SUMMARY
We used affinity-purification mass spectrometry to identify 747 candidate proteins that are complexed with Huntingtin (Htt) in distinct brain regions and ages in Huntington’s disease (HD) and wildtype mouse brains. To gain a systems-level view of the Htt interactome, we applied Weighted Gene Correlation Network Analysis (WGCNA) to the entire proteomic dataset to unveil a verifiable rank of Htt-correlated proteins and a network of Htt-interacting protein modules, with each module highlighting distinct aspects of Htt biology. Importantly, the Htt-containing module is highly enriched with proteins involved in 14-3-3 signaling, microtubule-based transport, and proteostasis. Top-ranked proteins in this module were validated as novel Htt interactors and genetic modifiers in an HD Drosophila model. Together, our study provides a compendium of spatiotemporal Htt-interacting proteins in the mammalian brain, and presents a conceptually novel approach to analyze proteomic interactome datasets to build in vivo protein networks in complex tissues such as the brain.
INTRODUCTION
Huntington’s disease (HD) is one of the most common dominantly inherited neurodegenerative disorders clinically characterized by a triad of movement disorder, cognitive dysfunction, and psychiatric impairment (Bates, 2002). HD neuropathology is characterized by selective and massive degeneration of the striatal medium spiny neurons (MSNs), and to a lesser extent, the deep layer cortical pyramidal neurons (Vonsattel and Difiglia, 1998). The disease is caused by a CAG repeat expansion resulting in an elongated polyglutamine (polyQ) stretch near the N-terminus of Huntingtin (Htt) (The Huntington's Disease Collaborative Research Group, 1993). HD is one of nine polyQ disorders with shared molecular genetic features such as an inverse relationship between the expanded repeat length and the age of disease onset, and evidence for toxic gain-of-function as a result of the polyQ expansion (Orr and Zoghbi, 2007). However, each of the polyQ disorders appears to target a distinct subset of neurons in the brain leading to disease-specific symptoms. Hence, it is postulated that molecular determinants beyond the polyQ repeat itself may be critical to disease pathogenesis (Orr and Zoghbi, 2007).
Protein interacting cis-domains (Lim et al., 2008) and post-translational modifications (PTMs) of polyQ proteins (Emamian et al., 2003; Gu et al., 2009) can significantly modify disease pathogenesis in vivo. Thus, studying the proteins that interact with domains beyond the polyQ region may provide important clues to disease mechanisms. In the case of HD, several hundred putative Htt interactors have been discovered using ex vivo methods such as yeast two-hybrid (Y2H) or in vitro affinity pull-down assays, utilizing only small, N-terminal fragments of Htt (Goehler et al., 2004; Kaltenbach et al., 2007). Such studies have provided insight into Htt’s normal function as a scaffolding protein involved in vesicular and axonal transport and nuclear transcription (Caviston and Holzbaur, 2009; Li and Li, 2006). The caveats of the prior Htt interactome studies include the exclusive use of small Htt N-terminal fragments as baits and the isolation of interactors ex vivo. Hence, it is not known which proteins can complex with full-length Htt (fl-Htt) in vivo within distinct brain regions and at different ages. Such information may shed light on age-dependent, selective neuropathogenesis in HD.
Immunoaffinity purification of native protein complexes followed by identification of its individual components using mass spectrometry (MS) has emerged as a powerful tool for deciphering in vivo neuronal signaling (Husi et al., 2000), synaptic (Fernández et al., 2009) and disease-related interactomes (Major et al., 2007). Although a “shotgun” proteomic approach is useful in creating a list of native interacting protein candidates from relevant mammalian tissues, formidable challenges exist in the unbiased bioinformatic analyses of such complex proteomic datasets to identify high-confidence interactors and to build accurate, endogenous protein interaction networks (Liao et al., 2009).
In this study, we performed a spatiotemporal in vivo proteomic interactome study of fl-Htt using dissected brain regions from a mouse model for HD and wildtype controls. The BACHD mouse model used in the study expresses full-length human mutant Htt (mHtt) with 97Q under the control of human Htt genomic regulatory elements on a BAC transgene (Gray et al., 2008). BACHD mice exhibit multiple disease-like phenotypes over the course of 12 months, including progressive motor, cognitive and psychiatric-like deficits and selective cortical and striatal atrophy (Gray et al., 2008; Menalled et al., 2009). Our multi-dimensional affinity purification-mass spectrometry (AP-MS) study uncovered a total of 747 candidate proteins complexed with fl-Htt in the mammalian brain. Moreover, we applied WGCNA to analyze the entire fl-Htt interactome dataset to define a verifiable rank of Htt-interacting proteins, and to uncover the organization of in vivo fl-Htt interacting protein networks in the mammalian brain.
RESULTS
Building an In Vivo Spatiotemporal Full-length Htt Interactome in BACHD and Wildtype Mouse Brains
To define the in vivo protein interactome for fl-Htt in BACHD and WT mouse brains, we performed immunoprecipitation (IP) of full-length mutant and WT Htt from BACHD and control mouse brains and identified the co-purified proteins by mass spectrometry. Since previous studies suggest that the majority of Htt interactors bind to Htt N-terminal fragments, with very few binding to the C-terminal region (Kaltenbach et al., 2007), we reasoned that IP with a Htt antibody against the C-terminal region of the protein should preserve the vast majority of in vivo Htt protein interactions. We identified a monoclonal antibody (clone HDB4E10) capable of preferentially pulling-down human Htt in BACHD brains, with lesser affinity for immunoprecipitating murine Htt in both BACHD and WT mice (Figure 1A). Considering the lack of suitable Htt antibodies that can immunoprecipitate only polyQ-expanded or WT Htt with equal efficiency, our AP-MS strategy of using HDB4E10 should be considered as a survey of in vivo Htt-complexed proteins regardless of Htt polyQ length. This is a reasonable strategy since full-length human mHtt can fully substitute the essential function of murine Htt in murine embryonic development (Gray et al., 2008), and hence both forms should share the majority of in vivo interacting proteins.
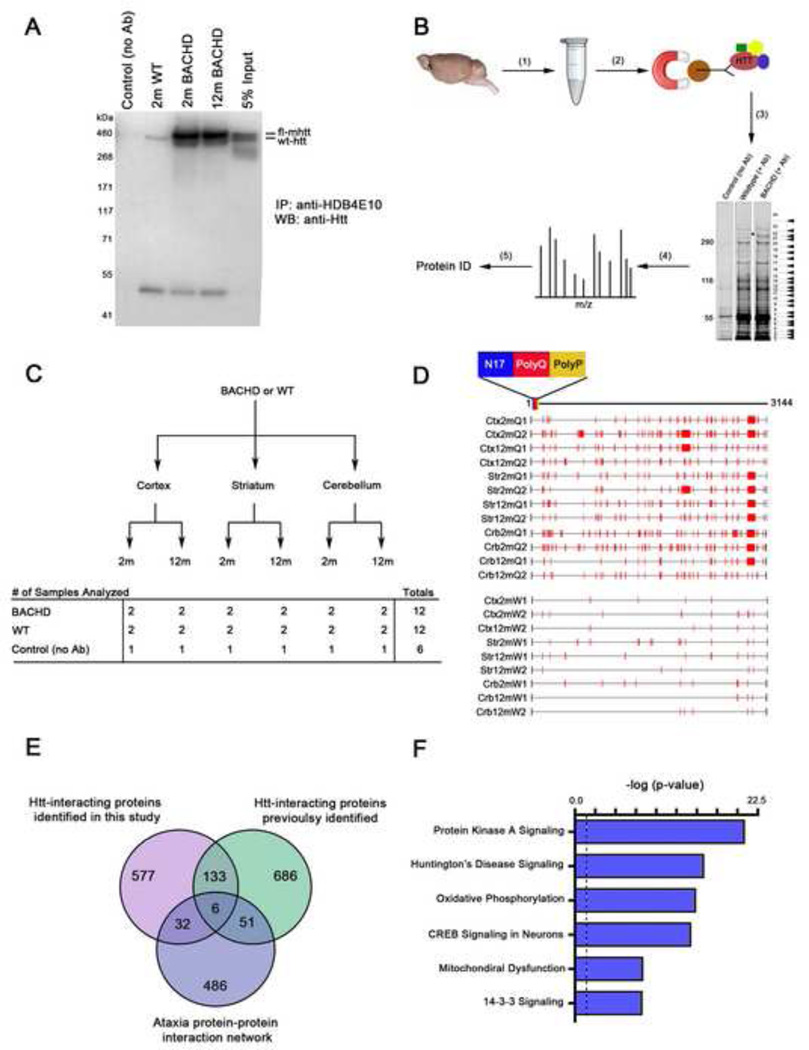
Figure 1. Generation and characterization of a full-length Htt in vivo proteomic interactome from BACHD and wildtype mouse brains.
(A) Immunoprecipitation using a human Htt-selective antibody (aa 1844–2131, clone HDB4E10) followed by Westernblot analysis using a human and mouse Htt antibody (MAB2166) confirms full-length Htt is pulled-down in both 2-month WT and 2- and 12-month BACHD soluble cortical extracts. A negative control (no Htt antibody) and 5% input loading control were also included. (B) The affinity purification followed by mass spectrometry (AP-MS) proteomic approach used to identify endogenous fl-Htt protein interactors: (1) The cortex, striatum, and cerebellum of 2- and 12-month WT and BACHD mice were homogenized, (2) The soluble lysates were subjected to anti-Htt IP using HDB4E10 and Protein G Dynabeads, (3) The immunoprecipitates were eluted with LDS loading buffer, separated on an SDS-PAGE gel, each gel was cut into 24–27 gel bands, and each gel band was subjected to in-gel trypsin digestion, (4) Approximately 750 independent LC-MS/MS analysis were performed, and (5) All mass spectra were searched using MASCOT (Matrix Science) to obtain a total of 747 high-scoring, putative Htt-interacting proteins. (C) The scheme of the spatiotemporal proteomic interactome study that includes 30 independent samples for the AP-MS experiments. (D) A schematic representation of the Htt tryptic peptides identified along the full-length Htt primary sequence (aa 1–3144). BACHD IP conditions labeled with Q and WT IP conditions labeled W. (E) Venn diagram comparison of Htt-interacting proteins identified in this study, literature-curated Htt-interacting proteins, and the published ataxia protein network. (F) Top IPA canonical signaling pathways enriched in the the Htt interactome, plotted as –log10 p-value significance. The black dotted line indicates canonical pathway genome-wide p-value significance (p < 0.05). See also Supplemental Tables S1–S3, and S5.
Human and/or murine fl-Htt complexes were immunoaffinity purified from the cortex, striatum, and cerebellum of BACHD and WT mice at 2- or 12-months of age using HDB4E10 (Figure 1B). As a negative control, a mock IP for each sample condition (defined by specific brain region, age and genotype) was performed in the absence of the Htt antibody. Therefore, a total of 30 independent IP experiments were performed, and approximately 765 trypsin-digested gel slices were subjected to LC-MS/MS analysis (Figure 1C). Using the MASCOT (Matrix Science) sequence database-searching tool, we identified a total of 747 high-scoring, putative Htt-interacting proteins from BACHD and WT mouse brains (Supplemental Table S1). Consistent with the claim that we are surveying fl-Htt interacting proteins, we identified Htt peptides spanning the entire sequence of Htt in both BACHD and WT samples (Figure 1D and Supplemental Table S2). As confirmation that HDB4E10 more efficiently immunoprecipitates human Htt than murine Htt, we observed more Htt peptides and more extensive sequence coverage of fl-Htt in BACHD mice compared to WT mice.
