Mimicking natural tissue structure is crucial for engineered tissues with intended applications ranging from regenerative medicine to biorobotics. Native tissues are highly organized at the microscale, thus making these natural characteristics an integral part of creating effective biomimetic tissue structures. There exists a growing appreciation that the incorporation of similar highly organized microscale structures in tissue engineering may yield a remedy for problems ranging from vascularization to cell function control/determination. In this review, we highlight the recent progress in the field of microscale tissue engineering in order to discuss the use of various biomaterials for generating engineered tissue structures with microscale features. In particular, we will discuss the use of microscale approaches to engineer the architecture of scaffolds, generate artificial vasculature, and control cellular orientation and differentiation. In addition, the emergence of microfabricated tissue units and the modular assembly to emulate hierarchical tissues will be discussed.
1. Introduction
Each year, millions of patients suffer from organ failure or damage. Many of these patients die while they are on organ transplantation waiting lists due to insufficient number of donors. The field of tissue engineering has emerged to address this need by creating transplantable tissues or organs in the laboratory. It is an interdisciplinary field which exploits living cells through integration of engineering, materials science, biological sciences and medicine to maintain, restore and enhance normal tissue and organ function.[1, 2]
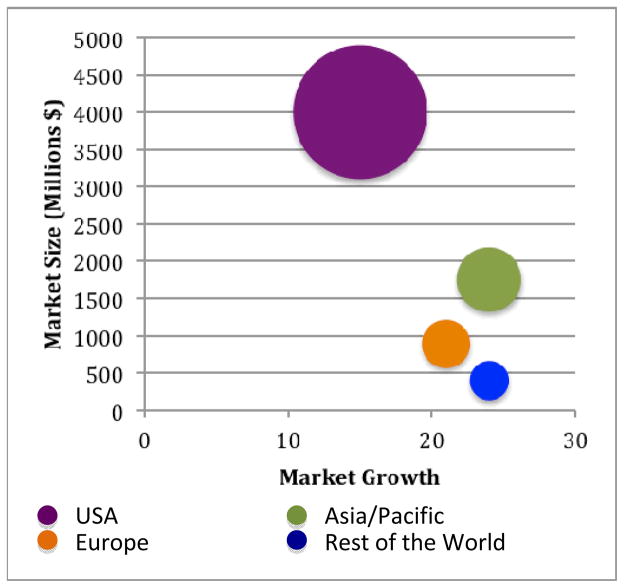
Almost two decades after Langer and Vacanti’s seminal article “Tissue Engineering” [3] in which they described the principles of tissue engineering, off-the-shelf engineered tissues are becoming more and more realistic. The market for tissue engineering and regenerative medicine in the United States has grown to $6.9 billion in 2009 (Fig. 1).[4] This market is estimated to further grow to about $32 billion in 2018. The potential market spans a wide range of tissues from degenerative or trauma caused orthopedic or nervous system therapies to cardiovascular and diabetes applications, and even to dental or ophthalmological problems. However, currently available engineered tissues are largely limited to skin epidermis, corneal epithelium and cartilage.[4] The common point of these tissues is their relatively simple architecture; in other words the simplicity in their tissue architecture and cellular organization. For example, there is less need for a vasculature in an engineered cartilage tissue or corneal epithelium compared to metabolically active and large tissues such as the heart or liver. Most tissues are composed of more than one cell type, usually with quite regular and microscale organization of these cells and extracellular matrix (ECM) components secreted by them, in order to perform a specific function. Since the functionality of a certain tissue is related with this complex architecture, tissue engineers have been trying to mimic and recapitulate this complexity in vitro. Although there are other approaches such as organizing cell sheets (which will not be covered in this review article), modulating the scaffold microarchitecture is one of the most potent ways of achieving biomimetic tissues. A three dimensional (3D) version of this biomimicry is a relatively recent phenomenon, and this research field is currently under intense exploration.[5] Another advantage of incorporating 3D microfeatures within tissue engineering scaffolds is the spatial organization of different types of cells in the same way as in the target tissue. Similarly, various ECM macromolecules or ECM-mimicking synthetic molecules can be spatially distributed in a predetermined manner to enhance biomimicry.
Figure 1.
Worldwide tissue engineering, cell therapy and transplantation market. Size and growth by region, 2009.[4]
Advances in microfabrication technologies have been exploited by an increasing number of research groups. Often technologies from other engineering disciplines have been translated and utilized in creating microfeatures in engineered scaffolds in a controlled manner. These include photolithographic approach of the electrical and electronic engineering, electrospinning tools of the textile industry, emulsification and fluid dynamics principles of chemical engineering and rapid prototyping methods of mechanical engineering. Currently, there are various 3D microfabrication techniques that have been explored as a potential remedy to the challenges facing tissue engineers. These challenges include controlling cell-cell and cell-material interactions for multicellular engineered tissues, vascularization of engineered tissues for proper oxygen/nutrient delivery, and directing stem cell fate using microengineered platforms. Here, we will first discuss the fabrication methods for creating 3D microscale tissue engineering constructs and then discuss the application of these approaches to the above mentioned problems.
2. Approaches for engineering 3D microfabricated tissues
2.1. Photomask-based methods
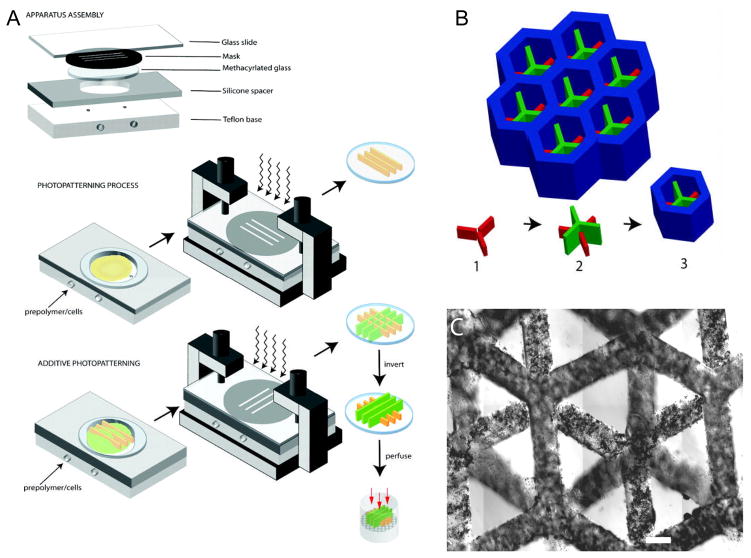
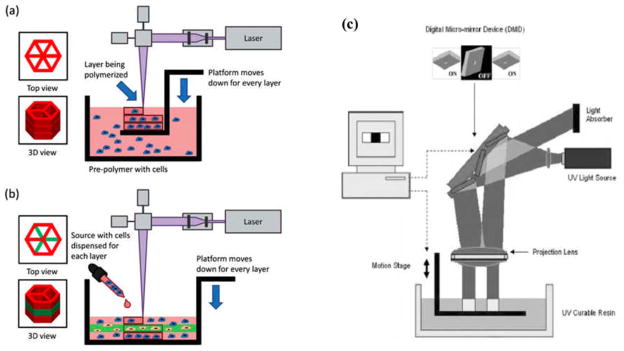
One commonly used technique to create micropatterned tissues and scaffolds is the use of direct polymerization through photomasks. A photomask is a two dimensional (2D) pattern printed onto a transparent sheet, designed such that light only passes through the mask in specific patterns.[6–12] The use of a photomask in conjunction with polymers that crosslink due to exposure to light of various wavelengths, most often, but not always ultraviolet (UV) wavelengths, restricts the crosslinking of the polymer to the regions where the light is permitted to pass through the photomask (Fig. 2A). As only the portions of the prepolymer residing below the transparent regions of the mask receive the necessary stimulus to initiate the polymerization reaction, only these areas are crosslinked. This technique has been used successfully for a number of hydrogels as well as other polymers, typically those containing acrylate groups in their backbone.
Figure 2.
Demonstration of photomask technique to create micropatterned structures. A) Prepolymer is loaded into a container of specified dimensions with a transparent top surface containing the photomask. UV light passing through the photomask to the photosensitive prepolymer causes polymerization only in the regions where UV light is able to pass through. B) Subsequent UV applications allows for layer-by-layer assembly of 3D structures. C) Functional 3D hepatic tissue created using this technique. Figure and legend reproduced with permission from.[6] Copyright 2007, Federation of American Societies for Experimental Biology.
The most common components required to create micropatterned polymer structures using photomask-based techniques are a light source, a photocurable polymer, a photomask, and a photoinitiator. As expected, there are several conditions, which must exist in order to create micropatterned polymers using photomask-based techniques. First, one needs a polymer with the appropriate chemistry that is amenable to polymerization caused by photosensitive free radicals. An appropriate photoinitiator must be used that not only initiates the reaction, but is stable enough to allow for rapid cessation of the reaction to avoid over polymerization and loss of pattern fidelity, while minimizing cytotoxicity. For acrylate-based polymer systems, the prepolymer backbone contains acrylate groups, which are involved in the crosslinking of polymer when exposed to light in the presence of the photoinitiator. There are many photoinitiators used in these processes, however, in general, all photoinitiators behave similarly in that light exposure causes the photoinitiators to decompose into free radicals. These free radicals then interact with the highly reactive double bond in the acrylate groups, leading to acrylate-acrylate permanent bonds when any two acrylate groups come in contact with each other following interaction with the free radicals. This reaction can occur rapidly, often within seconds or fractions of a second, dependent on the relative concentrations of the prepolymer and photoinitiator as well as the strength and duration of light exposure. Specific care must be taken to limit the exposure time to avoid this reaction from spreading outside the region of light exposure, as this will decrease the fidelity of the desired pattern. Careful modulation of the UV light exposure and the photoinitiator concentration must be observed to ensure that the cell viability is not affected by excess free radical generation or DNA damage due to UV light. In addition, the conditions of polymerization and the prepolymer characteristics, such as the solvent the prepolymer is dissolved in, can be cytotoxic. Typically, photomask based approaches are used with prepolymers which form hydrogels upon crosslinking or polymerization. Aqueous environment of a highly hydrophilic prepolymer solution provides sufficient mobility to the polymer chains and the acrylate groups, which are able to come into contact with one another as well as the free radical products of the photoinitiator. In addition, the use of hydrophilic polymer improves cell viability encapsulated within the hydrogels due to their diffusive properties.[13–15] Some examples of commonly used hydrogels for these applications are poly(ethylene glycol) (PEG),[16–22] methacrylated hyaluronic acid (meHA),[23–30] gelatin methacrylate (GelMA),[5,31–35] as well as co-polymer mixes containing these and other polymers.[24,31,33,36,37] PEG is one of the most hydrophilic polymers and its acrylated form (PEG-diacrylate or PEGDA) has been used extensively cell-laden micropatterned structures.[10,13] PEGDA hydrogels have demonstrated good early cell viability, structural integrity and micropattern fidelity below 100 μm feature size.[6,7,10] Many researchers have demonstrated using micropatterned PEG as the basis for creating macroscale engineered tissues with microscale features.[10,11,14,20,38] One example of such approach is the engineering of functional hepatic tissues.[6–8] In this process a three step photolithographic process was used to generate honeycomb structures which supported and surrounded the individual hepatic microtissues (Fig. 2). Primary hepatic cells were mixed with RGD functionalized PEGDA and placed into a chamber where the height was controlled by removable spacers. Overhang containing 3D structures were created through exposure to UV light using different photomasks in a layered fashion. After culturing these 3D hydrogel systems, the encapsulated cells were demonstrated to have good viability, as well as production of albumin and urea demonstrating the cell-laden constructs possessed properties similar to native tissues.
Common problems associated with PEGDA hydrogels include decreased cell viability over time and the lack of cell responsive characteristics, such as the ability for cells to attach, spread, migrate as well as polymer network degradation.[13] For some cell types, this lack of cell responsive characteristics is not an issue, and therefore loss of viability is less common, however for many engineered tissues, such as those requiring functional vasculature formation, this inability to allow for cell migration and alignment generates a major challenge. Researchers have sought to alleviate these shortcomings through conjugation of enzyme degradable motifs,[39] as well as cell binding motifs onto the PEG backbone.[6,20,21] These modifications allow for cell binding and migration through degradation of the polymer chains. Another solution to this problem is combining PEG with other polymers with cell responsive elements.[33] For example PEG and GelMA can be combined to generate a tunable, cell-initiated degradable gel with improved long-term cell viability compared to PEG alone. GelMA is a biocompatible and biodegradable material which has attracted much attention. Gelatin, or denaturated collagen, is an inexpensive material, which can be isolated from various animal sources relatively easily. Although it is denaturated, it maintains cell binding capacity. Modification of gelatin with methacrylate groups makes it light polymerizable which is mechanical robust and stable at 37 °C, and patternable into different geometries with desired dimensions through microfabrication technology.[34] Photopatterning of 3D cell-laden hydrogels has also proven to be an effective technique for creating microscale building blocks for constructing macroscale structures with microscale resolution and architecture.[10–12,38,40] Overall, the use of photopatterning to create microscale structures is a simple and straightforward manner to study tissue morphogenesis, cell behavior in response to microarchitectural cues, as well as the basis for creating macroscale engineered tissues with microscale resolution and controlled co-culture. As more photopolymerizable polymers are developed, this area of research will continue to expand and positively impact the field of tissue engineering.
2.2. Micromold based methods
Micromolding is a low expertise, robust, reproducible and inexpensive fabrication approach that enables patterning of both planar and non-planar substrates.[41–43] It is one of the widely used methods for generation of micropatterns in 3D tissue engineering constructs.[42,44] Shape and size of these patterns can be precisely controlled and they can range from 2–500 μm.[45,46] This approach involves elastomeric polymers, which are casted to form microstructures to be used as molds. Crosslinking of the prepolymer solution between a flat surface and a patterned elastomer provides the solid hydrogel in the shape of the pattern used (Fig. 3). Formation of molded patterns on the prepolymer solution is usually carried out by UV or temperature assisted crosslinking. Although micromolding is a popular technique to generate patterned cell-laden hydrogels, there are some drawbacks. For instance, hydrophobicity and swelling of the elastomeric molds are main concerns with this technique.[47] Polydimethylsiloxane (PDMS) is usually the elastomeric polymer of choice to produce the negative replica from silicon masters yielding molds for patterning.[43,47–49] PDMS is an inert polymer with high permeability possessing optical transparency and sufficient flexibility.[43,48] In addition to PDMS, polyurethanes, polyimines or Teflon™ are also utilized to prepare molds for patterning purposes.
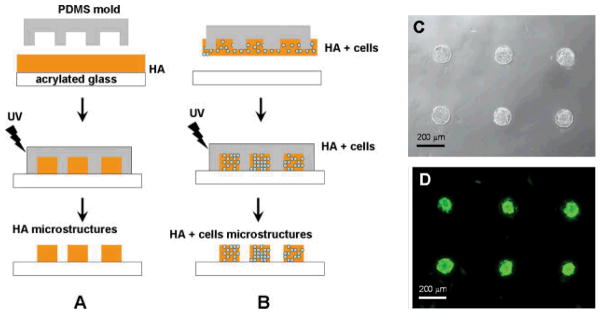
Figure 3.
Schematic diagram of the hyaluronic acid (HA) micromolding process. Poly(dimethylsiloxane) (PDMS) structures were used to mold a layer of HA into the void regions of the stamp. The polymer was then cured with exposure to UV light to fabricate HA microstructures. To fabricate HA microstructures without cells, a thin polymer film on the substrate was molded. A) To fabricate HA microstructures that encapsulated cells, B) the HA solution was transferred from the PDMS mold onto the substrate and subsequently crosslinked, C) Cell docking and viability within HA microwells after 24 h. Figure and legend reproduced with permission from.[133] Copyright 2006, John Wiley & Sons.
In a recent study, microgrooved molds fabricated from PDMS were chemically coated with a temperature responsive polymer, poly(N-isopropylacrylamide) (PNIPAAm), to generate size and shape controlled tissue constructs.[50] NIH-3T3 cells were seeded on PNIPAAm coated PDMS molds and cultured for three days to allow for the formation of longitudinal tissue modules. PDMS molds were then flipped on a glass slides to retrieve the tissue fibers at room temperature, which was facilitated by expansion of the PNIPAAm material. These final tissue units can be used to assemble and build larger constructs for tissue engineering and be useful in applications where alignment of cells is required. Similarly, sequentially patterned cells were encapsulated in agarose hydrogels on PNIPAAm patterned molds to create 3D mimics of microtissues.[51] In this study, cells were uniformly distributed within cubical, cylindrical or striped hydrogels. First, PNIPAAm patterns were made by UV-crosslinking and equilibrated at 24 °C to obtain their original shape. Second, cells were encapsulated in agarose gel precursor, then molded in patterns and crosslinked through incubation at 25 °C to achieve the mold patterns. After equilibrating at 37 °C, microgels were retrieved from the molds by taking advantage of shrinking ability of PNIPAAm at 37 °C. Furthermore, multicomponent hydrogels were generated using both 3T3s and human umbilical vein endothelial cells (HUVECs) or HepG2s and HUVECs in the same patterns. A second cell type was encapsulated in agarose and placed in the spaces created by the shrinking of PNIPAAm mold allowing two cell types patterned in the same gel structure. This micromolding technique can be used to produce microfabricated tissue constructs for tissue engineering applications.
