Abstract
Epigenetic changes are associated with the regulation of transcription of key cell regulatory genes [micro RNAs (miRNAs)] during different types of liver injury. This study evaluated the role of methylation-associated miRNA, miR-34a, in alcoholic liver diseases. We identified that ethanol feeding for 4 weeks significantly up-regulated 0.8% of known miRNA compared with controls, including miR-34a. Treatment of normal human hepatocytes (N-Heps) and cholangiocytes [human intrahepatic biliary epithelial cells (HiBECs)] with ethanol and lipopolysaccharide induced a significant increase of miR-34a expression. Overexpression of miR-34a decreased ethanol-induced apoptosis in both N-Heps and HiBECs. In support of the concept that the 5′-promoter region of miR-34a was noted to be embedded within a CpG island, the expression level of miR-34a was significantly increased after demethylation treatment in N-Heps and HiBECs. By methylation-specific PCR, we confirmed that miR-34a activation is associated with ethanol-linked hypomethylation of the miR-34a promoter. A combination of bioinformatics, dual-luciferase reporter assay, mass spectrometry, and Western blot analysis revealed that caspase-2 and sirtuin 1 are the direct targets of miR-34a. Furthermore, modulation of miR-34a also altered expression of matrix metalloproteases 1 and 2, the mediators involved in hepatic remodeling during alcoholic liver fibrosis. These findings provide the basis for an exciting field in which the epigenomic microRNAs of hepatic cells may be manipulated with potential therapeutic benefits in human alcoholic liver diseases.
Long-term alcohol consumption and the associated development of alcoholic liver disease (ALD) is a major health concern for the United States. Approximately 15% of individuals with alcoholism in the United States eventually develop ALD, one of the leading causes of liver diseases and liver-related deaths worldwide. ALDs encompass a broad spectrum of clinical features of alcoholic fatty liver, alcoholic steatohepatitis, alcoholic cirrhosis, and increased risk of hepatocellular carcinoma (HCC).1 The pathologic mechanisms of ALD involve complex interactions between the direct effects of alcohol and its toxic metabolites on various cell types in the liver, including induction of reactive oxygen species, up-regulation of the inflammatory cascade, and other cell-specific effects in the liver. Prominent features of ALD include ethanol-mediated cellular alterations, steatosis, and hepatic inflammation. However, a comprehensive understanding of the mechanisms involved in the pathogenesis of ALD remains incomplete. Thus, an understanding of the molecular mechanisms regulating hepatobiliary cell injury is important and may lead to more effective therapeutic approaches for ALD.
MicroRNAs (miRNAs) are a group of noncoding RNA that plays an important role in human liver diseases and have recently become of interest in the pathogenesis of ALD.2,3 In mammals, miRNAs can negatively regulate their targets by either binding to imperfect complementary sites within the 3′-untranslated region (UTR) of their mRNA targets or by targeting specific cleavage of homologous mRNAs.4 In our previous studies, we observed the increased expression of several miRNAs, including miR-181 and let-7 family members that are involved in hepatic cell survival, remodeling, and transformation.5 Similarly, altered expression of several miRNAs has been described in expression profiling of human liver diseases and in animal studies.6–8 We postulate that alterations in the expression of miRNAs influence cellular behavior, such as survival and remodeling by alteration of key cellular targets. In fact, aberrant expression of miRNAs, such as miR-181, alters the cellular expression of TIMP3 and Nemo-like kinase.5 However, the contribution of aberrantly expressed miRNAs to hepatic cell responses in ALD is unknown.
The regulation of miR-34a by the transcription factor p53 suggests a potential role for miR-34 in the modulation of hepatic cell behavior.9–12 Normally, p53 inhibits cell proliferation and stimulates cell death. However, disruption of the p53 pathway promotes liver injury. One pathway by which p53 regulates cell growth is through miRNA. Cellular stress stabilizes p53 that in turn regulates the expression of a set of miRNA, which control apoptosis and senescence.9–13 Recent studies show that the miRNA miR-34a is activated by p53.9–13 E2F3, a transcription factor involved in cell cycle progression, has also been identified as a target of miR-34a.13,14 Although derepression of E2F3 may promote neoplastic growth in tumors in which miR-34a is reduced, such as gliomas, neuroblastomas,14 and colorectal cancers,13 the overexpression of miR-34a during human liver regeneration suggests the presence of additional mechanisms by which miR-34a contributes to hepatic cell survival and regeneration.15 Thus, we assessed the role of aberrant expression of miR-34a in hepatic cell survival and remodeling during ALD by posing the following questions: i) Is miR-34a expression altered in ethanol-exposed mice and ALD human liver tissues? ii) Does modulation of miR-34a alter cell survival and remodeling? iii) Is miR-34a expression be epigenetically modulated? and iv) What downstream targets of miR-34a are involved in ALD?
Materials and Methods
Cells and Tissues
Normal human hepatocytes (N-Heps) and cholangiocytes [human intrahepatic biliary epithelial cells (HiBECs)] were obtained from Sciencell (San Diego, CA). The human hepatocellular cancer cell line, HepG2, was obtained from the American Type Culture Collection (Manassas, VA) and cultured as recommended by the supplier. Cells were grown to ∼75% confluency on 100-mm culture dishes. For studies on the effects of methylation inhibition, cells were incubated with either 10 μmol/L 5-aza-2′deoxycytidine (5-Aza-CdR) or diluent (acetic acid) for 24 hours at 37°C, after which cells were washed twice with cold 1× PBS and harvested for the isolation of genomic DNA or total protein. Six pairs of ALD human liver samples (adjacent liver tissues of HCC patients with heavy alcohol consumption history) and normal control tissues from surgical resections (distal normal liver tissue of patients with liver hemangioma) were analyzed in a masked manner from the Department of Hepatobiliary Surgery, Shengjing Hospital (see Supplemental Table S1 at http://ajp.amjpathol.org). The study protocol of human subjects for the collection of all human materials and data were approved by the Ethics Committee of Shengjing Hospital, China Medical University, Shenyang, China.
Transfections
Transfections were performed by nuclear electroporation using the Nucleofector system (Amaxa Biosystems, Koln, Germany). A total of 50 μL of 100 nmol/L microRNA precursor, antisense inhibitor, or controls (Ambion, Austin, TX) were added to 1 × 106 cells suspended in 50 μL of Nucleofector solution at room temperature. The sequences of the microRNA precursors and inhibitors used can be obtained from Ambion. After electroporation, transfected cells were resuspended in culture medium containing 10% fetal bovine serum for 48 to 72 hours before study. All studies were performed in quadruplicate unless otherwise specified.
Animal Experiments
For long-term alcohol administration, C57BL/6 mice (10 weeks old) were aseptically implanted with gastrostomy catheters as previously described.16,17 An increasing dose of ethanol (22.7 to 35 g/kg per day) or control solutions was infused for 4 weeks. All animal experiments were performed with age- and sex-matched mice from the same littermates and conducted in accordance with the approved Institutional Animal Care and Use Committee protocol at the University of Southern California.
miRNA Array Hybridization and Analysis
RNA was extracted using Trizol reagent (Invitrogen, Carlsbad, CA). Total RNA (5 μg) was reverse transcribed using biotin end-labeled random octamer oligonucleotide primers. Hybridization of biotin-labeled complementary DNA was performed using a custom miRNA microarray chip (Noncoding RNA Program at Center for Targeted Therapy, M.D. Anderson Cancer Center, Houston, TX), containing 627 probes for mature miRNA corresponding to 324 different human miRNAs spotted in quadruplicate. The images were scanned quantitated using an Axon 4000B scanner (Molecular Devices, Sunnyvale, CA). The scanned images were quantified using GenePix software version 6.0 (Molecular Devices). The data from three samples for each cell type were further analyzed by the BRB-ArrayTools software version 4.2.1 from the National Cancer Institute (Bethesda, MD).
Real-Time PCR for Mature miRNA
The expression of mature miRNAs in human hepatobiliary cell lines was analyzed by TaqMan miRNA Assay (Applied Biosystems, Foster City, CA). Briefly, single-stranded cDNA was synthesized from 10 ng of total RNA in a 15-μL reaction volume by using the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems). The reactions were incubated first at 16°C for 30 minutes and then at 42°C for 30 minutes. The reactions were inactivated by incubation at 85°C for 5 minutes. Each cDNA generated was amplified by quantitative PCR by using sequence-specific primers from the TaqMan microRNA Assays on a MX 3000P PCR Instrument (Stratagene, San Diego, CA). The 20-μL PCR included 10 μL of 2× Universal PCR Master Mix (No AmpErase UNG), 2 μL of each 10× TaqMan MicroRNA Assay Mix, and 1.5 μL of reverse transcription product. The reactions were incubated in a 96-well plate at 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. The CT is defined as the fractional cycle number at which the fluorescence passes the fixed threshold.
Plasmids and Oligonucleotides for miR-34a Methylation-Specific Luciferase Assay
Oligonucleotides were designed to the 5′-promoter region of has-miR-34a genomic sequence, and primers were obtained from Invitrogen. A fragment of 179 to 385 that contains the identified CpG island enriched elements of the 5′- promoter region of has-miR-34a was amplified by PCR and cloned into the MluI and BgIII sites of the pGL3-luciferase plasmid (Promega, Madison, WI) to form 5′-miR-34a-LUC; pCMV-Renilla was obtained from Promega. A 24-bp mutation of the hsa-miR-34a methylation site (206 bp) GCC to TAA was performed using oligonucleotides to the CpG island enriched region and a Quikchange site mutagenesis kit (Stratagene, San Diego, CA) according to the manufacturer's instructions to form miR-34a-MUT. Orientation and sequence identity were confirmed in all plasmids by sequencing.
SuperArray Quantitative PCR Assay and Real-Time PCR Analysis
RNA was isolated from colon tissues using TRIzol (Invitrogen) and cleaned with the Qiagen's RNeasy Kit (Qiagen, Valencia, CA) according to the manufacturer's protocol. The optional on-column DNase treatment was performed. Reverse transcription was performed using 1 μg of RNA with SABios-ciences' RT2 First Strand Kit (SABiosciences, Frederick, MD) according to the manufacturer's protocol. Mouse liver tissue cDNA was analyzed using SuperArray plates (Epigenetic Chromatin Modification Enzymes PCR Array, SABiosciences). To validate the translational significance of these gene expression findings, mice liver samples were analyzed using real-time PCR. RT2 qPCR Primer Assays were obtained from SABiosciences. Real-time PCR was performed using SABiosciences RT2 SYBR Green/ROX qPCR Master Mix for a Stratagene Mx3005P Real-Time PCR System according to the manufacturer protocol. ROX was used as an endogenous reference, and data were analyzed using SABiosciences PCR Array Data Analysis Template. The comparative CT method (ΔΔCT) was used for quantification of gene expression. All samples were tested in triplicate and mean values used for quantification.