The In Vivo fl-Htt Interactome Is Enriched with Known Htt Interactors and Proteins Involved in Pathways Relevant to HD
We next performed several standard bioinformatic analyses to determine the validity of our approach in isolating Htt interactors in vivo, and to define potential biological pathways that Htt may be engaged in at various ages within different brain regions. We compared our list of 747 candidates with previously reported Htt interactors. Among a curated list of 877 proteins previously identified as putative ex vivo Htt interactors, most of which were obtained from Y2H experiments (Goehler et al., 2004; Kaltenbach et al., 2007; HDbase.org), we found 139 proteins also present in our in vivo Htt interactome (Figures 1E and Supplemental Table S3). This represents a highly significant enrichment (p = 7.00 × 10−50; hypergeometric test). The disease specificity of our interactome is supported by the comparison of our dataset with that of another polyQ disease protein, Ataxin1 (Lim et al., 2006). Despite that one-third of our samples originated from the cerebellum, the same tissue source as the Ataxin1 interactome study, only 38 proteins were present in both datasets with relatively modest enrichment (p = 0.0005; Figure 1E and Supplemental Table S3). Thus, our in vivo fl-Htt interactome appears to be specific to Htt, and provides a valuable list of Htt-interacting proteins, both in vivo and ex vivo, for further investigation to determine their roles in novel Htt biology and HD pathogenesis.
Gene Ontology (GO) analysis (Huang et al., 2009a; Huang et al., 2009b) of our in vivo fl-Htt interactome dataset (Supplemental Table S4) showed significant overrepresentation of proteins involved in ‘Intracellular Transport’, ‘Synaptic Transmission’, and ‘Protein Folding’ (all previously implicated in HD pathogenesis), as well as proteins involved in pathways related to the ‘Generation of Precursor Metabolites and Energy’, ‘Cytoplasmic Membrane-Bound Organelles’, and ‘Nucleotide Binding’. This supports Htt’s involvement in multiple biological functions within several subcellular compartments in the brain (Li and Li, 2006).
We next probed the biological and disease pathways enriched in our in vivo fl-Htt interactome using Ingenuity Pathway Analyses (IPA, Ingenuity® Systems, www.ingenuity.com), a large curated database of published information on mammalian biology and disease (Figure 1F; Supplemental Table S5). As independent validation for the relevancy of our interactome to HD biology, the IPA ‘Huntington’s Disease Signaling’ pathway, based on published normal and disease-specific processes and pathways relevant to HD, was significantly enriched. Importantly, other top IPA signaling pathways enriched in our fl-Htt interactome include ‘Protein Kinase A Signaling’, ‘CREB Signaling in Neurons’, and ‘Mitochondrial Dysfunction’, which are pathways previously implicated in HD pathogenesis (Sugars et al., 2004; Kleiman et al., 2011).
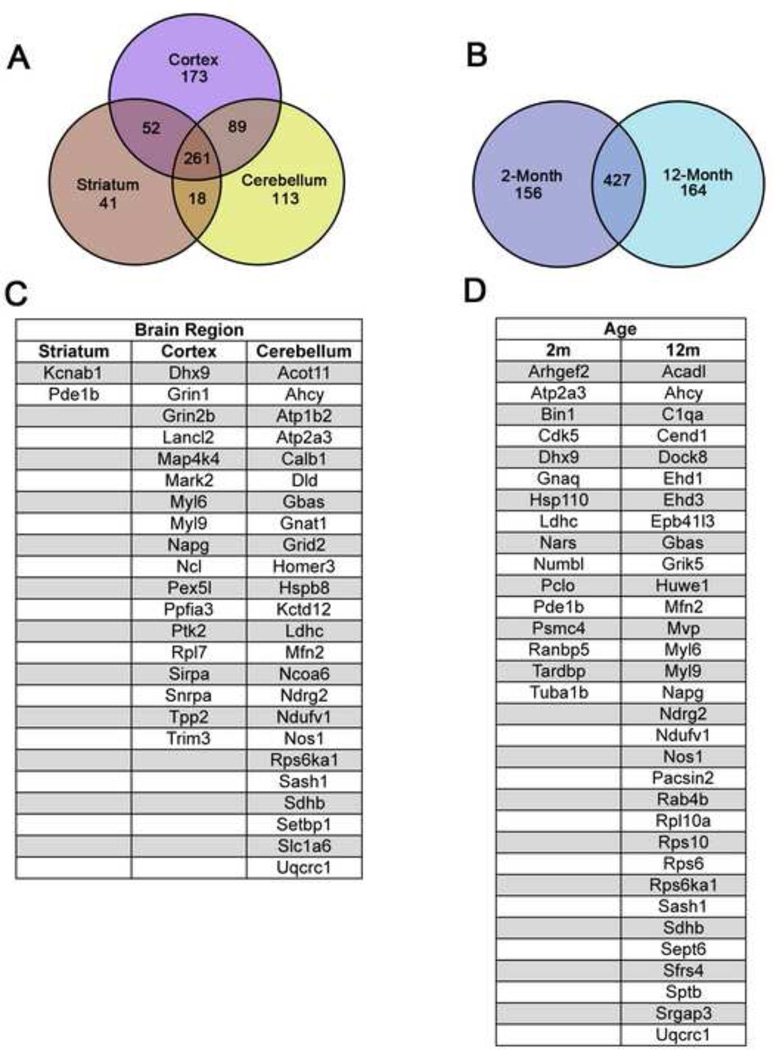
Our rationale for examining samples from three different brain regions at two different time points was to reveal dynamic in vivo differences between fl-Htt interactomes, which could possibly provide insight into selective, age-dependent disease processes. To this end, we identified candidate fl-Htt interactors exclusively from brain regions or ages relevant to HD (Figure 2, Supplemental Table S6), providing an interesting subset of proteins to further investigate their putative roles in selective neuronal vulnerability in HD. While the majority of proteins in our interactome are shared between all three brains regions (34.9%) and both age time points (57.3%), a subset of proteins were found to co-purify with Htt in specific brain regions (Cerebellum, 15.1%; Cortex, 23.1%; and Striatum 5.5%) and age time points (2m, 20.9%; and 12m, 22.0%). The proteins that appear reproducibly (at least 2 peptides in two IP conditions) and selectively complex with Htt at 12m, or in the striatum or cortex of our AP-MS dataset, are putative candidates for mediating age-dependent selective pathogenesis in HD, while those in complex with Htt only at 2m or in the cerebellum may be neuroprotective (Figure 2C and 2D).
Figure 2. Brain regional and age-specific protein interactions revealed in Htt interactome.
(A–B) Venn diagram comparison of the age (2- and 12-month) and brain region (cortex, striatum, and cerebellum) specific Htt protein interactions identified in this study. (C–D) A list of high confidence age and brain regional Htt protein interactions (protein identified in > 2 IP conditions, > 3 total peptides identified, no peptides identified in control IP). See also Supplemental Table S6.
Semi-Quantitative Measures of Protein Abundance Allow Unbiased Ranking of Candidate Interactors Based on Correlation with Fl-Htt
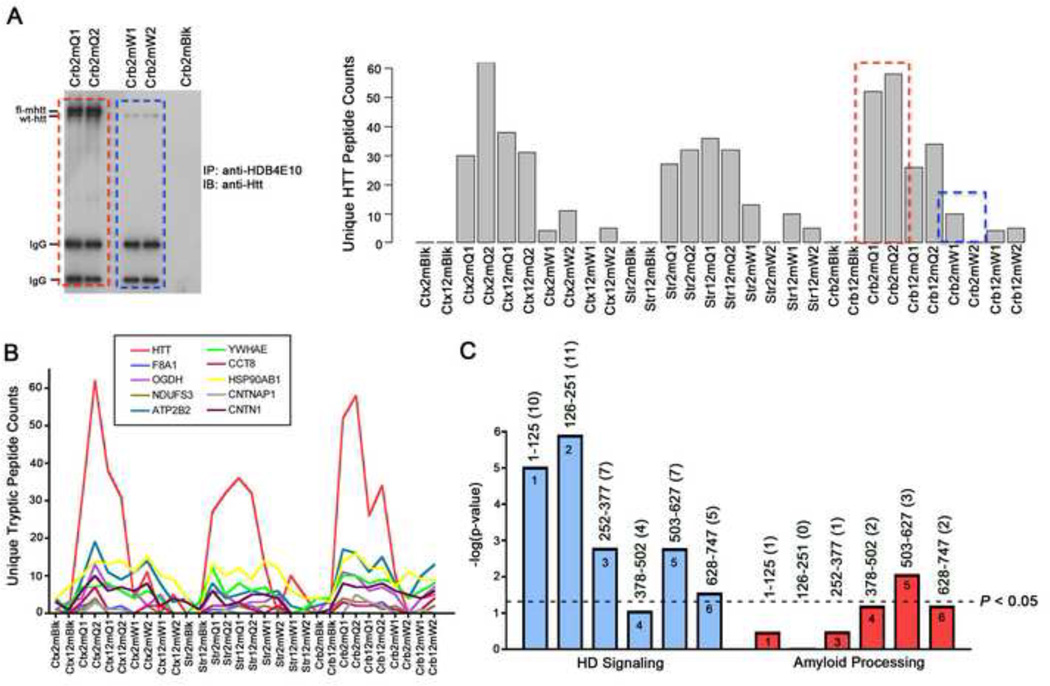
Although initial bioinformatics analyses indicated that our fl-Htt interactome was relevant to HD, we still needed to determine how to best prioritize the interacting proteins for biological validation. For this reason, we sought to explore whether the semi-quantitative MS information embedded within our dataset could be utilized to provide a system-level view of the interactome and enable a rationale prioritization of candidate interactors for functional studies. We performed a protein spiking experiment by adding increasing concentrations of bovine serum albumin (BSA) to our BACHD 2-month cortical extracts prior to LC-MS/MS (Supplemental Figure S1). Similar to a prior study (Liu et al., 2004), under our experimental conditions, the unique peptide counts for BSA were significantly correlated with the known BSA concentration in different samples (R2 = 0.822). To use such semi-quantitative information to uncover any novel relationships between candidate interactors and Htt, we took advantage of the observation that Htt peptide counts were consistently higher in BACHD samples than in WT control (or blank control) samples across all 30 IP experiments (Figure 3A). This is because there is approximately a two-fold increased level of fl-Htt in BACHD compared to WT control brains (Gray et al., 2008), and the selectivity of HDB4E10 for human Htt over murine Htt (Figure 3A). We reasoned that the broadly-expressed fl-Htt interacting proteins should have relative peptide abundances across our dataset similar to that of Htt (i.e., high in BACHD, low in WT, and none in the blank). We calculated pairwise correlations between fl-Htt and the 747 putative interacting proteins, using as input the unique tryptic peptide counts identified for each protein amongst the 30 sample conditions (Supplemental Tables S7 and S8). We found 163 proteins that are significantly correlated with Htt in our dataset (p < 0.05, Supplemental Table S8), and the top Htt-correlated proteins consisted of previously known Htt interactors such as F8a1 (Hap40), Ndufs3, Ywhae (14-3-3ε), Cct8, Hsp90ab1, and Cntn1 (Figure 3B; Supplemental Table S3). Importantly, F8a1/Hap40, a well-characterized fl-Htt interacting protein previously implicated in HD pathogenesis (Peters and Ross, 2001), was identified as being the most significantly correlated with Htt, despite having 3 or fewer peptides identified in any given sample (Supplemental Tables S7 and S8).
Figure 3. Unique peptide counts as a measure of relative protein abundance reveals proteins most correlated with Htt.
