Abstract
A haloalkaliphilic sulfate-respiring bacterium, strain SLSR-1, was isolated from a lactate-fed stable enrichment culture originally obtained from the extreme environment of Searles Lake, California. The isolate proved capable of growth via sulfate-reduction over a broad range of salinities (125–330 g/L), although growth was slowest at salt-saturation. Strain SLSR-1 was also capable of growth via dissimilatory arsenate-reduction and displayed an even broader range of salinity tolerance (50–330 g/L) when grown under these conditions. Strain SLSR-1 could also grow via dissimilatory nitrate reduction to ammonia. Growth experiments in the presence of high borate concentrations indicated a greater sensitivity of sulfate-reduction than arsenate-respiration to this naturally abundant anion in Searles Lake. Strain SLSR-1 contained genes involved in both sulfate-reduction (dsrAB) and arsenate respiration (arrA). Amplicons of 16S rRNA gene sequences obtained from DNA extracted from Searles Lake sediment revealed the presence of close relatives of strain SLSR-1 as part of the flora of this ecosystem despite the fact that sulfate-reduction activity could not be detected in situ. We conclude that strain SLSR-1 can only achieve growth via arsenate-reduction under the current chemical conditions prevalent at Searles Lake. Strain SLSR-1 is a deltaproteobacterium in the family Desulfohalobiacea of anaerobic, haloalkaliphilic bacteria, for which we propose the name Desulfohalophilus alkaliarsenatis gen. nov., sp. nov.
Keywords: Alkaliphile ecology, Systematics, Anaerobic bacteria, Metaloxidation and reduction, Halophiles, Ecology, Biotechnology, Phylogeny, Genetics, Taxonomy, Enzymes
Introduction
Environments characterized as “extremes” of temperature, pH, salinity, radiation, aridity, toxicants, or combinations of these factors have long held fascination to biologists. While some hypersaline environments can provide habitat to a selected few species of well-adapted metazoans (e.g., Artemia), they can also harbor dense and diverse populations of microorganisms (e.g., Bodaker et al. 2010). Yet even within the context of salinity extremes, there are gradations. The term “hyper-saline” is loosely defined as exceeding that of oceanic salt strength (~35 g/L). The upper range of hyper-salinity implies brines with high specific gravities (and low water activities); hence values of dissolved salts would significantly exceed 150 g/L and range up to and including dense, salt-saturated brines (~350 g/L). It is at this upper edge of extreme hyper-salinity that even microbial life becomes challenged. The Bacteria and Archaea that manage to inhabit dense brines are particularly adapted to tolerate low water activities by a variety of evolved molecular strategies to cope with osmotic stress. For anaerobes, which have an even more constrained bio-energetic regimen than aerobes, these physical challenges are even more difficult to bear (Oren 1999a, b).
Amongst the anaerobes, there is considerable scientific interest in sulfate-respiring prokaryotes owing in part to the ubiquity of these microbes in anoxic environments and the abundance of sulfate anions in saline systems, especially seawater (~28 mM). The biogeochemical process of dissimilatory sulfate-reduction is thought to be ancient, deriving from Archean times when the global oceans were much less saline and held much less sulfate content than today (Shen et al. 2001). Over the past 3.5 billion years, however, many sulfate-rich hyper-saline brines have appeared both on the Earth’s surface and buried within its crust. Oren (1999a) made the empirical observation that dissimilatory sulfate-reduction in natural waters may be constrained at only the very upper reaches of salinity, essentially reaching a cap at salt saturation (≥300 g/L), although no known pure cultures could achieve growth at the elevated salt levels that approached saturation. Hence, water bodies that are moderately hypersaline, such as Solar Lake (~150 g/L), Big Soda Lake (27–89 g/L), and Mono Lake (~90 g/L) have high primary productivity coupled with intensive sulfate-reduction (Cohen et al. 1977; Jørgensen and Cohen 1977; Cloern et al. 1983; Smith and Oremland 1987; Jellison and Melack 1993; Oremland et al. 2000). However, within sulfate-containing brines that are poised at salt saturation, clear evidence for the occurrence of bacterial sulfate-reduction is somewhat ambiguous. For example, in the case of the Dead Sea, the stable sulfur isotopic work of Nissenbaum and Kaplan (1976) was cited by Oren (1999a) as evidence for in situ sulfate-reduction. Nonetheless, direct evidence of sulfate reduction based on the use of 35S-radiotracer in Dead Sea sediments is lacking, although not for a want of trying (Oren 1999b). Similarly, sediment profiles of dissolved constituents in the Orca Brine suggested that the extreme salinity therein completely inhibited sulfate-reduction (Weisenburg et al. 1985). Contrasting with this is the report of active 35S-sulfate-reduction within the dense brines at the bottom of the eastern Mediterranean Sea (van den Wielen et al. 2005) and the amplification of dissimilatory sulfite reductase gene sequences (dsrA) in samples taken from the L’Atalante and Urania basins of those brines (van den Wielen and Heijs 2007).
The seminal review by Oren (1999a) proposing an upper salinity limit for sulfate reduction and other terminal electron accepting processes (such as methanogenesis) prompted others to more closely examine water bodies poised at the upper reaches of salinity for, amongst other microbial processes, the presence of detectable sulfate-reduction. Thus, 35S-sulfate-reduction was shown to be active in the moderately hyper-saline southern arm of the Great Salt Lake (salinity ~120 g/L) and was about tenfold lower but still detectable in the much more saline (~270 g/L) but still unsaturated northern arm (Kjeldsen et al. 2007). Sulfate-reducing bacteria could be both cultured as well as detected by culture-independent means (i.e., 16S rRNA and dissimilatory sulfite reductase, dsrAB, gene sequences) from the sediments of both arms of the Great Salt Lake (Brandt et al. 2001; Jakobsen et al. 2006; Kjeldsen et al. 2007). In a survey of 35S-sulfate-reduction assayed in sediments over salt concentration gradients in a series of coastal salt pans in South Africa, the highest detected rates were actually reported at the highest measured salinities, including super-saturation (e.g., 422 g/L; Porter et al. 2007). These results contrasted with an earlier survey conducted along South San Francisco Bay evaporation ponds which found greatly diminished activity at the highest salinities (Klug et al. 1985). The aforementioned investigations were performed in very saline water bodies that were poised at circum-neutral pH (~6.0–8.2). In a broad survey of hyper-saline and highly alkaline soda lake sediments located in Siberia, some of the highest 35S-sulfate-reduction rates occurred at the highest measured salinities, including super-saturation (e.g., 475 and 520 g/L), and the presence of sulfate-reducing bacteria in these systems was also confirmed by both culture-dependent and culture-independent techniques (Foti et al. 2007). These authors also had success in cultivating enrichment cultures of sulfate-reducers in a medium containing 4 M Na2CO3 (~424 g/L) with a mixture of lactate and butyrate serving as electron donors. Aspects of these above given efforts that transpired over the past decade were recently summarized by Oren (2011).
Enigmatically, in stark contrast to the above reports stand our results for Searles Lake, the alkaline salt-saturated brine located in the Mojave Desert of California. This lake is particularly rich in arsenic oxyanions (~4 mM) along with other toxic elements, most notably boron (0.46 molal; roughly equivalent to 0.62 M) (Oremland et al. 2005). Although the sediments contained some detectable free sulfide (~0.1 mM), all attempts to elicit 35S-sulfate-reduction from them failed (Kulp et al. 2006), including manipulations of incubated slurries designed to enhance activity by such stratagems as amendment with exogenous electron donors (i.e., lactate or H2), by lowering the sulfate-content of artificial brine matrix, or by raising the specific activity of the isotope applied (Kulp et al. 2007). We eventually were able to point to borate ions acting as direct inhibitors of sulfate-reduction as the primary reason for the absence of detectable sulfate-reduction in this system. Our subsequent investigations on the ecophysiology of the arsenate-respiring bacterium Halarsenatibacter silvermanii strain SLAS-1 isolated from this system clearly demonstrated the occurrence of sulfate-reduction within a stable, lactate-fed enrichment culture that included strain SLAS-1. Yet this sulfate-reducer was a separate, distinct entity from strain SLAS-1 (Switzer Blum et al. 2009). We now report isolation from this enrichment culture of Desulfohalophilus alkaliarsenatis strain SLSR-1; an isolate that is capable of sustaining anaerobic respiratory growth at near salt-saturation (~330 g/L), equivalent to a water activity (a w) of 0.81. The significance of this finding with regard to the search for microbial life beyond Earth will be discussed.