2.3. Rapid prototyping (RP) based methods
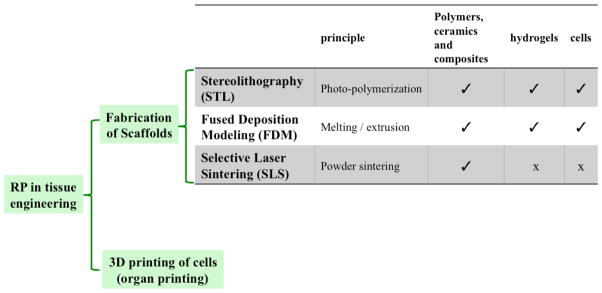
One of the requirements for translational applications of engineered tissues is a high throughput and automated method, which can also produce patient specific constructs. RP based methods are emerging fields in tissue engineering, which can potentially be used to fabricate such customized tissues. Besides being high throughput, fully automated and patient specific, these RP based approaches have excellent 3D microfabrication capacity.[52–54] RP is an automated additive manufacturing technique, which is a relatively recent fabrication method that was introduced around 1990s. Additive manufacturing is the “process of joining materials to make objects from 3D model data, usually layer upon layer, as opposed to subtractive manufacturing methodologies of traditional machining” as defined by American Society for Testing and Materials. Various additive fabrication methods have found applications in tissue engineering (Fig. 4). The phrase “rapid” is coined to emphasize the production time. It enabled fabrication of a construct within only few hours, which could have taken from several hours to several days using contemporary methods.
Figure 4.
Rapid Prototypoing (RP) techniques frequently employed in building microfabricated tissue engineering scaffold.
Although various rapid prototyping techniques are available, they all employ the same basic processes: 1) Computer-Aided Design (CAD) modeling of the construct, 2) conversion to a compatible format (i.e. STL), 3) slice the file into thin cross-sectional layers, 4) layer-by-layer fabrication of the construct. As the first step, pre-existing CAD files as well as custom made ones created using any solid modeler such as AutoCAD or Pro/ENGINEER can be used. This step is usually same in all of the RP techniques. As a second step, the CAD is converted into a consistent file format in order to standardize the rapid prototyping industry. To do so, the STL format has been used as a tribute to the format created by the first RP technique (i.e. stereolithography). The STL format approximates a 3D and curved surface as a set of planar triangles by storing the coordinates of the vertices and the normal vector for each triangle. Next, the 3D design is sliced into layers. The thickness of the layers can be varied from 0.01 mm to 0.7 mm depending on the RP technique. The program may also generate an auxiliary structure to support the model in case of delicate features such as thin-walled sections or internal cavities. The construct is fabricated one layer at a time.
Many competing additive technologies are available. Their main differences are found in the method how each layer is built. In some of them, thin layers are cut into desired shapes and consecutively joined together, while some requires liquid materials cured, and in others melting or softening of the material is used to produce the layers. One possible way of classifying RP techniques can be according to the nature of the material that is used for the fabrication. In broad sense, this can be a photolabile material (fabrication using light), or a material that requires high temperature to form an intact construct (fabrication using heat). Table 1 summarizes some of these techniques relevant to tissue engineering applications.
2.3.1. Photolabile RP Approaches
The work that initiated the revolution of Rapid Prototyping was Charles W Hull’s 1986 Patent on stereolithography (US Pat. 4,575,330). In this technique, 3D models are built, layer-by-layer, from liquid photolabile polymers (e.g. resin) that can be cured when exposed to UV light. In SLA, fabrication occurs on a platform that is immersed in a container of liquid resin, just below the surface. Highly focused UV laser (usually 325 nm wavelength) traces out each layer, solidifying the model’s cross sections one at a time, leaving excess areas in its liquid form. After each layer is fabricated, the platform is incrementally lowered by a motorized elevator; and a new layer is coated prior to laser tracing of the next slice. This process is repeated until the construct is complete. In its conventional form, SLA was used to fabricate micropatterned scaffolds for bone tissue engineering applications.[55] Cooke et al. used a blend of diethyl fumarate and poly(propylene fumarate), with SLA for scaffold fabrication. After this initial attempt to utilize SLA for hard tissue engineering applications, photolabile hydrogels were also microfabricated using this approach for possible soft tissue engineering applications.[56–59] Arcaute et al. characterized PEG solutions to control the crosslinking depth, which determines hydrogel thickness, which is crucial for fabricating structures with overhangs.[59] Characterization was done by measuring the thickness at different laser energies and photoinitiator concentrations. Two concentrations of PEG-DA 1000 Da (20% and 30% (w/v)) were characterized. By incorporation of cytocompatible photoinitiator Irgacure 2959, cells could be mixed into the polymer solution and cell-laden hydrogels could be fabricated. Similar approach was used to micropattern multiple materials (i.e. different molecular weigth PEGs) which can be useful for fabricating complex tissues (or tissue interfaces) that requires different material properties in a single construct.[56] These studies characterized the fabrication parameters required for incorporating cell compatible biomaterials with the SLA technology as a first step towards its utilization in tissue engineering approaches. However, viability and uniformity of the encapsulated cells was still an issue. Chan et al. modified the SLA process from its conventional top-down version to a bottom-up approach.[57] In the top-down approach, the elevator where the construct is fabricated moves inside a vat that is filled with a prepolymer solution. When cells are added to the prepolymer solution, they sit in the vat until all the layers are formed. This can decrease cell viability since the cells are continuously in contact with the photoinitiator. It may also result in cells settling towards the end layers, which leads to non-uniform cell encapsulation. In the bottom-up version, fresh cell containing prepolymer solution was added on each layer. This allows uniform cell distribution and viability up to 15 days (Fig. 5A and B).[57] This bottom-up approach also allowed the fabrication of multicellular constructs with varying cell type at each layer. In addition to the inherent z variability that comes with the layer-by-layer microfabrication approach, this method was further improved by incorporating multiple cell and material types in x and y directions.[58] Briefly, uncrosslinked polymer solution can be washed away in between the steps to add different cell and material types in different compartments in all 3 dimensions. SLA has a resolution of around 100 μm in step size (z resolution) and the x-y resolution depends on the laser beam diameter which is usually ca. 75 to 250 μm.[59]
Figure 5.
Schematic diagrams of modifications to stereolithography system for tissue engineering applications. A) In the top-down approach, the layout consists of a platform immersed just below the surface of a large tank of pre-polymer solution. After the layer is photopolymerized, the platform is lowered a specified distance to recoat the part with a new layer. B) In the bottoms-up approach, the pre- polymer solution is pipetted into the container one layer at a time from the bottom to the top. This setup was modified especially for cell encapsulation applications, which required: (1) reduction in total volume of photopolymer in use and (2) removal of photopolymer from static conditions that cause cells to settle. Figure and legend reproduced with permission from.[57] Copyright 2010, RSC Publishing. C) Digital Micromirror Device in a microstereolithography set-up. Figure reproduced with permission from.[62] Copyright 2005, John Wiley & Sons.
An approach that provides higher resolution with similar processing principles to a conventional SLA process, which is commonly referred as microstereolithography (μSL) uses a digital micromirror-array device (DMD).[60,61] The major difference is that instead of scanning each layer with a laser, DMD based approach uses a digital mask which is designed and sliced for each layer as usual, but this time as a series of PowerPoint slides. These slides are used to generate a dynamic mask through being executed on the DMD chip. This dynamic mask is used to create the micropatterns in each layer. The light source is illuminated on the polymer surface, resulting in the solidification of the exposed regions. Microfabricated complex 3D constructs can be built by sequential polymerization of the subsequent layers (Fig. 5C). This system uses a commercial projector and comprises of five main components: a dynamic mask created by the DMD chip embedded in the projector, a projection lens assembly, a translation stage with a micrometer, a vat containing macromer solution and a light source. Features as small as 20 μm can be fabricated using the μSL. This method has been employed in order to fabricate polymeric scaffolds containing pores and channels with wide variety of shapes, dimensions and layers.[60–62] Different geometries (hexagons, triangles, honeycombs with triangles, and squares) can be incorporated within a single scaffold (165–650 μm). Optical Diffraction and diffusion are considered to be the main limitation of this technique.[62]
2.3.2. High Temperature RP Approaches
High Temperature Approaches in Rapid Prototyping, also known as “melt–dissolution deposition system” are a class of technologies in which during the process of manufacturing, each layer is created by extrusion of a material through an aperture while it moves across the plane of the layer cross section. The material cools, solidifies itself and fixed to the previous layer. Successive formation of layers results in a complex 3D construct with a defined geometry in micron range. The basis of the process is either employing a powder form of a suitable material and apply high temp to melt/bond, which is called particle bonding, or using a material which is already in the molten state, which is called melt deposition.[63] Although these methods are useful in creating microstructured scaffolds, utilization of high temperature renders them undesirable for applications requiring incorporation of bioactive agents.
In particle bonding approach, particles are bonded selectively in successive thin layers of powder material. Since the object could be embedded, hence supported by the unprocessed powder, this technique enables the fabrication of overhanging features as well as through channels.[54] Major advantage of this approach is its ability to fabricate porous structures with controllable macro and micro-porosity.[64] In addition to that, the powder-based materials provide a rough surface to the fabricated scaffold, which can enhance the cell adhesion. A commonly used particle–bonding method in tissue engineering is selective laser sintering (SLS). The basic concept of SLS is similar to that of SLA. A laser is employed to merge powdered materials in a predetermined manner into a solid object. The system contains a platform similar to that of SLA, only with a heat-fusable powder instead of photolabile resin. The platform is lowered and fabrication continues until the part is complete. Finally, the excess powder, which serves as support during the process, will be washed off. Most biomaterials that do not deteriorate but can fuse under a laser beam can be used for fabrication by SLS. In addition to that, organic solvents are not required for SLS, which is usually the case with synthetic polymers for preparing solutions of them in order to cast them to certain shapes or create patterns. SLS was used to fabricate scaffolds from polycaprolactone (PCL) in order to successfully construct prototypes of mini-pig’s mandibular condyle scaffolds.[65] These constructs could be fabricated within three hours, and replicated the desired anatomy precisely. SLS was also used to microfabricate ceramic,[66,67] polymeric[68–70] and composite[71–73] scaffolds mostly for bone tissue engineering applications. Resolution of SLS is depend on the laser beam diameter similar to SLA.
Fused Deposition Modeling (FDM) is one of the most commonly employed melt deposition techniques for fabricating tissue engineering scaffolds. FDM employs thermoplastic materials that are liquefied and deposited by an extrusion head, based on a CAD plan. The materials are deposited in layers as fine as 125 μm thick. In general, the melt process is not desirable for many tissue engineering applications where bioactivity of the scaffolds is important. The major drawbacks of FDM are low resolution, limited choice of materials and high operating temperatures.[54] Furthermore its resolution is relatively low (ca. 250 μm). Since the material should be still usable/applicable after being melted and casted, only a limited range of materials can be used in FDM. This criterion almost completely excludes all natural polymers. Also, the operating temperature of the FDM system is high for many biomolecules; and this affects the biomimetic characteristic of the scaffolds. Despite these drawbacks, FDM has been used to fabricate a number of scaffolds with highly interconnecting and controllable pore structure mainly for bone tissue engineering applications.[74] These scaffold were fabricated using synthetic polymers such as PCL[75,76] and poly (lactic acid-co-glycolic acid) (PLGA),[77] and composites of them with ceramics.[78]
2.4. Approaches for controlled microporosity
2.4.1. Gas foaming
Gas foaming techniques have been employed to create biomaterials with uniform and controlled microporosity for generating engineered tissue structures. These techniques are based on the dispersion of gas bubbles as an internal phase through a continuous phase of a polymer solution.[79] The continuous phase is then solidified around the dispersed internal phase through the polymerization or fast gelation. The porous structure of polymer is formed when the gas bubbles diffuse out from the polymer matrix. The gas bubbles can be (a) generated in situ through chemical reaction (conventional gas foaming);[80–82] (b) formed through the addition of an external inert gas to the continuous phase of the polymer;[83,84] or (c) released from a presaturated dense gas CO2/polymer mixture at high pressure (gas foaming with dense gas CO2).[85,86]
Conventional gas foaming techniques have been utilized to create highly porous scaffolds from both natural and synthetic polymers. In this method, an inert gas such as N2 or CO2 is generated via a chemical reaction by the addition of a blowing/foaming agent (e.g. sodium bicarbonate or ammonium bicarbonate) to the prepolymer solution. The use of a surfactant is required to stabilize the foam and avoid liquid drainage and bubbles coalescence prior to solidification of the polymer phase.[80] The microstructures of resultant polymeric scaffolds can be controlled by the foaming agent concentration, surfactant type and its concentration, and the solidification process. Highly porous alginate scaffold was fabricated by the formation of CO2 bubbles through the reaction between tartaric acid and sodium bicarbonate in an aqueous solution of alginate.[80] The fabricated foam was stabilized by addition of Pluronic F-108 surfactant prior to crosslinking reaction which locked-in the structure of resultant scaffold.[80] It was found that the morphological characteristics of the fabricated scaffolds could be controlled by the concentration of surfactant. Increasing Pluronic F-108 concentration from 4 to 6% (w/v) enhanced the scaffold pore sizes from 180 μm to 260 μm that was due to the increased expansion of polymer foam at higher surfactant concentration.[80] A similar approach was used to fabricate porous gelatin scaffold by the formation of N2 gas through the reaction between sulfamic acid and sodium nitrite in gelation solution containing a foam stabilizer.[81] The foam was allowed for fast gelatin at low temperature after which the generated gas bubbles were released from the polymer matrix to form a porous gelatin scaffold. The fabricated construct had uniform porosity with average pore size in the range of 90–230 μm and 97% pore interconnectivity. Furthermore, It was shown that the resultant hydrogels supported human C3A cells adhesion and viability.[81] Biopolymeric scaffolds with controlled microarchitectures were also formed by the addition of an inert gas (e.g. argon), with controlled flow rate, into an aqueous solution of a polymer containing a surfactant. Following the foam formation, the polymer solution was solidified through physical gelation and subsequent chemical crosslinking to form a stable scaffold. This method was used to create uniform porosity in gelatin,[83] hyaluronic acid, chitosan, and alginate scaffolds.[84] It was reported that the volume of injected gas allowed the control over the microstructures of fabricated scaffolds. For example, the average pore size of gelatin scaffolds fabricated using this method was increased from 250 μm to 360 μm when the injected gas volume was enhanced from 80% (v/v) to 90% (v/v).[83] Similar approach was employed to form hyaluronic acid and chitosan scaffolds with uniform average pore size of 250 μm and 210 μm and pore interconnectivity of 88 % and 95%, respectively.[84]
Gas foaming using dense gas CO2 has been used widely as an effective means of producing porous biomaterials for various tissue engineering applications. This technique eliminates the use of organic solvents and allows for incorporation of temperature sensitive growth factors into the scaffold due to the low critical temperature of dense gas CO2 (31°C). Dense gas CO2 has been used as a plasticizer and foaming agent to create porosity in the structure of various hydrophobic polymers such as poly (lactic acid) (PLA), PLGA and PCL.[85–88] Using this technique, the microarchitectures of resultant scaffolds can be controlled by processing conditions such as pressure, temperature, soaking time, and depressurization rate.[86,88] Tai et al. used dense gas foaming process to fabricate PLA and PLGA scaffolds with controlled pore sizes and structures for tissue engineering applications.[86] It was reported that the pore sizes of fabricated scaffolds were increased when slower rate of depressurization was used as this allowed longer period for pore growth. However, higher pressure and longer soaking time allowed more CO2 to diffuse into the polymer matrices, leading to higher nucleation density and production of smaller pores.[86] Due to low solubility of dense gas CO2 in hydrophilic polymers, CO2-water emulsion templating techniques have been developed to increase CO2 diffusion into a biopolymer solution and create uniform porosity. These techniques have been used to produce highly porous hydrogel scaffolds from various biopolymers including dextran,[89] chitosan,[90] and alginate,[91] and synthetic polymers such as polyvinyl alcohol (PVA), blended PVA/PEG,[90] and CaCO3/polyacrylamide composites.[92] It has been shown that the morphologies of porous hydrogels, produced by CO2-water emulsion templating techniques, can be tailored by the polymer solution concentration, surfactant concentration, and volume fraction of CO2 as an internal phase. Palocci et al. used CO2-water emulsion templating method to produce highly porous dextran hydrogels with properties resembling natural tissues.[89] The pore architecture of fabricated dextran hydrogels was controlled by the concentration of surfactant; increasing the surfactant concentration from 3.5 to 5% (v/v) resulted in the formation of more interconnected pores. In one study, the variation of CO2 volume fraction had no significant effect on the hydrogel microstructure.[89] However, Lee et al. showed that the porous structures of emulsion-templated PVA hydrogels could be controlled by altering the CO2 volume fraction. It was found that the average pore size of resultant hydrogels increased about 2-folds when the volume fraction of CO2 enhanced from 74 % to 79%.[90] Similarly, Partap et al. demonstrated that the pore sizes of calcium alginate hydrogels fabricated by using CO2-water emulsion templating technique was significantly increased from 43.3 μm to 250 μm when the CO2 volume fraction was enhanced from 40% to 78%.[91] In this study, dense gas CO2 simultaneously served as a templating phase to generate porosity and a reagent to induce acidity that released calcium ions from their chelated form and initiated physical crosslinking of the polymer phase. The resultant alginate hydrogels exhibited uniform interconnected pores in the range of 24–250 μm depending on the surfactant concentration and CO2 fraction.[91]
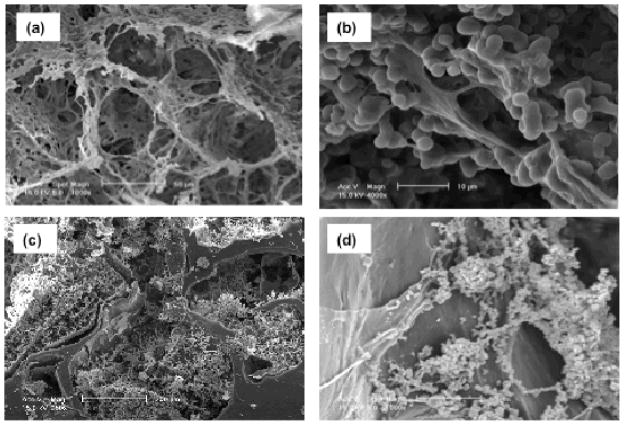
Dense gas CO2 was used to create porous tropoelastin/elastin composites with highly interconnected pores without the use of surfactant.[93] In this study, an aqueous solution of tropoelastin/elastin containing a crosslinking agent was pressurized with CO2 to dissolve the gas into the aqueous solution. The pores with ca. 78 μm size were then generated as a result of the release of CO2 from the aqueous solution during subsequent depressurization (Fig. 6A). The mechanical properties and microarchitectures of fabricated composites were controlled by processing parameters such as pressure, depressurization rate, crosslinker concentration, and biopolymer compositions.[93] The resultant composites were shown to support human skin fibroblast growth and migration within the 3D structures (Fig. 6B).[93] In a recent study, a combined gas foaming/salt leaching process was developed to produce porous 3D PCL/elastin composites.[88,94] In this process, PCL scaffolds with average pore size of 540 μm were first fabricated by melt-mixing of PCL with salt particles and subsequent gas foaming by using dense gas CO2.[88] The pore characteristics of PCL scaffolds were manipulated by gas foaming parameters such as pressure, temperature, depressurization rate, and salt concentration.[88] The PCL scaffolds were then impregnated with elastin solution and subsequently crosslinked under high pressure CO2 to form microporous structure of elastin within the macropores of PCL (Fig. 6C).[94] It was shown that the presence of crosslinked elastin within the pores of PCL promoted primary articular cartilage chondrocyte adhesion and proliferation within the 3D composites (Fig. 6D).[94]
Figure 6.
