Abstract
Environmentally induced epigenetic alterations are related to mental health. We investigated quantitative DNA methylation status before and after an acute psychosocial stressor in two stress-related genes: oxytocin receptor (OXTR) and brain-derived neurotrophic factor (BDNF ). The cross sectional study took place at the Division of Theoretical and Clinical Psychobiology, University of Trier, Germany and was conducted from February to August 2009. We included 83 participants aged 61–67 years. Thereof, 76 participants completed the full study procedure consisting of blood sampling before (pre-stress), 10 min after (post-stress) and 90 min after (follow-up) the Trier social stress test. We assessed quantitative DNA methylation of whole-blood cells using Sequenom EpiTYPER. Methylation status differed between sampling times in one target sequence of OXTR (P<0.001): methylation increased from pre- to post-stress (P=0.009) and decreased from post-stress to follow-up (P<0.001). This decrease was also found in a second target sequence of OXTR (P=0.034), where it lost statistical significance when blood cell count was statistically controlled. We did not detect any time-associated differences in methylation status of the examined BDNF region. The results suggest a dynamic regulation of DNA methylation in OXTR—which may in part reflect changes in blood cell composition—but not BDNF after acute psychosocial stress. This may enhance the understanding of how psychosocial events alter DNA methylation and could provide new insights into the etiology of mental disorders.
Keywords: acute psychosocial stress, brain-derived neurotrophic factor gene (BDNF ), dynamic DNA methylation, epigenetics, oxytocin receptor gene (OXTR)
Introduction
DNA methylation is an epigenetic mechanism related to mental and physical health and disease.1, 2, 3, 4 Aberrant DNA methylation has been implicated in the etiology of various mental disorders including, depression,5, 6, 7, 8, 9 psychotic disorders,10, 11, 12, 13, 14, 15 post-traumatic stress disorder,16, 17 autism,18, 19 eating disorders20, 21 and substance dependence (for review see22), but also has an important role in the pathology of physical illnesses, such as cancer.23 Thereby DNA methylation provides a biological basis for gene–environment interactions relevant to mental health24: animal and human studies have found that early life experiences can alter DNA methylation and affect gene expression and behavior.25, 26, 27, 28, 29, 30, 31, 32 Similarly, experiences later in life can modify the epigenome.33, 34 However, changes in DNA methylation immediately after adverse experiences, such as acute psychosocial stress, have not yet been investigated. Insight into how acute psychosocial stress affects DNA methylation may further elucidate our understanding of etiological mechanisms in mental health. Therefore, we investigated DNA methylation of two stress-related candidate genes—oxytocin receptor (OXTR)35 and brain-derived neurotrophic factor (BDNF)35, 36—before and after an acute psychosocial stressor.
We included the OXTR because the oxytocin system interacts with the hypothalamic-pituitary-adrenal axis35, 37, 38, 39, 40 and cardiovascular stress reactivity.41, 42 To the best of our knowledge, there have been no studies investigating methylation of OXTR with reference to stress in humans or animals. A study on patients suffering from autism spectrum disorder revealed aberrant DNA methylation in an OXTR region in peripheral mononuclear blood cells; similar results were found for brain tissue.43
BDNF, the second candidate gene, encodes a neuronal growth factor involved in neuronal development, cell differentiation and synaptic plasticity.44, 45 In addition to its pivotal role in the central nervous system, BDNF is also expressed in the periphery where it shows neuro-protective action.46 Peripheral BDNF concentration is decreased in various stress-related mental disorders47 including depression48 and post-traumatic stress disorder.49 Previous work has also shown that early life- and chronic stress resulted in a higher methylation status of Bdnf,32 and a decrease in Bdnf mRNA and BDNF protein levels in several rodent brain areas.32, 50, 51, 52, 53, 54 Animal studies have examined dynamic changes in DNA methylation of Bdnf associated with memory, learning and physical activity,55, 56, 57 but not following a psychosocial stressor.
The aim of this study was to investigate dynamic changes in DNA methylation in stress-related genes after an acute psychosocial stressor.
Materials and methods
Participants and procedure
The sample of this cross-sectional study consisted of 76 adults. All participants underwent three sequential study parts:
In the first study part (2006–2007), we contacted Trier inhabitants born between 1942 and 1947. This population experienced war adversities early in life and was chosen as the overall goal of the initial project was to assess long-term consequences of early adversities. Of 2117 contacted adults, 365 completed psychological and medical questionnaires. In the second study part (2007–2008), we invited participants from the first study part for a psychological interview. Thereby, exclusion criteria were medical conditions potentially interfering with planned biological measures: impaired general health status, signs of acute infection, untreated hypertension (blood pressure >160/95 mm Hg during unstimulated conditions) or diabetes mellitus, intake of glucocorticoid-, immunosuppressive-, anti-depressant- or antidiabetic-medication, current therapy for a mental disorder and previous participation in a study applying the Trier social stress test (TSST).58, 59 We invited 274 persons, of whom 179 completed the interview. The third study part (2009) consisted of a laboratory session at the Division of Theoretical and Clinical Psychobiology, University of Trier, Germany. Those participants of the second study part who did not suffer from clinically relevant symptoms of depression (assessed by the German Version of the Center for Epidemiological Studies Depression Scale60) and did not meet the above-described exclusion criteria were invited for the third study part. Of the 127 invited participants, 83 took part in the third study part. Blood samples of seven participants did not contain enough blood for analysis and had to be excluded from the statistical analyses. Thus the final sample consisted of 76 adults—43 women and 33 men—aged between 61 and 67 years (mean age: 64.11 years; s.d.: 1.65 years). Participants gave written informed consent in accordance with the Declaration of Helsinki and received financial compensation. The Chamber of Physicians (Landesärztekammer Rheinland-Pfalz, Germany) approved the study protocol.
For the laboratory session of the third study wave, we requested participants to abstain from heavy sports and alcohol the evening before and on the day of testing. In addition, they were asked to have a regular meal on the testing day and to avoid eating and drinking 2 hours before arriving at the laboratory. Upon arrival, we acquainted the participants with the staff and informed them about the general procedures. A study physician undertook a medical examination and placed a peripheral venous catheter into the antecubital vein of the nondominant arm for multiple blood draws. A study assistant then conducted two memory tests before starting with the TSST, which took place in a remote room, equipped with a standing microphone and a video camera in front of two desks. The TSST consisted of a 3-min anticipation period and a 10-min test period, during which the participants had to undergo a fictitious job interview and perform mental arithmetics in front of one male and one female expert, trained in behavioral observation techniques, as well as in abstaining from giving any positive or negative social cues. The experts were of about the same age as the participants themselves, wore white doctor coats and used stop watches in order to check time. Study participants were informed that they would be video- and speech-taped during the whole test period for later evaluation of his or her performance and behavior. After the TSST, we accompanied the participants back to a study room, where they were asked to complete two additional memory tests and to fill in several questionnaires. The whole study session took 3.5 h.
