SUMMARY
In Burkitt lymphoma (BL), a germinal center B-cell-derived tumor, the pro-apoptotic properties of c-MYC must be counterbalanced. Predicting that survival signals would be delivered by phosphoinositide-3-kinase (PI3K), a major survival determinant in mature B cells, we indeed found that combining constitutive c-MYC expression and PI3K activity in germinal center B cells of the mouse led to BL-like tumors, which fully phenocopy human BL with regard to histology, surface and other markers, and gene expression profile. The tumors also accumulate tertiary mutational events, some of which are recurrent in the human disease. These results and our finding of recurrent PI3K pathway activation in human BL indicate that deregulated c-MYC and PI3K activity cooperate in BL pathogenesis.
INTRODUCTION
While c-MYC (MYC) deregulation is a hallmark of BL (Jaffe and Pittaluga, 2011), an aggressive germinal center (GC)-derived B cell lymphoma characterized by immunoglobulin (IG)-MYC translocations, cooperating transforming events in BL are still poorly understood, despite the existence of MYC-induced murine lymphoma models (Adams and Cory, 1985; Kovalchuk et al., 2000; Park et al., 2005). MYC expression promotes malignancies by inhibiting cell differentiation and inducing proliferation, but also makes the cells prone to apoptosis. Since unlike other lymphoma entities BL typically do not exhibit constitutive activity of the pro-survival factor NF-κB (Dave et al., 2006; Klapproth et al., 2009), we considered a possible involvement of the PI3K pathway when we had identified PI3K signaling as the B cell receptor (BCR)-mediated survival signal in mature B cells (Srinivasan et al., 2009): MYC deregulation in BL is due to translocation of the MYC gene into one of the immunoglobulin loci of the cell, but exclusively non-productively rearranged immunoglobulin loci are affected, indicating that the cells are selected for BCR expression (Küppers et al., 1999). There is also evidence for a role of BCR signaling in MYC-driven lymphomagenesis from a transgenic mouse model in which the B cells express a BCR with specificity for a concomitantly expressed transgenic protein antigen (Refaeli et al., 2008). Although the polyclonal B cell proliferation seen in this model was in clear contrast to human BL and it remained unclear from which B cell differentiation stage it originated, we felt encouraged by the available evidence to try to better model BL pathogenesis.
RESULTS
Impact of MYC Overexpression and Constitutive PI3K Activation on the GC Reaction
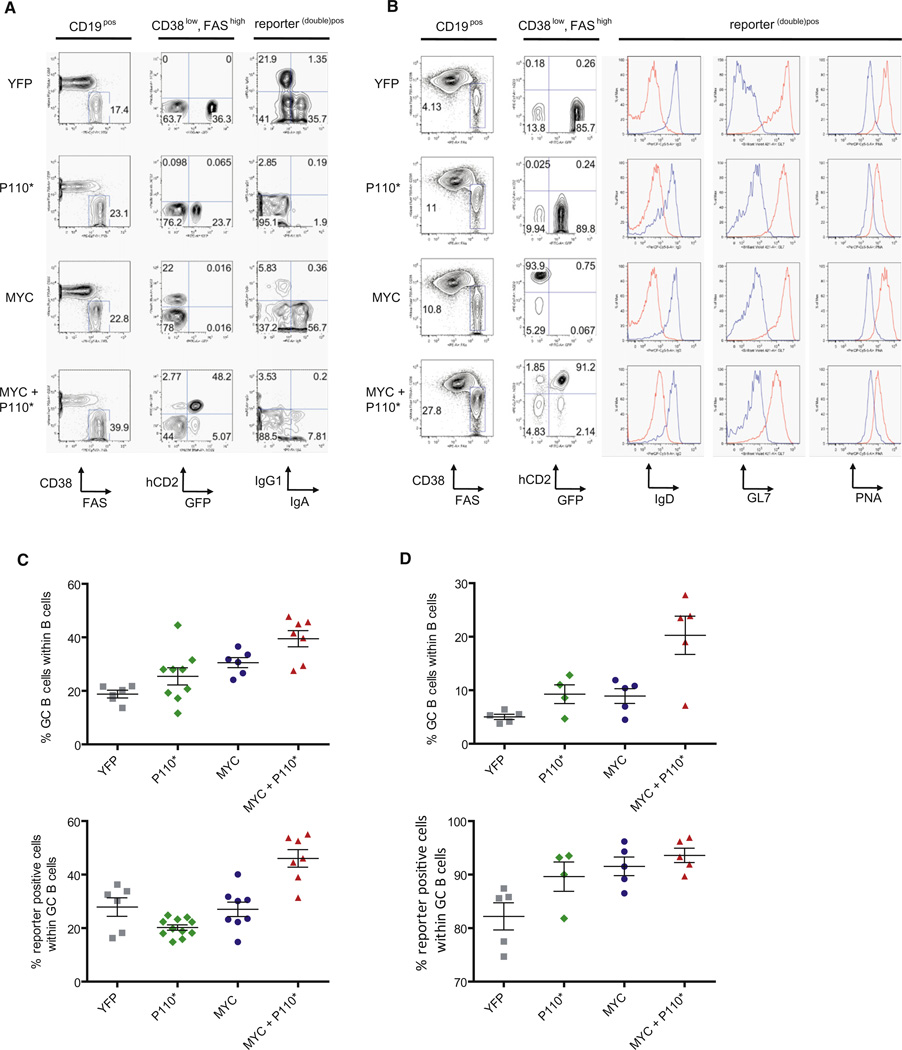
To determine the impact of MYC expression and PI3K pathway activation on GC B cells and lymphomagenesis, we generated mice expressing MYC and a constitutively active form of PI3K, here referred to as P110* (Srinivasan et al., 2009), specifically in B cells undergoing the GC reaction (Cγ1-cre,R26StopFLMYC,R26StopFLP110*; Figure S1 available online). Ten days after sheep red blood cell (SRBC) immunization, Peyer’s patches (PP) and spleens of transgenic mice were analyzed for reporter positive cells and their expression of GC B cell markers (Figures 1A and 1B). Transgenic expression of MYC and P110* was compatible with the formation of GCs. An increased proportion of GC B cells (CD38low, FAShigh) in the PP and the spleen of MYC and P110* co-expressing animals was detectable, accompanied by an increased proportion of reporter double-positive cells in comparison to the controls (Figures 1C and 1D). In addition to CD38 and FAS expression, MYC and P110* co-expressing GC B cells expressed less surface IgD than non-GC cells, at levels comparable to the controls (Figure 1B). The GC markers GL7 and PNA were also detectable on these cells although at lower levels than on GC B cells derived from Cγ1-cre,R26StopFLeYFP animals (Figure 1B). Class switch recombination (CSR) was impaired in MYC and P110* co-expressing cells (Figure 1A), presumably because of PI3K activation (Omori et al., 2006).
Figure 1. MYC and P110* Co-Expression Results in Increased GC B Cell Formation.
(A) Representative FACS analysis of PP isolated from Cγ1-cre, R26StopFLeYFP (YFP); Cγ1-cre, R26StopFLP110*(P110*); Cγ1-cre, R26StopFLMYC (MYC) and Cγ1-cre, R26StopFLMYC, R26StopFLP110* (MYC+P110*) animals. The sequential gating strategy is shown on top of each column.
(B) Representative FACS analysis in Rag2cgKO animals reconstituted with BM of the various genotypes and immunized with SRBC 10 days before analysis. The gating was performed according to (A). The histograms show expression of classical GC B cell markers in reporter (double) positive cells (red) and non-GC B cells (blue).
(C) Mean percentage (±SEM) of GC B cells (CD38low, FAShigh) and reporter (double) positive cells within PP of mice analyzed according to (A). At least six animals per genotype were analyzed.
(D) Mean percentage (±SEM) of GC B cells (CD38low, FAShigh) and reporter (double) positive cells within spleens of mice analyzed according to (B). At least 4 BM reconstituted animals per genotype were analyzed.
See also Figure S1.
