Abstract
In this review, we describe our current understanding of translation termination and pharmacological agents that influence the accuracy of this process. A number of drugs have been identified that induce suppression of translation termination at in-frame premature termination codons (PTCs; also known as nonsense mutations) in mammalian cells. We discuss efforts to utilize these drugs to suppress disease-causing PTCs that result in the loss of protein expression and function. In-frame PTCs represent a genotypic subset of mutations that make up ~11% of all known mutations that cause genetic diseases, and millions of patients have diseases attributable to PTCs. Current approaches aimed at reducing the efficiency of translation termination at PTCs (referred to as PTC suppression therapy) have the goal of alleviating the phenotypic consequences of a wide range of genetic diseases. Suppression therapy is currently in clinical trials for treatment of several genetic diseases caused by PTCs, and preliminary results suggest that some patients have shown clinical improvements. While current progress is promising, we discuss various approaches that may further enhance the efficiency of this novel therapeutic approach.
I. Overview of translation termination
The central dogma of molecular biology states that the genetic information stored in DNA is converted into proteins via mRNA intermediates. The process by which this information is transferred from the nucleotide code of mRNA into the amino acid code of proteins is called translation. Translation can be divided into four stages: initiation, elongation, termination, and recycling. During translation initiation and elongation, decoding of the genetic information is mediated by base pairing between codons in the mRNA and the complementary anticodons of aminoacyl-tRNAs as each successive codon enters the ribosomal A site. Because of stringent proofreading steps associated with the translational machinery, these codon-anticodon interactions are highly accurate.
Translation termination occurs when a stop codon enters the ribosomal A site. In contrast to initiation and elongation, no tRNA molecules are complementary to the three stop codons (UAA, UAG and UGA) in the genetic code. Instead, termination occurs through the action of release factor proteins. Most mechanistic details of translation termination were first delineated in prokaryotes, where stop codon recognition is mediated by one of two Class I release factors (Dunkle and Cate, 2010, Scolnick et al., 1968, Youngman et al., 2008). RF1 recognizes UAA and UAG codons, while RF2 recognizes UAA and UGA codons. Either of these factors alone is sufficient to mediate efficient release of the nascent polypeptide chain from the peptidyl-tRNA located in the ribosomal P site. Once release has occurred, the class II release factor RF3 binds and dissociates RF1 or RF2 from the post-termination complex. GTP hydrolysis by RF3 then facilitates its own dissociation (Zavialov et al., 2001). Subsequently, ribosome recycling is facilitated by ribosome recycling factor (RRF), EF-G, and initiation factor 3 (IF3) to dissociate the ribosomal subunits, tRNA, and mRNA in preparation for the next round of translation (Hirokawa et al., 2005, Peske et al., 2005, Zavialov et al., 2005).
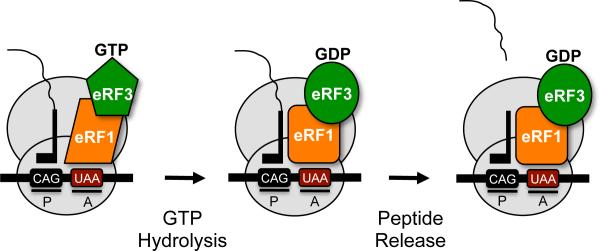
Translation termination in eukaryotes differs from the prokaryotic process in a number of ways. First, a single class I eukaryotic release factor, eRF1, recognizes all three stop codons (UAA, UAG and UGA) (Capecchi, 1967, Caskey et al., 1971, Frolova et al., 1994, Vogel et al., 1969) (for a review, see (Jackson et al., 2012)). eRF1 resides in a complex with the class II release factor, the GTPase eRF3 (Frolova et al., 1996). Unlike prokaryotic termination, GTP hydrolysis by eRF3 is an important feature of the eukaryotic termination process prior to peptide release. Mutations that compromised the GTPase activity of eRF3 were shown to reduce termination efficiency at some stop signals, but not others (Salas-Marco and Bedwell, 2004). This indicated that GTP hydrolysis by eRF3 influences stop codon recognition. In another study, Pestova and co-workers showed that GTP hydrolysis is a prerequisite for polypeptide release (Alkalaeva et al., 2006). Together, these results led to a model (Figure 1) in which GTP hydrolysis by eRF3 is required to alter both the conformation and position of eRF1 in the ribosomal A site in order to finalize stop codon recognition and trigger polypeptide release (Fan-Minogue et al., 2008, Jackson et al., 2012).
Figure 1.
Model of eukaryotic translation termination. A complex comprised of eRF1 and eRF3 mediate translation termination. eRF1 recognizes any of the three stop codons (UAA, UAG, UGA) in the ribosomal A site. GTP hydrolysis by eRF3 assists: 1) stop codon recognition by eRF1, and 2) eRF1 accommodation into the peptidyl transferase center so polypeptide release can occur. [Note: a color version of this figure is available online.]
II. Suppression of translation termination
A. Basal suppression of stop codons
The recognition of sense codons during translation elongation is mediated by aminoacyl-tRNAs bound in a complex with the GTPase EF-Tu in prokaryotes, or the GTPase eEF1A in eukaryotes. Codon recognition occurs when the aminoacyl-RNA enters the ribosomal A site and tests the base pairing to ensure that the proper codon-anticodon interaction occurs. Importantly, codon recognition is mediated by a sampling process where different aminoacyl-tRNAs (or eRF1) randomly enter the A site until the tRNA with the correct anticodon is selected.
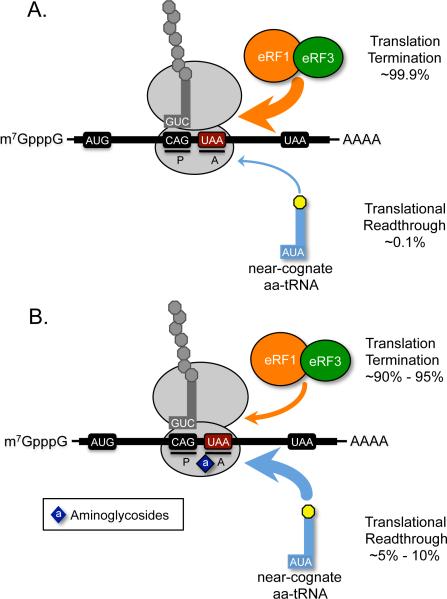
When a stop codon enters the ribosomal A site, aminoacyl-tRNAs and eRF1 randomly sample the A site until the stop codon is recognized by eRF1. While the translation termination process is generally accurate, a mistake can occur if a near-cognate aminoacyl-tRNA (whose anticodon is complementary to two of the three nucleotides of the codon) binds the stop codon. This mispairing may result in the incorporation of the amino acid attached to the near-cognate aminoacyl-tRNA into the nascent polypeptide chain, a process called stop codon suppression (or readthrough) (Figure 2). For reasons that will be discussed below, the error rate for suppression of normal stop codons at the end of genes is generally less than at premature stop codons (PTCs, or nonsense codons). Data from various studies have suggested that suppression generally occurs at a rate of 0.001-0.1% at normal stop codons (Parker, 1989) and 0.01 to 1% at PTCs (Bonetti et al., 1995, Cassan and Rousset, 2001, Manuvakhova et al., 2000). When an amino acid is incorporated at an in-frame PTC, synthesis of the protein resumes in the proper reading frame and results in production of a full-length protein. The only difference between the resulting translation product and the normal protein is the insertion of one of several possible amino acids at the position where suppression of the in-frame PTC occurred.
Figure 2.
Model of PTC suppression. A) The eRF1/eRF3 complex efficiently mediates translation termination in eukaryotes, where incorporation of an amino acid carried by a near-cognate aminoacyl-tRNA into the nascent polypeptide is rare. B) Drugs such as aminoglycosides bind to the small ribosomal subunit to stimulate misreading at stop codons, leading to an increased frequency of near-cognate aminoacyl-tRNA incorporation that allows continued translation elongation in the correct ribosomal reading frame. [Note: a color version of this figure is available online.]
As discussed above, suppression of stop codons is mediated by mispairing of a near-cognate aminoacyl-tRNA with a stop codon. By examining the codons (and encoded amino acids) that differ from stop codons at any one of the three nucleotide positions, the range of amino acids that could potentially be incorporated by the suppression event can be deduced (Table 1). It is often assumed that mispairing occurs only at the wobble position of the codon-anticodon pairing due to the lower discrimination of base pairing in the wobble position. However, this may be incorrect. In one study, an in-frame UAG PTC near the 5′-end of a ß-galactosidase reporter gene was suppressed and the resulting full-length protein was purified and subjected to N-terminal sequencing to determine the amino acid(s) incorporated (Fearon et al., 1994). It was found that three amino acids were primarily inserted at the PTC: tryptophan, tyrosine, and lysine. As shown in Table 1, these amino acids could be obtained when near-cognate mispairing occurs at the first nucleotide of the codon (lysine), the second (tryptophan), or the third (tyrosine). These results provide evidence that near-cognate mispairing can occur at any of the three positions of the stop codon during suppression. Overall, Table 1 shows that seven different amino acids could be incorporated at a UAG stop codon by near-cognate mispairing, while six different amino acids could be incorporated at either the UAA or UGA codons. In each case, one of these amino acids would represent the original amino acid found in the wild-type protein if the PTC arose by a single nucleotide change. Factors that are likely to affect aminoacyl-tRNA incorporation at PTCs include the PTC sequence as well as its surrounding mRNA sequence, the mode of action of the suppression drug used, the P-site tRNA and its bound polypeptide, and the relative abundance of the various near-cognate aminoacyl-tRNAs.
Table 1.
Amino acids that can be incorporated by mispairing of near-cognate aminoacyl-tRNAs at stop codonsa.
| UAA |
UAG |
UGA |
|||
|---|---|---|---|---|---|
| Near-cognate codon | Encoded amino acid | Near-cognate codon | Encoded amino acid | Near-cognate codon | Encoded amino acid |
| AAA | Lys | AAG | Lys | AGA | Arg |
| CAA | Gln | CAG | Gln | CGA | Arg |
| GAA | Glu | GAG | Glu | GGA | Gly |
| UCA | Ser | UCG | Ser | UAA | None |
| UGA | None | UGG | Trp | UCA | Ser |
| UUA | Leu | UUG | Leu | UUA | Leu |
| UAC | Tyr | UAA | None | UGC | Cys |
| UAG | None | UAC | Tyr | UGG | Tip |
| UAU | Tyr | UAU | Tyr | UGU | Cys |
During readthrough, stop codons are recognized by near-cognate aminoacyl-tRNAs that normally recognize the codons indicated above via mispairing of one of the three bases in the codon-anticodon interaction. For simplicity, we show all possible codons that can be deduced by a single mismatch at the first, second, or third position (including stop codons), rather than the anticodons of their cognate tRNAs.
B. Pharmacologically-induced suppression of stop codons
Aminoglycosides are the best-studied class of compounds that induce PTC suppression. These compounds inhibit translation in prokaryotes much more efficiently than in eukaryotes, an important difference that has been exploited for their broad clinical use as antibiotics. High doses of aminoglycosides block the initiation step of translation in prokaryotes. However, lower doses of these compounds alter the fidelity of translation, leading to an increased rate of amino acid misincorporation at sense codons during translation elongation and suppression of stop codons during translation termination (Gorini and Kataja, 1964, Lederberg et al., 1964). In the following sections, we will compare and contrast the mechanisms that promote fidelity during translation elongation and termination, and discuss how aminoglycosides disrupt these processes.
1) Aminoglycoside-induced misreading at sense codons
During translation elongation, selection of the correct aminoacyl-tRNA to pair with its corresponding codon occurs in the ribosomal A site. Structural and biochemical studies of prokaryotic ribosomes have provided important insights into how aminoacyl-tRNA selection and proofreading occurs during translation elongation. As discussed in the previous section, aminoacyl-tRNAs are delivered to the ribosome during translation as part of a ternary complex with the translation elongation factor EF-Tu and GTP (a similar ternary complex functions in eukaryotes, except the elongation factor is eEF1A). Translocation of each successive codon into the A site initiates a sampling process by ternary complexes carrying various aminoacyl-tRNAs. The choice of the aminoacyl-tRNA that yields the proper codon-anticodon interaction is determined through a three-stage process (Rodnina and Wintermeyer, 2001, Ruusala et al., 1982, Thompson and Stone, 1977, Wohlgemuth et al., 2011, Zaher and Green, 2009). 1) Incorrect ternary complexes containing a non-cognate aminoacyl-tRNA (capable of zero or one base pairs with the A site codon) are efficiently and immediately rejected prior to GTP hydrolysis by EF-Tu. 2) Incorrect ternary complexes containing mostly near-cognate aminoacyl-tRNA ternary complexes (capable of making two of the three possible base pairs with the A site codon) are rejected in a proofreading step following GTP hydrolysis by EF-Tu. 3) Hydrolysis of incorrect peptidyl tRNAs can occur following the subsequent translocation step.
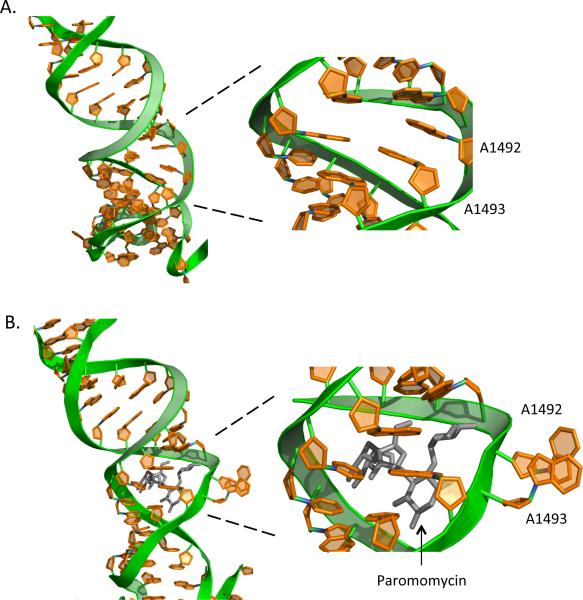
While each of the three steps described above contributes to the fidelity of translation elongation, the second (proofreading) step that discriminates against near-cognate aminoacyl-tRNAs is particularly relevant with regard to aminoglycoside-mediated misreading. Structural studies of prokaryotic ribosomes have shown that helix 44 of 16S rRNA, which contains the binding site for aminoglycosides, is located at the base of the ribosomal A site where codon recognition occurs. When a cognate aminoacyl-tRNA base pairs with the mRNA codon, three universally conserved nucleotides within the 16S rRNA, G530, A1492 and A1493, are positioned in close proximity to the codon-anticodon helix (Moazed and Noller, 1986). The bound cognate aminoacyl-tRNA adopts the A/T state on the ribosome, where the anticodon is stably associated with the codon in the A site but the aminoacyl end has not yet moved into proper position in the peptidyl transferase center. G530, A1492 and A1493 then undergo key structural rearrangements in the decoding center (Ogle et al., 2001). The bases of A1492 and A1493 flip out of the helix and interact directly with the minor groove of the codon-anticodon helix in the A site (Figure 3), while the base of G530 flips from a syn to an anti conformation. Together, these changes initiate a complex cascade of interactions, including a conversion of the 30S subunit from the open to the closed conformation and GTP hydrolysis by EF-Tu (Ogle et al., 2002). If the proper codon-anticodon interaction is present, EF-Tu rapidly dissociates from the ribosome and the aminoacyl-tRNA is accommodated into the peptidyl transferase center where peptide bond formation can occur. The presence of a single base pair mismatch in any position of the codon-anticodon complex results in a rapid dissociation of the entire ternary complex, thus preventing incorporation of an incorrect amino acid into the growing polypeptide chain.