Materials and methods
Enrichment culture and isolation
Strain SLSR-1 was isolated from a Searles Lake enrichment culture exhibiting clear evidence for the presence of sulfate-reduction, as determined by increases in sulfide with growth, and verified by the reduction of 35S-sulfate to 35S-sulfide (Switzer Blum et al. 2009). After isolation of strain SLAS-1, efforts were then re-oriented to isolate an extremely halophilic sulfate-reducer using the same anaerobic, high-salt lactate-based SearlesAb1 medium (salinity 340 g/L) described in Switzer Blum et al. (2009) but lacking any added arsenate, and thereby having sulfate as the sole electron acceptor. The composition of SearlesAb1 basal salts was (g/L): NaCl (180), Na2SO4 (100), K2SO4 (30), Na2CO3 (27), NaHCO3 (5.0), H3BO3 (4.0), (NH4)2SO4 (0.05), KH2PO4 (0.08), K2HPO4 (0.15), MgSO4·7H2O (0.025), Na2WO4 (0.075), Na2SeO4 (0.00001), and 3.0 ml of SL10 trace element solution (Widdel et al. 1983), yeast extract (0.2), cysteine-HCl (0.25), and 10 ml of a vitamin mix (Wolin et al. 1963; Oremland et al. 1994). The pH was adjusted to 9.5 with NaOH. The water activity (a w) of SearlesAb1 medium was 0.8 as determined analytically (see below). After three successive transfers at 38 °C, a serial dilution was conducted, with the 10−8 dilution level still showing growth. However, a microorganism similar in its odd motility to that displayed by Halarsensatibacter silvermanii strain SLAS-1 was also present (Switzer Blum et al. 2009). Hence, subsequent purification was achieved by conducting another dilution series at 20 °C, a temperature at which H. silvermanii strain SLAS-1 does not grow. In this case the 10−2 dilution demonstrated only one morphological type to be present. We were unable to cultivate this organism on solid agar plates composed of SearlesAb1 medium and thus could not pick isolated colonies. However, purity of the liquid cultures was confirmed by the presence of only a single band upon DGGE analysis of amplified 16S rRNA genes (see below) and designated strain SLSR-1. We subsequently determined that strain SLSR-1 did not require yeast extract for growth. Hence, other than in cases specified below to enhance growth rates, we eliminated it from the medium.
Growth experiments
The SearlesAb1 medium described above was used for growth experiments but modified with respect to electron acceptors added (sulfate or arsenate or both), the amount of electron donor (lactate) used, and the overall salinity employed. The salinity was lowered to 157 g/L (a w = 0.91) to improve growth. This was achieved by decreasing the amounts of NaCl (125 g/L), NaHCO3 (4 g/L), and Na2CO3 (24 g/L). Sulfate concentrations were manipulated for various media either by its total elimination or by adding incremental amounts of Na2SO4 (0.6–6 g/L), and K2SO4 (0.2–1.0 g/L). In order to be able to analytically discern lactate consumption in the presence of limited electron acceptor, we varied the amount of lactate employed. High concentrations (~60 mM) were used when conducting growth experiments with sulfate (30 mM), but lower concentrations of 15 mM lactate were used for sulfate (5 mM) plus arsenate (7 mM), and 3 mM lactate for arsenate alone (5 mM).
Effects of salinity, temperature, pH, and borate anions upon growth
The salinity range for growth was determined by varying the amount of NaCl added to SearlesAb1 medium (range 9–284 g/L). This was done individually for growth on sulfate and arsenate. Because growth in the basal salts medium was so slow during the above described salinity range experiments, the medium was modified by addition of 0.2 g/L of yeast extract for the pH, temperature, and borate concentration range experiments, which allowed us to shorten growth incubations from several months to several weeks. The pH range for growth was determined by employing the SearlesAb1 medium set at a salinity of 296 g/L (NaCl = 150 g/L), and the pH varied by changing the amounts of carbonate versus bicarbonate in the medium as previously described (Switzer Blum et al. 1998). The pH was re-checked in all tubes after the incubations and no deviations from the initial set of pH determinations occurred. Identical medium was employed for the temperature range experiments. The medium used to determine the effects of added borate anions upon growth under sulfate- or arsenate-reducing conditions was composed of SearlesAb1 as modified for a salinity of 180 g/L and a range of added borate (0–300 mM) and the pH was maintained at 8.8. In these long-term incubations (2–3 months) we used sulfide production or arsenate reduction in lieu of cell counts as proxies for cell growth
Experiments with washed cell suspensions
For the purpose of determining whether strain SLSR-1 achieved respiratory growth on nitrate by denitrification or by dissimilatory reduction to ammonia, we conducted incubations with washed cells suspensions. Cells were grown in lactate + nitrate Searles Ab1 medium (salinity 200 g/L), harvested by centrifugation, washed three times with a pH 9.5 buffer solution (g/L): NaCl (142), KH2PO4 (0.08), K2HPO4 (0.15), MgSO4·7H2O (0.025), H3BO3 (0.3), Na2CO3 (24), NaHCO3 (8.0), K2SO4 (0.87). All manipulations were carried out in an anaerobic glove box. Cells were re-suspended in buffer and supplemented with 15 mM Na lactate and 10 mM NaNO3 and 0.25 % cysteine-HCl as reducing agent and dispensed in 10 mL portions into serum bottles (37 mL volume) and crimp-sealed with butyl rubber stoppers and flushed with oxygen-free N2. Bottles were incubated statically at 38 °C, and selected samples were amended with a 15 % headspace addition of acetylene gas to inhibit N2O reductase (Balderston et al. 1976) using 63Ni electron-capture gas chromatography to detect headspace N2O and chemical analysis of liquid phase subsamples for their ammonia content (see below).
Analytical
Herein we have attempted to introduce the usage of water activity (a w) as a supplemental parameter to describe the experimental physiological constraints faced by extreme halophiles (Table 1). Activity of water (a w) was determined on stored samples and media using an Aqualab Water Activity Meter, Pawkit system (Decagon Devices, Pullman, WA). This device measures the humidity of an equilibrated enclosed air volume lying above the aqueous sample. Precision of triplicate measurements was 0.01 and accuracy was 0.02. Water activity measurements were conducted on SearlesAb1 medium, as well as on previously collected and archived (refrigerated at 6 °C) samples from Searles Lake, Mono Lake, and the Dead Sea. Organic acids, arsenic oxyanions, and sulfate were determined by high performance liquid chromatography (Hoeft et al. 2004), sulfide by the spectrophotometric method of Cline (1969), and direct cell counts by acridine orange staining (Hobbie et al. 1977). The acetylene-block technique of N2O-reductase (Balderston et al. 1976; Oremland et al. 1984) was used as a presumptive test for the presence of a full denitrification pathway leading to N2 formation from nitrate reduction. Ammonia in the aqueous phase was determined by the method of Solorzano (1969). Membrane-associated fatty acids (PLFAs: phospho-lipid fatty acids), a commonly used taxonomic tool, were obtained by harvesting lactate + sulfate-grown cells (Searles Ab1 medium; salinity 292 g/L), and the centrifuged frozen pellet was shipped to Microbial ID Inc. (Newark, DE). Analyses were performed on fatty acid methyl esters of the extracted PLFAs by flame ionization gas chromatography by methods detailed on-line (http://www.microbialid.com).
Table 1.
Estimated salinity composition of artificial media and natural brines compared with water activity measurements
| Component | Salinity (g/L) | Water activity (a w) |
|---|---|---|
| Searles Lake brine | >350 | 0.71 |
| Searles Lake medium | 346 | 0.79 |
| Searles Lake medium | 330 | 0.81 |
| Searles Lake medium | 296 | 0.83 |
| Searles Lake medium | 280 | 0.84 |
| Searles Lake medium | 230 | 0.87 |
| Searles Lake medium | 180 | 0.89 |
| Searles Lake medium | 160 | 0.91 |
| Searles Lake medium | 130 | 0.92 |
| Searles Lake medium | 80 | 0.95 |
| Searles Lake medium | 54 | 0.96 |
| Mono Lake brine | ~80 | 0.92 |
| Mono Lake medium (AML60)a | 75 | 0.94 |
| Dead Sea brine | ~337 | 0.67 |
| Dead Sea medium (high)b | 325 | 0.78 |
| Dead Sea medium (low) | 205 | 0.89 |
TEM/SEM
Sample preparation, thin section micro-toning, and scanning/transmission electron microscopy were carried out by procedures previously described (Switzer Blum et al. 2009).
Molecular analysis and alignments of strain SLSR-1 16S rRNA, dsrA, and arrA genes
DNA was extracted from a culture of SLSR-1 as previously described (Kulp et al. 2007). 16S rRNA genes and functional gene markers for dissimilatory sulfite reduction (dsrAB) were PCR amplified, cloned, screened, sequenced, and aligned as described previously (Kulp et al. 2007; Switzer Blum et al. 2009; Hoeft et al. 2010). For 16S rRNA and dsrAB genes, their evolutionary history was inferred using either the neighbor-joining method (16S rRNA genes; Saitou and Nei 1987) or the maximum likelihood method (dsrAB genes; Schwarz and Dayhoff 1979). The bootstrap consensus 16S rRNA gene tree inferred from 500 replicates was taken to represent the evolutionary history of the taxa analyzed for both 16S rRNA and dsrAB genes (Felsenstein 1985), and branches corresponding to partitions reproduced in <50 % bootstrap replicates were collapsed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) were aligned next to the branches displayed. The 16S rRNA gene tree was drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the 2-parameter method (Kimura 1980) and were given in the units of the number of base substitutions per site. The analysis involved 40 nucleotide sequences. All positions containing gaps and missing data were eliminated. There were a total of 490 positions in the final dataset, and evolutionary analyses were conducted in MEGA5 (Tamura et al. 2007). For the dsrAB genes, the initial tree(s) for the heuristic search were obtained automatically as follows: When the number of common sites was <100 or less than one fourth of the total number of sites, the maximum parsimony method was used; otherwise, BIONJ method with MCL distance matrix was used. The tree was drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 21 amino acid sequences. All positions containing gaps and missing data were eliminated. There were a total of 502 positions in the final dataset. Evolutionary analyses were conducted in MEGA5 (Tamura et al. 2007).