Isolation of RNA and Semiquantitative RT-PCR
The mRNA levels of DNMT3B and DNMT1 were analyzed by semiquantitative RT-PCR. A total of 1 μg of total RNA, isolated using an RNA isolation kit from Invitrogen, was used in reverse transcription reactions as described by the manufacturer. The resulting total cDNA was used in the PCR reaction to measure the mRNA levels of DNMT3B and DNMT1. The mRNA level of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as internal control. PCR was performed with Taq polymerase, and conditions were as follows: predenaturing at 94°C for 3 minutes and 94°C for 30 seconds, 55°C for 30 seconds, and 72°C for 60 seconds. The PCR cycle numbers were 26 for GAPDH, 30 for DNMT1, and 35 for DNMT3B. The primers used for RT-PCR were as follows: DNMT3B: sense primer, 5′-AATGTGAATCCAGCCAGGAAAGGC-3′; antisense primer, 5′-ACTGGATTACACTCCAGGAACCGT-3′; DNMT1: sense primer, 5′-ACCGCTTCTACTTCCTCGAGGCCTA-3′; antisense primer, 5′-GTTGCAGTCCTCTGTGAACACTGTGG-3′; GAPDH: sense primer, 5′-AAGGCTGAGAACGGGAAGCTTGTCATCAAT-3′; antisense primer, 5′-TTCCCGTTCAGCTCAGGGATGACCTTGCCC-3′. Under the conditions used, the cDNAs were exponentially amplified, and thus a semiquantitative estimation of the products was possible.
HPLC-Chip/MS Analysis
The cell lysate samples (500 μg) were reduced, alkylated, and double digested with trypsin to generate peptides. The digested peptides were dried in a SpeedVac and resuspended in 100 μL of 0.1% formic acid in 5% acetonitrile (mobile phase A). A total of 200 μg of peptides (40 μL) was directly loaded onto a 1 × 150-mm Poly-SEA strong cation exchange column (Michrom Bioresources, Auburn, CA) using an Agilent 1200 autosampler (Agilent Technologies, Santa Clara, CA). Peptides were eluted to 10 fractions using 0 to 100 mmol/L ammonium formate for 40 minutes (mobile phase B: 1 mmol/L ammonium formate, 10% formic acid in 5% acetonitrile) and five fractions in 100 to 1000 mmol/L ammonium formate for 10 minutes (on an Agilent 1200 Capillary LC and Analytical-Fraction Collector at a flow rate of 50 μL/min). Peptides were completely dried and reconstituted in 20 μL of 0.1% trifluoroacetic acid for liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) analysis using an LC/MS system consisting of an 1100 Series liquid chromatograph, HPLC-Chip Cube MS interface, and 1100 Series LC/MSD Trap XCT Ultra ion trap mass spectrometer (all Agilent Technologies). The system was equipped with an HPLC-Chip (Agilent Technologies) that incorporated a 40-nL enrichment column and a 43 × 75-μm analytical column packed with Zorbax 300SB-C18 5-μm particles. The gain control (intraclass correlation coefficient) was set to 500,000 with a maximum accumulation time of 150 milliseconds. The coefficient of intrinsic dependence was triggered on the six most abundant, not singly charged peptide ions in the m/z range of 450 to 1500. Precursors were set in an exclusion list for 1 minute after two MS/MS spectra. Coefficient of intrinsic dependence data were searched against the NCBInr human database, using the Agilent Spectrum Mill Server software version Rev A.03.03 (Agilent Technologies). A Spectrum Mill autovalidation was performed first in the protein details followed by peptide mode using default values [minimum scores, minimum scored peak intensity, forward minus reversed score threshold, and rank 1 minus rank 2 score threshold]. All protein hits found in a distinct database search by Spectrum Mill are nonredundant.
Western Blotting
Cells grown in 100-mm dishes were lysed and protein content quantitated using the Bradford protein assay. Equivalent amounts of protein were resolved by SDS-PAGE and transferred to nitrocellulose membranes. Membranes were blocked and incubated with the specific primary antibody overnight at 4°C, washed, and incubated with the appropriate IRDye700- and IRDye800-labeled secondary antibodies (Rockland, Gilbertsville, PA) (1:1000) for 1 hour. Blots were stripped and reprobed with mouse monoclonal antibodies for α-tubulin (Sigma Chemical, St. Louis, MO) (1:2000), which was used for normalization. Protein expression was visualized and quantified using the LI-COR Odyssey Infrared Imaging System (LI-COR Bioscience, Lincoln, NE).
MMP Gene Expression
Real-time PCR assays were performed as described.18,19 Briefly, cDNA was generated by reverse transcription using 1 μg of total RNA isolated using the RNA isolation kit (Bio-Rad, Hercules, CA) and the SuperScript III Reverse Transcription Kit (Invitrogen). Quantitative real-time PCR was performed on a MX 3005P PCR Instrument (Stratagene, San Diego, CA) as described.18,19 Each sample was tested in triplicate. The PCR products were verified by melting curve analysis and by 1.8% agarose gel electrophoresis of the PCR product. Matrix metalloproteinase (MMP) mRNA expression was normalized against expression of β-actin used as an internal control.
Cell Migration Assay
N-Heps, HiBECs, and HepG2 cells (5 × 104 cells) were placed into the top chamber of a BD Falcon HTS FluoroBlok insert with a membrane containing 8-μm pores (BD Biosciences, Rockville, MD) in 300 μL of serum-free Dulbecco's modified Eagle medium in triplicate. The inserts were placed into the bottom chamber wells of a 96-well plate containing Dulbecco's modified Eagle medium media and fetal bovine serum (5%) as a chemoattractant. Cells that migrated through the pores of the membrane to the bottom chamber were labeled with 8 μg/mL of calcein-AM (Molecular Probes, Eugene, OR) in PBS for 30 minutes at 37°C. The fluorescence of migrated cells was quantified using a fluorometer at excitation wavelengths of 485 nm and emission wavelengths of 530 nm and expressed as arbitrary fluorescence units.
Cell Proliferation Assay
Cell proliferation was measured using the CellTiter 96 AQueous Assay Kit (Promega). Transfected cells (10,000 per well) were plated in 96-well plates (BD Biosciences) and incubated at 37°C, and cell proliferation was assessed after 72 hours as previously described.5,19
Anchorage-Independent Hepatobiliary Cell Growth
N-Heps, HiBECs, and HepG2 cells were seeded in 96-well plates (10,000 per well) in modified Dulbecco's modified Eagle medium with 10% fetal bovine serum after miRNA transfection. The final concentration of the bottom and top feeder layers of the agar system was 0.6%, and the cell suspension layer was 0.4%. Anchorage-independent transformed cell growth was fluorometrically assayed after 7 days using Alamar Blue (Biosource International, Camarillo, CA), and the SpectraMax M5 Multi-Mode Microplate Reader (Molecular Devices Inc, Sunnyvale, CA; excitation, 530/25 nm; emission, 580/50 nm).
Luciferase Reporter Assay
Intact putative miR-34a recognition sequence from the 3′-UTR of caspase-2 (CASP2) and Sirtuin 1 (SIRT1) (pMIR-CASP2/SIRT1-wt-3′-UTR) or with random mutations (pMIR-CASP2/SIRT1-mut-3′-UTR) were cloned downstream of the firefly luciferase reporter gene. Cells were co-transfected with 1 μg of pMIR-CASP2/SIRT1-wt or mut-3′-UTR construct and 1 μg of pRL-TK Renilla luciferase expression construct with or without precursor miR-34a using TransIT-siQUEST transfection reagent (Mirus, Madison, WI). Luciferase assays were performed 72 hours after transfection using the Dual Luciferase Reporter Assay system (Promega).
Immunohistochemical Analysis
Lobular necrosis was evaluated in liver sections stained with H&E. Liver sections were incubated overnight at 4°C with the selected antibody (1:50), washed in 1× PBS, incubated for 20 minutes at room temperature with a secondary biotinylated antibody (Dako Cytomation LSAB Plus System-HRP, Glostrup, Denmark) and then with Dako ABC for 20 minutes, and developed with 3–3′ diaminobenzidine (Dako Cytomation Liquid DAB Plus Substrate Chromogen System). For all immunoreactions, negative controls were included. Immunohistochemical observations were taken by BX-51 light microscopy (Olympus, Tokyo, Japan) with a Videocam (Spot Insight; Diagnostic Instrument, Sterling Heights, MI) and processed with an Image Analysis System.
DNA Methylation Analysis
Genomic DNA was deaminated with sodium bisulfite using EZ DNA Methylation-Direct Kit (Zymo Research, Orange County, CA) according to the manufacturer's protocol. CpG islands within the target region for miR-34a promoter were determined using Promoter Scan (http://bimas.dcrt.nih.gov/molbio/proscan; Advanced Biosciences Computing Center, University of Minnesota, Duluth, MN) and CpG Island Searcher (http://ccnt.hsc.usc.edu/cpgislands2/cpg.aspx; University of Southern California, Los Angeles, CA). The regions were within several hundred base pairs upstream of miR-34a precursor transcription start point. Methylation-specific PCR (MSP) was performed using primer sequences for methylated or unmethylated DNA, which were designed using MethPrimer (http://www.urogene.org/methprimer; University of California At San Francisco, San Francisco, CA)20 (Table 1). The primer sets 1, 2, and 3 were used for detection of promoter methylation region, whereas primer sets 4 and 5 were for controls. Briefly, 3 μL of bisulfite-treated genomic DNA was amplified by fluorescence-based real-time MSP using SYBR Green ER qPCR SuperMix. Real-time PCR was performed under the following conditions: 95°C for 10 minutes, followed by 40 cycles of 95°C for 30 seconds, 52°C for 30 seconds, and 60°C for 30 seconds. A reaction tube without any DNA was used as a negative control. The specificity of products obtained was checked by dissociation curve analysis. Each sample was tested in triplicate. The relative methylated and unmethylated mRNA expression in normal or malignant cells was determined based on the mean and SE of the threshold (CT) values for each set of primers (promoter region and controls).
Table 1.
Primers Used for Methylation-Specific PCR
| Primer type | Methylated primer | Unmethylated primer |
|---|---|---|
| Target primer set 1 | F: −917 5′-GGTTGAAAGGTTTTAAGAGTAGAATC-3′ | F: −914 5′-TGAAAGGTTTTAAGAGTAGAATTGA-3′ |
| R: −747 5′-TTATTTCAAAAAATCGACTTACGTA-3′ | R: −747 5′-TTATTTCAAAAAATCAACTTACATA-3′ | |
| Target primer set 2 | F: −917 5′-GGTTGAAAGGTTTTAAGAGTAGAATC-3′ | F: −914 5′-TGAAAGGTTTTAAGAGTAGAATTGA-3′ |
| R: −747 5′- TTATTTCAAAAAATCGACTTACGTA-3′ | R: −746 5′-TTTATTTCAAAAAATCAACTTACATA-3′ | |
| Target primer set 3 | F: −914 5′-TGAAAGGTTTTAAGAGTAGAATCGA-3′ | F: −914 5′-TGAAAGGTTTTAAGAGTAGAATTGA-3′ |
| R: −747 5′-TTATTTCAAAAAATCGACTTACGTA-3′ | R: −747 5′-TTATTTCAAAAAATCAACTTACATA-3′ | |
| Control primer set 4 | F: −38 5′-TAGGTTTGTTTTTCGAGTTTTTTTC-3′ | F: −37 5′-AGGTTTGTTTTTTGAGTTTTTTTTG-3′ |
| R: 151 5′-CTCCCACTAATCTAAACATCTCTCG-3′ | R: 152 5′-TCTCCCACTAATCTAAACATCTCTCA-3′ | |
| Control primer set 5 | F: −38 5′-TAGGTTTGTTTTTCGAGTTTTTTTC-3′ | F: −38 5′-TAGGTTTGTTTTTTGAGTTTTTTTTG-3′ |
| R: 151 5′-CTCCCACTAATCTAAACATCTCTCG-3′ | R: 149 5′-CCCACTAATCTAAACATCTCTCACT-3′ |
Primer pairs were designed using MethPrimer software (http://www.urogene.org/methprimer).20 The positions relative to precursor miR-34a are shown for each pair. Sets 1 through 3 were used for detection of methylation within the promoter regions, whereas sets 4 and 5 were used for methylation within a control region.
