields of BrdU-DNA to drop precipitously after a double-IP. On the other hand, we have found that 2 hours does not detectably reduce resolution vs. 1 or 1.5 hour labeling times, and the increased yield of BrdU-substituted DNA eliminates replicate variability. We have not found the need to label longer than 2 hours, but with extremely limited S phase cell numbers, increasing the labeling time would be a reasonable approach, even though this could, in principle, come at a sacrifice of resolution.
3.4 Isolating Early and Late S phase Cells by Flow Cytometry
The quality of this assay critically depends upon clean separation of cells in early vs. late S phase. Hence a good quality FACS profile is crucial (Fig 1). To accomplish this, cells must be dissociated into a single cell suspension without introducing excessive variability in cell shape and size. The most common problem encountered with the standard protocol is cell clumping: some cell types are difficult to dissociate, while others aggregate during staining. An appropriate DNA staining method should be adopted to minimize cell clumping or cell breakage. A less common problem is cell sizes and shapes that confuse the scatter gates or cannot be distinguished easily from contaminating doublets. A third and surprisingly uncommon problem is a cell line with a karyotype so unstable that its S phase position is not homogeneously proportional to DNA content from cell to cell. In fact, in nearly 100 independent cell preparations, we have yet to encounter such a cell type.
Figure 1. Illustration of a successful FACS cell cycle profile and ideal gating.
Typical cell cycle FACS profile of proliferating cells. The early and late gates are positioned in such a way as to minimize cross contamination of early and late fractions, yet obtain maximum coverage of S-phase.
Once a clean cell cycle profile is obtained, gating the FACS profile becomes relatively easy for any trained FACS operator. Only two fractions are gated to simplify subsequent analyses and facilitate rapid cell type comparisons. Although intuitively one might imagine that higher resolution might be achieved by taking more fractions of S phase, we have not found any detectable differences between 2 and 6 fractions [9]. We attribute this to the likelihood that the limiting factor for resolution is the BrdU pulse, which labels hundreds of kilobases. Two fractions captures the entire S phase, due to slight overlap between sorting windows (Fig 1)
The primary problem encountered here is drifting in the gain of the machine, which needs to be checked from time to time. A second problem is nozzle clogging, which is a standard flow cytometry problem and should not be onerous with a good single cell suspension. Gates should be set to minimize cross contamination of early and late S phase fractions, although they can be set quite close to each other if the laser is stable and the operator is attentive throughout the sort. G1 or G2 phase contamination is less of a concern, since cells do not incorporate BrdU during these phases, although too much unlabeled contamination will reduce BrdU IP yield. To gather more cells in cases where material is limiting, the operator can set the left boundary of the early S phase gates to capture some G1 phase cells, and the right boundary of the late S phase window to capture G2/M cells, which legitimately contain DNA that was labeled during late S phase and completed replication by the end of the BrdU pulse label.
4. Adapting to different cell types
We have profiled nearly 100 different cell preparations, including primary patient samples, gene knockouts, and differentiation derivatives of stem cells (see www.replicationdomain.org) [20]. In theory the protocol is designed for any proliferating cell population that can be labeled and dissociated into a single cell suspension. However, some cell types pose particular challenges. Here we provide ways to address common problems encountered while extending the protocol to new cells types and available options to overcome them.
4.1 Types of Problems Most problems
Most problems occur at the FACS sorting step. A good FACS profile is the most reliable predictor of ultimate data quality and depends on obtaining a properly stained single cell suspension. The problems we encounter can be divided into three categories:
Difficulties dissociating cells. This situation usually applies to fresh tissue or biopsies and dispersal methods to solve this problem are highly sample specific and beyond the scope of this article. With cells such as mouse embryonic stem cells that grow in clumps and are sensitive to complete dissociation, one can use strong proteases such as trypsin without worrying about cell viability since the protocol is an end stage protocol.
