Abstract
Remote loading of liposomes by trans-membrane gradients is used to achieve therapeutically efficacious intra-liposome concentrations of drugs. We have developed Quantitative Structure Property Relationship (QSPR) models of remote liposome loading for a dataset including 60 drugs studied in 366 loading experiments internally or elsewhere. Both experimental conditions and computed chemical descriptors were employed as independent variables to predict the initial drug/lipid ratio (D/L) required to achieve high loading efficiency. Both binary (to distinguish high vs. low initial D/L) and continuous (to predict real D/L values) models were generated using advanced machine learning approaches and five-fold external validation. The external prediction accuracy for binary models was as high as 91–96%; for continuous models the mean coefficient R2 for regression between predicted versus observed values was 0.76–0.79. We conclude that QSPR models can be used to identify candidate drugs expected to have high remote loading capacity while simultaneously optimizing the design of formulation experiments.
Keywords: chemical descriptors, liposome, loading conditions, loading efficiency, QSPR, remote loading
Introduction
Liposomes are one of the most important and most successful drug delivery systems as illustrated by having already more than10 FDA approved liposomal drugs[1]. Liposomes are biocompatible, non toxic, can be formulated in small (nano) size, have long circulating life time and enhanced permeability and retention (EPR effect) thereby changing favorably drug bio-distribution [2–5]. Liposome-based formulations may also reduce the toxicity of drugs. In many cases delivering the drug via an optimized liposomal formulation improves the therapeutic index of the drug.
Remote / active loading of liposomes by trans-membrane gradients is one of the best approaches for achieving sufficiently high intra-liposome concentrations for drugs to be therapeutically efficacious. For the remote loading to be effective, the drug molecule should have structural features that allow the drug to permeate and diffuse via the lipid bilayer to accumulate within the liposome yet prevent permeation and diffusion from liposomes. Amphiphatic weak acids or bases fit this requirement very well [6–8] (see Fig. 1). Optimal remote loading by pH or ion gradients requires that the loaded molecules have a logD at pH 7 in the range of −2.5 to 2.0 and pKa of ≤11 for an amphipathic weak base or pKa of > 3 for an amphipathic weak acid [8].
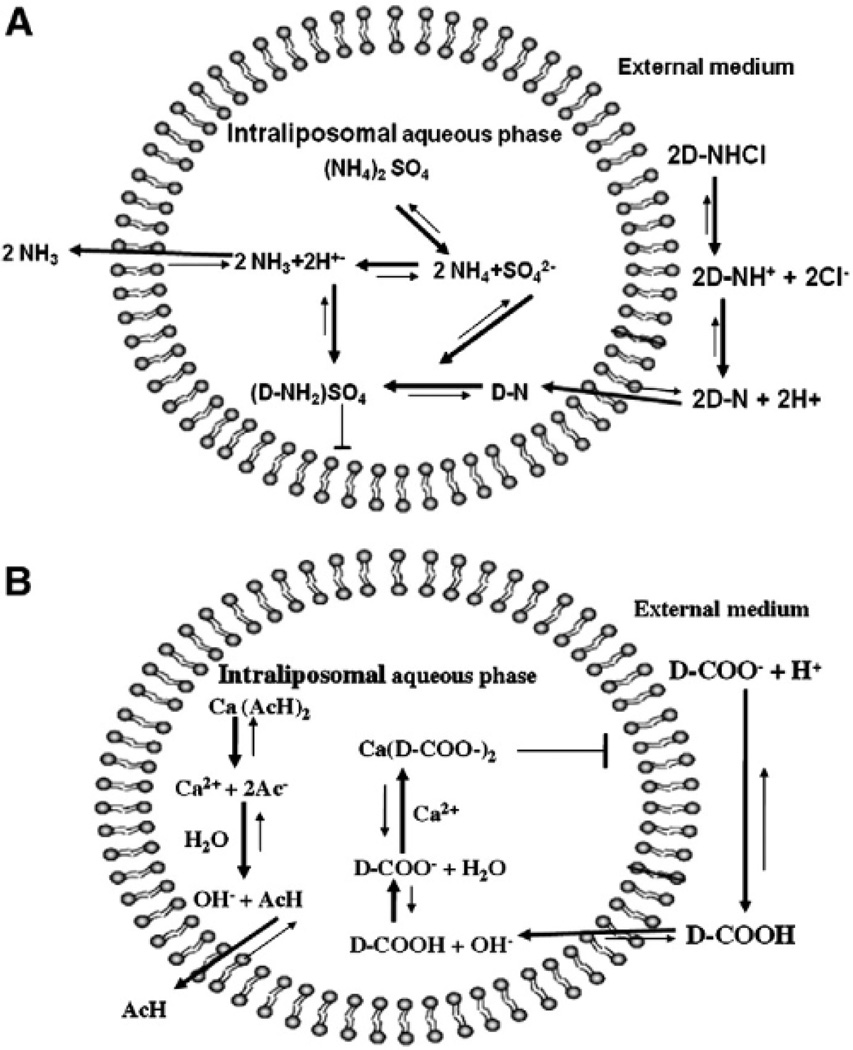
Figure 1.
Remote loading of amphipathic weak bases and acids into liposomes using an ion gradient. (A) loading of amphipathic weak base by transmembrane ammonium sulfate gradient.
(B) Loading of amphipathic weak acid by calcium acetate gradient.
Concentration of (NH4)2SO4 or Ca(C2H3O2)2 in the liposomes is 1000-fold greater than that in the extraliposomal medium. D = drug, AcH= acetic acid, Ac−= Acetate. Un-ionized drug base (D–N) or acid (D–COOH) crosses the liposomal membrane and is trapped inside after it is ionized; in some cases insoluble salt is formed with the counterion inside the liposome (sulfate and Ca ion respectively). This Figure was taken as is from[8] with permission.
In an attempt to better understand the drug characteristics and the corresponding loading conditions required to achieve high remote loading of such drugs, we have previously analyzed a unique data set generated during 15 years of research on drug remote loading into liposomes by the Barenholz laboratory. The dataset comprised of various parameters of loading experiments for nine drugs such as initial drug to lipid ratio (D/L), loading duration, loading temperature and more and physicochemical characteristics such as pKa, log P and solubility [8]. Using the decision tree method, the loading efficiency was correlated to the experimental conditions and physicochemical properties of the drugs with the accuracy of 77.5%, upgrading the tree through logical considerations increased the accuracy to 91.2%.
The loading efficiency is calculated by dividing the D/L mole ratio after the loading by the initial D/L ratio (used for loading). High loading efficiency is the desired outcome of liposome loading experiment; it enables good control of the experiment as the initial amount of the drug is almost entirely loaded, which is also important from pharmaceutical and cost perspectives. However, the initial D/L ratio resulting in high loading efficiency may be different for different drugs. Drug concentration in liposomes should be sufficiently high to enable the administration of a therapeutic dose, and hence low concentration of drugs in liposomes, even if the loading efficiency is high, will be suitable only for highly potent drugs. Therefore, it is generally accepted that a good candidate for remote loading to liposomes should be able to achieve high loading efficiency at all initial D/L ratios.