F, forward; R, reverse.
Gelatin Zymography
Twenty-five microliters of culture supernatant was subjected to electrophoresis under nondenaturing conditions on 7.5% SDS-polyacrylamide gel containing 0.1% bovine skin gelatin (Sigma Chemical) as substrate and analyzed. Bands indicated the presence of gelatinase activity.
Chemicals and Reagents
Specific miRNA precursors and inhibitors were obtained from Ambion. Rabbit anti-SIRT1, CASP2, Toll-like receptor-4, α-smooth muscle actin, survivin polyclonal antibodies, and DNMT3B and control small-interfering RNA plasmids were ordered from Santa Cruz Biotechnology Inc (Santa Cruz, CA). 5-aza-2′deoxycytidine and acetic acid were obtained from Sigma Chemical.
Statistical Analysis
Data are expressed as the mean ± SE from at least three separate experiments performed in triplicate, unless otherwise noted. The differences between groups were analyzed using a double-sided Student's t-test when only two groups were present and analysis of variance when more than two groups were present, and the null hypothesis was rejected at the 0.05 level unless otherwise specified.
Results
Moderate Steatohepatitis Is Induced by Long-Term Ethanol Exposure in Mice Liver
Four weeks of ethanol exposure caused significant increases in liver weight, liver/body weight ratio, and serum alanine aminotransferase level compared with control mice (Table 2). These results, along with liver histologic findings (Figure 1A) indicated that long-term ethanol feeding induced moderate steatosis. Ethanol exposure–induced liver injury was confirmed by analysis of total pathologic scores (Table 2).
Table 2.
Pathophysiologic and Biochemical Characterization of an Ethanol-Exposed Mouse Model
| Control | Ethanol exposed | |
|---|---|---|
| Final body weight, g | 23.75 ± 0.72 | 24.80 ± 0.66 |
| Liver weight, g | 1.02 ± 0.04 | 2.41 ± 0.19⁎ |
| Liver weight/body weight, % | 4.44 ± 0.09 | 10.15 ± 0.45⁎ |
| Alanine aminotransferase, U/L | 14.00 ± 1.15 | 253.60 ± 46.07⁎ |
| Blood alcohol level, mg/dL | 368.00 ± 0.84 | 309.78 ± 22.80⁎ |
| Adiponectin, μg/mL | 30.70 ± 0.07 | 73.40 ± 4.41⁎ |
| Total pathology score | 0.50 ± 0.29 | 6.40 ± 0.40⁎ |
Data are expressed as mean ± SE (n = 4 to 5).
P < 0.05 compared with the control.
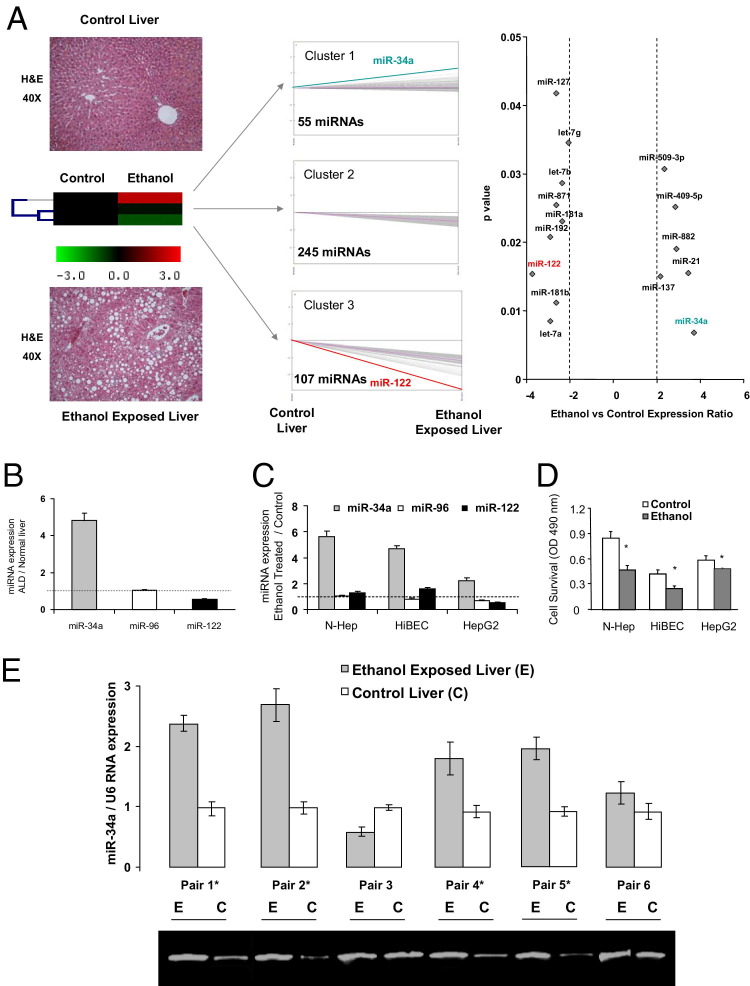
Figure 1.
Aberrant miR-34a expression in ethanol-exposed mouse liver. A: Left: Liver H&E staining of control (top) and ethanol-exposed (bottom) mouse liver that showed zonation of the liver, including portal and central veins, and miRNA expression patterns (middle) in ethanol-exposed mouse liver samples analyzed using a self-organizing tree algorithm using the Multiexperiment Viewer version 3.1 from the Institute for Genomic Research (Rockville, MD). A dendrogram showing three clusters was generated. miRNA expression in ethanol-exposed mouse liver tissues is shown on the right axis relative to normal mouse liver controls on the left axis. Cluster 1 is composed of a group of miRNAs, including miR-34a and miR-21, that were overexpressed after long-term alcohol exposure; cluster 2, 245 miRNAs that were not significantly altered after ethanol exposure; and cluster 3, 107 miRNAs that were decreased in expression. Right: Relative miRNA expression profile between ethanol-exposed livers versus normal control tissues. The expression of a panel of diverse updated miRNAs was evaluated by microarray analysis using Affymetrix GeneChip U133 Plus version 2.0 (Affymetrix Corporation, Santa Clara, CA). miRNA expression relative to U6 RNA was plotted, depicting the relative expression levels (Log2) for selected miRNAs in ethanol-exposed liver versus normal control panels (more than twofold change; P < 0.05). The relative expression levels and P values for each miRNA in the related samples were plotted against each other in the scatterplot. miR-34a and miR-122 are the most up- and down-regulated miRNAs among the 407 miRNAs detected in mice liver. Data represent the mean of three separate experiments. B and C: miRNA was isolated from either normal or ethanol-exposed mice liver (B) or from N-Heps, HiBECs, and HepG2 cell lines (C) with or without ethanol treatment (100 mmol/L) for 72 hours. The expression of a selected miRNA from each cluster (miR-34a from cluster 1, miR-96 from cluster 2, and miR-122a from cluster 3) was assessed using Taqman real-time PCR assay. Results represent the mean ± SE of miRNA expression from four separate determinations. D: Ethanol reduces hepatobiliary cell survival. Cell survival was assessed using a viable cell assay, and the survival index was assessed after the treatment of ethanol (100 mmol/L) or PBS for 72 hours. E: miR-34a is increased in human ALD liver. Total RNA was isolated from liver from control (C) or long-term ethanol-exposed patients (E). Real-time PCR analysis was performed, and the ratio of miR-34a to U6 small RNA expression in ethanol-exposed liver samples was determined. The PCR products were verified by 1.8% agarose gel electrophoresis. Data represent mean ± SE from three separate experiments. *P < 0.05 relative to controls.
miR-34a Is Aberrantly Expressed in Ethanol-Exposed Mice Tissues
Aberrant expression of selected miRNAs has been observed after alcoholic liver injury.8,21 To identify the miRNAs that are differentially increased in expression in ethanol-exposed liver tissues, we analyzed miRNA expression in three pairs of ethanol-treated mice and normal liver tissues using a miRNA microarray platform. Of the 697 human miRNAs represented in this microarray, the expression of 46 miRNAs was significantly altered relative to normal tissues. Of these, most aberrantly expressed miRNAs were decreased in expression. However, the expression of several miRNAs, including miR-34a, was markedly increased in expression (more than fourfold) in ethanol-exposed liver tissues compared with normal tissues (Figure 1, A and B). The expression of miR-34a was also increased in ethanol-treated N-Heps and HiBECs with a higher apoptotic rate compared with that of controls (Figure 1, C and D). The expression of miR-34a was also up-regulated in malignant hepatocytes (HepG2) relative to that of N-Heps. To further verify the expression of miR-34a in primary human ALD, real-time PCR analysis was performed using total RNA from ALD and normal human liver tissues obtained from six resected liver samples. miR-34a expression was increased by twofold or more in four of the six samples compared with the control liver tissues (Figure 1E and Table 2). These results indicate that aberrant expression of miR-34a is a frequent event in human primary ALDs.
Aberrant Expression of DNA Methyltransferases after Alcoholic Liver Injury
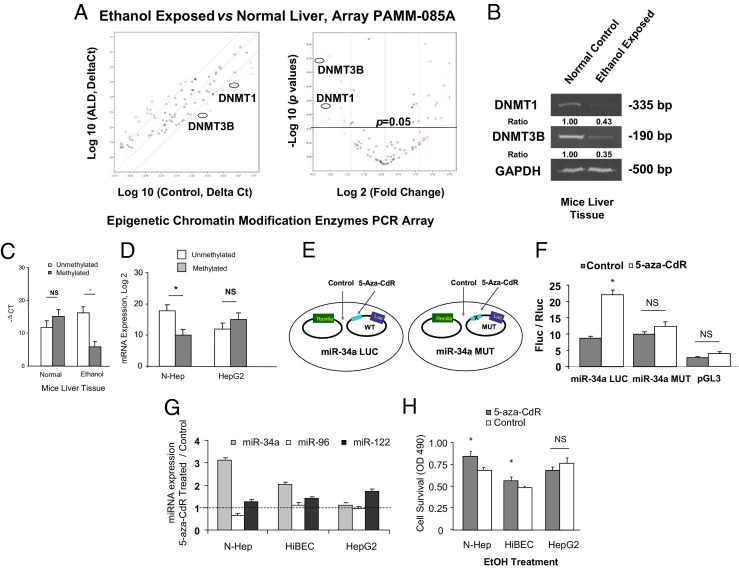
To investigate the mechanisms by which ethanol modulates methylation-dependent gene expression, we compared the genomic response of epigenetic modification enzymes to the treatment with ethanol in vivo using Epigenetic Chromatin Modification Enzymes PCR Array from SA Biosciences (Figure 2A and Table 3). In ethanol-exposed mouse liver samples, ethanol treatment significantly decreased the expression of the DNA methyltransferase enzymes, DNMT3B and DNMT1 (Figure 2, A and B; P < 0.05). Histone deacetylase, lysine (K)-specific demethylase, and ubiquitin-specific enzymes were also significantly altered after long-term ethanol in vivo exposure detected by PCR array analysis (Table 3). The functional role of DNMT3B was further studied because it is the most down-regulated gene on the list. In ethanol-treated N-Heps and HiBECs, reduced expression of DNMT3B was detected when compared with control cells (see Supplemental Figure S1 at http://ajp.amjpathol.org). DNMT3B mRNA expression was increased in N-Heps and HiBECs in response to 5-aza-CdR. On the basis of these observations, ethanol treatment decreases the overall methylation activity via a mechanism involving down-regulation of expression of DNMT3B methyltransferase.