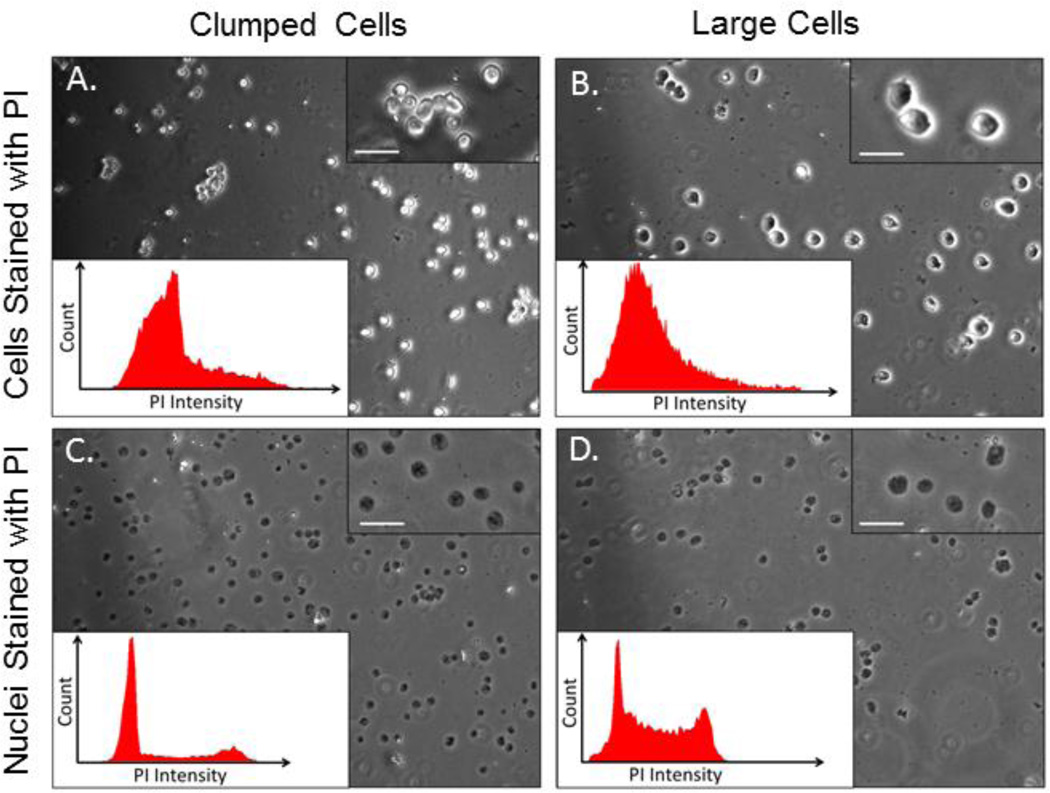
Cell clumping during ethanol fixation: some cell types spontaneously clump during ethanol fixation (Figure 2A).
Cell clumping during the staining protocol. It is not uncommon for cells to look well dissociated after ethanol fixation, but clump during steps of the staining protocol.
Cell morphology. We have also encountered problems during sorting due to large cell size (Figure 2B). These problems are pronounced when using certain models of cell sorters where the beam size or shape may be inadequate to illuminate the entire cell.
Figure 2. Common Problems Encountered With Certain Cell Types Can Be Resolved by Staining Nuclei Instead of Cells.
Panels A and B show cases where cells clump during the classical PI staining procedure (A), or are too large to be properly illuminated by the flow cytometer laser (B). In both cases, a well resolved cell cycle profile can be obtained by staining nuclei rather than cells (nuclei in C and D were derived from the same cells shown in A and B, respectively). Clumping can occur at different stages of the classical PI staining procedure, depending on cell type, but sorting nuclei has similarly resolved all cell types analyzed to date. Scale bar represents 5 µm.
4.2 Solutions
Clumping in ethanol can be avoided by being very gentle during ethanol fixation (gentle vortexing is essential) and effective trypsinzation during cell collection. Substituting trypsin with gentler proteases such as accutase might help in some cases. Some cells simply do not remain in a single cell suspension after ethanol fixation. In such cases a live cell sort can be performed using a dye such as DAPI.