Figure 3.
Structural changes that occur in the ribosomal decoding site. During proofreading of the codon-anticodon interaction, the bases of A1492 and A1493 of the 16S rRNA flip out and interact with the minor groove of the codon-anticodon helix. Shown are partial RNA structures of decoding sites from the 30S ribosomal subunit from Thermus thermophilus. These structures were obtained from 30S subunits in the (A) absence or (B) presence of paromomycin (shown in black). Structures were obtained using protein data bank numbers 1J5E (A) and 1IBK (B). [Note: a color version of this figure is available online.]
The mechanism described above is informative with regard to aminoglycoside-induced misreading. A1492 and A1493 are located in an internal loop of helix 44 that comprises the binding site for aminoglycosides. Aminoglycoside binding induces a similar rearrangement of A1492 and A1493 in the absence of a tRNA in the A site (Fourmy et al., 1996) (Figure 3). This drug-induced structural rearrangement in the decoding site increases the rate of GTPase activation, leading to an increased misincorporation rate (Pape et al., 2000). Much less is known about how fidelity is maintained during translation elongation in eukaryotes, but it is likely that most features of the process described above are conserved. However, the affinity of the eukaryotic decoding site for aminoglycoside binding is lower than the corresponding prokaryotic site (Recht et al., 1999), and studies in yeast have shown that aminoglycosides are more limited in their ability to induce misreading during translation elongation (Kramer et al., 2010, Salas-Marco and Bedwell, 2005). This suggests that aminoglycoside-induced effects on translation are largely restricted to the termination phase in eukaryotes.
2) Aminoglycoside-induced suppression of stop codons
As discussed above, the fidelity of translation elongation is maintained by a series of safeguards. In contrast, prokaryotic termination by RF1 and RF2 is thought to function in the absence of proofreading (Freistroffer et al., 2000). Structural studies have provided important insights into translation termination in prokaryotes (Jin et al., 2010, Korostelev et al., 2008, Korostelev et al., 2010, Laurberg et al., 2008, Weixlbaumer et al., 2008) (for a review, see (Korostelev, 2011)). RF1 or RF2 binding to a stop codon located in the ribosomal A site induces a conformational change that allows them to precisely span the distance from the decoding center in the 30S subunit to the peptidyl transferase center in the 50S subunit. This positioning allows the “decoding head” of the RF to recognize stop codons through a network of hydrogen bonds. Unlike the decoding of sense codons, the conserved nucleotides G530, A1492 and A1493 of the 16S rRNA are not directly involved in stop codon recognition. Instead, they stabilize the catalytically active conformation of the release factor, which allows the RF to recognize the stop codon through contacts with at least a dozen amino acids. Ultimately, the conformational changes in both the release factor and the ribosomal decoding site coordinate stop codon recognition with hydrolysis of the ester bond of the peptidyl-tRNA. While the mechanism of aminoglycoside-induced stop codon suppression has not been elucidated, the binding of these compounds to helix 44 and the resulting distortion of the decoding site (including A1492 and A1493) are likely to be involved.
Translation termination in eukaryotes differs significantly from the process in prokaryotes. As discussed earlier, a single class I release factor, eRF1, mediates polypeptide chain release at all three stop codons (UAA, UAG and UGA) (Capecchi, 1967, Caskey et al., 1971, Frolova et al., 1994, Vogel et al., 1969) (for a review, see (Jackson et al., 2012)), while the class II release factor eRF3 is a GTPase (Frolova et al., 1996). However, unlike the prokaryotic class I and class II factors, eRF1 and eRF3 can interact to form a stable complex. Formation of the eRF1/eRF3 complex significantly stabilizes GTP binding by eRF3, resulting in a stable eRF1/eRF3•GTP ternary complex. This ternary complex then participates in sampling the A site codon like the ternary complexes in translation elongation (aminoacyl-tRNA/eEF1A•GTP in eukaryotes, or aminoacyl-tRNA/EF-Tu•GTP in prokaryotes).
Overall, the class I release factors (RF1 and RF2 in prokaryotes and eRF1 in eukaryotes) carry out the conserved functions of stop codon recognition and polypeptide chain release. In contrast, the class II release factors (RF3 vs. eRF3) play completely different roles in the termination process. RF3•GTP binds after termination to facilitate release of the class I factors RF1 or RF2. In contrast, GTP hydrolysis by eRF3 is required for efficient stop codon recognition (Salas-Marco and Bedwell, 2004) and also facilitates the movement of eRF1's domain 2 into the peptidyl transferase center (in a manner analogous to tRNA accommodation) to mediate polypeptide chain release (Alkalaeva et al., 2006). After release, eRF1 remains associated with the post-termination complex to assist in recycling of the components of the post-termination complex by the recycling factor ABCE1 (Pisarev et al., 2010).
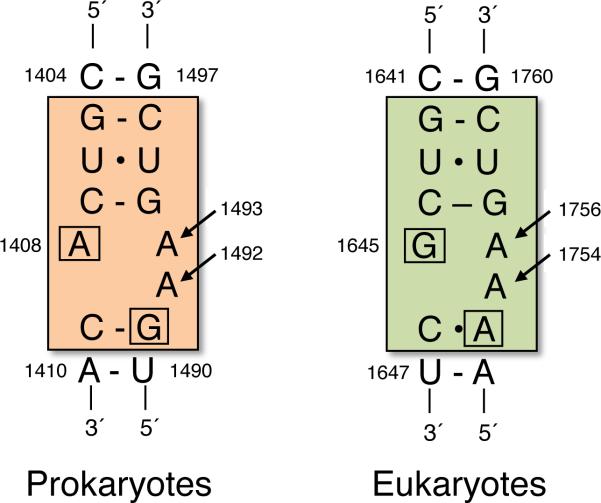
Structural studies have shown that aminoglycosides bind to the decoding site in the eukaryotic 18S rRNA, as they do in the corresponding structure within the prokaryotic 16S rRNA (Lynch and Puglisi, 2001). A critical difference is two base changes in the 18S rRNA that reduce the affinity of aminoglycoside binding to the eukaryotic decoding site, and correspondingly reduces the susceptibility of eukaryotic ribosomes to aminoglycoside-mediated inhibition (Recht et al., 1999) (Figure 4). Consistent with the importance of these nucleotide differences, conversion of these two residues in the eukaryotic structure to the prokaryotic sequence greatly enhanced aminoglycoside sensitivity in yeast (Fan-Minogue and Bedwell, 2007). While direct evidence is limited, these results suggest that conformational changes of the highly conserved residues G530, A1492 and A1493 (E. coli numbering) may also play an important role in aminoglycoside-induced suppression of translation termination in eukaryotes.
Figure 4.
Key differences between the prokaryotic and eukaryotic decoding sites. E. coli numbering is shown for prokaryotes (orange), while S. cerevisiae numbering is shown for eukaryotes (green). Arrows indicate nucleotides that are key in decoding correct base pair interactions within the A site. The boxed nucleotides indicate differing residues between prokaryotic and eukaryotic decoding sites that affect the affinity of aminoglycoside binding. [Note: a color version of this figure is available online.]
3) Suppression may occur primarily at PTCs
A concern of any therapeutic approach to suppress nonsense mutations is the potential to cause global readthrough of normal stop codons located at the end of genes. However, emerging evidence suggests that termination is more efficient at normal stop codons than PTCs, which could make normal stop codons less susceptible to suppression-inducing compounds. First, ribosomal toeprinting studies have found evidence of prolonged ribosomal pausing at PTCs, but not at normal stop codons (Amrani et al., 2004). It is possible that these prolonged pauses caused by a reduced efficiency of termination may make PTCs more susceptible to drug-induced suppression. In addition, previous studies have shown that poly(A) binding protein (PABP) interacts with eRF3 during termination and stimulates polypeptide chain release (Cosson et al., 2002, Hoshino et al., 1999). The position of normal stop codons near the poly(A) tail facilitates this association, thus ensuring that translation termination proceeds at an optimal rate. The random positioning of PTCs throughout the ORF frequently places them at a significant distance from the poly(A) tail, which could reduce the efficiency of translation termination. However, because mRNAs become circularized through an interaction between PABP and eIF4G (a component of the mRNA cap binding complex eIF4F), the linear distance between eRF3 and PABP is unlikely to be solely responsible for the efficiency of this interaction. In addition, the length of 3’ untranslated regions, which varies widely among mammalian transcripts, may also influence the association between eRF3 and PABP. Finally, the amount of PABP bound to the poly(A) tail of an mRNA may also influence the efficiency of its interaction with eRF3.
Some investigators have searched for evidence of suppression of normal stop codons under conditions where PTCs are efficiently suppressed. In one study, it was hypothesized that readthrough of normal stop codons would result in a range of proteins with C-terminal extensions that would induce a stress response due to accumulation of misfolded proteins (Keeling et al., 2001). To assess this possibility, the abundance of the inducible form of Hsp70 was compared in primary fibroblasts cultured in the presence or absence of gentamicin. Only a small change in the abundance of inducible Hsp70 was observed following growth in the presence of gentamicin, suggesting that suppression of natural stop codons did not occur to a significant extent. In another study, the relative suppression levels of a PTC vs. a normal stop codon within a luciferase gene were compared in HEK293 cells treated with the readthrough drug PTC124. A dose-dependent appearance of full-length protein was observed when cells containing the PTC reporter were grown in the presence of the drug. However, an extended form of the full-length protein was not observed in cells expressing the corresponding wild type reporter when grown under the same conditions. In addition, readthrough products could not be detected in multiple tissues from PTC124-treated human subjects, rats, and dogs (Welch et al., 2007). These results suggest that normal stop codons may be relatively resistant to readthrough. In addition, tandem stop codons have been shown to frequently occur at the end of ORFs, providing further protection against faulty termination at the end of mRNAs (Dalphin et al., 1999, Liang et al., 2005).
III. Therapeutic effects of suppression therapy
Dozens of published studies indicate that PTC suppression can restore partial levels of functional protein in a variety of disease models (Keeling and Bedwell, 2010, Keeling and Bedwell, 2011, Linde and Kerem, 2008). PTC suppression is a therapeutic approach that targets a class of mutation rather than a specific protein or disease pathway. As such, it can be considered a form of personalized medicine that could theoretically be applied to patients with a wide range of diseases attributable to nonsense mutations. Because of the large volume of suppression therapy data for many diverse diseases, we will limit our focus to two disease models, Duchenne muscular dystrophy and cystic fibrosis, that were used to establish this approach and have progressed to Phase 3 clinical trials. The following suppression therapy reviews may be viewed for more extensive disease coverage (Keeling and Bedwell, 2010, Keeling and Bedwell, 2011, Linde and Kerem, 2008).
A) Duchenne muscular dystrophy
Duchenne muscular dystrophy (DMD) is an X-linked neuromuscular disorder that results from a deficiency of functional dystrophin protein that is encoded by the DMD gene. The dystrophin protein plays a structural role in connecting the actin cytoskeleton of a muscle fiber to the extracellular matrix via an interaction with the dystrophin-associated protein complex (DAPC) (Nowak and Davies, 2004). The DAPC complex becomes destabilized in the absence of dystrophin, resulting in diminished levels of the DAPC complex and subsequent progressive muscle fiber damage.
DMD symptoms usually appear before age six and include muscle weakness that begins in the legs and pelvis. These symptoms progressively worsen, frequently leading to an inability to walk by age 12 (Fairclough et al., 2011). Other affected tissues include the cardiac and respiratory muscles, with most DMD patients succumbing to the disease by their late twenties due to respiratory distress. DMD occurs in 1 out of every 3,500 male births. Approximately 15% of DMD patients carry a nonsense mutation, suggesting that a significant number of DMD patients may benefit from a therapy that restores dystrophin by suppressing DMD nonsense mutations. Becker's muscular dystrophy (BMD), a much less common form of muscular dystrophy, is also caused by mutations in dystrophin that result in similar symptoms as DMD, but with a later age of onset and a much slower rate of progression.
1) DMD Mouse Studies
Suppression therapy was first investigated for DMD using the naturally occurring mdx mouse that contains a nonsense mutation at codon 3,195 (CAA to TAA) in exon 23 of the mouse Dmd locus (Sicinski et al., 1989). It has been estimated that approximately 20% of normal dystrophin protein must be restored in this mouse model to provide a significant correction of the muscle pathology (Chamberlain, 1997). A number of readthrough drugs have been shown to alleviate the DMD disease phenotype in this mouse model. When administered for two weeks at a dose of 34 mg/kg, gentamicin (Figure 6A) was shown to restore up to 20% of normal dystrophin protein levels in tibialis anterior, heart, and diaphragm muscle tissues and restored DAPC formation (Barton-Davis et al., 1999). Gentamicin-treated mdx mice also showed significant protection from contraction-induced damage and normalization of serum creatine kinase levels (an indicator of muscle damage). However, another group was unable to reproduce these results in the mdx mouse (Dunant et al., 2003). Possible reasons suggested for the discrepancy include differences in drug composition and administration protocols.
Figure 6.
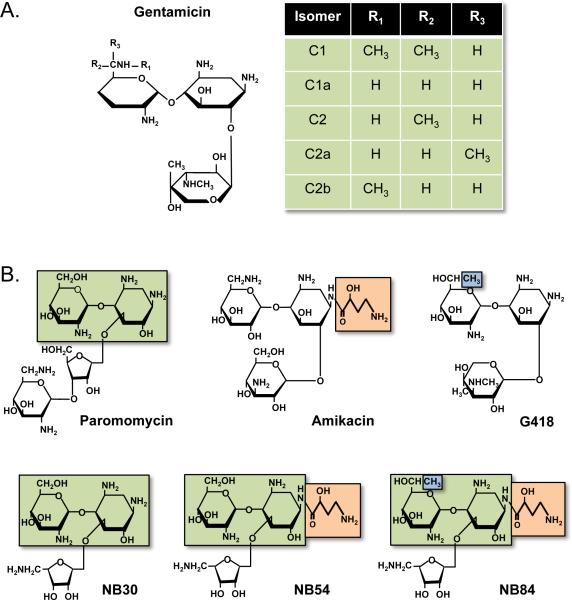
Structures of some aminoglycosides that induce PTC suppression in mammalian cells. A) Gentamicin isomers. B) Designer aminoglycoside derivatives. The boxed structures represent structural elements within conventional aminoglycosides that were used to generate the designer aminoglycosides NB30, NB54, and NB84. [Note: a color version of this figure is available online.]
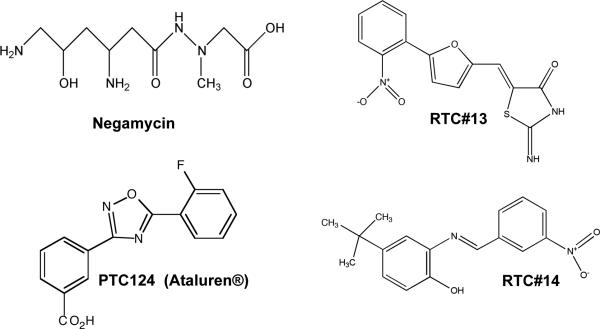
Administration of the dipeptide antibiotic negamycin (Figure 5) to mdx mice for two weeks at a daily dose of 120 millimoles/kg (a molar equivalent dose to 34 mg/kg gentamicin) also restored dystrophin protein and the DAPC in the tibialis anterior muscle tissues (Arakawa et al., 2003), while a four-week negamycin treatment restored dystrophin in cardiac muscle. This study also found that negamycin was significantly less toxic in mice than gentamicin.
Figure 5.
Structures of non-aminoglycosides that induce PTC suppression in mammalian cells.