Figure 2.
Epigenetic regulation of miR-34a in ethanol-exposed mouse liver. A: The expression levels of DNMT1 and DNMT3B are down-regulated in ethanol-exposed mouse liver. Relative gene expression profile between ethanol-exposed mouse livers versus control liver tissues is shown. The expression of a panel of diverse epigenetic-associated genes was evaluated by real-time PCR using Mouse Epigenetic Chromatin Modification Enzymes PCR Array (PAMM-085A) from SABiosciences Corporation (Valencia, CA). Gene expression relative to GAPDH was plotted as the Volcano Plots, depicting the relative expression levels (Log10) for selected genes in ALD versus control panels (left). The relative expression levels and P values for each gene in the related samples were also plotted against each other in the scatterplot (right). DNMT1 and DNMT3B are the greatest down-regulated genes among the six epigenetic signaling pathways in ethanol-exposed mouse liver. Data represent mean from three separate experiments. B: RT-PCR confirmed the reduced mRNA expression of DNMT1 and DNMT3B in ethanol-exposed mouse liver relative to controls. Expression of DNMTs was assayed by a standard RT-PCR method in control and ethanol-exposed mouse liver tissues. The PCR amplification was performed for 26 cycles for GAPDH (a housekeeping control), 30 cycles for DNMT1, and 35 cycles for DNMT3B. The ratios shown represent the mean value (relative to control) normalized with GAPDH from four independent experiments. C and D: Methylation-specific PCR analysis of the miR-34a upstream regulatory region was performed in ethanol-exposed mice and normal control liver, as well as ethanol-treated N-Heps and HepG2 cells (100 mmol/L, 7 days). The dark gray bar indicates hypermethylated miR-34a; the white bar indicates unmethylated miR-34a. Long-term alcohol exposure significantly demethylated CpG island enriched 5′-promoter region of miR-34a. miR-34a promoter was also hypomethylated in N-Heps but not in HepG2 cells after ethanol treatment. The results shown represent the mean ± SE from six independent experiments. **P < 0.01 compared with expression in control regions. E and F: Characterization of miR-34a promoter methylation activities by mutation site-specific luciferase assay. Experimental design strategy of miR-34a methylation-specific mutation is presented (E). A 24-bp mutation of the hsa-miR-34a methylation site (106 bp) converted from GCC to TAA was performed using oligonucleotides to the CpG island enriched region. Luciferase reporter constructs containing the sequence of potential methylation site (CpG island enriched region) from the 5′-promoter region of miR-34a inserted downstream of the luciferase gene were generated. miR-34a-LUC (wild type) contains the intact sequence whereas miR-34a-MUT contained the sequence with GCC to TAA nucleotide changes. Reporter constructs were transfected in normal human hepatocytes for 24 hours and then treated with 10 μmol/L 5-Aza-CdR or diluent control for 72 hours (F). The expression of firefly luciferase activity was normalized to that of Renilla luciferase activity for each sample. The increases in relative firefly luciferase activity in the presence of methylation inhibitor in miR-34a-LUC but not in miR-34a-MUT transfected cells indicate the presence of a methylation-modulated target sequence in the 5′-promoter region of miR-34a. The results shown represent the mean ± SE from eight independent experiments. G: N-Heps, HiBECs, and HepG2 cells were treated with 10 μmol/L 5-Aza-CdR for 72 hours. The expression of mature miR-34a, miR-96, and miR-122 was assessed using Taqman real-time PCR assay. 5-Aza-CdR increased miR-34a but not miR-96 expression in normal hepatocytes and cholangiocytes. Data represent mean ± SE from eight separate experiments. H: 5-Aza-CdR treated hepatobiliary cells were subjected to ethanol treatment for 72 hours simultaneously. Cell viability was measured by the MTS assay. Demethylation treatment significantly enhanced cell survival against ethanol in N-Heps and HiBECs but not HepG2 HCC cells. The results shown represent the mean ± SE from four independent experiments. *P < 0.05 relative to controls. NS, no significant difference.
Table 3.
Altered Epigenetic Chromatin Modification Enzymes on RT2 Profiler PCR Array in Ethanol-Exposed Mice Liver
| Refseq | Symbol | Description | Fold change | P value |
|---|---|---|---|---|
| NM_010068 | Dnmt3b | DNA methyltransferase 3B | 0.203 | 0.008 |
| NM_010066 | Dnmt1 | DNA methyltransferase (cytosine-5) 1 | 0.213 | 0.015 |
| NM_030241 | Setd8 | SET domain containing (lysine methyltransferase) 8 | 0.214 | 0.014 |
| NM_010413 | Hdac6 | Histone deacetylase 6 | 0.226 | 0.007 |
| NM_009458 | Ube2b | Ubiquitin-conjugating enzyme E2B, RAD6 homology (S. cerevisiae) | 0.240 | 0.037 |
| NM_009762 | Smyd1 | SET and MYND domain containing 1 | 0.340 | 0.068 |
| NM_178891 | Prmt6 | Protein arginine N-methyltransferase 6 | 0.379 | 0.012 |
| NM_145482 | Setd4 | SET domain containing 4 | 0.416 | 0.027 |
| XM_003085353 | Setd1b | SET domain containing 1B | 0.427 | 0.030 |
| NM_024258 | Usp16 | Ubiquitin specific peptidase 16 | 0.447 | 0.041 |
| NM_026984 | Mll5 | Myeloid/lymphoid or mixed-lineage leukemia 5 | 0.483 | 0.048 |
| NM_008084 | Gapdh | Glyceraldehyde-3-phosphate dehydrogenase | 0.967 | 0.138 |
| NM_007393 | Actb | Actin, beta | 1.018 | 0.885 |
| NM_020572 | Aurkc | Aurora kinase C | 2.212 | 0.007 |
| NM_017479 | Myst4 | MYST histone acetyltransferase monocytic leukemia 4 | 2.386 | 0.009 |
| NM_011035 | Pak1 | P21 protein (Cdc42/Rac)–activated kinase 1 | 2.492 | 0.010 |
| NM_145404 | Prmt7 | Protein arginine N-methyltransferase 7 | 2.578 | 0.014 |
| NM_201371 | Prmt8 | Protein arginine N-methyltransferase 8 | 2.726 | 0.036 |
| NM_080793 | Setd7 | SET domain containing (lysine methyltransferase) 7 | 2.791 | 0.035 |
| NM_024124 | Hdac9 | Histone deacetylase 9 | 2.808 | 0.013 |
| NM_028039 | Esco2 | Establishment of cohesion 1 homolog 2 (S. cerevisiae) | 2.876 | 0.038 |
| NM_144787 | Kdm4c | Lysine (K)–specific demethylase 4C | 3.008 | 0.011 |
| NM_008679 | Ncoa3 | Nuclear receptor co-activator 3 | 3.551 | 0.009 |
| NM_172382 | Kdm4a | Lysine (K)–specific demethylase 4A | 3.593 | 0.012 |
miR-34a Expression Is Epigenetically Regulated
CpG islands in human genomic DNA are GC-rich fragments whose aberrant methylation is associated with human disease development. To determine potential mechanisms by which microRNA expression was up-regulated in ALDs, the sequences containing 2000 bp upstream of the transcription start site of each up-regulated miRNA from ethanol-exposed mouse liver samples were entered and searched by MethPrimer for CpG islands.20 Analysis of the promoter region revealed the presence of CpG islands ∼300 bp upstream of the 5′-region of the mature miR-34a sequence (see Supplemental Figure S2 at http://ajp.amjpathol.org). These results suggest that the expression of miR-34a may be potentially regulated by modulation of promoter methylation. Of note, histone deacetylase inhibition has been demonstrated to activate the expression of miR-34a in human bladder cancer cells. Thus, we examined the methylation status of the miR-34a promoter by MSP, a bisulfite conversion-based PCR technique for the study of DNA CpG methylation. Using a real-time MSP assay, we found that the miR-34a promoter was hypomethylated in ethanol-exposed liver tissue (Figure 2C) and in normal hepatocytes but not in HepG2 cells after ethanol treatment (Figure 2D). To verify that miR-34a is translationally regulated by methylation through a CpG island enriched site of 5-promoter region, we performed studies using luciferase reporter constructs containing the sequence of CpG island enriched site from the 5′-promoter region of miR-34a inserted downstream of the luciferase gene (Figure 2, E and F). 5-aza-CdR treatment for 72 hours increased the reporter activity of miR-34a-LUC in N-Heps. However, when these studies were repeated with reporter constructs containing a 24-bp mutation in the recognition sequence, the effects of reporter activation by methylation inhibitor were abolished (Figure 2, E and F). Furthermore, a marked increase in miR-34a was also noted in N-Heps and HiBECs after 5-Aza-CdR, which was associated with enhanced cell survival during ethanol treatment (Figure 2, G and H). These results suggest that loss of the methylation mark in the miR-34a promoter may be associated with the reactivation of miR-34a expression in ethanol-exposed mouse liver. However, the effect of 5-Aza-CdR and the expression of pre-miR-34a on survival of alcohol-exposed HepG2 cells were minimal. Nevertheless, anchorage-independent growth and cell migration were affected, suggesting that other miRNAs or tumor suppressor genes may be involved in this event as supported by other studies.22
Overexpression of miR-34a Increases Cell Survival and Transformation in Vitro
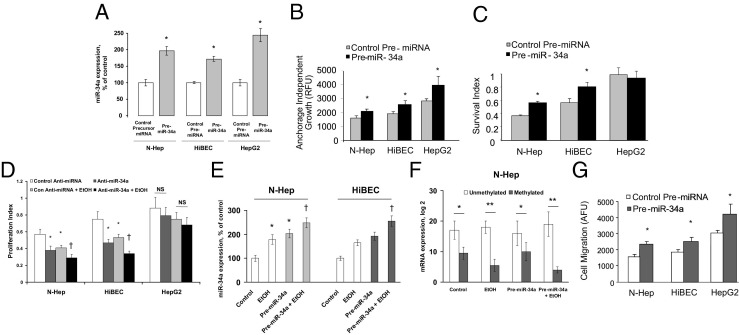
We have previously demonstrated a role for let-7 in hepatic cell behavior. However, the role of miR-34a in hepatobiliary injury remains unknown. Thus, we performed studies aimed to explore the possible biological significance of aberrant miR-34a by using a precursor specific to miR-34a. First, we verified the efficacy of transfection and target effects by assessing the expression of mature miR-34a by real-time PCR in N-Heps, HiBECs, and HepG2 cells transfected with miR-34a precursor (Figure 3A). Meanwhile, a significant change was seen in anchorage-independent growth after pre-miR-34a in all three cell lines tested (Figure 3B). Next, we assessed the cell survival after ethanol treatment in the cells lines of N-Heps, HiBECs, and HepG2 cells. In N-Heps and HiBECs transfected with pre-miR-34a or anti-miR-34a inhibitor, cell survival was significantly altered compared with cells transfected with controls with different dosage and duration of ethanol treatment (Figure 3, C and D). We also quantified miR-34a expression in miR-34a transfected cells before and after alcohol exposure and demonstrate that the levels of miR-34a significantly increased by approximately 1.3-fold and 1.4-fold in hepatocytes and cholangiocytes, respectively (Figure 3E). Meanwhile, relatively long-term ethanol treatment (1 week, without miR-34a overexpression) induced a 1.8-fold and 1.7-fold increase in miR-34a expression, respectively, which is higher than that of transfected cells. In addition, the degree of methylation was not altered in nontransfected hepatic cells compared with the miR-34a transfected cells with ethanol treatment (Figure 3F). These observations indicate a role for miR-34a in the regulation of survival and transformation of human hepatobiliary cells after alcoholic liver injury.