MYC and P110* Cooperate in Tumorigenesis
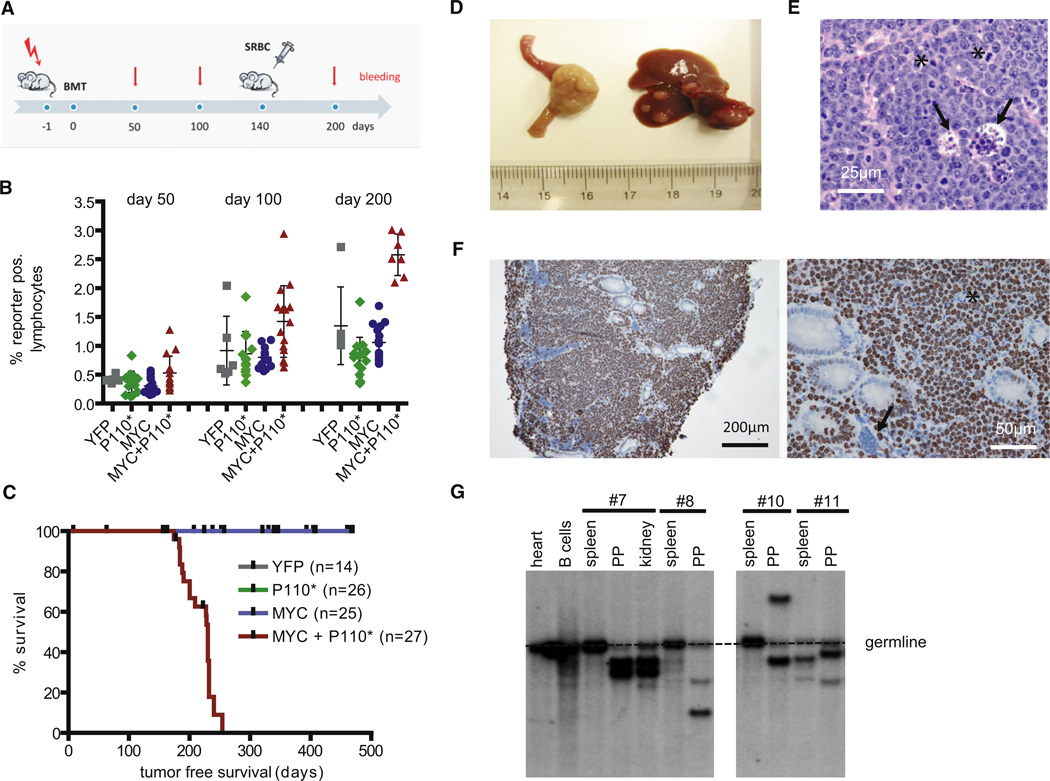
In order to obtain meaningful numbers of experimental animals in a timely fashion, bone marrow (BM) of individual triple transgenic animals (Cγ1-cre,R26StopFLMYC,R26StopFLP110*) and the corresponding controls was transferred to Rag2cgKO animals (Figure 2A). These animals lack a lymphatic system due to deficiency of the recombinase Rag2 and the cytokine receptor common subunit gamma (DiSanto et al., 1995; Shinkai et al., 1992). After BM transfer, the recipient mice generate lymphocytes that are genotypically identical to the donor BM cells. Blood analyses performed before and after a single boost of GC formation by SRBC demonstrated a steady increase of the percentage of lymphocytes co-expressing MYC and P110* over time, more so than in the case of lymphocytes expressing either transgene alone (Figure 2B). This correlated with lymphoma development and a reduced life span of the animals reconstituted with triple transgenic BM (median survival 227 days) (Figure 2C). In reconstituted animals expressing either MYC or P110* alone, tumor development was not detected within the period of observation.
Figure 2. MYC and PI3K Pathway Activation Cooperate in Tumorigenesis.
(A) Experimental protocol. Sublethally irradiated (day −1) Rag2cgKO mice were reconstituted with donor BM (from Cγ1-cre, R26StopFLeYFP; Cγ1-cre, R26StopFLP110*; Cγ1-cre, R26StopFLMYC; or Cγ1-cre, R26StopFLMYC, R26StopFLP110* animals) on day 0. Per genotype three individual BM donors were used. Transgene expression was enforced by a single SRBC immunization at day 140. Blood analyses were performed at days 50, 100, and 200 after BM transfer.
(B) Blood analysis of Rag2cgKO animals reconstituted with BM of the indicated genotypes. FACS analyses were performed at days 50, 100, and 200 after BM transfer. Mean percentage (±SEM) of reporter (double) positive lymphocytes is shown.
(C) Tumor-free survival of reconstituted Rag2cgKO animals. The total number of BM recipients is shown in parentheses. The ticks indicate non-tumor-related deaths.
(D) 12/21 tumors originated from the PP in the small intestine of MYC and P110* co-expressing animals (left). In 17/21 animals tumors disseminated to the liver (right).
(E) Representative HE staining in tumor no. 7. The asterisks mark mitotic figures within dividing cells. The arrowheads point to histiocytes clearing apoptotic cells. In total seven tumors were analyzed.
(F) Representative immunohistochemical staining for Ki67 in tumor no. 7. Interspersed nonmalignant (arrowhead) and dead cells (asterisk) are Ki67 negative. In total seven tumors were analyzed.
(G) Southern blot analysis for IgH gene rearrangements in PP derived tumors and potentially infiltrated organs using a JH4 probe. Monoclonal B cell expansion was seen in the PP, but not in the spleens of diseased animals (with exception of animal no. 11 showing expansion of an additional B cell clone in the spleen).
See also Table S1.
Macroscopically the animals reconstituted with triple transgenic BM displayed large tumors originating from the PP of the small intestine (12/21 tumors) or other lymphoid organs (spleen, lymph nodes) and infiltrating the liver and other nonlymphoid organs (e.g., kidney, lung) at an advanced stage (Figure 2D; Table S1). Histologic analysis revealed a characteristic BL morphology defined by the monotonous infiltration with medium-sized cells carrying uniform nuclei, prominent basophilic nucleoli, and frequent mitotic figures (Figure 2E). Like in human BL, the tumors displayed the typical “starry sky” pattern due to invading tissue macrophages that clear apoptotic tumor cells. In accordance with the diagnostic criteria of human BL, Ki67 staining demonstrated a proliferative index of nearly 100% in the tumors (Figure 2F).
The analysis of immunoglobulin heavy chain (IgH) gene rearrangements by Southern blot in the tumors and affected organs showed that the tumors were monoclonal (Figure 2G). The finding of distinct, unique VDJ rearrangements in B cell tumors of different recipient animals transferred with BM from a single donor argues against a transfer of tumor cells from the donor (Figure 2G). In three tumors derived from different recipient animals reconstituted with the same donor BM sequencing of the rearranged IgH variable (V) region genes confirmed unique VDJ rearrangements in the tumors (data not shown).
MYC and P110* Co-Expressing Tumors Originate from GC B Cells
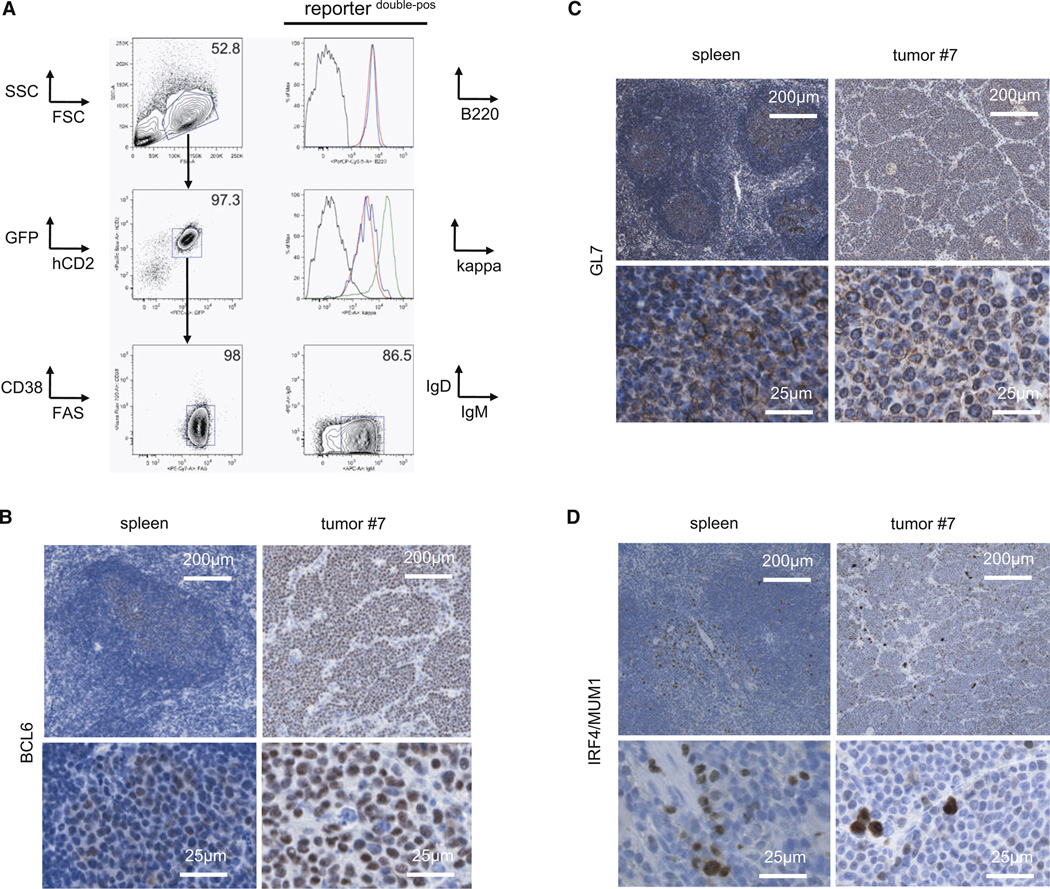
In accordance with our intention to generate a GC-derived MYC and PI3K induced tumor model, the tumor cells expressed both transgenes (GFPpos, hCD2pos) as well as mature GC B cell markers (B220pos, CD19pos, AA4.1low, CD38low, FAShigh, CD138neg, CD23neg, CD43neg, CD5neg; Figure 3A; Figure S2A). Similar to human BL, the tumors arose from non-switched GC B cells expressing surface IgM (Figure 3A). In addition, immunohistochemical analyses revealed expression of the GC B cell markers BCL6 and GL7 in the mouse tumors (Figures 3B and 3C) while PNA binding was not detectable in the tumors (Figure S2B). The latter might reflect its impaired binding on GC B cells upon P110* expression (see Figure 1B). The lack of IRF4/MUM1 expression, denoting B cell maturation toward plasma cells during late GC B cell differentiation, might indicate that the tumors arise from B cells at an early phase of the GC reaction (Figure 3D).
