Abstract
The intestinal microflora, typically equated with bacteria, influences diseases such as obesity and inflammatory bowel disease (IBD). Here we show that the mammalian gut contains a rich fungal community that interacts with the immune system through the innate immune receptor Dectin-1. Mice lacking Dectin-1 exhibited increased susceptibility t chemically-induced colitis, which was the result of altered responses to indigenous fungi. In humans we identified a polymorphism in the gene for Dectin-1 (CLEC7A) that is strongly linked to a severe form of ulcerative colitis. Together our findings reveal a novel eukaryotic fungal community in the gut (the “mycobiome”) that coexists with bacteria and substantially expands the repertoire of organisms interacting with the intestinal immune system to influence health and disease.
Interactions between the commensal microflora and the gut immune system are critical for establishing a balance between immunity and tissue health. Changes in gut bacteria described as “dysbiosis” have been associated with intestinal inflammation (1-3) and metabolic syndrome (4-6). The vast majority of studies on commensal microbiota have focused on gut bacteria, and the terms “intestinal microbiota” and “intestinal bacteria” are often used interchangeably. Recent studies have begun to note, however, that a fraction of gut microorganisms are not bacterial (7). Although a few studies have suggested the presence of commensal fungi in the gut (8, 9), whether they interact with the mucosal immune system or influence diseases is unknown.
Fungi are recognized by a number of immune receptors among which Dectin-1 has emerged as key for phagocytosis and killing by myeloid phagocytes. Dectin-1 is a C-type lectin receptor that recognizes β-1,3-glucans found in the cell walls of nearly all fungi. Dectin-1 activates intracellular signals through CARD9 leading to inflammatory cytokine production and induction of T helper 17 (Th17) immune responses (10-13). Deficiencies in either Dectin-1 or CARD9 result in enhanced susceptibility to pathogenic fungal infections in humans and mice (14-16). Polymorphic variants in the gene for CARD9 are strongly associated with Crohn’s disease and ulcerative colitis in humans (17, 18). Furthermore, anti-Saccharomyces cerevisiae antibodies (ASCA) against yeast mannan have been strongly associated with Crohn’s disease (19, 20). Together, these later findings suggest a possible link between immune responses to commensal fungi and intestinal disease.
We examined fungal distribution and detected fungal rDNA throughout the murine gastrointestinal tract with highest densities in the terminal colon of C57BL/6 (Fig. 1A) and 129S2/Sv (fig. S1A) mice. We stained colonic tissue sections and observed that fungi are abundant and in close proximity with commensal bacteria (Fig. 1B, fig. S1B, S2-S4). Furthermore, we found that a soluble Dectin-1 probe (21) binds to 5 to 7% of the fecal material consisting of fungal cells with various morphologies (Fig. 1C and fig. S5). Fungi were also present in rat, guinea pig, rabbit, pig, dog, and human feces (fig. S1C). Together the data demonstrate that commensal fungi contribute to the intestinal microbial community in many species.
Figure 1. Commensal fungi are present in the intestine and are recognized by Dectin-1.
(A) Prevalence of fungi in mucosa isolated from ileum, caecum, proximal (prox) and distal (dist) colon of C57BL/6J mice. ITS1-2 rDNA level was analyzed by qPCR and normalized to β-actin DNA. (B) Visualization of commensal fungi in the intestine. Colon sections were stained with a soluble Dectin-1 probe (sDEC-1) and counterstained with DAPI. The DAPI signal has been amplified in lower panels (B) to show that DAPI-stained bacteria and fungi are in close proximity to each other. (C) Intestinal fungi are recognized by Dectin-1. Fecal pellets were homogenized and labeled with sDEC-1 in presence (gray histogram) or absence (black histogram) of laminarin (a soluble β-glucan) to block specific binding. Binding was assessed by flow cytometry (left panels). Dectin-1-binding fungi were sorted (right panels) and visualized by confocal microscopy. (D) ASCA generation after DSS colitis. Mice were exposed twice to 2.5% DSS-supplemented water for 7 days each separated by two weeks of recovery. Serum samples were collected before DSS treatment (day 0) and 2 weeks after the last DSS cycle (42 days total) and ASCA IgM and IgG were measured by ELISA. Each symbol represents a mouse, all error bars indicate the s.d. *P < 0.05; unpaired t test. All data are representative of at least two independent experiments with similar results.