Cell clumping during FACS preparation can be reduced by adding FBS in the staining buffer throughout the sorting process, which reduces clumping by eliminating cell surface electrostatic effects. Finally, when conventional cell-staining methods fail, we have found that sorting PI-stained nuclei is a robust alternative that adds only a few steps to the protocol. We recently developed a nuclei sorting protocol that combines conditions used in several previously published protocols of nuclei isolation/sorting [21–23]. In our experience, this protocol effectively solves the clumping and morphology problems described above (Figure 2C and D).
4.3 Advantages and Disadvantages of different staining protocols
As described previously [12], live cells sorting using DAPI is a quick and easy method to overcome clumping during ethanol fixation. But a drawback of DAPI is the requirement for short-wave UV light for its excitation, since BrdU-substituted DNA is sensitive to UV-induced damage. Also, since the cells are live, long term storage is not an option.
The nuclei protocol has solved all the problems we have encountered to date. A major disadvantage of nuclei isolation is, since the cell membrane is lost, the cells cannot be sorted based on cell surface markers if needed. Also, the protocol is lengthier than conventional FACS protocol, but it obviates the need of filtering through nylon mesh (used to obtain single cell suspension), which is a cumbersome step in conventional FACS protocol.
5. BrdU labeling and Nuclei sorting protocol
5.1 Materials
Round-bottom polystyrene tube (5 ml; Falcon, Cat. no. 352054)
FACS Aria cell sorter (BD Biosciences, or a comparable sorter)
Hemocytometer
BrdU (5-bromo-2‟ -deoxyuridine, Sigma Aldrich, B5002) Make stock solutions of 10mg/mL and 1mg/mL in ddH2O and store at −20°C.
PBS (1×) To prepare 1 liter, dissolve 8 g NaCl, 0.2 g KCl, 1.44 g Na2HPO4 and 0.24 g KH2PO4 in 800 ml of ddH20. Adjust pH to 7.4 with HCl and adjust the final volume to 1 liter. Sterilize by autoclaving. Store at room temperature.
Fetal Bovine Serum (FBS)
Pepsin (Sigma, Cat.No: P6887-1G)
Propidium iodide (1 mg ml −1) To prepare 20 ml, dissolve 20 mg PI powder (Sigma P4179-100MG) in autoclaved ddH2O to obtain a final volume of 20 ml and filter by syringe. Store protected from light for up to 1 year at 4 °C.
RNase A (10 mg ml −1; Sigma, Cat. No: R6513)
-
0.2X Trypsin-EDTA To make 50mL, combine 10mL 1X Trypsin-EDTA (Mediatech 25-053-Cl) with 40mL 1X PBS. Store at 4°C for up to one month.
Warm to room temperature before each use.
5.2 BrdU labeling and fixing
Add BrdU to cells in culture medium at a final concentration of 50µM.
Incubate cells for two hours in a carbon dioxide incubator at 37°C, 5% CO2. For adherent cells, rinse gently with ice-cold PBS twice. For suspension cells, collect cells in a 15mL tube and proceed directly to step 6.
Detach adherent cells using 0.2X Trypsin-EDTA for 2–3 minutes or Accutase for 3–6 minutes. CRITICAL STEP Incubate cells at 37 °C with the enzyme treatment and/or use gentle trituration if necessary to achieve a single cell suspension, as this is essential for accurate FACS sorting.
Add 5mL of cell culture medium (containing FBS if trypsin has been used) to the cell culture dish or flask, pipette gently, and transfer contents to a 15mL round bottom tube.
Count the number of cells collected using a hemacytometer. Collect enough cells to obtain at least 20,000–30,000 (preferably >150,000) cells in each fraction after sorting; this will generally require 0.5–1×106 cells, with more required if few cells are in S-phase. For first-time users, we recommend starting with 4×106 – 8×106 cells.
Centrifuge at approximately 200 × g for 5 minutes at room temperature.