(A) The left panel is a Western blot analysis using anti-Htt (mAb2166) (left) of the anti-Htt immunoprecipitation (HDB4E10) of 2-month BACHD cerebellum biological duplicates (Crb2mQ1 and Crb2mQ2, red dashed box), 2-month WT cerebellum biological duplicates (Crb2mW1 and Crb2mW2, blue dashed box), and 2-month BACHD cerebellum no antibody control (Crb2mBlk). The right panel is a bar graph of unique tryptic Htt peptide counts identified across 30 experimental AP-MS conditions (right), which illustrate the reproducibility of the number of Htt peptides that is identified in duplicated sample conditions as well as the consistent variance in across sample conditions (i.e. age, brain region, genotype and with or without antibody in IP). (B) Unique peptide counts of the top ten proteins most correlated with Htt across the 30 sample conditions. (C) Htt correlation bins 1–6 (each consisting of 125 proteins, where bin 1 contains the proteins most correlated with Htt and bin 6 contains the proteins least correlated with Htt). IPA canonical pathway analysis of these bins showed their enrichment of ‘Huntington’s Disease Signaling’ (plotted as –log10 p-values values and the black dotted line indicates p < 0.05). The IPA ‘Amyloid Processing Pathway’, an Alzheimer’s disease related pathway, was used as a negative control. The number in parenthesis indicates the number of proteins identified in each category. See also Figures S1 and S2, and Supplemental Tables S7–S9.
To further increase the confidence in the relevancy of Htt-correlation ranking to HD biology, we divided the entire 747 candidate Htt-interacting protein pool into 6 bins of about 125 proteins, each based on ranking, and annotated each bin using IPA ‘Huntington’s Disease Signaling’ Pathway. Interestingly, the results showed a significant correlation between the fl-Htt correlation ranking and the IPA annotation of known Htt biology, with the top two bins being the most significantly enriched with ‘Huntington’s Disease Signaling’ proteins, while the bottom bins were only marginally or not significantly enriched at all (Figure 3C and Supplemental Table S9). As a control, the top bins of Htt-correlated proteins are not significantly enriched with an Alzheimer’s disease related IPA pathway (Figure 3C). This analysis provides independent support of the biological relevance of the Htt correlation ranking of the in vivo fl-Htt interactome.
Weighted Gene Correlation Network Analyses (WGCNA) Organizes the In Vivo fl-Htt Interactome into Network Modules
The construction of interacting protein networks based exclusively on AP-MS datasets has been largely limited to simple organisms (e.g., yeast). The use of AP-MS proteomic interactome information derived from complex biological systems to directly construct protein networks has been a challenge, but an important direction for the field (Vidal et al., 2011). WGCNA (Zhang and Horvath, 2005) was designed to provide a global analysis of microarray data across tissues, species, or disease conditions, and to build global gene networks based on co-expression relationships in order to identify gene modules that are tightly correlated across entire datasets (Miller et al., 2008; Oldham et al., 2008). However, to date, this method has never been applied to proteomics data.
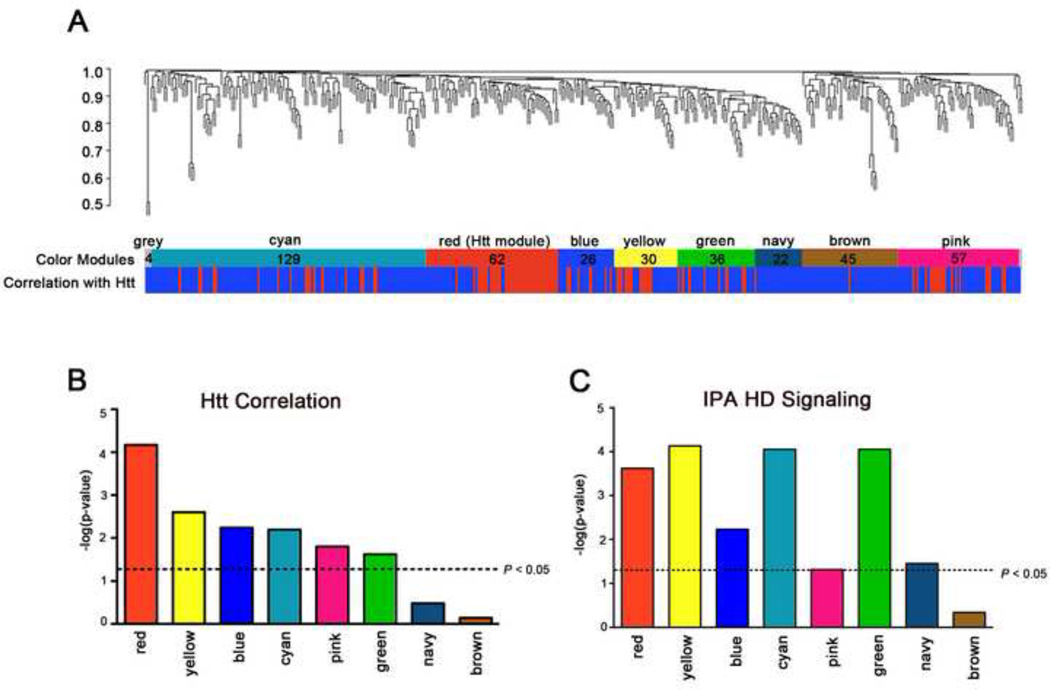
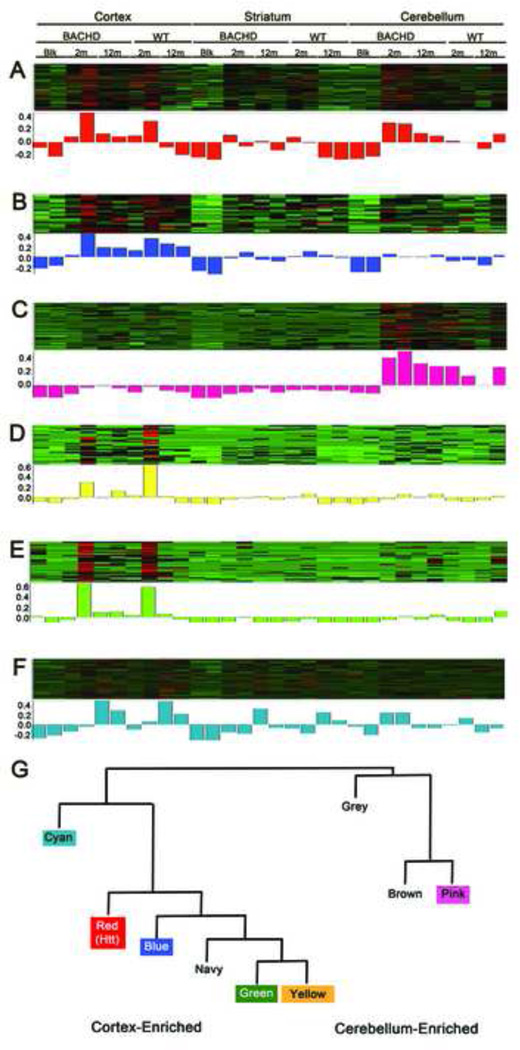
Because the semi-quantitative data provided by AP-MS provides a good proxy for relative protein abundance, we applied WGCNA to our proteomic dataset. We call this adapted application of the method to protein analysis, Weighted Gene Correlation Network Analyses (still abbreviated as WGCNA). Briefly, after selecting proteins present in at least three samples (n = 412), the pairwise correlation coefficients between one protein and every other detected protein were computed, weighted using a power function (Zhang and Horvath, 2005; Langfelder and Horvath, 2008), and used to determine the topological overlap, a measure of connection strength or ‘neighborhood sharing’ in the network. A pair of nodes in a network is said to have high topological overlap if they are both strongly connected to the same group of nodes. In WGCNA networks, genes with high topological overlap have been found to have an increased chance of being part of the same tissue, cell type, or biological pathway. Our analyses of the fl-Htt interactome produced 8 clusters of highly correlated proteins, or modules, with each including 22–145 proteins (Figure 4A, Supplemental Table S10). Based on the convention of WGCNA (ibid), the modules were named with different colors (Red, Yellow, Blue, Cyan, Pink, Green, Navy, Brown and Grey).
Figure 4. Weighted Gene Correlation Network Analysis (WGCNA) of Htt interactome.
(A) Cluster dendrogram generated by hierarchical clustering of proteins on the basis of topological overlap. Modules of correlated proteins were assigned colors and names (Cyan, Red, Blue, Yellow, Green, Navy, Brown, and Pink respectively) and are indicated by the horizontal bar beneath the dengrogram, where all unassigned proteins were placed in the Grey module (labeled Color Modules). The number in the color modules represents the number of proteins assigned to each module. Pairwise correlations were calculated for each protein to the Htt protein, and the p-value significance is colored below in the horizontal bar where a significant (p < 0.05, red) or non-significant correlation (p > 0.05, blue) with Htt protein levels is indicated. (B) Barplot depicting the –log10 p-values of the pairwise correlations between each module’s eigenprotein (see text) and Htt. Dotted black line: significance level at p < 0.05. (C) IPA canonical pathway analysis of each module to show their enrichment of ‘Huntington’s Disease Signaling’ (plotted as –log10 p-values values and the black dotted line indicates p < 0.05). See also Supplemental Tables S10 and S11.
To investigate the biological underpinning of the WGCNA modules, we addressed whether each module could have differential correlation strength with the central protein in our interactome, fl-Htt. We computed a Module Eigenprotein (MP) for each module, which is defined as the most representative protein member (i.e., a weighted summary) among all proteins in the module. We then calculated each MP correlation with fl-Htt (Figure 4B and Supplemental Table S11). The relationship between module membership (MM, defined as the correlation between each protein in the network and MP) and fl-Htt levels was determined (Supplemental Figure S2A–H). Both measures pointed to one module (Red) as the most correlated to fl-Htt across samples, with five other modules (Yellow, Blue, Cyan, Pink, and Green) also highly significantly correlated with fl-Htt. Importantly, the Red Module (comprised of 62 proteins, where 19 were previously known Htt interactors) includes Htt itself, thus giving further support that the proteins assigned to this module may have important biological relationships with Htt (Supplemental Table S12).
To further validate the Htt-correlated modules, we input the proteins in each module into IPA and analyzed for enrichment for ‘Huntington’s Disease Signaling Pathway’ proteins. The Red, Blue, Cyan, Yellow, Green, Pink, and Navy Modules were significantly enriched with proteins in this pathway (Figure 4C; Supplemental Table S13). The Brown Module was not significantly correlated with Htt, and is also not significantly enriched with IPA HD Signaling proteins (Figure 4C; Supplemental Table S13). Together, our analyses support the biological relevance to Htt of multiple WGCNA modules derived from our fl-Htt interactome.
Enrichment of Htt Interactome Modules and Meta-Networks in Specific Sample Conditions
We hypothesized that one of the underlying biological relationships driving the formation of different modules could be the differential enrichment of proteins within distinct AP-MS sample conditions (e.g., brain region, age, or genotype). To test this, we correlated the MPs for the six WGCNA modules to the 30 experimental conditions (Figure 5A–F). We found that Red Module is enriched in the cortical and cerebellar samples; The Blue, Yellow and Green Modules are enriched in the cortical samples; and Pink Module is enriched in the cerebellar samples. Interestingly, the Cyan Module appears to be an age-dependent module, with proteins consistently enriched in 12-month but not 2-month cortical samples in both BACHD and WT mice (Figure 5F).
Figure 5. Enrichment of Htt interactome modules and meta-networks in specific sample conditions.