We next examined whether gut fungi can be detected by the immune system upon intestinal insult. We utilized a mouse model of dextran sodium sulfate (DSS)-induced colitis extended to allow antibody responses to develop. We found that DSS-induced intestinal inflammation led to the development of circulating IgM and IgG antibodies to fungi (ASCA) (Fig. 1D), suggesting that fungal antigens indigenous to the gut might be responsible for the induction of ASCA during colitis.
Since we found that gut commensal fungi are recognized by Dectin-1, we tested whether Dectin-1-deficient mice (Clec7a−/−) are susceptible to DSS-induced colitis. Clec7a−/− mice experienced increased weight loss (Fig. 2A) and displayed altered histology characterized by increased mucosal erosion, crypt destruction, inflammatory cell infiltration, and TNF-α production in the colon (Fig. 2B-D) as compared to their wild type (WT) littermate controls. We further detected augmented production of interferon (IFN)-γ and interleukin (IL)-17 in intestines from Clec7a−/− mice (fig. S6). Similar results were obtained comparing co-housed animals (fig. S7). These results indicate that Dectin-1 deficiency leads to increased susceptibility to colitis.
Figure 2. Dectin-1 regulates severity of colitis.
Wild type (WT) and Clec7a−/− littermates were treated with 2.5% DSS for 7 days and kept on water for 4 additional days. Colitis progress and severity were assessed by measuring body weight during treatment (A) and histology (B, C), and TNF-α production in the colon (D) on day 11. (E, F) WT and Clec7a−/− mice were given an antibiotic cocktail including fluconazole for 3 weeks, transplanted as indicated (red) with fecal microflora from WT or Clec7a−/− mice and treated with DSS as in (A). Disease severity was accessed by histology score (E) and by cytokine production by anti-CD3/anti-CD28 stimulated LI-LP and MLN T cells (F). Each symbol represents a different mouse. One of four independent experiments is shown. Error bars, s.d., * P < 0.05, ** P < 0.01.
Because many studies have documented the importance of bacteria in intestinal inflammation, we examined whether bacteria could contribute to the susceptible phenotype. We observed no significant differences in major phyla of commensal bacteria between WT and Clec7a−/− mice (fig. S8). To directly determine whether microflora can transfer disease, we depleted intestinal bacteria and fungi with antibiotics, transplanted with fecal microflora from WT or Clec7a−/− mice, and exposed mice to DSS. Microflora from Clec7a−/− mice did not transfer susceptibility to disease (Fig. 2E, F, and fig. S9, S10). The data demonstrate that the disease phenotype in the Clec7a−/− mice is affected by the genotype of the mouse, not by initial differences in microflora.
We know very little about what commensal fungi populate the murine gut or how they contribute to colitis in Dectin-1 deficiency. To define the mouse intestinal fungal microbiome we isolated DNA from murine feces, amplified the internal transcribed spacer region (ITS1-2) of fungal rDNA, and performed high-throughput sequencing (22). Combining data from 23 mice, we obtained over 30 Mb of raw data from 454 pyrosequencing and over 2.2 GB of raw data from Illumina GA sequencing together containing over 1.3 million individual sequences which passed quality control. Detailed analysis identified over 100 different well-annotated fungal species representing at least 50 genera illustrating the fungal diversity. In addition, we identified over 100 novel/unannotated fungi representing the large uncharacterized nature of the mycobiome in the gut (fig. S11, S12). Interestingly, 97.3% of all the fungal sequences identified belonged to 10 fungal species with 65.2% of the sequences belonging to a single fungus: Candida tropicalis (Fig. 3A, and fig. S13). We found 7 of the 20 most common gut fungi also in mouse food (fig. S13, S14). These accounted, however, for only 1.5% of total fungi in the intestines, suggesting that highly represented fungal species are indigenous to the gut.
Figure 3. Defining the fungal microbiome and characterizing the specific role of Dectin-1-mediated host defense during colitis.