SEM images of scaffold with controlled microporosity. A) Tropoelastin/elastin composite fabricated by using dense gas CO2, B) the composite supported human skin fibroblast growth and migration. C) PCL/elastin scaffold produced by gas foaming/salt leaching process, D) the fabricated scaffold promoted primary articular cartilage chondrocyte adhesion and proliferation. Reproduced with permission from. [93, 94] Copyright 2010 & 2011, Elsevier.
Gas foaming processes are suitable techniques for the fabrication of porous scaffolds for various tissue engineering applications as they produce uniform porosity within materials and allow control over the microarchitecture of scaffolds. Biomaterials, fabricated using these techniques, can provide appropriate templates for cells seeded within their 3D structures. However, gas foaming techniques commonly involve conditions and chemicals that are detrimental for the cells, such as high pressure in dense gas foaming process and the use toxic surfactant in conventional gas foaming. Furthermore uniform cell distribution within the material might be prevented because of the inability to encapsulate the cells during the initial fabrication of scaffold.
2.4.2. Porogen leaching
Porogen leaching techniques have been widely used to generate porous scaffolds with controlled pore architectures. These techniques are based on the dispersion of a porogen within a polymer solution followed by solidification of the continuous polymer phase around the dispersed porogen particles. The porous structure is then formed by immersing the porogen/polymer construct in a suitable solvent to leach out the particles.[95] Various porogen materials such as salt,[88,96–99] sugar,[99] paraffin[100] and gelatin[101–103] have been used in this technique to generate highly porous biomaterials for different tissue engineering applications. The pore characteristics of resultant scaffolds including average pore size, porosity and pore interconnectivity can be controlled by the porogen geometry, size, and concentration.[88,99,104,105] For example, Horak et al. produced superporous poly(2-hydroxyethyl methacrylate) (pHEMA) scaffolds by combining a salt leaching technique with radical polymerization.[99] It was found that the pore sizes of pHEMA scaffolds increased from 28 μm to 69 μm by increasing the salt concentration from 40 to 41.4 vol.%. The porosity of resultant scaffolds was in the range of 81–91% depending on the volume of porogen.[99] In another variation, PLLA scaffolds, with controlled microstructures, were fabricated by using sugar sphere template leaching technique combined with thermally induced phase separation method.[106] In this study, a sugar template was first prepared by bonding sugar spheres with desirable sizes through hexane and heat treatment. The polymer solution was then cast onto the sugar template followed by thermally induced phase separation and leaching process. Using this techniques, porous PLLA scaffolds with pore sizes in the range of 180–250 μm and porosity of 97–98.2% were obtained.[106] In addition it was shown that the sugar spheres allowed control over the pore morphology and pore sizes of scaffolds; increasing the sugar particles enhanced the pore sizes and porosity of resultant scaffolds.[106] Following simulated body fluid incubation, bone-like apatite layer were grown uniformly throughout 3D scaffold. This demonstrated the bioactivity of the scaffold and its potential for bone repair.[106] A similar process by using paraffin spheres was employed to fabricate 3D nanofibrous gelatin scaffolds[107] and gelatin/apatite composites[108] with tunable physical properties, including fiber diameter and length, porosity, pore size, pore interconnectivity, and mechanical properties, for bone tissue engineering applications. Larger paraffin spheres led to larger scaffold pore size as controlled by the size of paraffin spheres, which in turn controls the pore sizes of gelatin scaffolds. In vitro studies demonstrated that the MC3T3-E1 osteoblasts seeded gelatin scaffolds had a greater dimensional stability compared to Gelfoam (a commercial gelatin foam) after four weeks of cell culture, demonstrating its ability to support tissue regeneration.[107] In another study, gelatin particles were utilized as porogen to produce porous PCL scaffold by using a combined gas foaming/porogen leaching technique.[103,109] In this process, PCL was first melt-mixed with gelatin particles at 60°C and then gas foamed by using a mixture of CO2/N2 as foaming agent. The porogen was subsequently removed by soaking the blend in water to form a porous structure of PCL containing both macropores (average pore diameter ~ 312 μm) and micropores (average pore diameter ~ 38 μm).[103,109] It was demonstrated that the weight ratio of gelatin and gas foaming parameters allowed the modulation of scaffold microstructures such as microporosity, average pore size, and pore interconnectivity.[109] The fabricated porous PCL scaffolds promoted hMSCs adhesion, proliferation, and 3D colonization.[103]
Particle leaching techniques allow for control over the overall porosity and average pore size of scaffolds. Control of biomaterial microstructures is important towards obtaining functional engineered tissues. Combining these processes with microfabrication technologies such as micropatterning, micromolding, and rapid prototyping techniques can provide advanced control over the microstructures, as well as macrostructures to create structurally biomimetic and functional engineered tissues.
3. Applications of microfabrication in 3D tissue engineering
3.1. Controlling cell-material and cell-cell interactions
The ability to create biomimetic microenvironments could potentially be the solution to the majority of the problems that tissue engineers are experiencing today.[110–116] The most prevalent among these problems is to fabricate multicellular complex tissues. The term biomimetic is coined to comprise precise, pre-designed, spatially patterned and temporally controlled biochemical and physical manipulation of the cellular microenvironment.[117–120] Biomimetic multicellular complex tissues can be achieved through 3D microfabrication approaches that are mentioned in the previous sections. Most recently, these technologies have started to be utilized as possible platforms to investigate cell-material and cell-cell interactions as a first step towards a functional engineered tissue. In particular, 3D microfeatures have been used to spatially control cell-cell interactions and to fabricate patterned 3D co-cultures of multiple cell types, to fabricate 3D scaffold with micropatterned materials or bioactive molecules, and to manipulate cell-material interactions via scaffold geometry.
The conventional approach to generate 3D co-cultures has been to seed cells on a biodegradable scaffold.[121] However, this approach has certain limitations since the cells in such tissues do not fully recreate tissue-like structures.[122,127] This may effect the final tissue function as adhesion, proliferation, differentiation and migration of cells are influenced by the nature of their homotypic and heterotypic cellular interactions.[25] Thus it may be difficult to fabricate fully functioning tissues if the spatial and temporal presentation of such biological signals are not taken into consideration. In their study, Hui and Bhatia demonstrated the importance of these multicellular interactions by using microfabrication techniques to achieve spatiotemporal control of the hepatocyte functionality. In particular they used a microfabricated movable culture device in which fibroblasts and hepatocytes were localized in a patterned manner to study the dynamics of cell-cell interaction. Their results showed that the hepatocyte phenotype was best retained when these cells were in direct contact with fibroblasts for a limited time (of the order of hours), and then separated for up to an effective range of 400 μm or less.[128]
Spatiotemporal presentation of bioactive factors can be detrimental for their action. They might be required to be presented in a sustained manner and their amounts might fluctuate depending on maturity of the cells. Furthermore, such bioactive molecules in native tissue may act in a bidirectional manner and with controlled feedback loops. Therefore, using conditioned media or adding bioactive molecules exogenously might not necessarily result in the expected outcome. In a recent study, microfabrication techniques were exploited to examine this question. It was found that skeletal muscle and neuron cells that were encapsulated in a spatially patterned manner resulted in enhanced functionality of the neurons compared to neurons encapsulated alone, while the conditioned media from the skeletal muscle cultures had no effect.[58]
Most of the spatially defined 3D microfabrication studies in literature have used hydrogels as the material of choice. There are two main reasons for this. First, hydrogels closely resemble the native tissue matrix due to their high water content and polymeric network structure.[129–131] A second factor is that cell-laden hydrogels enable the confinement of different cells or materials to certain compartments in 3D structures.[132] This property can be used to control cell behavior in a spatially regulated manner. For example, cell migration can be controlled by using different biomaterials, or tailored by adding peptide sequences that can render non-degradable hydrogels degradable via cellular enzymes.[120] Therefore, most of the examples in this section involve hydrogel-based scaffolds.
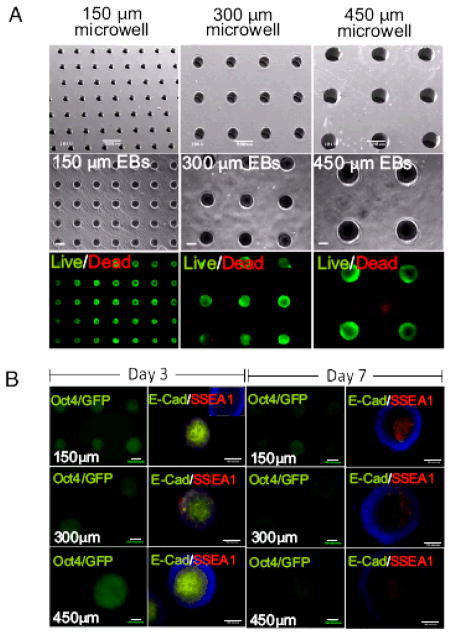
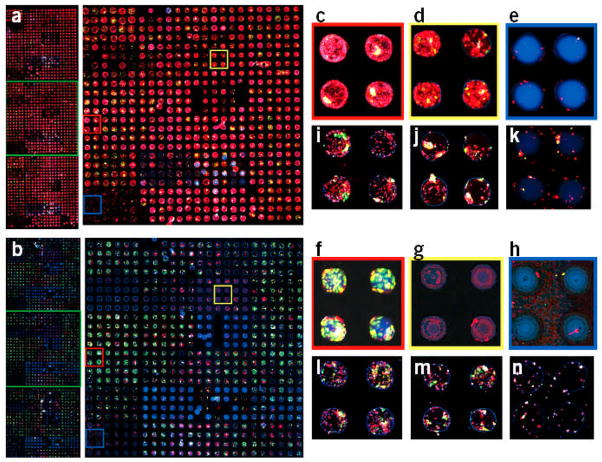
One approach in which cell-cell contact has been controlled by using microfabricated systems has been through the use of microwell templates. In this approach, photolithography or micromolding are used to fabricate platforms that can be used for controlled cell aggregation. [133–136] For example, polymers such as poly(acrylamide), PEG and HA have been micromolded to create microscale wells with different dimensions and geometries. Since these materials are non-adhesive, these microwells enhanced the degree of cell-cell interactions relative to cell-surface interactions and can be used to form cell aggregates, which can be further exploited to form 3D tissues. By varying the dimensions of these microwells the size and shape of the resulting cell aggregate can be controlled in a uniform manner.[133] Furthermore, these aggregates can be retrieved from microwells by mechanical agitation. A similar approach was used to co-culture fibroblasts and HUVECs, in polyacrylamide microwells to form multilayered aggregate structures.[137] In these aggregates fibroblasts occupied the inner core while HUVECs formed an outer layer. Furthermore, when fibroblasts were added onto already formed HUVEC spheroids, cells reorganized so that the fibroblasts again occupied the core of the structure. In addition, properly functioning tissues may be generated by providing sufficient cellular contact and managing diffusion related challenges. For example, in one study photocurable chitosan was used to fabricate microstructures for culturing spheroids in a spatially controlled manner.[138] Chitosan was first micromolded by a UV-crosslinking procedure to produce 50 μm deep and 200 μm diameter microwell shaped structures. Hepatocytes were then seeded and cultured within fabricated construct for three days to form hemispherical spheroid structures. In vitro studies showed the liver specific function of the hepatocytes by using an albumin secretion assay, demonstrating the potential application of the resultant hydrogels for liver tissue engineering.
Many native tissues consist of repeated functional units, such as the lobules in the liver, nephrons in the kidney, and muscle fibers. These “tissue modules” encompass the bulk of the function of the organs and tissues they comprise.[139] Given the ability to encapsulate cells in microscale gels, microfabrication techniques not only aim to accurately recreate engineered tissue components of specific shapes and microarchitecture,[139] but to assemble these structures into macroscale tissues.[140] Towards this end in the last decade many studies have investigated high throughput, automated and bottom-up fabrication of cell-laden microtissues. Some of the methods have exploited fabrication techniques from other engineering applications(i.e. bioplotting and stereolithography), [57,61,62,141] while others have found novel ways for directed assembly of microsized gels through modulating the polymer chemistry.[40,142]
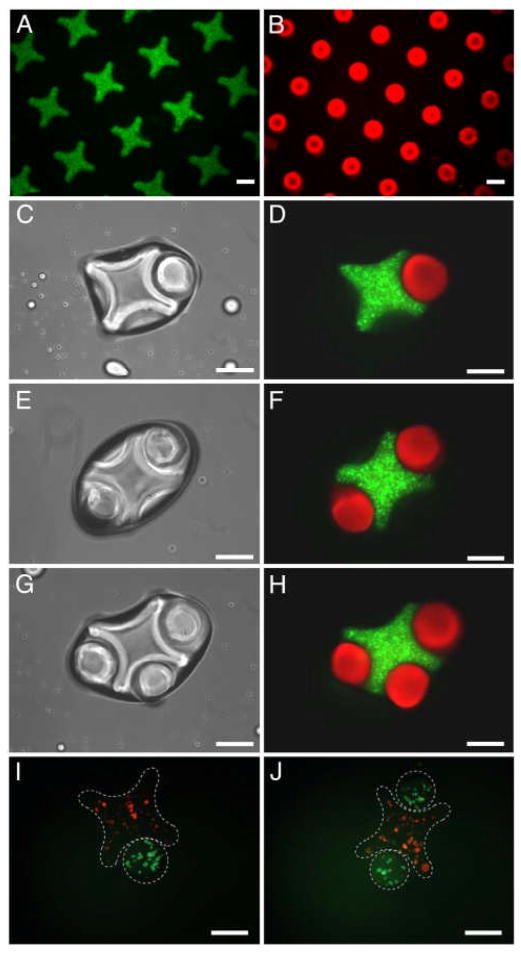
An early example in this process used directed assembly of cell-laden microgels by using hydrophobic and hydrophilic interactions (Fig. 7).[10] When microscale hydrogels were agitated in a hydrophobic medium, they assembled in an organized manner as a result of local minimization of the interaction free energy at the hydrophilic surface area exposed to the hydrophobic oil. These microscale hydrogel blocks (or microgels) were fabricated in specific geometries to favor particular assembled structures.[40,142] After directing the assembly of microgels into pre-defined shapes, a second crosslinking step was employed to stabilize the assembled microgels. Reducing the surface tension of the surrounding solution and increasing the hydrophobicity of the microgel have shown to improve this assembly process. This directed assembly technique can particularly be useful in fabricating multicomponent cell-laden constructs to achieve biomimetic tissue engineered constructs.[143] Du et al. also demonstrated that the secondary structures formed depend on the shape and aspect ratio of the rectangular microgel units. The concept of directed assembly was further demonstrated by creating complementary hydrogel structures, which naturally fit together in a lock-and-key mechanism. This showed that through careful design of the microgels, secondary and tertiary structures could be predictably controlled. This technique enables fabrication of centimeter scale tissues through directed assembly of microgels essentially floating on the surface of a dense, hydrophobic liquid. Expanding on the early work of the Whitesides and colleagues[144] which demonstrated similar mesoscale self assembly with PDMS structures, Zamanian et al. showed that PEG microgels would aggregate in predictable patterns (Fig. 7).[40] Similarly, with a brief secondary UV application, these centimeter scale single layer thick tissue-like sheets can be stabilized. Furthermore, a hierarchical assembly process was developed to create complex microgel blocks containing controlled co-cultures that could be used for creating centimeter scale structures with controlled cellular organization. Further advances in this technique have used wetting templates to generate 3D structures by using the liquid-air driven assembly process to assemble gels.[12]
Figure 7.
