Abstract
One of the main obstacles in quantitative interpretation of functional magnetic resonance imaging (fMRI) signal is that this signal is influenced by non‐neural factors such as vascular properties of the brain, which effectively increases signal variability. One approach to account for non‐neural components is to identify and measure these confounding factors and to include them as covariates in data analysis or interpretation. Previously, several research groups have independently identified four potential physiologic modulators of fMRI signals, including baseline venous oxygenation (Y v), cerebrovascular reactivity (CVR), resting state BOLD fluctuation amplitude (RSFA), and baseline cerebral blood flow (CBF). This study sought to directly compare the modulation effects of these indices in the same fMRI session. The physiologic parameters were measured with techniques comparable with those used in the previous studies except for CBF, which was determined globally with a velocity‐based phase‐contrast MRI (instead of arterial‐spin‐labeling MRI). Using an event‐related, scene‐categorization fMRI task, we showed that the fMRI signal amplitude was positively correlated with CVR (P < 0.0001) and RSFA (P = 0.002), while negatively correlated with baseline Y v (P < 0.0001). The fMRI‐CBF correlation did not reach significance, although the (negative) sign of the correlation was consistent with the earlier study. Furthermore, among the physiologic modulators themselves, significant correlations were observed between baseline Y v and baseline CBF (P = 0.01), and between CVR and RSFA (P = 0.05), suggesting that some of the modulators may partly be of similar physiologic origins. These observations as well as findings in recent literature suggest that additional measurement of physiologic modulator(s) in an fMRI session may provide a practical approach to control for inter‐subject variations and to improve the ability of fMRI in detecting disease or medication related differences. Hum Brain Mapp 34:2078–2088, 2013. © 2011 Wiley Periodicals, Inc.
Keywords: BOLD fMRI, normalization, venous oxygenation, cerebrovascular reactivity, resting state BOLD fluctuation, cerebral blood flow
INTRODUCTION
Functional magnetic resonance imaging (fMRI) provides a noninvasive approach to assess brain function and has the potential to be used in the diagnosis and treatment monitoring of neurological and psychiatric disorders. However, to date, this potential has not been turned into reality. One of the main reasons is that fMRI signal is an indirect measurement of neural activity and the signal is influenced by non‐neural factors such as vascular properties of the brain, which makes it difficult to interpret the results and also increases signal variability across individuals [D'Esposito et al., 1999; Jezzard and Buxton, 2006]. A possible approach to obtain a more accurate estimation of neural activity is to use a calibrated fMRI method [Chiarelli et al., 2007a, b; Davis et al., 1998; Hoge et al., 1999; Kim and Ugurbil, 1997; Sicard and Duong, 2005; Uludag et al., 2004]. Calibrated fMRI uses a biophysical model of the Blood‐Oxygenation‐Level‐Dependent (BOLD) effect, and by experimentally estimating various parameters of the model, one can separate the neural effect from vascular contributions. This method has shown great promises in quantitative evaluation of the BOLD effect and has been applied in studies of aging [Ances et al., 2009], Alzheimer's Disease [Fleisher et al., 2009], epilepsy [Stefanovic et al., 2005], and HIV patients [Ances et al., 2011]. However, a technical challenge of the calibrated fMRI approach is that the data acquisition requires the use of simultaneous BOLD and Arterial‐Spin‐Labeling (ASL) pulse sequence, which is often associated with lower signal‐to‐noise ratio (SNR) and reduced brain coverage.
An alternative approach to account for non‐neural components is to identify and measure these confounding factors and to include them as covariates in the data analysis [Bandettini and Wong, 1997; Birn et al., 2006; Handwerker et al., 2007; Kannurpatti and Biswal, 2008; Kannurpatti et al., 2011; Liau and Liu, 2009; Lu et al., 2008, 2010; Thomason et al., 2007]. With this approach, the fMRI acquisition will still use the routine BOLD sequence. But, additional sequences are performed during the same session to measure the sources of signal variability. Recently, much attention has been focused on identifying physiologic modulators of fMRI signals. To date, four markers have been proposed to explain (some) fMRI variance across individuals. First, it is well known that BOLD fMRI is based on the vasodilatation, thus the signal amplitude is correlated with the person's cerebrovascular reactivity (CVR), which can be measured by hypercapnia challenges [Bandettini and Wong, 1997; Handwerker et al., 2007; Thomason et al., 2007]. More recently, Lu et al. 2008 showed that fMRI response amplitude is negatively correlated with baseline venous oxygenation. That is, an individual with higher baseline venous oxygenation tends to have a smaller fMRI signal and vice versa. Kannurpatti and Biswal showed that stimulus‐induced fMRI signal amplitude is correlated with the amplitude of fluctuation in resting state BOLD time course [Kannurpatti and Biswal, 2008; Kannurpatti et al., 2011]. Finally, Liau and Liu 2009 showed that an individual's fMRI signal is correlated with his/her baseline cerebral blood flow (CBF) as measured with ASL MRI. It has also been shown that subject‐specific measurement of physiologic modulators was able to improve the statistical power in detecting group differences [Lu et al., 2010]. Therefore, addition of the physiologic measurement may provide a cost‐effective tool for normalizing fMRI signals and enhancing the use of fMRI in neurological and psychiatric research.
A remaining question is whether the various modulators described above are equivalent or complementary. This is an important and practical issue because if the modulators provide complementary information, they should be measured conjunctively to account for the maximum variability. On the other hand, if the modulators are of similar physiologic origins, one or a subset of them should be measured because extra measurements would not only increase scan time but also incur additional noise to the data set.
Here, we conducted an event‐related, scene‐categorization fMRI study in a group of young, healthy subjects and compared the above four fMRI modulators in explaining the inter‐subject variations in BOLD signals. The modulators were measured with techniques comparable with those used in the previous studies except for CBF, which was determined globally with a velocity‐based phase‐contrast MRI (instead of ASL MRI) [Haccke et al., 1999; Xu et al., 2009]. Potential correlations among the modulators themselves were examined, and the benefit of combining multiple physiologic modulators was assessed. Compared with previous studies which used simple sensory or motor tasks [Kannurpatti and Biswal, 2008; Liau and Liu, 2009; Lu et al., 2008], the cognitively oriented task allowed us to assess if the modulation effect can also be observed in higher order cortical areas.