Figure 3.
Overexpression of miR-34a increases cell survival and migration. A: miR-34a expression was assessed by real-time PCR in hepatobiliary cells transfected with either control or miR-34a precursors. The ability of these constructs to modulate miR-34a expression was verified in all three cell lines. Data represent mean ± SE from four separate experiments. B and C: Hepatobiliary cells were transfected with pre-miR-34a or control precursor. B: Cells were plated in agar wells in 96-well plates, and anchorage-independent growth assessed fluorometrically after 7 days. Pre-miR-34a increased anchorage independent growth in all three cell lines tested. The results shown represent the mean ± SE of four independent experiments. C: Cell survival against ethanol treatment (100 mmol/L for 72 hours) was assessed using a viable cell assay, and the survival index was assessed after 72 hours. Pre-miR-34a increased survival rate in cell lines from N-Heps and HiBECs but not from HepG2 cells. Data represent mean ± SE from four separate experiments. D: The hepatobiliary cells were transfected with either control or anti-miR-34a inhibitor for 48 hours. The transfected cells along with untransfected controls were plated in 96-well plates and treated with 20 mmol/L ethanol for 7 days. The proliferation index was measured by MTS assay in untransfected and transfected cells with or without ethanol treatment as indicted. Silencing of miR-34a has significantly further reduced cell survival in ethanol-treated human hepatocytes and HiBECs. Data represent mean ± SE from four separate experiments. E and F: N-Heps and HiBECs were transfected with either control or pre-miR-34a inhibitor for 48 hours. The transfected cells along with untransfected controls were further treated with 20 mmol/L ethanol for 7 days. Relative miR-34a expression (E) and methylation status of miR-34a upstream regulatory region (F) were detected by real-time PCR analysis, respectively. Ethanol-induced miR-34a up-regulation is more apparent in untransfected cells than that in miR-34a transfected cells (E). Furthermore, the degree of methylation is not altered in nontransfected hepatic cells compared with miR-34a transfected cell (both with ethanol treatment). The results shown represent the mean ± SE from four independent experiments. G: Cell migration was assessed as described in Materials and Methods and is expressed as arbitrary fluorescence units (AFU). The cell lines varied in their ability to migrate. Pre-miR-34a increased cell motility in all three cell lines. The results shown represent the mean ± SE from four independent experiments. *P < 0.05, **P < 0.01 relative to controls. †P < 0.05 relative to control miRNA precursor/inhibitor transfected cells.
Modulation of miR-34a Alters Cell Migration in Hepatobiliary Cell Lines
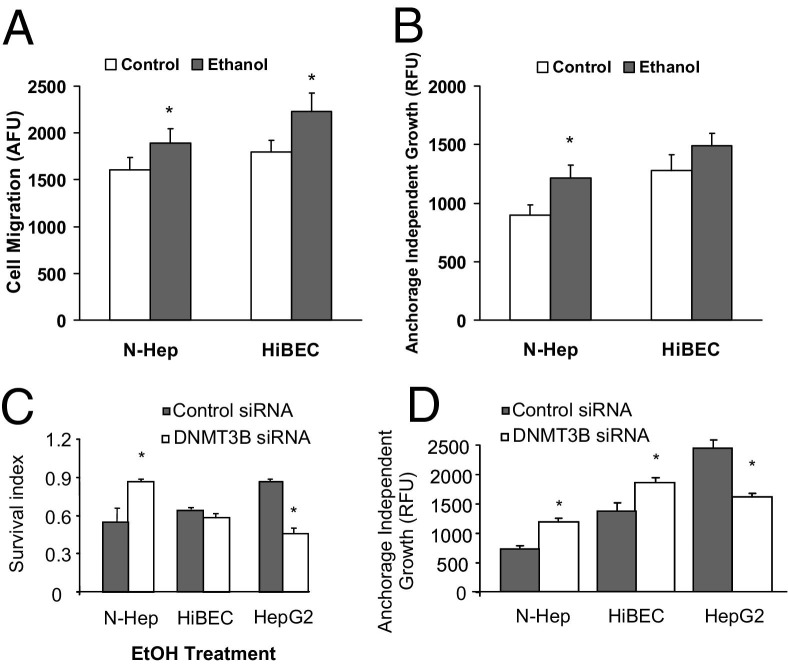
The ability of cells to migrate into adjacent tissues is a phenotypic characteristic of hepatobiliary cells and a key determinant of tissue remodeling and wound healing process. Hepatobiliary cell lines vary in their migratory potential, and among the cell lines studied, HepG2 cells exhibited the greatest migratory potential. To assess the effect of miR-34a on cell motility, N-Heps, HiBECs, and HepG2 cells were transfected with either control or miR-34a precursor, and we assessed vertical cell migration. Pre-miR-34a increased cell migration index in the three hepatobiliary cell lines studied compared with controls (Figure 3G). Moreover, neither cell migration nor survival was significantly altered by transfection of miR-96 precursor in normal human hepatocytes or HepG2 cells (not shown). Furthermore, cell motility and transformation were significantly altered after relatively long-term ethanol treatment along with a moderate increase in miR-34a expression (Figure 4, A and B). These results support a functional role for miR-34a and ethanol in mediating cell migration in hepatocytes and cholangiocytes and provide a mechanism by which overexpression of miR-34a induced by ethanol treatment may contribute to tissues remodeling and regeneration.
Figure 4.
Ethanol and DNMT3B silencing alter cell migration, survival, and transformation. A and B: N-Heps and HiBECs were treated with 20 mmol/L ethanol for 7 days. Cell migration (A) and transformation (B) were detected in treated cells by migration and soft agar assay. Relative longer-term ethanol treatment significantly increased cell motility and transformed survival potential in hepatobiliary cells tested. Data represent mean ± SE from six separate experiments. C and D: Cell survival against ethanol (C) and transformed cell growth (D) was assessed by MTS and soft agar assay in hepatobiliary cells transfected with either control or DNMT3B small-interfering RNA. Silencing DNMT3B increased the survival in normal hepatocytes but reduced HCC cell survival (C). In contrast to miR-34a, it decreased transformed cell growth in HepG2 cells (D). Data represent mean ± SE of eight separate experiments. *P < 0.05 relative to controls.
Effects of Specific DNMT3B Gene Depletion on Hepatobiliary Cell Survival, Transformation, and Migration
We then tested whether inhibition of the DNMT3B gene is sufficient to produce a significant effect of miR-34a in cell survival, transformation, and migration. miR-34a was significantly increased after silencing DNMT3B (with or without ethanol treatment) in all three cell lines tested (see Supplemental Figure S3 at http://ajp.amjpathol.org). However, silencing of DNMT3B increased the survival in normal hepatocytes but reduced HepG2 cell survival against ethanol treatment (Figure 4C). In contrast to miR-34a, DNMT3B silencing decreased transformed cell growth and migration in HepG2 cells, suggesting different epigenetic regulation mechanisms for normal and malignant hepatocytes (Figure 4D). These results demonstrated the distinct effects of specific DNMT3B promotion on the process of cell remodeling and tissue repair after ethanol exposure.
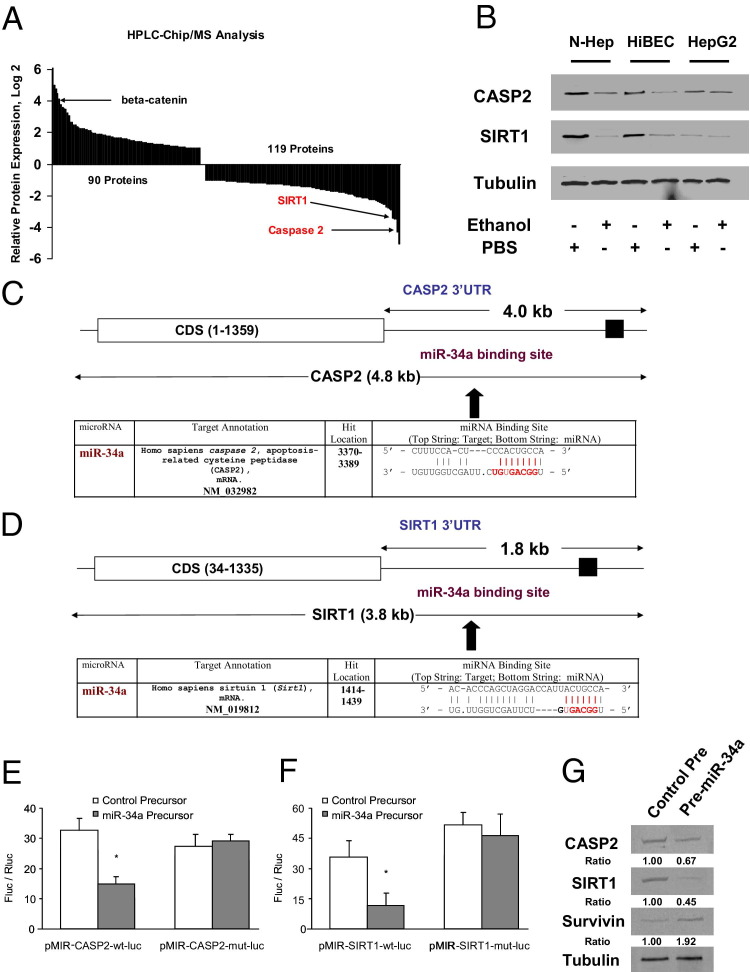
Identification of CASP2 and SIRT1 as Targets for miR-34a
Because miRNAs target mRNA stability and translation, we used HPLC-Chip/MS to identify specific target protein levels of miR-34a in pre-miR-34a treated N-Heps. Because the aim of our study is to correlate miR-34a with target transcript expression, we therefore used a cutoff of at least 30% difference in our expression analysis and a false discovery rate of 10% using the Benjamini-Hochberg correction for multiple testing. Nine genes were initially selected according to this strategy and further screened based on 3′-UTR sequence analysis and prediction algorithms; only two proteins can be potentially targeted by miR-34a (Figure 5A). Interestingly, the target prediction program Sanger miRBase database (http://microrna.sanger.ac.uk) indicated the presence of a highly conserved binding site for miR-34a in the 3′-UTR region of CASP2 and NAD+-dependent deacetylase SIRT1, the well-characterized regulator genes of apoptosis in liver biology. CASP2 and SIRT1 were significantly down-regulated in surviving N-Heps and HiBECs after ethanol treatment and are also silenced in HepG2 cells related to normal controls (Figure 5B). To verify that CASP2 and SIRT1 are bona fide targets of translational regulation by miR-34a in hepatocytes, we performed studies using luciferase reporter constructs containing the miR-34a recognition sequence from the 3′-UTR of CASP2 and SIRT1 inserted downstream of the luciferase gene (Figure 5, C and D). Transfection with miR-34a precursor decreased reporter activity of both CASP2 and SIRT1 in normal human hepatocytes. However, when these studies were repeated with reporter constructs that contained random mutations in the recognition sequence, the effects of reporter deactivation by miR-34a precursor were abolished (Figure 5, E and F). Moreover, a reduction in CASP2 and SIRT1 expression occurred after 3 days in cells incubated with pre-miR-34a. Concomitant with reduced SIRT1 expression, there was a significant increase of survivin expression, a confirmed downstream mediator of SIRT1 signaling (Figure 5G). In contrast, transfection with a precursor to miR-122a, which can also modulate cell survival in normal hepatocytes, did not alter the expression of CASP2 and SIRT1 with a relative expression of 0.8- ± 0.2-fold and 1.2- ± 0.3-fold of controls, respectively. Taken together, these findings indicate that CASP2 and SIRT1 are the biologically relevant targets of miR-34a in hepatobiliary cells.