In the previous study our aim was to define the conditions that could lead to high loading efficiency. In the present study we aimed to build models that may identify good candidates for remote loading among amphipathic drug molecules (Figure 1). For this purpose we have significantly expanded the original data set to include 60 drugs studied both in our laboratory and elsewhere in a total of 366 loading experiments with varied experimental conditions. Having as many as 60 drugs in the data set allowed us to develop predictive models employing not only the experimental conditions but also computed chemical descriptors of molecules forming a set of hybrid compound features. This approach, relying on hybrid descriptors was applied to achieve models that subsequently can be used for two main purposes: (1) virtual screening of large molecular libraries to find candidate drugs expected to have high remote loading capacity, and (2) designing compounds with improved remote loading characteristics while simultaneously optimizing the design of formulation experiments.
In the present study we have only considered a group of 60 compounds with high loading efficiency achieved at different initial D/L ratios and different experimental conditions. We have applied the Quantitative Structure Property Relationships (QSPR) modeling approach using several machine learning methods to develop two types of models: binary and continuous.. Binary models were built using decision tree, k Nearest Neighbor (kNN), Support Vector Machines (SVM) and Iterative Stochastic Elimination approaches (ISE)[9, 10]. For building binary models the loading experiments were classified into two groups: compounds achieving high loading efficiency with initial D/L of <0.3 and those achieving high loading efficiency with initial D/L of ≥0.3. Continuous models were generated using kNN and Support Vector Regression (SVR). In these models the target property was the actual initial D/L required for a drug to achieve high loading efficiency. Both binary and continuous models afforded high prediction accuracy.
Formulation scientists are always considering the physicochemical properties of drugs as the major guide for the formulation development. A QSPR study for the prediction of compound solubility in certain vehicle systems was recently published, suggesting routine implementation of pre-formulation screens before in vivo preclinical studies [11]. The approach presented in this study demonstrates that QSPR modeling may be successfully employed for rational design of drug delivery systems. The models generated in our studies may help in selecting drug candidates as well as in selecting the formulation conditions suitable for a specific molecule thereby aiding in optimizing the formulation conditions and accelerating the drug development process.
2. Methods
2.1 Experimental data
The data for liposome remote loading experiments were collected from the literature (presented in Supplementary Table 1) and from Prof. Barenholz’ lab (Supplementary Table 2). The experimental data was composed of very different incubation conditions that were generalized by the following variables:
-
▪
Tm- Phospholipid transition temperature
-
▪
% Cholesterol [mole fraction (mole% )] in liposome lipid composition
-
▪
Membrane rigidity scale (described in [8] or by an experimental approach, ref. no [12] and [13])
-
▪
Liposome size classified as small (S) for ≤100nm, medium (M) for 101–999nm and large (L) for ≥ 1000nm
-
▪
D/L initial mole ratio used for liposome preparation
-
▪
Loading duration (min)
-
▪
Loading temperature (°C)
-
▪
T to Tm is the ratio between loading temperature and Tm in Kelvin degrees
-
▪
Gradient type. Either pH gradient or ion gradient (the specific gradient ion was not considered)
-
▪
Extent of pH gradient
-
▪
Salt concentration (mM)
-
▪
Loading efficiency, i.e., the fraction of the drug used for loading that present in the liposomes. It is calculated by dividing the D/L ratio after the loading by the initial D/L ratio (used for loading). Loading efficiency was classified as low (0–29% loading), medium (30–69% loading) and high (70–100% loading).
2.2 Molecular descriptors
Molecular descriptors were calculated using MOE software[14]; complete list of descriptors is given in Supplementary Table 3. These descriptors were combined with the experimental conditions (described in the previous section) to assemble a hybrid set of descriptors for each entry in our dataset.
2.3 Data analysis
2.3.1 Data set preparation
The available data (Supplementary Tables 1 and 2) were processed to eliminate duplicates as well as to remove experiments that were performed under conditions not useful for future design, i.e., when salt concentrations were < 100mM, loading duration < 10min, and pH gradient <500. The final data set contained 60 drugs studied in 366 experiments of which 198 were classified as high loading (70–100% loading), 118 as medium loading (30–69% loading) and 50 as low loading (0–29% loading). Table 1 presents the chemical structures of the studied drugs.
Table 1.
Drugs for which remote loading experiments were collected in the data set
| Drug | Structure | Drug | Structure | Drug | Structure |
|---|---|---|---|---|---|
| Weak acids | |||||
| Diclofenac[16] |  |
Salicylic acid |  |
Methyl prednisolone hemisuccinate (MPS)[8] |  |
| Ferulic acid[17] |  |
Betamethasone succinate[8] |  |
||
| Weak bases | |||||
| Sildenafil citrate[18] |  |
Oxymatrine[19] |  |
Bupivacaine[8] |  |
| 4- hydroxyl tamoxifen (4HT)[20] |  |
Epirubicin[21] |  |
Pilocarpine[21] |  |
| Acridine orange[8] | Ethambutol[22] |  |
Primaquine[23] |  |
|
| Brucine[24] |  |
Fasudil[25] |  |
Propranolol[21] |  |
| Quinine[21]a |  |
Quinidine[21]a |  |
||
| Chlorpromazine[21] |  |
Gancyclovir[26] |  |
Quinacrine[21] |  |
| Cipro-glycine[27] |  |
Imipraine[21] |  |
Imatinib mesylate[28] |  |
| Carbendazim[29] |  |
Gemcitabine[30, 31] |  |
Idarubicin[32–34] |  |
| Chloroquine[21, 35] |  |
Hydrmorphone[36] |  |
S12363 sulfate[37] |  |
| Cipro-lysine[27] |  |
Irinotecan[38] |  |
Tas-103[39] |  |
| Citicholine[40] |  |
Levofloxacin[41] |  |
Tempamine[8] |  |
| CKD-602[42] |  |
Lidocaine[8, 21] |  |
Timolol[21] |  |
| Daunorubicin [21, 34] |  |
Mitoxantrone[21, 28, 43] |  |
Topotecan[8, 42] |  |
| Dibucaine[21] |  |
Codeine[21] |  |
Verapamil[44] |  |
| Serotonin[21] |  |
M-V-05[45] |  |
Vincristine[8] |  |
| Diphenhydramine[21] |  |
Oxmaniquine[46] |  |
Vinorelbine[47] |  |
| Docetaxel esterified at the 2′-hydroxyl group with 4-(4-methylpiperazin-1-yl)butanoic acid[48] |  |
Lurtotecan[42] |  |
Top 53[49] |  |
| Dopamine[21, 50] |  |
Oxymorphone[51] |  |
Ciprofloxacin[27, 52] |  |
| Doxorubicin[34, 44, 53, 54] |  |
Physostigmine[21] |  |
Stereoisomers
For the purpose of model building, only the group with high loading was used and the target property was the initial D/L. For binary modeling, initial D/L of <0.3 was classified as Ratio1 and initial D/L≥0.3 was classified as Ratio2. The dataset was further processed to eliminate entries for the same compounds with the same experimental conditions that had different initial D/L values. Our aim was to predict the highest initial D/L that will result in high loading efficiency and therefore the entry retained in the dataset was the one with the highest D/L. The dataset used for category model building was therefore composed of 143 experiments of which 112 experiments were classified as Ratio1 and 31 experiments were classified as Ratio2. For continuous model building, we used the same dataset but the actual D/L ratio was the target property; these values were distributed within the range of 0.004–0.569. One experiment with the D/L ratio of 0.8 was eliminated from the continuous data set because this value was an outlier compared to other points in the data set. Models were built under 5-fold external validations scheme: the dataset was randomly divided into five groups of nearly equal size; four groups were used systematically as training sets and the fifth group was employed as a test set.