MATERIALS AND METHODS
MRI Experiment
All MR imaging experiments were conducted on 3 T MR system (Philips Medical System, Best, The Netherlands). A total of 28 young healthy subjects (age range 20–35 years old, mean age 28.6 ± 5.2 years, 10 males and 18 females) were studied. Each subject gave informed written consent before participating in the study. The study protocol was approved by the Institutional Review Board of the University of Texas Southwestern Medical Center and the University of Texas at Dallas. All participants underwent extensive health screening and had no contraindications to MRI scanning (pacemaker, implanted metallic objects, claustrophobia) and were generally of good health, with no serious or unstable medical conditions such as neurological disease, brain injury, uncontrollable shaking, history of by‐pass surgery or chemotherapy, or use of medications that affect cognitive function. All participants were highly right‐handed, native English speakers with at least a high school education, and a Mini‐Mental State Exam [Folstein et al., 1975] score of 26 or greater.
The BOLD fMRI experiment used an event‐related, scene‐categorization task, during which the subject viewed full‐color photographs of outdoor scenes presented via a back‐projection system. Each subject received three fMRI runs with 32 photographs in each run. The paradigm started with a 20 s fixation period, during which the subject looked at a small white cross at the center of the black screen. Then, each photograph appeared for 3 s followed by a fixation period with randomized duration from 4 s to 17 s. The subject was instructed to determine if there is water in the scene and to press buttons in their right hand accordingly. Tasks of similar type are widely used in cognitive neuroscience and clinical fMRI studies [Gutchess et al., 2005; Ofen et al., 2007; Park et al., 2003]. The vision of the subjects was corrected using MR‐compatible corrective lenses, whenever necessary. The duration for each fMRI run was 5.75 min. Standard BOLD fMRI imaging parameters were used: TR/TE/flip angle = 2,000/25 ms/80°, field‐of‐view (FOV) = 220 × 220 mm2, matrix 64 × 64, voxel size 3.4 × 3.4 × 3.5 mm3, 43 axial slices with whole brain coverage, 171 brain volumes in each run.
The four physiologic modulators were measured as follows. Baseline venous oxygenation (Y v) was determined in sagittal sinus using a recently developed T2‐Relaxation‐Under‐Spin‐Tagging (TRUST) MRI technique [Lu and Ge, 2008]. The imaging parameters were: TR = 8,000 ms, TI = 1,200 ms, voxel size 3.44 × 3.44 × 5 mm3, four different T2‐weightings with TEs of 0, 40, 80, and 160 ms, duration 4 min and 16 s. The imaging slice was positioned axially to intersect the superior sagittal sinus at the level of ∼10 mm above the sinus congruence. The labeling slab was 80 mm in thickness and was positioned 17.5 mm above the imaging slice. These parameters yielded robust spin labeling signal to allow for accurate estimation of T2 of venous blood in the sagittal sinus [Lu and Ge, 2008]. Note that the TRUST MRI technique measures global baseline venous oxygenation, and no regional information is obtained.
CVR was measured using inhalation of 5% CO2 gas while simultaneously acquiring BOLD MR images. A number of previous studies have used a breathholding task to measure CVR [Handwerker et al., 2007; Thomason et al., 2007]. In this study, we used CO2 breathing as it allows us to monitor and record the end‐tidal CO2 (Et‐CO2) time course and to use it as an input signal in the general linear model (GLM) analysis [Yezhuvath et al., 2009], which may be useful to account for intersubject variations in respiratory responses to hypercapnia. The details of the CVR measurement have been described elsewhere [Yezhuvath et al., 2009]. Briefly, during the CVR scan, subjects were fitted with a nose clip and breathed through a mouthpiece. Hypercapnia was induced by breathing a gas mixture of 5% CO2, 74% N2, and 21% O2 contained within a Douglas bag through a two‐way non‐rebreathing valve and mouthpiece combination (Hans Rudolph, 2600 series, Shawnee, KS). The subject breathed room‐air and 5% CO2 in an interleaved fashion (switching every 1 min), while we continuously acquired BOLD MR images. Et‐CO2 was recorded continuously during the scan using a capnograph device (Capnogard, Model 1265, Novametrix Medical Systems, CT). The total duration for the CVR scan was 7 min. The imaging parameters were identical to those of the scene‐categorization fMRI. The CVR scan was always the last scan for each subject because of the additional setup of the CO2 breathing system.
Resting state BOLD fluctuation was quantified by acquiring BOLD MR images while the subject fixated on a white cross sign on the dark screen. The subject was instructed to stay awake but not to think of anything in particular. This procedure is similar to that of a functional connectivity MRI study. The imaging parameters were the same as those of the fMRI, and the duration of the scan was 5.4 min.
Baseline CBF was measured using a phase‐contrast quantitative flow technique [Haccke et al., 1999]. A previous study used ASL MRI to obtain regional CBF estimation [Liau and Liu, 2009]. In this study, we chose to use phase‐contrast MRI because of its simplicity in absolute flow quantification compared with ASL which may be sensitive to confounding factors associated with transit time [Yang et al., 2000], trailing time [Hendrikse et al., 2003], and labeling efficiency [Aslan et al., 2010]. Phase‐contrast MRI also tends to have a shorter scan time compared with ASL [Haccke et al., 1999]. Before conducting the phase‐contrast scan, a time‐of‐flight angiogram was performed to localize the internal carotid (ICA) and vertebral arteries (VA), which are the main feeding arteries to the brain. The imaging parameters for the angiogram were: TR/TE/flip angle = 23 ms/3.45 ms/18°, FOV = 160 × 160 × 70.5 mm3, voxel size 1.0 × 1.0 × 1.5 mm3, number of slices = 47, one saturation slab of 60 mm positioned above the imaging slab to suppress the venous vessels, duration 1 min 26 s. Based on the angiogram, a single‐slice phase‐contrast MRI was performed at the level of approximately cervical spine 2, intersecting both ICA and VA. The imaging parameters were: single slice, voxel size = 0.45 × 0.45 × 5 mm3, FOV = 230 × 230 × 5 mm3, maximum velocity encoding = 80 cm/s, scan duration 30 s. In addition, a T1‐weighted anatomic image (voxel size 1 × 1 × 1 mm3, duration 4 min) was acquired to provide an estimation of the intracranial volume, so that blood flow per unit mass of tissue can be calculated, which accounts for the variances in brain sizes across subjects.