The initial amplicon using an arrA primer set generated a ~200-bp product that was used to sequence flanking DNA by an inverse PCR approach as described in Switzer Blum et al. (2009) and Saltikov and Newman (2003). Inverse primers (SLSR1-iPCR-F1, 5′-CCT GGT CAA ATG GTG GAA T-3′ and SLSR1-iPCR-R1 5′-GCT CCG TCC TTG AAG TCT C-3′) were used in PCRs on EcoRI or HindIII digested and circularized SLSR-1 genomic DNA. The PCR amplicons were cloned, sequenced, and assembled into a ~1.8-kb DNA fragment. Phylogenetic analysis was performed on the translated ArrA of the partial arrA gene sequences from SLSR-1 and other bacteria as previously described (Switzer Blum et al. 2009; Zargar et al. 2012). Sequences were aligned using ClustalW (Thompson et al. 1994). PAUP* (Swofford 1999) was used to generate a neighbor-joining phylogeny on the resulting multi-sequence alignment where gaps in the alignment were ignored.
Nucleotide sequence accession numbers
The nucleotide sequences of 16S rRNA gene, dsrAB, and arrA genes of SLSR-1 submitted to GenBank were given the accession numbers JQ582408, JQ 582409, and JQ955737, respectively.
G + C content of strain SLSR-1
The molar G + C content of strain SLSR-1 was obtained from frozen cells shipped to the DSMZ using their standard extraction protocol as delineated elsewhere (http://www.dsmz.de/services/services-microorganisms/identification/gc-content.html).
Molecular analysis of 16S rRNA genes in Searles Lake sediments
16S rRNA gene sequences related to strain SLSR-1 were detected by DGGE of samples of Searles Lake sediment collected in 2005 (Kulp et al. 2006). To confirm relatedness to strain SLSR-1, we excised bands from Searles Lake 16S rRNA gene assemblages that migrated to the same position as strain SLSR-1 and sequenced them to confirm similarity as previously described (Kulp et al. 2007). Sequences with >97 % 16S rRNA gene similarities to strain SLSR-1 were considered to be highly related.
Results
Water activity measurements (aw)
The a w values for three naturally occurring brines and for our artificial media are shown in Table 1. Mono Lake surface water and our standard cultivation medium for that environment (AML60) were nearly identical and comparable to the values for Searles Lake media with the lowest applied salts. Searles Lake high salinity medium (346 g/L) yielded a much lower value of a w (0.79), but it was still higher than the natural brine sample from that environment (~0.71). By comparison, an archived Dead Sea sample exhibited the lowest value of all the natural brines and artificial media tested (~0.67), an a w that can only be achieved in brines containing divalent cations such as Ca2+ or Mg2+.
Morphological characteristics of strain SLSR-1
Strain SLSR-1 cells are motile vibrios (3.0 × 0.5 μm) (Fig. 1a) that stain Gram-negative. Thin sections exhibited numerous granular inclusions as well a system of internal membranes (Fig. 1b). In some instances fully formed, coccoid daughter cells having their own double outer membranes were observed within the structure of their parent cells (Fig. 1c). Transmission electron microscopy revealed that motility was conferred by a long (~10 μm) polar flagellum (Fig. 2), usually observed as a single array (Fig. 2a, c), but in rare instances, as paired polar flagella (Fig. 2b). The flagella of strain SLSR-1 proved highly fragile during the TEM processing, and many simply broke off the samples, leaving behind unadorned cells.
Fig. 1.
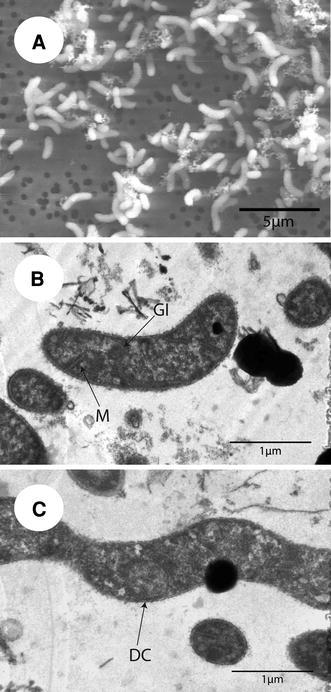
Electron micrographs of strain SLSR-1. a Scanning electron micrograph of sulfate-grown cells. b Thin section transmission micrograph of sulfate-grown cells showing presence of internal membranes (M-labeled arrow) and granular inclusions (G-labeled arrow). c Thin section of arsenate-grown cells showing presence of an internal daughter cell located near the node between larger cells (D-labeled arrow)
Fig. 2.
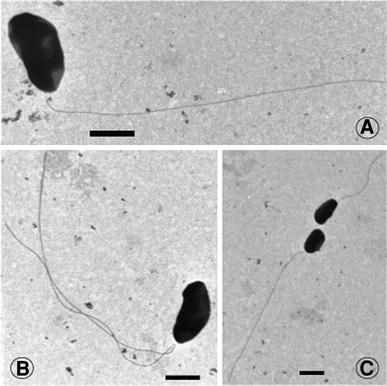
Transmission electron micrographs showing the polar flagella of strain SLSR-1, which most commonly were observed singly (a, c), but sometimes were paired (b). Bars indicate 1 μm
Growth characteristics of strain SLSR-1
Cell growth was evident on a lactate/sulfate medium with consumption of both over time, concomitant production of acetate and sulfide, and a cell density increase of 2 orders of magnitude, reaching a maximum (~1 × 108 cells/ml) by 36 days (Fig. 3a). There was a stoichiometric balance between lactate consumed (~33 mM) and acetate produced (~32 mM) indicating a decarboxylation with the production of CO2. The relationship between sulfate consumed (~18 mM) and sulfide formed (~25 mM), however, was only in very rough balance. Growth was slow (doubling time 82.5 h; μ = 0.008 h−1) at this salinity (160 g/L; a w = 0.9). In medium containing 5 mM As(V) but otherwise identical to the previous one (sulfate 30 mM), about 100 days elapsed before exponential growth was initiated, at which time As(V) reduction to As(III) briefly preceded sulfate reduction (Fig. 3b). There was reasonably good agreement between lactate and sulfate consumed, 43 and 18 mM, respectively, and the final accumulated amounts of acetate (40 mM) and sulfide (22 mM) produced. Arsenate was undetectable by ~140 days and was entirely replaced by As(III), which was also eventually removed, presumably as thioarsenite as a result of reaction with free sulfide in the medium. When sulfate and As(V) were both present at equivalent initial concentrations (5 mM) and the experiment conducted with much lower lactate (15 mM), a slow rate of growth and of As(V) reduction to As(III) occurred for the first 175 days, after which logarithmic phase was attained (Fig. 3c). Under these conditions, As(V) reduction clearly preceded that of sulfate-reduction, and the latter process did not commence until all the As(V) was reduced to As(III). By the incubation’s end there was a consumption of 5.75 mM lactate, which would have generated 23 mmol equiv. electrons. This source was balanced by the observed reduction of all the 5 mM As(V) plus 1.75 mM SO4 2−, thereby accounting for 24 mmol equiv. electrons as a cumulative respiratory sink. No growth or sulfate-reduction occurred in killed controls, or live samples incubated with lactate but without sulfate (data not shown). We did not observe growth when arsenate was employed as the sole electron acceptor unless some sulfate (~1 mM) was present in the medium (not shown).
Fig. 3.
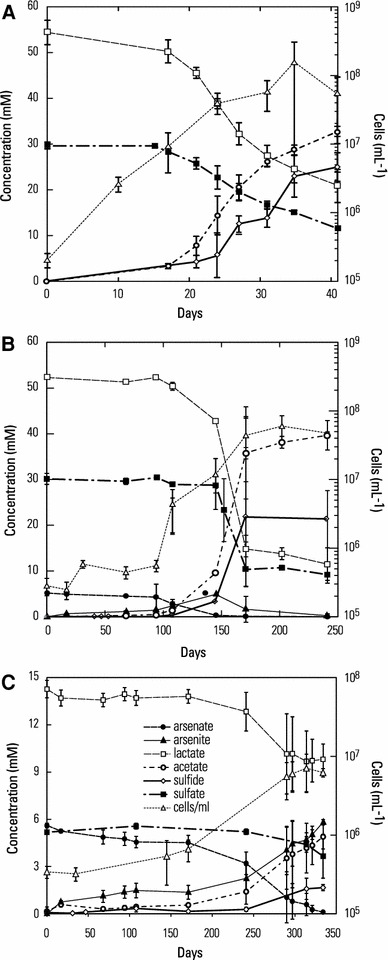
Growth curves of strain SLSR-1 achieved on defined media (salinity 157 g/L; a w = 0.91) with lactate as the electron donor. a Growth on high concentrations of sulfate (30 mM) and lactate (50 mM). b Growth on high concentrations of sulfate and lactate plus 5 mM arsenate. c Growth on lower concentrations of sulfate plus arsenate (~6 mM) with 15 mM lactate. Symbols represent the mean of three cultures. Bars indicate +1SD
Effects of salinity, pH, temperature, and borate
The salinity range for growth on sulfate was broad (125–330 g/L; a w = 0.93–0.8) with an optimum of ~200 g/L or a w = 0.84 (Fig. 4a). Curiously, the range for arsenate was even greater, with activity evident as low as 55 g/L (a w = 0.96), and an optimum between 125 and 175 g/L (a w = 0.92–0.89). Because we assumed a similar physiological response to pH with regard to available electron acceptor, we ran these salinity experiments at an alkaline pH of 9.5. However, an unexpected finding was that the pH optimum for growth on sulfate (~9.25) was higher than that for arsenate (8.75) (Fig. 4b). The response to borate additions is shown in Fig. 4c. Borate proved inhibitory to both sulfate reduction and arsenate reduction, but the former proved more sensitive and was completely blocked at 200 mM BO3 3−. In contrast, arsenate reduction was still evident at that concentration, and it was still in evidence at 300 mM BO3 3−, although it was strongly impaired (~78 % inhibition). The temperature range for growth was 20–50 °C, with an optimum at 44 °C and no growth evident at 16 °C, very slow growth at 20 °C, and no growth at 55 °C (Fig. 4d).