Figure 3. Lymphomas Arising upon MYC and PI3K Activation Express GC B-Cell-Specific Markers.
(A) Representative FACS analysis of GC B cell markers (B220, CD38, FAS, sIgD) and sIgM expression on tumor cells (defined as GFPpos and hCD2pos cells). The sequential gating strategy is indicated by arrows. Upper histogram: B220 expression on tumor cells (red) and normal splenic B cells (blue) in comparison to non-B cells (black). Lower histogram: Kappa light chain expression on tumor cells (red) and normal GC B cells (blue) in comparison to non-B cells (black) and normal follicular B cells (green).
(B–D) Representative immunohistochemical stainings for BCL6 (B), GL7 (C) and IRF4/MUM1 (D) in tumor no. 7 and control spleen (derived from a Cγ1-cre, R26StopFLeYFP animal 10 days after SRBC immunization). Per staining seven tumors were analyzed.
See also Figure S2.
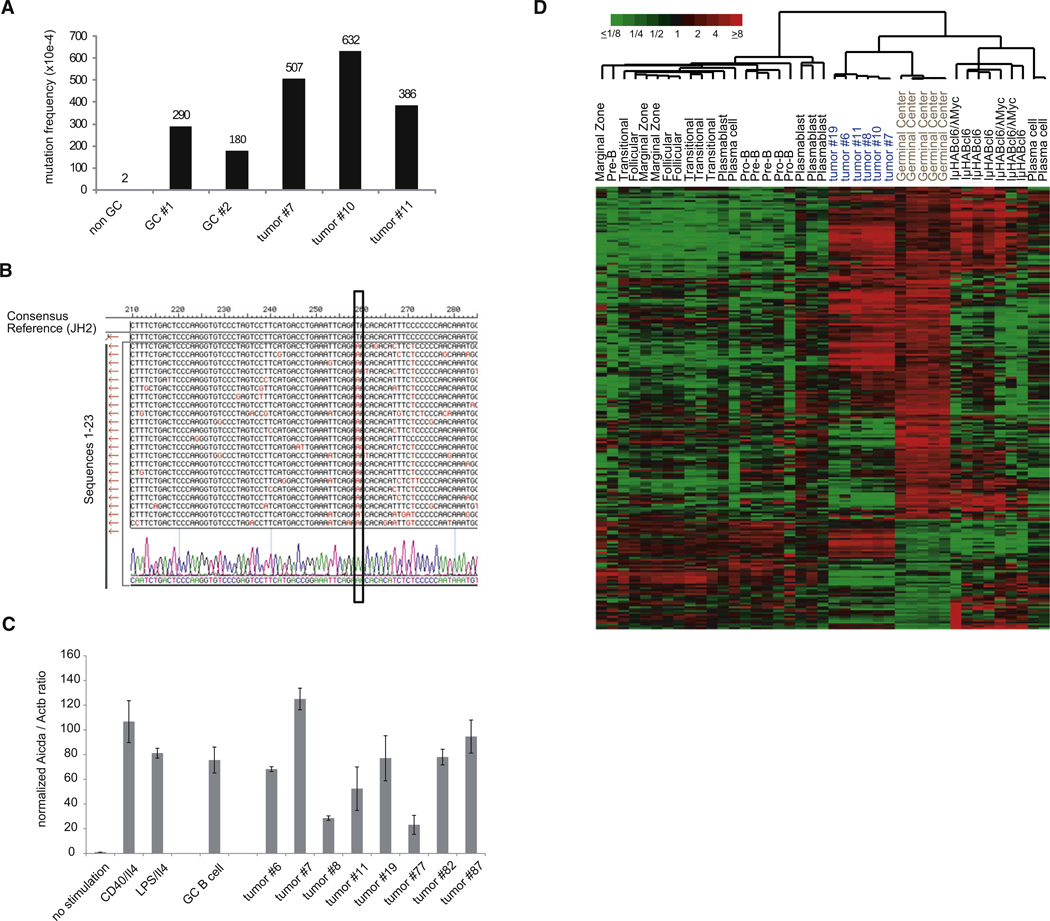
The finding of extensive ongoing somatic hypermutation (SHM) in the rearranged IgH-V region genes of the tumor cells (mean mutation frequency 508×10−4) confirmed the GC cell origin of the tumors (Figures 4A and 4B; Figure S3A). In agreement with this observation the tumors expressed cytidine deaminase AID at comparable transcript levels as GC and in vitro stimulated B cells (Figure 4C).
Figure 4. MYC and P110* Co-Expressing Tumors Show Ongoing SHM and Express GC B-Cell-Specific Genes.
(A) Mutation frequency in rearranged IgH-V region genes of non-GC B cells, GC B cells, and MYC+P110* co-expressing tumors (n = 3).
(B) SHM analysis in tumor no. 7. The rearranged IgH-V region genes of individual tumor cells were aligned to the JH2 reference sequence (n = 23). Mutations are labeled in red. The box marks a mutation shared by all sequences of this particular tumor.
(C) Aicda expression in stimulated B cells, GC B cells and MYC+P110* co-expressing tumors (n = 8) was analyzed by qRT-PCR. The ratio Aicda/Actb in non-stimulated cells was arbitrary defined as 1 and the values of the other samples were normalized to it. The mean expression from triplicate measurements (±SEM) was used for the calculations.
(D) Hierarchical cluster analysis based on relative transcript levels of 233 genes comprised in aGC B cell signature (Green et al., 2011) in normal B cell populations, MYC and P110* co-expressing tumor samples (n = 6) as well as other mouse lymphomas (IμHABcl6 (n = 4) and IμHABcl6/λMyc (n = 3); mean-centered log2 gene expression ratios are depicted by color scale). The meta-analysis was based on GEP data from Green et al. (GEO26408) and own experiments (GEO35219).
Additional information is provided in the Supplemental Experimental Procedures.
See also Figure S3.
The GC cell origin of the B cell tumors was also evident from gene expression profiling (GEP) data of purified MYC and P110* co-expressing tumor cells which we compared with published GEP data sets of various B cell subpopulations and mouse lymphoma models. Our tumors expressed a prominent GC B cell signature (Figures 4D and S3B), which was less pronounced in murine lymphomas resulting from transgenic expression of the GC B cell-specific transcriptional repressor BCL6 either alone or in combination with MYC (Green et al., 2011).
The Mouse Tumors Resemble Human BL
GC markers are typically associated with human BL, but are shared by a major subgroup of diffuse large cell B cell lymphomas (Alizadeh et al., 2000). We therefore performed a supervised comparison of global gene expression patterns established from MYC and P110* co-expressing tumors and BCL6 driven lymphomas, a mouse model recapitulating the pathogenesis of human DLBCL (Cattoretti et al., 2005), and identified a total of 2407 genes that were differentially expressed between these tumor entities (Figure 5A). We then looked among those genes for two sets of BL signature genes that had been identified in the human as differentially expressed between BL and DLBCL (Dave et al., 2006; Hummel et al., 2006). Comparing the expression of these BL signature genes between the two mouse lymphoma models, a clear positive association was detected between the MYC and P110* co-expressing mouse tumors and human BL (Figure 5B).
