Abstract
Bioluminescence resonance energy transfer (BRET) has become a widely used technique to monitor protein-protein interactions. It involves resonance energy transfer between a bioluminescent donor and a fluorescent acceptor. Because the donor emirs photons intrinsically, fluorescence excitation is unnecessary. Therefore, BRET avoids some of the problems inherent in fluorescence resonance energy transfer (FRET) approaches, such as photobleaching, autofluorescence, and undesirable stimulation of photo-biological processes. In the past, BRET signals have generally been too dim to be effectively imaged. Newly available cameras that are more sensitive coupled to image splitter now enable BRET imaging in plant and mammalian cells and tissues In addition, new substrates and enhanced luciferases enable brighter signals that allow even subcellular BRET imaging. Here, we report methods for BRET imaging or (I) localization of COP1 dimerization in plant cells and tissues and (2) subcellular distributions of interactions of the CCAAT/Enhancer Binding Protein α (C/EBPα) in single mammalian cells. We also discuss methods for the correction of BRET images for tissues that absorb light of different spectra. This progress should catalyze further applications of BRET for imaging and high-throughput assays.
Keywords: Bioluminescence resonance energy transfer (BRET), imaging assay, protein-protein interaction, Renilla luciferase, plant and mammalian cells
1. Introduction
The complicated network of protein interactions is pivotal to cellular “machinery.” Identifying the partners with whom a protein associates is a critical step in the elucidation of underlying mechanisms of action. Various approaches have been used to analyze protein–protein interactions, including the yeast two-hybrid assay, fluorescence resonance energy transfer (FRET), bioluminescence resonance energy transfer (BRET), protein mass Spectrometry, and evanescent wave methods (1). FRET and BRET are based on nonradiative energy transfer between a donor and an acceptor. In the case of FRET, two fluorophores with appropriately overlapping emission/absorption spectra (the “donor” and the “acceptor”) can transfer excited-state energy from donor to acceptor if they are within ~50 Å of each other (2). The orientation of the donor and acceptor can significantly influence the magnitude of the resonance transfer, as has been dramatically shown in a recent study using BRET fusion proteins (3). In the case of BRET, the donor is a luciferase enzyme that directly emits photons so that fluorescence excitation is unnecessary. This luciferase-catalyzed luminescence utilizes a substrate and can excite an acceptor fluorophore by resonance energy transfer if the luciferase and fluorophore are in close proximity (within a radius of ~50 Å) and have a luminescence emission spectrum for the luciferase that appropriately overlaps the absorption spectrum of the fluorophore. If candidate interacting proteins are fused to the luminescent “donor” and fluorescent “acceptor” molecules, BRET can be used as a gauge of interaction between the candidate proteins (4).
The disadvantages of fluorescence excitation limit the potential applications of FRET. These disadvantages include photo-bleaching, autofluorescence, direct excitation of the acceptor fluorophore, photoresponsivity of specialized tissues (e.g., retina), and phototoxicity. Because BRET allows the detection of interactions between fusion proteins without direct excitation of the acceptor fluorophore; therefore, it can be used in applications where those potential disadvantages are problematic (5). We initially developed BRET to investigate the oligomerization of cir-cadian clock proteins from cyanobacteria (4). During the past 8 years, the applications of BRET have multiplied (6–10), including new methods of analysis of BRET signals (11, 12), In addition, BRET has recently been coupled with its progenitor technique of FRET for detecting interaction in multi-protein complexes (13). Therefore, BRET has become a widely used technique to identify and monitor protein-protein interactions. BRET is potentially superior to FRET for high-throughput screening (HTS) because luminescence-monitoring HTS instruments are simpler and less expensive if fluorescence excitation is not involved. Moreover, low-resolution BRET imaging has shown in whole-animal analyses that BRET is advantageous for deep penetration of animal tissues (10, 14).
Nevertheless, BRET has not been used for high-resolution imaging of cells and tissues for two major reasons. First, BRET signals are very dim and cannot be increased by “turning up” the excitation, as with FRET (5, 15). Second, a wide range of ancillary techniques has been developed for fluorescence (e.g., FRET, FLIM, etc.) and many laboratories are equipped with microscopic setups that are designed for fluorescence. As we show herein, however, (i) new generation cameras can now detect the dim BRET signals, and (ii) many existing microscopic setups that were designed for FRET could be easily adapted for BRET by simply optimizing photon throughput and using the new cameras. We coupled a sensitive EB-CCD camera with a Dual-View™ image splitter to image BRET signals in two challenging applications: (i) subcellular imaging in single mammalian cells and (ii) tissue and cellular imaging in highly autofluorescent plant material (15, 16). The BRET fusion partners we used were the CCAAT/Enhancer Binding Protein α (C/EBPα) in isolated mammalian cells and COP1 (a regulator of the light signaling pathway) in plant seedlings.
This is an exciting time to use BRET technology for studying protein interactions because the application of (i) improved detection devices and (ii) new substrates that allow BRET signals to be easily detected now extends considerably the range of experiments that are feasible. This chapter discusses the use of an EB-CCD camera and Dual-View™ image splitter in addition to the first utilization of the ViviRen™ substrate for enhancing the luminescence intensity. Another publication has used a different but related substrate (EnduRen™) that extends the lifetime of the luminescence signal so that BRET signals can be measured over a long time course (17). In addition, mutagen-esis of the luciferase from Renilla (RLUC) has resulted in new versions of RLUC that can be of potential benefit, both in terms of brighter signals and in terms of the spectra of luminescence output (14, 18, 19). Finally, the crystal structure of RLUC has been determined (20), and this information can be used to make fusion proteins with RLUC in which the candidate proteins are fused into internal loops of RLUC rather than being limited to merely N- or C-terminal fusions of RLUC to the candidate inter-actors. We hope that the recent advances in detection devices, luciferase substrates, and luciferase structure will stimulate further applications of BRET for imaging and high-throughput assays.
2. Materials
2.1. Plant and Mammalian Cell Lines
Arabidopsis seedlings (Arabidopsis thaliana, ecotype Col-0).
Arabidopsis cell suspension culture line (derived from A. thaliana seedlings, ecotype Col-0).
Tobacco seedlings (Nicotiana tabacum cv. Xanthi).
Human embryonic kidney 293 (HEK293) cell lines.