Blood sampling, pre-analytics and blood cell count
At each blood sample collection, a study physician drew 5.5-ml blood from a peripheral venous catheter (Vasofix safety, Braun Melsungen AG, 18G, Melsungen, Germany) in EDTA-coated S-Monovettes (Sarstedt, Nuembrecht, Germany). Blood was taken 1 min before the TSST (pre-stress sampling), 10 min after the TSST (post-stress sampling) and 90 min after the TSST (follow-up sampling). To avoid acute orthostatic influences on pre-stress blood analyses, we asked persons to stand up and remain standing 10 min before the TSST until we collected the pre-stress blood sample.
EDTA samples collected for later blood counts were stored without centrifugation in a refrigerator until the end of the testing session. We delivered these samples to an external laboratory (SynLab Trier, Trier, Germany) the same day. Complete blood cell counts were obtained using an automated haematology analyzer (Sysmex XE2100i, Norderstedt, Germany).
Immediately after collection, EDTA samples for DNA methylation analysis were put on ice and centrifuged within 5 min (4000 rpm at +6 °C for 10 min) before freezing at –80 °C. The QIAamp DNA Blood Midi (Qiagen, Hilden, Germany) was used to extract DNA, following the manufacturer's protocol. Samples were stored at −20 °C for subsequent DNA methylation analysis.
DNA methylation analysis
Genomic DNA (540 ng) was treated with sodium bisulfite using the EZ-96 DNA Methylation Kit (Zymo Research, CA, USA) according to the manufacturers' standard protocol. Bisulfite PCR amplification of two target sequences in OXTR (OXTR1, OXTR2) and one target sequence in BDNF was conducted using Hot Star Taq DNA polymerase (Qiagen). The OXTR1 target sequence is located in the protein-coding region of OXTR exon III; the OXTR2 target sequence partly covers the noncoding and protein-coding promoter regions of OXTR exon III. Both target sequences were designed to cover the OXTR promoter region and the CpG island comprising exons I–III.61 The BDNF target sequence around the 3′ end of BDNF exon VI is situated mainly within a CpG island that covers BDNF exons V, Vh and VI.62 BDNF exon VI is frequently expressed especially in non-neuronal tissue of the periphery.62 PCR products were prepared according to the manufacturer's standard protocol for quantitative DNA methylation analysis using EpiTYPER 1.0 (Sequenom, CA, USA). For each run, a fully methylated positive control (New England BioLabs) and a blank control (distilled water) were included. The assays for the amplicons were designed using the Sequenom EpiDesigner software (for target sequences see Supplementary information).
Statistical analysis
The resolution of EpiTYPER yielded CpG units consisting of 1–6 individual CpG sites: 11 CpG units for OXTR1, 28 CpG units for OXTR2 and 12 CpG units for BDNF. Two CpG units in OXTR1, one CpG unit in OXTR2 and one CpG unit in BDNF could not be measured because of upper and lower detection limits of Sequenom EpiTYPER. CpG units with >20% missing data were excluded, which left eight CpGs units for OXTR1, 27 for OXTR2 and 10 for BDNF for statistical analyses. All sample CpG methylation values were compared with the values of the fully methylated positive control: If the value of the sample exceeded the value of the positive control, the value was set as missing data. All blank controls were negative. We identified suspicious samples by setting outlier values as missing data (⩾3 s.d.'s from mean methylation of the respective CpG unit). By conducting missing analyses separately for each gene, samples with >20% missing data were identified and excluded from the statistical analyses of the respective gene. Methylation values (OXTR2 and BDNF) were log-transformed to meet assumptions of normality and homoscedasticity. We analyzed time-associated changes in mean DNA methylation (averaged across CpG units) using multilevel models.63 The three hierarchical levels were subjects, CpGs within subjects, and time within CpGs within subjects. Methylation values were allowed to vary across time for individual CpGs within subjects as this improved model fit. In a first step, we examined overall effects of sampling time and analyzed differences between the three sampling time points using post-hoc contrasts. In a second step, we tested the same overall effect of sampling time while including blood cell counts as covariates.64 In both models, we included the identities of the bisulfite conversion plates and the Sequenom plates as covariates to negate laboratory batch effects. As we did not find any gender differences in DNA methylation, we did not include gender as potential confounder in the final models. We considered an alpha level of <0.05 as significant. All analyses were conducted using SPSS 20. DNA methylation values are presented as percent of cytosine methylation (%5MeC).
Results
Estimated means from the multilevel model of methylation (%5MeC) averaged across all CpG units of a target sequence and descriptive values of blood cell count are shown in Table 1.
Table 1. Estimated means and 95% confidence intervals (CI) from the multilevel model of methylation (%5MeC) averaged across CpG units of each target sequence and descriptive values of blood cell count for each sampling time (pre-stress, post-stress and follow-up).
|
Sampling time | ||||||
|---|---|---|---|---|---|---|
|
Pre-stress |
Post-stress |
Follow-up |
||||
| M | 95% CI | M | 95% CI | M | 95% CI | |
| Methylation (%5MeC) | ||||||
| OXTR1 | 17.64 | 16.48−18.81 | 18.02 | 16.85−19.19 | 16.98 | 15.79−18.16 |
| OXTR2 a | 5 | 4.67−5.35 | 5.06 | 4.73−5.42 | 4.76 | 4.42−5.11 |
| BDNF a | 6.61 | 6.10−7.15 | 6.56 | 6.05−7.10 | 6.31 | 5.80−6.84 |
| Blood cell count | ||||||
| Leukocytesb | 6.53 | 6.25−6.80 | 7.01 | 6.69−7.32 | 6.74 | 6.41−7.06 |
| Lymphocytesc | 30.59 | 28.98−32.21 | 32.61 | 30.85−34.36 | 26.74 | 25.09−28.40 |
| Monocytesc | 8 | 7.57−8.43 | 8.24 | 7.75−8.74 | 7.54 | 7.06−8.01 |
| Granulocytesc | 62.12 | 60.63−63.60 | 59.60 | 58.05−61.16 | 65.45 | 63.55−67.34 |
Abbreviations: BDNF, brain-derived neurotrophic factor; 95% CI, 95% confidence interval; M, mean; OXTR, oxytocin receptor.