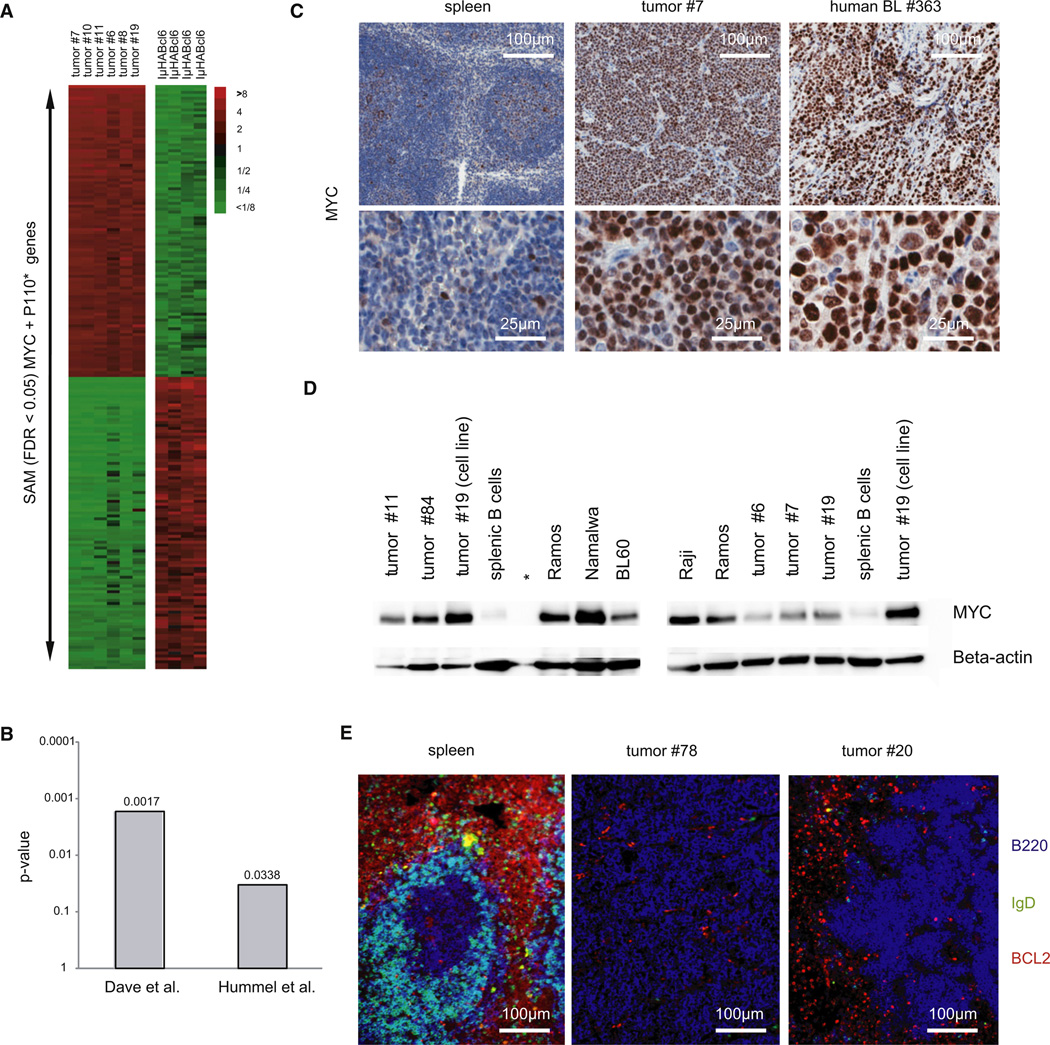
Figure 5. MYC and P110* Co-Expressing Tumors Resemble Human BL.
(A) Heat map showing relative transcript levels of the top 100 up- and downregulated genes distinguishing MYC and P110* co-expressing tumors (n = 6) from IμHABcl6 induced lymphomas (n = 4) as determined by SAM analysis (FDR < 0.05).
(B) p-values for the positive association of two human BL signatures (Dave et al., 2006; Hummel et al., 2006) with the MYC + P110* tumor signature as defined by SAM analysis.
(C) Representative immunohistochemical staining for MYC in tumor no. 7, primary human BL no. 363 and control spleen (derived from a Cγ1-cre, R26StopFLeYFP animal 10 days after SRBC immunization). In total seven mouse tumors and nine primary human BL were analyzed.
(D) Western blot analysis for MYC expression in five primary mouse tumors (tumor nos. 6, 7, 11, 19, and 84), one cell line derived from mouse tumor no. 19 (tumor no. 19 [cell line]), four human BL cell lines (BL60, Namalwa, Raji, Ramos), and splenic B cells. Beta-actin served as loading control. *Empty lane.
(E) Representative immunofluorescence analysis for BCL2 (red), B220 (blue), and IgD (green) in two tumors (tumor no. 78 and tumor no. 20) and control spleen (derived from a C57BL/6 animal 10 days after SRBC immunization).
To further distinguish our BL-like tumors from DLBCL we determined BL-typical proteins by immunohistochemistry, western blot, and immunofluorescence (Figures 5C–5E). Typically, human BL show elevated MYC levels due to translocations and mutations of the MYC gene. Similarly, MYC transgene expression in our tumor model resulted in abundant MYC protein as it is seen in primary BL samples and BL cell lines (Figures 5C and 5D). Besides MYC expression, histologic features and the high proliferation rate (Ki67 >95%), human BL cells as well as the mouse tumor cells are positive for BCL6 (Figure 3B) and lack BCL2 expression (Figure 5E).
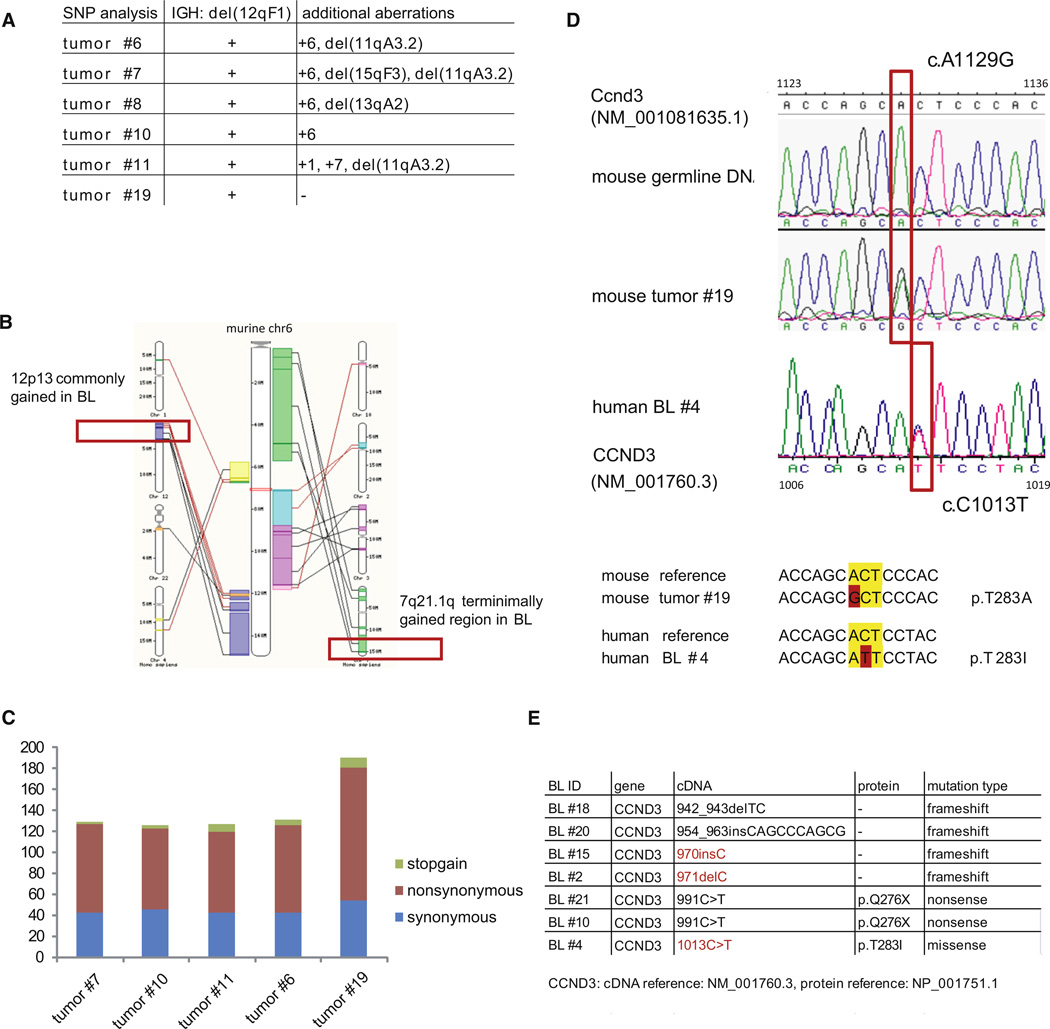
The MYC and P110* induced tumors did not only express BL characteristic markers and exhibit a GEP signature resembling that of human BL, but also displayed genomic aberrations reminiscent of aberrations previously reported in human BL (Mitelman et al., 2012). Overall SNP array analysis revealed a simple karyotype of the mouse tumors: beside the B cell specific rearrangements of the Ig loci the tumors displayed 1.8 aberrations per case (Figure 6A). The most frequent (4/6 tumors) DNA copy number alteration was a gain of chromosome 6, which comprises genomic regions gained in human BL such as 7q21.1qter and 12p13 (Figure 6B) (Boerma et al., 2009; Scholtysik et al., 2012). Exome sequencing of five murine MYC and P110* co-expressing lymphomas and the respective germline DNA revealed additional candidate oncogenic events (Table S2). In total, we observed on average 103 missense and/or nonsense mutations per murine BL tumor (Figure 6C). Although AID expression was clearly detectable in the tumor cells at the mRNA level, the mutations rarely coincided with classical AID hotspots (WRCY or the inverse RGYW; Figure S4).
Figure 6. Lymphomas Originating from MYC and P110* Co-Expressing GC B Cells Display Genetic Aberrations Commonly Found in Human BL.