Mouse pituitary GHFT1 cell lines.
2.2. Plasmids, Vectors, and Strains
2.2.1. For Plants
35 S::Rluc (pBIN19 binary vector with kanamycin marker) (Promega), which contains the Renilla luciferase (RLUC) coding region under the control of the 35 S promoter.
35 S::Rluc-Eyfp (pBIN19 binary vector with kanamycin marker), which encodes a fusion protein of RLUC and EYFP (EYFP = enhanced yellow fluorescence protein, which is a red-shifted mutant of the Aequorea victoria green fluorescent protein).
35 S::Rluc-COP1(N) (pPZP222 binary vector with gen-tamycin marker), which encodes a fusion protein of COP1 to N-terminus of RLUC.
35 S::Eyfp-COP1(N) (pBIN19 binary vector with Kanamycin marker), which encodes a fusion protein of COP1 to N-terminus of EYFP.
Agrobacterium tumefaciens strain GV3101, which is used for transformation of tobacco and Arabidopsis suspension cell culture lines. Escherichia coli DH5α is the host cloning strain.
2.2.2. For Mammalian Cells
hRlucC1 (= codon “humanized” pRlucC1 from Perkin-Elmer).
Venus, which is a yellow fluorescence protein variant inserted in the PCS2 vector under the control of the CMV promoter.
C/EBPalpha244, which is a nuclear transcription factor inserted in the EYFPC1r vector (this vector includes EYFP, but we replaced it with Venus as described below).
pcDNA 3.1 + vector (Invitrogen).
E. coli DH5a is the host cloning strain.
2.3. Media, Buffers, and Reagents
Dulbecco's Modified Eagle's Medium (DMEM).
Dulbecco's Modified Eagle's phenol-red free Medium (DMEM).
Phosphate buffered saline (PBS).
Opti-MEM: a reduced serum medium.
Fetal bovine serum (FBS).
Solution of trypsin (0.25%) and ethylenediamine tetraacetic acid (EDTA) (1 mM).
Lipofectamine 2000.
LB medium: 10 g/l bacto-tryptone, 5 g/l bacto-yeast extract, 10 g/l NaCl (pH 7.0).
Plant seedling growth medium: 1/2 MS salts with vitamins, 30 g/l sucrose, pH 5.8. Solid medium is the same except including 8 g/l agar.
Arabidopsis calli induction medium: MS medium with 0.5 mg/l 2,4-D (2,4-dichloro-phenoxyacetic acid), 2.0 mg/l NAA (α-naphthaleneacetic acid), 0.5 mg/l 6-BAP (6–benzylamino-purine), pH 5.8.
BRET assay buffer for plant seedlings: 1/2 MS salts with 2.5 µM coelenterazine (see Note 1).
BRET assay buffer for Arabidopsis suspension cell cultures: MS salts, 30 g/l sucrose, with 2.5 µM coelenterazine.
Coelenterazine (Native): 100 µM stock solution preparations dissolved in 95% EtOH. Prepare a fresh working solution before each use. Proper preparation and handling of coelenterazine stock and working solutions is critical (see Note 1).
Deep Blue C™ (BioSignal/PE): 100 µM stock solution, store desiccated and protected from light in deep freeze (−70°C). Deep Blue C™ was prepared the same as for native coelenterazine.
ViviRen™ (Promega): diluted in DMSO as a 10 mM stock solution, protected from light and stored at −20°C (see Note 2). From this 10 mM stock solution, a working solution of 10 µM was prepared on the day of the experiment.
PBI 1419 (Promega): diluted in DMSO to a 10 mM stock solution and stored at −20°C. From this 10 mM stock solution, a working solution of 30 µM was prepared on the day of the experiment.
2.4. Apparatus
EB-CCD Camera, a modified electron bombardment-charge coupled device camera (Hamamatsu Photonic Systems, Bridgewater NJ, USA) (see Note 3).
Dual-View™ micro-imager (Optical Insights, Tucson AZ, USA) (see Note 4).
Inverted microscope (IX-71, Olympus America Inc., Melville NY, USA), with Macro XLFLuor 2× objective, NA 0.14 (Olympus), UPlanFl 40× objective, NA 1.30 (oil immersion, Olympus), or a Plan Apo 60× objective, NA 1.45 (oil immersion, Olympus & 1-U2B616) (see Note 5). For measurement of fluorescence, an epifluorescence attachment (EX 500/20 nm, EM 520LP) was connected to the IX-71 inverted microscope.
Temperature controlled (22–37°C) light-tight box (see Note 6).
Spectrofluorimeter: QuantaMaster QM-7/SE (Photon Technology International, Birmingham NJ, USA) (see Note 7).
FB12 Luminometer (Zylux Corp., Maryville, TN).
3. Methods
The BRET technique has been widely used for bulk measurement of protein interaction and various methods have been described for bacterial cultures (4, 21), plant tissue (6, 9), and mammalian cell suspensions (7, 10, 22, 23). Nevertheless, BRET imaging of single cells has lagged behind fluorescence imaging because of the dim BRET signals. On the other hand, BRET is preferable to FRET for high-throughput screening because of its ease of measurement, exquisite sensitivity, and independence from excitation. We describe here our application of a modified EB-CCD camera coupled with a microimager to image BRET signals at tissue, cellular, and even subcellular levels. We used BRET imaging to monitor the protein interaction of COP1 in plant seedlings; these plant tissues absorb light of different colors differentially, but quantitative measurements of BRET are allowed by a simple correction using RLUC-EYFP emission profiles. Moreover, we detected CCAAT/Enhancer Binding Protein α (C/EBPa) interactions in the nuclei of isolated mammalian cells.
3.1. Construction of BRET Vectors
3.1.1. Constructs Used for Plants
The following constructs were made to express the fusion proteins under the control of the 35 S promoter:
Positive control: P35 S::Rluc-Eyfp for expression of the fusion protein RLUC-EYFP.
Negative control: P35S::Rluc for expression of RLUC.
P35 S::Rluc-COP1(N) for expression of COP1 fusions to the N-terminus of RLUC.
P35 S::Eyfp-COP1(N) for expression of COP1 fusions to the N-terminus of EYFP.