Fig. 4.
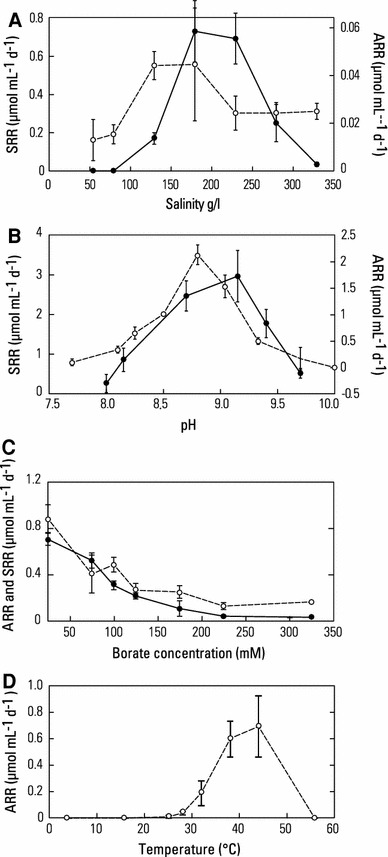
Physiological characterizations of strain SLSR-1 grown on either sulfate (filled circles) or arsenate (open circles). a Salinity. b pH. c Borate. d Temperature. Symbols represent the mean of three cultures and bars indicate +1SD. The medium for the pH, borate, and temperature experiments was supplemented with 0.2 % yeast extract to improve growth rates
Electron donors and acceptors
The substrate affinities and other phenotypic characterizations of strain SLSR-1 are listed in Table 2, and compared with those of 3 other species of anaerobic haloalkaliphiles capable of dissimilatory reduction of sulfate from the family Desulfohalobiaceae. Growth of strain SLSR-1 could be supported by the electron-donors lactate or pyruvate, but not by acetate, H2, H2 + acetate, formate or ethanol. Electron acceptors supporting growth on lactate besides sulfate and arsenate included nitrate, thiosulfate and sulfite, but not sulfur, fumarate, selenate, or Fe(III). In addition to heterotrophic growth on the listed organics, growth was also observed when sulfide was the e-donor and nitrate served as the e-acceptor. Growing cells consumed nitrate (~15.3 mM), produced sulfate (~4.0 mM), and traces of N2O were observed in the headspace of the culture tubes after several days’ incubation (69.6 ± 44.6 nmoles; n = 3). Growth could be sustained in this fashion upon repeated transfers, suggesting that growth was a result of chemolithoautotrophy and not the result of carry-over of an organic carbon source during inoculation. We also observed a capacity for oxidation of sulfide coupled to arsenate reduction, but growth could not be sustained in this fashion with repeated transfer. This implied a capacity for lithotrophy with the sulfide/arsenate coupling, but not for chemoautotrophic growth.
Table 2.
Phenotypic characteristics that distinguish Desulfohalophilus alkaliarsenatis strain SLSR-1 from other halo-alkaliphilic sulfate- or sulfur-reducers isolated from soda lakes
| Characteristic | SLSR-1 | Desulfonatronospira thiodismutans a | Desulfohalobium retbaense b | Desulfonatronovibrio hydrogenovorans c |
|---|---|---|---|---|
| Habitat | Searles Lake, CA, USA | Kulanda Steppe, Russia | Retba Lake, Senegal | Lake Magadi, Kenya |
| Morphology | Vibroid | Vibroid | Rod | Vibroid |
| Motility | + | + | + | + |
| Flagella | + | + | + | + |
| Size (μm) | 0.5 × 3.0 | 0.6–0.8 × 2–30 | 0.7–0.9 × 1.0–3.0 | 0.5 × 1.5–2.0 |
| pH range (optimum) | 7.75–9.75 (9.25) | 8.3–10.5 (10) | 5.5–8.0 (6.5–7.0) | 8.0–10.2 (9.6) |
| Salt range (g/L) | 55–330 | 87–232 | 29–240 | 10–120 |
| Temperature range (opt.; °C) | 20–50 (44) | “Mesophilic” (43) | 25–40 (37) | (37) |
| G + C content (mole%) | 45.1 | 50.4 | 57.1 | 48.6 |
| Chemolithotrophy | ||||
| H2 | − | + | − | +d |
| S2 − + NO3 − | + | NDe | ND | ND |
| S2 − + As(V)d | + | ND | ND | ND |
| Electron donors | ||||
| Lactate | + | + | + | − |
| Pyruvate | + | + | + | − |
| Acetate | − | − | − | − |
| H2 + acetate | − | − | + | + |
| Formate | − | + | + | − |
| Ethanol | − | + | + | − |
| Malate | − | ND | − | − |
| Succinate | − | ND | − | ND |
| Propionate | − | ND | − | − |
| Citrate | − | ND | ND | |
| Glycerol | − | ND | − | − |
| Glucose | − | ND | − | ND |
| Fructose | − | ND | − | ND |
| Fumarate | − | ND | − | − |
| Electron acceptors | ||||
| Sulfate | + | + | + | + |
| Arsenate | + | ND | ND | ND |
| Nitrate | + | ND | ND | ND |
| Nitrite | − | ND | ND | ND |
| DMSO | − | ND | ND | |
| Sulfite; thiosulfate | + | +f | + | +f |
| Oxygen | − | − | ND | |
| Sulfur | − | ND | + | − |
| Fumarate | − | ND | ND | ND |
| Fe(III) | − | ND | ND | ND |
| Mn(IV) | − | ND | ND | ND |
| Selenate | − | ND | ND | ND |
Incubation of washed cells with ~10 mM nitrate plus lactate produced N2O at a linear rate over a month-long incubation (data not shown) and accumulated 35.7 ± 2.4 nmoles N2O in the headspace by that time, which represented only ~0.07 % of the available nitrate. Samples (n = 3) incubated with C2H2 produced N2O, but at a ~fourfold slower rate than controls and accumulated only 10.6 ± 3.1 nmoles N2O in the headspace. During denitrification much more N2O would be expected in the presence of acetylene; hence these results indicated the lack of a functional N2O reductase. When compared with accumulated NH4 +, N2O was only a minor product. Samples incubated without acetylene accumulated more NH4 + (27.1 ± 1.4 μmoles) by the end of the incubation than those incubated in the presence of acetylene (14.1 ± 1.1 μmoles). By the end of the incubation, cells had consumed 52 μmoles nitrate and accumulated 14 μmoles nitrite plus 27 μmoles NH4 + as products, thereby accounting for 79 % of the nitrate removed, with the 21 % difference probably accountable via assimilatory metabolic pathways. Acetylene-inhibited samples by comparison demonstrated only 36 μmoles nitrate consumed, and were thus impaired by ~31 % in this regard, suggesting acetylene partially inhibited the nitrate reductase of strain SLSR-1. These results show that nitrate respiration by strain SLSR-1 proceeded via dissimilatory reduction to ammonia.
Phospho-lipid fatty acids
The major PLFAs, accounting for all those found in strain SLSR-1, can be seen in Table 3. Of the four molecular types determined, the group comprising normal, saturated fatty acids was the most prominent (44.8 %), with carbon chain lengths of 12, 16, and 18, respectively, having 6.7, 13.0, and 13.7 % abundances. There was one prominent (14 %) branched, saturated PLFA (15:0 iso) that was also a major PLFA found occurring in the haloalkaliphiles Desulfonatronospira thiodismutans and Dspira. delicata (Sorokin et al. 2008), but the other major PLFAs determined for these two species are different from those found in strain SLSR-1. Similarly, this (15:0 iso) branched PLFA occurs in Desulfonatronovibrio magnus but not in Dvibrio. thiodismutans (Sorokin et al. 2011a, b). Dvibio. magnus also contains 16:0 as a major normal, saturated fatty acid which is found in abundance in strain SLSR-1 (see above). Halarsenatibacter silvermanii strain SLAS-1 also contained abundant C16:0 and C18:0 as well as 15:0 iso PLFAs (Switzer Blum et al. 2009)
Table 3.