(A) Summary of genomic aberrations detected by SNP microarray analysis in 6 MYC and P110* co-expressing tumors.
(B) Schematic view of mouse chromosome 6 and its syntenic regions in human (based on Ensembl genome browser 65). Syntenic regions that have been described as gained in human BL (Boerma et al., 2009; Scholtysik et al., 2012) are marked in red.
(C) Number and classification of somatically acquired mutations based on exome sequencing in MYC and P110* co-expressing tumors (n = 5).
(D) Sanger sequencing of the Ccnd3 mutation (A1129G) in mouse tumor no. 19 and the corresponding germline DNA (upper and middle panel). In human BL no. 4 the CCND3 mutation (C1013T) affects the same codon as detected in the mouse tumor (lower panel).
(E) Summary of CCND3 mutations detected by Sanger Sequencing in 29 primary BL samples. Positions that are also affected in the mouse tumors are marked in red.
See also Figure S4 and Table S2.
Some of these additional mutations have recently also been identified in a parallel analysis of a large collection of human BL in the laboratory of L. Staudt (Schmitz et al., 2012). An example is the heterozygous mutation of cyclin D3 (Ccnd3) at codon 283 (A1129G) encoding a threonine residue which regulates cyclin D3 stability through phosphorylation (Figure 6D; Casanovas et al., 2004). An essential role of cyclin D3 in the control of GC B cell proliferation was recently reported (Cato et al., 2011; Peled et al., 2010).
To determine the incidence of Ccnd3 mutations in a larger tumor cohort we sequenced the 3′ end of the gene in 10 additional MYC and P110* co-expressing mouse tumors and identified two additional Ccnd3 mutations: a non-synonymous mutation at codon 239 (T997G) and a frameshift caused by a single base pair insertion (1087insC).
We also confirmed that cyclin D3 is recurrently mutated in human BL. Sequencing a 346-bp amplicon comprising part of CCND3 exon 5 in 29 primary BL samples, we found mutations in seven of the cases (Figure 6E). Strikingly, the mutation at codon 283 (C1013T) and the frameshift mutations at positions 970 and 971 (970insC, 971 delC) affected the very same conserved codons mutated in the mouse tumors (Figures 6D and 6E).
PI3K Pathway Activation in Human BL
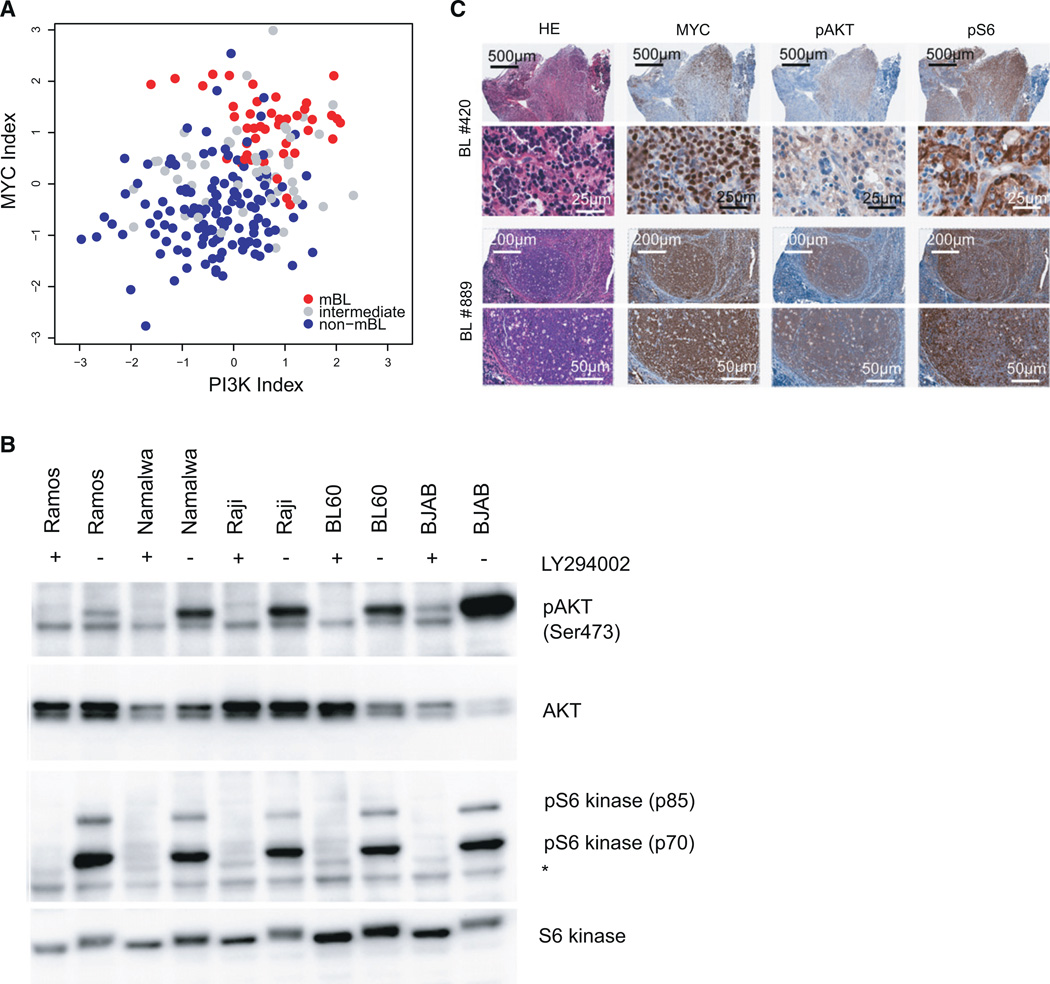
The present mouse model predicts PI3K pathway activation in human BL. To directly address this issue, we performed a bioinformatic analysis of GEP data of primary BL in comparison to other human lymphoma entities (Hummel et al., 2006), which revealed accompanying MYC activation (p < 2.2−16) and PI3K pathway activation (p = 2.3−8) in human BL (Figure 7A). In addition, western blot analysis indicated activation of the PI3K pathway as determined by phosphorylation of AKT at serine 473 and p70S6 kinase at threonine 389 in 5 human BL cell lines (Figure 7B). Phosphorylation of both kinases was reversible upon treatment with the PI3K inhibitor LY-294002 (Figure 7B). Finally, immunohistochemistry of nine primary human BL revealed phosphorylation of AKT and S6 in six of nine analyzed patients (Figure 7C), suggesting PI3K pathway inhibition as a therapeutic option in human BL.
Figure 7. PI3K Pathway Activation in Human BL.
(A) Scatter plot of the PI3K pathway activity (Gustafson et al., 2010) index against the MYC activation index (Bild et al., 2006) in human BL (GEO accession number GSE35219) (Hummel et al., 2006). Samples classified as molecular BL (mBL), intermediate, and non-mBL in the original study are shown as red, gray, and blue dots, respectively.
(B) Western blot analysis for Phospho-AKT (Ser473), AKT, phospho-S6 kinase (Thr389), and S6 kinase expression in five human BL cell lines (BJAB, BL60, Namalwa, Raji, Ramos). Cells were either treated with LY-294002 (+) or DMSO (−) 1 hr before protein extraction. *unspecific band.
(C) Representative immunohistochemical staining for MYC, phospho-AKT (Ser473), phospho-S6 (Ser235/236) in two primary human BL (human BL nos. 420 and 889). In total nine human BL were analyzed.
DISCUSSION
The present work describes the construction of a clinically relevant mouse model of a life-threatening human tumor by targeting constitutive expression of an oncogene known to be involved in tumor pathogenesis together with the activation of a suspected pathogenic signaling pathway specifically into the presumed cell of origin. The resulting tumors faithfully phenocopy their human counterparts, including strikingly similar gene expression profiles, expression of BL-typical markers like BCL6 and MYC at the protein level and tertiary transforming events, as discussed further below. The similarities between human BL and the mouse tumors also include histologic peculiarities like starry sky appearance, high proliferative activity with homogeneous Ki67 staining, and the conspicuous absence of the pro-survival protein BCL2, whose expression is observed in many other classes of B cell lymphomas. Like in human BL, the tumors predominantly originated from the PP (which are enriched in the ileocecal region in human where BL frequently arises), exhibit ongoing somatic hypermutation, and their monoclonality indicates a multistep pathogenesis with MYC and PI3K activation as initiating events. Consistent with the latter notion and underlining the relevance of the mouse model for the human disease we find PI3K pathway activation in human BL lines and a major fraction of primary Burkitt tumors.
While MYC deregulation is an established hallmark of human BL and was the basis of previous mouse models (Adams and Cory, 1985; Kovalchuk et al., 2000; Park et al., 2005), PI3K signaling had not been recognized as a critical element of BL pathogenesis. Although PI3K signaling can support MYC activity by blocking its degradation (Kumar et al., 2006) and inducing the degradation of the MYC antagonist MAD1 (Zhu et al., 2008), and MYC-mediated miR17–92 induction can increase PI3K signaling by targeting PTEN (Mu et al., 2009; Olive et al., 2009), combined MYC and PI3K pathway activation does not necessarily lead to malignant transformation of the targeted cells (Radziszewska et al., 2009).