3.1.2. Constructs Used for Mammalian Cells
The following constructs were made to express the fusion proteins under the control of the CMV promoter:
Positive control BRET construct PCMV::hRluc-Venus for expression of fusion protein hRLUC-Venus in cytoplasm (excluded from nucleus) (see Note 8 and Fig. 1.1f–j).
Negative-control BRET construct PCMV::hRluc.
PCMV::hRluc-C/EBP construct used for expression of the fusion protein hRLUC-C/EBP as a negative control for BRET in the nucleus (see Fig. 1.1z-c').
PCMV::Venus-C/EBP for expression of the fusion protein Venus-C/EBP in the nucleus (see Note 9 and Fig. 1.1u–y).
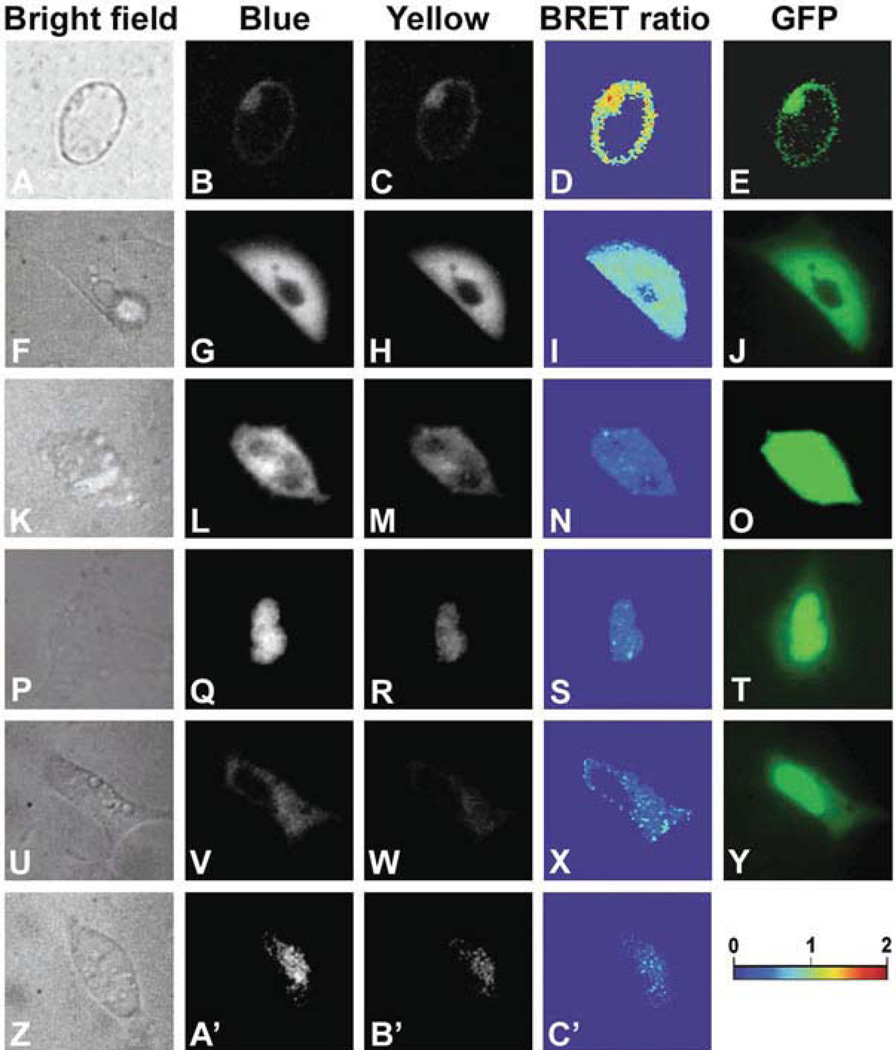
Fig. 1.1. Subcellular imaging of BRET in single plant and animal cells.
Arabidopsis suspension culture, HEK293, and mouse GHFT1 cells. a–e Isolated Arabidopsis cell from a suspension culture line that is expressing RLUC-EYFP. f–j HEK293 cells expressing hRLUC-Venus; Y ÷ B = 0.82 ± 0.07 SD over the luminescent portion of the cell. k–o HEK293 cell expressing unfused hRLUC and Venus; Y ÷ B = 0.36 over the luminescent portion of this cell. p–t Mouse GHFT1 cells expressing hRLUC-C/EBPα + Venus-C/EBPα; Y ÷ B = 0.39 ± 0.04 SD over the luminescent portion of the cell. u–y Mouse GHF1 cells expressing hRLUC + Venus-C/EBPα; Y ÷ B = 0.30 over the luminescent portion of this cell. z–c' Mouse GHF1 cells expressing hRLUC-C/EBPα; Y ÷ B = 0.24 ± 0.06 SD over the luminescent portion of the cell. Panels a, f, k, p, u, z are bright field, panels b, g, l, q, v, a' are Blue-range luminescence, panels c, h, m, r, w, b' are Yellow-range luminescence, panels d, i, n, s, x, c' are BRET ratios (Y ÷ B) over the entire images (pseudocolor scale shown below panel y), and panels e, j, o, t, y are fluorescence from EYFP or Venus in the fusion proteins. (Modified from Xu etal. (16).)
3.2. Transformation and Cotransfection of BRET Constructs
3.2.1. Induction of Arabidopsis Cell Suspension Culture Lines and Agrobacterium-Mediated Transformation
Prepare fresh Arabidopsis calli induction medium, 20-ml aliquot in a 100-ml flask.
Sterilize Arabidopsis seeds: sterilize seeds 30 s in 70% EtOH in a 1.5-ml microcentrifuge tube, followed by rinsing 3 times with sterile dH2O. Then add 20% Clorox bleach for 5 min, rinse 5 times with sterile dH2O.
Transfer sterilized seeds (~100) to a 100-ml volume flask containing 20 ml Arabidopsis calli induction medium, and place the flask on a shaker (120 rpm, 22°C, 12-h light/12-h dark cycle). Seeds will germinate in the liquid medium about 4 days later; calli and single cells will be induced and separated from the seedlings directly within 3–4 weeks (24). Subculture every 2 weeks by diluting and adding fresh induction medium (see Note 10).
Inoculate a single clone of Agrobacterium carrying the appropriate construct to 5 ml fresh LB medium with appropriate antibiotics and put on a rotating shaker (180 rpm, 28°C) overnight. Subculture once before transformation by transferring 100 µl to 10 ml fresh LB liquid medium and growing overnight.