PTC124 is a readthrough drug that has no antibiotic properties and has been shown to be nontoxic (Figure 5) (Hirawat et al., 2007, Welch et al., 2007). This compound was orally administered to mdx mice for two to eight weeks at 0.9-1.8 mg/ml doses in a liquid diet, and was further supplemented with intraperitoneal injections at doses of 33 mg/kg three times/day. Treated mice showed a restoration of up to 25% of normal dystrophin levels in tibialis anterior, heart, and diaphragm muscle tissues (Welch et al., 2007). In addition, PTC124 treatment improved protection against contraction-induced injury such that force decrement was not significantly different from wild-type control mice. Furthermore, serum creatine kinase levels were significantly reduced in PTC124-treated mice after two weeks of treatment and continued reductions were observed for up to eight weeks after treatment termination.
In summary, multiple drugs, including gentamicin, negamycin, and PTC124 were found to suppress the nonsense mutation in the mdx mouse at levels that restored a physiologically relevant amount of dystrophin protein.
2) DMD/BMD Clinical Trials
The promising results in the mdx mouse model led to the initiation of clinical trials of suppression therapy in DMD/BMD patients that carry nonsense mutations. Preliminary clinical trials using gentamicin administered intravenously produced mixed results. In one study, four DMD/BMD patients administered gentamicin once daily at a dose of 7.5 mg/kg for 2 weeks did not show significant changes in dystrophin levels or muscle strength (Wagner et al., 2001). In a separate study, three of four DMD patients treated with 6 to 7.5 mg/kg gentamicin in two 6-day intervals (separated by a 7 week period without treatment) showed a restoration of dystrophin protein, but no significant change in functional tests was observed (Politano et al., 2003). Recently, a more extensive trial was conducted in 26 DMD patients treated with 7.5 mg/kg gentamicin once daily for 14 days (Malik et al., 2010). A significant 50% decrease in serum creatine kinase activity was found after gentamicin treatment in the patients that carried nonsense mutations, but not in DMD patients that carried frameshift mutations. Additional DMD patients carrying nonsense mutations were subjected to a longer six-month gentamicin treatment regimen at a dose of 7.5 mg/kg that was administered once or twice weekly. Approximately 50% of the patients that provided muscle biopsies showed an increase in dystrophin protein, with levels ranging from 1% to 15% of normal. On average, a 54% decrease in serum creatine kinase levels was also found in patients post-treatment. While no other clinically meaningful endpoints showed statistically significant improvements at the end of the treatment period, muscle strength was stabilized in many of the gentamicin-treated patients, which is in contrast to the usual rate of steady functional decline observed in most DMD patients (Malik et al., 2010). In addition, no significant toxicity was associated with gentamicin administration in any of these studies.
Phase 2a clinical trials have also been carried out with PTC124 in 38 DMD/BMD patients with nonsense mutations (Finkel, 2010). PTC124 was administered orally for 28 days in 3 cohorts of patients at 16, 40, or 80 mg/kg/day. Comparison of primary cells from muscle biopsies obtained before vs. after treatment showed a dose-dependent restoration of dystrophin protein. During treatment, a reduction in serum creatine kinase was observed in most treated patients. This marker returned to the pre-treatment levels within a month of treatment cessation. In addition, dystrophin protein expression was restored in the majority of patients, but with no clear dose-dependence or correlation to the nonsense mutation sequence context.
A randomized, double-blinded, placebo-controlled phase 2b trial was conducted at multiple sites to evaluate the efficacy of PTC124 in 174 DMD/BMD patients administered either 40 or 80 mg/kg/day for 48 weeks [PTC Therapeutics, Inc., http://ptct.client.shareholder.com/releases.cfm]. A 6-minute walk distance (6MWD) test was used as the primary endpoint for evaluating the potential therapeutic benefit of treatment. Initially, failure to produce a significant improvement in the primary endpoint led to termination of the trial. However, subsequent stratification of the trial data revealed that patients who received 40 mg/kg/day PTC124 showed a significant improvement in the 6MWD test relative to placebo-treated patients, while a significant improvement was not found in patients treated with 80 mg/kg/day PTC124. In addition, it was found that patients who received the lower PTC124 dose also showed a significantly reduced disease progression than placebo-treated patients. While dystrophin protein expression was also analyzed, no correlation was found between dystrophin expression and results of the 6MWD test. Currently, Phase 3 trials are underway at 14 different centers to test the long-term effect of PTC124 administration in 110 DMD/BMD patients that carry nonsense mutations [ClinicalTrials.gov, identifier NCT01247207]. In these studies, PTC124 is being administered orally at a dose of 40 mg/kg/day.
In summary, the data compiled from preliminary clinical trials of suppression therapy in DMD/BMD patients is mixed. However, longer-term trials are warranted since a fraction of patients showed a therapeutic benefit from PTC suppression.
B) Cystic fibrosis
Cystic fibrosis (CF) is an autosomal recessive disorder caused by mutations in the CFTR (Cystic Fibrosis Transmembrane Regulator) gene. CF occurs in approximately 1 in 2,500 births. Approximately 30,000 CF patients reside in the United States, and roughly 1 in every 31 Americans is a carrier of a defective CF allele (Kim Chiaw et al., 2011, Wilschanski and Kerem, 2011). The CFTR protein functions as a cAMP-regulated chloride channel that resides in the apical membrane of epithelial cells that line the intestine, respiratory system, pancreas, gall bladder, and sweat glands. The CF phenotype includes dysfunction of the exocrine pancreas that results in pancreatic insufficiency and nutrient malabsorption, gastrointestinal disease including meconium ileus with obstruction, hepatobiliary disease, and absence of the vas deferens in male patients. However, the main pathologic feature of CF is respiratory disease manifested by recurrent episodes of infection, inflammation, and obstruction that lead to progressive lung damage and eventual respiratory failure. Since all states screen newborns for CF, the majority of CF patients are diagnosed as infants. Children with CF died in infancy sixty years ago, but improvements in diagnosis and treatments have extended the current average life expectancy of CF patients to 37 years (Kim Chiaw et al., 2011, Wilschanski and Kerem, 2011).
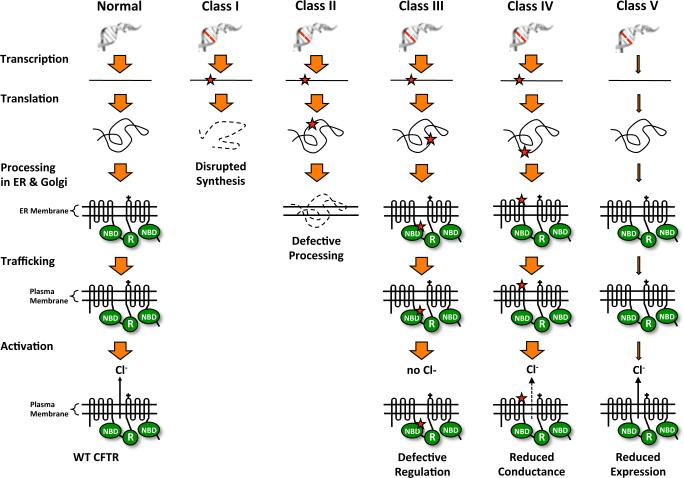
More than 1,900 different CFTR mutations have been reported and can be generally categorized into five classes according to their effect on CFTR biogenesis, processing, function, and expression (Rogan et al., 2011, Cuthbert, 2010) (Figure 7). Class I mutations disrupt CFTR protein synthesis, resulting in no CFTR protein production. This group includes nonsense, frameshift, and splicing mutations. Approximately 40% of CFTR mutations reside in this class. Specifically, nonsense mutations are carried by 10% of the general CF population, but in some populations, up to 50% of CF patients carry a nonsense mutation. The G542X mutation, found in 2% of Caucasian CF patients, is the most common CFTR nonsense mutation. Class II includes mutations that lead to faulty CFTR processing and subsequent protein misfolding that prohibits CFTR from properly localizing to the cell membrane. The F508del mutation (a deletion of the phenylalanine residue at position 508) is found in 75% of CF patients and represents a mutation from this class. Class III comprises mutations that allow the CFTR to be trafficked properly to the cell membrane, but the protein does not respond to cAMP stimulation and does not have chloride channel function. The G551D mutation, a CFTR gating mutant found in approximately 4% of CF patients, is an example of this mutation class. Class IV is made up of mutations that allow correct CFTR localization and activation by cAMP, but with reduced chloride conductance. Class V includes mutations that result in normal plasma membrane CFTR, but overall protein expression is reduced. Classes I-III result in severe reductions in CFTR function and lead to a severe CF phenotype, while Classes IV-V retain some level of residual CFTR protein function and confer a milder CF phenotype. It has been estimated that 5-35% of normal CFTR function may be required to alleviate the CF phenotype (Kerem, 2004, Zhang et al., 2009, Amaral, 2005).
Figure 7.
Classes of mutations that cause cystic fibrosis. CFTR mutations are categorized into five classes (I-V) depending on the effect of the mutation on CFTR protein abundance, processing, localization, and function. The mutation classes are as follows: Class I = no CFTR protein is produced (includes nonsense mutations); Class II = CFTR processing is defective, so CFTR is retained in the ER and subsequently degraded; Class III = CFTR localizes to the cell surface, but is nonfunctional; Class IV = CFTR localizes to the cell surface, but with reduced activity; Class V = CFTR localizes to the cell surface and is functional, but its expression is reduced. [Note: a color version of this figure is available online.]
1) CF Mouse studies
Previous studies have shown that a variety of readthrough drugs are able to suppress CFTR nonsense mutations and restore functional CFTR protein in both mouse models and in human clinical trials. However, the human CF phenotype varies from the mouse CF disease (Davidson and Rolfe, 2001, Scholte et al., 2004, Snouwaert et al., 1992). The most significant difference is that the lung pathology observed in the human CF disease does not occur in most CF mouse models. However, CF-associated intestinal defects are more prevalent. In some CF mouse strains, approximately 80% of CF mice die within seven days of weaning due to intestinal blockage. Maintenance on a liquid diet after weaning decreases the mortality rate to some extent.
Studies to examine suppression of a CFTR nonsense mutation utilized a transgenic mouse that carried a human CFTR cDNA containing the G542X nonsense mutation. This transgene was expressed under control of the rat fatty acid binding protein (FABP) promoter. The CFTR-G542X transgene was then crossed into a Cftrtm1Cam knockout mouse, resulting in a mouse that was suitable to evaluate the ability of suppression therapy to restore CFTR protein function (Du et al., 2002). Using this CFTR nonsense mouse model, a number of readthrough drugs were found to restore significant levels of CFTR function. Gentamicin administered at doses ranging from 5-34 mg/kg via once daily subcutaneous injections for 14 days resulted in a significant increase in intestinal cAMP-activated transepithelial chloride conductance, resulting in as much as 22% of wild-type CFTR function (Du et al., 2002, Du et al., 2006). In addition, CFTR protein could be detected at the apical surface of intestinal tissues following treatment. Similar results were observed when amikacin was administered using a similar administration protocol at doses ranging from 15-170 mg/kg. However, tobramycin was much less effective in restoring CFTR function than either gentamicin or amikacin (Du et al., 2002, Du et al., 2006). PTC124 administered orally for 14-21 days at a dose of 0.9 mg/ml in a liquid diet restored up to 24% of normal CFTR function, while once daily subcutaneous injections at doses ranging from 15-60 mg/kg restored up to 29% of CFTR function (Du et al., 2008). A partial restoration of CFTR protein at the apical surface of intestinal tissues was also observed.
In summary, a number of different drugs were found to suppress the common G542X mutation in a transgenic CF mouse model. These included gentamicin, amikacin, and PTC124. All of the drugs restored approximately 20-30% of wild-type CFTR levels, indicating the potential of these compounds to alleviate the CF phenotype.
2) CF Clinical Trials
Several clinical trials of suppression therapy using aminoglycosides have been conducted in CF patients carrying nonsense mutations. Notably, a range of results was obtained. In the first study, nine CF patients were administered 0.9 mgs of gentamicin daily via nasal drops for a total of 14 days (Wilschanski et al., 2003). Nasal potential difference (PD) measurements indicated a significant improvement in chloride conductance, and full-length CFTR protein was detected in nasal epithelia from treated subjects. No improvement in CFTR function or restoration of CFTR protein was detected in control patients homozygous for the F508del mutation. Another study administered gentamicin intravenously to ten CF patients at a dose of 2.5 mg/kg every 8 hours for one week (Clancy et al., 2001). This treatment protocol led to a three-fold increased incidence of nasal PD readings in the direction of chloride secretion in subjects with PTCs compared with CF controls. Airway chloride secretion was also stronger in the PTC group, with four of five subjects exhibiting a level of chloride secretion not seen in any of the control subjects. However, no improvement in sweat chloride measurements was observed in either treatment group. A more recent multi-center randomized, double-blinded, crossover study was conducted with eleven CF patients with nonsense mutations and eighteen CF patients that did not carry nonsense mutations (Clancy et al., 2007). Both groups of CF patients were administered 3.6 mg of gentamicin or tobramycin daily using a nasal spray device for 28 days. However, no improvement in chloride ion transport or CFTR localization was observed in either patient group treated with gentamicin or tobramycin.
Several CF suppression therapy clinical trials have also been conducted with PTC124. One Phase 2 trial was conducted with twenty-three CF patients carrying nonsense mutations who were administered PTC124 orally in two treatment phases (Kerem et al., 2008). Patients were administered 16 mg/kg PTC124 daily for 14 days in the first phase, followed by 14 days without treatment, and then PTC124 was administered at a dose of 40 mg/kg daily for 14 days in the second treatment phase. Nasal potential difference measurements after either treatment phase showed a substantial improvement in chloride conductance, where approximately half of the patients showed measurements within the normal range (Kerem et al., 2008). In addition, small improvements in lung forced vital capacity, neutrophil counts, and body weight were observed. Twelve-week treatments using the same treatment phases described above in nonsense-containing CF patients were also recently conducted (Wilschanski et al., 2011). Both treatment phases increased nasal chloride transport in approximately 60% of treated patients, with greater CFTR function restored with increased treatment time. In addition, trends toward improved pulmonary function and reduced coughing were observed. PTC124 has also been examined in thirty pediatric CF patients that carried nonsense mutations (Sermet-Gaudelus et al., 2010). PTC124 was administered in two fourteen-day cycles as described above. CFTR-mediated nasal epithelial chloride transport was at least partially restored in approximately 50% of the treated patients. In addition, localization of full-length CFTR protein was restored at the apical membrane of nasal epithelial cells from treated patients. Phase 3 clinical trials are currently underway to evaluate 48-week PTC124 treatment in pediatric and adult CF patients administered 40 mg/kg/day [ClinicalTrials.gov; identifier NCT00803205]. The clinical endpoint to evaluate the effectiveness will be lung forced vital capacity as well as secondary endpoints of nasal potential difference measurements, lung inflammation markers, and body weight.
In summary, the CF suppression therapy clinical trials indicate that both gentamicin and PTC124 are able to restore CFTR protein in a fraction of CF patients with PTCs. Longer-term studies will be required to determine whether the therapeutic effects found in patients upon short-term treatment can be sustained, and if enough CFTR can be restored to substantially alleviate manifestations of CF lung disease.
IV. Limitations of Suppression Therapy
The preliminary clinical data obtained with DMD and CF indicates that suppression therapy shows promise as a potential treatment; however, many of the studies produced highly variable results. While differences in administration protocols and technical differences may account for some of the variability, it appears that only a subset of patients elicits a robust response to the various suppression drugs. This indicates that substantial improvements must be made in suppression therapy before it can be generally applied as a treatment for the majority of patients that carry nonsense mutations. In the following section, we discuss possible reasons for the variable responses observed and ways that some of these limitations may be overcome.