Figure 5.
miR-34a regulates expression of SIRT1. A: Identification of protein targets modulated by miR-34a in N-Heps by HPLC-Chip/MS analysis. N-Heps were transfected with pre-miR-34a or control precursor (100 nmol/L) for 72 hours; HPLC-Chip/MS analysis was performed in protein lysates. Overexpressed or down-regulated proteins (more than twofold or <0.5-fold) from LC-MS have been ranked to their relative expression levels. Multifunctional oncogenic protein β-catenin is ranked in the up-regulated protein group, whereas CASP2 and SIRT1 are listed among the down-regulated targets. Data represent mean from three separate experiments. B: miR-34a target proteins are altered in normal hepatocytes after ethanol treatment. Hepatobiliary cells were treated with ethanol (100 mmol/L) or PBS controls. Cell lysates were obtained after 7 days and Western blots performed for CASP2, SIRT1, and α-tubulin. Treatment with ethanol down-regulated the expression of CASP2 and SIRT1 in all three cell lines. C and D: Schematic of predicted miR-34a site in the 3′UTR of human CASP2 and SIRT1. Positions 2 to 9 of the 5′ region of miRNA 34a are labeled in red. E and F: Luciferase reporter constructs containing the miR-34a recognition sequence from the 3′-UTR of CASP2 and SIRT1 inserted downstream of the luciferase gene were generated. pMIR-CASP2-wt-luc or pMIR-SIRT1-wt-luc contains the intact sequence, whereas pMIR-CASP2-mut-luc or pMIR-SIRT1-mut-luc contained the sequence with random nucleotide changes. Reporter constructs were co-transfected with either miR-34a precursor or control precursor in normal human hepatocytes. The expression of firefly luciferase activity was normalized to that of Renilla luciferase activity for each sample. The decreases in relative firefly luciferase activity in the presence of miR-34a indicate the presence of a miR-34a modulated target sequence in the 3′-UTR of CASP2 and SIRT1. Data represent the mean of eight separate experiments. *P < 0.05 relative to control precursor group. G: N-Heps were transfected with pre-miR-34a or control precursor. Cell lysates were obtained after 48 hours and Western blots performed for CASP2, SIRT1, survivin, and α-tubulin. Relative ratios normalized with α-tubulin and control group were displayed under the image. Overexpression of miR-34a decreased the expression of CASP2 and SIRT1 and subsequently increased the level of survivin, a downstream mediator of SIRT1 in N-Heps. The ratios shown represent the mean value (relative to control) normalized with α-tubulin from three independent experiments.
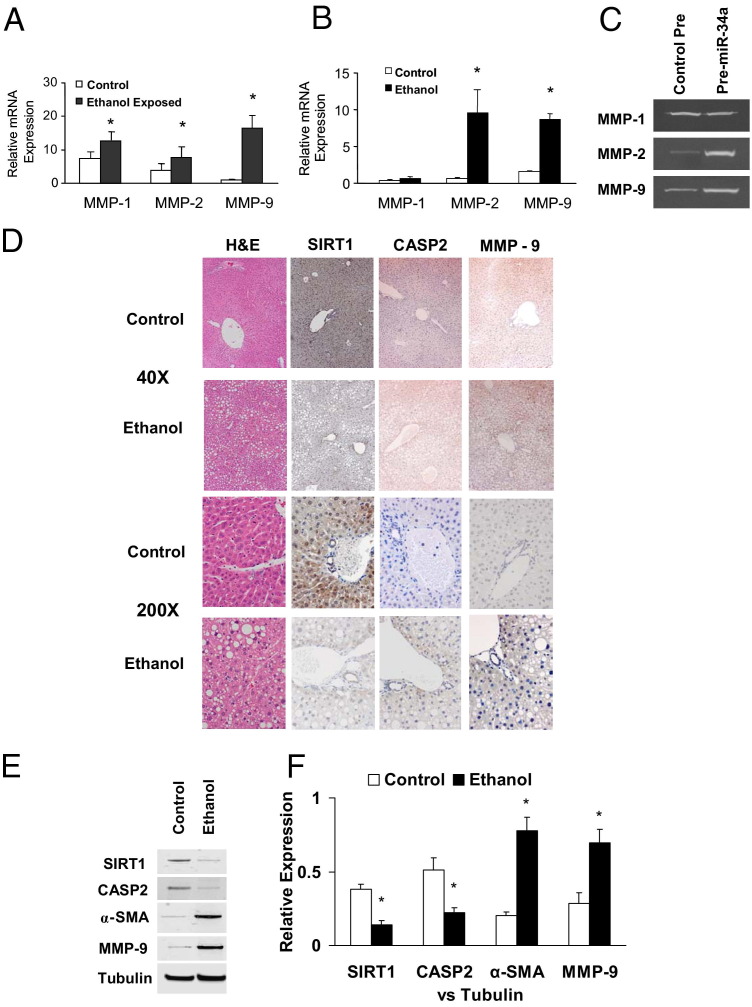
miR-34a Regulates MMP mRNA Expression
The cell remodeling and tissue repair process involves a series of proteolytic enzymes named MMPs. Alterations of the SIRT1 complexes have been mechanistically linked to decreased expression of MMPs and cell motility. Therefore, we examined the expression of selected MMPs involved in cell remodeling in normal and ethanol-exposed mouse liver tissues and cell lines. Compared with normal liver tissue, the expression of MMP-1, MMP-2, and MMP-9 increased in ethanol-exposed mouse liver tissue (Figure 6A). Increased expression of MMP-2 and MMP-9 was also observed in ethanol-treated N-Heps compared with controls (Figure 6B). Meanwhile, MMP-11 and MMP-13 were not altered in tissues and cells with ethanol exposure. To confirm the functional effect and relevance of miR-34a–dependent modulation of MMPs, we assessed the effect of modulation of miR-34a on MMPs expression. Transfection of normal human hepatocytes with miR-34a precursor increased MMP-2 and MMP-9 activity (Figure 6C). Furthermore, the expression of SIRT1 and CASP2 was reduced after long-term alcohol exposure, whereas MMP-9 and α-smooth muscle actin were increased in ethanol-exposed mouse liver sections and homogenates (Figure 6, D–F). These findings link miR-34a and putative mediators of tissue remodeling in ethanol-exposed mice and suggest that deregulated expression of miR-34a can contribute to liver reconstruction and fibrosis during alcoholic liver injury.
Figure 6.
miR-34a regulates the tissue remodeling molecules during ethanol exposure. A and B: Expression of MMP-1, MMP-2, and MMP-9 mRNA was assessed by quantitative real-time PCR and normalized to expression of β-actin in normal liver and ethanol-exposed mouse tissue (A) and in normal hepatocytes with or without 7 days of ethanol treatment (B). MMP-1, MMP-2, and MMP-9 are overexpressed in ethanol-exposed liver compared with normal liver. However, MMP-2 and MMP-9, but not MMP-1, were increased in ethanol-treated hepatocytes compared with PBS controls. The results shown represent the mea ± SE of four independent experiments. C: N-Heps were transfected with miR-34a precursors or controls. After 72 hours, MMP Zymogen Gel Assay was performed for MMP-1, MMP-2, and MMP-9 expression. Enhanced expression of miR-34a in N-Heps increases MMP-2 and MMP-9 expression. D: Steatohepatitis is induced by 4 weeks of intragastric ethanol infusion (ISI) in mice liver. Liver histologic analysis of regular feeding control mice and ISI mice is displayed. Enhanced expression of MMP-9 and reduced expression of CASP2 and SIRT1 were seen in ethanol-exposed mouse liver [H&E; original magnification: ×40 (top); ×200 (bottom)]. E and F: Liver tissue homogenates were obtained from ethanol-exposed and control mice. Increases of liver fibrotic marker α-SMA and MMP-9 along with the reduction of CASP2 and SIRT1 were verified by Western blot analysis. Representative immunoblots (E) and quantitative data (mean ± SE) from four separate blots (F) are shown.
Discussion
In this study, we demonstrated the role of altered expression of miR-34a in the processes that contribute to cellular phenotypic changes that are associated with ALD progression. We found that miR-34a is increased in ethanol-exposed mouse liver in vivo and overexpressed in ethanol-treated hepatobiliary cell lines compared with controls. We also demonstrated that miR-34a contributes to alcoholic liver injury and tissue repair by modulating cell proliferation, remodeling, and migration. Some of these effects are mediated through CASP2 and SIRT1, the well-characterized regulator genes of apoptosis that are also involved in tissue remodeling. Increased expression of miR-34a was found by in situ hybridization during liver regeneration, and a similar role for miR-34a has been postulated in renal injury.15,23 The concomitant miR-34a–dependent activation of metalloproteinases in hepatobiliary cells can facilitate tissue remodeling. Taken together, these findings support a functional role for miR-34a in promoting liver tissue repair and fibrosis during the development of ALD. However, the enhanced cell survival and migration by miR-34a may be important for the regression of alcohol-induced liver fibrosis and cirrhosis but may also mediate the higher potential for malignant transformation.
miRNA-mediated mechanisms are being increasingly implicated in liver injury. Deregulation of miR-34a can occur as a result of altered p53 expression. Likewise, ectopic expression of miR-34 can decrease expression of genes that are regulated by p53, such as cyclin E2, cyclin-dependent kinase 4, and hepatocyte growth factor receptor.24 These and other studies25 support the concept of a cell cycle regulation role for miR-34a. Variable expression of miR-34a has been reported in different organ systems and diseases states. Interestingly, although silencing of miR-34a was demonstrated in several human malignant neoplasms, up-regulation of miR-34a was demonstrated in liver injury and HCC.26–28 miR-34a was also demonstrated to be a critical link to disease progression from normal liver through cirrhosis to HCC.29 These observations necessitate a clear definition of liver tissue–specific expression and function of miR-34a expression. It is likely that targeted therapeutic approaches that involve miR-34a may result from defining tissue and hepatic disease state–specific roles of miR-34a.