Aspirate supernatant carefully and resuspend cells in 2.5 mL of ice-cold PBS containing 1% FBS.
Add 7.5 mL of ice-cold 100% ethanol dropwise while gently vortexing. CRITICAL STEP: Note that vortexing should be performed gently to avoid cell damage.
Seal the cap of the 15 mL tube with parafilm and mix gently but thoroughly.
Cells can be stored in the dark at −20°C indefinitely.
5.3 Staining protocol
Transfer 2 million fixed cells to a 5-ml polystyrene round-bottom tube.
Centrifuge at ~200g for 5 min at room temperature.
Decant supernatant carefully.
Resuspend the cell pellet in 2 ml of PBS with 1% (vol/vol) FBS. Mix well by tapping the tube.
Centrifuge at ~200g for 5 min at room temperature.
Decant supernatant carefully.
Incubate cells in 5 mL 0.015% pepsin (Sigma) w/v in 0.01N HCl for 45 minutes at 37°C with occasional tapping to resuspend.. Centrifuge at ~600g for 5 minutes and decant supernatant carefully.
Resuspend nuclei in 1 mL PBS.
Add 1 mg ml−1 of PI to a final concentration of 20 µg/ml.
Add 10 mg ml−1 of RNase A to a final concentration of 250 µg/ ml.
Incubate for 20–30 mins at RT.
Place samples on ice in the dark and proceed directly to FACS sorting.
Acknowledgements
We thank Dr. Takayo Sasaki for valuable inputs and suggestions. We thank the members of the Gilbert lab for their valuable input. DMG is supported by National Institute for General Medical Sciences awards GM083337 and GM085354.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- 1.Pope BD, Hiratani I, Gilbert DM. Domain-wide regulation of DNA replication timing during mammalian development. Chromosome Research : an International Journal on the Molecular, Supramolecular and Evolutionary Aspects of Chromosome Biology. 2010;18:127–136. doi: 10.1007/s10577-009-9100-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gilbert DM, Takebayashi S-I, Ryba T, Lu J, Pope BD, Wilson KA, et al. Space and time in the nucleus: developmental control of replication timing and chromosome architecture. Cold Spring Harbor Symposia on Quantitative Biology. 2010;75:143–153. doi: 10.1101/sqb.2010.75.011. [DOI] [PubMed] [Google Scholar]
- 3.Farkash-Amar S, Simon I. Genome-wide analysis of the replication program in mammals. Chromosome Research : an International Journal on the Molecular, Supramolecular and Evolutionary Aspects of Chromosome Biology. 2010;18:115–25. doi: 10.1007/s10577-009-9091-5. [DOI] [PubMed] [Google Scholar]
- 4.Hansen RS, Thomas S, Sandstrom R, Canfield TK, Thurman RE, Weaver M, et al. Sequencing newly replicated DNA reveals widespread plasticity in human replication timing. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:139–144. doi: 10.1073/pnas.0912402107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hiratani I, Ryba T, Itoh M, Rathjen J, Kulik M, Papp B, et al. Genome-wide dynamics of replication timing revealed by in vitro models of mouse embryogenesis. Genome Research. 2010;20:155–169. doi: 10.1101/gr.099796.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schübeler D, Scalzo D, Kooperberg C, van Steensel B, Delrow J, Groudine M. Genome-wide DNA replication profile for Drosophila melanogaster: a link between transcription and replication timing. Nature Genetics. 2002;32:438–442. doi: 10.1038/ng1005. [DOI] [PubMed] [Google Scholar]