Our initial motivation to study PI3K pathway activation in concert with MYC deregulation in GC B cells came from our previous identification of PI3K signaling as the “tonic” survival signal in B cells downstream of the BCR, for whose expression normal B cells as well as BL cells are known to be positively selected (Küppers et al., 1999; Srinivasan et al., 2009). Indeed, the present results indicate significant PI3K pathway activation in human BL, contrasting with the absence in these tumors of survival signals through the NF-κB pathway (Dave et al., 2006). Consistent with these data, recurrent mutations promoting PI3K signaling have recently been identified in human BL (Schmitz et al., 2012). The efficiency of MYC-PI3K cooperation in promoting the transformation of GC B cells to give rise to BL-like tumors in the mouse model indicates that these two factors indeed play a functional role in BL pathogenesis.
The induction of PI3K activation by the P110* transgene represents a limitation of the present mouse model in that the tumors arising in the mice cannot be expected to be selected for mutations promoting PI3K signaling. A similar argument can be made for mutations of tumor suppressor genes like TP53 or CDKN2A (recurrently mutated in human BL) (Bhatia et al., 1992; Sánchez-Beato et al., 2001), whose inactivation is considered to be an early event in tumorigenesis (Barrett et al., 1999) and may thus be upstream of the mutations deliberately introduced into the mouse model. The situation is different, however, for the tertiary mutations that are required for lymphomagenesis in the mice in addition to deregulated MYC and PI3K activity. Strikingly, these mutations include several genetic alterations that are recurrently seen in human BL. Thus, apart from shared copy number gains, the gene encoding cyclin D3, a critical cell cycle regulator in GC B cells, was mutated in both human BL and the mouse tumors. In both cases a point mutation affected a conserved codon critical for cyclin D3 stability, and frameshift mutations were found in the 3′ region of the gene. This further validates the mutant mice as a preclinical BL model, assigns functional significance to the mutations shared between the mouse and human tumors, and opens the way to in vivo analyses of their clinical relevance.
Taken together, we show that targeting MYC expression together with PI3K pathway activation into mouse GC B cells generates a faithful mouse model of human BL. The mouse tumors accumulate tertiary mutations at least some of which are recapitulated in the human disease; others may reflect genetic or epigenetic alterations that have not yet been uncovered in BL. The significance of the tertiary mutations for tumor progression can be assessed in the mouse model in functional terms, with clear perspectives for new therapeutic approaches. Already at this point the PI3K pathway has emerged as a promising therapeutic target in BL.
EXPERIMENTAL PROCEDURES
Mice, Immunization, and Tumor Cohorts
Cγ1-cre; R26StopFLP110*; and R26StopFLeYFP alleles have been described (Calado et al., 2010; Casola et al., 2006; Srinivasan et al., 2009). The R26StopFLMYC allele was generated following a strategy previously developed (Sasaki et al., 2006). Briefly, the ROSA26 allele was targeted with a construct containing human c-MYC cDNA preceded by a loxP flanked STOP cassette and marked by a signaling deficient truncated version of hCD2 under the control of an internal ribosomal entry site (IRES) downstream of the inserted cDNA. Transgene transcription is controlled by the CAG promoter. A detailed description of the mice will be given elsewhere (D.P.C., unpublished data).
Rag2cgKO animals were bred in our mouse colony or purchased from Taconic. 1 × 106 viable total BM cells were intravenously injected into sublethally irradiated (600 rad) 9- to 11-week-old Rag2cgKO animals. For each genotype BM of three different donor animals was individually transferred to at least five (or three in the case of Cγ1-cre, R26StopFLeYFP BM) recipients. At day 140 after BM transfer, mice were immunized once by i.v. injection of 1 × 109 sheep red blood cells (SRBCs; Cedarlane). Mouse cohorts were monitored twice a week for tumor development and euthanized if signs of tumor development were seen. All animal care and procedures were approved by the Institutional Animal Care and Use Committee (IACUC 03341) of Harvard University and the Immune Disease Institute as well as the governmental review board (Landesamt für Gesundheit und Soziales Berlin, LaGeSo G0273/11).
Flow Cytometry and Cell Sorting
Single-cell suspensions were stained with PNA (Vector Laboratories) and the following monoclonal antibodies from BD Biosciences, Biolegends or eBioscience: αCD19(ID3), αB220(RA3-6B2), αCD95(Jo2), αCD138(281-2), αCD38(90), αhCD2(TS1/8), αCD93(AA4.1), αIgM(FAB), αIgD(11-26c.2a), αIgG1(A85-1), αIgA(mA-6E1), αIgkappa(187.1), αCD23(B3B4), αCD43(S7), αCD5(53-7.3), αGL7(GL7). Topro3 or PI (Invitrogen) was used to exclude dead cells. Samples were acquired on a FACSCantoII (BD Biosciences), and analyzed using FlowJo software (Tree star). Viable (Topro3neg) GFP and hCD2 co-expressing tumor cells and GC B cells (CD19pos, CD38low, FAShigh, YFPpos) of Cγ1-cre, R26StopFLeYFP animals were sorted on a FACSAriaII (BD Biosciences) and used for RNA and DNA preparation.
Real-Time RT-PCR
Total RNA from sorted cells was extracted using the AllPrep DNA/RNA Kit (QIAGEN) and cDNA was synthesized using the Thermoscript RT-PCR system (Invitrogen). For qRT-PCR, we used Power SYBR Green, followed by analysis with the StepOnePlus system (Applied Biosystems). Samples were assayed in triplicate and messenger abundance was normalized to that of Actb.
Analysis of Tumor Clonality by Southern Blot
EcoRI digested genomic DNA from tumor bearing mice and normal splenic B cells (C57BL/6 animal) was probed with a JH probe spanning the JH4 exon and part of the downstream intronic sequence.
Histology and Immunohistochemistry
Tissues were fixed with 10% formalin (Sigma) and paraffin embedded sections were stained with H&E (Sigma), Ki67 (SP6; Vector Laboratories), PNA (Vector Laboratories), αGL7 (eBioscience), αMYC (N-terminal, Epitomics), αBCL6 (D65C10; Cell signaling), αpAKT (Ser473; D9E; Cell signaling), αpS6 (Ser235/236; D57.2.2E; Cell signaling), and αIRF4 (MUM1; Santa Cruz).
Immunofluorescence
Tissues were frozen in OCT (Sakura Finetek) in liquid nitrogen and staining was performed as described previously (Srinivasan et al., 2009) using B220-APC, IgD-FITC, and BCL2-PE (BD Biosciences) antibodies.
In Vitro Cell Culture of Splenic B Cells
Splenic B cells of C57BL/6 mice were purified by CD43 depletion (Miltenyi). Cells were cultured in the presence of 1 µg/ml of αCD40 (HM40-3, eBioscience) or 20 µg/ml of LPS (Sigma) and 25 ng/ml of IL-4 (R&D Systems) for 3 days. RNA was isolated using the RNeasy kit (QIAGEN) and cDNA was synthesized using the Thermoscript RT-PCR system (Invitrogen).
IgH Somatic Mutation Analysis
Genomic DNA was prepared from sorted tumor cells or GC B cells of Cγ1-cre, R26StopFLeYFP animals. IgH-V gene rearrangements were PCR amplified using the Expand High fidelity PCR system (Roche) combined with forward primers VHA, VHE, or VHG (Ehlich et al., 1994) and a reverse primer in the JH4 intron (5′-CTCCACCAGACCTCTCTAGACAGC-3′). Fragments were cloned, sequenced, and blasted against the NCBI database (http://www.ncbi.nlm.nih.gov/igblast/). Germline polymorphisms were excluded by blasting against the database of sequences generated in our laboratory.
Gene Expression Profiling and Data Analysis
GEP was performed on tumor samples and purified GC B cells and non-GC B cells from immunized Cγ1-cre, R26StopFLeYFP animals using Affymetrix GeneChip Mouse Genome 430 2.0 Arrays according to the manufacturer’s recommendations (Affymetrix). The complete microarray data are available at the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/projects/geo; accession number GSE35219). Further information is provided in Supplemental Experimental Procedures.
Pathway Activation Indices
The method to compute pathway activation indices is an extension of our previously published strategy (Läuter et al., 2009). Based on gene expression, we generated 50 clusters of highly correlated genes in the data set of Hummel et al. (GSE4475) (Hummel et al., 2006). We then mapped these clusters to interventional data sets from experiments in which either MYC (GSE3151) (Bild et al., 2006) or PI3K (GSE12815) (Gustafson et al., 2010) were activated. In all data sets, we summarized the clusters to 50 metagenes. To obtain an index of the relative activity of a pathway in a tumor sample, we computed the sum of the values of the 50 metagenes in this sample weighted by the correlations of the metagenes with the activation of this pathway in the corresponding interventional data set. We used preprocessed data as available at the Gene Expression Omnibus. Further information is provided in Supplemental Experimental Procedures.