Agrobacterium-mediated transformation: inoculate 2 ml Agrobacterium culture (OD600 ~ 1.0) to 20 ml of the cell suspension culture and co-culture for 48 h; rinse collected cells after a gentle centrifugation and replace medium with liquid calli induction medium that includes Cefotaxime (200 µg/ml, Sigma, to kill Agrobacterium) and culture for 48 h; after that, rinse the cells and add fresh medium that includes Cefotaxime and appropriate antibiotics (50 µg/ml Kanamycin or 100 µg/ml Gentamycin).
3.2.2. Agrobacterium-Mediated Tobacco Leaf Disk Double-Transformation
Take healthy fully expanded leaves from 3- to 4-week-old which grown in the sterilized MS medium since germination, cut into 0.25 cm2 leaf disks, and co-culture with the Agrobacterium suspension (see Section 3.2.1) for 10 min, then transfer to tobacco culture medium (MS medium with MES 0.59 g/l + NAA 0.1 mg/l + BAP 1 mg/l; 10 leaf disks per Petri dishes) for 2–3 days.
Rinse leaf disks three times with MS medium that includes Carbenicillin (Sigma; 500 mg/l, to kill Agrobacterium), transfer leaf disks to tobacco selective medium (MS medium with MES 0.59 g/l + NAA 0.1 mg/l + BAP 1 mg/l+Kanamycin 100 mg/l +Carbenicillin 500 mg/l), and subculture with fresh medium every 2 weeks.
Shoots should regenerate from the disks - if so, cut the regenerated young shoots from the basal disks, and transfer these shoots to 1/2 MS medium with selective antibiotics for rooting.
Prepare leaf disks from the regenerated and resistant seedlings for a second transformation with a second construct (if relevant), and use the same protocol as above, select then with two antibiotics (e.g., both Gentamycin and Kanamycin added to the selection medium).
3.2.3. Transient Transfection of Mammalian Cells
We have used human HEK293 and mouse pituitary GHFT1 cells for BRET detection. In general, HEK293 transfect well, but for studying the interactions of the C/EBP protein, we have used mouse GHFT1 cells because C/EBP takes up distinctive interaction patterns in the nuclei of GHFT1 cells.
Grow cells in DMEM medium supplemented with 10% FBS, 100 U/ml of penicillin, and 100 U/ml of streptomycin until they reach 80% confluence (see Note 11).
Dilute 4 µg of plasmid DNA in 250 µl OptiMEM medium without serum and mix gently.
Dilute 10 µl of Lipofectamine 2000 in 250 µl OptiMEM medium without serum. Mix gently and incubate for 5 min at room temperature (see Note 12).
Combine the diluted Lipofectamine 2000 with the diluted DNA and incubate for 20 min at room temperature (see Note 13).
Add the 500 µl of DNA-lipofectamine solution to cells in 35 mm dishes. Mix gently by rocking the dish back and forth.
Incubate the cells at 37°C in a CO2 incubator.
After 24–48 h, wash cells in PBS and replace the DMEM medium with phenol-red free DMEM supplemented with 10% FBS for imaging (phenol red absorbs light so it needs to be removed so that it does not absorb the luminescence signal).
3.3. Preparation of Plant Seedlings and Cell Suspension Lines
Sterilize Arabidopsisor tobacco seeds (see Section 3.2.1): for of Plant Seedlings tobacco seeds, 1 min for EtOH and 10 min for bleach.
Place seeds on plant seedling growth medium (1/2 MS + agar), and incubate in a 12-h light/12-h dark cycle, 120 µE m−2 s−1 (cool-white fluorescence lamps), 22°C.
5-day-old tobacco seedlings or 7-day-old Arabidopsis seedlings were used for BRET spectra acquisition and imaging analysis.
Newly grown Arabidopsis cell suspension lines (7 days after subculture, see Section 3.2.1 and Note 10) in appropriate selective liquid medium were used for single-cell imaging analysis.
3.4. BRET Luminescence Spectra Acquisition In Vivo
3.4.1. Plant Seedlings
Start spectrofluorimeter (QuantaMaster QM-7/SE) before luminescence measurement (see Note 7).
Transfer the transgenic seedlings (5–day-old Arabidopsis or 7-day-old tobacco) from solid medium directly to a 2-ml fluorescence cuvette containing 0.5 ml plant seedling BRET assay buffer without coelenterazine.
Add fresh coelenterazine to the assay buffer to a final concentration of 2.5 µM.
With the spectrofluorimeter's fluorescence excitation turned off, measure the luminescence emission spectrum between the wavelengths 440 and 580 nm in 2-nm steps with a 5-s integration for each step (see Note 14).
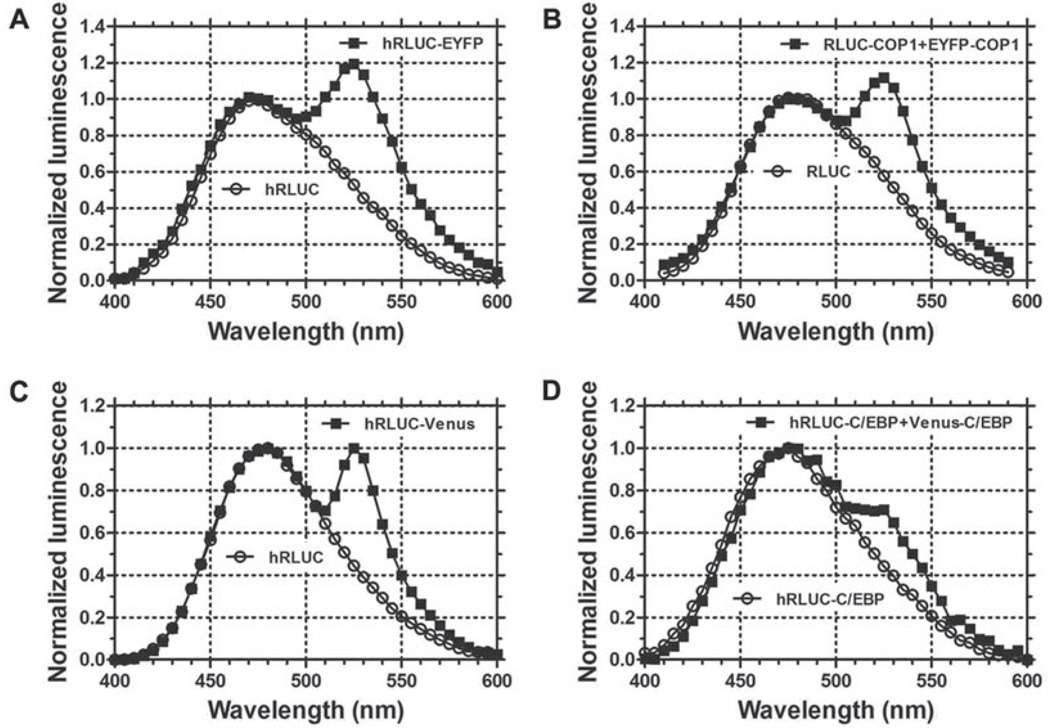
Analyze the BRET ratio by evaluating the emission spectra, especially the magnitude of the second peak at ~530 nm. Normalization of values is needed before analysis (see Section 3.8.2). Emission spectra of RLUC (negative control) and RLUC-EYFP (positive control) from whole Arabidopsis seedlings are shown in Fig. 1.2a. Spectra of RLUC-COP1 and RLUC-COP1 + EYFP-COP1 emission from whole Arabidopsis seedlings are shown in Fig. 1.2b.