Low and medium loading experiments were not used for model building (as previously explained). These experiments were therefore used as an additional test set for the models generated using the high loaded group. For this purpose duplicate experiments which differed only in their D/L ratio were eliminated. The overall low/medium test set contained 112 experiments.
2.3.2 QSPR Modeling
Two types of models were built: binary and continuous models. Binary models were built using one of four classification methods: J48 decision tree, kNN , SVM and ISE. Continuous models were built using kNN and SVR.
To characterize the predictive power of binary models, we used the following statistics.
Sensitivity = TP/(TP+FN) and Specificity=TN/(TN+FP), where TP and TN are true positives and true negatives, respectively, and FN and FP are false positives and false negatives, respectively. TP and TN are defined as instances predicted correctly as ratio2 and ratio1, respectively, while FN and FP are instances that were incorrectly classified as ratio1 (i.e., instances characterized experimentally as ratio2) and ratio2 (i.e., instances characterized experimentally as ratio1), respectively.
-
Correct classification rate (CCR) is the major determinant of success rate for each of the category models (as discussed later in the Results section and shown in Table 2). CCR was defined as follows:
CCR = 0.5*(Sensitivity+Specificity)
Table 2.
Mean 5-fold external validation results using four classification methods (mean±SD)
| ISE | kNN-GA | SVM | J48 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Confusion matrix | Pred ratio1 |
Pred ratio2 |
Pred ratio1 |
Pred ratio2 |
Pred ratio1 |
Pred ratio2 |
Pred ratio1 |
Pred ratio2 |
||||
| Obs ratio1 |
109 | 3 | Obs ratio1 |
108 | 4 | Obs ratio1 |
111 | 1 | Obs ratio1 |
105 | 7 | |
| Obs ratio2 |
7 | 24 | Obs ratio2 |
6 | 25 | Obs ratio2 |
5 | 26 | Obs ratio2 |
6 | 25 | |
| % Correctly classified | 93±5.6 | 93±5.0 | 96 ±2.9 | 91±5.4 | ||||||||
| CCR | 0.87±0.08 | 0.89±0.11 | 0.91 ±0.06 | 0.87±0.11 | ||||||||
| Specificity | 0.97±0.04 | 0.96±0.02 | 0.99 ±0.02 | 0.94±0.02 | ||||||||
| Sensitivity | 0.78±0.14 | 0.81±0.21 | 0.84 ±0.12 | 0.81±0.21 | ||||||||
Pred- Predicted, Obs- Observed
We have used also the Matthews correlation coefficient
| (3) |
For the continuous models the parameters calculated for the external testing were: squared correlation coefficient R (R2) for the best fit between the predicted and observed activities; coefficients of determination, i.e., squared correlation coefficients R20 for the best fit regression between predicted versus observed activities and R20’ for observed versus predicted activities forced to come through the origin and the slopes k and k' of the respective regression lines [15].
2.3.2.1 J48 decision tree using Weka 3.6 software(http://www.cs.waikato.ac.nz/~ml/index.html)
J48 is the implementation of the C4.5 algorithm [55, 56] in Weka 3.6 package. C4.5 builds a classification or regression pruned or non-pruned decision tree. Input data is a set of molecules (objects) with given descriptors (attributes) and activity values, which represent compound classes or a continuous response variable. The method makes use of the concept of information or entropy. Each node of the tree is characterized by a node’s entropy (information). Then, if possible (i.e. when the node includes compounds of different classes) the algorithm tries to find, a descriptor which provides the highest normalized information gain when the node is split into two child nodes. The splitting procedure stops when no one node can be split. Nodes that cannot be split are called leaf nodes. Pruning is used to avoid overfitting and chance correlation. The leaf node defines the predicted class of the molecule. Prediction of a query compound by the tree uses the descriptor values of this object which directs the object up the tree to a leaf node.
2.3.2.2 k Nearest Neighbor (kNN-GA) using ChemBench (http://chembench.mml.unc.edu/)
The kNN QSPR method [57] is based on the k nearest neighbors principle and the variable selection procedure. It employs the leave-one-out (LOO) cross-validation (CV) procedure and a genetic algorithm for the variable selection. The target functions used were the leave-one-out cross-validation CCR for binary QSPR or q2 for continuous QSPR(see section 2.3.2 above). The target function is the leave-one-out cross-validation correct classification rate (CCR) as defined above in section 2.3.2 for binary modeling or the mean accuracy calculated separately for each category in QSPR). The procedure starts with a random selection of a predefined number of descriptors. This step is repeated a predefined (500 by default) number of times and gives the first generation models. Each model is represented by a bit string with bits one corresponding to descriptors selected; other bits are zeros. For each model, a target function is calculated. A new generation of models is created by crossover and mutations of the first generation models. The procedure tries to maximize the target function, so 500 models with highest target function are retained. The procedure is repeated until one of the following stopping criteria is satisfied: no significant improvement between current and previous generation models; the maximum number of generations is reached, or all the models reach the threshold of CCR or q2 defined by the user. The additional details of variable selection kNN modeling approach are described elsewhere [57].
In case of the category (and binary) QSPR modeling, all models of the last generation are filtered using CCR statistics for prediction of the test set. In case of the continuous QSPR modeling, a few test set statistics are used to estimate the predictive power of the model[15]. LOO-CV q2; square of the correlation coefficient R (R2) between the predicted and observed activities; coefficients of determination (predicted versus observed activities, and observed versus predicted activities ); slopes k and k' of regression lines (predicted versus observed activities, and observed versus predicted activities) through the origin. We consider models acceptable, if they have q2>0.5; R2>0.6; (R2−R02)/R2<0.1 and 0.85≤k≤1.15 or (R2−R'02)/R2<0.1 and 0.85≤k'≤1.15; |R02−R'02|<0.3[15].