(A–F) The heat map correlation profiles (rows) across samples (columns) for proteins in the following color modules are shown: A) Red B) Blue C) Pink D) Yellow E) Green F) Cyan where green indicates areas of low correlation profiles and red indicates areas of high correlation profiles. The 30 sample conditions are indicated by brain regions, genotype (BACHD vs WT), and age (2m, 12m) or control IP lacking Htt antibody (Blank or Blk). The two duplicated conditions (two separate columns) are labeled only once. (G) Module eigenprotein meta-network reveals the cortical-enriched sub-network and the cerebellum-enriched sub-network.
Finally, the unbiased process of constructing WGCNA network modules also yields a higher-order meta-network called “module eigenprotein network”, which can be calculated based on pairwise correlation relationships of all possible pairs of MPs (Figure 5G). The two main branches of the network appear to represent either modules that are enriched with proteins in cortical samples (Red, Cyan, Blue, Green, and Yellow) or those enriched in the cerebellar samples (Pink). These analyses suggest that the hierarchical organization of the fl-Htt interactome modules and their meta-networks may reflect the tightly correlated group of proteins that preferentially complex with Htt in distinct sample conditions (brain regions and age).
WGCNA Modules Are Related to Distinct Aspects of Htt Biology
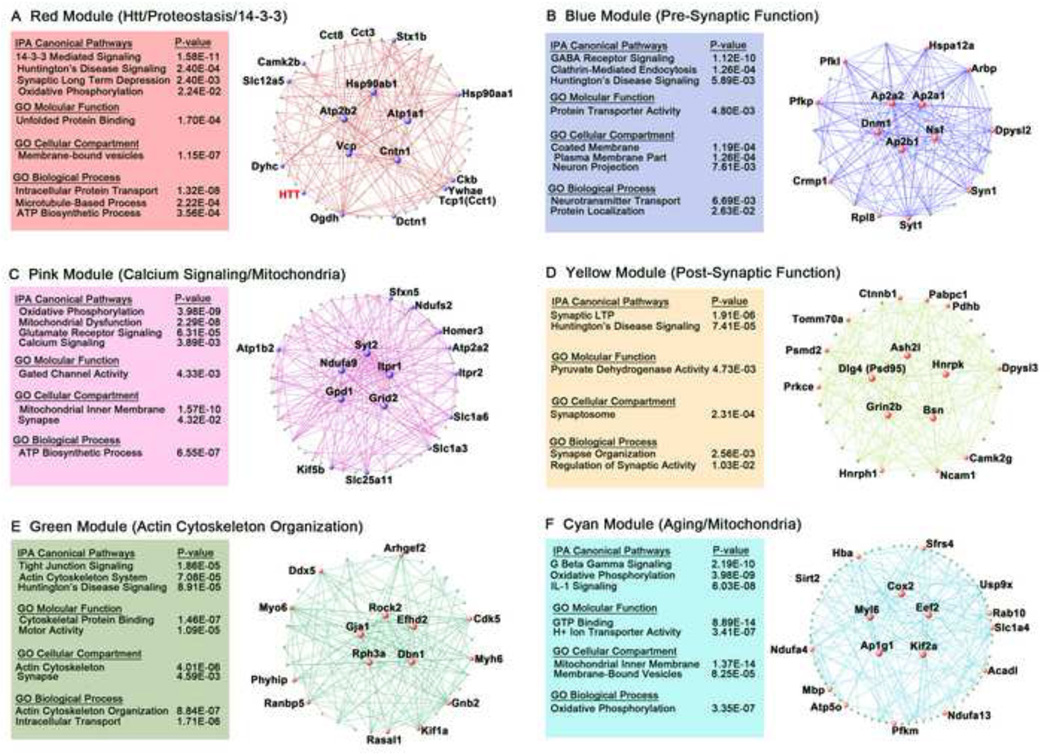
A key motivation for constructing an unbiased fl-Htt interactome network is to gain insights into different aspects of Htt molecular function in the intact mammalian brain. We analyzed the six Htt-correlated WGCNA modules using Gene Ontology and IPA (Supplemental Tables S13 and S14). HD-relevance and novel molecular characteristics of each module can be assessed based on their top module hub proteins, which are defined as the proteins with the highest correlation with each MP and can be ranked by the module connectivity values, kwithin (Figure 6 and Supplemental Table S10).
Figure 6. Functional annotation of top Htt-correlated modules.
(A–F) The Visant plots, IPA, and GO terms most enriched terms or signaling pathways in the top Htt-correlated modules are shown for: A) Red B) Blue C) Pink D) Yellow E) Green and F) Cyan Modules. Key characteristic describing each module is summarized in the parenthesis. Visant plots are shown representing the top 150 protein connections in each color module. The enlarged labeled nodes represent the top 15 proteins with the largest module connectivity values (kwithin) for each module, and the nodes in the middle of each Visant plot are the “hub genes” for each module. See also Supplemental Tables S10, S13, and S14
The Red Module, which is the most Htt-correlated module and contains Htt itself, is significantly enriched with hub proteins involved in unfolded protein binding (i.e., chaperones), 14-3-3 signaling, microtubule-based intracellular transport, and mitochondrial function (Figure 6A). Chaperones are key proteins involved in maintaining a healthy proteome (proteostasis) by preventing protein misfolding, a pathway directly implicated in the pathogenesis of neurodegenerative disorders including HD (Balch et al., 2008). Several chaperones in the Red Module (Tcp1, Hsp90s) have been shown to physically interact with Htt or modify mHtt toxicity in cell and invertebrate models of HD (Table 1). The roles of the others remain to be explored.
Table 1.
Summary of Top 50 Red Module Membership Proteins
| Protein | Module | Htt Correlation | MM.Red | IPA Function | IPA Subcellular Distribution | Interactors | Genetic Modifiers | IPA Drugs |
|---|---|---|---|---|---|---|---|---|
| Cntn1 | red | 1.000 | 0.911 | Cell adhesion | Membrane | Known | ||
| Atp2b2 | red | 1.000 | 0.907 | ATP biosynthesis | Membrane | This study | ||
| Vcp | red | 0.999 | 0.866 | DNA repair | Cytoplasm | f, w | ||
| Atp1a1 | red | 0.999 | 0.850 | ATP biosynthesis | Membrane | This study | f [This study] | Digoxin, Omeprazole |
| Hsp90ab1 | red | 1.000 | 0.848 | Molecular chaperone | Cytoplasm | f [This study] | 17-AAG | |
| Hsp90aa1 | red | 0.999 | 0.804 | Molecular chaperone | Cytoplasm | f | 17-AAG | |
| Ckb | red | 0.999 | 0.802 | Creatine metabolism | Cytoplasm | Creatine | ||
| Slc12a5 | red | 0.999 | 0.802 | Transporter | Membrane | |||
| Ogdh | red | 1.000 | 0.795 | Glycolysis | Cytoplasm | |||
| Dync1h1 | red | 0.999 | 0.774 | Microtubule movement | Cytoplasm | Known | f, w, m | |
| Stx1b | red | 0.999 | 0.771 | Neurotransmitter transport | Membrane | |||
| Dctn1 | red | 0.999 | 0.765 | Dynein retrograde movement | Cytoplasm | |||
| Ncam1 | yellow | 0.999 | 0.748 | Glycolysis | Membrane | BB-10901 | ||
| Camk2b | red | 0.999 | 0.746 | Kinase | Cytoplasm | f [This study] | ||
| Ywhae | red | 0.999 | 0.742 | Signal transduction | Cytoplasm | Known | f, f [This study] | |
| Cct8 | red | 0.999 | 0.733 | Molecular chaperone | Cytoplasm | Known | ||
| Tubb3 | red | 0.998 | 0.727 | Microtubule movement | Cytoplasm | Colchicine, Epothiolone B | ||
| Tubb4 | red | 0.998 | 0.726 | Microtubule movement | Cytoplasm | Colchicine, Epothiolone B | ||
| Vps35 | green | 0.999 | 0.723 | Transporter | Cytoplasm | This study | f [This study] | |
| Ndufs3 | red | 0.999 | 0.722 | Electron transport | Cytoplasm | Known | f [This study] | |
| HTT | red | 1.000 | 0.710 | Signaling, transport, etc. | Cytoplasm | |||
| Tcp1 | red | 0.997 | 0.707 | Molecular chaperone | Cytoplasm | Known | c, f, f [This study] | |
| Slc3a2 | red | 0.998 | 0.707 | Transporter | Membrane | |||
| Atp1b1 | yellow | 0.997 | 0.694 | ATP biosynthesis | Membrane | Known | f [This study] | |
| Ddx5 | green | 0.998 | 0.682 | mRNA processing | Nucleus | |||
| Tuba1b | red | 0.998 | 0.676 | Microtubule cytoskeleton | Cytoplasm | This study | ||
| Pdhb | yellow | 0.998 | 0.670 | Glycolysis | Cytoplasm | |||
| Atp2a2 | pink | 0.997 | 0.666 | ATP biosynthesis | Membrane | |||
| Pc | red | 0.997 | 0.664 | Gluconeogenesis | Cytoplasm | |||
| Nefm | yellow | 0.996 | 0.663 | Microtubule cytoskeleton | Cytoplasm | |||
| Sept3 | red | 0.998 | 0.662 | Cytokinesis | Cytoplasm | |||
| Slc25a3 | red | 0.998 | 0.657 | Transporter | Cytoplasm | Known | f [This study] | |
| Slc1a3 | pink | 0.998 | 0.657 | Transporter | Membrane | Riluzole | ||
| Ywhaz | red | 0.997 | 0.651 | Signal transduction | Cytoplasm | Known | c, f | |
| Rplp0 | blue | 0.998 | 0.650 | Protein synthesis | Cytoplasm | w | ||
| Nfasc | yellow | 0.998 | 0.650 | Cell adhesion | Membrane | |||
| Tomm70a | yellow | 0.996 | 0.648 | Transporter | Cytoplasm | |||
| Nsf | blue | 0.998 | 0.646 | Vesicle-mediated transport | Cytoplasm | w | ||
| Kif1a | green | 0.997 | 0.643 | Molecular movement | Cytoplasm | |||
| Syngr1 | red | 0.997 | 0.643 | Synaptic plasticity | Membrane | |||
| Rad23b | red | 0.998 | 0.632 | Nucleotide-excision repair | Nucleus | |||
| Ap1b1 | green | 0.997 | 0.630 | Protein transport | Cytoplasm | c, w | ||
| Cdk5 | green | 0.998 | 0.629 | Kinase | Nucleus | Flavopiridol | ||
| Hspa8 | green | 0.997 | 0.626 | Molecular chaperone | Cytoplasm | c, f, w | ||
| Ipo5 | green | 0.997 | 0.621 | GTPase inhibitor activity | Nucleus | f, w | ||
| Kif5c | pink | 0.998 | 0.621 | Molecular movement | Cytoplasm | |||
| Atp1a4 | red | 0.992 | 0.621 | ATP biosynthesis | Membrane | Digoxin, Omeprazole | ||
| Ppp2ca | yellow | 0.997 | 0.613 | Phosphatase | Cytoplasm | |||
| Gnao1 | cyan | 0.996 | 0.608 | Signal transduction | Membrane | Known | ||
| Stxbp1 | cyan | 0.997 | 0.607 | Exocytosis | Cytoplasm | Known | f |
Protein function, subcellular distribution, and drug annotation was based on Ingenuity Pathway Analysis (IPA). Protein interactors validated by co-IP/IB and fly genetic modifiers identified from ‘This Study’ are indicated. Previously identified Htt interactors are labeled as ‘Known’, and previously identified genetic modifiers (cells, c; fly, f, worm, w, and mouse, m) are indicated above. See Supplemental Material for complete list of references. See also Figure 7 and Supplemental Figure S4.