SNP Microarray Analysis
For genomic profiling we used Affymetrix Mouse Diversity Genotyping Arrays according to the manufacturer’s recommendations (Affymetrix). After generation of the raw data (CEL-files) using Command_Console software (Affymetrix) paired analysis of tumor and respective germline DNA samples was performed. Using R version 2.12.1 (http://www.r-project.org/), data were normalized using the aroma.affymetrix R package (Bengtsson et al., 2008) in combination with the R package DNAcopy (Olshen et al., 2004) for segmentation and detection of copy number aberrations. The complete SNP array data are available at the Gene Expression Omnibus (accession number GSE35219).
Exome Sequencing
Nonamplified genomic DNA (1µg) from sorted murine tumor cells and matched germline tissue (mouse tail) were used for exome sequencing using Illumina technology. Further information is provided in Supplemental Experimental Procedures.
Human BL Cell Lines
Human BL cell lines were cultured in RPMI medium 1640 with 10% fetal calf serum, 1% penicillin/streptomycin, 1% L-glutamine, 1% nonessential amino acids, and 0.1% beta-mercaptoethanol. For detection of PI3K pathway activation cell lines were treated with either 25 µM LY-294002 (Sigma) or DMSO for 1 hr.
Patient Samples
Primary BL samples [9 peripheral blood/BM (B cell acute lymphoblastic leukemia, B-ALL), 27 lymph node (BL), 1 jaw (BL), and 1 parotid gland (BL) specimens; all cases were translocation t(8;14) positive] were provided by the Department of Pathology, Brigham and Women’s Hospital (n = 9), and the Department of Internal Medicine III, University Hospital of Ulm (n = 29) with patient informed consent and institutional review board approval from all participating centers. One case represents endemic BL (1/38) whereas the others belong to the sporadic/HIV-associated subgroup (37/38). Sanger Sequencing was performed with genomic DNA obtained from frozen lymphoid tissue blocks (n = 20) and mononuclear cells isolated from the peripheral blood or BM (n = 9).
Cyclin D3 Mutation Analysis in Primary Human BL and Mouse Tumors
Genomic DNA of primary human BL samples (n = 29) was prepared using the QIAGEN DNeasy blood and tissue kit (QIAGEN). A 346bp amplicon within exon 5 of human CCND3 was PCR amplified (forward primer: 5′-GAAGCTGCACTCAGGGAGAG; reverse primer: 5′-AGCTTGACTAGCCACCGAAA) and sequenced (Sanger Sequencing). The 3′ end of Ccnd3 was amplified in genomic DNA of MYC and P110* co-expressing mouse tumors (n = 15) using 5′-CACCTGCTTGCTGTCAGTGCTGTGAG as forward primer and 5′- GCATGGATTGTTCTAGAGGCAGGGA as reverse primer.
Western Blot Analysis
RIPA extracts were fractionated on 10% sodium dodecyl sulfate polyacrylamide gels, electroblotted to polyvinylidene difluoride membranes and reacted with α-MYC (N-terminal, Epitomics), α-pAKT (Ser473) (D9E), α-AKT (5G3), α-pS6 kinase (Thr389) (108D2), α-S6 kinase (Cat #9202) (Cell Signaling), and α-beta-actin (Cat #A5316, Sigma) antibodies. Immunoreactivity was determined using the enhanced chemiluminescence method (Pierce Chemical).
Statistical Analysis
Data were analyzed using unpaired two-tailed Student’s t test and Fisher’s exact test, a p value % 0.05 was considered significant. Survival curves were compared using the Logrank test. Data in text and figures are represented as mean ± SEM (standard error of the mean).
Supplementary Material
Significance.
We describe a mouse model of a human lymphoma through targeting expression of an oncogene known to be involved in tumor pathogenesis together with the activation of a suspected pathogenic signaling pathway into the presumed cell of origin. The resulting tumors faithfully model their human counterparts and accumulate additional genetic alterations, with clear perspectives for an assessment of their clinical relevance. Our data establish a framework of Burkitt lymphoma pathogenesis by identifying PI3K pathway activation as a key element for the malignant transformation of c-MYC-expressing germinal center B cells and highlight this pathway as a potential therapeutic target.
ACKNOWLEDGMENTS
We thank J. Xia, C. Grosse, J. Grundy, X. Chen, D. Ghitza, C. Unitt, and M. Bamberg for technical assistance; M. Ottaviano for administrative assistance; T. Yasuda for contribution of reagents; E. Derudder and J. Seagal for assistance with irradiation; S. Koralov for help with SHM analyses; D. Bolgehn and T. Sommermann for technical support; and present and former Rajewsky lab members for critical comments and suggestions. We are grateful to L. Staudt and R. Schmitz for sharing unpublished results and primer sequences. This work was supported by grants from the National Institutes of Health to K.R. (P01 CA92625; R37 AI054636), the Leukemia & Lymphoma Society (Leukemia & Lymphoma Society SCOR) and the European Research Council (ERC advanced Grant ERC-AG-LS6) to K.R.; Leukemia & Lymphoma Society fellowships to S. Sander, B.Z., and D.P.C., and a Heisenberg scholarship by the Deutsche Forschungsgemeinschaft to L.B.
S. Sander, D.P.C., L.B., and K.R. designed research; S. Sander, D.P.C., L.S., B.Z., S.R.. and K.H. performed experiments; S. Sander, D.P.C., S.R., K.H., L.B., and K.R. analyzed data; S.R., S. Stilgenbauer, and R.S. provided primary human BL samples and human tumor data; K.K. and M.R. performed statistical analyses. S. Sander, L.B., and K.R. wrote the manuscript with contributions from all authors.
Footnotes
ACCESSION NUMBERS
The Gene Expression Omnibus accession number for the microarray data is GSE35219. The exome sequencing data are available at the Sequence Read Archive (SRA055727).
SUPPLEMENTAL INFORMATION
Supplemental Information includes four figures, two tables, Supplemental Experimental Procedures, and Supplemental References and can be found with this article online at http://dx.doi.org/10.1016/j.ccr.2012.06.012.
REFERENCES
- Adams JM, Cory S. Myc oncogene activation in B and T lymphoid tumours. Proc. R. Soc. Lond. B Biol. Sci. 1985;226:59–72. doi: 10.1098/rspb.1985.0079. [DOI] [PubMed] [Google Scholar]
- Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- Barrett MT, Sanchez CA, Prevo LJ, Wong DJ, Galipeau PC, Paulson TG, Rabinovitch PS, Reid BJ. Evolution of neoplastic cell lineages in Barrett oesophagus. Nat. Genet. 1999;22:106–109. doi: 10.1038/8816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtsson H, Irizarry R, Carvalho B, Speed TP. Estimation and assessment of raw copy numbers at the single locus level. Bioinformatics. 2008;24:759–767. doi: 10.1093/bioinformatics/btn016. [DOI] [PubMed] [Google Scholar]
- Bhatia KG, Gutiérrez MI, Huppi K, Siwarski D, Magrath IT. The pattern of p53 mutations in Burkitt’s lymphoma differs from that of solid tumors. Cancer Res. 1992;52:4273–4276. [PubMed] [Google Scholar]
- Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, Joshi MB, Harpole D, Lancaster JM, Berchuck A, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- Boerma EG, Siebert R, Kluin PM, Baudis M. Translocations involving 8q24 in Burkitt lymphoma and other malignant lymphomas: a historical review of cytogenetics in the light of todays knowledge. Leukemia. 2009;23:225–234. doi: 10.1038/leu.2008.281. [DOI] [PubMed] [Google Scholar]
- Calado DP, Zhang B, Srinivasan L, Sasaki Y, Seagal J, Unitt C, Rodig S, Kutok J, Tarakhovsky A, Schmidt-Supprian M, Rajewsky K. Constitutive canonical NF-κB activation cooperates with disruption of BLIMP1 in the pathogenesis of activated B cell-like diffuse large cell lymphoma. Cancer Cell. 2010;18:580–589. doi: 10.1016/j.ccr.2010.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanovas O, Jaumot M, Paules AB, Agell N, Bachs O. P38SAPK2 phosphorylates cyclin D3 at Thr-283 and targets it for proteasomal degradation. Oncogene. 2004;23:7537–7544. doi: 10.1038/sj.onc.1208040. [DOI] [PubMed] [Google Scholar]
