Exploring the “Anfinsen trail,” the pathway by which unstructured proteins spontaneously fold to their native functional form, has become a central goal of the protein chemists. In order to understand a biochemical pathway, one wants to isolate and describe the intermediate structures that lead from precursor to product. Unfortunately the intermediate forms that define protein folding pathways cannot be isolated or studied by the usual methodologies, however powerful, because folding intermediates are by definition unstable. Nevertheless, two reports in this issue, one by Miranker et al. on page 896, the other by Jennings and Wright on page 892, are able to present fairly detailed structural information on some kinetic folding intermediates in lysozyme and myoglobin, even though these ephemera only exist for less than 1 s during the folding process (1, 2). A related research article by Mayo and Baldwin on page 873 describes some characteristics of equilibrium unfolding intermediates in ribonuclease A that exist transiently at infinitesimal levels in equilibrium with the native state (3).
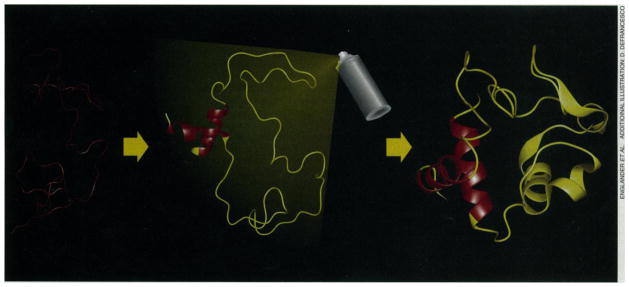
The folding experiments depend on a recently developed method (4) to obtain kinetic and structural information on partially folded protein forms that fly by on a millisecond time scale. The intermediates were labeled in-flight in a structurally sensitive way, in essence by writing on them with a brief pulse of chemical spray paint at any given point of time during folding. The painting technology (see figure) relies on the naturally occurring exchange of hydrogens between water and the peptide group NH hydrogens distributed uniformly throughout every protein molecule (5).
Figure. Stalking a folding protein.
The HX pulse labeling experiment. Folding proceeds from the random coil (left) through intermediate forms (middle) to the native state (right). Pulsed H-D exchange is used to label the protein in a time-resolved and structurally sensitive way while it refolds. Analysis of the refolded native protein by two-dimensional NMR or mass spectrometry (or both) can then provide kinetic and structural information on transient intermediates in the folding process.
The protein in D2O solution is initially unfolded with a denaturant, such as concentrated guanidinium chloride (GdmCl), so that the exchangeable amide NHs of all the peptide groups exist as ND (left side of figure). In a rapid-mixing experiment, the protein is diluted into H2O. Conditions are set so that ND-to-NH exchange of the freely exposed amides is slow (pH 6 and 10°C, for example, where the exchange half-time is several seconds). With the GdmCl now quite dilute, the protein begins to refold. At some time during the folding process, between 3 ms and several seconds, the refolding chains are labeled so that the parts of the protein that are already folded can be distinguished from the parts that are not (middle of figure). This is accomplished in a second mixing step simply by raising the pH of the refolding protein solution. High pH promotes the D-to-H exchange reaction. For example, at pH 10 and 10°C, amide NDs in parts of the protein backbone that are still unfolded will exchange to NH in about 1 ms. Other amides at positions that have already formed a hydrogen-bonded structure are protected from exchange and persist as ND. The high pH labeling pulse is maintained for some milliseconds and is then neutralized by a third mix, which drops pH to perhaps 5, effectively halting all further exchange. The protein molecules complete their run to the native state (right side of figure). The H-D exchange pattern imprinted on the protein by the labeling pulse is now locked in place by the native structure and can be analyzed at leisure, usually by two-dimensional nuclear magnetic resonance (NMR), to read out the degree of H labeling at many dozens of identifiable amides all through the protein. A series of such experiments with the labeling pulse imposed at different times during folding can reveal the time course for the formation of the various hydrogen bonds in the protein. From these data, the time course for forming each of the protein’s structural elements might be ascertained and thus the pathway for formation of the native structure.
Miranker et al. (1) used these methods to study the folding of hen egg lysozyme. Earlier hydrogen exchange (HX) labeling experiments showed that lysozyme shares a surprising and particularly frustrating aspect of most folding behavior so far observed. Folding is heterogeneous. The rigorously purified protein preparation in the chemist’s test tube splits into different sub-fractions that fold at different rates. This heterogeneity in folding leads to special problems of interpretation, which Miranker et al. solve by an ingenious applicaton of mass spectrometry. Mass spectrometry can separate the refolded proteins according to mass so that, unlike NMR analysis, the molecular distribution of H-D labeling obtained in an HX labeling experiment can be resolved.
The single chain lysozyme molecule contains two distinct lobes: an α domain rich in α helix and a β domain rich in β sheet. Earlier work (6) showed that protection against HX labeling develops in a fast phase (5 to 10 ms) for 40% of every measurable amide in the α domain and for 25% of every measurable amide in the β domain. The early folding phase might represent molecules with 40% having only their α domains folded and 25% only their β domains. In this case, a mass spectrometric analysis would show the early appearance of a partially deuterated fraction (40% + 25%) with intermediate mass. At the other extreme, 25% of the lysozyme molecules may fold both their α and β domains in the fast phase. Then, a mass spectrometric analysis would display an early forming heavy fraction (25%) with both domains protected and fully deuterated, an early forming intermediate mass fraction (15%) with only the α domain protected, and an unprotected light fraction (60%) that does not fold at all until later in the process. The mass spectrometry results clearly select the second case. Furthermore, α domains apparently can fold by themselves and then may or may not entrain the β domain, but β domains do not fold independently. The mass spectrometry experiment has general applicability. When any two structural elements fold on the same time scale, mass spectrometry can indicate whether the two elements fold together in the same molecule or independently in different molecules.
