Bortezomib (PS-341; Velcade) is a reversible inhibitor of the 26S proteasome that produces significant clinical responses in both newly diagnosed and advanced multiple myeloma (MM). However, only 40% of patients respond to bortezomib as a single-agent therapy, and most of those will become resistant. Consequently, understanding the mechanism of action of bortezomib is of great interest. Although the inhibitory activity of bortezomib is clearly defined, the downstream mechanisms of cytotoxity remain poorly understood and at times controversial.1 There is now emerging evidence that microRNAs (miRNAs) are important factors in anticancer drug activity and resistance.2 We therefore decided to investigate the potential involvement of miRNAs in the action of bortezomib on MM cells.
Three well-characterised MM cell lines (RPMI-8226, JJN3 and Thiel) were treated with bortezomib for 72 h (10 nℳ, LC laboratories, Woburn, MA, USA). Compared with diluent-only controls (0.008% dimethyl sulfoxide), levels of apoptosis (Cell Death Detection ELISAplus kit; Roche Diagnostics, Lewes, UK) in treated cell lines showed on average (n=3) an increase of 12.5- (JJN-3), 5.2- (Thiel) and 3.8-fold (RPMI-8226). Furthermore, cell proliferation (CellTiter 96 Aqueous One Solution Cell Proliferation Assay; Promega, WN, USA) levels were decreased 10.5- (JJN-3), 33.3- (Thiel) and 8.4-fold (RPMI-8226) (data not shown).
Total RNA was extracted by Trizol (Invitrogen, Paisley, UK) from biological triplicate experiments and the RNA used for whole transcriptome (Affymetrix U133plus2.0) and miRnome (Affymetrix Genechip miRNA v.1.0) microarray analyses (Affymetix, Santa Clara, CA, USA). Differences in cell line identity followed by bortezomib treatment were discriminatory features for both mRNA and miRNA expression levels as demonstrated by unsupervised cluster analyses (Supplementary Figure S1). Each treated sample was normalised against its' respective paired non-treated sample, and probes (mRNA or miRNA) filtered for a median±fold-change of >2 between triplicates. This resulted in 1516 and 4136 genes that were up- and downregulated, respectively, in JJN3 cells in response to bortezomib treatment; 824 and 1864 genes in RPMI-8226 cells; and 1292 and 5183 genes in Thiel cells (Supplementary Figure S2). There were 228 and 883 genes commonly upregulated and downregulated respectively in all three MM cell lines in response to treatment. Ontogeny analysis revealed that members of the protein ubiquitination canonical pathway were significantly enriched in the genes that were differentially expressed in response to bortezomib in each of the three MM cell lines individually, as well as for those genes that were commonly deregulated in all three lines (Supplementary Table S1). Interestingly, the most significant canonical pathway of differentially expressed genes in two of the cell lines (Thiel and JJN3) was the ‘role of BRCA1 in DNA damage response' recently it has been suggested that bortezomib induces ‘BRCAness' in MM cells.3
Many of the most deregulated genes that were common to all three cell lines (Supplementary Tables S2 and S3) had previously been associated with bortezomib treatment including upregulation of heat-shock proteins (HSP6, HSPA1), AP-1 complex (Jun), cell stress markers (ATF3),4 redox haemostasis genes (HMOX1, DDIT3),5 and the antiapoptotic protein BAG3,6 and downregulation of survivin (BIRC5), topoisomerase and MYB-oncogene.7