- Casola S, Cattoretti G, Uyttersprot N, Koralov SB, Seagal J, Hao Z, Waisman A, Egert A, Ghitza D, Rajewsky K. Tracking germinal center B cells expressing germ-line immunoglobulin gamma1 transcripts by conditional gene targeting. Proc. Natl. Acad. Sci. USA. 2006;103:7396–7401. doi: 10.1073/pnas.0602353103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cato MH, Chintalapati SK, Yau IW, Omori SA, Rickert RC. Cyclin D3 is selectively required for proliferative expansion of germinal center B cells. Mol. Cell. Biol. 2011;31:127–137. doi: 10.1128/MCB.00650-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattoretti G, Pasqualucci L, Ballon G, Tam W, Nandula SV, Shen Q, Mo T, Murty VV, Dalla-Favera R. Deregulated BCL6 expression recapitulates the pathogenesis of human diffuse large B cell lymphomas in mice. Cancer Cell. 2005;7:445–455. doi: 10.1016/j.ccr.2005.03.037. [DOI] [PubMed] [Google Scholar]
- Dave SS, Fu K, Wright GW, Lam LT, Kluin P, Boerma EJ, Greiner TC, Weisenburger DD, Rosenwald A, Ott G, et al. Lymphoma/Leukemia Molecular Profiling Project. Molecular diagnosis of Burkitt’s lymphoma. N. Engl. J. Med. 2006;354:2431–2442. doi: 10.1056/NEJMoa055759. [DOI] [PubMed] [Google Scholar]
- DiSanto JP, Müller W, Guy-Grand D, Fischer A, Rajewsky K. Lymphoid development in mice with a targeted deletion of the interleukin 2 receptor gamma chain. Proc. Natl. Acad. Sci. USA. 1995;92:377–381. doi: 10.1073/pnas.92.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlich A, Martin V, Müller W, Rajewsky K. Analysis of the B-cell progenitor compartment at the level of single cells. Curr. Biol. 1994;4:573–583. doi: 10.1016/s0960-9822(00)00129-9. [DOI] [PubMed] [Google Scholar]
- Green MR, Monti S, Dalla-Favera R, Pasqualucci L, Walsh NC, Schmidt-Supprian M, Kutok JL, Rodig SJ, Neuberg DS, Rajewsky K, et al. Signatures of murine B-cell development implicate Yy1 as a regulator of the germinal center-specific program. Proc. Natl. Acad. Sci. USA. 2011;108:2873–2878. doi: 10.1073/pnas.1019537108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson AM, Soldi R, Anderlind C, Scholand MB, Qian J, Zhang X, Cooper K, Walker D, McWilliams A, Liu G, et al. Airway PI3K pathway activation is an early and reversible event in lung cancer development. Sci. Transl. Med. 2010;2:26ra25. doi: 10.1126/scitranslmed.3000251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel M, Bentink S, Berger H, Klapper W, Wessendorf S, Barth TF, Bernd HW, Cogliatti SB, Dierlamm J, Feller AC, et al. Molecular Mechanisms in Malignant Lymphomas Network Project of the Deutsche Krebshilfe. A biologic definition of Burkitt’ lymphoma from transcriptional and genomic profiling. N. Engl. J. Med. 2006;354:2419–2430. doi: 10.1056/NEJMoa055351. [DOI] [PubMed] [Google Scholar]
- Jaffe ES, Pittaluga S. Aggressive B-cell lymphomas: a review of new and old entities in the WHO classification. Hematology (Am. Soc. Hematol. Educ. Program) 2011;2011:506–514. doi: 10.1182/asheducation-2011.1.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapproth K, Sander S, Marinkovic D, Baumann B, Wirth T. The IKK2/NF-kappaB pathway suppresses MYC-induced lymphomagenesis. Blood. 2009;114:2448–2458. doi: 10.1182/blood-2008-09-181008. [DOI] [PubMed] [Google Scholar]
- Kovalchuk AL, Qi CF, Torrey TA, Taddesse-Heath L, Feigenbaum L, Park SS, Gerbitz A, Klobeck G, Hoertnagel K, Polack A, et al. Burkitt lymphoma in the mouse. J. Exp. Med. 2000;192:1183–1190. doi: 10.1084/jem.192.8.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Marqués M, Carrera AC. Phosphoinositide 3-kinase activation in late G1 is required for c-Myc stabilization and S phase entry. Mol. Cell. Biol. 2006;26:9116–9125. doi: 10.1128/MCB.00783-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küppers R, Klein U, Hansmann ML, Rajewsky K. Cellular origin of human B-cell lymphomas. N. Engl. J. Med. 1999;341:1520–1529. doi: 10.1056/NEJM199911113412007. [DOI] [PubMed] [Google Scholar]
- Läuter J, Horn F, Roso1owski M, Glimm E. High-dimensional data analysis: selection of variables, data compression and graphics—application to gene expression. Biom. J. 2009;51:235–251. doi: 10.1002/bimj.200800207. [DOI] [PubMed] [Google Scholar]
- Mitelman F, Johansson B, Mertens FE. Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer. 2012 http://cgapncinihgov/Chromosomes/Mitelman.
- Mu P, Han YC, Betel D, Yao E, Squatrito M, Ogrodowski P, de Stanchina E, D’Andrea A, Sander C, Ventura A. Genetic dissection of the miR-17~92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 2009;23:2806–2811. doi: 10.1101/gad.1872909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive V, Bennett MJ, Walker JC, Ma C, Jiang I, Cordon-Cardo C, Li QJ, Lowe SW, Hannon GJ, He L. miR-19 is a key oncogenic component of mir-17–92. Genes Dev. 2009;23:2839–2849. doi: 10.1101/gad.1861409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- Omori SA, Cato MH, Anzelon-Mills A, Puri KD, Shapiro-Shelef M, Calame K, Rickert RC. Regulation of class-switch recombination and plasma cell differentiation by phosphatidylinositol 3-kinase signaling. Immunity. 2006;25:545–557. doi: 10.1016/j.immuni.2006.08.015. [DOI] [PubMed] [Google Scholar]
- Park SS, Kim JS, Tessarollo L, Owens JD, Peng L, Han SS, Tae Chung S, Torrey TA, Cheung WC, Polakiewicz RD, et al. Insertion of c-Myc into Igh induces B-cell and plasma-cell neoplasms in mice. Cancer Res. 2005;65:1306–1315. doi: 10.1158/0008-5472.CAN-04-0268. [DOI] [PubMed] [Google Scholar]
- Peled JU, Yu JJ, Venkatesh J, Bi E, Ding BB, Krupski-Downs M, Shaknovich R, Sicinski P, Diamond B, Scharff MD, Ye BH. Requirement for cyclin D3 in germinal center formation and function. Cell Res. 2010;20:631–646. doi: 10.1038/cr.2010.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radziszewska A, Choi D, Nguyen KT, Schroer SA, Tajmir P, Wang L, Suzuki A, Mak TW, Evan GI, Woo M. PTEN deletion and concomitant c-Myc activation do not lead to tumor formation in pancreatic beta cells. J. Biol. Chem. 2009;284:2917–2922. doi: 10.1074/jbc.M805183200. [DOI] [PubMed] [Google Scholar]
- Refaeli Y, Young RM, Turner BC, Duda J, Field KA, Bishop JM. The B cell antigen receptor and overexpression of MYC can cooperate in the genesis of B cell lymphomas. PLoS Biol. 2008;6:e152. doi: 10.1371/journal.pbio.0060152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Beato M, Sáez AI, Navas IC, Algara P, Sol Mateo M, Villuendas R, Camacho F, Sánchez-Aguilera A, Sánchez E, Piris MA. Overall survival in aggressive B-cell lymphomas is dependent on the accumulation of alterations in p53, p16, and p27. Am. J. Pathol. 2001;159:205–213. doi: 10.1016/S0002-9440(10)61686-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y, Derudder E, Hobeika E, Pelanda R, Reth M, Rajewsky K, Schmidt-Supprian M. Canonical NF-kappaB activity, dispensable for B cell development, replaces BAFF-receptor signals and promotes B cell proliferation upon activation. Immunity. 2006;24:729–739. doi: 10.1016/j.immuni.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Schmitz R, Young RM, Ceribelli M, Jhavar S, Xiao W, Zhang M, Wright G, Schaffer AL, Hodson DJ, Buras E, et al. Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature. 2012 doi: 10.1038/nature11378. http://dx.doi.org/10.1038/nature11378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholtysik R, Nagel I, Kreuz M, Vater I, Giefing M, Schwaenen C, Wessendorf S, Trümper L, Loeffler M, Siebert R, Küppers R. Recurrent deletions of the TNFSF7 and TNFSF9 genes in 19p13.3 in diffuse large B-cell and Burkitt lymphomas. Int. J. Cancer. 2012;131:E830–E835. doi: 10.1002/ijc.27416. [DOI] [PubMed] [Google Scholar]
- Shinkai Y, Rathbun G, Lam KP, Oltz EM, Stewart V, Mendelsohn M, Charron J, Datta M, Young F, Stall AM, et al. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- Srinivasan L, Sasaki Y, Calado DP, Zhang B, Paik JH, DePinho RA, Kutok JL, Kearney JF, Otipoby KL, Rajewsky K. PI3 kinase signals BCR-dependent mature B cell survival. Cell. 2009;139:573–586. doi: 10.1016/j.cell.2009.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Blenis J, Yuan J. Activation of PI3K/Akt and MAPK pathways regulates Myc-mediated transcription by phosphorylating and promoting the degradation of Mad1. Proc. Natl. Acad. Sci. USA. 2008;105:6584–6589. doi: 10.1073/pnas.0802785105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.