2.3.2.3 Support Vector Machines (SVM) using ChemBench (http://chembench.mml.unc.edu/)
The SVM method was developed by V. Vapnik[58]. The application of SVM to the binary classification problem was implemented as follows. Compounds can belong to two categories: active (activity equal to 1) and inactive (activity equal to −1). The SVM tries to separate active from inactive compounds of the training set by a hyperplane in the descriptor space in the best possible way (the hyperplane that has the largest distance to the nearest training data points of any class). If the solution of this problem is possible, the data set is referred to as separable. In most cases, the data set is nonseparable. In this case, a soft margin approach is implemented, in which the compounds with the mislabeled categories are allowed within the margins, and the distance from the dividing hyperplane to the margin (the support vectors) is maximized. Usually, the calculations are performed in the feature space defined by a kernel function. In our studies, the ν–SVM [59, 60] as applied within the LibSVM [61] was used as implemented in the ChemBench, and the radial basis function (RBF) kernel was used.
2.3.2.4 Support Vector Regression (SVR) using ChemBench (http://chembench.mml.unc.edu/)
The SVM approach can be applied to model datasets where the target property is a real value. In this case, the method is called support vector regression (SVR). Originally, it was developed by Vapnik. In SVM, the errors of classification were considered within the margin. In contrast, SVR ignores the errors within the margin, and takes into account errors outside of it. In these studies, the ν–SVR [59, 60] with the RBF kernel was used.
2.3.2.5 Iterative Stochastic Elimination (ISE)
ISE is a combinatorial optimization method developed in the Goldblum lab [9, 10]. ISE optimizes here the selection of filters that are able to distinguish between molecules with different activity classes. The ranking index of a certain molecule is determined by its ability to pass filters and by combining these filters' weights to give a score between −1 (negative) and +1 (positive). Molecules that pass more filters receive a higher index. The ranking model calculation is composed of the following steps:
Single range optimization- Finding the best ranges of each separate descriptor
Filters optimization –a filter is produced by combining ranges of a few descriptors, whose total number is given at the outset. The best sets are picked by a combinatorial search and optimization that is the core of the ISE algorithm. MCC (eq. 3 above) is used to score each randomly picked filter, and ISE eliminates ranges that contribute consistently to the worst MCC scores at the end of a huge sample of filters, which comprises a single iteration. Iterations continue to a point (usually < 106 combinations) from which it is possible to compute exhaustively all remaining combinations.
Constructing the model- It is now possible to sort all the combinations based on their MCC values, subsequently to cluster the top (~1000) filters and the remaining ones constitute the “Model”.
Virtual Screening: Molecular descriptors are calculated for any given molecule and are passed through the model, their final scores being a balance of filters passed successfully and those that were not successfully passed.
For more detailed description of ISE optimization tool see references [9, 10].
2.3.3 External testing
Models were validated by 5- fold external validation scheme: the data were randomly divided into five groups of nearly equal size; four groups were used systematically as training sets and the fifth group was employed as a test set. For the binary models, the parameters calculated for the external testing for each model were sensitivity, specificity and CCR as described below.
3. Results and Discussion
3.1 Binary models
For the purpose of binary in silico modeling of remote loading of drugs, the high loading group was classified into two groups: instances with initial D/L of <0.3 were classified as Ratio1 and instances with initial D/L≥0.3 were classified as Ratio2. Four classification methods were used for classification modeling of the data set; ISE, kNN, SVM and J48 decision tree.
Table 2 presents a summary of the 5-fold external validation results across the four methods used. SVM showed the highest specificity and sensitivity results with one false positive and 5 false negative instances. kNN and ISE showed similar specificity and sensitivity values. J48 showed slightly lower specificity results with seven false positive instances.
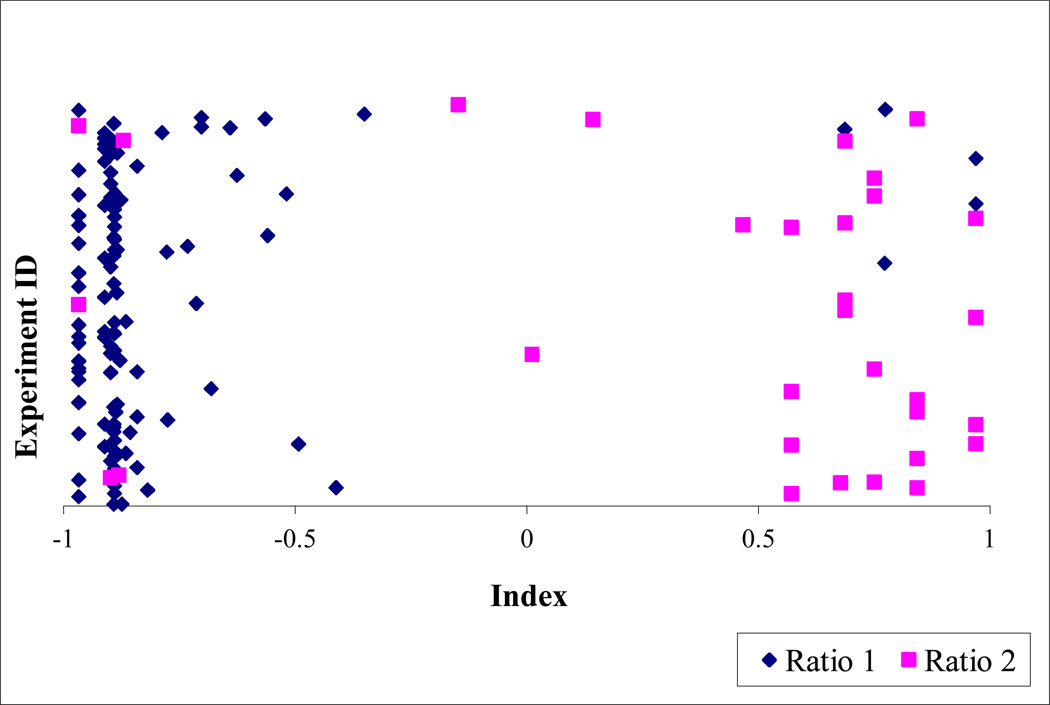
ISE generates indexing for each molecule probability to be in the ratio 2 group. Figure 2 presents the distribution of all instances in the external sets by the ISE indexing. Most of Ratio2 instances had ISE index in the range of 0.5 to 1 while most Ratio1 instances were in the range of −0.5 to −1. Thus, the ISE indexing resulted in a good discrimination between the two groups.
Figure 2.
External sets index distribution obtained using ISE
ISE provided sets of optimized filters in each of the five training fold rounds tested. Table 3 presents descriptors that were found in optimized filters of three or more of the training fold rounds tested and their percent of occurrence in the optimized filters. Most significant molecular descriptor included the total polar positive van der Waals (VDW) surface area (PEOE_VSA_PPOS), fractional polar VDW surface area (PEOE_VSA_FPOL), log P, SlogP and subdivided surface area and partial charges descriptors. The most frequent experimental conditions were Tm and T/ Tm ratio. A list of the most frequent descriptors in optimized ISE filters of each of the five training fold rounds is found in Supplementary Table 4.