1) Efficacy
While suppression therapy appears to be a promising treatment for diseases attributable to nonsense mutations, suppression therapy has not been successful in treating all diseases or in every PTC-bearing patient with a particular disease. As discussed above, PTC124 treatment was able to restore detectable CFTR function in only about half of treated patients (Kerem et al., 2008, Sermet-Gaudelus et al., 2008, Wilschanski et al., 2011). This indicates that the efficacy of suppression therapy must be improved in order to provide a therapeutic benefit to a majority of patients afflicted with diseases caused by PTCs. In the following sections, various factors that influence the efficiency of PTC suppression therapy will be discussed.
a) Sequence Context Effects
The ability of readthrough agents to suppress various PTCs may be influenced by a number of factors. The sequence of the stop codon as well as the surrounding upstream and downstream mRNA sequence have been shown to be important in mediating the efficiency of translation termination as well as the susceptibility to drug-induced PTC suppression (Cassan and Rousset, 2001, Manuvakhova et al., 2000, Martin, 1994, Phillips-Jones et al., 1995, Martin et al., 1989, Bonetti et al., 1995). The nucleotide immediately downstream of a stop codon appears to be the most influential of the surrounding sequence context in mediating termination efficiency, leading some investigators to propose that the eRF1 release factor may actually recognize a tetranucleotide termination signal (McCaughan et al., 1995, Brown et al., 1990). In general, it has been found that a purine (A or G) in the 4th nucleotide position of the termination signal resulted in more efficient termination than a pyrimidine (C or U). In mammalian in vitro translation systems, as much as a 20-fold difference in basal termination efficiency has been observed among different tetranucleotide termination signals (Manuvakhova et al., 2000). It has also been shown that various readthrough drugs have different abilities to suppress PTCs based on the PTC sequence context. As much as a 30-fold difference in suppression levels mediated by readthrough drugs has been observed among various tetranucleotide sequences (Manuvakhova et al., 2000). In addition, the extended sequence beyond the tetranucleotide can affect suppression efficiency, where as great as 150-fold differences in drug-mediated suppression levels can be observed among extended PTC contexts (Manuvakhova et al., 2000).
b) Nonfunctional amino acid incorporation at PTCs
Not all of the full-length protein restored by PTC suppression may be functional. As discussed earlier, several near-cognate aminoacyl-tRNAs that base pair with two of the three nucleotides of the PTC could potentially be utilized to suppress a PTC, including the 1st and 2nd nucleotides, the 2nd and 3rd nucleotides, or the 1st and 3rd nucleotides (Fearon et al., 1994). This could result in the incorporation of one of several amino acids at the PTC that may differentially affect protein function and/or stability (Table 1). However, unless a specific amino acid is required at the site of the PTC for protein function, at least a portion of the protein restored by PTC suppression is expected to be functional.
c) Nonsense-mediated mRNA decay (NMD)
In addition to producing truncated proteins that are often nonfunctional and/or unstable, PTCs also frequently reduce the stability of mRNAs, thus reducing their steady state abundance. Nonsense-mediated mRNA decay (NMD) is a conserved pathway in eukaryotes that degrades mRNAs containing PTCs (Isken and Maquat, 2007, Muhlemann et al., 2008, Bhuvanagiri et al., 2010). NMD decreases the efficiency of suppression therapy by reducing the pool of PTC-containing mRNAs available for translation and subsequent PTC suppression. Previous clinical trials of suppression therapy have provided evidence that NMD may influence the response to suppression therapy (Linde et al., 2007, Wilschanski et al., 2000, Wilschanski et al., 2003). It was shown that CF patients that responded most robustly to suppression therapy had higher levels of residual PTC-containing CFTR mRNA. Furthermore, it was shown that knocking down expression of NMD factors using RNA silencing to inhibit NMD in CF primary nasal epithelial cells increased both the level of PTC-containing CFTR transcripts and CFTR function (Linde et al., 2007). This strongly suggests that NMD efficiency can influence the efficacy of suppression therapy.
d) Toxicity
While PTC124 was found to be a nontoxic readthrough agent, many of the aminoglycosides that suppress PTCs pose significant risks of toxicity, including ototoxicity and nephrotoxicity. Approximately 2-25% of patients treated with aminoglycosides show signs of nephrotoxicity and/or ototoxicity (Lopez-Novoa et al., 2011, Huth et al., 2011). While kidney damage caused by aminoglycoside exposure can usually be reversed after treatment cessation, damage to the inner ear is permanent. Importantly, the toxicity associated with aminoglycosides is not due to their function in modulating translation termination efficiency at cytoplasmic ribosomes, but rather is due to off-target effects. Aminoglycosides can enter renal cells by binding an endocytic complex formed by the proteins megalin and cubilin that are enriched in the proximal tubules (Tauris et al., 2009). Within the low pH environment of lysosomes, aminoglycosides become positively charged and interact with the negatively charged phospholipids within the lysosomal membrane, which leads to inhibition of membrane phospholipases and phospholipidosis (De Broe et al., 1984, Laurent et al., 1982). Subsequent apoptosis and necrosis of the tubular epithelial cells occur, leading to renal damage. In the inner ear, aminoglycosides appear to enter the hair cells through both endocytosis and ion channels (Lopez-Novoa et al., 2011). Two main consequences result from aminoglycoside entry into hair cells: 1) production of reactive oxygen species that leads to apoptosis; and 2) apoptosis due to aminoglycoside interference with mitochondrial translation (Lopez-Novoa et al., 2011, Guthrie, 2008). Interestingly, around 1 in 500 individuals in the general population carries a mutation in their mitochondrial 12S rRNA that increases the structural similarity of the mitochondrial rRNA to the bacterial rRNA (Qian and Guan, 2009, Guan et al., 2000). This mutation promotes aminoglycoside binding to the rRNA, leading to greater susceptibility to inhibition of mitochondrial protein synthesis.
e) Immune Response
PTCs often lead to a complete absence of protein. Consequently, restoration of protein expression by suppression therapy may result in an immune response to the newly restored protein in some patients. Such a response was described in one DMD patient after a suppression therapy regimen with gentamicin (Malik et al., 2010). In that patient, no dystrophin was detected by western blotting following suppression therapy; however, a robust T-cell response was produced to dystrophin peptides downstream of the PTC. In two other patients that underwent the same suppression therapy protocol, a three- to five-fold increase in dystrophin protein was found using western blotting, but no T-cell response to dystrophin peptides was produced. It is unclear whether technical difficulties prevented an accurate quantitation of dystrophin protein in the patient where no dystrophin protein was detected, or if the restored dystrophin protein was cleared by the immune response in this patient. Further analyses will be required to determine whether an immune response to the protein restored by suppression therapy could reduce the therapy effectiveness in at least a subset of patients.
V. Efforts to overcome suppression therapy limitations
1) Develop Safer, More Effective Drugs
In order to find compounds that are more effective than earlier readthrough drugs such as gentamicin, a number of approaches have been explored. First, novel suppression agents have been identified through high throughput screens (HTS). PTC Therapeutics identified PTC124 (Figure 5A), which has been shown to be nontoxic, safe, and more effective than many aminoglycosides at suppressing PTCs (Du et al., 2008, Hirawat et al., 2004, Hirawat et al., 2007, Welch et al., 2007). Additional readthrough drugs have also been identified through HTS, including RTC#13 and RTC#14 (Figure 5), which have been shown to be effective in suppressing nonsense mutations in the ATM gene associated with ataxia telangiectasia (Jung et al., 2011, Du et al., 2009a). These novel readthrough compounds provide additional structural scaffolds to develop novel, safe readthrough drugs with increased efficacy.
Some controversy has been associated with the identification of readthrough drugs using HTS. In one example, PTC Therapeutics performed primary HTS of 800,000 compounds using PTC-containing firefly luciferase reporters to identify novel readthrough drugs (Welch et al., 2007). Candidate suppression drugs such as PTC124 were identified as a result of increased firefly luciferase activity (compared to controls treated with vehicle alone) due to PTC readthrough that restored full-length, functional firefly luciferase. However, Inglese and co-workers later found that the firefly luciferase protein binds PTC124 (Auld et al., 2009). This interaction was reported to stabilize the firefly luciferase protein, making it less susceptible to proteolysis, thereby increasing firefly luciferase protein steady state levels. Based on these results, it was suggested that PTC124 might have been inappropriately identified in the firefly-based HTS due to its ability to stabilize the small amount of firefly protein produced by basal readthrough, rather than by restoring functional firefly luciferase by PTC suppression. The discrepancies between the firefly luciferase results obtained by PTC Therapeutics and the Inglese group were suggested to have arisen from technical differences in the luciferase assay conditions (Peltz et al., 2009), although that explanation was disputed (Inglese et al., 2009). This controversy illustrates the need for extreme caution when designing HTS. At a minimum, a series of secondary screens should be used to verify initial hits. In this case, additional in vitro studies as well as multiple in vivo studies using both animal models and clinical trials have shown that PTC124 suppresses PTCs in mRNAs that encode a wide variety of proteins unrelated to firefly luciferase, including those associated with diseases such as DMD (Welch et al., 2007), CF (Du et al., 2008, Kerem et al., 2008, Sermet-Gaudelus et al., 2008, Wilschanski et al., 2008, Sermet-Gaudelus et al., 2010, Hirawat et al., 2004), Type 1C Usher syndrome (Goldmann et al., 2011), and carnitine palmitoyltransferase 1A deficiency (Tan et al., 2011).
Rational drug design has also been pursued to generate safer, more effective readthrough drugs. A number of aminoglycoside derivatives have been generated for the purpose of novel antibiotic development (Li et al., 2004, Elchert et al., 2004, Chang et al., 2002), and a subset of these drugs has been shown to suppress PTCs associated with spinal muscular atrophy (SMA) in mammalian cells (Mattis et al., 2006). From this group of drugs, the TC007 neomycin derivative was shown to significantly alleviate the disease phenotype in SMA nonsense mouse models (Mattis et al., 2009b, Mattis et al., 2009a). Recently, steps to develop aminoglycoside derivatives specifically for the purpose of suppressing PTCs have also been undertaken. Baasov and co-workers have used a medicinal chemistry approach to develop aminoglycoside derivatives with enhanced readthrough ability and reduced toxicity (Nudelman et al., 2010, Nudelman et al., 2006). They identified moieties within conventional aminoglycosides that are predicted to increase their association with the eukaryotic ribosome to enhance PTC suppression, while simultaneously reducing their association with bacterial and/or mitochondrial ribosomes to circumvent at least one aspect of aminoglycoside toxicity. Structural components of paromomycin were used to create the first generation of derivatives that included the designer aminoglycoside NB30 (Figure 6B), which was found to be 6 to 15 fold less toxic than paromomycin or gentamicin in mammalian cells. While NB30 suppressed PTCs associated with type 1 Usher syndrome using in vitro assays, it was not as effective as the conventional aminoglycosides paromomycin or gentamicin (Rebibo-Sabbah et al., 2007, Goldmann et al., 2010). However, the addition of a (S)-4-amino-2-hydroxybutanol (AHB) side group found in amikacin to NB30 generated the designer aminoglycoside NB54 (Figure 6B). While the toxicity of this new drug remained reduced compared to its parental aminoglycosides, NB54 produced more robust readthrough of PTCs than NB30. This was demonstrated in multiple in vitro assays (Nudelman et al., 2009, Vecsler et al., 2011) as well as in a nonsense CF mouse model (Rowe et al., 2011). The addition of a methyl group found in G418 to NB54 resulted in generation of NB84 (Figure 6B). While this designer aminoglycoside showed low toxicity like its precursors, NB84 more effectively promoted readthrough than NB54 in an in vitro model of Rett syndrome (Brendel et al., 2010) and a mouse model of mucopolysaccharidosis I-Hurler (MPS I-H) (Wang et al., 2012). This method of rational drug design has iteratively produced designer aminoglycosides with increasingly enhanced PTC suppression and reduced toxicity compared to traditional aminoglycosides. Further optimization of these designer aminoglycosides suggests that clinically relevant readthrough drugs may ultimately result from this approach (Kandasamy et al., 2011).
2) Reduce aminoglycoside toxicity
In addition to designing novel aminoglycosides that are safer for PTC suppression, other methods have also been investigated to offset aminoglycoside toxicity. Intravenous aminoglycoside administration is carefully monitored to maintain serum levels that result in maximal antimicrobial effects while minimizing toxicity. Aminoglycoside dosing regimens have been investigated that compared once daily dosing versus multiple daily dosing in order to find protocols that minimize toxicity risks. Recent data indicate that once daily treatment regimens are as effective as multiple smaller doses that cumulatively administer the same total daily dose. Most importantly, the once daily dose was shown to reduce nephrotoxicity in children (Smyth and Bhatt, 2012, Beauchamp and Labrecque, 2001).
Other methods of reducing aminoglycoside toxicity have also been pursued. For example, the administration of various antioxidants has been shown to alleviate aminoglycoside-induced ototoxicity (Sha and Schacht, 2000, Kawamoto et al., 2004, Reiter et al., 2011, Darrat et al., 2007) and nephrotoxicity (Ali et al., 2011). In addition, co-administration of polyanions such as daptomycin (Woodworth et al., 1994, Thibault et al., 1995) and poly-L-aspartic acid (PAA) (Kishore et al., 1992, Swan et al., 1991, Hulka et al., 1993) alleviate nephrotoxicity and ototoxicity by binding to aminoglycosides within the lysosomal compartment and preventing aminoglycoside association with phospholipids in the lysosomal membrane. Interestingly, PAA was found to increase cytoplasmic aminoglycoside concentrations in mammalian cells (Du et al., 2009b, Gilbert et al., 1989, Beauchamp et al., 1990, Kishore et al., 1992). Co-administration of PAA with aminoglycosides was also shown to enhance the efficacy of PTC suppression in human MPS I-H fibroblasts (Keeling, 2000) and in a nonsense CF mouse model (Du et al., 2009b).
Another approach to alleviate aminoglycoside toxicity is to encapsulate them into liposomes. Additional advantages of liposome-encapsulated aminoglycosides include improved pharmacokinetics and enhanced activity (Drulis-Kawa and Dorotkiewicz-Jach, 2010). However, tissue distribution has varied among different liposomal preparations. Encapsulation of amikacin or gentamicin into conventional liposomes resulted in their rapid clearing from the bloodstream by the mononuclear phagocyte system (Schiffelers et al., 2001). While acute toxicity was reduced, the drugs accumulated in the liver and spleen and their concentrations were low in other organs. Development of long-circulating liposomes (LCLs) resulted in a broader tissue exposure. The LCL-encapsulated drugs accumulated at similar concentrations as free aminoglycosides, but their localization within the kidney differed from free aminoglycosides, thus preventing nephrotoxicity. In addition, the LCL-aminoglycosides were found to have increased efficacy as antimicrobials. Arikace®, an inhaled form of liposome-encapsulated amikacin that can enter biofilms (Meers et al., 2008), will enter Phase 2 clinical trials for the treatment of lung infections [http://www.insmed.com/arikace.php#p2]; [ClinicalTrials.gov; NCT01315678].
Until recently, this method of aminoglycoside delivery was only tested for antibacterial purposes. However, a recent study evaluated the ability of liposome-encapsulated gentamicin to suppress a PTC in the mdx mouse model of Duchenne muscular dystrophy (Yukihara et al., 2011). The gentamicin-filled liposomes were found to accumulate in the mouse skeletal muscle and the liver, but not in the brain or kidney. A significant decrease in serum creatine kinase levels and an increase in dystrophin levels were observed in treated mice. In addition, a reduction in ototoxicity was observed. These observations suggest that liposomal aminoglycosides may be a way to improve their safety for suppression therapy.