As with the gene expression data the miRNA microarray data were normalised between paired samples. This resulted in 37 and 75 miRNAs that were up- and downregulated, respectively, in JJN3 cells (median fold-change >2); 23 and 26 miRNAs in RPMI-8226 cells; and 30 and 98 miRNAs in Thiel cells (Supplementary Figure S3). Nine and 14 miRNAs were commonly upregulated and downregulated, respectively, in all three MM cell lines (Supplementary Figure S3 and Supplementary Table S4). Four of these miRNAs (miR-92a-5p, let-7f, miR-27a-5p and miR-188 (Xp11.23)) were validated by qualitative reverse transcriptase-PCR (Supplementary Figure S4). Interestingly, the most downregulated miRNA was mir-92a-5p, which along with miR-18a (also down-regulated), are encoded by the miR-17∼92 cluster whose overexpression has been linked to tumorigenesis and poor prognosis in MM.8 Let-7f has been demonstrated to promote angiogenesis by targeting antiangiogenic genes.9 Administration of anti-Let-7f was found to increase apoptosis and decrease cell proliferation levels in MM cell lines and to significantly reduce size of tumours in an MM xenotransplant model.10
Four of the fourteen (29%) commonly downregulated miRNAs are encoded within a single cluster (Xp11.23) that contains a further four members, also downregulated in response to treatment (data not shown). The Xp11.23 cluster is encoded within intron 3 of the CLCN5 gene. Intriguingly, mutations in CLCN5 (including mutations involving intron 3) are frequently found in patients with Dent's disease, a rare renal tubular disorder characterised by progressive renal failure, that share many clinical manifestations with MM including presentation of Fanconi syndrome.11
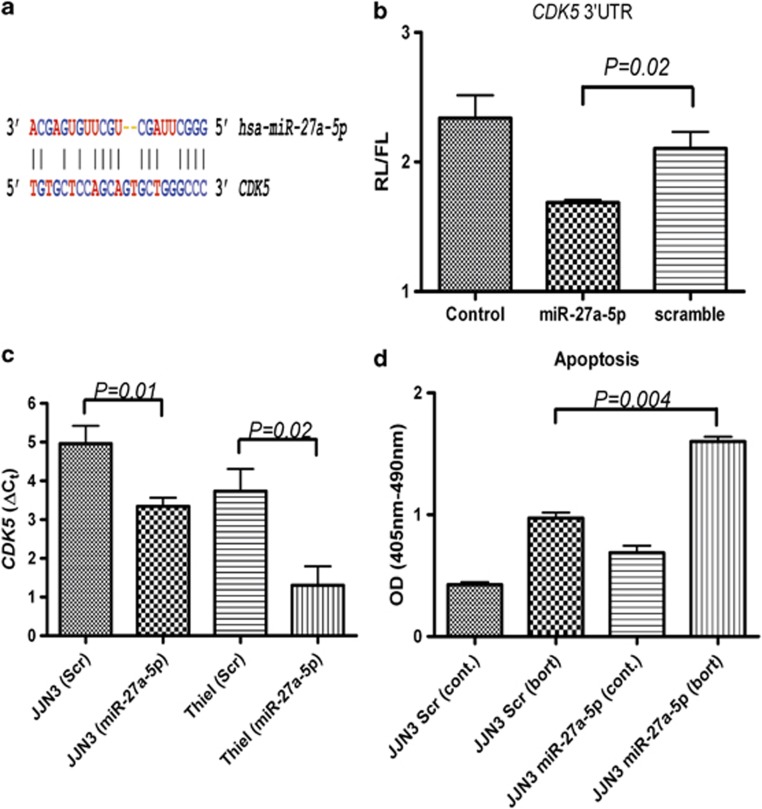
Recently cyclin-depedent kinase 5 (CDK5) was identified as being a major modulator of bortezomib sensitivity in both MM cell lines and patient tumour cells.12 We noted that miR-27a-5p, one of the miRNAs identified as being downregulated by all three MM cell lines in response to bortezomib treatment (Supplementary Table S4 and Supplementary Figure S4), was predicted to target CDK5 (Figure 1a). This putative interaction was tested by cloning the 3′-untranslated region sequence of CDK5 into a luciferase reporter vector. Transfection of miR-27a-5p significantly reduced luciferase output compared with a scrambled sequence (Figure 1b). Furthermore, we found that endogenous levels of CDK5 mRNA decreased significantly in MM cells transfected with miR-27a-5p (Figure 1c), but did not find a concordant change in CDK5 protein levels (data not shown). JJN3 cells transfected with miR-27a-5p appeared to have increased sensitivity to bortezomib activity as demonstrated by an increase in apoptosis levels (Figure 1d), although little or no difference was detected in proliferation levels. These findings are consistent with previous observations that downregulation of CDK5 (via siRNAs) increased the sensitivity of MM cell lines (and patient tumour cells) to bortezomib treatment.12 Consequently, it is tempting to suggest that miR-27a levels could be associated with the bortezomib-refractory status of MM patients; this is an area we are currently pursuing. Consistent with this hypothesis it has been observed that miR-27a levels are downregulated in bortezomib-resistant MM cell lines,13 and more generally downregulation of miR-27a has been associated with chemotherapy resistance.14
Figure 1.