(A) DNA was isolated from murine feces and mycobiome analysis was performed using Roche 454 and Illumina GA sequencing of ITS1-2 rDNA. The taxonomic distribution of the most abundant fungal genera is shown (large pie chart), and species breakdown for major groups are provided (small pie charts). (B) Quantitative analysis of the major intestinal fungal genera in wild type and Clec7a−/− mice before and after treatment with DSS. Illumina GA data were analyzed and presented as relative percentage of dominant fungal genera (n=16 mice). (C) Fungal invasion of colonic tissue in Clec7a−/− mice during colitis. Colon sections from WT and Clec7a−/− mice before and after colitis were stained with the sDEC-1 probe and counterstained with DAPI. (D) Intestinally conditioned dendritic cells were incubated with live C. tropicalis and killing was assessed after 6 and 18 hours. (E) Histology score of WT and Clec7a−/− mice supplemented or not with four doses of C. tropicalis or S. fibuligera every other day, and then treated with 2.5% DSS for 7 days and kept on water for 4 additional days. Data are representative of at least two independent experiments with similar results. Error bars, s.d., * P < 0.05, ** P < 0.01.
Many studies have shown that intestinal inflammation can lead to changes in commensal bacteria that affect the host (1, 2, 23). Whether colitis affects the makeup of the commensal mycobiome is unknown. One study has reported increased fungal burden in intestines of Crohn’s Disease patients (9), and another has shown increased colonization with exogenously added C. albicans during DSS colitis in mice (24). Notably, we found that during colitis in Clec7a−/− mice the proportion of opportunistic pathogenic fungi including Candida and Trichosporon increases whereas non-pathogenic Saccharomyces decreases (Fig. 3B and fig. S15). Examination of colons revealed that fungi invade inflamed tissues in DSS-treated Clec7a−/− mice but remain in the lumen of DSS-treated WT mice (Fig. 3C and fig. S16). These data are consistent with the observation that intestinally conditioned Clec7a−/− dendritic cells are less capable of killing C. tropicalis in vitro (Fig. 3D). Together the data suggest that Dectin-1 deficiency leads to altered immunity to commensal fungi in the gut.
Given that C. tropicalis is an opportunistic pathogen, we further analyzed its role during colitis. We supplemented mice with C. tropicalis and subjected them to DSS (See fig. S17A for dosing schedule). For comparison, another group of mice was supplemented with S. fibuligera, a non-pathogenic fungus that, like C. tropicalis, grows in yeast and filamentous forms and is recognized by Dectin-1 (fig. S18). Colitis symptoms including weight loss, crypt loss and inflammatory cell infiltration were more severe in Clec7a−/− mice supplemented with C. tropicalis compared to the Clec7a−/− controls (Fig. 3E and fig. S17B, C). In contrast, C. tropicalis supplementation did not aggravate colitis in WT mice. Consistent with the pathology, we detected increased IL-17 and IFN-γ production by T cells from the MLNs and colons of Clec7a−/− mice supplemented with C. tropicalis (fig. S17D, E) as well as increased message for TNF-α, IL-23p19, IL-17a, Cxcl2 and defensins in colons (fig. S19). This correlated with higher loads of C. tropicalis in the intestines of DSS-treated Clec7a−/− mice (fig. S20B). In contrast, S. fibuligera supplementation did not contribute to colitis pathology (Fig. 3E and fig. S17, S19), and fungal loads were unchanged (fig. S20C). The data suggest that an inability of Clec7a−/− mice to mount effective immune responses to specific intestinal fungi creates conditions that promote inflammation.