Major membrane fatty acid (PLFA) composition of strain SLSR-1
| Fatty Acida | Percent |
|---|---|
| Branched/saturated | |
| 11:0 iso | 3.4 |
| 13:0 iso | 3.2 |
| 15:0 iso | 14.0 |
| 15:0 anteiso | 0.3 |
| 15:0 iso DMA | 3.0 |
| 17:0 iso | 3.9 |
| 17:0 anteiso | 1.7 |
| Total = 29.5 % | |
| Branched/unsaturated | |
| 15:1 iso w9c | 0.8 |
| Total = 0.8 % | |
| Normal/saturated | |
| 12:0 | 6.7 |
| 12:0 DMA | 0.5 |
| 14:0 | 1.4 |
| 14:0 DMA | 0.7 |
| 16:0 | 13.0 |
| 16:0 DMA | 2.8 |
| 16:0 aldehyde | 1.0 |
| 17:0 | 1.5 |
| 18:0 | 13.7 |
| 18:0 DMA | 3.5 |
| Total = 44.8 % | |
| Normal/unsaturated | |
| 16:1 w7c alcohol | 0.4 |
| 16:2 DMA | 0.9 |
| 17:1 w7c | 0.7 |
| 17:1 w6c | 1.4 |
| 18:3 w3c | 1.0 |
| 18:2 w6c | 4.2 |
| 18:1 w9c | 7.7 |
| 18:1 w7c | 1.7 |
| 18:1 w9c DMA | 3.7 |
| 18:1 w7c DMA | 1.2 |
| 19:1 w6c | 0.6 |
| 22:1 w9c | 1.7 |
| Total = 24.9 % | |
| Grand total = 100 % | |
aCarbon number for double bond positions (w) relative to the methyl carbon end of the hydrocarbon chain; DMA refers to dimethyl acetal group occurring at the carboxyl (#1 carbon) end
Taxonomic alignment of strain SLSR-1
Phylogenetic analysis of the 16S rRNA gene sequences shows that strain SLSR-1 and other sequences obtained from Searles Lake sediments (SLAB, SL, and SRC) form a distinct clade in the family Desulfohalobiaceae near the root of the genus Desulfonatronovibrio (Fig. 5). Members of this family of sulfidogenic haloakaliphiles exhibit diverse substrate affinities and salt tolerances. Desulfonatronovibrio hydrogenovorans is a sulfate reducer that can use H2 or formate as electron donors for reduction of sulfate, sulfite, or thiosulfate (Zhilina et al. 1997). Other members of this family include the genus Desulfonatronum which in addition to their use of H2 or formate, can use ethanol or in one case lactate (Pikuta et al. 2003; Zhilina et al. 2005). Members of the genus Desulfonatrospira have a more versatile metabolism, in that they can use H2 for the reduction of sulfate or sulfite along with electron donors like lactate and alcohols, but can also ferment pyruvate or grow chemoautotrophically via disproportionation of either sulfite or thiosulfate (Sorokin et al. 2008). The moderate halophile Desulfovermiculus halophilus not only has metabolic versatility in being a chemo-organotroph with a broad array of organic electron donors, but also harbors the ability to grow as an autotroph on H2 + CO2 (Beliakova et al. 2006). Another denizen of anoxic soda lake mud worth mentioning is Desulfurispira natronophila. But while this bacterium is a moderate haloalkaliphile that employs a diverse array of organic electron donors to achieve growth, it uses elemental sulfur or arsenate as electron acceptors but cannot use sulfate, thiosulfate, or nitrate. Indeed, it belongs to the family Crysiogenetes rather than the Desulfohalobiaceae (Sorokin and Muyzer 2010).
Fig. 5.
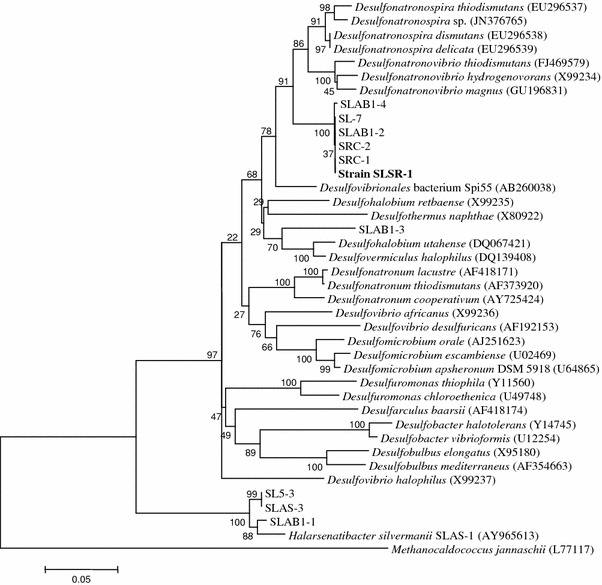
Neighbor-joining tree of 16S rRNA genes showing the phylogenetic position of strain SLSR-1 and other amplicon sequences obtained from Searles Lake (prefix SL). Genbank accession numbers are included next to the corresponding species names
Despite being displayed in Fig. 5 at the base of the Desulfonatronovibrio genus, Desulfonatronovibrio hydrogenovorans shows only 90 % homology with strain SLSR-1 based on 16S rRNA gene sequence BLAST alignments. The closest phylogenetic relatives of strain SLSR-1 are from the genus Desulfonatronospira, with highest homologies for D.spira thiodismutans (95 %), D.spira delicata (94 %) and D.spira dismutans (94 %) (Fig. 5). The DsrAB phylogeny also places SLSR-1 within the Desulfohalobiaceae (Fig. 6), being most similar to Desulfonatronospira thiodismutans (Sorokin et al. 2008). A similar case can be made for the respiratory arsenate reductase of strain SLSR-1 (see below). But there are substrate disparities between strain SLSR-1 and these diverse haloalkaliphiles. As mentioned above, chemoautotrophic growth via disproportionation of sulfite or thiosulfate is a distinguishing characteristic of Desulfonatronospira thiodismutans and Desulfonatronospira delicata in addition to heterotrophic growth by oxidation of low-molecular-weight organics (e.g., lactate, ethanol, butanol) linked to respiratory reduction of sulfate, thiosulfate, or sulfite (Table 2; Sorokin et al. 2008). The ability to metabolize H2 was observed for Desulfonatronum thiodismutans, Desulfonatronum cooperativum, Desulfonatronovibrio hydrogenovorans (Table 2), and Desulfurispira natronophila (Sorokin and Muyzer 2010) However, strain SLSR-1 was not capable of H2 metabolism, and although it could grow via dissimilatory reduction of thiosulfate or sulfite, it did not display a clear capacity for chemoautotrophic growth via disproportionation of these sulfur compounds. Moreover, although strain SLSR-1 could use pyruvate as an electron donor, it could not grow via pyruvate fermentation in the absence of an electron acceptor, nor could it employ acetate as an electron donor for growth on sulfate. Finally, strain SLSR-1 is clearly morphologically distinct from the Desulfonatronospira by having a distinct vibroid rather than a spiraled shape, which when taken together with its lower G + C abundance (e.g., 45.1 vs. 50.4 %) argues for its taxonomic classification in a separate genus.
Fig. 6.
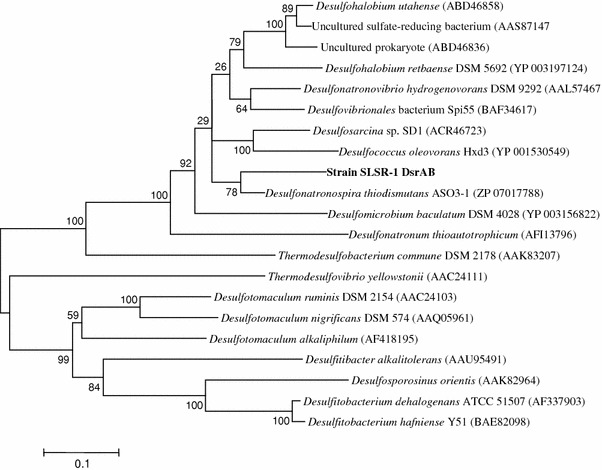
Maximum likelihood tree of predicted amino acid translation of the dsrAB genes showing the phylogenetic position of strain SLSR-1. Genbank accession numbers are included next to the corresponding species names
Strain SLSR-1did exhibit a mode of sustained growth with sulfide as the electron donor and nitrate as the electron acceptor which would suggest a capacity for chemolithotrophy. However, when the sulfide/arsenate couple was tested cell lines could not be sustained after one or two transfers, indicating a requirement for a carbon skeleton (e.g., lactate, acetate) rather than via autotrophic fixation of inorganic carbon. Nonetheless, nitrate- or arsenate-linked lithotrophic oxidation of sulfide is analogous to modes of chemoautotrophy displayed by two isolates from Mono Lake, Alkalilimnicola ehrlichii strain MLHE-1 and strain MLMS-1 (Oremland et al. 2002; Hoeft et al. 2004, 2007), and by Halarsenatibacter silvermanii from Searles Lake (Oremland et al. 2005).
Phylogenetic analysis of the predicted protein of the arrA-like gene sequence of SLSR-1 revealed that it is part of the ArrA, arsenate respiratory reductase clade of the DMSO reductase family of molybdenum enzymes (Saltikov and Newman 2003). The SLSR-1 partial ArrA was most similar to an ArrA-like encoding gene found in the genome of Desulfonatronospira thiodismutans (71 and 82 % amino acid identity and similarity, respectively) although it is not known if this organism can grow via dissimilatory arsenate reduction (Fig. 7). Strain SLSR-1 ArrA formed a cluster with ArrAs of D.spira thiodismutans and the Mono Lake sulfide-oxidizing arsenate reducer, MLMS-1. Additional partial arrA sequences from uncultured bacteria previously retrieved from Searles Lake using a different primer set (Kulp et al. 2006) shared only 70–80 % amino sequence similarities to the corresponding regions of the SLSR-1 ArrA. These observations indicated that the strain SLSR-1 arsenate respiratory reductase is most likely a unique member of the ArrA clade. These collective considerations argue that strain SLSR-1 represents the first described member species of a new genus of extremely haloalkaliphilic sulfate-respiring bacteria, for which we propose the name Desulfohalophilus alkaliarsenatis gen. nov., sp. nov.
Fig. 7.