The second molecular feature of the Red Module is the presence of six 14-3-3 family proteins (Ywhab, Ywhae, Ywhag, Ywhah, Ywhaq, Ywhaz), with Ywhae as a top hub protein (Figure 6A). Impressively, ‘14-3-3-Mediated Signaling’ in the Red Module is the most significantly enriched IPA Canonical Pathway for all modules in the fl-Htt interactome network (Supplemental Table S13). The 14-3-3 pathway has been implicated in the pathogenesis of a variety of neurodegenerative disorders, and four 14-3-3 members have been shown to physically or genetically interact with Htt N-terminal fragments (Kaltenbach et al., 2007; Omi et al., 2008) (Supplemental Table S3). Since 14-3-3 proteins are phospho-serine/phospho-threonine binding proteins (Morrison, 2009), and Htt phosphorylation at several serine residues has been shown to modify HD pathogenesis (Humbert et al., 2002; Gu et al., 2009; Thompson et al., 2009), it could be a promising direction to investigate whether 14-3-3 proteins in the Red Module could directly interact with relevant phospho-Htt species to affect the disease process.
The third molecular pathway enriched in the Red Module is ‘Intracellular Protein Transport’ (Dynactin, Dynein, Vcp, and Ran) consistent with the convergent evidence supporting the role of Htt function in the microtubule-based transport process (Caviston and Holzbaur, 2009) and the disruption of such function in HD (Gauthier et al., 2004).
Although the Red Module appears to be enriched with proteins from divergent molecular processes, several lines of evidence suggest these proteins indeed have close biological connectivity. First, 26 out of the 61 Red Module proteins are included in the same IPA network, which is constructed based on the archived IPA Knowledge Base derived from published studies. This network has the highest IPA network score among all of the networks constructed from Htt interactome modules (Supplemental Table S13), suggesting that the proteins in Red Module already have a close functional link based on existing knowledge. Second, the Red Module has a marked enrichment for proteins implicated in other neurological and genetic disorders. Using another IPA core analysis (IPA Function), the Red Module has dramatically higher enrichment for proteins in the categories of Neurological Disorders and Genetic Disorders compared to the other modules (Supplemental Figure S3A–B), which cannot be accounted for by enrichment of the HD Signaling Pathway alone (Figure 4C). Furthermore, 16 Red Module proteins (Supplemental Figure S3C–D) are mutated in neurological disorders ranging from Frontotemporal dementia (Vcp) to Parkinson’s disease (Vps35). Finally, among the top 50 Red Module proteins based on module memberships, there are 19 proteins that are known to interact with Htt that can modify mHtt toxicity in cell or invertebrate models of HD (Table 1), and several genes are also known therapeutic targets for HD (e.g., creatine targeting Ckb; and 17-AAG targeting Hsp90s) (Herbst and Wanker, 2007; Dorsey and Shoulson, 2012). In summary, the Red Module contains proteins that are highly correlated with Htt (including Htt itself), and is enriched in a highly connected group of proteins involved in proteostasis, 14-3-3 signaling, microtubule-based transport and mitochondria function.
The second most Htt-correlated module is the Blue Module, with its member proteins enriched in the cortex and playing roles in presynaptic function. The most significant GO terms enriched in Blue Module are ‘Coated Membrane’ and ‘Neurotransmitter Transport’ (Supplemental Table S14). The top IPA Canonical Signaling Pathways enriched in Blue Module are ‘GABA Receptor Signaling’, ‘Clathrin-Mediated Endocytosis’ and ‘Huntington’s Disease Signaling’. The hub proteins in Blue Module (Ap2a2, Dnm1, and Syt1) are members of a Htt protein network previously established based on ex vivo interactions with mHtt fragments and are validated as genetic modifiers in an HD fly model (Kaltenbach et al., 2007). Together, this evidence supports the notion that Blue Module contains cortex-enriched Htt interactors that preferentially function in pre-synaptic terminals, and hence may influence corticostriatal neurotransmission that is known to be affected in HD (Raymond et al., 2011).
The Pink Module is a cerebellum-enriched module with HD-relevant hub proteins functioning in calcium signaling (Itpr1 and Itpr2), mitochondria function (Ndufa9, Ndufs2 and Uqcrc2), and glutamate receptor function (Grid2 and Slc1a3). Not surprisingly, several hub proteins are either selectively expressed (Grid2 and Slc1a3) or highly enriched in the cerebellum (Itpr1, Syt2 and Gpd1; see Allen Brain Atlas). Consistent with the idea that cerebellar-enriched Htt interactors may confer beneficial neuroprotective function, one interesting Pink Module protein, Ucqrc2, was shown to be one of nine core modulators of the proteostasis network (e.g., mHtt polyQ fragment and endogenous metastable proteins) in a genome-wide C. elegans screen (Silva et al., 2011). Since our interactome also identified more Ucqrc2 peptides in brain tissues (cerebellum) and at ages (2m) relatively unaffected in HD mice (Supplemental Table S7), this evidence strongly encourages further investigation in the role of Ucqrc2 and its interaction with Htt in HD selective pathogenesis.
The Yellow Module is driven by top hub proteins involved in excitatory post-synaptic function (Supplemental Table 14). Two of the top hub proteins (Grin2b/NR2b and Dlg4/PSD95) have been implicated in pathogenesis in HD mice (Zeron et al., 2002; Fan et al., 2009). Another interesting member, beta-catenin (Ctnnb1), is a known modifier of mHtt-induced toxicity in HD cell and fly models (Godin et al., 2010; Dupont et al, 2012). Ctnnb1 is also a key member of the Wnt signaling pathway, which has been implicated in multiple neurodegenerative disorders (Wexler et al., 2011) and is being explored for neuroprotective therapies (Toledo et al., 2008). Thus, further study of Yellow Module hub proteins may yield new therapeutic targets for mitigating post-synaptic dysfunction in HD.
The Green Module is highly enriched with proteins involved in actin cytoskeleton organization (Figure 6E). The annotation of this module is consistent with the emerging role of Htt as a direct actin binding protein (Angeli et al., 2010), with an evolutionarily-ancient function in regulating the actin-binding protein, myosin, in chemotaxis and cytokinesia (Wang et al., 2011). Green Module hub proteins, including Cdk5 (Anne et al., 2007), Rph3A (Smith et al., 2007), Rock2 (Shao et al., 2008), and Gja1/Connexin-43 (Vis et al., 1998), have previously been implicated in HD. Therefore, this module provides a novel set of actin binding candidates impetus to study in the context of normal Htt function and HD pathogenesis.
Finally, the Cyan Module is the only age-dependent module and is also highly enriched with proteins residing in mitochondria, or functioning in inflammation, G-protein signaling and as modifiers of mHtt aggregation. Several of these proteins (Usp9x, Rock1, and Sirt2) appear to modify mHtt aggregation or toxicity in cellular models and are known drug targets (Kaltenbach et al., 2007; Shao et al., 2008; Pallos et al., 2008; Luthi-Carter et al., 2010). The role of Cyan Module proteins in influencing mHtt aggregation gained further support after a genome-wide RNA interference screen for modifiers of mHtt aggregation identified a number of similar genes, including Aldoa, Csnk2a1, Hspa9, Pfkm, and Rab1a in Drosophila cells (Zhang et al, 2010). Thus, the Cyan Module is enriched with proteins complexed with Htt in an age-dependent manner, which may help to probe the poorly understood role of aging in the pathogenesis of HD.
Experimental Validation of Red Module Proteins as Huntingtin Interactors and Genetic Modifiers of Functional Deficit in a Fly Model of HD
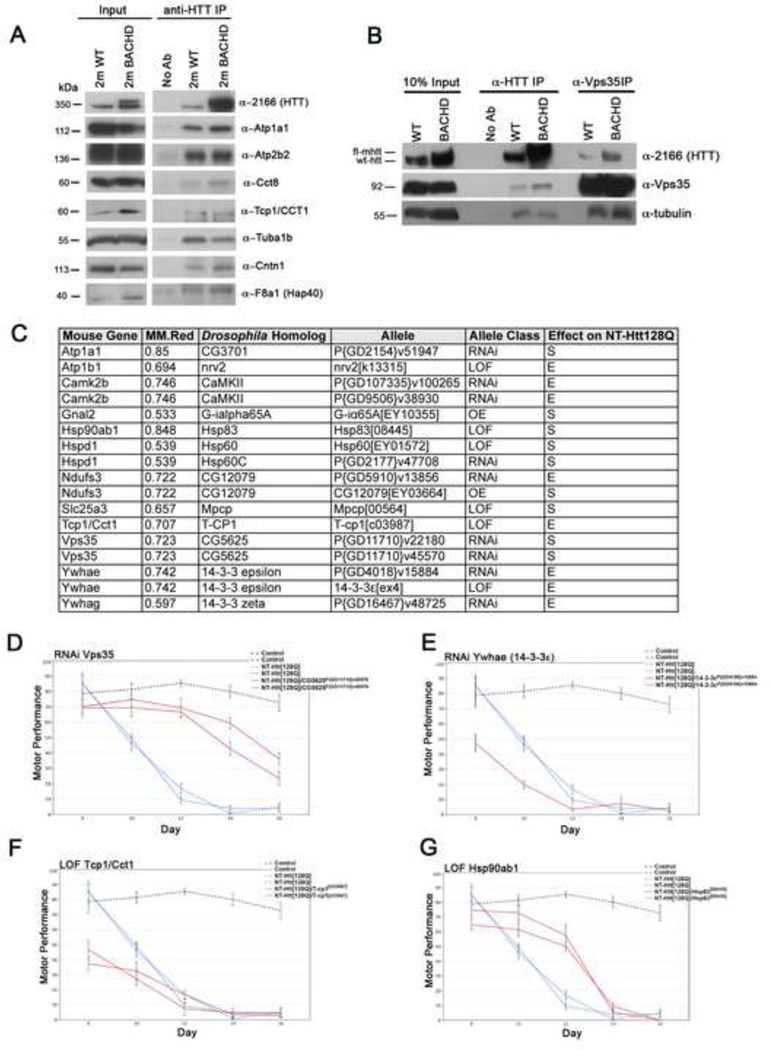
We next sought to validate those proteins not previously implicated in HD, but with a high Red Module connectivity (hub proteins) as novel Htt physical interactors and/or genetic modifiers (Table 1). We performed an anti-Htt co-IP from BACHD and WT mouse brains followed by Western blot analysis for the specific candidate proteins to confirm interaction (Figure 7A). We confirmed six novel proteins (Atp1a1, Atp2b2, Cct8, Cct1, Tuba1b, Cntn1) and one positive control Red Module protein (Hap40/F8A1) as co-immunoprecipitating with Htt. Furthermore, Vps35, a protein in the retromer complex that functions in the retrieval of certain membrane proteins from the endosome to the plasma membrane (Attar and Cullen, 2010), was found in both the IP of Htt in both the BACHD and WT mouse brain extracts (Figure 7B). Moreover, Htt is also present in the reciprocal co-IP of Vps35 from these brain extracts (Figure 7B). Thus, the top Red Module proteins are indeed complexed with Htt in the mouse brain.
Figure 7. Validation of Red Module proteins in Htt interactome uncovers Htt-interacting proteins in WT and BACHD mice and novel genetic modifiers of disease in a fly model of HD.