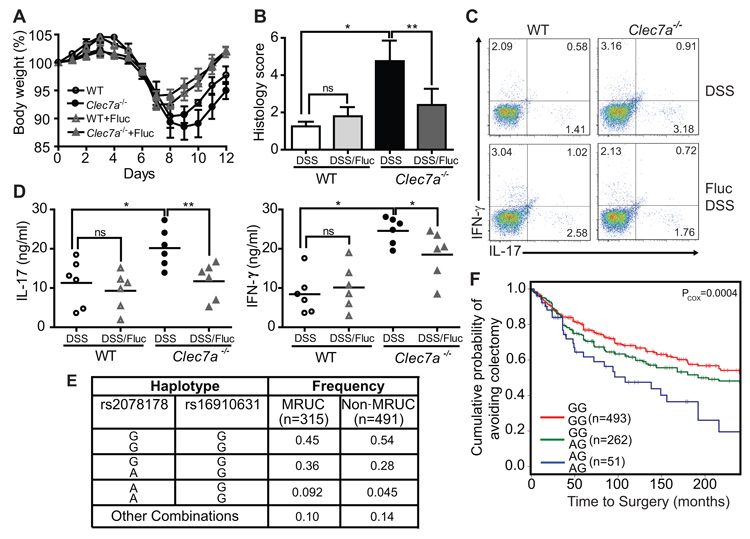
To determine whether the altered fungal burden during colitis contributes to disease severity in the absence of Dectin-1, we suppressed fungal growth with fluconazole, a specific anti-fungal drug (fig. S3). Fluconazole treatment during colitis led to reduced weight loss (Fig. 4A), and milder histological disease characteristics (Fig. 4B) specifically in Clec7a−/− mice. We similarly observed decreased Th1 and Th17 responses (Fig. 4C, D and fig. S21A, B) and decreased production of inflammatory cytokines (fig. S21C, D). Taken together, these results further support the conclusion that an inability to control fungi in the gut leads to more severe colitis in Dectin-1 knockout mice.
Figure 4. Anti-fungal therapy ameliorates colitis in Clec7a−/− mice and CLEC7A associates with ulcerative colitis severity in humans.
(A) WT and Clec7a−/− mice were given fluconazole in their drinking water for total of 14 days (starting 2 days prior the induction of DSS colitis), and body weight was measured. Weight loss is shown in (A) (p<0.05). Histology score (B), the percentage of IL-17 and IFN-γ producing CD4+ T cells in LI-LP (C), and IL-17 and IFN-γ production in MLNs (D) were determined 4 days after the 7 days of DSS treatment. Each symbol represents a different mouse. One of three independent experiments with similar results is shown. Error bars, s.d., * P < 0.05, ** P < 0.01. (E) Specific CLEC7A haplotypes associate with medically refractory ulcerative colitis (MRUC). Haplotypes were formed from rs2078178 and rs16910631 using PHASE v2.3. Haplotypes listed as “Other Combinations” were those that could not be reliably determined (posterior p<0.95). (F) The CLEC7A “AG/AG” haplotype associates with severity of disease as indicated by earlier progression to colectomy. Haplotypes were tested for association with time to surgery by fitting the MRUC/non-MRUC and time to surgery with a Cox proportional hazards model.
Having established a role for Dectin-1 in fungal control during colitis in mice, we next explored whether there is an association between inflammatory bowel disease and genetic variation of the human Dectin-1 gene (CLEC7A). Because the mouse model suggested that Dectin-1 is involved in the severity of colonic disease, we focused our human studies on ulcerative colitis (UC), a disease of the colon, and in particular on severe UC. Up to 30% of patients with UC require colectomy usually for severe disease that will not respond to medical therapy including systemic corticosteroids, cyclosporine and biological therapies (medically refractory UC (MRUC)). We compared CLEC7A alleles in an MRUC group to a group of patients with UC who had not required colectomy (non-MRUC) (25). We identified an association of CLEC7A SNP rs2078178 in patients with MRUC (logistic regression p=0.007). Notably, a two marker haplotype, rs2078178-rs16910631, was more strongly associated with MRUC (AG haplotype; p logistic regression = 0.00013/p fisher = 0.0005; Fig. 4E and fig. S22 and table S1), shorter time to surgery and thus with a more severe UC (Fig. 4F). Compared to healthy controls, the haplotype is strongly associated with MRUC and not with non-MRUC, further consistent with the idea that the haplotype is associated with severe disease (table S2). CLEC7A has not been identified in any GWAS study yet as an IBD susceptibility gene. Unlike susceptibility genes that predispose to disease, severity gene variants aggravate disease that is initially established through other mechanisms. The CLEC7A risk haplotype we report here fits this latter situation and agrees with our observation that Clec7a−/− mice do not develop spontaneous colitis. These findings suggest more in-depth studies on the role of the CLEC7A gene and pathway on the natural history of UC, and will require further validation in independent cohorts.