Table 3.
Descriptors appeared in optimized ISE filters in three or more of the five training fold rounds.
| Descriptor | % Occurrence |
|---|---|
| PEOE_VSA_PPOS | 91.1 |
| Tm | 73.5 |
| SMR_VSA2 | 52.6 |
| SlogP | 49.3 |
| TtoTm | 38.8 |
| PEOE_VSA-3 | 38.2 |
| logP(o/w) | 23.6 |
| PEOE_VSA+4 | 21.5 |
| PEOE_VSA_FPOL | 14.1 |
| GCUT_SMR_3 | 13.6 |
Table 4 summarizes 20 most frequent descriptors identified by kNN models and their percent of occurrence in the models. The most important experimental conditions were Tm, size, rigidity, salt concentration, T/Tm ratio and % Cholesterol. Most important molecular descriptors included logP, subdivided surface area and partial charges descriptors, VDW basic surface area (vsa_base), VDW donor surface area (vsa_don), radius and diameter. In J48 decision tree the most significant experimental conditions were Tm and loading duration. The important chemical descriptors included radius, number of sulfur atoms (a_nS), adjacency matrix descriptors (BCUT_SlogP_3 and GCUT_SMR_1) and total polar negative VDW surface area (Q_VSA_PNEG). For a complete list of MOE descriptors see Supplementary Table 3. At the same time, implemented nonlinear SVM method is not a variable selection method, therefore descriptors analysis was not done for SVM models.
Table 4.
Twenty most frequent descriptors in kNN models
| Descriptor | % Occurrence | Descriptor | % Occurrence |
|---|---|---|---|
| Tm | 42.9 | % Cholesterol | 17.5 |
| Size | 34.4 | vsa_base | 17.3 |
| rigidity | 25.9 | radius | 16.9 |
| reactive | 22.5 | PEOE_VSA+6 | 16.5 |
| Salt concentration | 20.0 | BCUT_PEOE_2 | 15.9 |
| logP(o/w) | 20.0 | chiral | 15.3 |
| T/ Tm ratio | 19.7 | GCUT_SMR_0 | 15.1 |
| PEOE_VSA-0 | 19.3 | diameter | 14.7 |
| SMR_VSA2 | 18.6 | SlogP | 14.2 |
| SMR_VSA3 | 17.7 | vsa_don | 13.9 |
Models obtained by the four classification methods were further applied to the low and medium categorized instances, combined as an additional external test set (defined in sec. 2.3.1). Using SVM and ISE models, 6 and 8 instances respectively, were classified as Ratio2; kNN and J48 models classified 13 instances in this dataset as Ratio2. Many of these instances in the low/medium group were actually tested with D/L ratio of ≥0.3 and their loading efficiency was medium. The Ratio2 classified instances in the low/medium group may be therefore considered as borderline instances. The total number of Ratio2 classified instances in all four models tested did not exceed 12% of all low/medium instances.
3.2.2 Continuous models
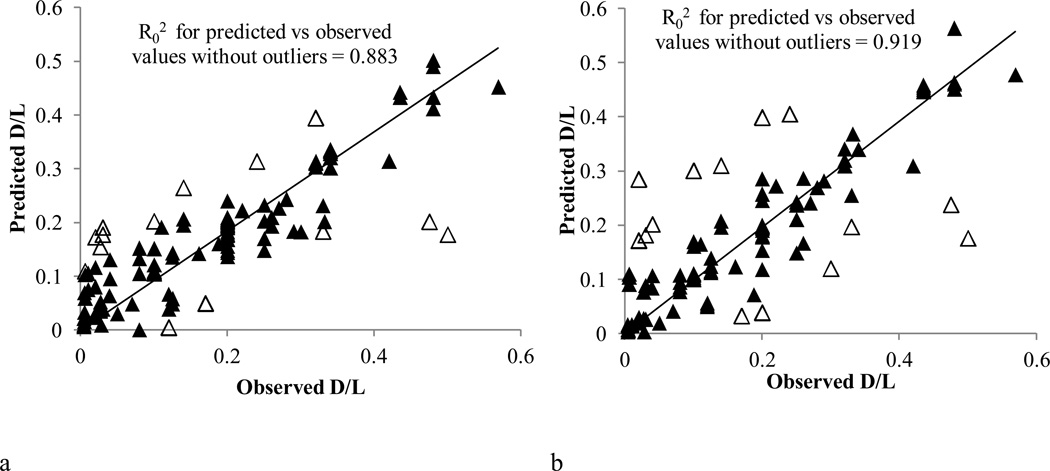
Continuous models for the prediction of the initial D/L required for a drug, under certain experimental conditions, to achieve high loading efficiency were built using SVR and kNN implemented within ChemBench. Table 5 summarizes the mean external validation results for SVR and kNN. The mean values for all figures of merit were indeed satisfactory. Figure 3 presents the comparison between predicted vs observed D/L values obtained for the external validation tests in both models. The empty triangles in the figures present the outliers in the predictions. Outliers were defined as the instances having the highest absolute distances from the overall line (top 10%). In both models most of the outliers were above the line. In these cases it may be that testing higher D/L ratios for the specific drugs would have also resulted in high loading efficiency. The maximal distance from the line was 0.27 and 0.30 for SVR and kNN respectively. Two points in both cases showed such a difference: Imatinib and Topotecan which were reported to have high loading efficiency with the D/L ratio of 0.5 and 0.48, respectively, whereas both models predicted these values to be 0.3 and 0.2 respectively. Excluding the outlier points, the predicted vs observed plots showed high R02 of 0.88 for SVR and 0.92 for kNN.
Table 5.
Mean (± SD) 5-fold external validation results of SVR and kNN continuous models
| R2 | R02 | R'02 | k | k' | |
|---|---|---|---|---|---|
| SVR | 0.789±0.084 | 0.734±0.093 | 0.781±0.084 | 0.894±0.072 | 1.033±0.062 |
| kNN | 0.758±0.105 | 0.732±0.120 | 0.731±0.136 | 0.945±0.093 | 0.970±0.098 |
Figure 3.
Comparison between predicted vs observed D/L values obtained for the external validation tests a. based on SVR models, b. based on kNN models. The empty triangles represent outlier points
Continuous models were also applied to the combined low/medium dataset. Most of the D/L ratios predicted for these instances were below the D/L actually tested implying that these compounds would be expected to have low/medium loading efficiency as observed. This means that using lower D/L ratios for these compounds would have made these instances to achieve the high loading efficiency. These results additionally inllustrate the predictive ability of the obtained models.