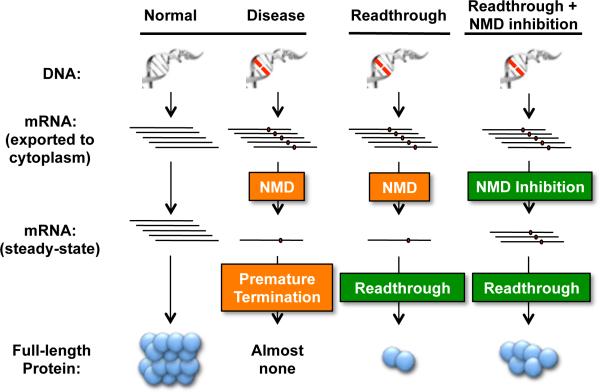
3) Decrease NMD Efficiency
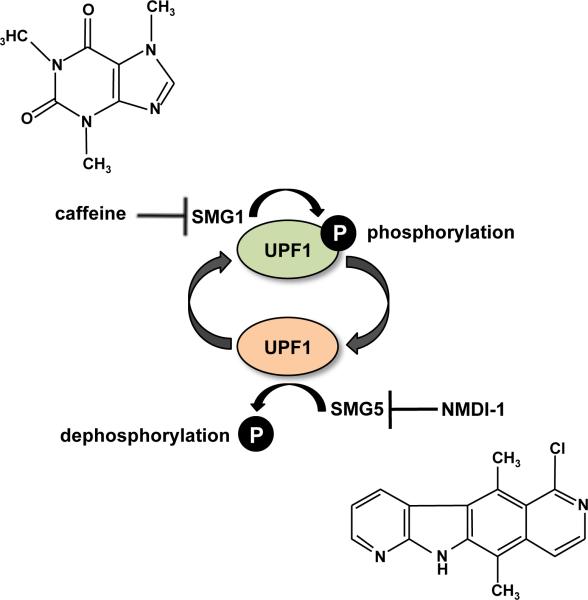
Another way to possibly increase nonsense suppression efficacy is to partially inhibit NMD to increase the pool of PTC-containing mRNA available for translation and readthrough (Figure 8). It was previously demonstrated that siRNA-mediated knockdown of NMD factor expression to reduce NMD efficiency increases the efficacy of suppression therapy in nasal epithelia derived from CF patients (Linde et al., 2007). A possible target for pharmacological NMD inhibition is the UPF1 protein (Figure 9). The NMD factor UPF1 is a phosphoprotein that must undergo a cycle of phosphorylation and dephosphorylation in order for efficient NMD to occur (Kashima et al., 2006, Ohnishi et al., 2003, Yamashita et al., 2006). Several pharmacological agents have been shown to target the UPF1 phosphorylation cycle. Phosphatidylinositol-3-kinase inhibitors such as caffeine, wortmannin, and LY294002SMG1 inhibit SMG1, a member of the phosphatidylinositol-3-kinase-like kinases that phosphorylates UPF1 (Pal et al., 2001, Usuki et al., 2004). In addition, another NMD inhibitor called NMDI-1 was recently identified from a high throughput drug screen to inhibit UPF1 dephosphorylation by blocking the interaction between UPF1 and SMG5, which recruits the PP2A phosphatase to dephosphorylate UPF1 (Durand et al., 2007).
Figure 8.
Rationale of combining suppression therapy with NMD inhibition. The presence of a nonsense mutation within an mRNA leads to premature translation termination, resulting in a truncated polypeptide that is nonfunctional and/or unstable. However, nonsense mutations often also trigger nonsense-mediated mRNA decay of the transcript that severely reduces its steady-state levels. The combination of these PTC-induced events contributes to the near complete loss of protein expression that often results in a disease state. Suppression therapy targets premature translation termination by inducing readthrough at PTCs. NMD inhibition could increase the pool of PTC-containing mRNAs available for translation and subsequent PTC suppression. The combination of suppression therapy and NMD inhibition simultaneously targets both PTC-mediated events to enhance protein restoration. [Note: a color version of this figure is available online.]
Figure 9.
NMD inhibitors. UPF1 is a critical NMD factor that undergoes a cycle of phosphorylation and de-phosphorylation, which represents a pharmacological target for NMD inhibitors. SMG1, the kinase that phosphorylates UPF1, is inhibited by caffeine. The interaction between UPF1 and SMG5, which recruits PP2A phosphatase to dephosphorylate UPF1, is blocked by NMDI-1. [Note: a color version of this figure is available online.]
In some instances, NMD inhibition alone may be beneficial in alleviating a disease phenotype. One example was documented for a collagen deficiency caused by a frameshift mutation in the COL IV α2 gene (Usuki et al., 2004). It was found that the truncated collagen VI α2 protein produced by this mutant gene retained partial function, but NMD reduced the abundance of the COL IV α2 mRNA so severely that the resulting expression of the mutant, partially functional collagen VI α2 protein was not sufficient to prevent the onset of Ullrich's syndrome. However, it was found that NMD inhibition mediated by caffeine or wortmannin restored enough truncated protein expression to alleviate the disease phenotype in fibroblasts from an Ullrich's syndrome patient. In addition, NMD factor knockdowns using RNA silencing to decrease NMD efficiency resulted in a therapeutic benefit within in vitro model systems for Ullrich's syndrome (Usuki et al., 2006) and type 2 long QT syndrome (Zarraga et al., 2011). However, NMD inhibition must be approached with caution since several NMD factors have been shown to be essential for embryonic development (Hwang and Maquat, 2011). NMD also regulates the abundance of a large number of physiological transcripts, and NMD and/or NMD factors appear to have additional roles in other cellular pathways (Isken and Maquat, 2008).
4) Combine Suppression Therapy with Other Therapeutic Approaches
The threshold of functional protein required for a therapeutic benefit dictates the effectiveness of suppression therapy. This threshold not only varies for different diseases, but also among different patient populations with the same disease. For some disorders, even enhanced forms of suppression therapy may not be capable of restoring enough functional protein to surpass the therapeutic threshold. However, suppression therapy may be combined with other therapeutic approaches to enhance the overall therapeutic benefits. For example, current treatments for the lysosomal storage disease MPS I-H, such as hematopoietic stem cell transplantation (Aldenhoven et al., 2008) and enzyme replacement therapy (ERT) (Brooks, 2002, Wraith, 2005) are unable to alleviate abnormal lysosomal glycosaminoglycan storage in all tissues. In particular, ERT is unable to treat MPS I-H neurological defects because the exogenously supplied recombinant protein is unable to cross the blood-brain barrier. Since PTC suppression agents are small molecules that cross the blood-brain barrier to some extent (McCracken et al., 1971, Riff and Jackson, 1971, Smith et al., 1988), suppression therapy may be a suitable approach to alleviate the neurological disease associated with MPS I-H. In addition, numerous drugs are under development to treat cystic fibrosis (CF) by targeting different aspects of CFTR protein localization and function depending upon the type of mutation in the affected individuals (Rogan et al., 2011, Cuthbert, 2010). For example, several drugs have been developed by Vertex Pharmaceuticals to treat CF according to the specific class of CFTR mutation (see Figure 7). VX-809 was developed for CF patients with the Class II mutation F508del that affects CFTR trafficking (Van Goor et al., 2011), and VX-770 (Kalydeco®) was developed for the Class III mutation G551D that produces correctly localized CFTR, but causes channel gating defects (Van Goor et al., 2009). In particular, drugs like VX-770 that increase CFTR gating could potentially be combined with PTC suppression therapy in patients with nonsense mutations to further enhance the activity of the CFTR protein restored by suppression.
Summary
Due to improvements in genomic analysis and genetic testing, mutation detection is becoming routine for many genetic diseases. This has led to an increase in the development of mutation-specific drugs, thus making personalized medicine a reality. Nonsense suppression is an example of personalized medicine that alters how a specific class of mutation is recognized by the translation machinery. Thus, it is not restricted to any specific disease. The need for such a broad therapeutic class of drugs is dire, since approximately 2.5 million patients in the U.S. alone have genetic disorders attributable to nonsense mutations. Notably, progress is being made in the development of PTC suppression as a clinical therapy. PTC124 is currently in Phase 3 clinical trials for the treatment of cystic fibrosis and Duchenne muscular dystrophy. However, suppression therapy in its current form may be effective in only a subset of patients with nonsense mutations. Accordingly, improvements in the efficacy of this approach are needed in order to broaden its applicability to a larger number of patients with diverse disorders. Active research investigating the mechanisms of translation termination, PTC suppression, and NMD is likely to provide additional pharmaceutical targets for PTC suppression that will enhance the effectiveness of this novel therapeutic approach.
Acknowledgments
DECLARATION OF INTERESTS: The authors acknowledge grant support from the NIH (P30 DK072482, RO1 NS057412, RO1 GM0688554 and RO1 GM094792) and the University of Pennsylvania. DMB is a paid consultant of PTC Therapeutics, Inc.
REFERENCES
- ALDENHOVEN M, BOELENS JJ, DE KONING TJ. The clinical outcome of Hurler syndrome after stem cell transplantation. Biol Blood Marrow Transplant. 2008;14:485–98. doi: 10.1016/j.bbmt.2008.01.009. [DOI] [PubMed] [Google Scholar]
- ALI BH, AL ZA'ABI M, BLUNDEN G, NEMMAR A. Experimental gentamicin nephrotoxicity and agents that modify it: a mini-review of recent research. Basic Clin Pharmacol Toxicol. 2011;109:225–32. doi: 10.1111/j.1742-7843.2011.00728.x. [DOI] [PubMed] [Google Scholar]
- ALKALAEVA EZ, PISAREV AV, FROLOVA LY, KISSELEV LL, PESTOVA TV. In Vitro Reconstitution of Eukaryotic Translation Reveals Cooperativity between Release Factors eRF1 and eRF3. Cell. 2006;125:1125–36. doi: 10.1016/j.cell.2006.04.035. [DOI] [PubMed] [Google Scholar]
- AMARAL MD. Processing of CFTR: traversing the cellular maze--how much CFTR needs to go through to avoid cystic fibrosis? Pediatr Pulmonol. 2005;39:479–91. doi: 10.1002/ppul.20168. [DOI] [PubMed] [Google Scholar]
- AMRANI N, GANESAN R, KERVESTIN S, MANGUS DA, GHOSH S, JACOBSON A. A faux 3'-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature. 2004;432:112–8. doi: 10.1038/nature03060. [DOI] [PubMed] [Google Scholar]
- ARAKAWA M, SHIOZUKA M, NAKAYAMA Y, HARA T, HAMADA M, KONDO S, IKEDA D, TAKAHASHI Y, SAWA R, NONOMURA Y, SHEYKHOLESLAMI K, KONDO K, KAGA K, KITAMURA T, SUZUKI-MIYAGOE Y, TAKEDA S, MATSUDA R. Negamycin restores dystrophin expression in skeletal and cardiac muscles of mdx mice. J Biochem (Tokyo) 2003;134:751–8. doi: 10.1093/jb/mvg203. [DOI] [PubMed] [Google Scholar]
- AULD DS, THORNE N, MAGUIRE WF, INGLESE J. Mechanism of PTC124 activity in cell-based luciferase assays of nonsense codon suppression. Proc Natl Acad Sci U S A. 2009;106:3585–90. doi: 10.1073/pnas.0813345106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BARTON-DAVIS ER, CORDIER L, SHOTURMA DI, LELAND SE, SWEENEY HL. Aminoglycoside antibiotics restore dystrophin function to skeletal muscles of mdx mice. J Clin Invest. 1999;104:375–81. doi: 10.1172/JCI7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BEAUCHAMP D, LABRECQUE G. Aminoglycoside nephrotoxicity: do time and frequency of administration matter? Curr Opin Crit Care. 2001;7:401–8. doi: 10.1097/00075198-200112000-00006. [DOI] [PubMed] [Google Scholar]
- BEAUCHAMP D, LAURENT G, MALDAGUE P, ABID S, KISHORE BK, TULKENS PM. Protection against gentamicin-induced early renal alterations (phospholipidosis and increased DNA synthesis) by coadministration of poly-L-aspartic acid. J Pharmacol Exp Ther. 1990;255:858–66. [PubMed] [Google Scholar]
- BHUVANAGIRI M, SCHLITTER AM, HENTZE MW, KULOZIK AE. NMD: RNA biology meets human genetic medicine. Biochem J. 2010;430:365–77. doi: 10.1042/BJ20100699. [DOI] [PubMed] [Google Scholar]
- BONETTI B, FU L, MOON J, BEDWELL DM. The efficiency of translation termination is determined by a synergistic interplay between upstream and downstream sequences in Saccharomyces cerevisiae. J Mol Biol. 1995;251:334–45. doi: 10.1006/jmbi.1995.0438. [DOI] [PubMed] [Google Scholar]
- BRENDEL C, BELAKHOV V, WERNER H, WEGENER E, GARTNER J, NUDELMAN I, BAASOV T, HUPPKE P. Readthrough of nonsense mutations in Rett syndrome: evaluation of novel aminoglycosides and generation of a new mouse model. J Mol Med. 2010;89:389–98. doi: 10.1007/s00109-010-0704-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BROOKS DA. Alpha-L-iduronidase and enzyme replacement therapy for mucopolysaccharidosis I. Expert Opin Biol Ther. 2002;2:967–76. doi: 10.1517/14712598.2.8.967. [DOI] [PubMed] [Google Scholar]
- BROWN CM, STOCKWELL PA, TROTMAN CN, TATE WP. Sequence analysis suggests that tetra-nucleotides signal the termination of protein synthesis in eukaryotes. Nucleic Acids Res. 1990;18:6339–45. doi: 10.1093/nar/18.21.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CAPECCHI MR. Polypeptide chain termination in vitro: isolation of a release factor. Proc Natl Acad Sci U S A. 1967;58:1144–51. doi: 10.1073/pnas.58.3.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CASKEY CT, BEAUDET AL, SCOLNICK EM, ROSMAN M. Hydrolysis of fMettRNA by peptidyl transferase. Proc Natl Acad Sci U S A. 1971;68:3163–7. doi: 10.1073/pnas.68.12.3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CASSAN M, ROUSSET JP. UAG readthrough in mammalian cells: effect of upstream and downstream stop codon contexts reveal different signals. BMC Mol Biol. 2001;2:3. doi: 10.1186/1471-2199-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHAMBERLAIN J. Dystrophin levels required for genetic correction of Duchenne Muscular Dystrophy. Basic Appl Myol. 1997;7:251–5. [Google Scholar]
- CHANG CW, HUI Y, ELCHERT B, WANG J, LI J, RAI R. Pyranmycins, a novel class of aminoglycosides with improved acid stability: the SAR of D-pyranoses on ring III of pyranmycin. Org Lett. 2002;4:4603–6. doi: 10.1021/ol0269042. [DOI] [PubMed] [Google Scholar]