(A) Co-immunoprecipitation of top MM.Red proteins from 2-month WT and BACHD mouse brains. Htt was immunoprecipitated with anti-Htt (clone HDB4E10) and probed with mouse monoclonal antibody MAB2166. The 10% input for each protein (left panels) and resulting immunoprecipitations are shown (right panels). A no Htt antibody negative control was performed in parallel to confirm specific protein interaction. F8a1 (Hap40) served as the positive control for Htt protein interaction. (B) Reciprocal co-immunoprecipitation of Htt and Vps35 in 2-month WT and BACHD mouse brains. Anti-alpha tubulin was used as a loading control. (C) Table summarizing the effects caused by modulating the levels of the indicated Red Module proteins on NT-Htt[128Q]-induced motor performance in a HD Drosophila model. (D–G) Charts represent motor performance as a function of age. Each chart includes controls (elav-GAL4) and animals expressing NT-Htt[128Q] either alone or together with a loss of function allele targeting the Drosophila homolog of the indicated gene (red continuous lines). Control flies show robust motor performance for the duration of the experiment (black dashed line). Animals expressing NT-Htt[128Q] in the nervous system using elav-GAL4 show a progressive decline in their motor performance (blue discontinuous lines). D and G show the suppressor effect of decreasing the levels of the Drosophila homologs of Vps35 and Hsp90ab1. E and F show the enhancer effect of decreasing the levels of the Drosophila homologs of Ywhae and Tcp1/Cct1. Error bars represent standard error of the mean (SEM). Two experimental replicates are shown for the NT-Htt128Q and the NT-Htt128Q/modifier animals except for G. See also Figure S4.
To test the hypothesis that Red Module proteins are also modifiers of mHtt toxicity in vivo, we performed a genetic modifier study using an established Drosophila model of HD expressing a human Htt fragment (amino acids 1–336) with an expanded polyQ repeat of 128 glutamines (NT-Htt[128Q]) (Kaltenbach et al., 2007). Directed expression of NT-Htt[128Q] to all neurons of the CNS results in a robust and progressive motor deficit that can be quantified in a climbing assay. We used this behavioral assay to test 32 Red Module genes for which there were available mutants in the corresponding Drosophila ortholog genes, and we were able to validate 12 Red Module hub proteins as novel modifiers of neuronal dysfunction (Figure 7C–7G; Supplemental Figure S4A–J). Among the genetic enhancers of the HD motor deficits are Atp1b1, Camk2b, Ndufs3, Tcp1/Cct1, Ywhae, and Ywhag. The genetic suppressors are Atp1a1, Gnai2, Hsp90ab1, Hspd1, Ndufs3, Vps35, and Slc25a3. Interestingly, Ndufs3 is both a suppressor when over-expressed, and an enhancer by partial loss of function demonstrating dosage sensitive modulation of mHtt-induced motor deficits. In summary, our validation studies confirmed six Red Module proteins as novel Htt-complexed proteins in vivo and 12 Red Module proteins as novel genetic modifiers in HD fly. By integrating our validation studies with the existing HD literature, we found a total of 25 out of the top 50 Red Module proteins (based on MMred) to physically or genetically capable of interacting with Htt in various HD model systems (Table 1), lending further support that the Red Module is a central Htt in vivo protein network, mediating critical aspects of normal Htt function and HD pathogenesis in the brain.
DISCUSSION
We have used a spatiotemporal AP-MS approach to obtain the first compendium of in vivo full-length Htt interacting proteins in the mammalian brain, with the identification of 747 candidate proteins that complex with fl-Htt in vivo, creating one of the largest in vivo proteomic interactome datasets to date and directly validating more than 100 previously identified ex vivo interactors shown to associate with small N-terminal Htt fragments. We have also provided information on the context (age or brain regions) in which these proteins associate with fl-Htt. Moreover, we were able unbiasedly rank the interacting proteins, based on their correlation strength with Htt, and to construct a WGCNA network that describes this interactome. Proteins in several WGCNA network modules are highly correlated with Htt itself, and appear to reflect distinct biological contexts in their interactions with Htt. Finally, we were able to validate 18 Red Module proteins as in vivo physical interactors or genetic modifiers in an HD fly model. In summary, our study provides the first comprehensive survey of proteins complexed with fl-Htt in distinct brain regions at different ages, and demonstrates a conceptually novel but simple approach to building unbiased in vivo interactomes based on AP-MS data from tissues as complex as the mammalian brain.
This study highlights core HD-relevant molecules and pathways via integration of our spatiotemporal fl-Htt interactome with previously generated Htt ex vivo interactome and cell- or invertebrate-based genetic modifier datasets, many of which are archived in IPA ‘Huntington’s Disease Signaling’ pathway. We show that 139 previously identified ex vivo Htt interactors (e.g., Y2H) also complex with fl-Htt in the mammalian brain (Goehler et al., 2004; Kaltenbach et al., 2007). Moreover, comparison of our interactome with datasets derived from genetic modifier screens in Drosophila and C. elegans models of HD may also help identify proteins that can interact with mHtt in the mammalian brain and possibly modify its toxicity, and could be prioritized for further validation in mammalian models of HD. For example, comparison of our dataset with data obtained from genetic screens in yeast, C. elegans, and fly models of HD (Nollen et al., 2004; Wang et al., 2009; Zhang et al., 2010; Silva et al., 2011) reveal several Red Module (CCTs, Hsp90s, 14-3-3s, and Vcp; Table 1) and Pink Module (Uqcrc2) proteins in common, which could possibly represent evolutionarily conserved modifiers of mHtt-induced toxicity. Therefore, their disease-modifying role should be fully explored in HD mammalian models.
A key motivation for this work was to obtain an unbiased global view of complex biological function or disease processes related to fl-Htt protein in the intact brain, and to formulate novel, testable hypotheses. The first crucial insight obtained from our WGCNA analyses is that distinct Htt-correlated modules represent proteins preferentially complexed with Htt in specific sample conditions that reflect key biological processes: cortical Htt IP samples (Red, Blue, Yellow and Green Modules), cerebellar Htt IP samples (Pink Module), and 12-month but not 2-month cortical samples (Cyan Module). The second important insight is that each of the six significant modules provides critical aspects of known Htt and HD biology. All are significantly enriched with ‘Huntington’s Disease Signaling’ in IPA (Figure 6C). Knowing the architecture and Htt-relevance of each network module can seed novel hypotheses based on important hubs and/or molecular or pathogenic processes defined by the module, e.g., Cntn1 and Vcp in Red Module mediating mHtt toxicity; Rad23b in Red Module implicating specific DNA repair and ubiquitin/proteasome pathway in Htt biology; Sirt2, Cox2 and Usp9x in the Cyan Module in age-dependent pathogenesis; and Itpr1 and Grid2 in cerebellar neuroprotection in HD. Testing such hypotheses will constitute a crucial next step towards unraveling the complex biology of Htt in healthy and diseased brains, but also further deciphering the biological significance of the in vivo Htt protein network.
One potentially exciting area of data integration would be to combine proteomic interactome data with large-scale genetic modifier/gene expression profiling studies from HD patients. The increasing capacity of DNA sequencing provides an unprecedented opportunity for such large-scale studies using patient samples, and our fl-Htt brain interactome may provide converging information on candidates that may have both a genetic and proteomic link to Htt. Thus, our study lends strong support to a systems biology strategy of vertically integrating large genetic, genomic, and interactome datasets (Geschwind and Konopka, 2009) derived from HD models of different organismal complexity to unravel the conserved mechanism related to Htt biology and HD pathogenesis.
Our study supports the view that the majority of Htt interacting proteins are relatively stable across brain tissue and age (Figure 2A–B), while a portion of the Htt interactome is quite dynamic. The latter group of proteins, particularly those that consistently complex with mHtt in a brain-regional-specific or age-specific manner (Figure 2C–D), could be interesting candidates to study for their contribution to selective neuronal or regional vulnerability and age-dependent pathogenesis in HD. Our studies also raised the intriguing possibility that age-dependent changes in the normal brain proteome (e.g. Sirt2) may alter Htt interactions, which could in turn contribute to the presently unexplained role of aging in disease pathogenesis (Maxwell et al., 2011).
A major advance in this study is the use of a systems biology approach to construct in vivo protein interaction networks exclusively using proteomic interactome datasets generated from complex tissue, such as the mammalian brain. We applied, for the first time, WGCNA to analyze all the peptide count information for an entire group of Htt complexed proteins in our spatiotemporal AP-MS dataset. WGCNA provides an unbiased systems level organization of gene expression modules in both normal and diseased brains (Voineagu et al., 2011) and has been demonstrated to be among the most powerful methods for global network construction (Allen et al., 2012). Several lines of evidence support the validity and value of WGCNA analyses of our in vivo Htt interactome dataset. We were able to show that the pairwise correlation measure leads to a meaningful ranking of Htt-related proteins with respect to the external annotated knowledge of HD related proteins (Huntington’s Disease Signaling in IPA; Figure 3C). WGCNA identified six significant Htt-correlated modules with distinct tissue- or age-specific over-representation, and significant enrichment of distinct biological function previously implicated in Htt biology (Figure 6), effectively providing an in silico dissection of the molecular processes related to fl-Htt biology.
Several experimental factors were instrumental to the construction of WGCNA networks based on our AP-MS dataset. First, the relative level of bait protein (fl-Htt) brought down by IP is markedly, but reproducibly, variable across all samples. Such variation is due to our experimental design of using samples from distinct brain regions, ages, genotypes, and no-antibody controls, and also from the use of an anti-Htt antibody that preferentially binds to human over murine Htt. The quantitative difference in the amount of Htt precipitated in each sample results in a similar quantitative variation for those proteins that were tightly associated with Htt (i.e, highly correlated with Htt), while background proteins (false positives) in the sample are less likely to vary in a similar manner as Htt. Hence, rather than being weakened by experimental variance, WGCNA was able to extract the quantitative correlation relationships among the proteins identified in our study. The second important factor for WGCNA analyses was the large-scale and multi-dimensional nature (e.g., brain region, age, and genotype) of our study. We estimated that one would need at least 24 independent AP-MS experiments (at least one biological replicates per sample condition), with systematic changes in the sample conditions to create differential pull-down of the bait protein and its complexes, in order to construct a robust WGCNA protein interaction network. One caveat of the current study is our use of MS unique tryptic peptide counts as a semi-quantitative readout of relative protein abundance. Such limitation could have been resolved by using stable isotope labeling in intact animals for a quantitative AP-MS study (Krüger et al., 2008).
Finally, our analysis provides a central molecular network, the Red Module, which is likely to contain proteins crucial to Htt biology and may constitute novel molecular targets to study for HD pathogenesis and therapeutics. The Red Module has Htt as its member, and is highly enriched with Htt interactors and genetic modifiers (Table 1). We were able to validate 6 Red Module proteins as novel in vivo Htt interactors by co-IP (Figure 7) and 12 as novel modifiers of Htt-induced neuronal dysfunction in a fly model (Figure 7; Supplemental Figure S4A–J). Moreover, Red Module proteins are targets for small molecules that are in HD clinical trials (i.e., creatine targeting Ckb; Hersch et al., 2006), or show effectiveness in preclinical studies in HD or other polyglutamine disorders in mice (Waza et al., 2005; Masuda et al., 2008). Considering several other proteins in this module can also be targeted by small molecules (Table 1), it would be interesting to explore whether pharmacological targeting of these proteins could be therapeutic in HD preclinical models.
In conclusion, we have constructed the first compendium of in vivo fl-Htt interacting proteins in distinct brain regions and ages, thereby providing a valuable resource for further exploration of the normal function of Htt in several disease-relevant biological context, and for identification of novel molecular targets critical to selective pathogenesis for HD and to develop new therapy. Moreover, we have demonstrated an innovative approach utilizing WGCNA to analyze multidimensional AP-MS datasets to produce in vivo protein networks for a given bait protein, and validated key proteins in the network using a HD fly model. This powerful application of systems biology to proteomics can be readily applied to decipher in vivo protein networks for other complex biological systems or disease processes in tissues as complex as the mammalian brain.
EXPERIMENTAL PROCEDURES
Please see Supplemental Data for complete details of experimental procedures.