4. Conclusions
QSPR modeling is widely used for correlating chemical structure with biological activity or chemical reactivity. In this study we used QSPR methods to correlate both chemical structural features and experimental conditions with the classification of drugs as good or poor candidates for liposomes remote loading. A good candidate for remote loading to liposomes was defined as a drug that can achieve high loading efficiency at all initial drug to lipid ratios. However, high loading efficiency at low D/L ratio is impractical as it would not allow to achieve sufficiently high intra-liposome concentrations. Therefore, we have focused on predicting the intial D/L ratio to guide the selection of viable candidates for remote loading.
Two types of models were generated: binary and continuous models. The binary models showed very high specificity values with few false negative instances while sensitivity values were lower having more false negative instances. The continuous models were also found to be reliable. The methods used for constructing the two types of models showed high predictability using the low/medium loading test set. The approach presented in this study allows rational selection of new candidates for liposomes remote loading, and it can also aid in the design of formulation experiments thereby accelerating the drug development process.
Supplementary Material
Acknowledgements
The partial support by the Barenholz Fund is gratefully acknowledged. The authors would like to thank Dr. David Marcus for the development and implementation of the ISE algorithm. Mr. Theo Walker is acknowledged for his technical support of the Chembench portal. The UNC group acknowledges support from the NIH grant GM066940.
References
- 1.Wagner V, Dullaart A, Bock AK, Zweck A. The emerging nanomedicine landscape. Nat Biotechnol. 2006;24:1211–1217. doi: 10.1038/nbt1006-1211. [DOI] [PubMed] [Google Scholar]
- 2.Gabizon A, Catane R, Uziely B, Kaufman B, Safra T, Cohen R, Martin F, Huang A, Barenholz Y. Prolonged Circulation Time and Enhanced Accumulation in Malignant Exudates of Doxorubicin Encapsulated in Polyethylene-Glycol Coated Liposomes. Cancer Res. 1994;54:987–992. [PubMed] [Google Scholar]
- 3.Gabizon A, Shmeeda H, Barenholz Y. Pharmacokinetics of pegylated liposomal doxorubicin - Review of animal and human studies. Clin Pharmacokinet. 2003;42:419–436. doi: 10.2165/00003088-200342050-00002. [DOI] [PubMed] [Google Scholar]
- 4.Barenholz Y. Harnessing biomaterials for Nanomedicine: Preparation, toxicity and applications. Pan Stanford Publishing Pte Ltd; 2011. Doxil®— The First FDA-Approved Nano-Drug: From an Idea to a Product. in Press. www.panstanford.com. [Google Scholar]
- 5.Naparstek Y, Avnir Y, Ulmansky R, Wasserman V, Even-Chen S, Broyer M, Barenholz Y. Amphipathic weak acid glucocorticoid Prodrugs remote-loaded into sterically stabilized nanoliposomes evaluated in arthritic rats and in a beagle dog. Arthritis Rheum. 2008;58:119–129. doi: 10.1002/art.23230. [DOI] [PubMed] [Google Scholar]
- 6.Barenholz Y. Liposome application: problems and prospects. Current Opinion in Colloid & Interface Science. 2001;6:66–77. [Google Scholar]
- 7.Barenholz Y. Relevancy of drug loading to liposomal formulation therapeutic efficacy. J Liposome Res. 2003;13:1–8. doi: 10.1081/lpr-120017482. [DOI] [PubMed] [Google Scholar]
- 8.Zucker D, Marcus D, Barenholz Y, Goldblum A. Liposome drugs' loading efficiency: a working model based on loading conditions and drug's physicochemical properties. J Control Release. 2009;139:73–80. doi: 10.1016/j.jconrel.2009.05.036. [DOI] [PubMed] [Google Scholar]
- 9.Glick M, Rayan A, Goldblum A. A stochastic algorithm for global optimization and for best populations: a test case of side chains in proteins. Proc Natl Acad Sci U S A. 2002;99:703–708. doi: 10.1073/pnas.022418199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rayan A, Marcus D, Goldblum A. Predicting oral druglikeness by iterative stochastic elimination. J Chem Inf Model. 2010;50:437–445. doi: 10.1021/ci9004354. [DOI] [PubMed] [Google Scholar]
- 11.Crivori P, Morelli A, Pezzetta D, Rocchetti M, Poggesi I. Development and validation of in silico models for estimating drug preformulation risk in PEG400/water and Tween80/water systems. Eur J Pharm Sci. 2007;32:169–181. doi: 10.1016/j.ejps.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 12.Shinitzky M, Barenholz Y. Fluidity parameters of lipid regions determined by fluorescence polarization. Biochim Biophys Acta. 1978;515:367–394. doi: 10.1016/0304-4157(78)90010-2. [DOI] [PubMed] [Google Scholar]
- 13.Shinitzky M, Barenholz Y. Dynamics of the hydrocarbon layer in liposomes of lecithin and sphingomyelin containing dicetylphosphate. J Biol Chem. 1974;249:2652–2657. [PubMed] [Google Scholar]
- 14.Molecular Operating Environment (MOE) http://www.chemcomp.com/index.htm. 2010.10.