For clarity, we summarize the four parameters and their respective acquisition techniques in Table 1.
Table 1.
Summary of the acquisition techniques of the four physiologic parameters
| Physiologic parameters | Imaging technique | Acquisition duration | Task for subjects |
|---|---|---|---|
| Baseline venous oxygenation (%) | TRUST | 4 min 16 s | Rest |
| Cerebrovascular reactivity (%BOLD/mm Hg CO2) | BOLD | 7 min | Breathing 5% CO2 |
| Resting state fluctuation (SD/mean × 100%) | BOLD | 5 min | Fixation on a cross |
| Baseline CBF (ml/100 g/min) | Time‐of‐flight angiogram, Phase contrast, T1w MPRAGE | 6 min 30 s | Rest |
Data Processing
The data analysis was conducted using the software Statistical Parametric Mapping (SPM) (University College London, UK) and in‐house MATLAB scripts. The image volumes of fMRI, hypercapnia MRI, and resting state MRI were registered to their respective first volume within each scan. The mean image of the hypercapnia MRI was then coregistered to the mean image of fMRI data, and the resulting transformation was applied to each image volume in hypercapnia MRI. Similar procedures were conducted for the resting state MRI data. Next, all BOLD images were coregistered to the image template of Montreal Neurological Institute (MNI) with a resampled voxel size of 2 × 2 × 2 mm3. Finally, the BOLD images were smoothed using a Gaussian filter with a full‐width half‐maximum of 8 mm.
Detection of activated voxels in the fMRI data was conducted using a general linear analysis between the signal time course and the stimulus paradigm convoluted with a SPM‐defined hemodynamic response. Activation patterns comparing photograph viewing to fixation was determined using a one‐sample t‐test with a voxel‐wise P value of 0.001 and cluster size of 200 voxels, which corresponded to a family‐wise‐error (FWE) rate of <0.002 for the smoothness of our data. Region of interest (ROI) analysis was based on the following anatomic regions defined in WFU PickAtlas Tool (Wake Forest Univ., http://fmri.wfubmc.edu/cms/software), early visual areas (including occipital lobe and fusiform gyrus), medial temporal lobe (MTL), right inferior frontal gyrus (IFG), and left IFG. For each PickAtlas region, the top 5% most activated voxels were identified on a subject‐by‐subject basis, and these voxels were included in the final mask for signal averaging. This criterion resulted in 1,191, 300, 178, and 178 voxels (resolution 2 × 2 × 2 mm3) for visual areas, MTL, right IFG, and left IFG, respectively. The subject‐specific voxel mask as opposed to a fixed mask was used to allow for slight variations in brain anatomy across individuals. We used a fixed number of voxels for every subject so that the intersubject variations in activation volume do not present a confounding factor. The threshold value of 5% was based on a tradeoff between false positive and false negative voxels. If the threshold was chosen to be too low (e.g., top 10% voxels), many voxels included would not be truly activated and thus only add noise to the analysis. If the threshold was chosen to be too high (e.g., top 1% voxels), very few voxels will be included in the ROI, and this also results in poor stability in signal estimation. As a confirmation, we examined the cutoff t value associated with this selection criteria, which was found to be 17.0 ± 6.1 (mean ± SD, min 5.0, max 28.5), 10.4 ± 4.2 (min 1.45, max 19.0), 5.9 ± 3.0 (min 1.5, max 12.7), and 5.0 ± 2.7 (min 1.1, max 11.7) for visual areas, MTL, right IFG, and left IFG, respectively.
The physiologic modulators for each subject were estimated using their respective MRI data. TRUST MRI data were used to estimate venous blood T2, which was in turn converted to venous oxygenation (Y v) using a calibration plot [Lu and Ge, 2008; Lu et al., 2012]. This value in the sagittal sinus was used as the baseline venous oxygenation, which is thought to be homogenous across the brain at resting state [An and Lin, 2000; Fox and Raichle, 1986; He and Yablonskiy, 2007].
CVR data were processed using a GLM similar to a typical fMRI data set, except that the regressor was the Et‐CO2 time course rather than the “fMRI paradigm.” In‐house MATLAB scripts were used to obtain Et‐CO2 time courses that were synchronized with MRI acquisitions [Yezhuvath et al., 2009]. The synchronization between the Et‐CO2 time course and the BOLD time course was achieved by introducing a delay to the recorded Et‐CO2 time course, at which the Et‐CO2 provided the maximal cross‐correlation coefficient (CC) with the BOLD time course. By applying the GLM analysis on the synchronized Et‐CO2 and BOLD time courses, the absolute CVR was calculated in units of %BOLD signal change per mm Hg of Et‐CO2 change (%BOLD/mm Hg CO2). CVR value in each fMRI ROI was determined.
For resting state fluctuation, the BOLD data in the ROI were averaged, and the ratio between the temporal standard deviation and the temporal mean was calculated [Kannurpatti and Biswal, 2008], referred to as resting state fluctuation amplitude (RSFA).
Whole‐brain CBF was estimated from the phase‐contrast MRI data [Xu et al., 2009]. A mask was drawn on each of the four arteries (left and right ICA, left and right VA) based on the magnitude image. The operator was instructed to trace the boundary of the targeted vessel without including adjacent vessels. The phase signals, i.e., velocity values, within the mask were summed to yield the whole brain blood flow. To account for brain size differences, the unit volume CBF (ml/100 g/min) was then obtained by normalizing the total CBF (ml/min) to the intracranial mass (g), which was estimated from the high‐resolution T1‐weighted image using the software FSL (FMRIB Software Library, Oxford University).
We note that, of the four modulators measured in our study, CVR and RSFA can provide region‐specific value whereas Y v and CBF provide whole‐brain values only.
Statistical Analysis
Relationships between fMRI signals and the physiologic modulators were evaluated. For each modulator, a mixed‐effect analysis accounting for multiple subjects and multiple regions was used to determine whether there is a correlation between them. A multiple‐comparison‐corrected P value of <0.05 was considered significant.