Estimates were re-transformed from natural logarithm to %5MeC;
Number × 103/μl;
% of leukocytes.
Methylation of OXTR1
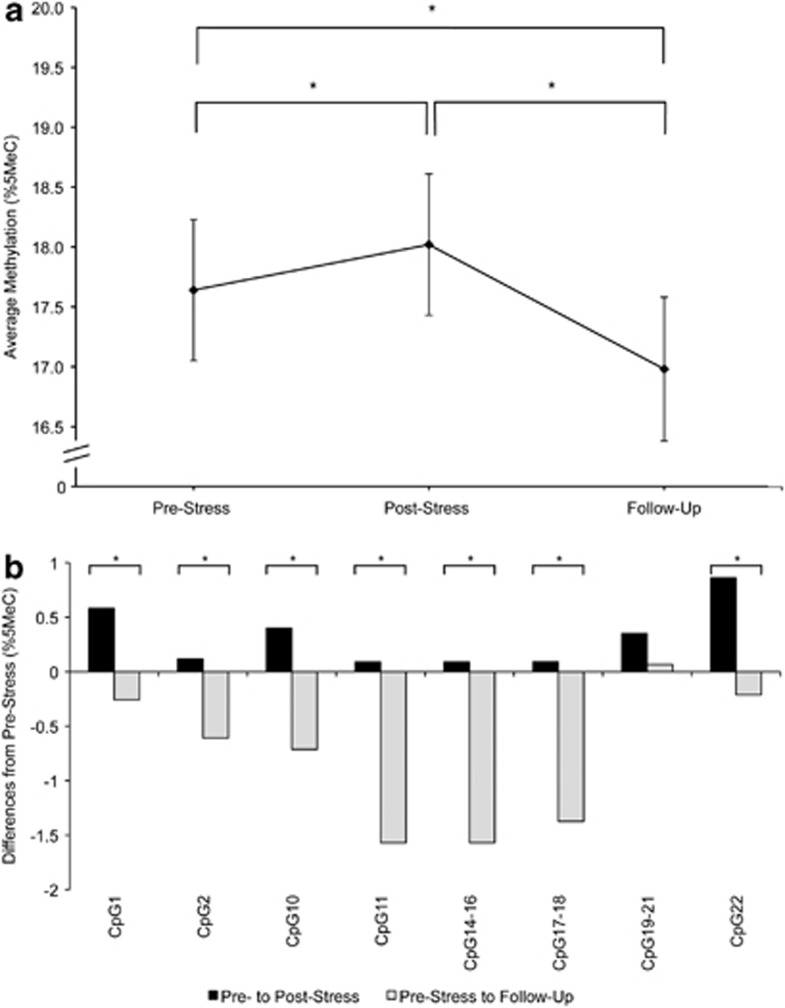
We found an overall effect for sampling time on OXTR1 mean methylation status. All post-hoc contrasts between sampling times were significant, with the greatest difference in mean methylation between post-stress and follow-up (Figure 1a, Table 2). Moreover, seven of eight individual CpG units within OXTR1 revealed significant time effects (Figure 1b). Notably, when adjusting for blood cell counts, the overall effect of sampling time on OXTR1 methylation averaged across CpG units remained significant. However, of the three post-hoc contrasts, the one between pre-stress and post-stress was no longer significant.
Figure 1.
(a) Estimated mean DNA methylation level (%5MeC) in OXTR1 amplicons averaged across CpGs at pre-stress, post-stress, and 90 min follow-up stress assessments. Error bars are s.e. of the estimated mean. (b) Differences in individual CpG mean methylation (%5MeC) from pre-stress to post-stress and from pre-stress to follow-up. All estimates obtained from the unadjusted model. *P<0.05.
Table 2. Overall effects of sampling time on mean methylation of OXTR 1, OXTR 2 a and BDNF a without and with adjustment for blood cell count as covariates; post-hoc contrasts between sampling times pre-stress, post-stress and follow-up. Results based on multilevel analysis.
|
Main effects |
Contrasts |
Number of observations |
|||||
|---|---|---|---|---|---|---|---|
| df b | F | p | C1 | C2 | C3 | N | |
| p | p | p | |||||
| OXTR1 | |||||||
| Overall model | 2; 802 | 25.84 | <0.001 | 0.009 | <0.001 | <0.001 | 600 |
| Adjusted for blood cell count | 2; 1133 | 10.70 | <0.001 | 0.278 | <0.001 | <0.001 | 600 |
| OXTR2a | |||||||
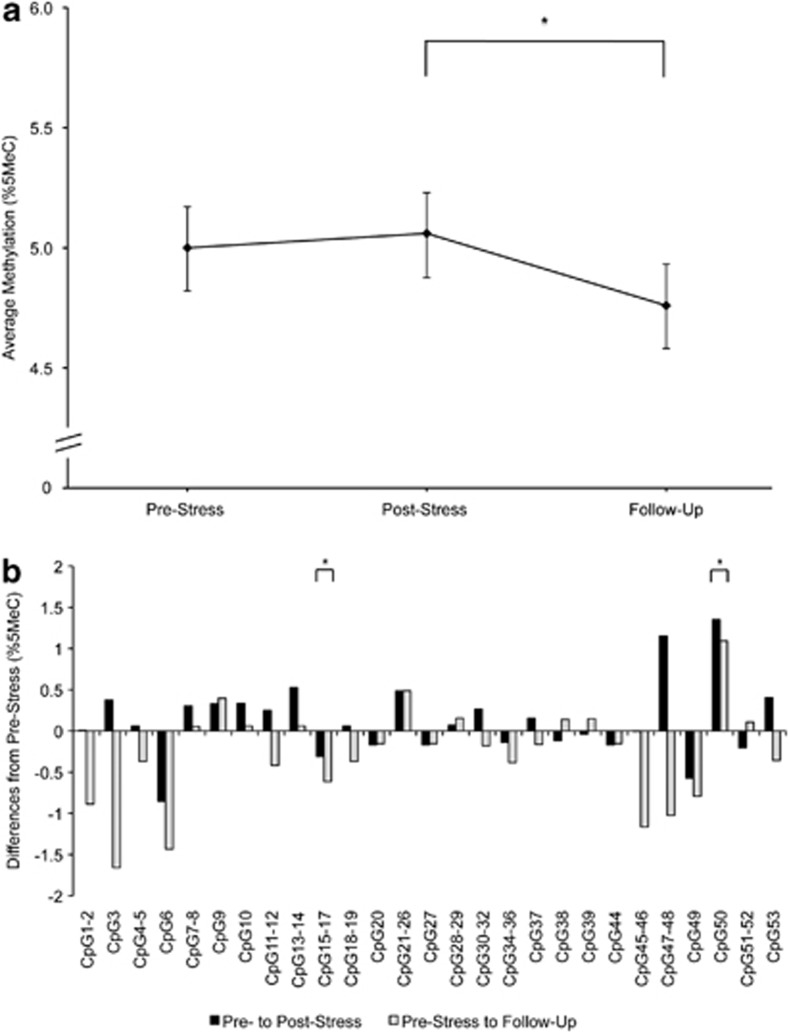
| Overall model | 2; 2998 | 2.46 | 0.086 | 0.672 | 0.034 | 0.099 | 2045 |
| Adjusted for blood cell count | 2; 1368 | 1.92 | 0.146 | 0.536 | 0.058 | 0.137 | 2044 |
| BDNF a | |||||||
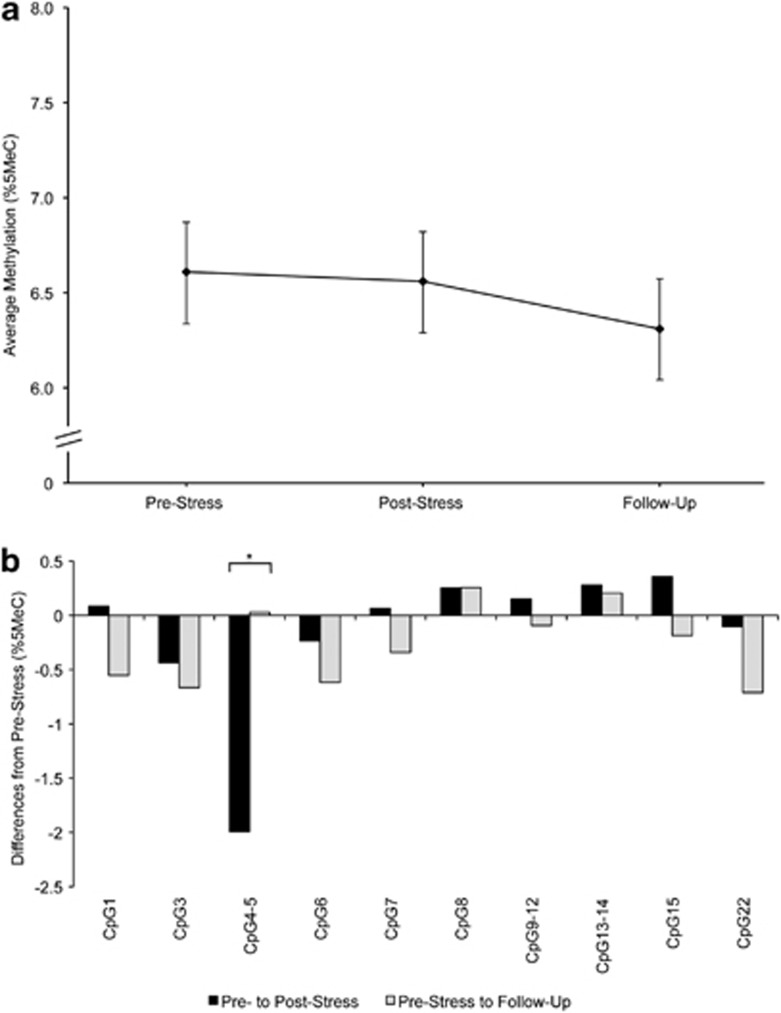