- 15.Tropsha A, Golbraikh A. Predictive Quantitative Structure–Activity Relationships Modeling. Development and Validation of QSAR Models. In: Faulon J-L, Bender A, editors. Handbook of Chemoinformatics Algorithms. London: Taylor and Francis; 2009. [Google Scholar]
- 16.Sung Hee Hwang YM, Qi Xian-Rong, Takayama Kozo, Nagai Tsuneji. Remote loading of diclofenac, insulin and fluorescein isothiocyanate labeled insulin into liposomes by pH and acetate gradient methods. International Journal of Pharmaceutics. 1999;179:85–95. doi: 10.1016/s0378-5173(98)00392-5. [DOI] [PubMed] [Google Scholar]
- 17.Qin J, Chen D, Lu W, Xu H, Yan C, Hu H, Chen B, Qiao M, Zhao X. Preparation, characterization, and evaluation of liposomal ferulic acid in vitro and in vivo. Drug Dev Ind Pharm. 2008;34:602–608. doi: 10.1080/03639040701833559. [DOI] [PubMed] [Google Scholar]
- 18.Wagner A, Vorauer-Uhl K, Katinger H. Liposomal composition comprising an active ingredient for relaxing smooth muscle, the production of this composition and the therapeutic use thereof. 2009/0324698 A1. USA Patent. 2009
- 19.Du S, Deng Y. Studies on the encapsulation of oxymatrine into liposomes by ethanol injection and pH gradient method. Drug Dev Ind Pharm. 2006;32:791–797. doi: 10.1080/03639040600760556. [DOI] [PubMed] [Google Scholar]
- 20.Urbinati G, Audisio D, Marsaud V, Plassat V, Arpicco S, Sola B, Fattal E, Renoir JM. Therapeutic potential of new 4-hydroxy-tamoxifen-loaded pH-gradient liposomes in a multiple myeloma experimental model. Pharm Res. 2010;27:327–339. doi: 10.1007/s11095-009-0023-z. [DOI] [PubMed] [Google Scholar]
- 21.Madden TD, Harrigan PR, Tai LC, Bally MB, Mayer LD, Redelmeier TE, Loughrey HC, Tilcock CP, Reinish LW, Cullis PR. The accumulation of drugs within large unilamellar vesicles exhibiting a proton gradient: a survey. Chem Phys Lipids. 1990;53:37–46. doi: 10.1016/0009-3084(90)90131-a. [DOI] [PubMed] [Google Scholar]
- 22.Wiens T, Redelmeier T, Av-Gay Y. Development of a Liposome Formulation of Ethambutol. Antimicrobial Agents and Chemotherapy. 2004;48:1887–1888. doi: 10.1128/AAC.48.5.1887-1888.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stensrud G, Sande SA, Kristensen S, Smistad G. Formulation and characterisation of primaquine loaded liposomes prepared by a pH gradient using experimental design. Int J Pharm. 2000;198:213–228. doi: 10.1016/s0378-5173(00)00338-0. [DOI] [PubMed] [Google Scholar]
- 24.Chen J, Lin A, Chen Z, Wang W, Zhang T, Cai H, Cai B. Ammonium sulfate gradient loading of brucine into liposomes: effect of phospholipid composition on entrapment efficiency and physicochemical properties in vitro. Drug Dev Ind Pharm. 2010;36:245–253. doi: 10.1080/03639040903099736. [DOI] [PubMed] [Google Scholar]
- 25.Ishida T, Takanashi Y, Doi H, Yamamoto I, Kiwada H. Encapsulation of an antivasospastic drug, fasudil, into liposomes, and in vitro stability of the fasudil-loaded liposomes. Int J Pharm. 2002;232:59–67. doi: 10.1016/s0378-5173(01)00896-1. [DOI] [PubMed] [Google Scholar]
- 26.Engelmann C, Panis Y, Bolard J, Diquet B, Fabre M, Nagy H, Soubrane O, Houssin D, Klatzmann D. Liposomal encapsulation of ganciclovir enhances the efficacy of herpes simplex virus type 1 thymidine kinase suicide gene therapy against hepatic tumors in rats. Hum Gene Ther. 1999;10:1545–1551. doi: 10.1089/10430349950017879. [DOI] [PubMed] [Google Scholar]
- 27.Guo SSL, Gittelman J, Zalipsky S, Martin FJ. Liposome composition and method for administrating a quinolone. WO 99/51202. PCT. 1999
- 28.Pinto AC, Moreira JN, Simoes S. Liposomal imatinib-mitoxantrone combination: formulation development and therapeutic evaluation in an animal model of prostate cancer. Prostate. 2011;71:81–90. doi: 10.1002/pros.21224. [DOI] [PubMed] [Google Scholar]
- 29.Jia L, Garza M, Wong H, Reimer D, Redelmeier T, Camden JB, Weitman SD. Pharmacokinetic comparison of intravenous carbendazim and remote loaded carbendazim liposomes in nude mice. J Pharm Biomed Anal. 2002;28:65–72. doi: 10.1016/s0731-7085(01)00702-6. [DOI] [PubMed] [Google Scholar]
- 30.Celano M, Calvagno MG, Bulotta S, Paolino D, Arturi F, Rotiroti D, Filetti S, Fresta M, Russo D. Cytotoxic effects of gemcitabine-loaded liposomes in human anaplastic thyroid carcinoma cells. BMC Cancer. 2004;4:63. doi: 10.1186/1471-2407-4-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Immordino ML, Brusa P, Rocco F, Arpicco S, Ceruti M, Cattel L. Preparation, characterization, cytotoxicity and pharmacokinetics of liposomes containing lipophilic gemcitabine prodrugs. J Control Release. 2004;100:331–346. doi: 10.1016/j.jconrel.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 32.Dos Santos N, Mayer LD, Abraham SA, Gallagher RC, Cox KA, Tardi PG, Bally MB. Improved retention of idarubicin after intravenous injection obtained for cholesterol-free liposomes. Biochim Biophys Acta. 2002;1561:188–201. doi: 10.1016/s0005-2736(02)00345-0. [DOI] [PubMed] [Google Scholar]
- 33.Gubernator J, Chwastek G, Korycinska M, Stasiuk M, Grynkiewicz G, Lewrick F, Suss R, Kozubek A. The encapsulation of idarubicin within liposomes using the novel EDTA ion gradient method ensures improved drug retention in vitro and in vivo. J Control Release. 2010;146:68–75. doi: 10.1016/j.jconrel.2010.05.021. [DOI] [PubMed] [Google Scholar]
- 34.Dos Santos N, Cox KA, McKenzie CA, van Baarda F, Gallagher RC, Karlsson G, Edwards K, Mayer LD, Allen C, Bally MB. pH gradient loading of anthracyclines into cholesterol-free liposomes: enhancing drug loading rates through use of ethanol. Biochim Biophys Acta. 2004;1661:47–60. doi: 10.1016/j.bbamem.2003.11.016. [DOI] [PubMed] [Google Scholar]
- 35.Qiu L, Jing N, Jin Y. Preparation and in vitro evaluation of liposomal chloroquine diphosphate loaded by a transmembrane pH-gradient method. Int J Pharm. 2008;361:56–63. doi: 10.1016/j.ijpharm.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 36.Tu S, McGinnis T, Krugner-Higby L, Heath TD. A mathematical relationship for hydromorphone loading into liposomes with trans-membrane ammonium sulfate gradients. J Pharm Sci. 2010;99:2672–2680. doi: 10.1002/jps.22017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chemin C, Pean JM, Bourgaux C, Pabst G, Wuthrich P, Couvreur P, Ollivon M. Supramolecular organization of S12363-liposomes prepared with two different remote loading processes. Biochim Biophys Acta. 2009;1788:926–935. doi: 10.1016/j.bbamem.2008.11.017. [DOI] [PubMed] [Google Scholar]