CC were also computed on a region‐by‐region and modulator‐by‐modulator basis. P values associated with the CCs (uncorrected) are reported.
General linear regression analysis was used to examine the effect of combining two or more modulators in explaining fMRI variations. For example, in the case of all four modulators, the BOLD fMRI signal was written as:
| (1) |
where Y v, CVR, RSFA, and CBF are the regressors, and a i is the coefficient associated with each regressor. a Neural represents the fMRI signal after removing effects from all physiologic modulators, which may be a better marker for neural activity. Separate analyses were conducted for all possible combinations of physiologic modulators.
To evaluate whether some of the modulators may have partly overlapping physiologic origins, relationships among Y v, CVR, RSFA, and CBF were also assessed.
RESULTS
Robust fMRI activations were detected in all subjects. Group‐level activation maps are shown in Figure 1a. Based on these patterns, quantitative analysis was conducted on the following regions defined in PickAtlas (Fig. 1b): early visual areas (including occipital lobe and fusiform gyrus), MTL, right IFG, and left IFG. The BOLD percentage changes, S BOLD, in the final masks (i.e., top 5% voxels inside the anatomic ROI as described above) were 1.39 ± 0.46% (mean ± standard deviation, N = 28), 0.71% ± 0.22%, 0.47% ± 0.21%, and 0.39% ± 0.18% for these regions, respectively.
Figure 1.
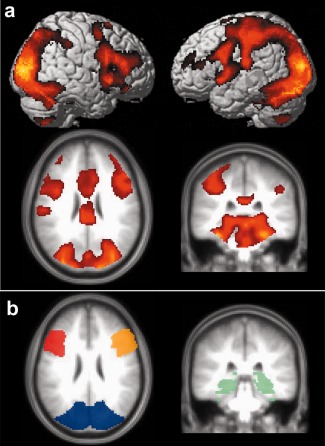
Group‐level activation results comparing photograph viewing to fixation. (a) Activation maps. Threshold: P value < 0.001 in one‐sample t test, cluster > 200 voxels. The top panels show rendered views and the bottom panels show cross‐sectional views. The activated brain regions included early visual areas (including occipital lobe and fusiform gyrus), MTL, left IFG, right IFG, left pre/postcentral gyrus, insula, and anterior cingulated gyrus. (b) Anatomic ROIs defined in PickAtlas encompassing representative activated regions: early visual areas (blue), MTL (green), right IFG (orange), left IFG (red). For each individual, the final mask used for averaging consists of the most activated voxels in these ROIs. [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
Figure 2 shows a representative data set for the four physiologic measurements. TRUST MRI data yielded baseline venous oxygenation, Y v, of 63.6 ± 4.4% (N = 28). Whole brain CVR value was 0.19% ± 0.03%/mm Hg CO2, while regional mean CVR ranged from 0.16%–0.23%/mm Hg CO2. Whole brain RSFA value was 0.23% ± 0.07%, while regional mean RSFA varied between 0.37% and 0.55% among all areas studied. Whole brain CBF value was 60.6 ± 7.0 ml/100 g/min.
Figure 2.
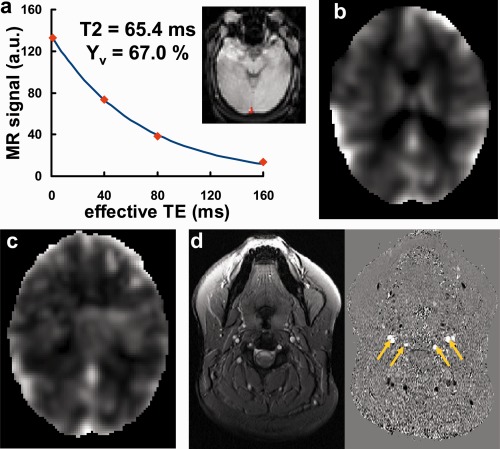
Results of the four physiological modulators from a representative subject. (a) T2 fitting of the TRUST data. Inset: ROI in the sagittal sinus. The fitted T2 was 65.4 ms, corresponding to a Y v of 67.0%. (b) CVR map from hypercapnia challenge using 5% CO2. (c) Map of resting state BOLD fluctuation (SD/mean × 100%). (d) Anatomic (left) and phase (right) images from phase‐contrast MRI. Yellow arrows indicate the four feeding arteries of the brain. [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
Comparison between fMRI and each of the physiologic modulators revealed that, across individuals, the BOLD signal had a significantly negative correlation with Y v (mixed effect model using all brain regions studied, P < 0.0001), while it was positively correlated with CVR (P < 0.0001) and RSFA (P = 0.002). These findings are consistent with previous reports on individual modulators [Handwerker et al., 2007; Kannurpatti and Biswal, 2008; Lu et al., 2008; Thomason et al., 2007]. Figure 3a,b show scatter plots comparing fMRI signal to Y v and CVR, respectively, where the brain regions of interest, i.e., early visual areas, MTL, right IFG, and left IFG, are shown in separate panels. We did not find a significant correlation between fMRI signals and whole‐brain CBF as measured by phase‐contrast MRI (P = 0.28 using the mixed effect model), although the slopes between them were found to be negative for all brain regions studied (−0.0034% ± 0.0020%/ml/100 g/min, N = 4, one‐sample t test, P = 0.04), consistent with the relationship reported in a previous study [Liau and Liu, 2009].
Figure 3.