A deeper understanding of the mechanisms by which fungi stimulate inflammatory immune responses in the gut may lead to better therapies for IBD, and may be especially beneficial to patients with particularly severe forms of ulcerative colitis carrying the risk haplotype of the gene for Dectin-1. Overall, the idea that fungi are present in the gut and that they interact strongly with the immune system will fundamentally alter how we think about the gut microflora and inflammatory bowel diseases.
Supplementary Material
Acknowledgments
This study was supported in part by the NIH (D.M.U. – AI071116) and the Janis and William Wetsman Family Chair in Inflammatory Bowel Disease Research (D.M.U.). I.D.I. held a Research Fellowship Award from the Crohn’s and Colitis Foundation of America (#3064). Further support came from NIH/NIDDK grant P01-DK046763; CTSI Grant UL1RR033176; Cedars-Sinai Medical Center Inflammatory Bowel & Immunobiology Research Institute Funds; The Feintech Family Chair in IBD (S.R. Targan); The Cedars-Sinai Board of Governors’ Chair in Medical Genetics (J.I.R.); The Abe and Claire Levine Chair in Pediatric IBD (M.D.), Joshua L and Lisa Z. Greer Chair in IBD Genetics (D.P.B.M). G.D.B. is supported by the Wellcome Trust. Clec7a−/− mice are available through an MTA with the University of Aberdeen. The data presented in this paper are tabulated in the main paper and in the supplementary materials. All sequences generated in this study have been deposited in the NCBI Short Read Archive (http://www.ncbi.nlm.nih.gov/Traces/sra, Accession # SRA051853.1). G.D.B. is an advisory board member of MiniVax and Fiberbiotics. M.D. is a consultant for Prometheus Labs.
References
- 1.Lupp C, et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2:204. doi: 10.1016/j.chom.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 2.Willing BP, et al. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology. 2010;139:1844. doi: 10.1053/j.gastro.2010.08.049. [DOI] [PubMed] [Google Scholar]
- 3.Elinav E, et al. NLRP6 Inflammasome Regulates Colonic Microbial Ecology and Risk for Colitis. Cell. 2011 doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arumugam M, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henao-Mejia J, et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 2012;482:179. doi: 10.1038/nature10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vijay-Kumar M, et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scupham AJ, et al. Abundant and diverse fungal microbiota in the murine intestine. Appl Environ Microbiol. 2006;72:793. doi: 10.1128/AEM.72.1.793-801.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ott SJ, et al. Fungi and inflammatory bowel diseases: Alterations of composition and diversity. Scand J Gastroenterol. 2008;43:831. doi: 10.1080/00365520801935434. [DOI] [PubMed] [Google Scholar]
- 10.Cheng SC, et al. The dectin-1/inflammasome pathway is responsible for the induction of protective T-helper 17 responses that discriminate between yeasts and hyphae of Candida albicans. J Leukoc Biol. 2011;90:357. doi: 10.1189/jlb.1210702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gringhuis SI, et al. Dectin-1 is an extracellular pathogen sensor for the induction and processing of IL-1beta via a noncanonical caspase-8 inflammasome. Nat Immunol. 2012 doi: 10.1038/ni.2222. [DOI] [PubMed] [Google Scholar]
- 12.LeibundGut-Landmann S, et al. Syk- and CARD9-dependent coupling of innate immunity to the induction of T helper cells that produce interleukin 17. Nat Immunol. 2007;8:630. doi: 10.1038/ni1460. [DOI] [PubMed] [Google Scholar]
- 13.Conti HR, et al. Th17 cells and IL-17 receptor signaling are essential for mucosal host defense against oral candidiasis. J Exp Med. 2009;206:299. doi: 10.1084/jem.20081463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferwerda B, et al. Human dectin-1 deficiency and mucocutaneous fungal infections. N Engl J Med. 2009;361:1760. doi: 10.1056/NEJMoa0901053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glocker EO, et al. A homozygous CARD9 mutation in a family with susceptibility to fungal infections. N Engl J Med. 2009;361:1727. doi: 10.1056/NEJMoa0810719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taylor PR, et al. Dectin-1 is required for beta-glucan recognition and control of fungal infection. Nat Immunol. 2007;8:31. doi: 10.1038/ni1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franke A, et al. Genome-wide association study for ulcerative colitis identifies risk loci at 7q22 and 22q13 (IL17REL) Nat Genet. 2010;42:292. doi: 10.1038/ng.553. [DOI] [PubMed] [Google Scholar]