Use of 3D microgels as building blocks for higher order macroscale tissue-like structures. A) Directed assembly of lock-and-key-shaped microgels. B) Fluorescence images of cross-shaped microgels stained with FITC-dextran. C – H) Rod-shaped microgels stained with Nile red. I – J) Phase-contrast and fluorescence images of lock-and-key assemblies with one to three rods per cross. Fluorescence images of microgel assembly composed of cross-shaped microgels containing red-stained cells, and rod-shaped microgels containing green-stained cells. (Scale bars, 200 μm.) Reproduced with permission from.[10] Copyright 2008, National Academy of Sciences.
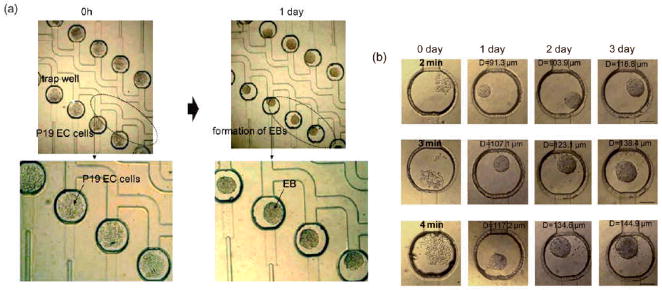
Micromolding is another approach used for assembly of small building blocks to make larger tissue-like units. By using this technique, free toroid units made from aggregated rat hepatoma (H35) cells were generated by utilizing agarose molds with different shapes (toroid and spheroid).[145] Due to the non-adhesive nature of agarose, cells seeded in wells interacted with each other to form the desired aggregate sizes (diameter: 400–1000 μm; height: 400–800 μm) and shapes in a uniform manner. The toroid aggregates were then allowed to assemble to form larger units. The toroids were found to be intact once they were retrieved from the molds. In addition, fusion of toroids was also observed and viability was found to be higher in the toroid shaped samples compared to spheroids. This approach can be conveniently used to generate multilayered scaffold-free tissue constructs in particular where formation of lumen structures are required.
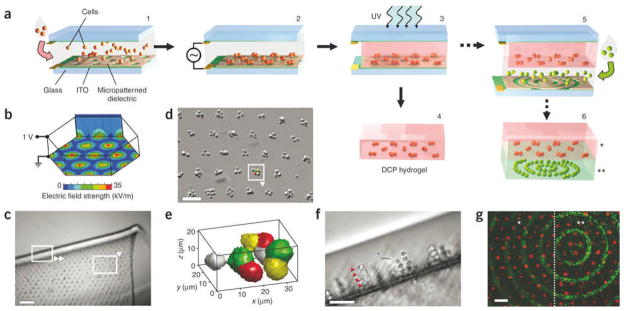
Spatial organization of cells can also be achieved through dielectrophoretic forces exerted on cells within photolabile polymers and subsequent crosslinking. This approach has been applied on various cell types including hepatocytes, fibroblasts, chondrocytes and embryonic liver precursors (Fig. 8).[146] In one example, up to 20,000 chondrocyte cell clusters with precise size and shape were formed in parallel within a thin 2 cm2 hydrogel. These chondrocytes not only remained viable, but also were able to remain functional up to 2 weeks. Interestingly, bovine articular chondrocyte biosynthesis was affected by the microarchitecture of these cell-laden hydrogels, which demonstrated cell function can potentially be controlled by the form of the engineered construct.
Figure 8.
Fabrication method and examples of DCP hydrogels. A) Cells in prepolymer solution are introduced into the transparent chamber (1) and localize via dielectrophoretic forces to micropatterned gaps in the 1.8 μm-thick dielectric layer upon application of the AC. chamber bias (2). Next UV light exposure polymerizes the hydrogel (3), embedding cells in a stable microorganization. The ‘DCP hydrogel’ can then be removed and cultured (4) or incorporated into multilayer constructs by expanding the chamber height and repeating steps 1–3; each layer may contain distinct cell organization, cell type and hydrogel formulation (5, 6). B) Electric field strength is high above circular gaps in the dielectric layer arranged in a hexagonal array (100 μm spacing), as shown by finite element modeling (CFD Research Corp.). C–F) The 100 μm thick DCP hydrogels contain a microarray of embedded fibroblast clusters. In C, the free-floating hydrogel is shown folded; boxes indicate orientation of panels D and F. In E, the 3D shape of an 8-cell cluster from D is rendered from confocal data (MATLAB; pseudocolored to depict individual cells). Cross-sectional view of linear columns 4–5 cells long (arrowheads) demonstrates cluster shape versatility (F). G) A bilayered hydrogel (200 μm total thickness) contains distinct fluorescently labeled fibroblasts in a cluster array above (*) and in concentric rings below (**), as depicted in a (step 6). The same microscope field is shown focused on the upper or lower cell pattern, as delineated by the dotted line. Scale bars: c, 250 μm; d,g, 100 μm; f, 50 μm. Reproduced with permission from.[146] Copyright 2006, Nature Publishing Group.
Spatial patterning of ECM components and growth factors is another crucial aspect of biomimicry. Such 3D tissue engineering constructs are important for fabricating multicellular complex tissues, since each cell type in the target tissue requires different microenvironmental cues.[2,147,148] Traditional tissue engineering methods, however are incapable of regulating various features at the length scales of a few microns to control cells in specific niches.[149–152] Approaches to create precise, spatially distributed microenvironments within a single scaffold therefore would be a significant advancement towards engineering complex tissues and develop concepts to ultimately engineer highly sophisticated organ structures. This can be achieved through the development of novel microfabrication techniques. One of the most potent techniques towards this end is stereolithography-based systems. These microfabrication methods not only allow for generation of complex layer-by-layer architectures but also enable precise, patterning of multiple material types or bioactive molecules.[62] Furthermore, bioactive molecules or controlled-released particles can be incorporated in different layers creating spatially distributed environments with micron size resolution. This technique was used to fabricate patterns of two different bioactive molecules using PEGDA solutions containing either Cy-5 or FITC-labeled polystyrene particles. Similar approaches were adapted to create 3D micropatterned scaffolds made of multiple materials including different molecular weight PEGs,[56,153] and PEG-acrylated alginate composite.[58] Controlling the scaffold geometry and shape in microscale is another essential aspect of biomimicry.[154–156] As in the case of employing multiple cells, material types and bioactive molecules that locally control the material properties (pore size, shape and distribution) can modulate cell-material interactions in a spatially patterned manner. Specifically, directing nuclear and cytoskeletal alignment of the cells have been regarded as an essential component of biomimicry.[157,158] Such studies have been performed on 2D surfaces fabricated from different materials, with diverse cell types, in a range of patterns.[159–163] For example, it has been shown that, actin, microtubules, and other cytoskeletal elements can be regulated by nano/microscale grooves on the substrates that resulted in the controlled alignment of cells parallel to the direction of the grooves.[164–168] As microarchitecture has been shown to direct cell behavior, for example in stem cell niches,[122,169–171] researchers have recently focused on using photopatterning techniques to investigate 3D cell behavior on the microscale which is more relevant to native tissues. Cell alignment in 3D is a rather new phenomena, which can be critical in the fabrication of tissue-like structures[44] and mimicking native microenvironments.[172] 3D cell-laden channels fabricated using GelMA and photomasks have been shown to result in 3D alignment of the encapsulated cells depending on the channel dimensions (Fig. 9).[5] Direct-write base approaches were also used to guide cell orientation. In one example micropatterns in 3D hydrogel were created using two photon laser scanning lithography (TP-LSL).[141] In this approach, PEG-diacrylate (PEGDA) hydrogels were selectively photocrosslinked to include micropatterns of cell attachment sequence (i.e. RGDS) in the polymer backbone to guide cellular orientation and the cell migration in 3D. Control over spatial presentation and concentration of biomolecules within the scaffolds was proven to be effective by showing the guidance of fibroblast migration. Using the same techniques, more than one type of cell attachment sequences were micropatterned in a single construct, which can be valuable in studies towards multicellular complex tissues.[173] Bryant et al. combined a sphere templating technique with photolithography process to fabricate pHEMA hydrogels with well-defined architectures.[174] In this study, polymer solution was poured over a poly(methyl methacrylate) (PMMA) microsphere template and subsequently photopatterned to create microchannels within the hydrogels. Using this technique, vertical microchannels ranging from 360 μm to 730 μm were patterned into porous pHEMA hydrogels with pore sizes in the range of 62–147 μm.[174] This sphere templating process allowed the control of the pore structure, size and interconnectivity while the photopatterning process enabled controlling the macroarchitecture of scaffold. The resultant hydrogels were shown to support elongation, spreading and fibrillar formation of C2C12 myoblasts.[174]
Figure 9.
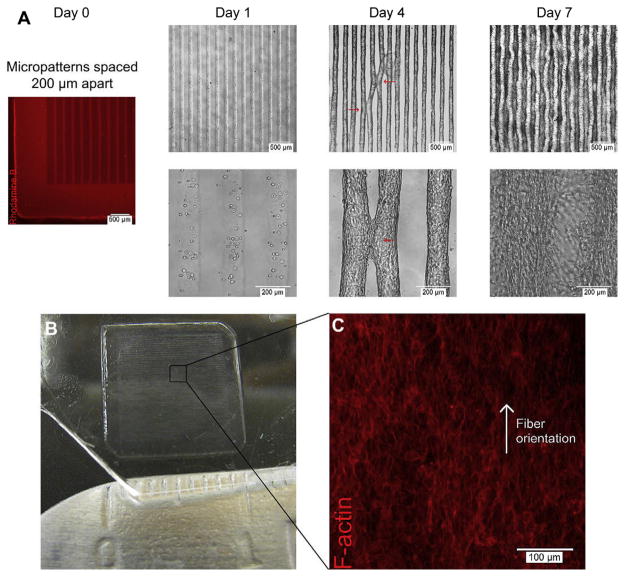
Self assembly of multiple aligned microconstructs into a macroscale and aligned 3D tissue construct. 3T3-fibroblast-laden 5% methacrylated gelatin (GelMA) hydrogels patterned into rectangular microconstructs (50 μm (w) & 800 μm (l) & 150 μm (h)) spaced 200 μm apart self-assembled into macroscale and aligned 3D tissue constructs after 7 days of culture through convergence of multiple, aligned microconstructs. A) Rhodamine B stained hydrogel shows initial microconstruct spacing of 200 μm at Day 0; representative phase contrast images of cell-laden microconstructs at Day 1, 4 and 7 respectively showing focal points of contact between neighboring aligned microconstructs at Day 4 (red arrows) and convergence into a macroscale 3D tissue construct at Day 7. B) Image of a 1 cm _ 1 cm, self-assembled 3D tissue construct at Day 7. C) Representative F-Actin staining of middle xy-plane of macroscale 3D tissue construct shows orientated actin fiber organization in single direction. Reproduced with permission from.[5] Copyright 2010, Elsevier.
Micromolding methods are also used to guide cell alignment. For example, skeletal muscle cells were aligned and differentiated in the pores of micromolded fibrin gels, forming tissue constructs relevant for in vitro models.[175] This was achieved by patterning C2C12 cell loaded collagen and fibrinogen hydrogels with PDMS molds that were previously treated with oxygen plasma. After gelation with the addition of sodium hydroxide (collagen) or thrombin (fibrinogen), the patterned cell-laden hydrogels were removed from the PDMS mold and fixed on a frame to generate myotubes with uniform alignment.
3.2. Vascularization of the 3D engineered tissues
Despite significant progresses in tissue engineering, a number of challenges remain towards the aim of developing fully functional off-the-shelf engineered tissue constructs. One engineering bottleneck is vascularization of the tissue constructs to sufficiently deliver nutrition and oxygen and remove the waste products from the cells.[176–178] Therefore, most of the success in this field has been made in developing thin and avascular tissues such as skin, bladder and cartilage. In the past few years, there has been a tremendous effort in developing strategies to address this challenge and engineer thick and complex tissues or organs such as the heart, muscle, kidney, liver and lung. Recently microtechnology has been proven to be a potentially useful tool in addressing the current vascularization challenge in tissue engineering.[179]
A number of studies have used micropatterning of natural or synthetic hydrogels to enhance endothelial cell organization to consequently promote vasculogenesis.[5,34,180–182] Commonly used biomaterials to this end include collagen, hyaluronic acid, alginate, PEG and PVA.[183] Through the integration of microtechnology and hydrogels, attempts have been made to tailor these hydrogels to mimic native tissue environment.[184] For example West and colleagues have used PEG hydrogels to generate microfluidic hydrogels for vascularization applications.[181,182,185,186] PEG can be modified by using cell adhesive ligands as well as other bioactive molecules and growth factors to support several cell functions such as cell adhesion, migration and proliferation. This tailorability of PEG has been adapted to direct the angiogenesis and vascularization via patterned angiogenic and/or vasculogenic bioactive molecules. Bioactive patterning of PEG was achieved by using multi-step photolithography. In this process a layer of the PEGDA was initially crosslinked to generate a substrate and then polymer solutions containing PEG hydrogel and cell adhesive ligands such as Arg-Gly-Asp-Ser (RGDS), or vascular endothelial growth factor (VEGF) were poured on the base layer and spatially patterned through standard photolithography or laser scanning lithography techniques.[181,182] The pattern geometries generated using this process consisted of features with variable widths ranging from 10 μm to 200 μm. Preliminary studies demonstrated that HUVECs underwent morphogenesis on intermediate RGDS concentrations (20 μg/cm2) in which the cells assembled on top of each other and formed cord-like structures along the stripes.[182] In addition, the cord formation was enhanced on the stripes with smaller widths compared to the wider ones. As expected, the addition of VEGF to RGDS ligands on the patterned layout further enhanced the lumen formation of the cells.[181] Furthermore, significantly higher expression of angiogenic markers VEGFR1, VEGFR2, EphA 7 and laminin, was detected on narrow stripes (10 μm) compared to the wide stripes. These results demonstrate that the cell patterning on the hydrogel surface promoted tissue organization and endothelial cells tubule formation while modification of the PEG hydrogel with cell adhesive ligands and active molecules promoted the expression of angiogenic markers. GelMA has also been employed for the endothelial cell alignment and organization studies.[5,34] Notably, the HUVECs were able to form lumen-like structures on GelMA with 5%, 10% and 15% gel concentrations.[34] In addition, alignment of HUVEC was significantly increased within patterned microchannels (50 μm width) compared to unpatterned regions confirming the potential of micropatterned GelMA to create 3D vascularized networks.[5]
Patterning proteins on 2D surfaces has also been found to promote alignment and organization of endothelial cells along the patterned regions.[187,188] This approach has resulted in 2D endothelial cell organization which may alter the cell behavior from their behavior in natural microenvironment. Gerecht et al. used this 2D approach and followed it by the addition of hydrogel to promote tubulogenesis of endothelial progenitor cells (hEPC) in a 3D microenvironment.[189] Fibronectin was patterned on glass substrates to guide and align hEPC on strips with variable width ranging from 2.5 μm to 70 μm. Preliminary studies showed optimal cell attachment, alignment and organization on 50 μm width patterns after 5 days of culture. Von Willebrand factor (vWF) expression was significantly increased, confirming the ability of the cells to differentiate toward endothelial lineage and form vasculature. Furthermore, E-selectin and ICAM-1 expression was significantly enhanced in response to tumor necrosis factor-α (TNF-α) showing the angiogenic ability of the cells. Finally, the patterned cells were inverted on cured fibrin gel after one day of the culture time. Within 24 hours after the addition of the gel to the patterned cells, the cells were able to from 3D tubular structures. This study demonstrated the importance of a 3D microenvironment for cellular support and vascularization as opposed to micropatterning the cells on 2D surfaces. Micromolding has also been shown to be an effective method to spatially control the organization of endothelial cell and enhance in vitro tubulogenesis.[190] For example, a PDMS mold consisting of channels with the desired geometries was used to generate microfabricated vascular structure. In this process endothelial cells were encapsulated inside microfabricated collagen gels and induced to form tubules. Tubule formation of endothelial cells, which was triggered by basic fibroblast growth factor (bFGF) and VEGF, started as early as 24 hours after the cell encapsulation within the channels. Increasing the collagen concentration as well as the channel width resulted in formation of tubules with larger diameter. The major advantage of this method was the precise control over the tubule geometry such as tubule diameter and tubule branching toward the desired direction and angle. In these approaches, 3D vascularized networks were mainly relied on encapsulation/seeding of endothelial cells within hydrogels and stimulating the process of tubulogenesis by using active molecules such as bFGF and VGEF. The enhancement of the process of tubulogenesis was quantified through tubule length measurement, lumen formation and angiogenic markers expressions.