- CLANCY JP, BEBOK Z, RUIZ F, KING C, JONES J, WALKER L, GREER H, HONG J, WING L, MACALUSO M, LYRENE R, SORSCHER EJ, BEDWELL DM. Evidence that systemic gentamicin suppresses premature stop mutations in patients with cystic fibrosis. Am J Respir Crit Care Med. 2001;163:1683–92. doi: 10.1164/ajrccm.163.7.2004001. [DOI] [PubMed] [Google Scholar]
- CLANCY JP, ROWE SM, BEBOK Z, AITKEN ML, GIBSON R, ZEITLIN P, BERCLAZ P, MOSS R, KNOWLES MR, OSTER RA, MAYER-HAMBLETT N, RAMSEY B. No Detectable Improvements in CFTR by Nasal Aminoglycosides in CF Patients with Stop Mutations. Am J Respir Cell Mol Biol. 2007;37:57–66. doi: 10.1165/rcmb.2006-0173OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COSSON B, COUTURIER A, CHABELSKAYA S, KIKTEV D, INGE-VECHTOMOV S, PHILIPPE M, ZHOURAVLEVA G. Poly(A)-binding protein acts in translation termination via eukaryotic release factor 3 interaction and does not influence [PSI(+)] propagation. Mol Cell Biol. 2002;22:3301–15. doi: 10.1128/MCB.22.10.3301-3315.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CUTHBERT A. New horizons in the treatment of cystic fibrosis. Br J Pharmacol. 2010;163:173–83. doi: 10.1111/j.1476-5381.2010.01137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DALPHIN ME, STOCKWELL PA, TATE WP, BROWN CM. TransTerm, the translational signal database, extended to include full coding sequences and untranslated regions. Nucleic Acids Res. 1999;27:293–4. doi: 10.1093/nar/27.1.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DARRAT I, AHMAD N, SEIDMAN K, SEIDMAN MD. Auditory research involving antioxidants. Curr Opin Otolaryngol Head Neck Surg. 2007;15:358–63. doi: 10.1097/MOO.0b013e3282efa641. [DOI] [PubMed] [Google Scholar]
- DAVIDSON DJ, ROLFE M. Mouse models of cystic fibrosis. Trends Genet. 2001;17:S29–37. doi: 10.1016/s0168-9525(01)02452-0. [DOI] [PubMed] [Google Scholar]
- DE BROE ME, PAULUS GJ, VERPOOTEN GA, ROELS F, BUYSSENS N, WEDEEN R, VAN HOOF F, TULKENS PM. Early effects of gentamicin, tobramycin, and amikacin on the human kidney. Kidney Int. 1984;25:643–52. doi: 10.1038/ki.1984.69. [DOI] [PubMed] [Google Scholar]
- DRULIS-KAWA Z, DOROTKIEWICZ-JACH A. Liposomes as delivery systems for antibiotics. Int J Pharm. 2010;387:187–98. doi: 10.1016/j.ijpharm.2009.11.033. [DOI] [PubMed] [Google Scholar]
- DU L, DAMOISEAUX R, NAHAS S, GAO K, HU H, POLLARD JM, GOLDSTINE J, JUNG ME, HENNING SM, BERTONI C, GATTI RA. Nonaminoglycoside compounds induce readthrough of nonsense mutations. J Exp Med. 2009a;206:2285–97. doi: 10.1084/jem.20081940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DU M, JONES JR, LANIER J, KEELING KM, LINDSEY JR, TOUSSON A, BEBOK Z, WHITSETT JA, DEY CR, COLLEDGE WH, EVANS MJ, SORSCHER EJ, BEDWELL DM. Aminoglycoside suppression of a premature stop mutation in a Cftr-/- mouse carrying a human CFTR-G542X transgene. J Mol Med. 2002;80:595–604. doi: 10.1007/s00109-002-0363-1. [DOI] [PubMed] [Google Scholar]
- DU M, KEELING KM, FAN L, LIU X, BEDWELL DM. Poly-L-aspartic Acid Enhances and Prolongs Gentamicin-mediated Suppression of the CFTR-G542X Mutation in a Cystic Fibrosis Mouse Model. J Biol Chem. 2009b;284:6885–92. doi: 10.1074/jbc.M806728200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DU M, KEELING KM, FAN L, LIU X, KOVACS T, SORSCHER E, BEDWELL DM. Clinical doses of amikacin provide more effective suppression of the human CFTR-G542X stop mutation than gentamicin in a transgenic CF mouse model. J Mol Med. 2006;84:573–82. doi: 10.1007/s00109-006-0045-5. [DOI] [PubMed] [Google Scholar]
- DU M, LIU X, WELCH EM, HIRAWAT S, PELTZ SW, BEDWELL DM. PTC124 is an orally bioavailable compound that promotes suppression of the human CFTR-G542X nonsense allele in a CF mouse model. Proc Natl Acad Sci U S A. 2008;105:2064–9. doi: 10.1073/pnas.0711795105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DUNANT P, WALTER MC, KARPATI G, LOCHMULLER H. Gentamicin fails to increase dystrophin expression in dystrophin-deficient muscle. Muscle Nerve. 2003;27:624–7. doi: 10.1002/mus.10341. [DOI] [PubMed] [Google Scholar]
- DUNKLE JA, CATE JH. Ribosome structure and dynamics during translocation and termination. Annu Rev Biophys. 2010;39:227–44. doi: 10.1146/annurev.biophys.37.032807.125954. [DOI] [PubMed] [Google Scholar]
- DURAND S, COUGOT N, MAHUTEAU-BETZER F, NGUYEN CH, GRIERSON DS, BERTRAND E, TAZI J, LEJEUNE F. Inhibition of nonsense-mediated mRNA decay (NMD) by a new chemical molecule reveals the dynamic of NMD factors in P-bodies. J Cell Biol. 2007;178:1145–60. doi: 10.1083/jcb.200611086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ELCHERT B, LI J, WANG J, HUI Y, RAI R, PTAK R, WARD P, TAKEMOTO JY, BENSACI M, CHANG CW. Application of the synthetic aminosugars for glycodiversification: synthesis and antimicrobial studies of pyranmycin. J Org Chem. 2004;69:1513–23. doi: 10.1021/jo035290r. [DOI] [PubMed] [Google Scholar]
- FAIRCLOUGH RJ, BAREJA A, DAVIES KE. Progress in therapy for Duchenne muscular dystrophy. Exp Physiol. 2011;96:1101–13. doi: 10.1113/expphysiol.2010.053025. [DOI] [PubMed] [Google Scholar]
- FAN-MINOGUE H, BEDWELL DM. Eukaryotic ribosomal RNA determinants of aminoglycoside resistance and their role in translational fidelity. RNA. 2007;14:148–57. doi: 10.1261/rna.805208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAN-MINOGUE H, DU M, PISAREV AV, KALLMEYER AK, SALAS-MARCO J, KEELING KM, THOMPSON SR, PESTOVA TV, BEDWELL DM. Distinct eRF3 requirements suggest alternate eRF1 conformations mediate peptide release during eukaryotic translation termination. Mol Cell. 2008;30:599–609. doi: 10.1016/j.molcel.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FEARON K, MCCLENDON V, BONETTI B, BEDWELL DM. Premature translation termination mutations are efficiently suppressed in a highly conserved region of yeast Ste6p, a member of the ATP-binding cassette (ABC) transporter family. J Biol Chem. 1994;269:17802–8. [PubMed] [Google Scholar]
- FINKEL RS. Read-through strategies for suppression of nonsense mutations in Duchenne/ Becker muscular dystrophy: aminoglycosides and ataluren (PTC124). J Child Neurol. 2010;25:1158–64. doi: 10.1177/0883073810371129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FOURMY D, RECHT MI, BLANCHARD SC, PUGLISI JD. Structure of the A site of Escherichia coli 16S ribosomal RNA complexed with an aminoglycoside antibiotic. Science. 1996;274:1367–71. doi: 10.1126/science.274.5291.1367. [DOI] [PubMed] [Google Scholar]
- FREISTROFFER DV, KWIATKOWSKI M, BUCKINGHAM RH, EHRENBERG M. The accuracy of codon recognition by polypeptide release factors. Proc Natl Acad Sci U S A. 2000;97:2046–51. doi: 10.1073/pnas.030541097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FROLOVA L, LE GOFF X, RASMUSSEN HH, CHEPEREGIN S, DRUGEON G, KRESS M, ARMAN I, HAENNI AL, CELIS JE, PHILIPPE M, JUSTESEN J, KISSELEV L. A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature. 1994;372:701–3. doi: 10.1038/372701a0. [DOI] [PubMed] [Google Scholar]
- FROLOVA L, LE GOFF X, ZHOURAVLEVA G, DAVYDOVA E, PHILIPPE M, KISSELEV L. Eukaryotic polypeptide chain release factor eRF3 is an eRF1- and ribosome-dependent guanosine triphosphatase. RNA. 1996;2:334–41. [PMC free article] [PubMed] [Google Scholar]
- GILBERT DN, WOOD CA, KOHLHEPP SJ, KOHNEN PW, HOUGHTON DC, FINKBEINER HC, LINDSLEY J, BENNETT WM. Polyaspartic acid prevents experimental aminoglycoside nephrotoxicity. J Infect Dis. 1989;159:945–53. doi: 10.1093/infdis/159.5.945. [DOI] [PubMed] [Google Scholar]
- GOLDMANN T, OVERLACK N, WOLFRUM U, NAGEL-WOLFRUM K. PTC124 mediated translational read-through of a nonsense mutation causing Usher type 1C. Hum Gene Ther. 2011;22:537–47. doi: 10.1089/hum.2010.067. [DOI] [PubMed] [Google Scholar]
- GOLDMANN T, REBIBO-SABBAH A, OVERLACK N, NUDELMAN I, BELAKHOV V, BAASOV T, BEN-YOSEF T, WOLFRUM U, NAGEL-WOLFRUM K. Beneficial read-through of a USH1C nonsense mutation by designed aminoglycoside NB30 in the retina. Invest Ophthalmol Vis Sci. 2010;51:6671–80. doi: 10.1167/iovs.10-5741. [DOI] [PubMed] [Google Scholar]
- GORINI L, KATAJA E. Phenotypic Repair by Streptomycin of Defective Genotypes in E. Coli. Proc Natl Acad Sci U S A. 1964;51:487–93. doi: 10.1073/pnas.51.3.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GUAN MX, FISCHEL-GHODSIAN N, ATTARDI G. A biochemical basis for the inherited susceptibility to aminoglycoside ototoxicity. Hum Mol Genet. 2000;9:1787–93. doi: 10.1093/hmg/9.12.1787. [DOI] [PubMed] [Google Scholar]
- GUTHRIE OW. Aminoglycoside induced ototoxicity. Toxicology. 2008;249:91–6. doi: 10.1016/j.tox.2008.04.015. [DOI] [PubMed] [Google Scholar]
- HIRAWAT S, NORTHCUTT VJ, WELCH EM, ELFRING GL, HWANG S, ALMSTEAD NG, JU W, MILLER LL. Phase 1 safety and PK study of PTC124 for nonsense-mutation suppression therapy of cystic fibrosis. Pediatric Pulmonology. 2004;38:248. [Google Scholar]
- HIRAWAT S, WELCH EM, ELFRING GL, NORTHCUTT VJ, PAUSHKIN S, HWANG S, LEONARD EM, ALMSTEAD NG, JU W, PELTZ SW, MILLER LL. Safety, tolerability, and pharmacokinetics of PTC124, a nonaminoglycoside nonsense mutation suppressor, following single- and multiple-dose administration to healthy male and female adult volunteers. J Clin Pharmacol. 2007;47:430–44. doi: 10.1177/0091270006297140. [DOI] [PubMed] [Google Scholar]
- HIROKAWA G, NIJMAN RM, RAJ VS, KAJI H, IGARASHI K, KAJI A. The role of ribosome recycling factor in dissociation of 70S ribosomes into subunits. RNA. 2005;11:1317–28. doi: 10.1261/rna.2520405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOSHINO S, IMAI M, KOBAYASHI T, UCHIDA N, KATADA T. The eukaryotic polypeptide chain releasing factor (eRF3/GSPT) carrying the translation termination signal to the 3'-Poly(A) tail of mRNA. Direct association of erf3/GSPT with polyadenylate-binding protein. J Biol Chem. 1999;274:16677–80. doi: 10.1074/jbc.274.24.16677. [DOI] [PubMed] [Google Scholar]
- HULKA GF, PRAZMA J, BROWNLEE RE, PULVER S, PILLSBURY HC. Use of poly-l-aspartic acid to inhibit aminoglycoside cochlear ototoxicity. Am J Otol. 1993;14:352–6. [PubMed] [Google Scholar]
- HUTH ME, RICCI AJ, CHENG AG. Mechanisms of aminoglycoside ototoxicity and targets of hair cell protection. Int J Otolaryngol. 2011;2011:937861. doi: 10.1155/2011/937861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HWANG J, MAQUAT LE. Nonsense-mediated mRNA decay (NMD) in animal embryogenesis: to die or not to die, that is the question. Curr Opin Genet Dev. 2011 doi: 10.1016/j.gde.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- INGLESE J, THORNE N, AULD DS. Reply to Peltz et al: Post-translational stabilization of the firefly luciferase reporter by PTC124 (Ataluren). Proc Natl Acad Sci U S A. 2009;106:E65. [Google Scholar]
- ISKEN O, MAQUAT LE. Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev. 2007;21:1833–3856. doi: 10.1101/gad.1566807. [DOI] [PubMed] [Google Scholar]
- ISKEN O, MAQUAT LE. The multiple lives of NMD factors: balancing roles in gene and genome regulation. Nat Rev Genet. 2008;9:699–712. doi: 10.1038/nrg2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JACKSON RJ, HELLEN CU, PESTOVA TV. Termination and post-termination events in eukaryotic translation. Adv Protein Chem Struct Biol. 2012;86:45–93. doi: 10.1016/B978-0-12-386497-0.00002-5. [DOI] [PubMed] [Google Scholar]
- JIN H, KELLEY AC, LOAKES D, RAMAKRISHNAN V. Structure of the 70S ribosome bound to release factor 2 and a substrate analog provides insights into catalysis of peptide release. Proc Natl Acad Sci U S A. 2010;107:8593–8. doi: 10.1073/pnas.1003995107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JUNG ME, KU JM, DU L, HU H, GATTI RA. Synthesis and evaluation of compounds that induce readthrough of premature termination codons. Bioorg Med Chem Lett. 2011;21:5842–8. doi: 10.1016/j.bmcl.2011.07.107. [DOI] [PubMed] [Google Scholar]
- KANDASAMY J, ATIA-GLIKIN D, BELAKHOV V, BAASOV T. Repairing faulty genes by aminoglycosides: Identification of new pharmacophore with enhanced suppression of disease-causing nonsense mutations. Med Chem Commun. 2011;2:165–71. doi: 10.1016/j.bmc.2010.03.060. [DOI] [PubMed] [Google Scholar]
- KASHIMA I, YAMASHITA A, IZUMI N, KATAOKA N, MORISHITA R, HOSHINO S, OHNO M, DREYFUSS G, OHNO S. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 2006;20:355–67. doi: 10.1101/gad.1389006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KAWAMOTO K, SHA SH, MINODA R, IZUMIKAWA M, KURIYAMA H, SCHACHT J, RAPHAEL Y. Antioxidant gene therapy can protect hearing and hair cells from ototoxicity. Mol Ther. 2004;9:173–81. doi: 10.1016/j.ymthe.2003.11.020. [DOI] [PubMed] [Google Scholar]
- KEELING KM. PhD. University of Alabama at Birmingham; 2000. Suppression of premature stop mutations using aminoglycosides. [Google Scholar]
- KEELING KM, BEDWELL DM. Recoding therapies for genetic diseases. In: ATKINS JF, GESTELAND RF, editors. Recoding: Expansion of Decoding Rules Enriches Gene Expression. Springer Publishing; New York: 2010. [Google Scholar]