| Overall model | 2; 1098 | 1.31 | 0.271 | 0.780 | 0.184 | 0.139 | 747 |
| Adjusted for blood cell count | 2; 1523 | 0.87 | 0.418 | 0.536 | 0.518 | 0.191 | 737 |
Abbreviations: BDNF, brain-derived neurotrophic factor; C1, contrast pre-stress versus post-stress; C2, contrast post-stress versus 90 min after stress; C3, contrast pre-stress versus 90 min after stress; OXTR, oxytocin receptor.
Natural logarithm transformed;
Numerator; denominator.
Methylation of OXTR2
We found a trend effect for sampling time in OXTR2. Post-hoc contrast analyses indicated a difference in OXTR2 mean methylation between post-stress and follow-up (Figure 2a, Table 2). Time effects were significant in two of 27 CpG units (Figure 2b). After adjustment for blood cell count, the overall effect for sampling time remained nonsignificant; the contrast between post-stress and follow-up was no longer significant.
Figure 2.
(a) Estimated mean DNA methylation level (%5MeC) in OXTR2 amplicons averaged across CpGs at pre-stress, post-stress and 90 min follow-up stress assessments. Error bars are s.e. of the estimated mean. (b) Differences in individual CpG mean methylation (%5MeC) from pre-stress to post-stress and from pre-stress to follow-up. All estimates obtained from the unadjusted model. *P<0.05.
Methylation of BDNF (Figure 3, Table 2)
Figure 3.
(a) Estimated mean DNA methylation level (%5MeC) in BDNF amplicons averaged across CpGs at pre-stress, post-stress and 90 min follow-up stress assessments. Error bars are s.e. of the estimated mean. (b) Differences in individual CpG mean methylation (%5MeC) from pre-stress to post-stress and from pre-stress to follow-up. All estimates obtained from the unadjusted model. *P<0.05.
The analysis revealed no overall effect for sampling time on BDNF mean methylation and no post-hoc contrast was significant (Figure 3a, Table 2). However, analysis of individual CpG units revealed a significant time effect in 1 of 10 CpG units (Figure 3b). The inclusion of blood cell count as covariates did not change these results.