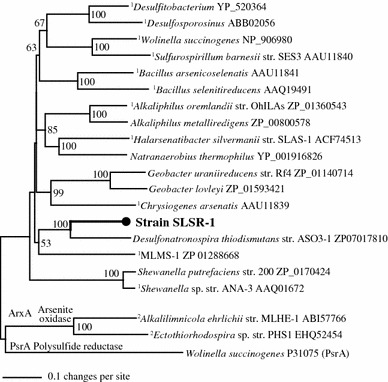
Phylogenetic relationship of the ArrA sequence of strain SLSR-1 in relationship to the ArrA enzymes of other known arsenate respiring bacteria or predicted ArrA identified in sequenced microbial genomes. The bootstrap values 50 % and greater are indicated at the corresponding nodes. Superscript 1 and 2 correspond to organisms known to respire arsenate or oxidize arsenite, respectively
Presence of SLSR-1-like sequences in Searles Lake
Although strain SLSR-1 was isolated from a stable enrichment culture from Searles Lake, its significance to the Searles Lake ecosystem is unclear. DGGE analysis indicates that sequences closely related to SLSR-1 are present in sediments of Searles Lake and are predominant at 7.5 cm depth in the sediments. The presence of strain SLSR-1 relatives in Searles Lake sediments is supported by our previous finding of sequences highly related to SLSR-1 (Fig. 5; Kulp et al. 2006). However, the arrA gene from strain SLSR-1 was distinct from those found in Searles Lake samples. Thus, we cannot directly confirm that this isolate is a dominant member of the Searles Lake microbial community. Nonetheless, the physiology of this isolate, i.e. its capacity to reduce As(V) under conditions restrictive for sulfate reduction, is likely a common mechanism in Searles Lake and other As(V) and SO4 2− containing environments.
Discussion
Strain SLSR-1 demonstrated slow but obvious growth as an anaerobic heterotroph when cultivated in defined mineral medium with sulfate or in combinations of sulfate plus arsenate serving as electron acceptors under conditions of moderate hypersalinity (at an a w of 0.9, Fig. 3). We noted that growth on arsenate required the presence of sulfate. This could have been due to the latter anion serving as a nutritional source of sulfur in this chemically defined medium which lacked any complex nutritional supplements (e.g., yeast extract, peptone) and contained only a small quantity of cysteine serving as a reductant. When both arsenate and sulfate were present at equimolar concentrations, dissimilatory reduction of the former preceded that of the latter (Fig. 3b, c), a condition first noted for Desulfotomaculum auripigmentum (renamed: Desulfosporosinus auripigmenti) (Newman et al. 1997a), and explained by the much greater energy yield associated with respiration of arsenate as opposed to sulfate (Newman et al. 1997b). Supplementing the medium with 0.2 % yeast extract and incubating at a salinity closer to optimal (a w = 0.86) decreased doubling times from 82.5 to 18.4 h, and increasing μ from 0.008 to 0.038 h−1.
Despite the extraordinarily high concentration of sulfate in the brines of Searles Lake (0.730 molal; roughly equivalent to 1 M), in situ sulfate-reduction was undetectable in its sediments (Oremland et al. 2005; Kulp et al. 2006, 2007). Sulfate-reduction by strain SLSR-1 was strongly inhibited by the imposed salinity gradients (Fig. 4a), being almost completely eliminated at a salinity (~330 g/L; a w = 0.79) that approached the actual strength of the lake’s brine (a w = 0.71; Table 1). Hence, these results are in general accordance with the original observations of Oren (1999a) with regard to the bioenergetic-constraints upon sulfate-reduction at high salinities. The survival of strain SLSR-1 under such ambient conditions in Searles Lake is assured by its ability to grow via respiration of the abundant arsenate ions in the brine (~4 mM; Oremland et al. 2005) in lieu of sulfate. Arsenate reduction by strain SLSR-1 was impeded comparatively modestly (~50 %) at the highest salinity tested (Fig. 4a).
The natural abundance of borate ions in the brinewater of Searles Lake (0.43 molal) was postulated as a selective cause of inhibition of sulfate-reduction in its sediments (Kulp et al. 2007). High borate concentrations strongly inhibited sulfate-reduction by strain SLSR-1 (93 % inhibition), while its effect on arsenate-reduction was less pronounced (78 % inhibition) (Fig. 4c) leaving this variable as a contributing complication to the original salinity conundrum. Faced with the “double whammy” of Searles Lake brine’s low water activity combined with its high borate concentrations, it appears that strain SLAS-1 is operating in situ at the margin of its physiological survival range. Nonetheless, vigorous in situ 73As(V)-reduction was measured in the anoxic, shallow sediments of Searles Lake that lay beneath its thick salt crust and overlying brine (Kulp et al. 2006). The results prove that the bacterial community, if not strain SLSR-1 itself, is functional under these extreme conditions. Examples of poly-extremophiles, namely the ability to tolerate multiple severe environmental stresses, can be found in other microorganisms isolated from evaporative soda lakes (Bowers and Wiegel 2011).
Low but detectable free sulfide (~0.1–0.3 mM) is prevalent within the anoxic sediment lying below the dense brinewater of Searles Lake (Oremland et al. 2005; Kulp et al. 2006). In the absence of sulfate-reduction, the origin of the sulfide could arise from the dissimilatory reduction or disproportionation of less oxidized intermediates of the sulfur cycle such as thiosulfate, sulfite, or elemental sulfur. The bacteria that carry out these types of sulfidogenesis occur commonly in soda lakes (Sorokin et al. 2008, 2011a; Sorokin and Muyzer 2010), as do the bacteria that regenerate these substances by oxidative processes (Sorokin et al. 2011b). Strain SLSR-1 was capable of growth upon thiosulfate and sulfite via dissimilatory reduction, although notably not elemental sulfur, as were its closest phylogenetic relatives listed in Table 2. Desulfurispira natronophila can grow on sulfur in lieu of sulfate, thiosulfate, or sulfite (Table 2), as can Halarsenatibacter silvermanii an arsenate-respirer isolated from Searles Lake (Switzer Blum et al. 2009). Collectively, a sediment flora consisting of substrate affinities represented by these haloalkaliphiles would assure modes of in situ sulfide production via intermediates of the sulfur cycle. Supporting this is the detection of 16S rDNA amplicons of strains taxonomically related to strain SLSR-1 from extracted DNA from Searles Lake mud (Fig. 5). Yet that leaves unanswered the question of how intermediates of the sulfur cycle arose in the first place without the occurrence of some initial sulfate-reduction.
One possibility is that the free sulfide currently detected in the lake’s sediments could have arisen from the degradation of organic sulfur compounds, such as the common osmolyte dimethylsulfoniopropionate, and perhaps from sulfur-containing proteins as well. The question of the origin of the sulfide detected in Searles Lake is ecologically relevant because both strain SLSR-1 and H. silvermanii also have the ability to oxidize sulfide using either arsenate or nitrate as the oxidant (Table 2; Oremland et al. 2005; Switzer Blum et al. 2009).
The search for extant microbial life in the Solar System beyond Earth’s orbit holds the likelihood of someday encountering and testing highly dense brines formed either by cryo-concentration (Europa) or evapo-concentration (Mars). Planetary scientists and geochemists employ a w, rather than salinity, as a means to describe the density of such brines. Specifically, in the case of Mars the residual fluvial salt deposits encountered by the Rover Opportunity at Meridiani Planum have been proposed to be derived from brines having such strong ionic strengths as to severely impede or even preclude the possibility of life. Tosca et al. (2008) calculated a w values of 0.78–0.86 of the brines that once flowed over these sites, roughly 3.0 billion years ago (Andrews-Hanna et al. 2007). Strain SLSR-1 can clearly grow at these low water activities (Table 1; Fig. 4), and in situ As(V)-reductase activity occurs in the sediments of Searles Lake at a w = 0.71 (Kulp et al. 2006). Hence, the consideration of salinity per se as an absolute impediment to anaerobic microbial life at these a w values can be challenged, although the brine’s highly acidic rather than alkaline nature would add another strong physiological constraint. Another severe physiological constraint would include considerations of chaotropicity (high MgCl2 content; Hallsworth et al. 2007). However, the gypsum veins of the early Noachian age recently encountered by the Rover Opportunity at the Cape York site (Endeavor Crater) gave “evidence for relatively dilute [a w = ~0.98] water at moderate temperature, perhaps supporting locally and transiently habitable environments” (Squyres et al. 2012). Such a scenario could conceivably allow time for the adaptation to and natural selection of a microbial flora able to tolerate the extreme conditions that subsequently ensued over the Meridiani Planum of the late Noachian and early Hesperian epochs of Mars. Regardless of these events, adopting a general usage of a w within the extremophile community would make it better conversant with planetary scientists searching for extant (or extinct) life at the margins of known terrestrial physiology encountered in possible biomes lying beyond Earth.
Description of Desulfohalophilus, gen. nov.
De.sul.fo.ha.lo’phi.lus. L. pref. de-, from; L. n. sulfur, sulfur; N.L. pref. desulfo-, desulfuricating (prefix used to characterize a dissimilatory sulfate-reducing prokaryote); Gr. n. hals halos, salt; N.L. adj. philus (from Gr. adj. philos -ê -on), friend, loving; N.L. masc. n. Desulfohalophilus, sulfate-reducing salt-loving bacterium; Gram negative motile vibrios. Obligately anaerobic with a respiratory metabolism of dissimilatory reduction of sulfate. Chemoorganotroph, incompletely oxidizes electron donors like lactate and pyruvate. Alkaliphilic and halophilic. Habitats are soda lakes. Belongs to the class deltaproteobacteria. The type species is Desulfohalophilus alkaliarsenatis.
Description of Desulfohalophilus alkaliarsenatis gen. nov., sp. nov.