Fig. 1.2. Luminescence emission spectra from whole plant seedlings and mammalian cells.
a The hRLUC vs. hRLUC-EYFP emission spectra from 7-day-old transgenic Arabidopsis seedlings. b The spectra of RLUC vs. RLUC-COP1+EYFP-COP1 emission from 5-day-old transgenic tobacco seedlings. c Luminescence spectra of hRLUC and hRLUC-Venus emission from HEK 293 cells. d The spectra of hRLUC-C/EBPα vs. hRLUC-C/EBPα + Venus-C/EBPα emission from single mouse GHFT1 cells. Luminescence spectra were normalized to the emission at 480 nm. (Modified from Xu et al. (16).)
3.4.2. Mammalian Cells
Wash cells twice with phosphate buffered saline (PBS).
Trypsinize the adherent cells with 0.05% trypsin-EDTA solution at 37°C for 5~10 min.
Add 900 µl of serum-containing DMEM to inactivate the trypsin. Transfer the cells to a 1.5-Ml microcentrifuge tube.
Centrifuge the cells at 300×g for 5 min and wash twice with PBS.
Resuspend cells with phenol-red free DMEM supplemented with 10% FBS.
Add ViviRen™ to the assay buffer to a final concentration of 10µM.
Check the total luminescence using the F12 Luminometer.
With the spectrofluorimeter's (QuantaMaster QM-7/SE) fluorescence excitation turned off, record the luminescence emission spectrum between the wavelengths 440 and 580 nm in 2-nm steps with a 5-s integration for each step (see Fig. 2c, d).
3.5. Comparison of Substrates for BRET Imaging (Native Coelenterazine, Deep Blue C™, and ViviRen™)
3.5.1. Autoluminescence, Brightness, and Stability
For assessing autoluminescence, we tested different substrates in DMEM phenol-red free medium with or without 10% FBS. For assessing enzyme-catalyzed brightness and stability, we used transfected HEK293 cells in DMEM with or without serum (see Note 15).
24–48 h after transfection, cells were washed twice with phosphate buffered saline (PBS).
Release adherent cells by adding trypsin to a final concentration of 0.05% trypsin-EDTA (incubate at 37°C for 5~10 min).
Add 900 µl DMEM supplemented with 10% FBS to inactivate the trypsin and collect the cells in a 1.5-ml microcen-trifuge tube.
Gently spin down the cells and wash them twice with PBS.
Gently resuspend the cell pellet in 1 ml of phenol-red free DMEM with or without 10%FBS.
Add substrate solution (100 µl) to cells to reach a final concentration of 5–10 µM. For the stability assay, add 30 µM of stabilizer to the ViviRen™-treated samples (see Note 15).
Record the time dependence of total luminescence in the FB12 luminometer (see Fig. 1.3a–c).
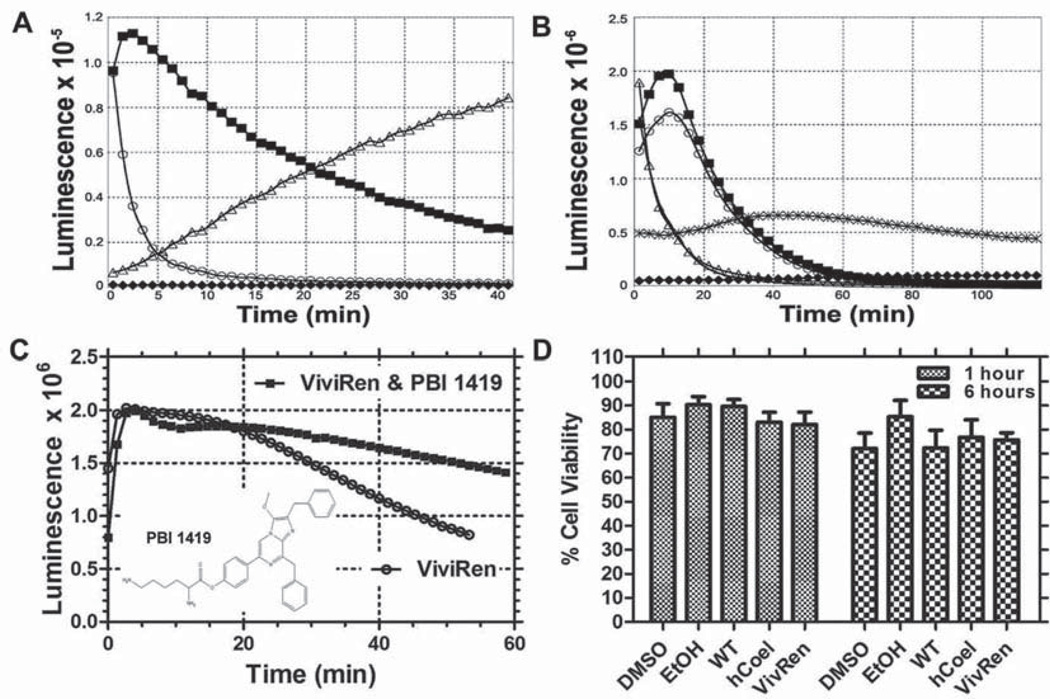
Fig. 1.3. Comparison of coelenterazine substrates.