Discussion
The aim of this study was to investigate immediate changes in DNA methylation in stress-related genes after acute psychosocial stress. We found stress-associated DNA methylation changes in one of two OXTR target sequences but not in the assessed target sequence of BDNF, suggesting a considerable variation in the sensitivity of short-term DNA methylation responses among different stress-related genes. For OXTR1, we found an increase in DNA methylation from pre-stress to post-stress and a decrease from post-stress to follow-up. In OXTR2, methylation decreased from post-stress to follow-up only. Notably, in OXTR1 the time-associated changes, as well as the difference from post-stress to follow-up, remained significant even after controlling for blood cell count. The changes from pre-stress to post-stress in OXTR1 and from post-stress to follow-up in OXTR2 may have been secondary to stress-associated changes in blood cell composition.65
Although (i) methylation increase in OXTR is associated with decreased OXTR expression61 and (ii) the oxytocin system antagonizes the short-term stress response,37, 41 methylation increase from pre- to post-stress in OXTR1 could constitute a part of the immediate stress response, which relies on rapid autonomic sympathetic activation to mobilize resources and increase performance.66 After the stressor had passed, DNA methylation of the OXTR not only receded back to pre-stress baseline, but also fell below pre-stress levels. This could indicate an overcompensating mechanism in OXTR methylation after acute psychosocial stress, allowing for an upregulation of the oxytocin system as a middle-term physiological buffer of the acute stress response. Previous studies have shown that the oxytocin system has an essential role in the regulation of blood pressure and volume, heart rate and cardiovascular homeostasis, as well as in the cardiovascular response to stress.42, 67, 68 Therefore, a decrease in DNA methylation of the OXTR and the subsequent increase in expression43 may indeed be a potential mechanism to support physiological recovery after acute stress on an epigenetic level.
Regarding BDNF, our results suggest that in the periphery, DNA methylation in BDNF remains stable after a short and non-recurring psychosocial stressor. Previous studies found lifelong and transgenerational perpetuation of changes in BDNF methylation after early-life adversity.32 Fuchikami et al.69 recently suggested DNA methylation of BDNF in peripheral blood as a diagnostic biomarker of major depression. These results and our finding implicate that BDNF methylation has a long-term, rather than a short-term, role in stress adaptation.
This study has several strengths: First, the TSST is a highly established and robust standardized protocol to induce psychosocial stress and a robust hypothalamic-pituitary-adrenal axis activation.70 Various biological markers of acute stress have been investigated in relation to the TSST. Here we extend previous findings, by adding DNA methylation changes in OXTR as an additional biomarker of acute psychosocial stress, especially from post-stress to 90 min after the stressor. Second, we included blood cell count as a time-varying covariate into the analyses to ensure that DNA methylation changes were not the result of alterations in blood cell composition in response to stress.64 Indeed, our results highlight the necessity to consider blood cell count in the analyses while investigating DNA methylation in the periphery. Third, DNA methylation was not only assessed at pre- and post-stress, but also after a time interval of 90 min, which provided insight into methylation changes after stress recovery. Fourth, the focus on not only one, but on multiple genes (OXTR and BDNF) and target sequences revealed remarkable specificity of the short-term DNA methylation response of individual stress-related genes.
Several limitations of this study should also be noted: First, we measured DNA methylation in peripheral blood, which does not allow us to directly draw conclusions about processes in the central nervous system. To what degree DNA methylation in the periphery corresponds to DNA methylation in the brain remains to be elucidated, although some studies suggest certain consistency across tissues.43, 71, 72 Second, we did not apply an unstressed control group and can therefore not completely exclude that DNA methylation changes were due to factors unrelated to the psychosocial stress experience. Third, we analyzed DNA methylation changes after acute psychosocial stress in a study population with high likelihood of early experiences of war-related adversities, who may have been sensitized to stress. As a consequence, study subjects might have been especially susceptible to changes in OXTR DNA methylation after acute psychosocial stress. Therefore, generalizability of our results to populations without early adversities may be limited. Fourth, it should be noted that differences in mean DNA methylation between time points were small, and the functional effects of such modest alterations are not known. In this context, however, the following should be considered: (i) We did not compare different study groups, but assessed changes in DNA methylation over time within the same individuals. Therefore mean values in methylation are not independent of each other and differences are expected to be smaller in contrast to between-group comparison. (ii) Changes in DNA methylation were larger for several individual CpG units than for averaged target sequences (Figures 1, 2, 3). (iii) The absolute change of 1% in methylation of OXTR1 (%5MeC) from post-stress to follow-up represents a relative change of 5–6%. (iv) DNA methylation changes may accumulate and increase in magnitude in case of repeated psychosocial stress experience.
Future studies should replicate our findings for OXTR and BDNF, but also include additional stress-related candidate genes. Furthermore, we suggest shortening blood-sampling intervals to identify the time point of greatest DNA methylation changes. In addition, future studies could assess DNA methylation not only after different stressors, but also after positive experiences to determine whether DNA methylation is sensitive not only to aversive but also to positive psychosocial experiences. Moreover, subjects from other populations (such as cohorts without increased likelihood of early adversities) should be studied to scrutinize the generalizability of our results. Finally, future studies should assess DNA methylation after repeated psychosocial experiences to elucidate possible long-term modifications in DNA methylation. Identifying and studying short- and long-term effects of psychosocial experiences—which for example could reverse aberrant DNA methylation—could become an important goal in the development of new treatment approaches.
Conclusion
To the best of our knowledge, this is the first study in humans investigating dynamic short-term changes in DNA methylation related to a specific life event, namely a psychosocial stressor. We found different DNA methylation states in the OXTR when comparing pre-stress, post-stress and 90-min follow-up stress measurement. These findings contribute to the understanding of epigenetic mechanisms in general, but may also have clinical significance in the future: We found that psychosocial experiences are linked to immediate epigenetic modifications in a sample of subjects with early adverse experiences. This could have clinical implications regarding the etiology of mental and stress-related disorders, as well as of general medical conditions.
Acknowledgments
This work was supported by a grant of the German Research Foundation to DH (DFG grant number HE 1013/21-1 and HE 1013/21-2) and by grants of the Swiss National Science Foundation to GM and RL (project number 100014-135328 and project number 51A240-104890). We thank all people involved in data collection and entry, especially Malgorzata Kaszynska, Stephanie Hartmann, Dr Andrea Gierens and Celine Pleimling from the Trier lab and research team, everyone supporting the biological analyses in Basel, as well as the staff in the laboratories of the ‘MRC Social, Genetics and Developmental Psychiatry center' of the King's College in London.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Translational Psychiatry website (http://www.nature.com/tp)
Supplementary Material
References
- Docherty S, Mill J. Epigenetic mechanisms as mediators of environmental risks for psychiatric disorders. Psychiatry. 2008;7:500–506. [Google Scholar]
- Toyokawa S, Uddin M, Koenen KC, Galea S. How does the social environment ‘get into the mind'? Epigenetics at the intersection of social and psychiatric epidemiology. Soc Sci Med. 2012;74:67–74. doi: 10.1016/j.socscimed.2011.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Institute of Mental Health. National Institute of Mental Health Strategic Plan. 2008.