miR-27a-5p regulates levels of CDK5 and increases sensitivity of MM cells to bortezomib. (a) Predicted binding site of miR-27a-5p within the 3′-untranslated region of CDK5 gene (source MicroCosm target database (http://www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5/)). (b) miR-27a-5p inhibits in vitro translation of luciferase reporter gene containing 3′-untranslated region of CDK5 gene. A luciferase reporter vector (HmiT000261-MT01) was constructed containing the 3′-untranslated region sequence of the CDK5 gene (197 bp) immediately downstream of a firefly luciferase reporter gene (Genecopeia; Rockville, MD, USA). HeLa cells were cotransfected with 2.5 μg of plasmid and either 20 nℳ of miR-27a-5p or a scrambled negative control (miRIDIAN mimics, Dharmacon, Lafayette, CO, USA). Cells were harvested 48 h post transfection and firefly luminescence and internal control Renilla luminescence measured using Luc-Pair luciferase assay (Genecopeia) according to the manufacturer's instructions and read using an Appliskan plate reader (Thermo Scientific, Waltham, MA, USA). Experiments were carried out in biological triplicate and P-values calculated by Mann–Whitney independent t-test (Graphpad Prism v.4, La Jolla, CA, USA). (c) miR-27a-5p inhibits endogenous levels of CDK5 mRNA in MM cells. JJN3 and Thiel cell lines were transfected with either 20 nℳ of miR-27a-5p or a scrambled negative control (miRIDIAN mimics, Dharmacon). Cells were harvested 72 h post transfection and total RNA purified by Trizol according to the manufacturer's instructions (Invitrogen). Levels of B2M (control gene) and CDK5 gene were measured in triplicate using Taqman probes to carry out quantitative reverse transcriptase-PCR as described by the manufacturer's (Applied biosytems, Carlsbad, CA, USA). CDK5 levels were not affected by bortezomib treatment (data not shown). (d) miR-27a-5p increases levels of apoptosis induced by bortezomib in MM cells. The JJN3 cell line was transfected with either 20 nℳ of miR-27a-5p or a scrambled negative control. After 48 h, either bortezomib 10 nℳ or diluent-only (0.008% dimethyl sulfoxide) was added to cells, and 24 h later levels of apoptosis were measured using the Cell Death Detection ELISAplus kit (Roche Diagnostics) according to the manufacturer's instructions. Experiments were carried out in triplicate. Cell proliferation levels were not found to vary significantly in response to miR-27a-5p transfection (data not shown).
In order to ascertain whether deregulation of miRNAs by bortezomib could explain the pattern of differential gene expression observed, we used the TargetScan predictive algorithm15 (as implemented in Ingenuity Pathway Analysis software suite (Ingenuity, Redwood City, CA, USA)) to identify putative target genes that were also deregulated >2-fold in the opposite sense to their respective miRNA regulator. Of the Affymetrix miRNA v.1.0 human probe set (n=1101), 393 miRNAs are included in the Targetscan database, along with 16 245 genes of the Affymetrix U133plus2.0 probe set (n=42661). In JJN3 cells, 96% (67/70 deregulated miRNAs in Targetscan) of miRNAs were predicted to target 1 or more of 2558 genes (51% of 5010 deregulated genes in database); RPMI-8226 cells, 100% (32/32) miRNAs were predicted to target 1 or more of 943 genes (38% of 2455 deregulated genes); and for Thiel cells 94% (78/79) were predicted to target 1 or more of 2447 (42% of 5764 deregulated genes). Of the 12 miRNAs in the Targetscan database that were commonly differentially expressed by all three cell lines in response to bortezomib treatment, all were predicted to target 1 or more of 302 genes (30% of 1000 deregulated genes).
Surprisingly, ontogeny analysis of these potentially miRNA-regulated genes once again showed a significant enrichment for genes that form part of the protein ubiquitination pathway (Supplementary Table S5). These results suggest that in addition to targeting the proteasome directly, bortezomib alters the expression of specific miRNAs that target downstream components of the protein ubiquitination pathway presumably acting in a synergistic fashion. How the miRNAs are themselves deregulated by bortezomib remains unclear, but nevertheless represents a hitherto unknown function for miRNAs and provides additional information, and targets for improving the efficacy of this promising treatment for MM patients.