Motile vibrio with one or two polar flagella. Cells are 0.5 × 3.0 μm, occur singly or in pairs. Multiplication is by binary fission, and cells have a Gram negative cell wall structure. Strictly anaerobic, growth is chemohetereotrophic or chemoautotrophic. Slow growth occurs in defined mineral medium, but is stimulated by the inclusion of yeast extract. Utilizes lactate or pyruvate as electron donors when grown with sulfate or arsenate as the electron acceptor, the products include acetate, sulfide or arsenite. Other electron acceptors supporting heterotrophic growth include nitrate, sulfite and thiosulfate. Chemoautotrophic growth occurs with sulfide as the electron donor and nitrate as the electron acceptor. Lithotrophic oxidation of sulfide with arsenate, but requires a carbon skeleton (e.g., lactate) for growth. Obligate alkaliphile that does not grow at pH below 7.75 or above 9.75 and has an optimum at 9.25. Salinity range for growth ranges from 55 to 330 g/L (a w = 0.96–0.8) with an optimum at 200 g L−1 (0.84 a w). The optimum temperature for growth is 44 °C and growth occurs over the range of 20–55 °C. The G + C content is 45.1 mol% (as determined by the thermal denaturation method). Habitat: bottom deposits of alkaline athlassic and extremely hypersaline soda lakes. The type strain is strain SLSR-1, which was isolated from the sediments of Searles Lake, located in the Mojave Desert of southeastern California, USA. The strain has been deposited in the American Type Culture Collection (accession number BAA-2432) and the DSMZ (DSM accession number 25765)
Acknowledgments
We are extremely grateful to A. Oren and D. Sorokin for helpful comments and guidance on an earlier draft of this manuscript. This work was supported by a NASA Exobiology grant to RSO and JFS, and by the Toxics Hydrology Program of the US Geological Survey
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
References
- Andrews-Hanna JC, Phillips RJ, Zuber MT. Meridiani Planum and the global hydrology of Mars. Nature. 2007;446:163–166. doi: 10.1038/nature05594. [DOI] [PubMed] [Google Scholar]
- Balderston WL, Sherr B, Payne WJ. Blockage by acetylene of nitrous oxide reduction in Pseudomonas perfectomarinus. Appl Environ Microbiol. 1976;31:504–508. doi: 10.1128/aem.31.4.504-508.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beliakova EV, Rozanova EP, Borzenkov IA, Turova TP, Pusheva MA, Lysenko AM, Kolganov TV. The new facultatively chemolithoautotrophic, moderately halophilic, sulfate-reducing bacterium Desulfovermiculus halophilus gen. nov., sp. nov., isolated from an oil field. Mikrobiologia. 2006;75:201–211. [PubMed] [Google Scholar]
- Bodaker I, Sharon I, Suzuki MT, Feingersch R, Shmoish M, Andreshcheva E, Sogin ML, Rosenberg M, Maguire ME, Belkin S, Oren A, Béjà O. Comparative community genomics on the Dead Sea: an increasingly extreme environment. ISME J. 2010;4:399–407. doi: 10.1038/ismej.2009.141. [DOI] [PubMed] [Google Scholar]
- Bowers KJ, Wiegel J. Temperature and pH optima of extremely halophilic Archaea. A minireview. Extremophiles. 2011;15:119–128. doi: 10.1007/s00792-010-0347-y. [DOI] [PubMed] [Google Scholar]
- Brandt KK, Vester F, Jensen AN, Ingvorsen JK. Sulfate reduction dynamics and enumeration of sulfate-reducing bacteria in hypersaline sediments of the Great Salt Lake (Utah, USA) Microbial Ecol. 2001;41:1–11. doi: 10.1007/s002480000059. [DOI] [PubMed] [Google Scholar]
- Cline JD. Spectrophotometric determination of hydrogen sulfide in natural waters. Limnol Oceanogr. 1969;14:454–459. [Google Scholar]
- Cloern JE, Cole BE, Oremland RS. Autotrophic processes in meromictic Big Soda Lake, Nevada. Limnol Oceanogr. 1983;28:1049–1061. [Google Scholar]
- Cohen Y, Krumbein WE, Shilo M. Solar Lake (Sinai). 2. Distribution of photosynthetic microorganisms and primary production. Limnol Oceanogr. 1977;22:609–620. [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Foti M, Sorokin DY, Lomans B, Mussman M, Zacharova EE, Pimenov NV, Kuenen JG, Muyzer G. Diversity, activity, and abundance of sulfate-reducing bacteria in saline and hypersaline soda lakes. Appl Environ Microbiol. 2007;73:2093–2100. doi: 10.1128/AEM.02622-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallsworth JE, Yakimov MM, Golyshin PN, Gillion JLM, D’Auria G, de Lima Alves F, La Cono V, Genovese M, McKew BA, Hayes SL, Harris G, Giuliano L, Timmis KN, McGenity TJ. Limits of life in MgCl2-containing environments: chaotropicity defines the window. Environ Microbiol. 2007;9:801–813. doi: 10.1111/j.1462-2920.2006.01212.x. [DOI] [PubMed] [Google Scholar]
- Hobbie JE, Daley RL, Jaspar S. Use of nuclepore filters for counting bacteria for fluorescence microscopy. Appl Environ Microbiol. 1977;33:1225–1228. doi: 10.1128/aem.33.5.1225-1228.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeft SE, Kulp TR, Stolz JF, Hollibaugh JT, Oremland RS. Dissimilatory arsenate reduction with sulfide as the electron donor: experiments with Mono Lake water and isolation of strain MLMS-1, a chemoautotrophic arsenate respirer. Appl Environ Microbiol. 2004;70:2043–2049. doi: 10.1128/AEM.70.5.2741-2747.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeft SE, Switzer Blum J, Stolz JF, Tabita JR, Witte B, King GM, Santini JM, Oremland RS. Alkalilimnicola ehlichii, sp. nov., a novel, arsenite-oxidizing haloalkaliphilic γ-proteobacterium capable of chemoautotrophic or heterotrophic growth with nitrate or oxygen as the electron acceptor. Int J Syst Evol Microbiol. 2007;57:504–512. doi: 10.1099/ijs.0.64576-0. [DOI] [PubMed] [Google Scholar]
- Hoeft SE, Kulp TR, Han S, Lanoil B, Oremland RS. Coupled arsenotrophy in a photosynthetic hot spring biofilm from Mono Lake, California. Appl Environ Microbiol. 2010;76:463–4633. doi: 10.1128/AEM.00545-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsen TF, Kjeldsen KU, Ingvorsen K. Desulfohalobium utahense sp. nov., a moderately halophilic, sulfate-reducing bacterium isolated from Great Salt Lake. Int J Syst Evol Microbiol. 2006;56:2063–2069. doi: 10.1099/ijs.0.64323-0. [DOI] [PubMed] [Google Scholar]
- Jellison R, Melack JM. Algal photosynthetic activity in response to meromixis in hypersaline Mono Lake, California. Limnol Oceanogr. 1993;38:818–837. [Google Scholar]
- Jørgensen BB, Cohen Y. Solar Lake (Sinai). 5. The sulfur cycle of the benthic cyanobacterial mats. Limnol Oceanogr. 1977;22:657–666. [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Kjeldsen KU, Loy A, Jakobsen TF, Thompson TR, Wagner M, Ingvorsen K. Diversity of sulfate-reducing bacteria from an extreme hypersaline sediment, Great Salt Lake (Utah) FEMS Microbiol Ecol. 2007;60:287–298. doi: 10.1111/j.1574-6941.2007.00288.x. [DOI] [PubMed] [Google Scholar]
- Klug MJ, Boston P, Francois R, Gyure R, Javor B, Tribble G, Vairavamurthy A (1985) Sulfur reduction in sediments of marine and evaporate environments. In: Sagan D (ed) The global sulfur cycle. NASA Technical Memorandum 87570. National Aeronautics and Space Administration, Washington, DC, pp 128–157
- Kulp TR, Hoeft SE, Miller LG, Saltikov C, Murphy JN, Han S, Lanoil B, Oremland RS. Dissimilatory arsenate and sulfate reduction in the sediments of two hypersaline, arsenic-rich soda lakes: Mono and Searles Lakes, California. Appl Environ Microbiol. 2006;72:6514–6526. doi: 10.1128/AEM.01066-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulp TR, Han S, Saltikov CW, Lanoil BD, Zargar K, Oremland RS. Effects of imposed salinity gradients on dissimilatory arsenate reduction, sulfate reduction, and other microbial processes in sediments from two California soda lakes. Appl Environ Microbiol. 2007;73:5130–5137. doi: 10.1128/AEM.00771-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman DK, Kennedy EK, Coates JD, Ahmann D, Ellis DJ, Lovley DR, Morel FMM. Dissimilatory arsenate and sulfate reduction in Desulfotomaculum auripigmentum sp. nov. Arch Microbiol. 1997;168:380–388. doi: 10.1007/s002030050512. [DOI] [PubMed] [Google Scholar]
- Newman DK, Ahamnn D, Morel FMM. A brief review of microbial arsenate respiration. Geomicrobiol J. 1997;15:255–268. [Google Scholar]
- Nissenbaum A, Kaplan IR (1976) Sulfur and carbon isotopic evidence for biogeochemical processes in the Dead Sea ecosystem. In: Nriagu JO (ed) Environmental biogeochemistry, vol 1. Ann Arbor Scientific, MI, pp 309–325
- Ollivier B, Hatchikian CE, Prensier G, Guezennec J, Garcia JL. Desulfohalobium retbaense gen. nov., sp. nov., a halophilic sulfate-reducing bacterium from sediments of a hypersaline lake in Senegal. Int J Syst Bacteriol. 1991;41:74–81. [Google Scholar]