Mouse Breeding, Maintenance, and Genotyping
BACHD mice were bred, maintained in the FvB/NJ background, and genotyped as previously described (Gray et al., 2008). BACHD mice were maintained under standard conditions consistent with National Institutes of Health guidelines and approved by the University of California, Los Angeles, Institutional Animal Care and Use Committees.
Brain Tissue Lysate Preparation and Anti-Htt Immunoprecipitation
Protein was prepared as previously described (Gu et al., 2009). Briefly, BACHD and WT mouse brains were dissected in ice-cold 100 mM PBS and homogenized in modified RIPA buffer supplemented with Complete Protease Inhibitor Mixture tablets (Roche) using 10 strokes from a Potter-Elvehjem homogenizer followed by centrifugation at 4 °C for 15 min at 16,000g. The resultant supernatant is the soluble fraction and protein concentrations were determined using the Bio-Rad Protein Assay (Bio-Rad). Brain lysates (2.5-mg) were subjected to immunoprecipitation with anti-huntingtin clone HDB4E10 (MCA2050, AbD Serotec, 1:500) using Protein G Dynabeads (Invitrogen). Immunoprecipitated proteins (500-ug) were washed, eluted with NuPAGE LDS loading buffer and subjected to Western blot analysis.
In-gel Trypsin Digestion
Immunoprecipitated protein samples were separated on NuPAGE 3–8% Tris-Acetate gels (Invitrogen), stained using GelCode Blue stain reagent (Pierce), destained in ddH2O, and then cut into approximately 24–27 gel slices. The gel slices were washed 3× in alternating solutions of a 50:50 mix of 100mM NaHCO3 buffer/CH3CN and 100% CH3CN. Disulfide bonds were reduced by incubation in 10mM dithiothreitol (DTT) at 60°C for 1 hour. Free sulfhydryl bonds were blocked by incubating in 50mM iodoactamide at 45 °C for 45 minutes in the dark, followed by washing 3× in alternating solutions of 100mM NaHCO3 and CH3CN. The slices were dried, and then incubated in a 20-ng/µL solution of porcine trypsin (Promega) for 45 minutes at 4 °C, followed by incubation at 37 °C for 4 to 6 hours. Afterwards, the supernatant was transferred into a fresh collection tube. The gels were incubated for 10 minutes in a solution of 50% CH3CN/1% trifluoroacetic acid (TFA), in which the supernatant was removed and combined with the previously removed supernatants. This step was repeated a total of 3 times. The supernatant samples containing the peptides were then spun to dryness and prepared for LC-MS/MS analysis by resuspension in 10-µL of 0.1% formic acid.
Mass Spectrometry Analysis
Peptide sequencing was accomplished by nanoLC-MS/MS with a QqTOF-MS (QSTAR Pulsar XL, Applied Biosystems) equipped with nanoelectrospray interface (Protana, Odense, Denmark) and LC Packings (Sunnyvale, CA) nano-LC system. The nano-LC was equipped with homemade precolumn (150 mm × 5 mm) and analytical column (75 mm × 150 mm) packed with Jupiter Proteo C12 resin (particle size 4 mm, Phenomenex, Torrance, CA). The dried peptides were resuspended in 1% formic acid (FA) solution. Six mL of sample solution was loaded to the precolumn for each LC-MS/MS run. The precolumn was washed with the loading solvent (0.1% FA) for 4 min before the sample was injected onto the LC column. The eluents used for the LC were 0.1% FA (solvent A) and 95% CAN containing 0.1% FA (solvent B). The flow rate was 200 nL/min, and the following gradient was used: 3% B to 35% B in 72 min, 35% B to 80% B in 18 min, and maintained at 80% B for 9 min. The column was finally equilibrated with 3% B for 15 min prior to the next run. Electrospray ionization was performed using a 30 mm (i.d.) nano-bore stainless steel online emitter (Proxeon, Odense, Denmark) and a voltage set at 1900 V. Sequences were searched against Swiss-Prot mouse and mammalian genomes using MASCOT software versions 2.1.0 and 2.1.04 (Matrix Science, London, UK). Peptides were required to have a rank =1 and a score >18.
Weighted Gene Correlation Network Analysis and Module Detection
We constructed a weighted correlation network as previously described using the R software package (Langfelder and Horvath, 2008). We used proteins with unique tryptic peptide counts coming from at least 3 IP conditions as an input (n = 412). See Supplemental Material for complete WGCNA procedure.
Drosophila Motor Performance Assay
Using a population of 15 age-matched (+/− 4 hours) virgin female flies, we estimated the percentage of animals able to climb past a line set at 9 cm in 15 sec. These tests were repeated 10 consecutive times for each replicate per experimental day. The experiment was carried out in duplicate (2 populations of 15 animals) in flies that were 8, 10, 12, 14, and 16 days old. Tests were always performed in the same time of the day and in the same place, to avoid circadian rhythm and environmental variability. The average number of flies climbing per day is averaged and plotted independently for each replicate.
Statistical Analysis
All values are presented as the mean ± SEM, and p < 0.05 was considered statistically significant.
Supplementary Material
ACKNOWLEDGMENTS
X.W.Y. is supported by grants from NINDS/NIH grants (R01NS049501; and R01NS074312), CHDI Foundation, the Hereditary Disease Foundation (HDF), David Weil Fund to the Semel Institute at University of California, Los Angeles, and Neuroscience of Brain Disorders Award from The McKnight Endowment Fund for Neuroscience. D.S. was supported by the NIH Chemistry-Biology Interface Research Training Program at UCLA (T32GM008496). D.S. and E.G., were supported by the UCLA Dissertation Year Fellowship Program. J.A.L. is supported by an NIH grant (R01RR20004), and by the W. M. Keck Foundation for the establishment of the UCLA Functional Proteomics Center. S.H. is supported by the NIH grant (1R01DA030913-01). UCLA Informatics Center for Neurogenomics and Neurogenetics (NINDS P30NS062691) provided bioinformatics analyses. We would like to thank advice and help from Dr. Rachel Ogorzalek Loo and helpful discussion from members and Yang and Loo labs.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Allen JD, Xie Y, Chen M, Girard L, Xiao G. Comparing statistical methods for constructing large scale gene networks. PLoS ONE. 2012;7:e29348. doi: 10.1371/journal.pone.0029348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angeli S, Shao J, Diamond MI. F-actin binding regions on the androgen receptor and huntingtin increase aggregation and alter aggregate characteristics. PLoS ONE. 2010;5:e9053. doi: 10.1371/journal.pone.0009053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anne SL, Saudou F, Humbert S. Phosphorylation of huntingtin by cyclin-dependent kinase 5 is induced by DNA damage and regulates wild-type and mutant huntingtin toxicity in neurons. JNeurosci. 2007;27:7318–7328. doi: 10.1523/JNEUROSCI.1831-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attar N, Cullen PJ. The retromer complex. Adv. Enzyme Regul. 2010;50:216–236. doi: 10.1016/j.advenzreg.2009.10.002. [DOI] [PubMed] [Google Scholar]
- Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- Bates G. Bates: Huntington’s Disease, Ed 3 (Bates GP, Harper…- Google Scholar. 2002. [Google Scholar]
- Caviston JP, Holzbaur ELF. Huntingtin as an essential integrator of intracellular vesicular trafficking. Trends Cell Biol. 2009;19:147–155. doi: 10.1016/j.tcb.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HK, Fernandez-Funez P, Acevedo SF, Lam YC, Kaytor MD, Fernandez MH, Aitken A, Skoulakis EM, Orr HT, Botas J, et al. Interaction of Akt-phosphorylated ataxin-1 with 14-3-3 mediates neurodegeneration in spinocerebellar ataxia type 1. Cell. 2003;113:457–468. doi: 10.1016/s0092-8674(03)00349-0. [DOI] [PubMed] [Google Scholar]
- Dorsey RE, Shoulson I. Neurobiology of Huntington's Disease: Applications to Drug Discovery. Boca Raton, FL: CRC Press; 2012. [PubMed] [Google Scholar]
- Dupont P, Besson MT, Devaux J, Liévens JC. Reducing canonical Wingless/Wnt signaling pathway confers protection against mutant Huntingtin toxicity in Drosophila. Neurobiol Dis. 2012 doi: 10.1016/j.nbd.2012.04.007. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Emamian E, Kaytor M, Duvick L, Zu T, Tousey S, Zoghbi H, Clark H, Orr H. Serine 776 of ataxin-1 is critical for polyglutamine-induced disease in SCA1 transgenic mice. Neuron. 2003;38:375–387. doi: 10.1016/s0896-6273(03)00258-7. [DOI] [PubMed] [Google Scholar]
- Fan J, Cowan CM, Zhang LYJ, Hayden MR, Raymond LA. Interaction of postsynaptic density protein-95 with NMDA receptors influences excitotoxicity in the yeast artificial chromosome mouse model of Huntington's disease. J. Neurosci. 2009;29:10928–10938. doi: 10.1523/JNEUROSCI.2491-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández E, Collins MO, Uren RT, Kopanitsa MV, Komiyama NH, Croning MDR, Zografos L, Armstrong JD, Choudhary JS, Grant SGN. Targeted tandem affinity purification of PSD-95 recovers core postsynaptic complexes and schizophrenia susceptibility proteins. Mol. Syst. Biol. 2009;5:269. doi: 10.1038/msb.2009.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier LR, Charrin BC, Borrell-Pagès M, Dompierre JP, Rangone H, Cordelières FP, De Mey J, MacDonald ME, Lessmann V, Humbert S, et al. Huntingtin controls neurotrophic support and survival of neurons by enhancing BDNF vesicular transport along microtubules. Cell. 2004;118:127–138. doi: 10.1016/j.cell.2004.06.018. [DOI] [PubMed] [Google Scholar]
- Godin JD, Poizat G, Hickey MA, Maschat F, Humbert S. Mutant huntingtin-impaired degradation of beta-catenin causes neurotoxicity in Huntington's disease. Embo J. 2010;29:2433–2445. doi: 10.1038/emboj.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goehler H, Lalowski M, Stelzl U, Waelter S, Stroedicke M, Worm U, Droege A, Lindenberg KS, Knoblich M, Haenig C, et al. A protein interaction network links GIT1, an enhancer of huntingtin aggregation, to Huntington's disease. Mol Cell. 2004;15:853–865. doi: 10.1016/j.molcel.2004.09.016. [DOI] [PubMed] [Google Scholar]
- Gray M, Shirasaki DI, Cepeda C, Andre VM, Wilburn B, Lu XH, Tao J, Yamazaki I, Li SH, Sun YE, et al. Full-length human mutant huntingtin with a stable polyglutamine repeat can elicit progressive and selective neuropathogenesis in BACHD mice. J. Neurosci. 2008;28:6182–6195. doi: 10.1523/JNEUROSCI.0857-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Greiner E, Mishra R, Kodali R, Osmand A, Finkbeiner S, Steffan J, Tompson L, Wetzel R, Yang X. Serines 13 and 16 are critical determinants of full-length human mutant huntingtin induced disease pathogenesis in HD mice. Neuron. 2009;64:824–840. doi: 10.1016/j.neuron.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbst M, Wanker EE. Small molecule inducers of heat-shock response reduce polyQmediated huntingtin aggregation A possible therapeutic strategy. Neurodegener Dis. 2007;4:254–260. doi: 10.1159/000101849. [DOI] [PubMed] [Google Scholar]