Acknowledgments
This study was supported by funding from the Starmer-Smith memorial lymphoma fund, Leukaemia and Lymphoma Research and IKERBASQUE, Basque Foundation for Science.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Blood Cancer Journal website (http://www.nature.com/bcj)
Supplementary Material
References
- Hideshima T, Ikeda H, Chauhan D, Okawa Y, Raje N, Podar K, et al. Bortezomib induces canonical nuclear factor-kappaB activation in multiple myeloma cells. Blood. 2009;114:1046–1052. doi: 10.1182/blood-2009-01-199604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng T, Wang J, Chen X, Liu L. Role of microRNA in anticancer drug resistance. Int J Cancer. 2010;126:2–10. doi: 10.1002/ijc.24782. [DOI] [PubMed] [Google Scholar]
- Neri P, Ren L, Gratton K, Stebner E, Johnson J, Klimowicz A, et al. Bortezomib-induced ‘BRCAness' sensitizes multiple myeloma cells to PARP inhibitors. Blood. 2011;118:6368–6379. doi: 10.1182/blood-2011-06-363911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shringarpure R, Catley L, Bhole D, Burger R, Podar K, Tai YT, et al. Gene expression analysis of B-lymphoma cells resistant and sensitive to bortezomib. Br J Haematol. 2006;134:145–156. doi: 10.1111/j.1365-2141.2006.06132.x. [DOI] [PubMed] [Google Scholar]
- Nerini-Molteni S, Ferrarini M, Cozza S, Caligaris-Cappio F, Sitia R. Redox homeostasis modulates the sensitivity of myeloma cells to bortezomib. Br J Haematol. 2008;141:494–503. doi: 10.1111/j.1365-2141.2008.07066.x. [DOI] [PubMed] [Google Scholar]
- Liu P, Xu B, Li J, Lu H. BAG3 gene silencing sensitizes leukemic cells to Bortezomib-induced apoptosis. FEBS Lett. 2009;583:401–406. doi: 10.1016/j.febslet.2008.12.032. [DOI] [PubMed] [Google Scholar]
- Ling X, Calinski D, Chanan-Khan AA, Zhou M, Li F. Cancer cell sensitivity to bortezomib is associated with survivin expression and p53 status but not cancer cell types. J Exp Clin Cancer Res. 2010;29:8. doi: 10.1186/1756-9966-29-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Li C, Zhang R, Gao X, Qu X, Zhao M, et al. miR-17-92 cluster microRNAs confers tumorigenicity in multiple myeloma. Cancer Lett. 2011;309:62–70. doi: 10.1016/j.canlet.2011.05.017. [DOI] [PubMed] [Google Scholar]
- Kuehbacher A, Urbich C, Zeiher AM, Dimmeler S. Role of Dicer and Drosha for endothelial microRNA expression and angiogenesis. Circ Res. 2007;101:59–68. doi: 10.1161/CIRCRESAHA.107.153916. [DOI] [PubMed] [Google Scholar]
- Adamia S, Fulciniti M, Avet-Loiseau H, Amin SB, Shah P, Carrasco DR, et al. Biological and therapeutic potential of Mir-155, 585 and Let-7f in myeloma in vitro and in vivo [abstract] Blood. 2009;114:883. [Google Scholar]
- Devuyst O, Thakker RV. Dent's disease. Orphanet J Rare Dis. 2010;5:28. doi: 10.1186/1750-1172-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu YX, Tiedemann R, Shi CX, Yin H, Schmidt JE, Bruins LA, et al. RNAi screen of the druggable genome identifies modulators of proteasome inhibitor sensitivity in myeloma including CDK5. Blood. 2011;117:3847–3857. doi: 10.1182/blood-2010-08-304022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neri P, Gratton K, Ren L, Mansoor A, Duggan P, Stewart DA, et al. miRNA expression in multiple myeloma as predictive model of response to bortezomib [abstract] Blood. 2009;114:4918. [Google Scholar]
- Feng DD, Zhang H, Zhang P, Zheng YS, Zhang XJ, Han BW, et al. Down-regulated miR-331-5p and miR-27a are associated with chemotherapy resistance and relapse in leukaemia. J Cell Mol Med. 2011;15:2164–2175. doi: 10.1111/j.1582-4934.2010.01213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.