- Scarano MI, Strazzullo M, Matarazzo MR, D'Esposito M. DNA methylation 40 years later: Its role in human health and disease. J Cell Physiol. 2005;204:21–35. doi: 10.1002/jcp.20280. [DOI] [PubMed] [Google Scholar]
- Fuchikami M, Morinobu S, Segawa M, Yamawaki S. DNA methylation profiles of the BDNF gene in patients with major depression. Int J Neuropsychopharmacol. 2010;13:147. [Google Scholar]
- Olsson CA, Foley DL, Parkinson-Bates M, Byrnes G, McKenzie M, Patton GC, et al. Prospects for epigenetic research within cohort studies of psychological disorder: a pilot investigation of a peripheral cell marker of epigenetic risk for depression. Biol Psychol. 2010;83:159–165. doi: 10.1016/j.biopsycho.2009.12.003. [DOI] [PubMed] [Google Scholar]
- Rotter A, Asemann R, Decker A, Kornhuber J, Biermann T. Orexin expression and promoter-methylation in peripheral blood of patients suffering from major depressive disorder. J Affect Disord. 2011;131:186–192. doi: 10.1016/j.jad.2010.12.004. [DOI] [PubMed] [Google Scholar]
- Uddin M, Koenen KC, Aiello AE, Wildman DE. de los Santos R, Galea S. Epigenetic and inflammatory marker profiles associated with depression in a community-based epidemiologic sample. Psychol Med. 2011;41:997–1007. doi: 10.1017/S0033291710001674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mill J, Petronis A. Molecular studies of major depressive disorder: the epigenetic perspective. Mol Psychiatry. 2007;12:799–814. doi: 10.1038/sj.mp.4001992. [DOI] [PubMed] [Google Scholar]
- Abdolmaleky HM. Reelin hypermethylation in schizophrenia and beyond. Int J Neuropsychopharmacol. 2006;9:S37. [Google Scholar]
- Ghadirivasfi M, Nohesara S, Ahmadkhaniha HR, Eskandari MR, Mostafavi S, Thiagalingam S, et al. Hypomethylation of the serotonin receptor type-2A gene (HTR2A) at T102C polymorphic site in DNA derived from the saliva of patients with schizophrenia and bipolar disorder. Am J Med Genet Part B. 2011;156B:536–545. doi: 10.1002/ajmg.b.31192. [DOI] [PubMed] [Google Scholar]
- Kuratomi G, Iwamoto K, Bundo M, Kusumi I, Kato N, Iwata N, et al. Aberrant DNA methylation associated with bipolar disorder identified from discordant monozygotic twins. Mol Psychiatry. 2008;13:429–441. doi: 10.1038/sj.mp.4002001. [DOI] [PubMed] [Google Scholar]
- Mill J, Tang T, Kaminsky Z, Khare T, Yazdanpanah S, Bouchard L, et al. Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. Am J Hum Genet. 2008;82:696–711. doi: 10.1016/j.ajhg.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimabukuro M, Sasaki T, Imamura A, Tsujita T, Fuke C, Umekage T, et al. Global hypomethylation of peripheral leukocyte DNA in male patients with schizophrenia: a potential link between epigenetics and schizophrenia. J Psychiatr Res. 2007;41:1042–1046. doi: 10.1016/j.jpsychires.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Dempster EL, Pidsley R, Schalkwyk LC, Owens S, Georgiades A, Kane F, et al. Disease-associated epigenetic changes in monozygotic twins discordant for schizophrenia and bipolar disorder. Hum Mol Genet. 2011;20:4786–4796. doi: 10.1093/hmg/ddr416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AK, Conneely KN, Kilaru V, Mercer KB, Weiss TE, Bradley B, et al. Differential immune system DNA methylation and cytokine regulation in post-traumatic stress disorder. Am J Med Genet B Neuropsychiatr Genet. 2011;156B:700–708. doi: 10.1002/ajmg.b.31212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin M, Aiello AE, Wildman DE, Koenen KC, Pawelec G, de Los Santos R, et al. Epigenetic and immune function profiles associated with posttraumatic stress disorder. Proc Natl Acad Sci U S A. 2010;107:9470–9475. doi: 10.1073/pnas.0910794107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schanen NC. Epigenetics of autism spectrum disorders. Hum Mol Genet. 2006;2:R138–R150. doi: 10.1093/hmg/ddl213. [DOI] [PubMed] [Google Scholar]
- Nguyen A, Rauch TA, Pfeifer GP, Hu VW. Global methylation profiling of lymphoblastoid cell lines reveals epigenetic contributions to autism spectrum disorders and a novel autism candidate gene, RORA, whose protein product is reduced in autistic brain. FASEB J. 2010;24:3036–3051. doi: 10.1096/fj.10-154484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieling H, Romer KD, Scholz S, Mittelbach F, Wilhelm J, De Zwaan M, et al. Epigenetic dysregulation of dopaminergic genes in eating disorders. Int J Eat Disord. 2010;43:577–583. doi: 10.1002/eat.20745. [DOI] [PubMed] [Google Scholar]
- Frieling H, Bleich S, Otten J, Romer KD, Kornhuber J, de Zwaan M, et al. Epigenetic downregulation of atrial natriuretic peptide but not vasopressin mRNA expression in females with eating disorders is related to impulsivity. Neuropsychopharmacology. 2008;33:2605–2609. doi: 10.1038/sj.npp.1301662. [DOI] [PubMed] [Google Scholar]
- Wong CC, Mill J, Fernandes C. Drugs and addiction: an introduction to epigenetics. Addiction. 2011;106:480–489. doi: 10.1111/j.1360-0443.2010.03321.x. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nat Med. 2011;17:330–339. doi: 10.1038/nm.2305. [DOI] [PubMed] [Google Scholar]
- Meaney MJ. Epigenetics and the biological definition of gene x environment interactions. Child Dev. 2010;81:41–79. doi: 10.1111/j.1467-8624.2009.01381.x. [DOI] [PubMed] [Google Scholar]
- Liu D, Diorio J, Tannenbaum B, Caldji C, Francis D, Freedman A, et al. Maternal care, hippocampal glucocorticoid receptors, and hypothalamic-pituitary-adrenal responses to stress. Science. 1997;277:1659–1662. doi: 10.1126/science.277.5332.1659. [DOI] [PubMed] [Google Scholar]