SIRT1, an NAD+-dependent class III protein deacetylase, plays a role in a wide variety of processes, including stress resistance, metabolism, differentiation, and aging.30–33 Overexpression of SIRT1 orthologs leads to increased life span of organisms34–36 and mammals.37 A recent study has shown that livers from older animals have reduced expression of SIRT1 and have lost proper control of the regulation of SIRT1 after partial hepatectomy.38 Long-term ethanol administration inhibits activities of hepatic SIRT1 in mice and rats.39–43 SIRT1 is partially localized in the cytoplasm in certain cell lines, and the cytoplasm-localized SIRT1 is associated with apoptosis, leading to increased sensitivity to apoptosis.44 The apoptosis enhanced by cytoplasm-localized SIRT1 is dependent on caspases.45 Overexpression of SIRT1 has been shown to inhibit cell proliferation and differentiation of pig preadipocytes.46 Down-regulation of SIRT1 may be an important mechanism for hepatobiliary cells to cope with unfavorable growing conditions with the reduction of related caspase molecules. Moreover, another verified target gene of miR-34a, CASP2, plays a key role in apoptosis by trophic factor deprivation, β-amyloid cytotoxicity, and granzyme B; CASP2-deficient mice lack a discernible phenotype. Nonetheless, CASP2 is involved in stress and ethanol-induced apoptosis, most likely upstream of the mitochondria.47,48 Involvement of CASP2 and Bcl-2 family proteins has been found in oxidative stress-induced apoptosis related to ALD.47,49
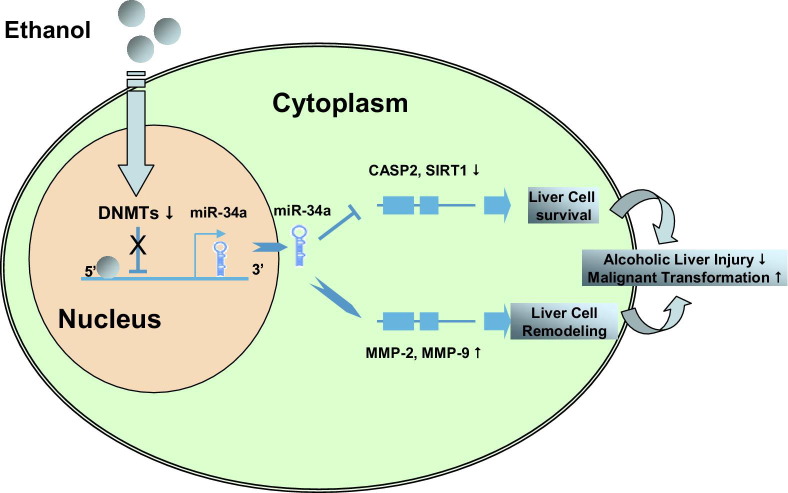
Our findings identify a previously unrecognized mechanism for direct regulation of SIRT1 and CASP2, involving noncoding RNA in ALD. Aberrant DNA methylation has been implicated in many human diseases, including ALDs. Recent studies indicate that ethanol induces epigenetic alterations, particularly acetylation, methylation of histones, and hypomethylation and hypermethylation of DNA.47,50–52 This has opened up a new area of interest in ethanol research and is providing novel insight into actions of ethanol at the nucleosomal level in relation to gene expression and pathophysiologic consequences. Although DNA methylation has been tightly linked to liver injury and poor outcomes for many hepatic disorders, including human ALDs, its application to ethanol-dependent noncoding RNA expression is novel. In summary, the current study has revealed an epigenetic mechanism that highlights the relationship of ethanol-induced aberrant methylation enzymes on the epigenetic regulation of expression of selected miRNA genes that are relevant to cell survival and remodeling during alcoholic liver injury (Figure 7). A better understanding of how ethanol interacts with specific DNA methyltransferases to contribute to aberrant noncoding RNA expression will advance the field and increase our understanding of the mechanisms involved in the development of ALDs. Besides methylation enzymes, histone deacetylase, lysine (K)–specific demethylase, and ubiquitin-specific enzymes were also significantly altered after long-term ethanol exposure in mouse liver (Table 3), suggesting the importance of other epigenetic and translational regulation mechanisms during alcoholic liver injury. Genomic scanning approaches to identify epigenetically and translationally modified targets in ALD are lacking, but such strategies could identify other novel targets that could be epigenetically modified in ALDs.
Figure 7.
Summary diagram. Ethanol modulates liver parenchymal cell survival, remodeling, and transformation by activation of miR-34a–dependent signaling pathways.
Until recently, only a few therapeutic agents were available for ALD. We postulate that therapeutic strategies based on targeting miR-34a may be useful to consider in the adjuvant setting to human ALDs. Studies undertaken to identify the interplay of epigenetic events and therapeutic effects of anti-miR-34a inhibition in vivo will provide new insights into translational mechanisms of ALD. The availability of therapeutic strategies targeting miRNA will enable translation of our observations to potential strategies for the treatment of alcohol-induced liver fibrosis in ALD patients.
Footnotes
Supported by the Dr. Nicholas C. Hightower Centennial Chair of Gastroenterology from Scott & White; NIH grants R01DK054811 and DK76898 (G.A.), R01-DK081442 (S.G.), and P50AA011999 and U01 AA018663 (H.T.); VA Merit Review (F.M. and H.T.); and Scott & White Research Grants Program Project110536 (F.M.).
Supplemental material for this article can be found at http://ajp.amjpathol.org or at http://dx.doi.org/10.1016/j.ajpath.2012.06.010.
Contributor Information
Fanyin Meng, Email: fmeng@tamu.edu.
Gianfranco Alpini, Email: galpini@tamu.edu.
Supplementary data
Silencing DNMT3B by ethanol in human hepatobiliary cells. N-Heps (A) and HiBECs (B) were treated with 20 mmol/L ethanol along with the diluents control for 7 days. Total RNA was extracted from treated hepatobiliary cells, and DNMT1 and DNMT3B expressions were assessed by quantitative real-time PCR. The relative mRNA expression in the ethanol-treated group was normalized to expression of the control group as a percentage of the control. The results shown represent the mean ± SE of four independent experiments. *P < 0.05 when compared with mRNA expression of control group of cells.
Analysis of the CpG island enriched sites of the miR-34a regulatory region using the MethPrimer program. Analysis of the promoter region using MethPrimer software revealed the presence of CpG islands ∼300 bp upstream of the 5′-region of the mature miR-34a sequence. The positions of five pair primer sets relative to precursor miR-34a are shown for each pair. Sets 1 through 3 were used for detection of methylation within the promoter regions, whereas sets 4 and 5 were used for methylation within a control region.
Silencing DNMT3B enhanced miR-34a expression in hepatobiliary cells with or without ethanol exposure. N-Heps, HiBECs, and HepG2 cells were transfected with either control or DNMT3B small-interfering RNA for 48 hours. The transfected cells were further treated with 20 mmol/L ethanol or PBS for 7 days. Relative miR-34a expression was detected by real-time PCR analysis. miR-34a was significantly increased after silencing DNMT3B (with or without ethanol treatment) in all three cell lines tested. Data represent mean ± SE from four separate experiments. *P < 0.05 versus PBS control; #P < 0.05 versus control small-interfering RNA group.
References
- 1.Mathurin P., Louvet A., Dharancy S. [Acute alcoholic hepatitis: management practices for 2007] French. Gastroenterol Clin Biol. 2008;32:S179–S181. doi: 10.1016/j.gcb.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 2.Lakner A.M., Bonkovsky H.L., Schrum L.W. microRNAs: fad or future of liver disease. World J Gastroenterol. 2011;17:2536–2542. doi: 10.3748/wjg.v17.i20.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kerr T.A., Korenblat K.M., Davidson N.O. MicroRNAs and liver disease. Transl Res. 2011;157:241–252. doi: 10.1016/j.trsl.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Meng F., Glaser S.S., Francis H., Demorrow S., Han Y., Passarini J.D., Stokes A., Cleary J.P., Liu X., Venter J., Kumar P., Priester S., Hubble L., Stoloch D., Sharma J., Liu C.G., Alpini G. Functional analysis of microRNAs in human hepatocellular cancer stem cells. J Cell Mol Med. 2012;16:160–173. doi: 10.1111/j.1582-4934.2011.01282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roderburg C., Urban G.W., Bettermann K., Vucur M., Zimmermann H., Schmidt S., Janssen J., Koppe C., Knolle P., Castoldi M., Tacke F., Trautwein C., Luedde T. Micro-RNA profiling reveals a role for miR-29 in human and murine liver fibrosis. Hepatology. 2011;53:209–218. doi: 10.1002/hep.23922. [DOI] [PubMed] [Google Scholar]
- 7.Mann J., Chu D.C., Maxwell A., Oakley F., Zhu N.L., Tsukamoto H., Mann D.A. MeCP2 controls an epigenetic pathway that promotes myofibroblast transdifferentiation and fibrosis. Gastroenterology. 2010;138:705–714. doi: 10.1053/j.gastro.2009.10.002. e701–e704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bala S., Marcos M., Kodys K., Csak T., Catalano D., Mandrekar P., Szabo G. Up-regulation of microRNA-155 in macrophages contributes to increased tumor necrosis factor α (TNFα) production via increased mRNA half-life in alcoholic liver disease. J Biol Chem. 2011;286:1436–1444. doi: 10.1074/jbc.M110.145870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He L., He X., Lim L.P., de Stanchina E., Xuan Z., Liang Y., Xue W., Zender L., Magnus J., Ridzon D., Jackson A.L., Linsley P.S., Chen C., Lowe S.W., Cleary M.A., Hannon G.J. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang T.C., Wentzel E.A., Kent O.A., Ramachandran K., Mullendore M., Lee K.H., Feldmann G., Yamakuchi M., Ferlito M., Lowenstein C.J., Arking D.E., Beer M.A., Maitra A., Mendell J.T. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tarasov V., Jung P., Verdoodt B., Lodygin D., Epanchintsev A., Menssen A., Meister G., Hermeking H. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle. 2007;6:1586–1593. doi: 10.4161/cc.6.13.4436. [DOI] [PubMed] [Google Scholar]
- 12.Raver-Shapira N., Marciano E., Meiri E., Spector Y., Rosenfeld N., Moskovits N., Bentwich Z., Oren M. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 13.Tazawa H., Tsuchiya N., Izumiya M., Nakagama H. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc Natl Acad Sci U S A. 2007;104:15472–15477. doi: 10.1073/pnas.0707351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Welch C., Chen Y., Stallings R.L. MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene. 2007;26:5017–5022. doi: 10.1038/sj.onc.1210293. [DOI] [PubMed] [Google Scholar]
- 15.Chen H., Sun Y., Dong R., Yang S., Pan C., Xiang D., Miao M., Jiao B. Mir-34a is upregulated during liver regeneration in rats and is associated with the suppression of hepatocyte proliferation. PLoS One. 2011;6:e20238. doi: 10.1371/journal.pone.0020238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsukamoto H., Mkrtchyan H., Dynnyk A. Intragastric ethanol infusion model in rodents. Methods Mol Biol. 2008;447:33–48. doi: 10.1007/978-1-59745-242-7_3. [DOI] [PubMed] [Google Scholar]