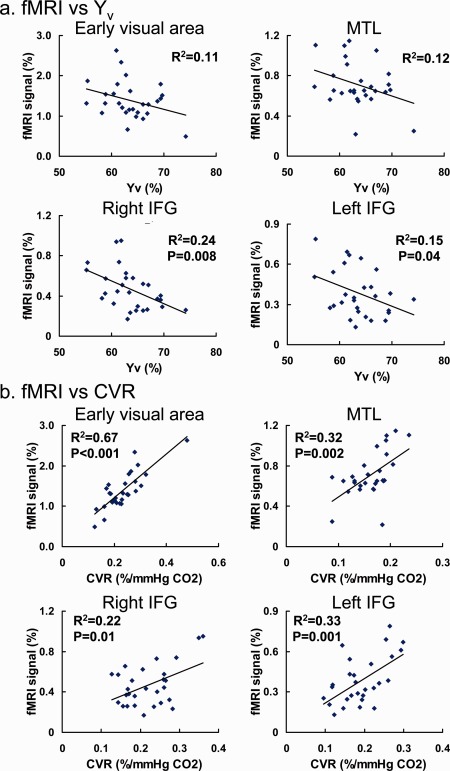
Scatter plots between fMRI signals and the physiologic modulators in activated brain regions. Each symbol in the plot represents data from one subject. The solid curves show the linear fitting of the experimental data. (a) Relationship between fMRI signal and Y v. (b) Relationship between fMRI signal and CVR. [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
Next, we investigated the potential benefit of combing multiple modulators in explaining fMRI variations. Table 2 shows the fraction of fMRI variance explained by combinations of modulators. The inclusion of all four physiologic parameters in the multilinear regression model explained 42%–74% of the total variance (Table 2), with early visual areas having a much bigger value than the higher order cortical areas. This may be attributed to the fact that, in higher order cortical areas with smaller fMRI signals, the relative contribution of random noise to the intersubject variation is greater and cannot be corrected by the physiologic modulators. Further evaluations of the intercorrelations among the modulators themselves revealed that Y v is significantly correlated with CBF (P = 0.01, Fig. 4a), and that CVR is significantly correlated with RSFA (P = 0.05, Fig. 4b). Thus, a subset of these parameters may be sufficient to explain the majority of the variance. For example, the pair of (Y v, CVR) was able to explain almost as much as that explained by all four modulators (0.49 vs. 0.52 in Table 2).
Table 2.
Fraction of inter‐subject fMRI variance explained by different combinations of physiologic modulators (calculated as R 2 from multiple regression analysis)
| ROI | Y v | CVR | RSFA | CBF | Y v CVR | CBF CVR | Y v RSFA | CBF RSFA | Y v CBF | CVR RSFA | Y v CVR CBF | Y v CVR RSFA | CBF CVR RSFA | Y v CBF RSFA | All four |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Early visual areas | 0.11 | 0.67 | 0.16 | <0.01 | 0.74 | 0.70 | 0.23 | 0.16 | 0.12 | 0.67 | 0.74 | 0.74 | 0.70 | 0.24 | 0.74 |
| MTL | 0.12 | 0.32 | 0.09 | <0.01 | 0.41 | 0.34 | 0.21 | 0.09 | 0.14 | 0.33 | 0.41 | 0.42 | 0.33 | 0.24 | 0.42 |
| Right IFG | 0.24 | 0.22 | 0.03 | 0.04 | 0.44 | 0.24 | 0.25 | 0.07 | 0.24 | 0.07 | 0.44 | 0.45 | 0.27 | 0.25 | 0.46 |
| Left IFG | 0.15 | 0.33 | 0.23 | 0.01 | 0.38 | 0.33 | 0.37 | 0.24 | 0.16 | 0.37 | 0.39 | 0.43 | 0.37 | 0.39 | 0.45 |
| Mean | 0.15 | 0.39 | 0.13 | 0.01 | 0.49 | 0.40 | 0.26 | 0.14 | 0.16 | 0.36 | 0.50 | 0.51 | 0.42 | 0.28 | 0.52 |
| SD | 0.06 | 0.20 | 0.09 | 0.02 | 0.16 | 0.20 | 0.07 | 0.08 | 0.05 | 0.25 | 0.16 | 0.15 | 0.19 | 0.07 | 0.15 |
Figure 4.
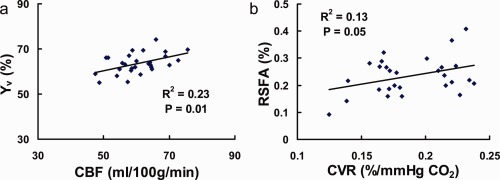
Relationship among the physiologic modulators. Each symbol in the plot represents data from one subject. The solid curves show the linear fitting of the experimental data. (a) Scatter plot between baseline venous oxygenation (Y v) and global CBF, P = 0.01. (b) Scatter plot between whole brain CVR and RSFA, P = 0.05. [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
DISCUSSION
The main findings from this study can be summarized by the following four observations. (1) We reproduced the effects of three physiologic modulators on fMRI signals simultaneously in one session. (2) Using a cognitively relevant paradigm, we demonstrated that the modulation effect appeared to be present throughout the brain, extending previous findings in primary sensory or motor cortices. (3) The four reported modulators (including CBF) showed correlations among themselves, thus a subset of these parameters may be sufficient to explain the majority of the variance. (4) We demonstrated that physiologic modulation of fMRI signals can also be measured in event‐related design, a feature that has been previously demonstrated for CVR [Handwerker et al., 2007; Thomason et al., 2007] but not for the other modulators. The potential of using physiologic modulators in event‐related design may present an advantage over the calibrated fMRI approach for which virtually all studies conducted have used a block design.
FMRI data suffer from two types of noise: thermal noise associated with equipment sensitivity and stability, and physiologic noise associated with variations or fluctuations of living systems [Hu and Kim, 1994; Kruger and Glover, 2001]. With recent advances in high‐field MR systems and improved hardware performance, the thermal noise is expected to be a manageable component and the ultimate sensitivity and specificity of fMRI is likely to be determined by physiologic noise of the data [Triantafyllou et al., 2005]. FMRI modulation by physiologic parameters in practice presents a significant source of noise in clinical studies [D'Esposito et al., 1999]. Physicians in psychiatry and neurology have high expectations for fMRI, as it can potentially provide a useful marker for diagnosis of disease on an individual level. However, different individuals (even in healthy controls) may have vastly different physiologic states, thus the ability to separate abnormal fMRI responses from normal ones is considerably diminished by physiologic modulator effects. Only after these physiologic modulators are measured and accounted for can fMRI signal become a “personalized” marker. That is, the physician can interpret the fMRI signal in the context of each patient's physiologic state. At present, a typical fMRI session consists of localizers, anatomic scans, and functional scans. Based on our observations as well as findings in recent literature [Handwerker et al., 2007; Kannurpatti and Biswal, 2008; Liau and Liu, 2009; Lu et al., 2008, 2010; Thomason et al., 2007], we recommend that a measurement of physiologic modulator(s) be added to the standard protocol.