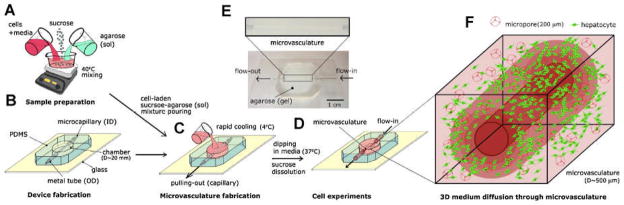
Another approach uses microfluidic systems to create vascularized network within the tissue constructs. Both hydrogels[191–193] and other biocompatible, biodegradable polymers[194,195] have been used to create microfluidic networks for vascularization purposes. For example calcium alginate hydrogels have been used to provide an appropriate microenvironment for cellular support and for creating an embedded microfluidic network within the cell seeded hydrogel.[192] In another study, microfluidic networks were created in agarose hydrogels and highly porous channels with variable dimensions was fabricated.[191] Cells mostly remained viable near the channels confirming proper diffusion of nutrient and waste exchange within the proximity of the channels. Cellular viability can be further improved by using a combination of micromolding technique and sucrose crystal leaching process to generate porous cell-laden agarose hydrogels containing microchannels (Fig. 10).[196] In this approach, the mechanical properties and the pores architecture (pore sizes and porosity) of hydrogel were controlled by varying the concentration of sucrose. Furthermore, the viability of human hepatic carcinoma cells encapsulated within the porous hydrogels containing microchannels was 10–20% higher compared to non-porous hydrogels.[196]
Figure 10.
Microfabrication scheme to generate cell encapsulated agarose based hydrogel channels. A) Cells, agarose and sucrose were combined under agitated conditions. B) PDMS mold with a microneedle was cured. C) The cell and porogen loaded hydogel precursor was placed in the PDMS mold to obtain the microchannel. D) The microneedle was removed once the cell-laden polymer is cured in the PDMS mold. E) The microconstruct was cultured in the media under perfusion conditions upto 5 days. F) Encapsulated hepatocytes in the 3D porous hydrogels mimicking microvasculature geometry. Reproduced with permission from.[196] Copyright 2010, John Wiley & Sons.
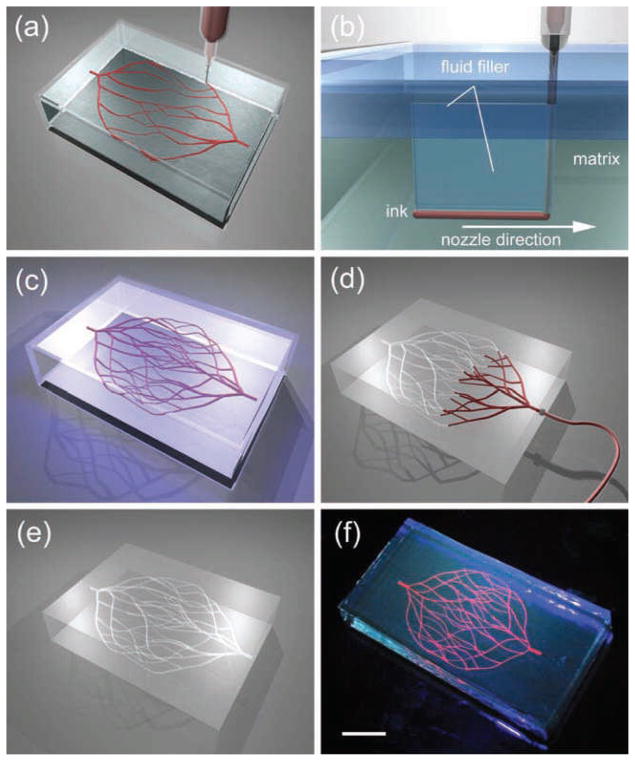
In another example of this approach Golden et. al. developed microfluidic networks in collagen and fibrin hydrogels.[197] In their approach, geometrical features of the channels were primarily defined through using a PDMS stamp, which was treated with PluronicR. Then, a sacrificial gelatin layer, was filled inside the channels and induced to gel upon cool. Subsequently a solution of the precursor hydrogel (collagen I and fibrin) was poured on top of the gelatin layer and polymerized at room temperature. To remove the gelatin layer the entire construct was heated to 37 °C to induce the formation of a liquid gelatin, which was removed by a flushing process to create the microfluidic channels. Through this method, it was possible to produce microfluidic channels as small as 6 μm in diameter. Further studies demonstrated spreading, proliferation and viability of human microvascular endothelial cells, seeded within the hydrogel, after 5 days of culture. A potential limitation of this technique was the generation of channels that were slightly wider than the original PDMS stamps due to distortion and swelling of the gelatin layer. Figure 11 shows another attempt to create microvasculature network by using bioprinter technology. In this approach 3D biomimetic microvascular networks were embedded within a hydrogel matrix through omnidirectional printing.[198] Tailoring the chemical and rheological properties of the ink controlled the printing process.
Figure 11.
a–e) Schematics of omnidirectional printing of 3D microvascular networks within a hydrogel reservoir. A) Deposition of a fugitive ink into a physical gel reservoir allows hierarchical, branching networks to be patterned. B) Voids induced by nozzle translation are filled with liquid that migrates from the fluid capping layer. C) Subsequent photopolymerization of the reservoir yields a chemically cross-linked, hydrogel matrix. D,E) The ink is liquified and removed under a modest vacuum to expose the microvascular channels. f) Fluorescent image of a 3D microvascular network fabricated via omnidirectional printing of a fugitive ink (dyed red) within a photopolymerized Pluronic F127-diacrylate matrix. (scale bar = 10 mm). Reproduced with permission from.[198] Copyright 2011, John Wiley & Sons.
In addition to hydrogels, biodegradable polymers such as PLGA[194] and poly(glycerol sebacate) (PGS)[195] have been utilized to create highly branched multi-layer microfluidic networks that resemble in vivo microvasculature. In one approach, a PDMS stamp was used to create the desired architecture out of PLGA through a melt molding process.[194] In addition, it was demonstrated that multiple layers of patterned PLGA could be bonded together through thermal bonding process to form 3D multilayer microfluidic networks. In another study, replica-molding technique was employed to generate a multilayered microfluidic network in PGS. [195] A unique feature of their architecture was the uniform distribution of shear stress throughout the fluidic network, making it a suitable platform for vascularization purposes. Although significant progress has been made in creating 3D vascularized network through the integration of microtechnology and tissue engineering, there are still numerous challenges in building a fully functional vascularized tissue constructs for clinical applications. Some of these challenges include: a) Understanding the biological processes governing angiogenesis; b) Mimicking the functionality and complexity of the in vivo vascularized network in a scalable fashion; c) Optimal selection of biomaterials to incorporate the bioactive and growth factors to successfully support the cellular function and; d) Implantation of the vascularized tissue construct inside the patient’s body.
3.3. Directing stem cell fate using microengineered platforms
In using stem cells for therapeutic applications, microscale approaches have been increasingly utilized to address various challenges associated with the inability of the current technologies to suitable regulate the stem cell fate decisions. Such platforms are fairly compatible with biological phenomena[199–201] and have been used to study and/or regulate cellular responses in a tightly controlled microenvironment. For example, the ability to fabricate microscale units with spatially controlled material properties has been used to the aid investigation of cellular behaviors through interaction in different microenvironments.[14,138,202] These advanced microengineering techniques have advanced the field of stem cell engineering through specific control of stem cell behaviors by mimicking the complex in vivo cellular microenvironments.[122–127]
There have been many attempts to control the spatial organizations of individual cells or cellular colonies in vitro by using a variety of microscale technologies to study and characterize complex cell-cell interactions. For instance, microarrays have been fabricated from many types of biomaterials for various cell culture applications. In one such microarray platform, the dynamics in the fate decisions of individual stem cell aggregates was investigated in high-throughput manner by using microfabricated adhesive stencils to control the sizes of ESC-aggregates that ranged from 100 μm to 500 μm in diameter.[203] In another study, Bauwens et al. formed size-controlled EBs using a microcontact printing approach, showing size-dependent differentiation of embryoid bodies (EBs).[204] Moreover, Lee et al. studied the effects of regulating ESC-colony sizes along with the supplementation of growth factors on mesodermal and endodermal lineage developments.[205] In our previous studies, a microwell culture system was developed using PEG as templates for directing the formation of EBs that could offer homogeneity in the size and shape as well as retrievability for further biological analyses.[206] Using this PEG microwell system, it was demonstrated that the different sizes of EBs directed the differentiation of stem cells to endothelial and cardiomyogenic lineages respectively, in a size-dependent manner (Fig. 12).[200] In addition, the inner surface of this microarray system can be functionalized with ECM molecules which mimic essential features of the native 3D extracellular milieu, providing quasi-3D single cell microenvironments.[207,208] In another study, the spatially organized human ESC expansion was successfully achieved through patterning of murine feeder cells within microwells.[209]
Figure 12.
Arrays of hydrogel microwells for culturing embryonic stem cells (ESCs). A) Analysis of embryoid bodies (EBs) cultured within microwells for 7 days. Scanning electron microscopy (SEM) (Top) and phase contrast (Middle) images show the formation of uniform arrays of PEG microwells with different diameters (150 μm, 300 μm, and 450 μm) and live and dead cells were presented by fluorescent images (Bottom) of EBs cultured within microwells after 7 days. (Scale bar, 100 μm), B) The molecular expression of ESC pluripotency markers (Oct4, E-cadherin, and SSEA1) after 3 and 7 days (Scale bar, 100 μm). Reproduced with permission from.[200] Copyright 2009, National Academy of Sciences.
The development of combinatorial microarrays of ECMs and/or synthetic biomaterials could also be used to elucidate the mechanisms responsible for the interactions between stem cells and extracellular environments and directed cell fate decisions in a more predictive manner. For example, a microarray of various natural ECM protein combinations has been developed to study ESC differentiation.[210,211] These studies allowed small amounts of biosignaling molecules to be effectively used to control and regulate stem cell differentiation in a cost-effective manner.[212] Also, MSCs were induced to differentiate to osteoblasts or adipocytes depending on the sizes of adhesive ECM-islands on which the stem cells were attached and grown.[213] Furthermore, the responses of human ESCs to various extracellular signals have been studied by a nanoliter-scale synthesis technique for different synthetic polymers (Fig. 13).[214] In another study, human ESCs were micropatterned by microcontact printing techniques to study their differentiation as a function of changes to ESC-colony sizes.[204] The micropatterned human ESC-colonies responded differently to the varying sizes such that endodermal and neuronal expression increased inversely with the colony size, whereas greater mesoderm and cardiac induction was observed in the larger colonies. This suggested that the size-regulated microprinted human ESC colonies could specify the subsets of appropriate differentiation conditions for a certain lineage. A 3D approach of microarray system has also been introduced. For example, murine ESCs encapsulated in alginate hydrogels were micropatterned for studying the interactions between stem cells and soluble factors in a 3D environment.[215] Furthermore, various methodologies of co-cultures could be achieved by micropatterning technologies in order to control the degree of homotypic and heterotypic cell-cell contacts. For instance, a dynamic co-culture environment was created using microscale stencils made from parylene-C for investigating temporal changes of murine ESCs in co-cultures with other cell types.[216] More complex combinations of co-culture system was generated such that various proteins and cells were micropatterned using reversible sealing microfabricated parylene-C stencils.[217]
Figure 13.
Human embryonic stem cells (hESCs) grown on microarrays. A,B) Four million hESC embryoid body Day 6 cells were grown on the microarray in the presence (B) and absence (A) of retinoic acid for 6 days and then stained for cytokeratin 7 (green) and vimentin (red). C–N) The blue channel can be used to identify the location of polymer spots (b,e,h) or cells lacking other signal by nuclear staining (blue in A,C,D,F,G,I–N). Reproduced with permission from.[214] Copyright 2004, Nature Publishing Group.
In a developing organism, tissue organization is made within a dynamic microenvironment involving coordinated sequences of stem cell renewal, differentiation, and spontaneous assembly into sub-tissues that are regulated by spatial and temporal influences of multiple factors. Most of the biological events in vivo progress through a series of cellular mechanisms activated by growth factors that bind to specific receptors on the cell surfaces. Indeed, a variety of studies about using various growth factors for biochemically mimicking in vivo environments have been conducted for directing the lineage specific differentiation of stem cells. However, this complex interplay of various factors present that the in vivo environments are difficult to recreate in vitro by traditional culture techniques.[218] Furthermore, 3D interactions as they normally exist in vivo between cells and either ECMs or other neighboring cells would also be disrupted by a 2D culture environment.[219] Even though stem cells theoretically can be expanded indefinitely, traditional culture methods still suffer from their labor-intensive and time-consuming nature as well as restricted surface area available for culture that results in a limited amount of therapeutically applicable cells. Therefore, the use of stem cells for therapeutic applications require a novel culture methodology with natural 3D settings that can support the recreation of the appropriate microenvironment in vitro.[220] Bioreactors are culture devices with the ability to control cell or tissue specific environmental conditions in automatable and reproducible manners. The development of bioreactors can provide a means to quantitatively study cellular responses to various physical and/or biochemical stimuli. By using microengineering techniques one can facilitate the designs of bioreactor to be modular, miniscaled, and multiparametric while increasing the number of samples that can be simultaneously analyzed. For example, the advent of microfluidic culture platforms showed the feasibility to deliver precise concentrations of various culturing factors such as oxygen, nutrients, and other molecular and physical regulatory factors to the desired locations with minimized batch to batch variability.[221] Furthermore, the media volume required to run the system is much smaller than the traditional culture systems, which in turn reduces the cost associated with expensive soluble factors.[222] Moreover, laminar flow generated in the microfluidic system can offer many advantages in that the iterations can be reduced due to its predictable environment through precise control of media mixing and shear stress over the cells.[223,224] To date, microfluidic technologies have been used in a number of different applications for studying cellular behaviors, such as cell patterning under microfluidic systems, analysis of cellular behaviors on controlled shear stress, and creation of soluble factor gradients.[225–228] Microfluidic devices, through precise control of critical culture parameters, ensure optimal conditions for studies that aim to recapitulate in vivo environment.[229,230] Another application where precise submicroliter control of culture parameters can be benefited is regulating stem cell differentiation using soluble factors.[225,231] For example, microfluidic devices were shown to produce a logarithmic scale of flow rates and concentration gradients that could influence cell morphogenesis and proliferation of ESCs.[232] By controlling the flow rate and shear stress within the microfluidic system, self-renewal and proliferation of ESCs were regulated, demonstrating that high flow rates resulted in increased proliferation.[233] In another study, neural stem cell differentiation and proliferation has been regulated by microfluidic system, such that the effective screening of different growth factor combinations consisting of epidermal growth factor (EGF), fibroblast growth factor 2 (FGF2) and platelet-derived growth factor (PDGF) could be performed in the laminar flow range for differentiation towards astrocyte lineage.[234]
Current designs of bioreactor platform incorporate the cascades of biological and physical stimuli to exert greater influence over cellular differentiation into tissue constructs with enhanced functionalities.[235] For example, Park et al. implemented a mechanical stimulation module in the microfluidic system, demonstrating that the osteogenic differentiation of human MSCs was enhanced by compressive cyclic loading in a microfluidic network.[236] In another study, shear stress gradients were generated in the microfluidic channels in order to study the responses of human ESC-derived endothelial cells upon the shear stress changes through changing a variety of gene expressions.[237] However, one potential drawback is that the complex microplumbing system to generate various gradients increase device complexity which in turn leads to low reproducibility and reliability. Therefore, device simplicity and reliability are considered essential in the integration processes of multiple microfluidic devices. To this end, a simplified bioreactor array has been developed with a capability of different gradient generation and continuous perfusion inside bioreactors for a long-term culture.[238,239] In addition, a microfluidic device was further developed for automated EB formation within a microchip containing microwell arrays that are amenable to 3D culture of ESCs, size controlled EB formation, and perfused biochemical treatment for directing EB differentiation (Fig. 14).[240] Although non-uniform cell loading and cross contamination between channels were still problematic, developed microfluidic devices demonstrated versatile functions including continuous medium perfusion, generation of various conditions within bioreactors, and post-analytical processing. Since each type of tissue constructs requires an individualized culture system design due to their specific functional, physicochemical characteristics in vivo, tissue-specific bioreactors are to be designed on the basis of comprehensive understanding of biological and engineering aspects.[241] Moreover, the advances in optical technologies and their combinations with microfluidic platform support the development of non-invasive monitoring system in real time. The development of such bioreactor systems is believed to accelerate the transfer of laboratory-based practices to the clinic. In particular, the specific directions of stem cell differentiation can greatly benefit from these advanced bioreactor systems. It is expected that the continuous development of microbioreactors, will lead to cost-effective manufacture of tissue engineered products that could generate therapeutic procedures with sufficient donor cells/tissues.[242]
Figure 14.