- Hersch SM, Gevorkian S, Marder K, Moskowitz C, Feigin A, Cox M, Como P, Zimmerman C, Lin M, Zhang L, et al. Creatine in Huntington disease is safe, tolerable, bioavailable in brain and reduces serum 8OH2'dG. Neurology. 2006;66:250–252. doi: 10.1212/01.wnl.0000194318.74946.b6. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009a;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nature Protoc. 2009b;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Humbert S, Bryson EA, Cordelieres FP, Connors NC, Datta SR, Finkbeiner S, Greenberg ME, Saudou F. The IGF-1/Akt pathway is neuroprotective in Huntington's disease and involves Huntingtin phosphorylation by Akt. Dev Cell. 2002;2:831–837. doi: 10.1016/s1534-5807(02)00188-0. [DOI] [PubMed] [Google Scholar]
- Husi H, Ward MA, Choudhary JS, Blackstock WP, Grant SG. Proteomic analysis of NMDA receptor-adhesion protein signaling complexes. Nat. Neurosci. 2000;3:661–669. doi: 10.1038/76615. [DOI] [PubMed] [Google Scholar]
- Kaltenbach LS, Romero E, Becklin RR, Chettier R, Bell R, Phansalkar A, Strand A, Torcassi C, Savage J, Hurlburt A, et al. Huntingtin interacting proteins are genetic modifiers of neurodegeneration. PLoS Genet. 2007;3:e82. doi: 10.1371/journal.pgen.0030082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiman RJ, Kimmel LH, Bove SE, Lanz TA, Harms JF, Romegialli A, Miller KS, Willis A, Etages des S, Kuhn M, et al. Chronic suppression of phosphodiesterase 10A alters striatal expression of genes responsible for neurotransmitter synthesis, neurotransmission, and signaling pathways implicated in Huntington's disease. J. Pharmacol Exp. Ther. 2011;336:64–76. doi: 10.1124/jpet.110.173294. [DOI] [PubMed] [Google Scholar]
- Krüger M, Moser M, Ussar S, Thievessen I, Luber CA, Forner F, Schmidt S, Zanivan S, Fässler R, Mann M. SILAC mouse for quantitative proteomics uncovers kindling-3 as an essential factor for red blood cell function. Cell. 2008;134:353–364. doi: 10.1016/j.cell.2008.05.033. [DOI] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Li X-J. Multiple pathways contribute to the pathogenesis of Huntington disease. Mol Neurodegener. 2006;1:19. doi: 10.1186/1750-1326-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao L, McClatchy DB, Yates JR. Shotgun proteomics in neuroscience. Neuron. 2009;63:12–26. doi: 10.1016/j.neuron.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, Crespo-Barreto J, Jafar-Nejad P, Bowman AB, Richman R, Hill DE, Orr HT, Zoghbi HY. Opposing effects of polyglutamine expansion on native protein complexes contribute to SCA1. Nature. 2008;452:713–718. doi: 10.1038/nature06731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, Hao T, Shaw C, Patel AJ, Szabó G, Rual JF, Fisk CJ, Li N, Smolyar A, Hill DE, et al. A protein-protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell. 2006;125:801–814. doi: 10.1016/j.cell.2006.03.032. [DOI] [PubMed] [Google Scholar]
- Liu H, Sadygov RG, Yates JR. A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- Luthi-Carter R, Taylor DM, Pallos J, Lambert E, Amore A, Parker A, Moffitt H, Smith DL, Runne H, Gokce O, et al. SIRT2 inhibition achieves neuroprotection by decreasing sterol biosynthesis. Proc. Nat. Acad Sci. U.S. A. 2010;107:7927–7932. doi: 10.1073/pnas.1002924107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Major MB, Camp ND, Berndt JD, Yi X, Goldenberg SJ, Hubbert C, Biechele TL, Gingras AC, Zheng N, Maccoss MJ, et al. Wilms tumor suppressor WTX negatively regulates WNT/beta-catenin signaling. Science. 2007;316:1043–1046. doi: 10.1126/science/1141515. [DOI] [PubMed] [Google Scholar]
- Masuda N, Peng Q, Li Q, Jiang M, Liang Y, Wang X, Zhao M, Wang W, Ross CA, Duan W. Tiagabine is neuroprotective in the N171-82Q and R6/2 mouse models of Huntington's disease. Neurobiol Dis. 2008;30:293–302. doi: 10.1016/j.nbd.2008.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell MM, Tomkinson EM, Nobles J, Wizeman JW, Amore AM, Quinti L, Chopra V, Hersch SM, Kazantsev AG. The Sirtuin 2 microtubule deacetylase is an abundant neuronal protein that accumulates in the aging CNS. Hum Mol Genet. 2011;20:3986–3996. doi: 10.1093/hmg/ddr326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menalled L, El-Khodor B, M P, Suarez-Farinas M, Orenstein S, B Z, Leahy C, Wheeler V, Yang X, MacDonald M, et al. Systematic behavioral evaluation of Huntington's disease transgenic and knock-in mouse models. Neurobiology of Disease. 2009;35:319–336. doi: 10.1016/j.nbd.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JA, Oldham MC, Geschwind DH. A systems level analysis of transcriptional changes in Alzheimer's disease and normal aging. J. Neurosci. 2008;28:1410–1420. doi: 10.1523/JNEUROSCI.4098-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison DK. The 14-3-3 proteins: integrators of diverse signaling cues that impact cell fate and cancer development. Trends Cell Biol. 2009;19:16–23. doi: 10.1016/j.tcb.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nollen EAA, Garcia SM, van Haaften G, Kim S, Chavez A, Morimoto RI, Plasterk RHA. Genome-wide RNA interference screen identifies previously undescribed regulators of polyglutamine aggregation. Proc. Natl. Acad. Sci. U.S.A. 2004;101:6403–6408. doi: 10.1073/pnas.0307697101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham MC, Konopka G, Iwamoto K, Langfelder P, Kato T, Horvath S, Geschwind DH. Functional organization of the transcriptome in human brain. Nat. Neurosci. 2008;11:1271–1282. doi: 10.1038/nn.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omi K, Hachiya NS, Tanaka M, Tokunaga K, Kaneko K. 14-3-3zeta is indispensable for aggregate formation of polyglutamine-expanded huntingtin protein. Neurosci Lett. 2008;431:45–50. doi: 10.1016/j.neulet.2007.11.018. [DOI] [PubMed] [Google Scholar]
- Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu Rev Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- Pallos J, Bodai L, Lukacsovich T, Purcell JM, Steffan JS, Thompson LM, Marsh JL. Inhibition of specific HDACs and sirtuins suppresses pathogenesis in a Drosophila model of Huntington's disease. Hum Mol Genet. 2008;17:3767–3775. doi: 10.1093/hmg/ddn273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters MF, Ross CA. Isolation of a 40-kDa Huntingtin-associated protein. J. Biol. Chem. 2001;276:3188–3194. doi: 10.1074/jbc.M008099200. [DOI] [PubMed] [Google Scholar]
- Raymond LA, Andre VM, Cepeda C, Gladding CM, Milnerwood AJ, Levine MS. Pathophysiology of Huntington's disease: time-dependent alterations in synaptic and receptor function. Neuroscience. 2011;198:252–273. doi: 10.1016/j.neuroscience.2011.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao J, Welch WJ, Diamond MI. ROCK and PRK-2 mediate the inhibitory effect of Y-27632 on polyglutamine aggregation. FEBS Lett. 2008;582:1637–1642. doi: 10.1016/j.febslet.2008.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva MC, Fox S, Beam M, Thakkar H, Amaral MD, Morimoto RI. A genetic screening strategy identifies novel regulators of the proteostasis network. PLoS Genet. 2011;7:e1002438. doi: 10.1371/journal.pgen.1002438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R, Klein P, Koc-Schmitz Y, Waldvogel HJ, Faull RL, Brundin P, Plomann M, Li JY. Loss of SNAP-25 and rabphilin 3a in sensory-motor cortex in Huntington's disease. J Neurochem. 2007;103:115–123. doi: 10.1111/j.1471-4159.2007.04703.x. [DOI] [PubMed] [Google Scholar]
- Sugars KL, Brown R, Cook LJ, Swartz J, Rubinsztein DC. Decreased cAMP response element-mediated transcription: an early event in exon 1 and full-length cell models of Huntington's disease that contributes to polyglutamine pathogenesis. J. Biol. Chem. 2004;279:4988–4999. doi: 10.1074/jbc.M310226200. [DOI] [PubMed] [Google Scholar]
- The Huntington's Disease Collaborative Research Group. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- Thompson L, Aiken C, Kaltenbach L, Agrawal N, Illes K, Khoshnan A, Martinez-Vicente M, Arrasate M, O'Rourke J, Khashwji H, et al. IKK phosphorylates Huntingtin and targets it for degradation by the proteasome and lysosome. The Journal of Cell Biology. 2009;187:1083–1099. doi: 10.1083/jcb.200909067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo EM, Colombres M, Inestrosa NC. Wnt signaling in neuroprotection and stem cell differentiation. Prog. Neurobiol. 2008;86:281–296. doi: 10.1016/j.pneurobio.2008.08.001. [DOI] [PubMed] [Google Scholar]
- Vidal M, Cusick ME, Barabási AL. Interactome networks and human disease. Cell. 2011;144:986–998. doi: 10.1016/j.cell.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vis JC, Nicholson LF, Faull RL, Evans WH, Severs NJ, Green CR. Connexin expression in Huntington's diseased human brain. Cell Biol Int. 1998;22:837–847. doi: 10.1006/cbir.1998.0388. [DOI] [PubMed] [Google Scholar]
- Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S, Mill J, Cantor RM, Blencowe BJ, Geschwind DH. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonsattel JP, Difiglia M. Huntington disease. J. Neuropathol. Exp. Neurol. 1998;57:369–384. doi: 10.1097/00005072-199805000-00001. [DOI] [PubMed] [Google Scholar]
- Wang Y, Meriin AB, Zaarur N, Romanova NV, Chernoff YO, Costello CE, Sherman MY. Abnormal proteins can form aggresome in yeast: aggresome-targeting signals and components of the machinery. Faseb J. 2009;23:451–463. doi: 10.1096/fj.08-117614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Steimle PA, Ren Y, Ross CA, Robinson DN, Egelhoff TT, Sesaki H, Iijima M. Dictyostelium huntingtin controls chemotaxis and cytokinesis through the regulation of myosin II phosphorylation. Mol. Biol. Cell. 2011;22:2270–2281. doi: 10.1091/mbc.E10-11-0926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waza M, Adachi H, Katsuno M, Minamiyama M, Sang C, Tanaka F, Inukai A, Doyu M, Sobue G. 17-AAG, an Hsp90 inhibitor, ameliorates polyglutamine-mediated motor neuron degeneration. Nat. Med. 2005;11:1088–1095. doi: 10.1038/nm1298. [DOI] [PubMed] [Google Scholar]
- Wexler EM, Rosen E, Lu D, Osborn GE, Martin E, Raybould H, Geschwind DH. Genome-wide analysis of a Wnt1-regulated transcriptional network implicates neurodegenerative pathways. Sci Signal. 2011;4:ra65. doi: 10.1126/scisignal.2002282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeron MM, Hansson O, Chen N, Wellington CL, Leavitt BR, Brundin P, Hayden MR, Raymond LA. Increased sensitivity to N-methyl-D-aspartate receptor-mediated excitotoxicity in a mouse model of Huntington's disease. Neuron. 2002;33:849–860. doi: 10.1016/s0896-6273(02)00615-3. [DOI] [PubMed] [Google Scholar]
- Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4 doi: 10.2202/1544-6115.1128. Article17. [DOI] [PubMed] [Google Scholar]
- Zhang S, Binari R, Zhou R, Perrimon N. A genomewide RNA interference screen for modifiers of aggregates formation by mutant Huntingtin in Drosophila. Genetics. 2010;184:1165–1179. doi: 10.1534/genetics.109.112516. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.