The modulation effects observed in this study are in general agreement with literature reports, where the modulators were examined individually. Specifically, the fMRI signal was greater in subjects with lower baseline oxygenation [Lu et al., 2008, 2010], higher vascular reactivity [Handwerker et al., 2007; Liau and Liu, 2009; Thomason et al., 2007], and higher resting state fluctuation [Kannurpatti and Biswal, 2008; Kannurpatti et al., 2011]. The correlation between fMRI signal and whole‐brain CBF was not significant (although all regions studied showed a consistent slope). One difference between the CBF technique used in this study and that used in the previous studies was that we have used a phase‐contrast MRI instead of ASL [Liau and Liu, 2009]. Phase‐contrast MRI was tested because this technique requires relatively short scan time, which is an advantage in patient studies and because of its simplicity in flow quantification compared with ASL (without the need of measuring arterial transit time, tissue T1, tissue M0, etc.). However, a limitation of this technique is the lack of regional values, which may be more relevant to local BOLD signal. Regional measures of a modulator also automatically account for gray/white partial volume effects in BOLD signal. It is probably for these reasons that the phase‐contrast CBF used in this study only showed the correct trend but did not show statistically significant correlation with fMRI. It is also interesting to note that the use of whole brain Y v did not appear to suffer from this issue, possibly because oxygen extraction fraction is more homogeneous across the brain compared with CBF or cerebral metabolic rate of oxygen (CMRO2) [An and Lin, 2000; Fox and Raichle, 1986; He and Yablonskiy, 2007].
Among the four physiological modulators, baseline venous oxygenation and CBF were correlated across subjects (Fig. 4a). One possible mechanism is that CMRO2 is relatively constant across individuals. However, blood supply could vary considerably across subjects due to a number of vascular factors such as breathing pattern, consumption of vasoactive substance (e.g., caffeine), and blood pressure [Liau et al., 2008; Wise et al., 2004]. Then, given the relationship CMRO2 = CBF × (1 − Y v) [Kety and Schmidt, 1948], it can be readily expected that an individual with higher CBF tends to have a higher Y v. We also observed a marginal correlation between CVR and RSFA (Fig. 4b). This observation is comparable with those reported by Kannurpatti and Biswal 2008. A possible explanation is that both CVR and RSFA reflect to some extent the vessel's ability to expand in response to CO2 content in the blood. The difference between these two parameters is that the CO2 change in CVR measurement was induced by hypercapnia challenge whereas that in RSFA was due to spontaneous variations in breathing depth resulting in small fluctuations in CO2 levels [Birn et al., 2006; Kannurpatti and Biswal, 2008; Wise et al., 2004]. However, the correlation between CVR and RSFA was relatively weak, possibly because RSFA is also affected by other factors such as spontaneous neural activity, bulk motion, and the extent of breathing variations.
No correlation was observed between Y v and CVR even though both were found to be significant modulators of fMRI. Therefore, it seems that these two parameters affect the fMRI signals via different mechanisms. One way to understand this is to examine the classic equation for the BOLD signal [Davis et al., 1998; Hoge et al., 1999]:
| (2) |
where M reflects baseline physiological state and is given by M = TE · A · CBV0 · B β · (1 − Y v)β, A and B are constants, and β is related to vascular geometry. In Eq. (2), ΔCMRO2/CMRO20 is thought to be directly related to neural activity. The other terms can be considered to be modulators of fMRI signals. Therefore, it seems that Y v could affect fMRI signals by modulating M. On the other hand, CVR may affect the signals by modulating the term ΔCBF/CBF0 in Eq. (2). They indeed seem to influence the signal in independent manners.
A systematic investigation to compare these physiologic modulators is useful for determining optimal acquisition and processing strategies in an fMRI session. Given the above findings that the modulators showed potential correlations among themselves, it is reasonable to expect that the acquisition of two, rather than four, of the modulators may be adequate to account for the majority of physiology‐related variance in fMRI signals. Indeed, our data suggest that about 94% (= 0.49/0.52 in Table 2) of the total variance explained by the four modulators can be accounted for by the combination of Y v and CVR. Another factor to consider in designing a study protocol is complexity and subject burden associated with the measurements. Among the four physiologic parameters investigated, three of them, Y v, CBF, and RSFA, are baseline measures and do not involve any tasks. CVR measurement, on the other hand, requires a hypercapnia challenge in the form of either CO2 inhalation or breath‐holding. This is sometimes considered inconvenient or impractical. If hypercapnia is not desired, an alternative set of physiologic parameters is Y v and RSFA, which can explain 50% (= 0.26/0.52 in Table 2) of the total variance explained by the four modulators. If scan duration is a significant constraint, one can consider including only one of the modulators. The time it takes to acquire each of these parameters ranges from 4 to 7 min (Table 1). Note, however, that further technical development may reduce the duration of these techniques. For example, the Y v method used in this study took 4 min and 16 s. A recent improvement of the TRUST technique has reduced the scan duration to 1 min and 12 s without sacrificing measurement accuracy [Xu et al., 2011].
The fMRI modulation model described in this study can be extended to clinical studies to compare fMRI signals between two subject populations or between two time points. This can be done by including an additional term in Eq. (1), a Group · GroupIndex, where GroupIndex represents whether the subject is a patient or control, or whether it is Time point #1 or Time point #2. a Group is the coefficient associated with the GroupIndex. The statistical test is then to evaluate if a Group is significantly different from zero. If so, the data would suggest that there is a group difference or time dependence in fMRI signals. The presence of the physiologic modulators serves to reduce intragroup variance and to improve intergroup separation. A demonstration of such a utility accounting for only Y v‐related variance was presented previously [Lu et al., 2010].