- McGowan PO, Sasaki A, D'Alessio AC, Dymov S, Labonte B, Szyf M, et al. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci. 2009;12:342–348. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller BR, Bale TL. Sex-specific programming of offspring emotionality after stress early in pregnancy. J Neurosci. 2008;28:9055–9065. doi: 10.1523/JNEUROSCI.1424-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radtke KM, Ruf M, Gunter HM, Dohrmann K, Schauer M, Meyer A, et al. Transgenerational impact of intimate partner violence on methylation in the promoter of the glucocorticoid receptor. Transl Psychiatry. 2011;1 doi: 10.1038/tp.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver ICG, Cervoni N, Champagne FA, D'Alessio AC, Sharma SS, et al. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- Oberlander TF, Weinberg J, Papsdorf M, Grunau R, Misri S, Devlin AM. Prenatal exposure to maternal depression, neonatal methylation of human glucocorticoid receptor gene (NR3C1) and infant cortisol stress responses. Epigenetics. 2008;3:97–106. doi: 10.4161/epi.3.2.6034. [DOI] [PubMed] [Google Scholar]
- Spengler D, Murgatroyd C, Patchev AV, Wu Y, Micale V, Bockmuhl Y, et al. Dynamic DNA methylation programs persistent adverse effects of early-life stress. Nat Neurosci. 2009;12:1559–U1108. doi: 10.1038/nn.2436. [DOI] [PubMed] [Google Scholar]
- Roth TL, Lubin FD, Funk AJ, Sweatt JD. Lasting epigenetic influence of early-life adversity on the BDNF gene. Biol Psychiatry. 2009;65:760–769. doi: 10.1016/j.biopsych.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestart ML, et al. Epigenetic differences arise during the lifetime of monozygotic twins. P Natl Acad Sci USA. 2005;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CC, Caspi A, Williams B, Craig IW, Houts R, Ambler A, et al. A longitudinal study of epigenetic variation in twins. Epigenetics. 2010;5:516–526. doi: 10.4161/epi.5.6.12226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann ID. Involvement of the brain oxytocin system in stress coping: interactions with the hypothalamo-pituitary-adrenal axis. Prog Brain Res. 2002;139:147–162. doi: 10.1016/s0079-6123(02)39014-9. [DOI] [PubMed] [Google Scholar]
- Vaidya VA, Nair A, Vadodaria KC, Banerjee SB, Benekareddy M, Dias BG, et al. Stressor-specific regulation of distinct brain-derived neurotrophic factor transcripts and cyclic AMP response element-binding protein expression in the postnatal and adult rat hippocampus. Neuropsychopharmacology. 2007;32:1504–1519. doi: 10.1038/sj.npp.1301276. [DOI] [PubMed] [Google Scholar]
- Heinrichs M, Baumgartner T, Kirschbaum C, Ehlert U. Social support and oxytocin interact to suppress cortisol and subjective responses to psychosocial stress. Biol Psychiatry. 2003;54:1389–1398. doi: 10.1016/s0006-3223(03)00465-7. [DOI] [PubMed] [Google Scholar]
- Rodrigues SM, Saslow LR, Garcia N, John OP, Keltner D. Oxytocin receptor genetic variation relates to empathy and stress reactivity in humans. Proc Natl Acad Sci USA. 2009;106:21437–21441. doi: 10.1073/pnas.0909579106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon I, Young EA. Effects of stress and glucocorticoids on CNS oxytocin receptor binding. Psychoneuroendocrinology. 1997;22:411–422. doi: 10.1016/s0306-4530(97)00045-0. [DOI] [PubMed] [Google Scholar]
- Meinlschmidt G, Heim C. Sensitivity to intranasal oxytocin in adult men with early parental separation. Biol Psychiat. 2007;61:1109–1111. doi: 10.1016/j.biopsych.2006.09.007. [DOI] [PubMed] [Google Scholar]
- Grewen KM, Light KC. Plasma oxytocin is related to lower cardiovascular and sympathetic reactivity to stress. Biol Psychol. 2011;87:340–349. doi: 10.1016/j.biopsycho.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutkowska J, Jankowski M, Mukaddam-Daher S, McCann SM. Oxytocin is a cardiovascular hormone. Braz J Med Biol Res. 2000;33:625–633. doi: 10.1590/s0100-879x2000000600003. [DOI] [PubMed] [Google Scholar]
- Gregory SG, Connelly JJ, Towers AJ, Johnson J, Biscocho D, Markunas CA, et al. Genomic and epigenetic evidence for oxytocin receptor deficiency in autism. BMC Med. 2009;7 doi: 10.1186/1741-7015-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder DK, Scharfman HE. Brain-derived neurotrophic factor. Growth Factors. 2004;22:123–131. doi: 10.1080/08977190410001723308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoenen H. Neurotrophins and neuronal plasticity. Science. 1995;270:593–598. doi: 10.1126/science.270.5236.593. [DOI] [PubMed] [Google Scholar]
- Kerschensteiner M, Gallmeier E, Behrens L, Leal VV, Misgeld T, Klinkert WEF, et al. Activated human T cells, B cells, and monocytes produce brain-derived neurotrophic factor in vitro and in inflammatory brain lesions: A neuroprotective role of inflammation. J Exp Med. 1999;189:865–870. doi: 10.1084/jem.189.5.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulle F, van den Hove DL, Jakob SB, Rutten BP, Hamon M, van Os J, et al. Epigenetic regulation of the BDNF gene: implications for psychiatric disorders. Mol Psychiatry. 2011. [DOI] [PubMed]
- Karege F, Perret G, Bondolfi G, Schwald M, Bertschy G, Aubry JM. Decreased serum brain-derived neurotrophic factor levels in major depressed patients. Psychiat Res. 2002;109:143–148. doi: 10.1016/s0165-1781(02)00005-7. [DOI] [PubMed] [Google Scholar]
- Dell'Osso L, Carmassi C, Del Debbio A, Dell'Osso MC, Bianchi C, da Pozzo E, et al. Brain-derived neurotrophic factor plasma levels in patients suffering from post-traumatic stress disorder. Prog Neuro-Psychoph. 2009;33:899–902. doi: 10.1016/j.pnpbp.2009.04.018. [DOI] [PubMed] [Google Scholar]