- KEELING KM, BEDWELL DM. Suppression of nonsense mutations as a therapeutic approach to treat genetic diseases. WIREs RNA. 2011 doi: 10.1002/wrna.95. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KEELING KM, BROOKS DA, HOPWOOD JJ, LI P, THOMPSON JN, BEDWELL DM. Gentamicin-mediated suppression of Hurler syndrome stop mutations restores a low level of alpha-L-iduronidase activity and reduces lysosomal glycosaminoglycan accumulation. Hum Mol Genet. 2001;10:291–9. doi: 10.1093/hmg/10.3.291. [DOI] [PubMed] [Google Scholar]
- KEREM E. Pharmacologic therapy for stop mutations: how much CFTR activity is enough? Curr Opin Pulm Med. 2004;10:547–52. doi: 10.1097/01.mcp.0000141247.22078.46. [DOI] [PubMed] [Google Scholar]
- KEREM E, HIRAWAT S, ARMONI S, YAAKOV Y, SHOSEYOV D, COHEN M, NISSIMRAFINIA M, BLAU H, RIVLIN J, AVIRAM M, ELFRING GL, NORTHCUTT VJ, MILLER LL, KEREM B, WILSCHANSKI M. Effectiveness of PTC124 treatment of cystic fibrosis caused by nonsense mutations: a prospective phase II trial. Lancet. 2008;372:719–27. doi: 10.1016/S0140-6736(08)61168-X. [DOI] [PubMed] [Google Scholar]
- KIM CHIAW P, ECKFORD PD, BEAR CE. Insights into the mechanisms underlying CFTR channel activity, the molecular basis for cystic fibrosis and strategies for therapy. Essays Biochem. 2011;50:233–48. doi: 10.1042/bse0500233. [DOI] [PubMed] [Google Scholar]
- KISHORE BK, IBRAHIM S, LAMBRICHT P, LAURENT G, MALDAGUE P, TULKENS PM. Comparative assessment of poly-L-aspartic and poly-L-glutamic acids as protectants against gentamicin-induced renal lysosomal phospholipidosis, phospholipiduria and cell proliferation in rats. J Pharmacol Exp Ther. 1992;262:424–32. [PubMed] [Google Scholar]
- KOROSTELEV A, ASAHARA H, LANCASTER L, LAURBERG M, HIRSCHI A, ZHU J, TRAKHANOV S, SCOTT WG, NOLLER HF. Crystal structure of a translation termination complex formed with release factor RF2. Proc Natl Acad Sci U S A. 2008;105:19684–9. doi: 10.1073/pnas.0810953105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KOROSTELEV A, ZHU J, ASAHARA H, NOLLER HF. Recognition of the amber UAG stop codon by release factor RF1. EMBO J. 2010;29:2577–85. doi: 10.1038/emboj.2010.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KOROSTELEV AA. Structural aspects of translation termination on the ribosome. RNA. 2011;17:1409–21. doi: 10.1261/rna.2733411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KRAMER EB, VALLABHANENI H, MAYER LM, FARABAUGH PJ. A comprehensive analysis of translational missense errors in the yeast Saccharomyces cerevisiae. RNA. 2010;16:1797–808. doi: 10.1261/rna.2201210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAURBERG M, ASAHARA H, KOROSTELEV A, ZHU J, TRAKHANOV S, NOLLER HF. Structural basis for translation termination on the 70S ribosome. Nature. 2008;454:852–7. doi: 10.1038/nature07115. [DOI] [PubMed] [Google Scholar]
- LAURENT G, CARLIER MB, ROLLMAN B, VAN HOOF F, TULKENS P. Mechanism of aminoglycoside-induced lysosomal phospholipidosis: in vitro and in vivo studies with gentamicin and amikacin. Biochem Pharmacol. 1982;31:3861–70. doi: 10.1016/0006-2952(82)90303-3. [DOI] [PubMed] [Google Scholar]
- LEDERBERG EM, CAVALLI-SFORZA L, LEDERBERG J. Interaction of Streptomycin and a Suppressor for Galactose Fermentation in E. Coli K-12. Proc Natl Acad Sci U S A. 1964;51:678–82. doi: 10.1073/pnas.51.4.678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI J, WANG J, CZYRYCA PG, CHANG H, ORSAK TW, EVANSON R, CHANG CW. Application of glycodiversification: expedient synthesis and antibacterial evaluation of a library of kanamycin B analogues. Org Lett. 2004;6:1381–4. doi: 10.1021/ol0497685. [DOI] [PubMed] [Google Scholar]
- LIANG H, CAVALCANTI AR, LANDWEBER LF. Conservation of tandem stop codons in yeasts. Genome Biol. 2005;6:R31. doi: 10.1186/gb-2005-6-4-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LINDE L, BOELZ S, NISSIM-RAFINIA M, OREN YS, WILSCHANSKI M, YAACOV Y, VIRGILIS D, NEU-YILIK G, KULOZIK AE, KEREM E, KEREM B. Nonsense-mediated mRNA decay affects nonsense transcript levels and governs response of cystic fibrosis patients to gentamicin. J Clin Invest. 2007;117:683–92. doi: 10.1172/JCI28523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LINDE L, KEREM B. Introducing sense into nonsense in treatments of human genetic diseases. Trends Genet. 2008;24:552–63. doi: 10.1016/j.tig.2008.08.010. [DOI] [PubMed] [Google Scholar]
- LOPEZ-NOVOA JM, QUIROS Y, VICENTE L, MORALES AI, LOPEZ-HERNANDEZ FJ. New insights into the mechanism of aminoglycoside nephrotoxicity: an integrative point of view. Kidney Int. 2011;79:33–45. doi: 10.1038/ki.2010.337. [DOI] [PubMed] [Google Scholar]
- LYNCH SR, PUGLISI JD. Structure of a eukaryotic decoding region A-site RNA. J Mol Biol. 2001;306:1023–35. doi: 10.1006/jmbi.2000.4419. [DOI] [PubMed] [Google Scholar]
- MALIK V, RODINO-KLAPAC LR, VIOLLET L, WALL C, KING W, AL-DAHHAK R, LEWIS S, SHILLING CJ, KOTA J, SERRANO-MUNUERA C, HAYES J, MAHAN JD, CAMPBELL KJ, BANWELL B, DASOUKI M, WATTS V, SIVAKUMAR K, BIEN-WILLNER R, FLANIGAN KM, SAHENK Z, BAROHN RJ, WALKER CM, MENDELL JR. Gentamicin-induced readthrough of stop codons in Duchenne muscular dystrophy. Ann Neurol. 2010;67:771–80. doi: 10.1002/ana.22024. [DOI] [PubMed] [Google Scholar]
- MANUVAKHOVA M, KEELING K, BEDWELL DM. Aminoglycoside antibiotics mediate context-dependent suppression of termination codons in a mammalian translation system. RNA. 2000;6:1044–55. doi: 10.1017/s1355838200000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARTIN R. On the relationship between preferred termination codon contexts and nonsense suppression in human cells. Nucleic Acids Res. 1994;22:15–9. doi: 10.1093/nar/22.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARTIN R, MOGG AE, HEYWOOD LA, NITSCHKE L, BURKE JF. Aminoglycoside suppression at UAG, UAA and UGA codons in Escherichia coli and human tissue culture cells. Mol Gen Genet. 1989;217:411–8. doi: 10.1007/BF02464911. [DOI] [PubMed] [Google Scholar]
- MATTIS VB, EBERT AD, FOSSO MY, CHANG CW, LORSON CL. Delivery of a read-through inducing compound, TC007, lessens the severity of a spinal muscular atrophy animal model. Hum Mol Genet. 2009a;18:3906–13. doi: 10.1093/hmg/ddp333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MATTIS VB, FOSSO MY, CHANG CW, LORSON CL. Subcutaneous administration of TC007 reduces disease severity in an animal model of SMA. BMC Neurosci. 2009b;10:142. doi: 10.1186/1471-2202-10-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MATTIS VB, RAI R, WANG J, CHANG CW, COADY T, LORSON CL. Novel aminoglycosides increase SMN levels in spinal muscular atrophy fibroblasts. Hum Genet. 2006;120:589–601. doi: 10.1007/s00439-006-0245-7. [DOI] [PubMed] [Google Scholar]
- MCCAUGHAN KK, BROWN CM, DALPHIN ME, BERRY MJ, TATE WP. Translational termination efficiency in mammals is influenced by the base following the stop codon. Proc Natl Acad Sci U S A. 1995;92:5431–5. doi: 10.1073/pnas.92.12.5431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MCCRACKEN GH, JR., CHRANE DF, THOMAS ML. Pharmacologic evaluation of gentamicin in newborn infants. J Infect Dis. 1971;124(Suppl 124):214. doi: 10.1093/infdis/124.supplement_1.s214. [DOI] [PubMed] [Google Scholar]
- MEERS P, NEVILLE M, MALININ V, SCOTTO AW, SARDARYAN G, KURUMUNDA R, MACKINSON C, JAMES G, FISHER S, PERKINS WR. Biofilm penetration, triggered release and in vivo activity of inhaled liposomal amikacin in chronic Pseudomonas aeruginosa lung infections. J Antimicrob Chemother. 2008;61:859–68. doi: 10.1093/jac/dkn059. [DOI] [PubMed] [Google Scholar]
- MOAZED D, NOLLER HF. Transfer RNA shields specific nucleotides in 16S ribosomal RNA from attack by chemical probes. Cell. 1986;47:985–94. doi: 10.1016/0092-8674(86)90813-5. [DOI] [PubMed] [Google Scholar]
- MUHLEMANN O, EBERLE AB, STALDER L, ZAMUDIO OROZCO R. Recognition and elimination of nonsense mRNA. Biochim Biophys Acta. 2008;1779:538–49. doi: 10.1016/j.bbagrm.2008.06.012. [DOI] [PubMed] [Google Scholar]
- NOWAK KJ, DAVIES KE. Duchenne muscular dystrophy and dystrophin: pathogenesis and opportunities for treatment. EMBO Rep. 2004;5:872–6. doi: 10.1038/sj.embor.7400221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NUDELMAN I, GLIKIN D, SMOLKIN B, HAINRICHSON M, BELAKHOV V, BAASOV T. Repairing faulty genes by aminoglycosides: development of new derivatives of geneticin (G418) with enhanced suppression of diseases-causing nonsense mutations. Bioorg Med Chem. 2010;18:3735–46. doi: 10.1016/j.bmc.2010.03.060. [DOI] [PubMed] [Google Scholar]
- NUDELMAN I, REBIBO-SABBAH A, CHERNIAVSKY M, BELAKHOV V, HAINRICHSON M, CHEN F, SCHACHT J, PILCH DS, BEN-YOSEF T, BAASOV T. Development of Novel Aminoglycoside (NB54) with Reduced Toxicity and Enhanced Suppression of Disease-Causing Premature Stop Mutations. J Med Chem. 2009;52:2836–45. doi: 10.1021/jm801640k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NUDELMAN I, REBIBO-SABBAH A, SHALLOM-SHEZIFI D, HAINRICHSON M, STAHL I, BEN-YOSEF T, BAASOV T. Redesign of aminoglycosides for treatment of human genetic diseases caused by premature stop mutations. Bioorg Med Chem Lett. 2006;16:6310–5. doi: 10.1016/j.bmcl.2006.09.013. [DOI] [PubMed] [Google Scholar]
- OGLE JM, BRODERSEN DE, CLEMONS WM, JR., TARRY MJ, CARTER AP, RAMAKRISHNAN V. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- OGLE JM, MURPHY FV, TARRY MJ, RAMAKRISHNAN V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002;111:721–32. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- OHNISHI T, YAMASHITA A, KASHIMA I, SCHELL T, ANDERS KR, GRIMSON A, HACHIYA T, HENTZE MW, ANDERSON P, OHNO S. Phosphorylation of hUPF1 induces formation of mRNA surveillance complexes containing hSMG-5 and hSMG-7. Mol Cell. 2003;12:1187–200. doi: 10.1016/s1097-2765(03)00443-x. [DOI] [PubMed] [Google Scholar]
- PAL M, ISHIGAKI Y, NAGY E, MAQUAT LE. Evidence that phosphorylation of human Upfl protein varies with intracellular location and is mediated by a wortmannin-sensitive and rapamycin-sensitive PI 3-kinase-related kinase signaling pathway. RNA. 2001;7:5–15. doi: 10.1017/s1355838201000127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PAPE T, WINTERMEYER W, RODNINA MV. Conformational switch in the decoding region of 16S rRNA during aminoacyl-tRNA selection on the ribosome. Nat Struct Biol. 2000;7:104–7. doi: 10.1038/72364. [DOI] [PubMed] [Google Scholar]
- PARKER J. Errors and alternatives in reading the universal genetic code. Microbiol Rev. 1989;53:273–98. doi: 10.1128/mr.53.3.273-298.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PELTZ SW, WELCH EM, JACOBSON A, TROTTA CR, NARYSHKIN N, SWEENEY HL, BEDWELL DM. Nonsense suppression activity of PTC124 (ataluren). Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0901936106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PESKE F, RODNINA MV, WINTERMEYER W. Sequence of steps in ribosome recycling as defined by kinetic analysis. Mol Cell. 2005;18:403–12. doi: 10.1016/j.molcel.2005.04.009. [DOI] [PubMed] [Google Scholar]
- PHILLIPS-JONES MK, HILL LS, ATKINSON J, MARTIN R. Context effects on misreading and suppression at UAG codons in human cells. Mol Cell Biol. 1995;15:6593–600. doi: 10.1128/mcb.15.12.6593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PISAREV AV, SKABKIN MA, PISAREVA VP, SKABKINA OV, RAKOTONDRAFARA AM, HENTZE MW, HELLEN CU, PESTOVA TV. The role of ABCE1 in eukaryotic posttermination ribosomal recycling. Mol Cell. 2010;37:196–210. doi: 10.1016/j.molcel.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- POLITANO L, NIGRO G, NIGRO V, PILUSO G, PAPPARELLA S, PACIELLO O, COMI LI. Gentamicin administration in Duchenne patients with premature stop codon. Preliminary results. Acta Myol. 2003;22:15–21. [PubMed] [Google Scholar]
- QIAN Y, GUAN MX. Interaction of aminoglycosides with human mitochondrial 12S rRNA carrying the deafness-associated mutation. Antimicrob Agents Chemother. 2009;53:4612–8. doi: 10.1128/AAC.00965-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- REBIBO-SABBAH A, NUDELMAN I, AHMED ZM, BAASOV T, BEN-YOSEF T. In vitro and ex vivo suppression by aminoglycosides of PCDH15 nonsense mutations underlying type 1 Usher syndrome. Hum Genet. 2007;122:373–81. doi: 10.1007/s00439-007-0410-7. [DOI] [PubMed] [Google Scholar]
- RECHT MI, DOUTHWAITE S, PUGLISI JD. Basis for prokaryotic specificity of action of aminoglycoside antibiotics. EMBO J. 1999;18:3133–8. doi: 10.1093/emboj/18.11.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- REITER RJ, TAN DX, KORKMAZ A, FUENTES-BROTO L. Drug-mediated ototoxicity and tinnitus: alleviation with melatonin. J Physiol Pharmacol. 2011;62:151–7. [PubMed] [Google Scholar]
- RIFF LJ, JACKSON GG. Pharmacology of gentamicin in man. J Infect Dis. 1971;124(Suppl 124):98–1. doi: 10.1093/infdis/124.supplement_1.s98. [DOI] [PubMed] [Google Scholar]