Compared with the calibrated fMRI approach [Chiarelli et al., 2007a, b; Davis et al., 1998; Hoge et al., 1999; Sicard and Duong, 2005; Uludag et al., 2004], this normalization method does not provide a quantitative assessment of brain metabolism, thus the interpretation of the data is still based on BOLD fMRI signal itself, which does not have a direct link to neural activity. On the other hand, it provides a more practical approach to control for non‐neural factors in fMRI signals and may improve the sensitivity and specificity of fMRI in distinguishing abnormal neural activity. Importantly, the proposed method does not require the alteration of existing fMRI acquisition protocol, unlike the calibrated fMRI in which simultaneous ASL and BOLD acquisitions are necessary [Chiarelli et al., 2007a, b; Sicard and Duong, 2005]. Simultaneous ASL and BOLD acquisitions often result in smaller brain coverage, lower SNR, and poorer temporal resolutions. The proposed method functions by including additional measurements of physiologic parameters and, while it modestly increases the total scan duration, the researchers maintain the options to examine the original data.
The physiologic modulators measured in this study were able to explain some (42%–74%), but not all, of the variance in fMRI signals. Note that even a slight reduction in intersubject variations may be useful in improving statistical power in clinical applications. We performed a simple Monte Carlo simulation in which the fMRI signal of the control and patient groups were assumed to be 1.0 ± 0.3 and 0.8 ± 0.3, respectively. The relationship between detection power and sample size is shown by the black curve in Supporting Information Figure S1. Then, we reduced the variance by 15%, 30%, 45%, and 60% (from a full variance of 0.3), it was found that, at a power of 80%, the required sample size decreased by 26%, 47%, 66%, and 79%, respectively (colored curves in Supporting Information Fig. S1). That is, if our modulator(s) can explain 30% of the variance, the sample size can be reduced to about half to achieve the same power.
A limitation of this study is that the correlations between fMRI signals and individual modulators were only modest and many would not survive a Bonferroni correction given the relatively large number of regions and modulators examined. This could be attributed to a number of possible reasons. One is that an event‐related fMRI design was used and the reliability of the fMRI signal is lower than that of a block‐design. Second, even if the measurement noise is negligible, the correlation between fMRI signal and each modulator is not expected to be very high, because the modulation effect from one modulator will become “noise” in the correlation analysis between fMRI and another modulator. Potential intersubject variability in neural signal further adds “noise” to the correlation analysis.
CONCLUSIONS
FMRI signal of an individual is simultaneously modulated by baseline venous oxygenation, CVR to CO2, resting state BOLD fluctuations, and possibly baseline CBF. These effects seem to be present throughout the brain. The modulators showed certain correlations among themselves, thus the measurement of two may be sufficient to explain most of the physiologic variance. These findings suggest that additional measurement of physiologic modulator(s) in an fMRI session may provide a practical approach to control for intersubject variations and to improve the ability of fMRI in detecting disease or medication effects.
Supporting information
Supporting Information Figures
REFERENCES
- An H, Lin W (2000): Quantitative measurements of cerebral blood oxygen saturation using magnetic resonance imaging. J Cereb Blood Flow Metab 20:1225–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ances B, Vaida F, Ellis R, Buxton R (2011): Test‐retest stability of calibrated BOLD‐fMRI in HIV‐ and HIV+ subjects. Neuroimage 54:2156–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ances BM, Liang CL, Leontiev O, Perthen JE, Fleisher AS, Lansing AE, Buxton RB (2009): Effects of aging on cerebral blood flow, oxygen metabolism, and blood oxygenation level dependent responses to visual stimulation. Hum Brain Mapp 30:1120–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslan S, Xu F, Wang PL, Uh J, Yezhuvath US, van Osch M, Lu H (2010): Estimation of labeling efficiency in pseudocontinuous arterial spin labeling. Magn Reson Med 63:765–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandettini PA, Wong EC (1997): A hypercapnia‐based normalization method for improved spatial localization of human brain activation with fMRI. NMR Biomed 10:197–203. [DOI] [PubMed] [Google Scholar]
- Birn RM, Diamond JB, Smith MA, Bandettini PA (2006): Separating respiratory‐variation‐related fluctuations from neuronal‐activity‐related fluctuations in fMRI. Neuroimage 31:1536–1548. [DOI] [PubMed] [Google Scholar]
- Chiarelli PA, Bulte DP, Piechnik S, Jezzard P (2007a): Sources of systematic bias in hypercapnia‐calibrated functional MRI estimation of oxygen metabolism. Neuroimage 34:35–43. [DOI] [PubMed] [Google Scholar]
- Chiarelli PA, Bulte DP, Wise R, Gallichan D, Jezzard P (2007b): A calibration method for quantitative BOLD fMRI based on hyperoxia. Neuroimage 37:808–820. [DOI] [PubMed] [Google Scholar]
- D'Esposito M, Zarahn E, Aguirre GK, Rypma B (1999): The effect of normal aging on the coupling of neural activity to the bold hemodynamic response. Neuroimage 10:6–14. [DOI] [PubMed] [Google Scholar]
- Davis TL, Kwong KK, Weisskoff RM, Rosen BR (1998): Calibrated functional MRI: Mapping the dynamics of oxidative metabolism. Proc Natl Acad Sci USA 95:1834–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleisher AS, Podraza KM, Bangen KJ, Taylor C, Sherzai A, Sidhar K, Liu TT, Dale AM, Buxton RB (2009): Cerebral perfusion and oxygenation differences in Alzheimer's disease risk. Neurobiol Aging 30:1737–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folstein MF, Folstein SE, McHugh PR (1975): “Mini‐mental state”. A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res 12:189–198. [DOI] [PubMed] [Google Scholar]
- Fox PT, Raichle ME (1986): Focal physiological uncoupling of cerebral blood flow and oxidative metabolism during somatosensory stimulation in human subjects. Proc Natl Acad Sci USA 83:1140–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutchess AH, Welsh RC, Hedden T, Bangert A, Minear M, Liu LL, Park DC (2005): Aging and the neural correlates of successful picture encoding: Frontal activations compensate for decreased medial‐temporal activity. J Cogn Neurosci 17:84–96. [DOI] [PubMed] [Google Scholar]