- Branchi I. The mouse communal nest: Investigating the epigenetic influences of the early social environment on brain and behavior development. Neurosci Biobehav Rev. 2009;33:551–559. doi: 10.1016/j.neubiorev.2008.03.011. [DOI] [PubMed] [Google Scholar]
- Branchi I, D'Andrea I, Fiore M, Di Fausto V, Aloe L, Alleva E. Early social enrichment shapes social behavior and nerve growth factor and brain-derived neurotrophic factor levels in the adult mouse brain. Biol Psychiatry. 2006;60:690–696. doi: 10.1016/j.biopsych.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Lippmann M, Bress A, Nemeroff CB, Plotsky PM, Monteggia LM. Long-term behavioural and molecular alterations associated with maternal separation in rats. Eur J Neurosci. 2007;25:3091–3098. doi: 10.1111/j.1460-9568.2007.05522.x. [DOI] [PubMed] [Google Scholar]
- Meaney MJ, Liu D, Diorio J, Day JC, Francis DD. Maternal care, hippocampal synaptogenesis and cognitive development in rats. Nat Neurosci. 2000;3:799–806. doi: 10.1038/77702. [DOI] [PubMed] [Google Scholar]
- Gronli J, Bramham C, Murison R, Kanhema T, Fiske E, Bjorvatn B, et al. Chronic mild stress inhibits BDNF protein expression and CREB activation in the dentate gyrus but not in the hippocampus proper. Pharmacol Biochem Behav. 2006;85:842–849. doi: 10.1016/j.pbb.2006.11.021. [DOI] [PubMed] [Google Scholar]
- Gomez-Pinilla F, Zhuang Y, Feng J, Ying Z, Fan G. Exercise impacts brain-derived neurotrophic factor plasticity by engaging mechanisms of epigenetic regulation. Eur J Neurosci. 2011;33:383–390. doi: 10.1111/j.1460-9568.2010.07508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubin FD, Roth TL, Sweatt JD. Epigenetic regulation of BDNF gene transcription in the consolidation of fear memory. J Neurosci. 2008;28:10576–10586. doi: 10.1523/JNEUROSCI.1786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Nabeshima T. Brain-derived neurotrophic factor/TrkB signaling in memory processes. J Pharmacol Sci. 2003;91:267–270. doi: 10.1254/jphs.91.267. [DOI] [PubMed] [Google Scholar]
- Kirschbaum C, Pirke KM, Hellhammer DH. The Trier social stress test—a tool for investigating psychobiological stress responses in a laboratory setting. Neuropsychobiology. 1993;28:76–81. doi: 10.1159/000119004. [DOI] [PubMed] [Google Scholar]
- Dickerson SS, Kemeny ME. Acute stressors and cortisol reactivity: a meta-analytic review. Psychosom Med. 2002;64:105–105. [Google Scholar]
- Hautzinger M, Bailer M.ADS: Allgemeine Depressions Skala Beltz Test: Weinheim 19933Einheitenpp.
- Kusui C, Kimura T, Ogita K, Nakamura H, Matsumura Y, Koyama M, et al. DNA methylation of the human oxytocin receptor gene promoter regulates tissue-specific gene suppression. Biochem Biophys Res Commun. 2001;289:681–686. doi: 10.1006/bbrc.2001.6024. [DOI] [PubMed] [Google Scholar]
- Pruunsild P, Kazantseva A, Aid T, Palm K, Timmusk T. Dissecting the human BDNF locus: Bidirectional transcription, complex splicing, and multiple promoters. Genomics. 2007;90:397–406. doi: 10.1016/j.ygeno.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck RH, Thomas SL, Tabata LN.Multilevel and Longitudinal Modeling with IBM SPSS Routledge: New York; 2010. 343 pp. [Google Scholar]
- Zhu ZZ, Hou L, Bollati V, Tarantini L, Marinelli B, Cantone L, et al. Predictors of global methylation levels in blood DNA of healthy subjects: a combined analysis. Int J Epidemiol. 2010;41:126–139. doi: 10.1093/ije/dyq154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhabhar FS, Miller AH, McEwen BS, Spencer RL. Effects of stress on immune cell distribution. Dynamics and hormonal mechanisms. J Immunol. 1995;154:5511–5527. [PubMed] [Google Scholar]
- Eriksen HR, Olff M, Murison R, Ursin H. The time dimension in stress responses: relevance for survival and health. Psychiatry Res. 1999;85:39–50. doi: 10.1016/s0165-1781(98)00141-3. [DOI] [PubMed] [Google Scholar]
- Favaretto AIV, Ballejo GO, AlbuquerqueAraujo WIC, Gutkowska J, AntunesRodrigues J, McCann SM. Oxytocin releases atrial natriuretic peptide from rat atria in vitro that exerts negative inotropic and chronotropic action. Peptides. 1997;18:1377–1381. doi: 10.1016/s0196-9781(97)00209-x. [DOI] [PubMed] [Google Scholar]
- Jankowski M, Wang DH, Hajjar F, Mukaddam-Daher S, McCann SM, Gutkowska J. Oxytocin and its receptors ar synthesized in the rat vasculature. Proc Natl Acad Sci USA. 2000;97:6207–6211. doi: 10.1073/pnas.110137497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchikami M, Morinobu S, Segawa M, Okamoto Y, Yamawaki S, Ozaki N, et al. DNA methylation profiles of the brain-derived neurotrophic factor (BDNF) gene as a potent diagnostic biomarker in major depression. Plos One. 2011;6:8. doi: 10.1371/journal.pone.0023881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley P, Kirschbaum C. Human hypothalamus-pituitary-adrenal axis responses to acute psychosocial stress in laboratory settings. Neurosci Biobehav Rev. 2010;35:91–96. doi: 10.1016/j.neubiorev.2010.01.010. [DOI] [PubMed] [Google Scholar]
- Pidsley R, Mill J. Epigenetic studies of psychosis: current findings, methodological approaches, and implications for postmortem research. Biol Psychiatry. 2011;69:146–156. doi: 10.1016/j.biopsych.2010.03.029. [DOI] [PubMed] [Google Scholar]
- Yuferov V, Nielsen DA, Levran O, Randesi M, Hamon S, Ho A, et al. Tissue-specific DNA methylation of the human prodynorphin gene in post-mortem brain tissues and PBMCs. Pharmacogenet Genomics. 2011;21:185–196. doi: 10.1097/FPC.0b013e32833eecbc. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.