a Autoluminescence of substrates in DMEM with and without 10% FBS (no cells present): for 10 µM native coelenterazine, closed squares = DMEM + 10% FBS and open circles = DMEM without FBS; for 10 µM ViviRen™, open triangles = DMEM + 10% FBS and closed diamonds = DMEM without FBS. b Brightness of enzyme-catalyzed luminescence with native vs. ViviRen™ coelenterazine. For HEK293 cells in DMEM + 10% FBS and transfected with PCMV::hRLUC-Venus: line without symbols = 5 µM native coelenterazine, open triangles = 10 µM native coelenterazine, open circles = 5 µM ViviRen™, closed squares = 10 µM ViviRen™. For tobacco seedlings transfected with P35S::RLUC-EYFP in 1/2 MS medium: "x" = 10 µM native coelenterazine, closed diamonds = 10 µM ViviRen™. c Stability of enzyme-catalyzed luminescence with ViviRen™ vs. ViviRen™ and the stabilizer PBI 1419. For HEK293 cells in DMEM + 10% FBS and transfected with PCMV::hRluc-Venus: open circles = 10 µM ViviRen™, closed squares = 10 µM ViviRen™ + 30 µM PBI 1419. Inset: structure of PBI 1419, a stabilizer molecule for ViviRen™ (obtainable from Promega by special request). d HEK293 cell viability after exposure to three different substrates. Cell viability was assayed by Trypan Blue exclusion after exposure to substrates and/or solvents for 1 or 6 h. HEK293 cells were in DMEM + 10% FBS. Data are shown as % viability (± S.E.M.) as compared with untreated cells. Treatments were as follows: 0.1 % DMSO, 0.1 % ethanol, 10 µM native coelenterazine (0.1 % ethanol final concentration), 10 µM coelenterazine-h (0.1% ethanol final concentration), and 10 µM ViviRen™ (0.1% DMSO final concentration).
3.5.2. Cell Viability Assay
Cell viability was assayed to test the possibility of toxicity of the different substrates.
Grow HEK293cells in 24-well plates overnight until they reach 80% confluence.
Wash cells with PBS.
Add medium with or without serum to the cells.
Incubate cells for 1 h or 6 h with the different substrates to a final concentration of 5 µM (native coelenterazine and coelenterazine-h) or 10 µM (ViviRen™).
Count viable cells by using 0.4% (w/v) Trypan Blue in a hemacytometer under the microscope (see Note 16, Fig. 1.3d). Viable cells exclude the Trypan Blue whereas the dye stains dead cells.
3.6. BRET Imaging
3.6.1. Apparatus for Whole Organism and Microscopic Imaging
Microscope setup: for plant cell and mammalian cell imaging, the EB-CCD camera was coupled to the Dual-View™ image splitter, which was connected to the bottom port of the IX-71 microscope. The inverted microscope (but not the EB-CCD camera or Dual-View™) resided inside a temperature controlled light-tight box (see Note 6).
“Box” setup for whole organism imaging: non-infinity-corrected objectives were attached directly to the Dual-View™ inside a light-tight box (at room temperature ~ 22°C). The EB-CCD camera was coupled to the Dual-View™, but the camera resided outside the box (see Note 17). Dual-View™ requires precise alignment before use (see Note 18).
3.6.2. “Box” Setup for Imaging of Plant Seedlings
Plant seedlings were placed into a drop of BRET assay buffer on a slide without coelenterazine, room temperature 22°C.
Put the slide onto a support, then fine-tune and focus using the non-infinity-corrected objectives (e.g., Plan 4×, NA=0.13 DL, 160/-; and Plan 10×, NA=0.30 DL, 160/0.17; Nikon).
Gently add the coelenterazine to the samples (final concentration of 10 µM).
Fine-tune and focus the samples precisely in brightfield with the Dual-View™ in “Bypass Mode.” Then turn the Dual-View™ off “Bypass Mode” and push the filter holder inside the tube. Close the light-tight box carefully (see Note 19).
Capture the BRET images of short-pass (“Blue”) and long-pass (“Yellow”) emission simultaneously from the whole seedlings (~7- to 10-min exposure time) (see Fig. 1.4).
Take a brightfield image of the above same sample with the door of the light-tight box open (see Note 19, Fig. 1.4, panels a, f, k).
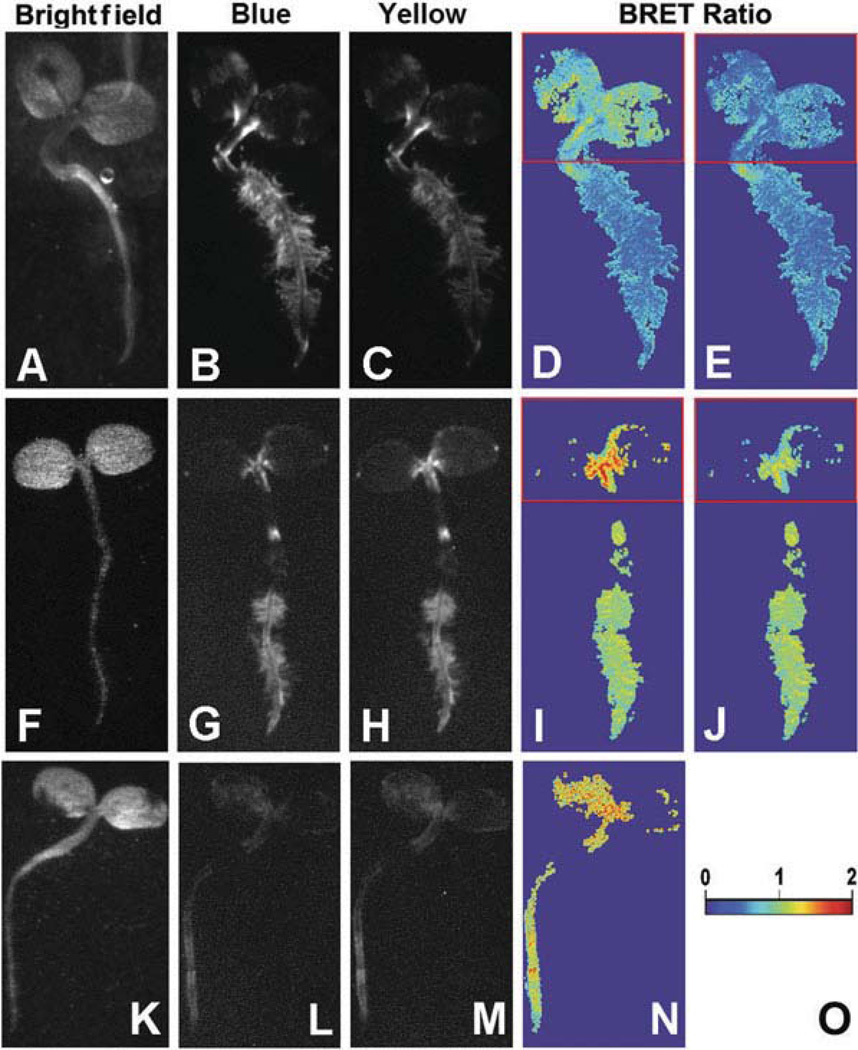
Fig. 1.4. BRET macro-imaging of tobacco seedlings.