- Haccke EM, Brown RW, Thompson MR, Venkatesan R (1999): MR Angiography and Flow Quantification. Magnetic Resonance Imaging: Physical Principles and Sequence Design. New York: Wiley Periodicals. [Google Scholar]
- Handwerker DA, Gazzaley A, Inglis BA, D'Esposito M (2007): Reducing vascular variability of fMRI data across aging populations using a breathholding task. Hum Brain Mapp 28:846–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Yablonskiy DA (2007): Quantitative BOLD: Mapping of human cerebral deoxygenated blood volume and oxygen extraction fraction: Default state. Magn Reson Med 57:115–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrikse J, Lu H, van der Grond J, Van Zijl PC, Golay X (2003): Measurements of cerebral perfusion and arterial hemodynamics during visual stimulation using TURBO‐TILT. Magn Reson Med 50:429–433. [DOI] [PubMed] [Google Scholar]
- Hoge RD, Atkinson J, Gill B, Crelier GR, Marrett S, Pike GB (1999): Linear coupling between cerebral blood flow and oxygen consumption in activated human cortex. Proc Natl Acad Sci USA 96:9403–9408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Kim SG (1994): Reduction of signal fluctuation in functional MRI using navigator echoes. Magn Reson Med 31:495–503. [DOI] [PubMed] [Google Scholar]
- Jezzard P, Buxton RB (2006): The clinical potential of functional magnetic resonance imaging. J Magn Reson Imaging 23:787–793. [DOI] [PubMed] [Google Scholar]
- Kannurpatti SS, Biswal BB (2008): Detection and scaling of task‐induced fMRI‐BOLD response using resting state fluctuations. Neuroimage 40:1567–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannurpatti SS, Motes MA, Rypma B, Biswal BB (2011): Non‐neural BOLD variability in block and event‐related paradigms. Magn Reson Imaging 29:140–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kety SS, Schmidt CF (1948): The effects of altered arterial tensions of carbon dioxide and oxygen on cerebral blood flow and cerebral oxygen consumption of normal young men. J Clin Invest 27:484–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SG, Ugurbil K (1997): Comparison of blood oxygenation and cerebral blood flow effects in fMRI: Estimation of relative oxygen consumption change. Magn Reson Med 38:59–65. [DOI] [PubMed] [Google Scholar]
- Kruger G, Glover GH (2001): Physiological noise in oxygenation‐sensitive magnetic resonance imaging. Magn Reson Med 46:631–637. [DOI] [PubMed] [Google Scholar]
- Liau J, Liu TT (2009): Inter‐subject variability in hypercapnic normalization of the BOLD fMRI response. Neuroimage 45:420–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liau J, Perthen JE, Liu TT (2008): Caffeine reduces the activation extent and contrast‐to‐noise ratio of the functional cerebral blood flow response but not the BOLD response. Neuroimage 42:296–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Ge Y (2008): Quantitative evaluation of oxygenation in venous vessels using T2‐Relaxation‐Under‐Spin‐Tagging MRI. Magn Reson Med 60:357–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Xu F, Grgac K, Liu P, Qin Q, van Zijl P (2012): Calibration and validation of TRUST MRI for the estimation of cerebral blood oxygenation. Magn Reson Med 67:42–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Yezhuvath US, Xiao G (2010): Improving fMRI sensitivity by normalization of basal physiologic state. Hum Brain Mapp 31:80–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Zhao C, Ge Y, Lewis‐Amezcua K (2008): Baseline blood oxygenation modulates response amplitude: Physiologic basis for intersubject variations in functional MRI signals. Magn Reson Med 60:364–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofen N, Kao YC, Sokol‐Hessner P, Kim H, Whitfield‐Gabrieli S, Gabrieli JD (2007): Development of the declarative memory system in the human brain. Nat Neurosci 10:1198–1205. [DOI] [PubMed] [Google Scholar]
- Park DC, Welsh RC, Marshuetz C, Gutchess AH, Mikels J, Polk TA, Noll DC, Taylor SF (2003): Working memory for complex scenes: Age differences in frontal and hippocampal activations. J Cogn Neurosci 15:1122–1134. [DOI] [PubMed] [Google Scholar]
- Sicard KM, Duong TQ (2005): Effects of hypoxia, hyperoxia, and hypercapnia on baseline and stimulus‐evoked BOLD, CBF, and CMRO2 in spontaneously breathing animals. Neuroimage 25:850–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovic B, Warnking JM, Kobayashi E, Bagshaw AP, Hawco C, Dubeau F, Gotman J, Pike GB (2005): Hemodynamic and metabolic responses to activation, deactivation and epileptic discharges. Neuroimage 28:205–215. [DOI] [PubMed] [Google Scholar]
- Thomason ME, Foland LC, Glover GH (2007): Calibration of BOLD fMRI using breath holding reduces group variance during a cognitive task. Hum Brain Mapp 28:59–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triantafyllou C, Hoge RD, Krueger G, Wiggins CJ, Potthast A, Wiggins GC, Wald LL (2005): Comparison of physiological noise at 1.5 T, 3 T and 7 T and optimization of fMRI acquisition parameters. Neuroimage 26:243–250. [DOI] [PubMed] [Google Scholar]
- Uludag K, Dubowitz DJ, Yoder EJ, Restom K, Liu TT, Buxton RB (2004): Coupling of cerebral blood flow and oxygen consumption during physiological activation and deactivation measured with fMRI. Neuroimage 23:148–155. [DOI] [PubMed] [Google Scholar]
- Wise RG, Ide K, Poulin MJ, Tracey I (2004): Resting fluctuations in arterial carbon dioxide induce significant low frequency variations in BOLD signal. Neuroimage 21:1652–1664. [DOI] [PubMed] [Google Scholar]
- Xu F, Ge Y, Lu H (2009): Noninvasive quantification of whole‐brain cerebral metabolic rate of oxygen (CMRO2) by MRI. Magn Reson Med 62:141–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Uh J, Liu P, Lu H (2011): On improving the speed and reliability of T2‐Relaxation‐Under‐Spin‐Tagging (TRUST) MRI. Magn Reson Med (in press). DOI: 10.1002/mrm.23207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Engelien W, Xu S, Gu H, Silbersweig DA, Stern E (2000): Transit time, trailing time, and cerebral blood flow during brain activation: Measurement using multislice, pulsed spin‐labeling perfusion imaging. Magn Reson Med 44:680–685. [DOI] [PubMed] [Google Scholar]
- Yezhuvath US, Lewis‐Amezcua K, Varghese R, Xiao G, Lu H (2009): On the assessment of cerebrovascular reactivity using hypercapnia BOLD MRI. NMR Biomed 22:779–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Figures