- Oremland RS, Umberger C, Culbertson CW, Smith RL. Denitrification in San Francisco bay intertidal sediments. Appl Environ Microbiol. 1984;47:1106–1112. doi: 10.1128/aem.47.5.1106-1112.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oremland RS, Switzer Blum J, Culbertson CW, Visscher PT, Miller LG, Dowdle P, Strohmaier FE. Isolation, growth, and metabolism of an obligately anaerobic, selenate-respiring bacterium, strain SES-3. Appl Environ Microbiol. 1994;60:3011–3019. doi: 10.1128/aem.60.8.3011-3019.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oremland RS, Dowdle PR, Hoeft S, Sharp JO, Schaefer JK, Miller LG, Switzer Blum J, Smith RL, Bloom NS, Wallschlaeger D. Bacterial dissimilatory reduction of arsenate and sulfate in meromictic Mono Lake, California. Geochim Cosmochim Acta. 2000;64:3073–3084. [Google Scholar]
- Oremland RS, Hoeft SE, Bano N, Hollibaugh RA, Hollibaugh JT. Anaerobic oxidation of arsenite in Mono Lake water and by a facultative, arsenite-oxidizing chemoautotroph, strain MLHE-1. Appl Environ Microbiol. 2002;68:4795–4802. doi: 10.1128/AEM.68.10.4795-4802.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oremland RS, Kulp TR, Switzer Blum J, Hoeft SE, Baesman S, Miller LG, Stolz JF. A microbial arsenic cycle in a salt-saturated, extreme environment. Science. 2005;308:1305–1308. doi: 10.1126/science.1110832. [DOI] [PubMed] [Google Scholar]
- Oren A. Bioenergetic aspects of halophilism. Microbiol Mol Biol Rev. 1999;63:334–348. doi: 10.1128/mmbr.63.2.334-348.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren A. Microbiological studies in the Dead Sea: future challenges toward the understanding of life at the limit of salt concentrations. Hydrobiologia. 1999;405:1–9. [Google Scholar]
- Oren A. Thermodynamic limits to microbial life at high salt concentrations. Environ Microbiol. 2011;13:1908–1923. doi: 10.1111/j.1462-2920.2010.02365.x. [DOI] [PubMed] [Google Scholar]
- Pikuta EV, Hoover RB, Bej AK, Marsic D, Whitman WB, Cleland D, Krader P. Desulfonatronum thiodismutans sp. nov., a novel alkaliphilic, sulfate-reducing bacterium capable of lithoautotrophic growth. Int J Syst Evol Microbiol. 2003;53:1327–1332. doi: 10.1099/ijs.0.02598-0. [DOI] [PubMed] [Google Scholar]
- Porter D, Roychoudhury AN, Cowan D. Dissimilatory sulfate reduction in hypersaline coastal pans: activity across a salinity gradient. Geochim Cosmochim Acta. 2007;71:5102–5116. [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Saltikov CW, Newman DK. Genetic identification of a respiratory arsenate reductase. Proc Natl Acad Sci USA. 2003;100:10983–10988. doi: 10.1073/pnas.1834303100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz R, Dayhoff M (1979) Matrices for detecting distant relationships. In: Dayhoff M (ed) Atlas of protein sequences. National Biomedical Research Foundation, pp 353–58
- Shen Y, Buick R, Canfield DE. Isotopic evidence for microbial sulphate reduction in the early Archean. Nature. 2001;410:77–81. doi: 10.1038/35065071. [DOI] [PubMed] [Google Scholar]
- Smith RL, Oremland RS. Big Soda Lake (Nevada). 2. Pelagic sulfate reduction. Limnol Oceanogr. 1987;32:794–803. [Google Scholar]
- Solorzano L. Determination of ammonia in natural waters by the phenol hypochlorite method. Limnol Oceanogr. 1969;14:799–801. [Google Scholar]
- Sorokin DY, Muyzer G. Desulfurispira natronophila gen. nov. sp. nov.: an obligately anaerobic dissimilatory sulfur-reducing bacterium from soda lakes. Extremophiles. 2010;14:349–355. doi: 10.1007/s00792-010-0314-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin DY, Tourova TP, Henstra AM, Stams AJM, Galinski EA, Muyzer G. Sulfidogenesis under extremely haloalkaline conditions by Desulfonatronospira thiodismutans gen. nov., sp. nov., and Desulfonatronospira delicata sp. nov.—a novel lineage of Deltaproteobacteria from hypersaline soda lakes. Microbiology. 2008;154:1444–1453. doi: 10.1099/mic.0.2007/015628-0. [DOI] [PubMed] [Google Scholar]
- Sorokin DY, Tourova TP, Detkova EN, Kolganova TV, Galinski EA, Muyzer G. Culturable diversity of lithotrophic haloalkaliphilic sulfate-reducing bacteria in soda lakes and the description of Desulfonatronum thioautotrophicum sp. nov., Desulfonatronum thiosulfatophilum sp. nov., Desulfonatrovibrio thiodismutans sp. nov., and Desulfonatrovibrio magnus sp. nov. Extremophiles. 2011;15:391–401. doi: 10.1007/s00792-011-0370-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin DY, Kuenen JG, Muyzer G. The microbial sulfur cycle at extremely haloalkaline conditions of soda lakes. Frontiers Microbiol. 2011;2:1–16. doi: 10.3389/fmicb.2011.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squyres SW, Arvidson RE, Bell JF, III, Calef F, III, Clark BC, Cohen BA, Crumpler LA, de Souza Jr PA, Farrand WH, Gellert R, Grant J, Herkenhoff KE, Hurowitz JA, Johnson JR, Jolliff BL, Knoll AH, Li R, McLennan SM, Ming DW, Mittlefehldt DW, Parker TJ, Paulsen G, Rice MS, Ruff SW, Schröder C, Yen AS, Zacny K. Ancient impact and aqueous processes at Endeavor Crater Mars. Science. 2012;336:570–576. doi: 10.1126/science.1220476. [DOI] [PubMed] [Google Scholar]
- Switzer Blum J, Burns Bindi A, Buzzelli J, Stolz JF, Oremland RS. Bacillus arsenicoselenatis sp. nov., and Bacillus selenitireducens sp. nov.: two haloalkaliphiles from Mono Lake, California which respire oxyanions of selenium and arsenic. Arch Microbiol. 1998;171:19–30. doi: 10.1007/s002030050673. [DOI] [PubMed] [Google Scholar]
- Switzer Blum J, Stolz JF, Oren A, Oremland RS. Selenihalanaerobacter shriftii gen. nov., sp. nov., a halophilic anaerobe from Dead Sea sediments that respires selenate. Arch Microbiol. 2001;175:208–219. doi: 10.1007/s002030100257. [DOI] [PubMed] [Google Scholar]
- Switzer Blum J, Han S, Lanoil B, Saltikov C, Witte B, Tabita FR, Langley S, Beveridge TJ, Jahnke L, Oremland RS. Ecophysiology of “Halarsenatibacter silvermanii” strain SLAS-1, gen. nov., sp. nov., a facultative chemoautotrophic arsenate respirer from salt-saturated Searles Lake, California. Appl Environ Microbiol. 2009;75:1950–1960. doi: 10.1128/AEM.02614-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL (1999) Phylogenetic analysis using parsimony (and other methods), version 4.0b10b. Sinauer Associates, Sunderland
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosca NJ, Knoll AH, McLennan SM. Water activity and the challenge for life on early Mars. Science. 2008;320:1204–1207. doi: 10.1126/science.1155432. [DOI] [PubMed] [Google Scholar]
- van den Wielen PW, Heijs SK. Sulfate-reducing prokaryotic communities in two deep hypersaline anoxic basins in the Eastern Mediterranean deep sea. Environ Microbiol. 2007;9:1335–1340. doi: 10.1111/j.1462-2920.2006.01210.x. [DOI] [PubMed] [Google Scholar]
- van den Wielen PW, Bolhuis H, Borin S, Daffonchio D, Corselli C, Giuliano L, et al. The enigma of prokaryotic life in deep hypersaline anoxic basins. Science. 2005;307:121–123. doi: 10.1126/science.1103569. [DOI] [PubMed] [Google Scholar]
- Weisenburg DA, Brooks JM, Bernard BB. Biogenic hydrocarbon gases and sulfate reduction in the Orca Basin brine. Geochim Cosmochim Acta. 1985;49:2069–2080. [Google Scholar]
- Widdel F, Kohring GW, Mayer F. Studies on dissimilatory sulfate-reducing bacteria that decompose fatty acids. III. Characterization of the filamentous gliding Desulfonema limicola gen. nov., sp. nov., and Desulfonema magnum sp. nov. Arch Microbiol. 1983;134:286–294. [Google Scholar]
- Wolin EA, Wolin MJ, Wolfe RS. Formation of methane by bacterial extracts. J Biol Chem. 1963;238:2882–2886. [PubMed] [Google Scholar]
- Zargar K, Conrad A, Bernick DL, Lowe TM, Stolc V, Hoeft S, Oremland RS, Stolz J, Saltikov CW (2012) ArxA, a new clade of arsenite oxidase within the DMSO reductase family of molybdenum oxidoreductases. Environ Microbiol (in press) [DOI] [PubMed]
- Zhilina TN, Zavarzin GA, Rainey FA, Pikuta EN, Osipov GA, Kostrikina NA. Desulfonatronovibrio hydrogenovorans gen. nov., sp. nov., an alkaliphilic, sulfate-reducing bacterium. Int J Syst Bacteriol. 1997;47:144–149. doi: 10.1099/00207713-47-1-144. [DOI] [PubMed] [Google Scholar]
- Zhilina TN, Zavarzina DG, Kuever J, Lysenko AM, Zavarzin GA. Desulfonatronum cooperativum sp. nov., a novel hydrogenotrophic, alkaliphilic, sulfate-reducing bacterium, from a syntrophic culture growing on acetate. Int J Syst Evol Microbiol. 2005;55:1001–1006. doi: 10.1099/ijs.0.63490-0. [DOI] [PubMed] [Google Scholar]