Seven-day-old tobacco seedlings were transformed with (i) P35 s::Rluc (a–e), (ii) P35s::Rluc-EYFP (f–j), or (iii) P35 s::Rluc-COP1+ P35 s::Eyfp-COP1 (k–n). Panels a, f, k are bright field images, panels b, g, l are images of short-pass luminescence (Blue), panels c, h, m are images of long-pass luminescence (Yellow), panels d, i, n are BRET ratios (Y ÷ B) over the entire luminescent portion of the image (pseudocolor scale shown in panel o), panels e and j are corrected images of panel d and i, respectively, shown with a red box encasing the pigmented (cotyledon) portion of the seedlings (correction factor for boxed regions of panels e and j was 1.27). (Modified from Xu et al. (16).)
3.6.3. Microscope Setup for Imaging of Plant Cell Culture
Let the flask of cell culture stand for several seconds, then remove suspension cells from the middle of the flask gently and quickly place a drop onto the slide.
Mix the sample with the coelenterazine working solution (for Arabidopsis suspension cell use, see Section 2.3) and place a cover glass atop the sample.
In brightfield, focus on a single cell using the inverted microscope with the 40× oil immersion objective (NA 1.30). Close the box for imaging (exclude ALL incidental light by using a light-tight box!).
Capture the BRET images of short-pass (“Blue”) and long-pass (“Yellow”) simultaneously using the Dual-View™ and EB-CCD (~5- 15-min exposure time).
Take fluorescence images (EX 500/20 nm, EM 520LP) and/or brightfield images (see Fig. 1.1a–e). It is better to take the luminescence image first while the substrate is active and avoid any photobleaching and/or phototoxicity of the sample caused by fluorescence excitation or brightfield.
3.6.4. Mammalian Cells Imaged in the Microscope Setup
Wash cells with PBS that have been transfected and grown in 35-mm petri dishes with cover-glass bottoms (MatTek Corporation).
Add 1 ml of phenol-red free DMEM supplemented with 10%FBS.
Add ViviRen™ substrate directly to the medium to a final concentration of 10 µM.
Gently agitate the petri dish by hand to mix the substrate and medium.
Check the total luminescence in the FB12 luminometer before looking at the sample in the microscope. The brightness of luminescence as measured by the luminometer gives an approximation of the transfection efficiency of the luminescence constructs.
In brightfield, focus on a single cell using the inverted microscope with the Plan Apo 60× objective, NA 1.45 (oil immersion, Olympus – 1-U2B616) using the Dual-View™ in “Bypass Mode.”
After focusing in brightfield, turn off the brightfield excitation and switch the Dual-View™ out of “Bypass Mode” by pushing the filter holder inside the Dual-View™. Close the box for imaging (exclude ALL incidental light by using a light-tight box!).
Capture the BRET images of short-pass (“Blue”) and long-pass (“Yellow”) simultaneously for sequential exposures of 100 ms (see Note 20). See Fig. 1.1, panels g–i, l–n, q–S, v–x, a'–c'.
Take fluorescence images (EX 500/20 nm, EM 520LP, see Fig. 1.1, panels j, o, t, y), and/or brightfield images (see Fig. 1.1, panels f, k, p, u, z). It is usually better to take the luminescence image first while the substrate is active and before fluorescence or brightfield excitation causes any pho-tobleaching and/or phototoxicity of the sample.
3.7. Set Up Appropriate “Controls”
“Negative control”: RLUC alone. It is used for normalization (see Fig. 1.1a)
“Positive control”: RLUC-EYFP or hRLUC-Venus fusion proteins. These are commonly used to test the whole system, e.g., the setup of the spectrofluorimeter or imaging apparatus, especially since they tend to be the brightest construct in our experience with plant and mammalian cells. The RLUC-EYFP (or hRLUC-Venus) spectrum can also be used to calculate the absorption ratio for differentially absorbing tissues, and for other image corrections (see Fig. 1.1a and Note 21). For examples of images using these positive control fusion proteins, see Figs. 1. 1f—j and 1.4f–j.
Unfused luciferase + unfused yellow fluorescent protein: this combination is to confirm that the luciferase and yellow fluorescent protein will not spuriously interact by themselves, as shown in Fig. 1.1 k–o for unfused hRLUC + unfused Venus (in bacterial, plant, and mammalian cells, we have found that these proteins do not interact, but this should be tested for any new cell type).
Measure the spectra of relevant fusion proteins alone (e.g., RLUC vs. RLUC-COP1; or EYFP vs. EYFP-COP1) to make sure that the fusion proteins do not alter the spectra of RLUC emission or EYFP fluorescence. For an example of an image with hRLUC-C/EBPα, see Fig. 1.1z-c'.