A) Trapped embryonic stem cells (ESCs) and P19 cells were aggregated in each microwell and formed a spherical cell body after 1 day. B) The trapped ESCs were cultured to form the embryoid bodies for 3 days. The number of trapped cells increased monotonically with longer cell loading duration time as well as the cell sphere diameter. Reproduced with permission from.[240] Copyright 2011, RSC Publishing.
4. Conclusion
Although engineered tissues are being considered as a potential remedy for organ failure, major challenges such as lack of sufficient vascularization and suitable tissue functionality as well as lack of a suitable cell source remain to be addressed. Despite being at an early developmental stage, the combination of microengineering approaches with novel biomaterials will lead to more thorough understanding of cell biology and substantially contribute to the therapeutic potential of engineered tissues. Consolidation of these fields holds great promise for the advancement of regenerative medicine which in long term will add in realizing the dream of reproducing degenerated or defective tissues.
Acknowledgments
The authors acknowledge funding from the National Science Foundation CAREER Award (DMR 0847287), the office of Naval Research Young National Investigator Award and the National Institutes of Health (HL092836, DE021468, AR05837, EB012597, HL099073).
Contributor Information
Dr. Pinar Zorlutuna, Center for Biomedical Engineering, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Cambridge, MA 02139 (USA), Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge, MA, 02139 (USA)
Dr. Nasim Annabi, Center for Biomedical Engineering, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Cambridge, MA 02139 (USA), Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge, MA, 02139 (USA)
Dr. Gulden Camci-Unal, Center for Biomedical Engineering, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Cambridge, MA 02139 (USA), Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge, MA, 02139 (USA)
Dr. Mehdi Nikkah, Center for Biomedical Engineering, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Cambridge, MA 02139 (USA), Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge, MA, 02139 (USA)
Dr. Jae Min Cha, Center for Biomedical Engineering, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Cambridge, MA 02139 (USA), Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge, MA, 02139 (USA)
Dr. Jason Nichol, Center for Biomedical Engineering, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Cambridge, MA 02139 (USA), Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge, MA, 02139 (USA)
Amir Manbachi, Institute for Biomaterials and Biomedical Engineering, University of Toronto, Toronto (Canada).
Dr. Hojae Bae, Center for Biomedical Engineering, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Cambridge, MA 02139 (USA), Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge, MA, 02139 (USA)
Prof. Shaochen Chen, Department of Nanoengineering, University of California, San Diego, 9500 Gilman Drive, La Jolla, CA, 92093-0448 (USA)
Prof. Ali Khademhosseini, Email: alik@rics.bwh.harvard.edu, Wyss Institute for Biologically Inspired Engineering, Harvard University, Boston, MA 02115 (USA)
References
- 1.Khademhosseini A, Langer R, Borenstein J, Vacanti JP. Proc Natl Acad Sci U S A. 2006;103:2480. doi: 10.1073/pnas.0507681102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Griffith LG. Ann NY Acad Sci. 2002;961:83. doi: 10.1111/j.1749-6632.2002.tb03056.x. [DOI] [PubMed] [Google Scholar]
- 3.Langer R, Vacanti JP. Science. 1993;260:920. doi: 10.1126/science.8493529. [DOI] [PubMed] [Google Scholar]
- 4.Report #S520. 2009. MedMarket Diligence, Tissue Engineering & Cell Therapy. [Google Scholar]
- 5.Aubin H, Nichol JW, Hutson CB, Bae H, Sieminski AL, Cropek DM, Akhyari P, Khademhosseini A. Biomaterials. 2010;31:6941. doi: 10.1016/j.biomaterials.2010.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsang VL, Chen AA, Cho LM, Jadin KD, Sah RL, DeLong S, West JL, Bhatia SN. Faseb J. 2007;21:790. doi: 10.1096/fj.06-7117com. [DOI] [PubMed] [Google Scholar]
- 7.Tsang VL, Bhatia SN. Adv Biochem Eng Biotechnol. 2007;103:189. doi: 10.1007/10_010. [DOI] [PubMed] [Google Scholar]
- 8.Tsang VL, Bhatia SN. Adv Drug Deliv Rev. 2004;56:1635. doi: 10.1016/j.addr.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 9.Du Y, Hancock MJ, He J, Villa-Uribe JL, Wang B, Cropek DM, Khademhosseini A. Biomaterials. 2010;31:2686. doi: 10.1016/j.biomaterials.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du Y, Lo E, Ali S, Khademhosseini A. Proc Natl Acad Sci U S A. 2008;105:9522. doi: 10.1073/pnas.0801866105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Du Y, Lo E, Vidula MK, Khabiry M, Khademhosseini A. Cell Mol Bioeng. 2008;1:157. doi: 10.1007/s12195-008-0020-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernandez JG, Khademhosseini A. Adv Mater. 2010;22:2538. doi: 10.1002/adma.200903893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Slaughter BV, Khurshid SS, Fisher OZ, Khademhosseini A, Peppas NA. Adv Mater. 2009;21:3307. doi: 10.1002/adma.200802106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peppas NA, Hilt JZ, Khademhosseini A, Langer R. Adv Mater. 2006;18:1345. [Google Scholar]
- 15.Khademhosseini A, Langer R. Biomaterials. 2007;28:5087. doi: 10.1016/j.biomaterials.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 16.Burdick JA, Anseth KS. Biomaterials. 2002;23:4315. doi: 10.1016/s0142-9612(02)00176-x. [DOI] [PubMed] [Google Scholar]
- 17.Dikovsky D, Bianco-Peled H, Seliktar D. Biomaterials. 2006;27:1496. doi: 10.1016/j.biomaterials.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 18.Khademhosseini A, Yeh J, Jon S, Eng G, Suh KY, Burdick JA, Langer R. Lab Chip. 2004;4:425. doi: 10.1039/b404842c. [DOI] [PubMed] [Google Scholar]
- 19.Kim P, Jeong HE, Khademhosseini A, Suh KY. Lab Chip. 2006;6:1432. doi: 10.1039/b610503c. [DOI] [PubMed] [Google Scholar]
- 20.Lee HJ, Lee JS, Chansakul T, Yu C, Elisseeff JH, Yu SM. Biomaterials. 2006;27:5268. doi: 10.1016/j.biomaterials.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 21.Raeber GP, Lutolf MP, Hubbell JA. Biophys J. 2005;89:1374. doi: 10.1529/biophysj.104.050682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weber LM, He J, Bradley B, Haskins K, Anseth KS. Acta Biomater. 2006;2:1. doi: 10.1016/j.actbio.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 23.Brigham MD, Bick A, Lo E, Bendali A, Burdick JA, Khademhosseini A. Tissue Eng. 2009;15:1645. doi: 10.1089/ten.tea.2008.0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Camci-Unal G, Aubin H, Ahari AF, Bae H, Nichol JW, Khademhosseini A. Soft Matter. 2010;6:5120. doi: 10.1039/c0sm00508h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fukuda J, Khademhosseini A, Yeh J, Eng G, Cheng J, Farokhzad OC, Langer R. Biomaterials. 2006;27:1479. doi: 10.1016/j.biomaterials.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 26.Gerecht S, Burdick JA, Ferreira LS, Townsend SA, Langer R, Vunjak-Novakovic G. Proc Natl Acad Sci U S A. 2007;104:11298. doi: 10.1073/pnas.0703723104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim J, Park Y, Tae G, Lee KB, Hwang SJ, Kim IS, Noh I, Sun K. J Mater Sci. 2008;19:3311. doi: 10.1007/s10856-008-3469-3. [DOI] [PubMed] [Google Scholar]
- 28.Radice M, Brun P, Cortivo R, Scapinelli R, Battaliard C, Abatangelo G. J Biomed Mater Res. 2000;50:101. doi: 10.1002/(sici)1097-4636(200005)50:2<101::aid-jbm2>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 29.Sahoo S, Chung C, Khetan S, Burdick JA. Biomacromolecules. 2008;9:1088. doi: 10.1021/bm800051m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Solchaga LA, Dennis JE, Goldberg VM, Caplan AI. J Orthop Res. 1999;17:205. doi: 10.1002/jor.1100170209. [DOI] [PubMed] [Google Scholar]
- 31.Bae H, Ahari AF, Shin H, Nichol JW, Hutson CB, Masaeli M, Kim SH, Aubin H, Yamanlar S, Khademhosseini A. Soft Matter. 2011;7:1903. doi: 10.1039/C0SM00697A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benton JA, DeForest CA, Vivekanandan V, Anseth KS. Tissue Eng. 2009;15:3221. doi: 10.1089/ten.tea.2008.0545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hutson CB, Nichol JW, Aubin H, Bae H, Yamanlar S, Al-Haque S, Koshy ST, Khademhosseini A. Tissue Eng. 2011;17:1713. doi: 10.1089/ten.tea.2010.0666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nichol JW, Koshy ST, Bae H, Hwang CM, Yamanlar S, Khademhosseini A. Biomaterials. 2010;31:5536. doi: 10.1016/j.biomaterials.2010.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Den Bulcke AI, Bogdanov B, De Rooze N, Schacht EH, Cornelissen M, Berghmans H. Biomacromolecules. 2000;1:31. doi: 10.1021/bm990017d. [DOI] [PubMed] [Google Scholar]
- 36.Xiao W, He J, Nichol JW, Wang L, Hutson CB, Wang B, Du Y, Fan H, Khademhosseini A. Acta Biomater. 2011;7:2384. doi: 10.1016/j.actbio.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shin H, Nichol JW, Khademhosseini A. Acta Biomater. 2011;7:106. doi: 10.1016/j.actbio.2010.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Du Y, Ghodousi M, Lo E, Vidula MK, Emiroglu O, Khademhosseini A. Biotechnol Bioeng. 2010;105:655. doi: 10.1002/bit.22552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gobin AS, West JL. Faseb J. 2002;16:751. doi: 10.1096/fj.01-0759fje. [DOI] [PubMed] [Google Scholar]
- 40.Zamanian B, Masaeli M, Nichol JW, Khabiry M, Hancock MJ, Bae H, Khademhosseini A. Small. 2010;6:937. doi: 10.1002/smll.200902326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Folch A, Toner M. Annu Rev Biomed Eng. Vol. 2. United States: 2000. p. 227. [DOI] [PubMed] [Google Scholar]
- 42.Oh JK, Drumright R, Siegwart DJ, Matyjaszewski K. Prog in Polym Sci. 2008;33:448. doi: 10.1016/j.progpolymsci.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Napolitano AP, Dean DM, Man AJ, Youssef J, Ho DN, Rago AP, Lech MP, Morgan JR. Biotechniques. 2007;43:494. doi: 10.2144/000112591. [DOI] [PubMed] [Google Scholar]
- 44.Kane RS, Takayama S, Ostuni E, Ingber DE, Whitesides GM. Biomaterials. 1999;20:2363. doi: 10.1016/s0142-9612(99)00165-9. [DOI] [PubMed] [Google Scholar]
- 45.Rivest C, Morrison DWG, Ni B, Rubin J, Yadav V, Mahdavi A, Karp JM, Khademhosseini A. J Mech Mater Struct. 2007;2:1103. [Google Scholar]
- 46.Yeh J, Ling Y, Karp JM, Gantz J, Chandawarkar A, Eng G, Blumling J, 3rd, Langer R, Khademhosseini A. Biomaterials. Vol. 27. England: 2006. p. 5391. [DOI] [PubMed] [Google Scholar]
- 47.Quist AP, Pavlovic E, Oscarsson S. Anal Bioanal Chem. 2005;381:591. doi: 10.1007/s00216-004-2847-z. [DOI] [PubMed] [Google Scholar]
- 48.Suh K-Y, Park MC, Kim P. Adv Funct Mater. 2009;19:2699. doi: 10.1002/adfm.200990041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oh JK, Lee DI, Park JM. Prog Polym Sci. 2009;34:1261. [Google Scholar]
- 50.Tekin H, Ozaydin-Ince G, Tsinman T, Gleason KK, Langer R, Khademhosseini A, Demirel MC. Langmuir. 2011;27:5671. doi: 10.1021/la200183x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tekin H, Tsinman T, Sanchez JG, Jones BJ, Camci-Unal G, Nichol JW, Langer R, Khademhosseini A. JACS. 2011 doi: 10.1021/ja204266a. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Melchels FPW, Bertoldi K, Gabbrielli R, Velders AH, Feijen J, Grijpma DW. Biomaterials. 2010;31:6909. doi: 10.1016/j.biomaterials.2010.05.068. [DOI] [PubMed] [Google Scholar]
- 53.Melchels FPW, Feijen J, Grijpma DW. Biomaterials. 2010;31:6121. doi: 10.1016/j.biomaterials.2010.04.050. [DOI] [PubMed] [Google Scholar]
- 54.Peltola SMMF, Grijpma DW, Kellomäki M. Ann Med. 2008;40:12. doi: 10.1080/07853890701881788. [DOI] [PubMed] [Google Scholar]
- 55.Cooke MN, Fisher JP, Dean D, Rimnac C, Mikos AG. J Biomed Mater Res B Appl Biomater. 2003;64B:65. doi: 10.1002/jbm.b.10485. [DOI] [PubMed] [Google Scholar]
- 56.Arcaute K, Mann B, Wicker R. Acta Biomater. 2010;6:1047. doi: 10.1016/j.actbio.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 57.Chan V, Zorlutuna P, Jeong JH, Kong H, Bashir R. Lab Chip. 2010;10:2062. doi: 10.1039/c004285d. [DOI] [PubMed] [Google Scholar]
- 58.Zorlutuna P, Jeong JH, Kong HJ, Bashir R. Adv Funct Mater. 2011;21:3642. [Google Scholar]
- 59.Arcaute K, Mann BK, Wicker RB. Ann Biomed Eng. 2006;34:1429. doi: 10.1007/s10439-006-9156-y. [DOI] [PubMed] [Google Scholar]
- 60.Han L-H, Suri S, Schmidt CE, Chen S. Biomed Microdevices. 2010;12:721. doi: 10.1007/s10544-010-9425-2. [DOI] [PubMed] [Google Scholar]
- 61.Mapili G, Lu Y, Chen S, Roy K. J Biomed Mater Res B Appl Biomater. 2005;75:414. doi: 10.1002/jbm.b.30325. [DOI] [PubMed] [Google Scholar]
- 62.Lu Y, Mapili G, Suhali G, Chen SC, Roy K. J Biomed Mater Res A. 2006;77A:396. doi: 10.1002/jbm.a.30601. [DOI] [PubMed] [Google Scholar]
- 63.Antonov EN, Bagratashvili VN, Whitaker MJ, Barry JJA, Shakesheff KM, Konovalov AN, Popov VK, Howdle SM. Adv Mater. 2005;17:327. doi: 10.1002/adma.200400838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sudarmadji N, Tan JY, Leong KF, Chua CK, Loh YT. Acta Biomater. 2011;7:530. doi: 10.1016/j.actbio.2010.09.024. [DOI] [PubMed] [Google Scholar]