Jennings and Wright (2) used the HX pulse labeling experiment with NMR analysis to study the folding of apomyoglobin, the myoglobin molecule with its heme group removed. The results define a folding intermediate that forms in less than 5 ms and strongly resembles a previously known (7) equilibrium folding intermediate, the so-called molten globule form of apomyoglobin. This result will provide comfort and joy to a growing group of investigators who have attempted to sidestep the problems of studying kinetic folding by studying folding intermediates that can be obtained and examined directly in an equilibrium form. Equilibrium molten globule forms, it has been suggested, provide easily accessible analogs of true kinetic intermediates.
The molten globule has an interesting history. After some early skepticism, protein chemists have now embraced the idea that proteins can exist not only in their native and fully unfolded states but also as intermediate forms referred to as molten globules. The molten globule in its early incarnation was a strictly defined construct (8), but a growing zoo of intermediate protein forms has broadened the definition to include any protein form that is less than fully native but not yet fully unfolded. Examples include the equilibrium molten globule of apomyoglobin, which has three of the normal myoglobin helices, helices A, G, and H, identified by equilibrium HX labeling experiments (7). This motif, together with the early part of helix B, is copied in the newly found kinetic molten globule. The lysozyme form discussed before, with only the α domain folded, may provide another example.
In their research article, Mayo and Baldwin (3) deal with a series of transient, partially unfolded forms of ribonuclease A that exist under native protein conditions only about one-millionth of the time. These forms represent conformationally excited states in dynamic equilibrium with the native state. Having safely traversed the complex energy landscape that led them to fold to their lowest energy, native conformation, protein molecules are fated to spend their lifetime restlessly reexploring that same Boltzmann landscape. Just as the Oxford and Scripps groups used HX-based methods to learn how proteins descend through this terrain in folding, Mayo and Baldwin exploited the naturally occurring HX behavior of native protein molecules to study the equilibrium reexploration process.
The investigation of these rare forms is possible because, according to the unfolding model for protein-hydrogen exchange (5), H-bonded NHs can exchange with solvent only during the small fraction of time when their protecting H bonds are broken in transient, high-energy, conformational unfolding reactions. The unfolding model relates HX rate to an unfolding equilibrium constant and therefore quantitatively connects HX rates with the Gibbs energies for unfolding. In this view, the pattern of exchanging hydrogens can reveal the conformation of the partially unfolded form that mediates their exchange, and their exchange rate reveals the energy level of the partially unfolded state.
Mayo and Baldwin set out to test the proposition that the naturally occurring HX behavior of structurally blocked protein NHs depends on global and local unfolding reactions. They measured the response of the slowest exchanging NHs of ribonuclease A to low levels of the denaturant, GdmCl, reasoning that the energy of each unfolding reaction, measured by the HX rates of the appropriate NHs, should correlate with the size of the unfolding unit, measured by the sensitivity of HX rates to GdmCl concentration. For most of the NHs, this behavior is found. However, when the global denaturation energy for ribonuclease is considered, some surprises appear. Unfolding energies calculated from the HX rates appear to be displaced upward from the denaturation energy by several kilocalories, perhaps implying residual protecting structure in the globally unfolded state. The HX pathway for all of the slowest NHs appears to involve a common “globally unlocked” state and, in addition, further energy steps that vary from one NH to another. The unlocked state, seen here as a transiently populated high-energy form, may represent the transition state that normally determines the rate of protein unfolding reactions. It appears to resemble a “dry molten globule.” These experiments begin to show how hydrogen exchange can be used as a tool for surveying the energy landscape for protein unfolding.
The three papers on protein folding and unfolding in this issue illustrate the new avenues opened by hydrogen exchange, NMR, mass spectrometry, and other powerful techniques now available for studies of protein structure and behavior. These approaches provide the bright promise that we can ultimately understand how the factors encoded in the amino acid sequences of proteins lead to the range of protein forms that govern all the processes of living organisms.
References
- 1.Miranker A, Robinson CV, Radford SE, Aplin RT, Dobson CM. Science. 1993;262:896. doi: 10.1126/science.8235611. [DOI] [PubMed] [Google Scholar]
- 2.Jennings PA, Wright PE. :892. ibid. [Google Scholar]
- 3.Mayo SL, Baldwin RL. :873. ibid. [Google Scholar]
- 4.Baldwin RL. Curr Opin Struct Biol. 1993;3:84. [Google Scholar]; Englander SW, Mayne L. Annu Rev Biophys Biomol Struct. 1992;21:243. doi: 10.1146/annurev.bb.21.060192.001331. [DOI] [PubMed] [Google Scholar]; Elove GA, Roder H. Frontiers in Protein Folding. In: Georgiou G, editor. ACS Symposium Series 470. American Chemical Society; Washington, DC: 1991. pp. 50–63. [Google Scholar]
- 5.Englander SW, Kallenbach NR. Q Rev Biophys. 1984;16:521. doi: 10.1017/s0033583500005217. [DOI] [PubMed] [Google Scholar]
- 6.Radford SE, Dobson CM, Evans PA. Nature. 1992;358:302. doi: 10.1038/358302a0. [DOI] [PubMed] [Google Scholar]
- 7.Hughson FM, Wright PE, Baldwin RL. Science. 1990;249:1544. doi: 10.1126/science.2218495. [DOI] [PubMed] [Google Scholar]
- 8.Ptitsyn OB, Pain RH, Semisotnov GV, Zerovnik E, Razgulyaev OI. FEBS Lett. 1990;262:20. doi: 10.1016/0014-5793(90)80143-7. [DOI] [PubMed] [Google Scholar]