- 17.Xiong S., She H., Zhang A.S., Wang J., Mkrtchyan H., Dynnyk A., Gordeuk V.R., French S.W., Enns C.A., Tsukamoto H. Hepatic macrophage iron aggravates experimental alcoholic steatohepatitis. Am J Physiol Gastrointest Liver Physiol. 2008;295:G512–G521. doi: 10.1152/ajpgi.90327.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Francis H., Franchitto A., Ueno Y., Glaser S., DeMorrow S., Venter J., Gaudio E., Alvaro D., Fava G., Marzioni M., Vaculin B., Alpini G. H3 histamine receptor agonist inhibits biliary growth of BDL rats by downregulation of the cAMP-dependent PKA/ERK1/2/ELK-1 pathway. Lab Invest. 2007;87:473–487. doi: 10.1038/labinvest.3700533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alpini G., Franchitto A., Demorrow S., Onori P., Gaudio E., Wise C., Francis H., Venter J., Kopriva S., Mancinelli R., Carpino G., Stagnitti F., Ueno Y., Han Y., Meng F., Glaser S. Activation of alpha(1) -adrenergic receptors stimulate the growth of small mouse cholangiocytes via calcium-dependent activation of nuclear factor of activated T cells 2 and specificity protein 1. Hepatology. 2011;53:628–639. doi: 10.1002/hep.24041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li L.C., Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 21.Miranda R.C., Pietrzykowski A.Z., Tang Y., Sathyan P., Mayfield D., Keshavarzian A., Sampson W., Hereld D. MicroRNAs: master regulators of ethanol abuse and toxicity. Alcohol Clin Exp Res. 2010;34:575–587. doi: 10.1111/j.1530-0277.2009.01126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goeppert B., Schmezer P., Dutruel C., Oakes C., Renner M., Breinig M., Warth A., Vogel M.N., Mittelbronn M., Mehrabi A., Gdynia G., Penzel R., Longerich T., Breuhahn K., Popanda O., Plass C., Schirmacher P., Kern M.A. Down-regulation of tumor suppressor A kinase anchor protein 12 in human hepatocarcinogenesis by epigenetic mechanisms. Hepatology. 2010;52:2023–2033. doi: 10.1002/hep.23939. [DOI] [PubMed] [Google Scholar]
- 23.Bhatt K., Zhou L., Mi Q.S., Huang S., She J.X., Dong Z. MicroRNA-34a is induced via p53 during cisplatin nephrotoxicity and contributes to cell survival. Mol Med. 2010;16:409–416. doi: 10.2119/molmed.2010.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Corney D.C., Flesken-Nikitin A., Godwin A.K., Wang W., Nikitin A.Y. MicroRNA-34b and MicroRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth. Cancer Res. 2007;67:8433–8438. doi: 10.1158/0008-5472.CAN-07-1585. [DOI] [PubMed] [Google Scholar]
- 25.He L., He X., Lowe S.W., Hannon G.J. microRNAs join the p53 network–another piece in the tumour-suppression puzzle. Nat Rev Cancer. 2007;7:819–822. doi: 10.1038/nrc2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mizuguchi Y., Mishima T., Yokomuro S., Arima Y., Kawahigashi Y., Shigehara K., Kanda T., Yoshida H., Uchida E., Tajiri T., Takizawa T. Sequencing and bioinformatics-based analyses of the microRNA transcriptome in hepatitis B-related hepatocellular carcinoma. PLoS One. 2011;6:e15304. doi: 10.1371/journal.pone.0015304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pogribny I.P., Starlard-Davenport A., Tryndyak V.P., Han T., Ross S.A., Rusyn I., Beland F.A. Difference in expression of hepatic microRNAs miR-29c, miR-34a, miR-155, and miR-200b is associated with strain-specific susceptibility to dietary nonalcoholic steatohepatitis in mice. Lab Invest. 2010;90:1437–1446. doi: 10.1038/labinvest.2010.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cermelli S., Ruggieri A., Marrero J.A., Ioannou G.N., Beretta L. Circulating microRNAs in patients with chronic hepatitis C and non-alcoholic fatty liver disease. PLoS One. 2011;6:e23937. doi: 10.1371/journal.pone.0023937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pineau P., Volinia S., McJunkin K., Marchio A., Battiston C., Terris B., Mazzaferro V., Lowe S.W., Croce C.M., Dejean A. miR-221 overexpression contributes to liver tumorigenesis. Proc Natl Acad Sci U S A. 2010;107:264–269. doi: 10.1073/pnas.0907904107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blander G., Guarente L. The Sir2 family of protein deacetylases. Annu Rev Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- 31.Buck S.W., Gallo C.M., Smith J.S. Diversity in the Sir2 family of protein deacetylases. J Leukoc Biol. 2004;75:939–950. doi: 10.1189/jlb.0903424. [DOI] [PubMed] [Google Scholar]
- 32.Smith J.S., Avalos J., Celic I., Muhammad S., Wolberger C., Boeke J.D. SIR2 family of NAD(+)-dependent protein deacetylases. Methods Enzymol. 2002;353:282–300. doi: 10.1016/s0076-6879(02)53056-1. [DOI] [PubMed] [Google Scholar]
- 33.Solomon J.M., Pasupuleti R., Xu L., McDonagh T., Curtis R., DiStefano P.S., Huber L.J. Inhibition of SIRT1 catalytic activity increases p53 acetylation but does not alter cell survival following DNA damage. Mol Cell Biol. 2006;26:28–38. doi: 10.1128/MCB.26.1.28-38.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaeberlein M., McVey M., Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rogina B., Helfand S.L. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci U S A. 2004;101:15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tissenbaum H.A., Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- 37.Bordone L., Guarente L. Calorie restriction. SIRT1 and metabolism: understanding longevity. Nat Rev Mol Cell Biol. 2005;6:298–305. doi: 10.1038/nrm1616. [DOI] [PubMed] [Google Scholar]
- 38.Jin J., Iakova P., Jiang Y., Medrano E.E., Timchenko N.A. The reduction of SIRT1 in livers of old mice leads to impaired body homeostasis and to inhibition of liver proliferation. Hepatology. 2011;54:989–998. doi: 10.1002/hep.24471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ajmo J.M., Liang X., Rogers C.Q., Pennock B., You M. Resveratrol alleviates alcoholic fatty liver in mice. Am J Physiol Gastrointest Liver Physiol. 2008;295:G833–G842. doi: 10.1152/ajpgi.90358.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lieber C.S., Leo M.A., Wang X., Decarli L.M. Alcohol alters hepatic FoxO1, p53, and mitochondrial SIRT5 deacetylation function. Biochem Biophys Res Commun. 2008;373:246–252. doi: 10.1016/j.bbrc.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 41.Lieber C.S., Leo M.A., Wang X., Decarli L.M. Effect of chronic alcohol consumption on Hepatic SIRT1 and PGC-1alpha in rats. Biochem Biophys Res Commun. 2008;370:44–48. doi: 10.1016/j.bbrc.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 42.You M., Cao Q., Liang X., Ajmo J.M., Ness G.C. Mammalian sirtuin 1 is involved in the protective action of dietary saturated fat against alcoholic fatty liver in mice. J Nutr. 2008;138:497–501. doi: 10.1093/jn/138.3.497. [DOI] [PubMed] [Google Scholar]
- 43.You M., Liang X., Ajmo J.M., Ness G.C. Involvement of mammalian sirtuin 1 in the action of ethanol in the liver. Am J Physiol Gastrointest Liver Physiol. 2008;294:G892–G898. doi: 10.1152/ajpgi.00575.2007. [DOI] [PubMed] [Google Scholar]
- 44.Chen S., Xiao X., Feng X., Li W., Zhou N., Zheng L., Sun Y., Zhang Z., Zhu W. Resveratrol induces Sirt1-dependent apoptosis in 3T3-L1 preadipocytes by activating AMPK and suppressing AKT activity and survivin expression. J Nutr Biochem. 2011 doi: 10.1016/j.jnutbio.2011.06.003. [DOI] [PubMed] [Google Scholar]; [Epub ahead of press]
- 45.Jin Q., Yan T., Ge X., Sun C., Shi X., Zhai Q. Cytoplasm-localized SIRT1 enhances apoptosis. J Cell Physiol. 2007;213:88–97. doi: 10.1002/jcp.21091. [DOI] [PubMed] [Google Scholar]
- 46.Bai L., Pang W.J., Yang Y.J., Yang G.S. Modulation of Sirt1 by resveratrol and nicotinamide alters proliferation and differentiation of pig preadipocytes. Mol Cell Biochem. 2008;307:129–140. doi: 10.1007/s11010-007-9592-5. [DOI] [PubMed] [Google Scholar]
- 47.Madesh M., Zong W.X., Hawkins B.J., Ramasamy S., Venkatachalam T., Mukhopadhyay P., Doonan P.J., Irrinki K.M., Rajesh M., Pacher P., Thompson C.B. Execution of superoxide-induced cell death by the proapoptotic Bcl-2-related proteins Bid and Bak. Mol Cell Biol. 2009;29:3099–3112. doi: 10.1128/MCB.01845-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McVicker B.L., Tuma D.J., Kubik J.L., Tuma P.L., Casey C.A. Ethanol-induced apoptosis in polarized hepatic cells possibly through regulation of the Fas pathway. Alcohol Clin Exp Res. 2006;30:1906–1915. doi: 10.1111/j.1530-0277.2006.00235.x. [DOI] [PubMed] [Google Scholar]
- 49.Brandl A., Meyer M., Bechmann V., Nerlich M., Angele P. Oxidative stress induces senescence in human mesenchymal stem cells. Exp Cell Res. 2011;317:1541–1547. doi: 10.1016/j.yexcr.2011.02.015. [DOI] [PubMed] [Google Scholar]
- 50.Shukla S.D., Aroor A.R. Epigenetic effects of ethanol on liver and gastrointestinal injury. World J Gastroenterol. 2006;12:5265–5271. doi: 10.3748/wjg.v12.i33.5265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bricambert J., Miranda J., Benhamed F., Girard J., Postic C., Dentin R. Salt-inducible kinase 2 links transcriptional coactivator p300 phosphorylation to the prevention of ChREBP-dependent hepatic steatosis in mice. J Clin Invest. 2010;120:4316–4331. doi: 10.1172/JCI41624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bardag-Gorce F., Li J., Oliva J., Lu S.C., French B.A., French S.W. The cyclic pattern of blood alcohol levels during continuous ethanol feeding in rats: the effect of feeding S-adenosylmethionine. Exp Mol Pathol. 2010;88:380–387. doi: 10.1016/j.yexmp.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Silencing DNMT3B by ethanol in human hepatobiliary cells. N-Heps (A) and HiBECs (B) were treated with 20 mmol/L ethanol along with the diluents control for 7 days. Total RNA was extracted from treated hepatobiliary cells, and DNMT1 and DNMT3B expressions were assessed by quantitative real-time PCR. The relative mRNA expression in the ethanol-treated group was normalized to expression of the control group as a percentage of the control. The results shown represent the mean ± SE of four independent experiments. *P < 0.05 when compared with mRNA expression of control group of cells.
Analysis of the CpG island enriched sites of the miR-34a regulatory region using the MethPrimer program. Analysis of the promoter region using MethPrimer software revealed the presence of CpG islands ∼300 bp upstream of the 5′-region of the mature miR-34a sequence. The positions of five pair primer sets relative to precursor miR-34a are shown for each pair. Sets 1 through 3 were used for detection of methylation within the promoter regions, whereas sets 4 and 5 were used for methylation within a control region.
Silencing DNMT3B enhanced miR-34a expression in hepatobiliary cells with or without ethanol exposure. N-Heps, HiBECs, and HepG2 cells were transfected with either control or DNMT3B small-interfering RNA for 48 hours. The transfected cells were further treated with 20 mmol/L ethanol or PBS for 7 days. Relative miR-34a expression was detected by real-time PCR analysis. miR-34a was significantly increased after silencing DNMT3B (with or without ethanol treatment) in all three cell lines tested. Data represent mean ± SE from four separate experiments. *P < 0.05 versus PBS control; #P < 0.05 versus control small-interfering RNA group.