- 65.Williams JM, Adewunmi A, Schek RM, Flanagan CL, Krebsbach PH, Feinberg SE, Hollister SJ, Das S. Biomaterials. 2005;26:4817. doi: 10.1016/j.biomaterials.2004.11.057. [DOI] [PubMed] [Google Scholar]
- 66.Shuai C, Gao C, Nie Y, Hu H, Zhou Y, Peng S. Nanotechnology. 2011:22. doi: 10.1088/0957-4484/22/28/285703. [DOI] [PubMed] [Google Scholar]
- 67.Liu FH, Chen SH, Lee RT, Lin WS, Liao YS. Proc Wrld Acad Sci E. 2011;76:214. [Google Scholar]
- 68.Fu Y, Cheng C, Zhang BB, Hu CB, Fu CH, Wang YL. Gongneng Cailiao/Journal of Functional Materials. 2010;41:1667. [Google Scholar]
- 69.Bukharova TB, Antonov EN, Popov VK, Fatkhudinov TK, Popova AV, Volkov AV, Bochkova SA, Bagratashvili VN, Gol’Dshtein DV. Bull Exp Biol Med. 2010;149:148. doi: 10.1007/s10517-010-0895-2. [DOI] [PubMed] [Google Scholar]
- 70.Eshraghi S, Das S. Acta Biomater. 2010;6:2467. doi: 10.1016/j.actbio.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Duan B, Cheung WL, Wang M. Biofabrication. 2011;3:015001. doi: 10.1088/1758-5082/3/1/015001. [DOI] [PubMed] [Google Scholar]
- 72.Duan B, Wang M, Zhou WY, Cheung WL, Li ZY, Lu WW. Acta Biomater. 2010;6:4495. doi: 10.1016/j.actbio.2010.06.024. [DOI] [PubMed] [Google Scholar]
- 73.Eosoly S, Brabazon D, Lohfeld S, Looney L. Acta Biomater. 2010;6:2511. doi: 10.1016/j.actbio.2009.07.018. [DOI] [PubMed] [Google Scholar]
- 74.Masood SH, Alamara K. Manama. 2010;83–86:269. [Google Scholar]
- 75.Zein I, Hutmacher DW, Tan KC, Teoh SH. Biomaterials. 2002;23:1169. doi: 10.1016/s0142-9612(01)00232-0. [DOI] [PubMed] [Google Scholar]
- 76.Ramanath HS, Chua CK, Leong KF, Shah KD. J Mater Sci Mater Med. 2008;19:2541. doi: 10.1007/s10856-007-3203-6. [DOI] [PubMed] [Google Scholar]
- 77.Yen HJ, Tseng CS, Hsu SH, Tsai CL. Biomed Microdevices. 2009;11:615. doi: 10.1007/s10544-008-9271-7. [DOI] [PubMed] [Google Scholar]
- 78.Endres M, Hutmacher DW, Salgado AJ, Kaps C, Ringe J, Reis RL, Sittinger M, Brandwood A, Schantz JT. Tissue Eng. 2003;9:689. doi: 10.1089/107632703768247386. [DOI] [PubMed] [Google Scholar]
- 79.Lips PAM, Velthoen IW, Dijkstra PJ, Wessling M, Feijen J. Polymer. 2005;46:9396. [Google Scholar]
- 80.Barbetta A, Barigelli E, Dentini M. Biomacromolecules. 2009;10:2328. doi: 10.1021/bm900517q. [DOI] [PubMed] [Google Scholar]
- 81.Barbetta A, Gumiero A, Pecci R, Bedini R, Dentini M. Biomacromolecules. 2009;10:3188. doi: 10.1021/bm901051c. [DOI] [PubMed] [Google Scholar]
- 82.Keskar V, Marion NW, Mao JJ, Gemeinhart RA. Tissue Eng A. 2009;15:1695. doi: 10.1089/ten.tea.2008.0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Barbetta A, Rizzitelli G, Bedini R, Pecci R, Dentini M. Soft Matter. 2010;6:1785. [Google Scholar]
- 84.Barbetta A, Carrino A, Costantini M, Dentini M. Soft Matter. 2010;6:5213. [Google Scholar]
- 85.Tai H, Popov VK, Shakesheff KM, Howdle SM. Biochem Soc Trans. 2007;35:516. doi: 10.1042/BST0350516. [DOI] [PubMed] [Google Scholar]
- 86.Tai H, Mather ML, Howard D, Wang W, White LJ, Crowe JA, Morgan SP, Chandra A, Williams DJ, Howdle SM, Shakesheff KM. Eur Cells Mater. 2007;14:64. doi: 10.22203/ecm.v014a07. [DOI] [PubMed] [Google Scholar]
- 87.Kazarian SG. Polymer Sci, Ser C. 2000;42:78. [Google Scholar]
- 88.Annabi N, Fathi A, Mithieux SM, Weiss AS, Dehghani F. J Supercrit Fluids. 2011 In press. [Google Scholar]
- 89.Palocci C, Barbetta A, La Grotta A, Dentini M. Langmuir. 2007;23:8243. doi: 10.1021/la700947g. [DOI] [PubMed] [Google Scholar]
- 90.Lee J-Y, Tan B, Cooper AI. Macromolecules. 2007;40:1955. [Google Scholar]
- 91.Partap S, Rehman I, Jones JR, Darr JA. Adv Mater. 2006;18:501. [Google Scholar]
- 92.Bing Z, Lee JY, Choi SW, Kim JH. Eur Polym J. 2007;43:4814. [Google Scholar]
- 93.Annabi N, Mithieux SM, Weiss AS, Dehghani F. Biomaterials. 2010;31:1655. doi: 10.1016/j.biomaterials.2009.11.051. [DOI] [PubMed] [Google Scholar]
- 94.Annabi N, Fathi A, Mithieux SM, Martens P, Weiss AS, Dehghani F. Biomaterials. 2011;32:1517. doi: 10.1016/j.biomaterials.2010.10.024. [DOI] [PubMed] [Google Scholar]
- 95.Annabi N, Nichol JW, Zhong X, Ji C, Koshy S, Khademhosseini A, Dehghani F. Tissue Eng B. 2010;16:371. doi: 10.1089/ten.teb.2009.0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Park JS, Woo DG, Sun BK, Chung H-M, Im SJ, Choi YM, Park K, Huh KM, Park K-H. J Control Release. 2007;124:51. doi: 10.1016/j.jconrel.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 97.Dadsetan M, Hefferan TE, Szatkowski JP, Mishra PK, Macura SI, Lu L, Yaszemski MJ. Biomaterials. 2008;29:2193. doi: 10.1016/j.biomaterials.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kim JH, Lee SB, Kim SJ, Lee YM. Polymer. 2002;43:7549. [Google Scholar]
- 99.Horak D, Hlidkova H, Hradil J, Lapcikova M, Slouf M. Polymer. 2008;49:2046. [Google Scholar]
- 100.Draghi L, Resta S, Pirozzolo M, Tanzi M. J Mater Sci Mater Med. 2005;16:1093. doi: 10.1007/s10856-005-4711-x. [DOI] [PubMed] [Google Scholar]
- 101.Gong Y, Zhou Q, Gao C, Shen J. Acta Biomater. 2007;3:531. doi: 10.1016/j.actbio.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 102.Salerno A, Guarnieri D, Iannone M, Zeppetelli S, Netti PA. Tissue Eng. 2010;16:2661. doi: 10.1089/ten.tea.2009.0494. [DOI] [PubMed] [Google Scholar]
- 103.Salerno A, Guarnieri D, Iannone M, Zeppetelli S, Di Maio E, Iannace S, Netti PA. Acta Biomater. 2009;5:1082. doi: 10.1016/j.actbio.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 104.Zhang J, Wu L, Jing D, Ding JA. Polymer. 2005;46:4979. [Google Scholar]
- 105.Suh SW, Shin JY, Kim J, Kim J, Beak CH, Kim D-I, Kim H, Jeon SS, Choo I-W. ASAIO J. 2002:460. doi: 10.1097/00002480-200209000-00003. [DOI] [PubMed] [Google Scholar]
- 106.Wei G, Ma PX. J Biomed Mater Res A. 2006;78:306. doi: 10.1002/jbm.a.30704. [DOI] [PubMed] [Google Scholar]
- 107.Liu X, Ma PX. Biomaterials. 2009;30:4094. doi: 10.1016/j.biomaterials.2009.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Liu X, Smith LA, Hu J, Ma PX. Biomaterials. 2009;30:2252. doi: 10.1016/j.biomaterials.2008.12.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Salerno A, Oliviero M, Di Maio E, Iannace S, Netti PA. J Appl Polym Sci. 2007;106:3335. [Google Scholar]
- 110.Alves NM, Pashkuleva I, Reis RL, Mano JF. Small. 2010;6:2208. doi: 10.1002/smll.201000233. [DOI] [PubMed] [Google Scholar]
- 111.Barber RW, Emerson DR. Altern Lab Anim. 2010;38:67. doi: 10.1177/026119291003801S02. [DOI] [PubMed] [Google Scholar]
- 112.Kaji H, Camci-Unal G, Langer R, Khademhosseini A. Biochim Biophys Acta. 2011;1810:239. doi: 10.1016/j.bbagen.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tay CY, Irvine SA, Boey FYC, Tan LP, Venkatraman S. Small. 2011;7:1361. doi: 10.1002/smll.201100046. [DOI] [PubMed] [Google Scholar]
- 114.Shekaran A, Garcia AJ. Biochim Biophys Acta. 2011;1810:350. doi: 10.1016/j.bbagen.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sreejalekshmi KG, Nair PD. J Biomed Mater Res A. 2011;96 A:477. doi: 10.1002/jbm.a.32980. [DOI] [PubMed] [Google Scholar]
- 116.Pradhan S, Farach-Carson MC. Regen Med. 2010;5:961. doi: 10.2217/rme.10.61. [DOI] [PubMed] [Google Scholar]
- 117.Shin H, Jo S, Mikos AG. Biomaterials. 2003;24:4353. doi: 10.1016/s0142-9612(03)00339-9. [DOI] [PubMed] [Google Scholar]
- 118.Grayson WL, Martens TP, Eng GM, Radisic M, Vunjak-Novakovic G. Semin Cell Dev Biol. 2009;20:665. doi: 10.1016/j.semcdb.2008.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ma PX. Adv Drug Deliv Rev. 2008;60:184. doi: 10.1016/j.addr.2007.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Patterson J, Martino MM, Hubbell JA. Mater Today. 2010;13:14. [Google Scholar]
- 121.Kaji H, Camci-Unal G, Langer R, Khademhosseini A. Biochim Biophys Acta. Vol. 1810. Elsevier B.V; Netherlands: 2010. p. 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Murtuza B, Nichol JW, Khademhosseini A. Tissue Eng B. 2009;15:443. doi: 10.1089/ten.teb.2009.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bissell MJ, Radisky DC, Rizki A, Weaver VM, Petersen OW. Differentiation. 2002;70:537. doi: 10.1046/j.1432-0436.2002.700907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bissell MJ, Rizki A, Mian IS. Curr Opin Cell Bio. 2003;15:753. doi: 10.1016/j.ceb.2003.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Engler AJ, Humbert PO, Wehrle-Haller B, Weaver VM. Science. 2009;324:208. doi: 10.1126/science.1170107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ingber DE. Proc Natl Acad Sci U S A. 2005;102:11571. doi: 10.1073/pnas.0505939102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Rivron NC, Rouwkema J, Truckenmuller R, Karperien M, De Boer J, Van Blitterswijk CA. Biomaterials. 2009;30:4851. doi: 10.1016/j.biomaterials.2009.06.037. [DOI] [PubMed] [Google Scholar]
- 128.Hui EE, Bhatia SN. Proc Natl Acad Sci U S A. 2007;104:5722. doi: 10.1073/pnas.0608660104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Tibbitt MW, Anseth KS. Biotechnol Bioeng. 2009;103:655. doi: 10.1002/bit.22361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Drury JL, Mooney DJ. Biomaterials. 2003;24:4337. doi: 10.1016/s0142-9612(03)00340-5. [DOI] [PubMed] [Google Scholar]
- 131.Lee KY, Mooney DJ. Chem Rev. 2001;101:1869. doi: 10.1021/cr000108x. [DOI] [PubMed] [Google Scholar]
- 132.Owen SC, Shoichet MS. Journal of Biomedical Materials Research - Part A. 2010;94:1321. doi: 10.1002/jbm.a.32834. [DOI] [PubMed] [Google Scholar]
- 133.Khademhosseini A, Eng G, Yeh J, Fukuda J, Blumling J, III, Langer R, Burdick JA. J Biomed Mater Res A. 2006;79:522. doi: 10.1002/jbm.a.30821. [DOI] [PubMed] [Google Scholar]
- 134.Suh KY, Khademhosseini A, Yang JM, Eng G, Langer R. Adv Mater. 2004;16:584. [Google Scholar]
- 135.Revzin A, Russell RJ, Yadavalli VK, Koh W-G, Deister C, Hile DD, Mellott MB, Pishko MV. Langmuir. 2001;17:5440. doi: 10.1021/la010075w. [DOI] [PubMed] [Google Scholar]
- 136.Albrecht DR, Tsang VL, Sah RL, Bhatia SN. Lab Chip. 2005;5:111. doi: 10.1039/b406953f. [DOI] [PubMed] [Google Scholar]
- 137.Napolitano AP, Chai P, Dean DM, Morgan JR. Tissue Eng. 2007;13:2087. doi: 10.1089/ten.2006.0190. [DOI] [PubMed] [Google Scholar]
- 138.Fukuda J, Khademhosseini A, Yeo Y, Yang X, Yeh J, Eng G, Blumling J, Wang C-F, Kohane DS, Langer R. Biomaterials. 2006;27:5259. doi: 10.1016/j.biomaterials.2006.05.044. [DOI] [PubMed] [Google Scholar]
- 139.Annabi N, Nichol JW, Zhong X, Ji C, Koshy S, Khademhosseini A, Dehghani F. Tissue Eng B. 2010;16:371. doi: 10.1089/ten.teb.2009.0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Nichol JW, Khademhosseini A. Soft Matter. 2009;5:1312. doi: 10.1039/b814285h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lee SH, Moon JJ, West JL. Biomaterials. 2008;29:2962. doi: 10.1016/j.biomaterials.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Du Y, Lo E, Vidula MK, Khabiry M, Khademhosseini A. Cell Mol Bioeng. 2008;1:157. doi: 10.1007/s12195-008-0020-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Du Y, Ghodousi M, Lo E, Vidula MK, Emiroglu O, Khademhosseini A. Biotechnol Bioeng. 2010;105:655. doi: 10.1002/bit.22552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bowden N, Terfort A, Carbeck J, Whitesides GM. Science. 1997;276:233. doi: 10.1126/science.276.5310.233. [DOI] [PubMed] [Google Scholar]
- 145.Livoti CM, Morgan JR. Tissue Eng A. 2010;16:2051. doi: 10.1089/ten.tea.2009.0607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Albrecht DR, Underhill GH, Wassermann TB, Sah RL, Bhatia SN. Nat Methods. 2006;3:369. doi: 10.1038/nmeth873. [DOI] [PubMed] [Google Scholar]
- 147.Orban JM, Marra KG, Hollinger JO. Tissue Eng. 2002;8:529. doi: 10.1089/107632702760240454. [DOI] [PubMed] [Google Scholar]
- 148.Sharma B, Elisseeff JH. Ann Biomed Eng. 2004;32:148. doi: 10.1023/b:abme.0000007799.60142.78. [DOI] [PubMed] [Google Scholar]
- 149.Richardson TP, Peters MC, Ennett AB, Mooney DJ. Nat Biotechnol. 2001;19:1029. doi: 10.1038/nbt1101-1029. [DOI] [PubMed] [Google Scholar]
- 150.Johnstone B, Hering TM, Caplan AI, Goldberg VM, Yoo JU. Exp Cell Res. 1998;238:265. doi: 10.1006/excr.1997.3858. [DOI] [PubMed] [Google Scholar]
- 151.Nuttelman CR, Tripodi MC, Anseth KS. J Biomed Mater Res A. 2004;68A:773. doi: 10.1002/jbm.a.20112. [DOI] [PubMed] [Google Scholar]
- 152.Williams CG, Kim TK, Taboas A, Malik A, Manson P, Elisseeff J. Tissue Eng. 2003;9:679. doi: 10.1089/107632703768247377. [DOI] [PubMed] [Google Scholar]
- 153.Choi JW, MacDonald E, Wicker R. Int J Adv Manuf Tech. 2010;49:543. [Google Scholar]
- 154.Khademhosseini A, Langer R, Borenstein J, Vacanti JP. Proc Natl Acad Sci U S A. 2006;103:2480. doi: 10.1073/pnas.0507681102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Chung BG, Kang L, Khademhosseini A. Expt Opin Drug Dis. 2007;2:1653. doi: 10.1517/17460441.2.12.1653. [DOI] [PubMed] [Google Scholar]
- 156.Khademhosseini A, Eng G, Yeh J, Kucharczyk PA, Langer R, Vunjak-Novakovic G, Radisic M. Biomed Microdevices. 2007;9:149. doi: 10.1007/s10544-006-9013-7. [DOI] [PubMed] [Google Scholar]