- 38.Chou TH, Chen SC, Chu IM. Effect of composition on the stability of liposomal irinotecan prepared by a pH gradient method. J Biosci Bioeng. 2003;95:405–408. [PubMed] [Google Scholar]
- 39.Shimizu K, Takada M, Asai T, Kuromi K, Baba K, Oku N. Cancer chemotherapy by liposomal 6-[12-(dimethylamino)ethyl]aminol-3-hydroxy-7H-indeno[2,1-clquinolin-7-one dihydrochloride (TAS-103), a novel anti-cancer agent. Biol Pharm Bull. 2002;25:1385–1387. doi: 10.1248/bpb.25.1385. [DOI] [PubMed] [Google Scholar]
- 40.Rajnarayan MA, Ishwarlal GN, Ramgopal BM, Babulal SB, Shantaram SR, Pinjari JAS. Liposomal Citicoline Injection. WO 2010/092597 A2. PCT. 2010
- 41.Zhang X, Sun P, Bi R, Wang J, Zhang N, Huang G. Targeted delivery of levofloxacin-liposomes for the treatment of pulmonary inflammation. J Drug Target. 2009;17:399–407. doi: 10.1080/10611860902795407. [DOI] [PubMed] [Google Scholar]
- 42.Slater JL, Colbern GT, Working PK. Liposome-entrapped topoisomerase inhibitors. 7,244,449 B2. USA Patent. 2007
- 43.Li C, Cui J, Wang C, Li Y, Zhang H, Wang J, Zhang L, Guo W, Wang Y. Encapsulation of mitoxantrone into pegylated SUVs enhances its antineoplastic efficacy. Eur J Pharm Biopharm. 2008;70:657–665. doi: 10.1016/j.ejpb.2008.05.019. [DOI] [PubMed] [Google Scholar]
- 44.Wang J, Goh B, Lu W, Zhang Q, Chang A, Liu XY, Tan TM, Lee H. In vitro cytotoxicity of Stealth liposomes co-encapsulating doxorubicin and verapamil on doxorubicin-resistant tumor cells. Biol Pharm Bull. 2005;28:822–828. doi: 10.1248/bpb.28.822. [DOI] [PubMed] [Google Scholar]
- 45.Tan BJ, Quek KS, Wong MY, Chui WK, Chiu GN. Liposomal M-V-05: formulation development and activity testing of a novel dihydrofolate reductase inhibitor for breast cancer therapy. Int J Oncol. 2010;37:211–218. doi: 10.3892/ijo_00000669. [DOI] [PubMed] [Google Scholar]
- 46.Frezard F, Silva-Barcellos NM, Melo ALd. Schistosomicidal activity of oxamniquine encapsulated in sterically stabilized liposomes: influence of the route of administration. XVI CONGRESSO BRASILEIRO DE PARASITOLOGIA; Poços de Caldas, MG. 1999. [Google Scholar]
- 47.Li CL, Cui JX, Wang CX, Zhang L, Li YH, Xiu X, Li YF, Wei N. Development of pegylated liposomal vinorelbine formulation using "post-insertion" technology. Int J Pharm. 2010;391:230–236. doi: 10.1016/j.ijpharm.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 48.Zhigaltsev IV, Winters G, Srinivasulu M, Crawford J, Wong M, Amankwa L, Waterhouse D, Masin D, Webb M, Harasym N, Heller L, Bally MB, Ciufolini MA, Cullis PR, Maurer N. Development of a weak-base docetaxel derivative that can be loaded into lipid nanoparticles. J Control Release. 2010;144:332–340. doi: 10.1016/j.jconrel.2010.02.029. [DOI] [PubMed] [Google Scholar]
- 49.Shimizu K, Takada M, Asai T, Irimura K, Baba K, Oku N. Potential usage of liposomal 4betaaminoalkyl-4'-O-demethyl-4-desoxypodophyllotoxin (TOP-53) for cancer chemotherapy. Biol Pharm Bull. 2002;25:783–786. doi: 10.1248/bpb.25.783. [DOI] [PubMed] [Google Scholar]
- 50.Zhigaltsev IV, Kaplun AP, Kucheryanu VG, Kryzhanovsky GN, Kolomeichuk SN, Shvets VI, Yurasov VV. Liposomes containing dopamine entrapped in response to transmembrane ammonium sulfate gradient as carrier system for dopamine delivery into the brain of parkinsonian mice. J Liposome Res. 2001;11:55–71. doi: 10.1081/LPR-100103170. [DOI] [PubMed] [Google Scholar]
- 51.Krugner-Higby L, KuKanich B, Schmidt B, Heath TD, Brown C, Smith LJ. Pharmacokinetics and behavioral effects of an extended-release, liposome-encapsulated preparation of oxymorphone in rhesus macaques. J Pharmacol Exp Ther. 2009;330:135–141. doi: 10.1124/jpet.108.150052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.OH YK, Nix DE, Straubinger RM. Formulation and efficacy of liposome-encapsulated antibiotics for therapy of intracellular mycobacterium avium infection . Antimicrobial Agents and Chemotherapy. 1995:2104–2111. doi: 10.1128/aac.39.9.2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mayer LD, Bally MB, Cullis PR. Uptake of adriamycin into large unilamellar vesicles in response to a pH gradient. Biochim Biophys Acta. 1986;857:123–126. doi: 10.1016/0005-2736(86)90105-7. [DOI] [PubMed] [Google Scholar]
- 54.Fritze A, Hens F, Kimpfler A, Schubert R, Peschka-Suss R. Remote loading of doxorubicin into liposomes driven by a transmembrane phosphate gradient. Biochim Biophys Acta. 2006;1758:1633–1640. doi: 10.1016/j.bbamem.2006.05.028. [DOI] [PubMed] [Google Scholar]
- 55.Quinlan JR. Improved use of continuous attributes in c4.5. Journal of Artificial Intelligence Research. 1996;4:77–90. [Google Scholar]
- 56.Kotsiantis SB. Supervised Machine Learning: A Review of Classification Techniques. Informatica. 2007;31:249–268. [Google Scholar]
- 57.Zheng W, Tropsha A. Novel variable selection quantitative structure--property relationship approach based on the k-nearest-neighbor principle. J Chem Inf Comput Sci. 2000;40:185–194. doi: 10.1021/ci980033m. [DOI] [PubMed] [Google Scholar]
- 58.Vapnik VN. The Nature of Statistical Learning Theory. New York: Springer; 2000. [Google Scholar]
- 59.Sch¨olkopf B, Smola AJ, Williamson R. Shrinking the tube: A new support vector regression algorithm. In: Kearns MS, Solla SA, Cohn DA, editors. Advances in Neural Information Processing Systems. Cambridge, MA: MIT Press; 1999. [Google Scholar]
- 60.Scholkopf B, Smola A, Williamson RC, Bartlett PL. New support vector algorithms. Neural Computation. 2000;12:1207–1245. doi: 10.1162/089976600300015565. [DOI] [PubMed] [Google Scholar]
- 61.Chang C-C, Lin C-J. LIBSVM : a library for support vector machines, in, Software available at http://www.csie.ntu.edu.tw/~cjlin/libsvm. 2001
- 62.Frezard GSA, Demicheli C F. Encapsulation of Mithramycin in Liposomes in Response to a Transmembrane Gradient of Calcium Ions. Journal of Inclusion Phenomena and Molecular Recognition in Chemistry. 1997;28:51–62. [Google Scholar]
- 63.Gaspar MM, Cruz A, Penha AF, Reymao J, Sousa AC, Eleuterio CV, Domingues SA, Fraga AG, Filho AL, Cruz ME, Pedrosa J. Rifabutin encapsulated in liposomes exhibits increased therapeutic activity in a model of disseminated tuberculosis. Int J Antimicrob Agents. 2008;31:37–45. doi: 10.1016/j.ijantimicag.2007.08.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.