- 7.Desprat R, Thierry-Mieg D, Lailler N, Lajugie J, Schildkraut C, Thierry-Mieg J, et al. Predictable dynamic program of timing of DNA replication in human cells. Genome Research. 2009;19:2288–2299. doi: 10.1101/gr.094060.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hiratani I, Ryba T, Itoh M, Yokochi T, Schwaiger M, Chang C-W, et al. Global reorganization of replication domains during embryonic stem cell differentiation. PLoS Biology. 2008;6:e245. doi: 10.1371/journal.pbio.0060245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ryba T, Hiratani I, Lu J, Itoh M, Kulik M, Zhang J, et al. Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Research. 2010;20:761–770. doi: 10.1101/gr.099655.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science (New York, N.Y.) 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilbert DM. Evaluating genome-scale approaches to eukaryotic DNA replication. Nature Reviews. Genetics. 2010;11:673–684. doi: 10.1038/nrg2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ryba T, Battaglia D, Pope BD, Hiratani I, Gilbert DM. Genome-scale analysis of replication timing: from bench to bioinformatics. Nature Protocols. 2011;6:870–895. doi: 10.1038/nprot.2011.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farkash-Amar S, Lipson D, Polten A, Goren A, Helmstetter C, Yakhini Z, et al. Global organization of replication time zones of the mouse genome. Genome Research. 2008;18:1562–1570. doi: 10.1101/gr.079566.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilbert DM. Temporal order of replication of Xenopus laevis 5S ribosomal RNA genes in somatic cells. Proceedings of the National Academy of Sciences of the United States of America. 1986;83:2924–2928. doi: 10.1073/pnas.83.9.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilbert DM, Cohen SN. Bovine papilloma virus plasmids replicate randomly in mouse fibroblasts throughout S phase of the cell cycle. Cell. 1987;50:59–68. doi: 10.1016/0092-8674(87)90662-3. [DOI] [PubMed] [Google Scholar]
- 16.Yokochi T, Gilbert DM. Replication labeling with halogenated thymidine analogs. Current Protocols in Cell Biology / Editorial Board, Juan S. Bonifacino … [et Al.] 2007;Chapter 22 doi: 10.1002/0471143030.cb2210s35. Unit 22.10. [DOI] [PubMed] [Google Scholar]
- 17.Ryba T, Hiratani I, Sasaki T, Battaglia D, Kulik M, Zhang J, et al. Replication Timing: A Fingerprint for Cell Identity and Pluripotency. PLoS Computational Biology. 2011;7 doi: 10.1371/journal.pcbi.1002225. e1002225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pope BD, Tsumagari K, Battaglia D, Ryba T, Hiratani I, Ehrlich M, et al. DNA Replication Timing Is Maintained Genome-Wide in Primary Human Myoblasts Independent of D4Z4 Contraction in FSH Muscular Dystrophy. PloS One. 2011;6:e27413. doi: 10.1371/journal.pone.0027413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu J, Li F, Murphy CS, Davidson MW, Gilbert DM. G2 phase chromatin lacks determinants of replication timing. The Journal of Cell Biology. 2010;189:967–980. doi: 10.1083/jcb.201002002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weddington N, Stuy A, Hiratani I, Ryba T, Yokochi T, Gilbert DM. Replication Domain: a visualization tool and comparative database for genome-wide replication timing data. BMC Bioinformatics. 2008;9:530. doi: 10.1186/1471-2105-9-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larsen JK, Munch-Petersen B, Christiansen J, Jørgensen K. Flow cytometric discrimination of mitotic cells: resolution of M, as well as G1, S, and G2 phase nuclei with mithramycin, propidium iodide, and ethidium bromide after fixation with formaldehyde. Cytometry. 1986;7:54–63. doi: 10.1002/cyto.990070108. [DOI] [PubMed] [Google Scholar]
- 22.Schutte B, Reynders MM, van Assche CL, Hupperets PS, Bosman FT, Blijham GH. An improved method for the immunocytochemical detection of bromodeoxyuridine labeled nuclei using flow cytometry. Cytometry. 1987;8:372–376. doi: 10.1002/cyto.990080405. [DOI] [PubMed] [Google Scholar]
- 23.Tennenbaum T, Giloh H, Fusenig NE, Kapitulnik J. A rapid procedure for flow cytometric DNA analysis in cultures of normal and transformed epidermal cells. The Journal of Investigative Dermatology. 1988;90:857–860. doi: 10.1111/1523-1747.ep12462098. [DOI] [PubMed] [Google Scholar]