- RODNINA MV, WINTERMEYER W. Fidelity of aminoacyl-tRNA selection on the ribosome: kinetic and structural mechanisms. Annu Rev Biochem. 2001;70:415–35. doi: 10.1146/annurev.biochem.70.1.415. [DOI] [PubMed] [Google Scholar]
- ROGAN MP, STOLTZ DA, HORNICK DB. Cystic fibrosis transmembrane conductance regulator intracellular processing, trafficking, and opportunities for mutation-specific treatment. Chest. 2011;139:1480–90. doi: 10.1378/chest.10-2077. [DOI] [PubMed] [Google Scholar]
- ROWE SM, SLOANE P, TANG LP, BACKER K, MAZUR M, BUCKLEY-LANIER J, NUDELMAN I, BELAKHOV V, BEBOK Z, SCHWIEBERT E, BAASOV T, BEDWELL DM. Suppression of CFTR premature termination codons and rescue of CFTR protein and function by the synthetic aminoglycoside NB54. J Mol Med (Berl) 2011 doi: 10.1007/s00109-011-0787-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RUUSALA T, EHRENBERG M, KURLAND CG. Is there proofreading during polypeptide synthesis? EMBO J. 1982;1:741–5. doi: 10.1002/j.1460-2075.1982.tb01240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SALAS-MARCO J, BEDWELL DM. GTP Hydrolysis by eRF3 Facilitates Stop Codon Decoding during Eukaryotic Translation Termination. Mol Cell Biol. 2004;24:7769–78. doi: 10.1128/MCB.24.17.7769-7778.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SALAS-MARCO J, BEDWELL DM. Discrimination between defects in elongation fidelity and termination efficiency provides mechanistic insights into translational readthrough. J Mol Biol. 2005;348:801–15. doi: 10.1016/j.jmb.2005.03.025. [DOI] [PubMed] [Google Scholar]
- SCHIFFELERS R, STORM G, BAKKER-WOUDENBERG I. Liposome-encapsulated aminoglycosides in pre-clinical and clinical studies. J Antimicrob Chemother. 2001;48:333–44. doi: 10.1093/jac/48.3.333. [DOI] [PubMed] [Google Scholar]
- SCHOLTE BJ, DAVIDSON DJ, WILKE M, DE JONGE HR. Animal models of cystic fibrosis. J Cyst Fibros. 2004;3(Suppl 2):183–90. doi: 10.1016/j.jcf.2004.05.039. [DOI] [PubMed] [Google Scholar]
- SCOLNICK E, TOMPKINS R, CASKEY T, NIRENBERG M. Release factors differing in specificity for terminator codons. Proc Natl Acad Sci U S A. 1968;61:768–74. doi: 10.1073/pnas.61.2.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SERMET-GAUDELUS I, BOECK KD, CASIMIR GJ, VERMEULEN F, LEAL T, MOGENET A, ROUSSEL D, FRITSCH J, HANSSENS L, HIRAWAT S, MILLER NL, CONSTANTINE S, REHA A, AJAYI T, ELFRING GL, MILLER LL. Ataluren (PTC124) induces cystic fibrosis transmembrane conductance regulator protein expression and activity in children with nonsense mutation cystic fibrosis. Am J Respir Crit Care Med. 2010;182:1262–72. doi: 10.1164/rccm.201001-0137OC. [DOI] [PubMed] [Google Scholar]
- SERMET-GAUDELUS I, LEAL T, DEBOECK CEA. PTC124 induces CFTR full-length production and activity in children with nonsense-mutation-mediated CF. J Cyst Fibros. 2008;7:S22. [Google Scholar]
- SHA SH, SCHACHT J. Antioxidants attenuate gentamicin-induced free radical formation in vitro and ototoxicity in vivo: D-methionine is a potential protectant. Hear Res. 2000;142:34–40. doi: 10.1016/s0378-5955(00)00003-4. [DOI] [PubMed] [Google Scholar]
- SICINSKI P, GENG Y, RYDER-COOK AS, BARNARD EA, DARLISON MG, BARNARD PJ. The molecular basis of muscular dystrophy in the mdx mouse: a point mutation. Science. 1989;244:1578–80. doi: 10.1126/science.2662404. [DOI] [PubMed] [Google Scholar]
- SMITH AL, DAUM RS, SIBER GR, SCHEIFELE DW, SYRIOPOULOU VP. Gentamicin penetration into cerebrospinal fluid in experimental Haemophilus influenzae meningitis. Antimicrob Agents Chemother. 1988;32:1034–9. doi: 10.1128/aac.32.7.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SMYTH AR, BHATT J. Once-daily versus multiple-daily dosing with intravenous aminoglycosides for cystic fibrosis. Cochrane Database Syst Rev. 2012;2:CD002009. doi: 10.1002/14651858.CD002009.pub4. [DOI] [PubMed] [Google Scholar]
- SNOUWAERT JN, BRIGMAN KK, LATOUR AM, MALOUF NN, BOUCHER RC, SMITHIES O, KOLLER BH. An animal model for cystic fibrosis made by gene targeting. Science. 1992;257:1083–8. doi: 10.1126/science.257.5073.1083. [DOI] [PubMed] [Google Scholar]
- SWAN SK, KOHLHEPP SJ, KOHNEN PW, GILBERT DN, BENNETT WM. Long-term protection of polyaspartic acid in experimental gentamicin nephrotoxicity. Antimicrob Agents Chemother. 1991;35:2591–5. doi: 10.1128/aac.35.12.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TAN L, NARAYAN SB, CHEN J, MEYERS GD, BENNETT MJ. PTC124 improves readthrough and increases enzymatic activity of the CPT1A R160X nonsense mutation. J Inherit Metab Dis. 2011;34:443–7. doi: 10.1007/s10545-010-9265-5. [DOI] [PubMed] [Google Scholar]
- TAURIS J, CHRISTENSEN EI, NYKJAER A, JACOBSEN C, PETERSEN CM, OVESEN T. Cubilin and megalin co-localize in the neonatal inner ear. Audiol Neurootol. 2009;14:267–78. doi: 10.1159/000199446. [DOI] [PubMed] [Google Scholar]
- THIBAULT N, GRENIER L, SIMARD M, BERGERON MG, BEAUCHAMP D. Protection against gentamicin nephrotoxicity by daptomycin in nephrectomized rats. Life Sci. 1995;56:1877–87. doi: 10.1016/0024-3205(95)00162-y. [DOI] [PubMed] [Google Scholar]
- THOMPSON RC, STONE PJ. Proofreading of the codon-anticodon interaction on ribosomes. Proc Natl Acad Sci U S A. 1977;74:198–202. doi: 10.1073/pnas.74.1.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- USUKI F, YAMASHITA A, HIGUCHI I, OHNISHI T, SHIRAISHI T, OSAME M, OHNO S. Inhibition of nonsense-mediated mRNA decay rescues the phenotype in Ullrich's disease. Ann Neurol. 2004;55:740–4. doi: 10.1002/ana.20107. [DOI] [PubMed] [Google Scholar]
- USUKI F, YAMASHITA A, KASHIMA I, HIGUCHI I, OSAME M, OHNO S. Specific inhibition of nonsense-mediated mRNA decay components, SMG-1 or Upf1, rescues the phenotype of ullrich disease fibroblasts. Mol Ther. 2006;14:351–60. doi: 10.1016/j.ymthe.2006.04.011. [DOI] [PubMed] [Google Scholar]
- VAN GOOR F, HADIDA S, GROOTENHUIS PD, BURTON B, CAO D, NEUBERGER T, TURNBULL A, SINGH A, JOUBRAN J, HAZLEWOOD A, ZHOU J, MCCARTNEY J, ARUMUGAM V, DECKER C, YANG J, YOUNG C, OLSON ER, WINE JJ, FRIZZELL RA, ASHLOCK M, NEGULESCU P. Rescue of CF airway epithelial cell function in vitro by a CFTR potentiator, VX-770. Proc Natl Acad Sci U S A. 2009;106:18825–30. doi: 10.1073/pnas.0904709106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VAN GOOR F, HADIDA S, GROOTENHUIS PD, BURTON B, STACK JH, STRALEY KS, DECKER CJ, MILLER M, MCCARTNEY J, OLSON ER, WINE JJ, FRIZZELL RA, ASHLOCK M, NEGULESCU PA. Correction of the F508del-CFTR protein processing defect in vitro by the investigational drug VX-809. Proc Natl Acad Sci U S A. 2011;108:18843–8. doi: 10.1073/pnas.1105787108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VECSLER M, BEN ZEEV B, NUDELMAN I, ANIKSTER Y, SIMON AJ, AMARIGLIO N, RECHAVI G, BAASOV T, GAK E. Ex vivo treatment with a novel synthetic aminoglycoside NB54 in primary fibroblasts from Rett syndrome patients suppresses MECP2 nonsense mutations. PLoS One. 2011;6:e20733. doi: 10.1371/journal.pone.0020733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VOGEL Z, ZAMIR A, ELSON D. The possible involvement of peptidyl transferase in the termination step of protein biosynthesis. Biochemistry. 1969;8:5161–8. doi: 10.1021/bi00840a070. [DOI] [PubMed] [Google Scholar]
- WAGNER KR, HAMED S, HADLEY DW, GROPMAN AL, BURSTEIN AH, ESCOLAR DM, HOFFMAN EP, FISCHBECK KH. Gentamicin treatment of Duchenne and Becker muscular dystrophy due to nonsense mutations. Ann Neurol. 2001;49:706–11. [PubMed] [Google Scholar]
- WANG D, BELAKHOV V, KANDASAMY J, BAASOV T, LI SC, LI YT, BEDWELL DM, KEELING KM. The designer aminoglycoside NB84 significantly reduces glycosaminoglycan accumulation associated with MPS I-H in the Idua-W392X mouse. Mol Genet Metab. 2012;105:116–125. doi: 10.1016/j.ymgme.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WEIXLBAUMER A, JIN H, NEUBAUER C, VOORHEES RM, PETRY S, KELLEY AC, RAMAKRISHNAN V. Insights into translational termination from the structure of RF2 bound to the ribosome. Science. 2008;322:953–6. doi: 10.1126/science.1164840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WELCH EM, BARTON ER, ZHUO J, TOMIZAWA Y, FRIESEN WJ, TRIFILLIS P, PAUSHKIN S, PATEL M, TROTTA CR, HWANG S, WILDE RG, KARP G, TAKASUGI J, CHEN G, JONES S, REN H, MOON YC, CORSON D, TURPOFF AA, CAMPBELL JA, CONN MM, KHAN A, ALMSTEAD NG, HEDRICK J, MOLLIN A, RISHER N, WEETALL M, YEH S, BRANSTROM AA, COLACINO JM, BABIAK J, JU WD, HIRAWAT S, NORTHCUTT VJ, MILLER LL, SPATRICK P, HE F, KAWANA M, FENG H, JACOBSON A, PELTZ SW, SWEENEY HL. PTC124 targets genetic disorders caused by nonsense mutations. Nature. 2007;447:87–91. doi: 10.1038/nature05756. [DOI] [PubMed] [Google Scholar]
- WILSCHANSKI M, ARMONI S, YAACOV YEA. PTC124 treatment over 3 months improves pharmacodynamic and clinical parameters in patients with nonsense-mutation-mediated CF. J Cyst Fibros. 2008;7:S22. [Google Scholar]
- WILSCHANSKI M, FAMINI C, BLAU H, RIVLIN J, AUGARTEN A, AVITAL A, KEREM B, KEREM E. A pilot study of the effect of gentamicin on nasal potential difference measurements in cystic fibrosis patients carrying stop mutations. Am J Respir Crit Care Med. 2000;161:860–5. doi: 10.1164/ajrccm.161.3.9904116. [DOI] [PubMed] [Google Scholar]
- WILSCHANSKI M, KEREM E. New drugs for cystic fibrosis. Expert Opin Investig Drugs. 2011;20:1285–92. doi: 10.1517/13543784.2011.600304. [DOI] [PubMed] [Google Scholar]
- WILSCHANSKI M, MILLER LL, SHOSEYOV D, BLAU H, RIVLIN J, AVIRAM M, COHEN M, ARMONI S, YAAKOV Y, PUGATCH T, COHEN-CYMBERKNOH M, MILLER NL, REHA A, NORTHCUTT VJ, HIRAWAT S, DONNELLY K, ELFRING GL, AJAYI T, KEREM E. Chronic ataluren (PTC124) treatment of nonsense mutation cystic fibrosis. Eur Respir J. 2011;38:59–69. doi: 10.1183/09031936.00120910. [DOI] [PubMed] [Google Scholar]
- WILSCHANSKI M, YAHAV Y, YAACOV Y, BLAU H, BENTUR L, RIVLIN J, AVIRAM M, BDOLAH-ABRAM T, BEBOK Z, SHUSHI L, KEREM B, KEREM E. Gentamicin-induced correction of CFTR function in patients with cystic fibrosis and CFTR stop mutations. N Engl J Med. 2003;349:1433–41. doi: 10.1056/NEJMoa022170. [DOI] [PubMed] [Google Scholar]
- WOHLGEMUTH I, POHL C, MITTELSTAET J, KONEVEGA AL, RODNINA MV. Evolutionary optimization of speed and accuracy of decoding on the ribosome. Philos Trans R Soc Lond B Biol Sci. 2011;366:2979–86. doi: 10.1098/rstb.2011.0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WOODWORTH JR, NYHART EH, WOLNY JD, BRIER GL, BLACK HR. Tobramycin and daptomycin disposition when co-administered to healthy volunteers. J Antimicrob Chemother. 1994;33:655–9. doi: 10.1093/jac/33.3.655. [DOI] [PubMed] [Google Scholar]
- WRAITH JE. The first 5 years of clinical experience with laronidase enzyme replacement therapy for mucopolysaccharidosis I. Expert Opin Pharmacother. 2005;6:489–506. doi: 10.1517/14656566.6.3.489. [DOI] [PubMed] [Google Scholar]
- YAMASHITA A, KASHIMA I, OHNO S. Role of SMG-1-mediated phosphorylation of Upf1 in NMD. In: MAQUAT LE, editor. Nonsense-Mediated mRNA Decay. Landes Bioscience; Georgetown, Texas: 2006. [Google Scholar]
- YOUNGMAN EM, MCDONALD ME, GREEN R. Peptide release on the ribosome: mechanism and implications for translational control. Annu Rev Microbiol. 2008;62:353–73. doi: 10.1146/annurev.micro.61.080706.093323. [DOI] [PubMed] [Google Scholar]
- YUKIHARA M, ITO K, TANOUE O, GOTO K, MATSUSHITA T, MATSUMOTO Y, MASUDA M, KIMURA S, UEOKA R. Effective drug delivery system for duchenne muscular dystrophy using hybrid liposomes including gentamicin along with reduced toxicity. Biol Pharm Bull. 2011;34:712–6. doi: 10.1248/bpb.34.712. [DOI] [PubMed] [Google Scholar]
- ZAHER HS, GREEN R. Quality control by the ribosome following peptide bond formation. Nature. 2009;457:161–6. doi: 10.1038/nature07582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZARRAGA IG, ZHANG L, STUMP MR, GONG Q, VINCENT GM, ZHOU Z. Nonsense-mediated mRNA decay caused by a frameshift mutation in a large kindred of type 2 long QT syndrome. Heart Rhythm. 2011;8:1200–6. doi: 10.1016/j.hrthm.2011.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZAVIALOV AV, BUCKINGHAM RH, EHRENBERG M. A posttermination ribosomal complex is the guanine nucleotide exchange factor for peptide release factor RF3. Cell. 2001;107:115–24. doi: 10.1016/s0092-8674(01)00508-6. [DOI] [PubMed] [Google Scholar]
- ZAVIALOV AV, HAURYLIUK VV, EHRENBERG M. Splitting of the Posttermination Ribosome into Subunits by the Concerted Action of RRF and EF-G. Mol Cell. 2005;18:675–86. doi: 10.1016/j.molcel.2005.05.016. [DOI] [PubMed] [Google Scholar]
- ZHANG L, BUTTON B, GABRIEL SE, BURKETT S, YAN Y, SKIADOPOULOS MH, DANG YL, VOGEL LN, MCKAY T, MENGOS A, BOUCHER RC, COLLINS PL, PICKLES RJ. CFTR delivery to 25% of surface epithelial cells restores normal rates of mucus transport to human cystic fibrosis airway epithelium. PLoS Biol. 2009;7:e1000155. doi: 10.1371/journal.pbio.1000155. [DOI] [PMC free article] [PubMed] [Google Scholar]