- 157.Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Science. 1997;276:1425. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- 158.Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Biotechnol Prog. 1998;14:356. doi: 10.1021/bp980031m. [DOI] [PubMed] [Google Scholar]
- 159.Hasirci V, Vrana E, Zorlutuna P, Ndreu A, Yilgor P, Basmanav FB, Aydin E. J Biomater Sci Polym Ed. 2006;17:1241. doi: 10.1163/156856206778667442. [DOI] [PubMed] [Google Scholar]
- 160.Ito Y. Biomaterials. 1999;20:2333. doi: 10.1016/s0142-9612(99)00162-3. [DOI] [PubMed] [Google Scholar]
- 161.Itoga K, Okano T. Nippon rinsho. Jpn J Clin Med. 2008;66:887. [PubMed] [Google Scholar]
- 162.Yap FL, Zhang Y. Biosens Bioelectron. 2007;22:775. doi: 10.1016/j.bios.2006.03.016. [DOI] [PubMed] [Google Scholar]
- 163.Nakanishi J, Takarada T, Yamaguchi K, Maeda M. Anal Sci. 2008;24:67. doi: 10.2116/analsci.24.67. [DOI] [PubMed] [Google Scholar]
- 164.Chou L, Firth JD, Uitto V-J, Brunette DM. J Biomed Mater Res A. 1998;39:437. doi: 10.1002/(sici)1097-4636(19980305)39:3<437::aid-jbm13>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 165.Rajnicek A, Britland S, McCaig C. J Cell Sci. 1997;110:2905. doi: 10.1242/jcs.110.23.2905. [DOI] [PubMed] [Google Scholar]
- 166.Wójciak-Stothard B, Curtis A, Monaghan W, Macdonald K, Wilkinson C. Exp Cell Res. 1996;223:426. doi: 10.1006/excr.1996.0098. [DOI] [PubMed] [Google Scholar]
- 167.Meyle J, Gültig K, Wolburg H, Recum AFv. J Biomed Mater Res. 1993;27:1553. doi: 10.1002/jbm.820271212. [DOI] [PubMed] [Google Scholar]
- 168.Meyle J, Gültig K, Nisch W. J Biomed Mater Res. 1995;29:81. doi: 10.1002/jbm.820290112. [DOI] [PubMed] [Google Scholar]
- 169.Boni A, Urbanek K, Nascimbene A, Hosoda T, Zheng H, Delucchi F, Amano K, Gonzalez A, Vitale S, Ojaimi C, Rizzi R, Bolli R, Yutzey KE, Rota M, Kajstura J, Anversa P, Leri A. Proc Natl Acad Sci U S A. 2008;105:15529. doi: 10.1073/pnas.0808357105. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 170.Dellatore SM, Garcia AS, Miller WM. Curr Opin Biotechnol. 2008;19:534. doi: 10.1016/j.copbio.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Urbanek K, Cesselli D, Rota M, Nascimbene A, De Angelis A, Hosoda T, Bearzi C, Boni A, Bolli R, Kajstura J, Anversa P, Leri A. Proc Natl Acad Sci U S A. 2006;103:9226. doi: 10.1073/pnas.0600635103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Cheng C-M, LeDuc PR. Mol Biosyst. 2006;2:299. doi: 10.1039/b606496p. [DOI] [PubMed] [Google Scholar]
- 173.Hoffmann JC, West JL. Soft Matter. 2010;6:5056. [Google Scholar]
- 174.Bryant SJ, Cuy JL, Hauch KD, Ratner BD. Biomaterials. 2007;28:2978. doi: 10.1016/j.biomaterials.2006.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Bian W, Bursac N. Biomaterials. 2009;30:1401. doi: 10.1016/j.biomaterials.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Lovett M, Lee K, Edwards A, Kaplan DL. Tissue Eng B. 2009;15:353. doi: 10.1089/ten.teb.2009.0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Kaully T, Kaufman-Francis K, Lesman A, Levenberg S. Tissue Eng B. 2009;15:159. doi: 10.1089/ten.teb.2008.0193. [DOI] [PubMed] [Google Scholar]
- 178.Moon JJ, West JL. Curr Top Med Chem. 2008;8:300. doi: 10.2174/156802608783790983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Whitesides GM, Ostuni E, Takayama S, Jiang XY, Ingber DE. Annu Rev Biomed Eng. 2001;3:335. doi: 10.1146/annurev.bioeng.3.1.335. [DOI] [PubMed] [Google Scholar]
- 180.Hahn MS, Miller JS, West JL. Adv Mater. 2006;18:2679. [Google Scholar]
- 181.Leslie-Barbick JE, Shen C, Chen C, West JL. Tissue Eng A. 2011;17:221. doi: 10.1089/ten.tea.2010.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Moon JJ, Hahn MS, Kim I, Nsiah BA, West JL. Tissue Eng A. 2009;15:579. doi: 10.1089/ten.tea.2008.0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Bettinger CJ, Cyr KM, Matsumoto A, Langer R, Borenstein JT, Kaplan DL. Adv Mater. 2007;19:2847. doi: 10.1002/adma.200602487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Khademhosseini A, Langer R. Biomaterials. 2007;28:5087. doi: 10.1016/j.biomaterials.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 185.Hahn MS, Taite LJ, Moon JJ, Rowland MC, Ruffino KA, West JL. Biomaterials. 2006;27:2519. doi: 10.1016/j.biomaterials.2005.11.045. [DOI] [PubMed] [Google Scholar]
- 186.Moon JJ, Saik JE, Poche RA, Leslie-Barbick JE, Lee S-H, Smith AA, Dickinson ME, West JL. Biomaterials. 2010:31. doi: 10.1016/j.biomaterials.2010.01.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Dike LE, Chen CS, Mrksich M, Tien J, Whitesides GM, Ingber DE. In Vitro Cell Dev Biol Anim. 1999;35:441. doi: 10.1007/s11626-999-0050-4. [DOI] [PubMed] [Google Scholar]
- 188.Li S, Bhatia S, Hu YL, Shin YT, Li YS, Usami S, Chien S. Biorheology. 2001;38:101. [PubMed] [Google Scholar]
- 189.Dickinson LE, Moura ME, Gerecht S. Soft Matter. 2010;6:5109. [Google Scholar]
- 190.Raghavan S, Nelson CM, Baranski JD, Lim E, Chen CS. Tissue Eng A. 2010;16:2255. doi: 10.1089/ten.tea.2009.0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Ling Y, Rubin J, Deng Y, Huang C, Demirci U, Karp JM, Khademhosseini A. Lab Chip. 2007;7:756. doi: 10.1039/b615486g. [DOI] [PubMed] [Google Scholar]
- 192.Choi NW, Cabodi M, Held B, Gleghorn JP, Bonassar LJ, Stroock AD. Nature Mater. 2007;6:908. doi: 10.1038/nmat2022. [DOI] [PubMed] [Google Scholar]
- 193.Karp JM, Yeh J, Eng G, Fukuda J, Blumling J, Suh KY, Cheng J, Mahdavi A, Borenstein J, Langer R, Khademhosseini A. Lab Chip. 2007;7:786. doi: 10.1039/b705085m. [DOI] [PubMed] [Google Scholar]
- 194.King KR, Wang CCJ, Kaazempur-Mofrad MR, Vacanti JP, Borenstein JT. Adv Mater. 2004;16:2007. [Google Scholar]
- 195.Bettinger CJ, Weinberg EJ, Kulig KM, Vacanti JP, Wang YD, Borenstein JT, Langer R. Adv Mater. 2006;18:165. doi: 10.1002/adma.200500438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Park JH, Chung BG, Lee WG, Kim J, Brigham MD, Shim J, Lee S, Hwang CM, Durmus NG, Demirci U, Khademhosseini A. Biotechnol Bioeng. 2010;106:138. doi: 10.1002/bit.22667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Golden AP, Tien J. Lab Chip. 2007;7:720. doi: 10.1039/b618409j. [DOI] [PubMed] [Google Scholar]
- 198.Wu W, Deconinck A, Lewis JA. Adv Mater. 2011;23:H178. doi: 10.1002/adma.201004625. [DOI] [PubMed] [Google Scholar]
- 199.Andersson H, van den Berg A. Lab Chip. 2004;4:98. doi: 10.1039/b314469k. [DOI] [PubMed] [Google Scholar]
- 200.Hwang YS, Chung BG, Ortmann D, Hattori N, Moeller HC, Khademhosseini A. Proc Natl Acad Sci U S A. 2009;106:16978. doi: 10.1073/pnas.0905550106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Whitesides GM, Ostuni E, Takayama S, Jiang X, Ingber DE. Annu Rev Biomed Eng. 2001;3:335. doi: 10.1146/annurev.bioeng.3.1.335. [DOI] [PubMed] [Google Scholar]
- 202.Nichol JW, Khademhosseini A. Soft Matter. 2009;5:1312. doi: 10.1039/b814285h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Park J, Cho CH, Parashurama N, Li Y, Berthiaume F, Toner M, Tilles AW, Yarmush ML. Lab Chip. 2007;7:1018. doi: 10.1039/b704739h. [DOI] [PubMed] [Google Scholar]
- 204.Bauwens CL, Peerani R, Niebruegge S, Woodhouse KA, Kumacheva E, Husain M, Zandstra PW. Stem Cells. 2008;26:2300. doi: 10.1634/stemcells.2008-0183. [DOI] [PubMed] [Google Scholar]
- 205.Lee LH, Peerani R, Ungrin M, Joshi C, Kumacheva E, Zandstra P. Stem Cell Res. 2009;2:155. doi: 10.1016/j.scr.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 206.Moeller HC, Mian MK, Shrivastava S, Chung BG, Khademhosseini A. Biomaterials. 2008;29:752. doi: 10.1016/j.biomaterials.2007.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Dusseiller MR, Schlaepfer D, Koch M, Kroschewski R, Textor M. Biomaterials. 2005;26:5917. doi: 10.1016/j.biomaterials.2005.02.032. [DOI] [PubMed] [Google Scholar]
- 208.Ochsner M, Dusseiller MR, Grandin HM, Luna-Morris S, Textor M, Vogel V, Smith ML. Lab Chip. 2007;7:1074. doi: 10.1039/b704449f. [DOI] [PubMed] [Google Scholar]
- 209.Khademhosseini A, Ferreira L, Blumling J, 3rd, Yeh J, Karp JM, Fukuda J, Langer R. Biomaterials. 2006;27:5968. doi: 10.1016/j.biomaterials.2006.06.035. [DOI] [PubMed] [Google Scholar]
- 210.Xu Y, Shi Y, Ding S. Nature. 2008;453:338. doi: 10.1038/nature07042. [DOI] [PubMed] [Google Scholar]
- 211.Flaim CJ, Teng D, Chien S, Bhatia SN. Stem Cells Dev. 2008;17:29. doi: 10.1089/scd.2007.0085. [DOI] [PubMed] [Google Scholar]
- 212.Fernandes TG, Diogo MM, Clark DS, Dordick JS, Cabral JM. Trends Biotechnol. 2009;27:342. doi: 10.1016/j.tibtech.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Dev Cell. 2004;6:483. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 214.Anderson DG, Levenberg S, Langer R. Nat Biotechnol. 2004;22:863. doi: 10.1038/nbt981. [DOI] [PubMed] [Google Scholar]
- 215.Fernandes TG, Kwon SJ, Bale SS, Lee MY, Diogo MM, Clark DS, Cabral JM, Dordick JS. Biotechnol Bioeng. 2010;106:106. doi: 10.1002/bit.22661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Jinno S, Moeller HC, Chen CL, Rajalingam B, Chung BG, Dokmeci MR, Khademhosseini A. J Biomed Mater Res A. 2008;86:278. doi: 10.1002/jbm.a.32030. [DOI] [PubMed] [Google Scholar]
- 217.Wright D, Rajalingam B, Karp JM, Selvarasah S, Ling Y, Yeh J, Langer R, Dokmeci MR, Khademhosseini A. J Biomed Mater Res A. 2008;85:530. doi: 10.1002/jbm.a.31281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Abbott A. Nature. 2003;424:870. doi: 10.1038/424870a. [DOI] [PubMed] [Google Scholar]
- 219.Polak JM, Mantalaris S. Pediatr Res. 2008;63:461. doi: 10.1203/pdr.0b013e31816a8c1c. [DOI] [PubMed] [Google Scholar]
- 220.Zhang S. Nat Biotech. 2004;22:151. [Google Scholar]
- 221.Burdick JA, Vunjak-Novakovic G. Tissue Eng. 2009;15:205. doi: 10.1089/ten.tea.2008.0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Zhong JF, Chen Y, Marcus JS, Scherer A, Quake SR, Taylor CR, Weiner LP. Lab Chip. 2008;8:68. doi: 10.1039/b712116d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.McDonald JC, Duffy DC, Anderson JR, Chiu DT, Wu H, Schueller OJ, Whitesides GM. Electrophoresis. 2000;21:27. doi: 10.1002/(SICI)1522-2683(20000101)21:1<27::AID-ELPS27>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 224.Park TH, Shuler ML. Biotechnol Prog. 2003;19:243. doi: 10.1021/bp020143k. [DOI] [PubMed] [Google Scholar]
- 225.Rhee SW, Taylor AM, Tu CH, Cribbs DH, Cotman CW, Jeon NL. Lab Chip. 2005;5:102. doi: 10.1039/b403091e. [DOI] [PubMed] [Google Scholar]
- 226.Tan W, Desai TA. Tissue Eng. 2003;9:255. doi: 10.1089/107632703764664729. [DOI] [PubMed] [Google Scholar]
- 227.Chiu DT, Jeon NL, Huang S, Kane RS, Wargo CJ, Choi IS, Ingber DE, Whitesides GM. Proc Natl Acad Sci U S A. 2000;97:2408. doi: 10.1073/pnas.040562297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Takayama S, Ostuni E, LeDuc P, Naruse K, Ingber DE, Whitesides GM. Nature. 2001;411:1016. doi: 10.1038/35082637. [DOI] [PubMed] [Google Scholar]
- 229.Aref A, Horvath R, McColl J, Ramsden JJ. J Biomed Opt. 2009:14. doi: 10.1117/1.3065541. [DOI] [PubMed] [Google Scholar]
- 230.Kirouac DC, Zandstra PW. Cell Stem Cell. 2008;3:369. doi: 10.1016/j.stem.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 231.Tourovskaia A, Figueroa-Masot X, Folch A. Lab Chip. 2005;5:14. doi: 10.1039/b405719h. [DOI] [PubMed] [Google Scholar]
- 232.Kim L, Vahey MD, Lee HY, Voldman J. Lab Chip. 2006;6:394. doi: 10.1039/b511718f. [DOI] [PubMed] [Google Scholar]
- 233.Meinel L, Karageorgiou V, Fajardo R, Snyder B, Shinde-Patil V, Zichner L, Kaplan D, Langer R, Vunjak-Novakovic G. Ann Biomed Eng. 2004;32:112. doi: 10.1023/b:abme.0000007796.48329.b4. [DOI] [PubMed] [Google Scholar]
- 234.Chung BG, Flanagan LA, Rhee SW, Schwartz PH, Lee AP, Monuki ES, Jeon NL. Lab Chip. 2005;5:401. doi: 10.1039/b417651k. [DOI] [PubMed] [Google Scholar]
- 235.Freed LE, Langer R, Martin I, Pellis NR, Vunjak-Novakovic G. Proc Natl Acad Sci U S A. 1997;94:13885. doi: 10.1073/pnas.94.25.13885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Park SH, Sim WY, Park SW, Yang SS, Choi BH, Park SR, Park K, Min BH. Tissue Eng. 2006;12:3107. doi: 10.1089/ten.2006.12.3107. [DOI] [PubMed] [Google Scholar]
- 237.Metallo CM, Vodyanik MA, de Pablo JJ, Slukvin II, Palecek SP. Biotechnol Bioeng. 2008;100:830. doi: 10.1002/bit.21809. [DOI] [PubMed] [Google Scholar]
- 238.Hung P, Lee P, Sabounchi P, Lin R, Lee L. Biotechnol Bioeng. 2005;89:1. doi: 10.1002/bit.20289. [DOI] [PubMed] [Google Scholar]
- 239.Hung PJ, Lee PJ, Sabounchi P, Aghdam N, Lin R, Lee LP. Lab Chip. 2005;5:44. doi: 10.1039/b410743h. [DOI] [PubMed] [Google Scholar]
- 240.Kim C, Lee KS, Bang JH, Kim YE, Kim MC, Oh KW, Lee SH, Kang JY. Lab Chip. 2011;11:874. doi: 10.1039/c0lc00516a. [DOI] [PubMed] [Google Scholar]
- 241.Ratcliffe A, Niklason LE. Ann NY Acad Sci. 2002;961:210. doi: 10.1111/j.1749-6632.2002.tb03087.x. [DOI] [PubMed] [Google Scholar]
- 242.Pei M, Solchaga LA, Seidel J, Zeng L, Vunjak-Novakovic G, Caplan AI, Freed LE. Faseb J. 2002;16:1691. doi: 10.1096/fj.02-0083fje. [DOI] [PubMed] [Google Scholar]