- 18.McGovern DP, et al. Genome-wide association identifies multiple ulcerative colitis susceptibility loci. Nat Genet. 2010;42:332. doi: 10.1038/ng.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seow CH, et al. Novel anti-glycan antibodies related to inflammatory bowel disease diagnosis and phenotype. Am J Gastroenterol. 2009;104:1426. doi: 10.1038/ajg.2009.79. [DOI] [PubMed] [Google Scholar]
- 20.Joossens S, et al. The value of serologic markers in indeterminate colitis: a prospective follow-up study. Gastroenterology. 2002;122:1242. doi: 10.1053/gast.2002.32980. [DOI] [PubMed] [Google Scholar]
- 21.Gantner BN, Simmons RM, Underhill DM. Dectin-1 mediates macrophage recognition of Candida albicans yeast but not filaments. EMBO J. 2005;24:1277. doi: 10.1038/sj.emboj.7600594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghannoum MA, et al. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog. 2010;6:e1000713. doi: 10.1371/journal.ppat.1000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garrett WS, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8:292. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jawhara S, et al. Colonization of mice by Candida albicans is promoted by chemically induced colitis and augments inflammatory responses through galectin-3. J Infect Dis. 2008;197:972. doi: 10.1086/528990. [DOI] [PubMed] [Google Scholar]
- 25.Haritunians T, et al. Genetic predictors of medically refractory ulcerative colitis. Inflamm Bowel Dis. 2010;16:1830. doi: 10.1002/ibd.21293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Landers CJ, et al. Selected loss of tolerance evidenced by Crohn’s disease-associated immune responses to auto- and microbial antigens. Gastroenterology. 2002;123:689. doi: 10.1053/gast.2002.35379. [DOI] [PubMed] [Google Scholar]
- 27.Iliev ID, Mileti E, Matteoli G, Chieppa M, Rescigno M. Intestinal epithelial cells promote colitis-protective regulatory T-cell differentiation through dendritic cell conditioning. Mucosal Immunol. 2009;2:340. doi: 10.1038/mi.2009.13. [DOI] [PubMed] [Google Scholar]
- 28.Kelsall B. Recent progress in understanding the phenotype and function of intestinal dendritic cells and macrophages. Mucosal Immunol. 2008;1:460. doi: 10.1038/mi.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matteoli G, et al. Gut CD103+ dendritic cells express indoleamine 2,3-dioxygenase which influences T regulatory/T effector cell balance and oral tolerance induction. Gut. 2010;59:595. doi: 10.1136/gut.2009.185108. [DOI] [PubMed] [Google Scholar]
- 30.Liu Z, DeSantis TZ, Andersen GL, Knight R. Accurate taxonomy assignments from 16S rRNA sequences produced by highly parallel pyrosequencers. Nucleic Acids Res. 2008;36:e120. doi: 10.1093/nar/gkn491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nilsson RH, Bok G, Ryberg M, Kristiansson E, Hallenberg N. A software pipeline for processing and identification of fungal ITS sequences. Source Code Biol Med. 2009;4:1. doi: 10.1186/1751-0473-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kent WJ. BLAT--the BLAST-like alignment tool. Genome Res. 2002;12:656. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chenna R, et al. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fried LP, et al. The Cardiovascular Health Study: design and rationale. Ann Epidemiol. 1991;1:263. doi: 10.1016/1047-2797(91)90005-w. [DOI] [PubMed] [Google Scholar]
- 37.Psaty BM, et al. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: Design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ Cardiovasc Genet. 2009;2:73. doi: 10.1161/CIRCGENETICS.108.829747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stephens M, Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73:1162. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.R. D. C. Team R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. 2005 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.