Control for RLUC and EYFP concentrations: This kind of control is important to confirm that the BRET ratio is independent of the concentration of RLUC/EYFP molecules. In some cases, energy transfer can arise from random interactions of the luciferase and fluorophore in a cellular compartment (e.g., the membrane), and testing a range of concentrations of the RLUC and EYFP fusion proteins to assess which concentrations of those proteins lead to a constant BRET ratio is an important control (8). Also, in situations where RLUC and EYFP fusion protein are produced independently or expressed at different concentrations, this control is important for quantitative measurement of protein interaction (25). In the case of transient transfections, the experimenter can vary the intracellular concentration of the BRET proteins by using different amounts of plasmids in the transfection reaction.
Make sure your imaging setup totally excludes all incidental light. BRET signals are very dim and any incidental light will ruin image quality and/or generate misleading images.
3.8. Calculation of BRET Ratio
3.8.1. Correction of BRET Signal in Differentially Absorbing Tissues
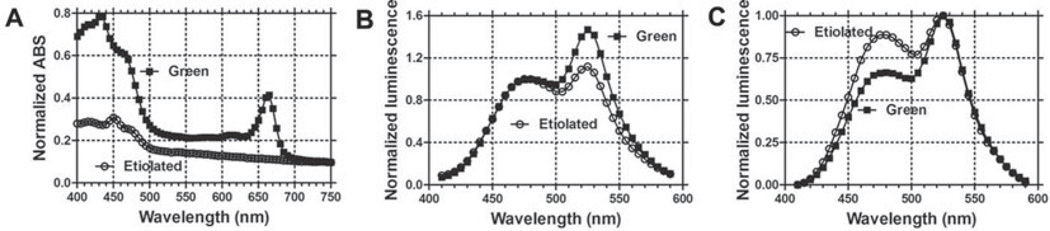
Many types of tissue contain pigments that differentially absorb light, and this property can interfere with an accurate measurement of BRET or FRET spectra. An example of this problem is green plant tissue, which contains many pigments that differentially absorb luminescence of different wavelengths. Green plant tissue absorbs more strongly at 480 nm than at 530 nm (see Fig. 1.5a), and this can be visualized by the difference in RLUC-EYFP spectrum from green plant tissue as compared with etiolated (= non-pigmented) plant tissue (see Fig. 1.5b, c). Therefore, to obtain an accurate BRET spectrum from pigmented tissue, the BRET signal should be corrected for differential absorption (see Note 21).
Prepare light-grown (“green”) and dark-grown (“etiolated”) seedlings (e.g., 5-day-old tobacco seedlings). Measure the emission spectrum of RLUC-EYFP in etiolated and green seedlings using the spectrofluorimeter.
Extract the total pigments from tobacco cotyledons (both green and etiolated seedlings) using ethanol.
Measure the absorption of above two kinds of ethanol extracts with spectrophotometer. Calculate the absorption ratio 480:530 for green seedlings (e.g., 2.07) and etiolated seedlings (e.g., 1.64) to get the correction factor, 1.27 (e.g., 1.27 = 2.07 ÷ 1.64) (see Fig. 1.5a).
The factor can be used to correct the image for the differential absorption using the image calculation function of ImageJ (or an equivalent function in other imaging software) (compare Fig. 1.4d with 1.4e and 1.4i with 1.4j). In the case of the plant seedlings, the areas which are to be corrected in the “Blue image” are multiplied by the correction factor to produce a “corrected Blue image,” then the BRET ratio image can be produced by re-calculating the ratio between the “Yellow image” and the “corrected Blue image.”
We describe the correction process for plant tissue here, but this correction could be applied to any kind of tissue, plant, or animal, by simply assuming the spectrum of RLUC-EYFP emission from the tissue in question should have roughly equivalent peaks at 480 nm and at 530 nm. Deviations from equal 480/530 nm peaks can be assumed to be due to differential tissue absorption (as in Fig. 1.5b, c) and a correction factor can be derived from those deviations.
Fig. 1.5. Correction of BRET signals from tobacco seedlings for differential absorption of luminescence.
a Absorption spectra of an ethanol extraction of pigments from light-grown (“green,” filled squares) and dark-grown (“etiolated,” open circles) seedlings. b RLUC-EYFP emission spectra for green and etiolated seedlings, normalized to the emission at 480 nm. c RLUC-EYFP emission spectra for green and etiolated seedlings, normalized to the emission at 530 nm. (Modified from Xuetal. (16).)
3.8.2. Normalization and Ratio Calculation
To compare the BRET luminescence spectra traces between RLUC-EYFP and RLUC (negative control), we normalized the first peak (480 nm) to 1.0 (as in all panels of Fig. 1.2).
Calculate BRET ratio (530 nm ÷ 480 nm) of individual traces.
Take the average of replicate samples and calculate standard deviations (see Note 22).
3.9. Methods of BRET Imaging Analysis
Background subtraction: first, select a region to serve as background and calculate the average optical density, then use the image calculator function to subtract the background (ImageJ has a plug-in for background subtraction).
Image alignment: In some situations where the two images from the Dual-View™ (i.e., the short-pass “Blue” and the long-pass “Yellow”) do not align well (e.g., which might be caused by poor alignment within the Dual-View™), imaging alignment should be processed before image ratio calculation. (ImageJ and some other software packages have this function and can align two images automatically by calculating an optimal alignment. Image alignment can also be accomplished by manual translation, rotation, and scaling.)
Derivation of BRET ratio image: divide “Yellow” by “Blue” ({Yellow range}÷{Blue range}= Y ÷ B), to produce a ratio-metric image with pseudo-color (as in Figs. 1.1, panels d, i, n, s, x, c' and 1.4, panels d, e, i, j, n).
Using ImageJ's ROI (Region Of Interest) and measurement functions, select the same ROI in both Blue and Yellow images; calculate the densities inside the ROIs that have been selected.
Calculate the ratio of density of objects between images (Y ÷ B). Take the average and standard deviation on parallel experiments (see Note 23).
Acknowledgements
We thank Dr. R.N. Day for mouse GHFT1 cells and C/EBP244 fused to EYFP, Dr. Roger Tsien for Venus YFP, Drs. Yao Xu, Michael Geusz, David Piston & Shin Yamazaki for advice concerning BRET techniques, imaging techniques, and image analysis, and Drs. Keith Wood & Erika Hawkins of Promega for making ViviRen™ and PBI 1419 available to us prior to its commercial release. Stein Servick contributed technical expertise to the images shown in Figure 4. This work was supported by NSF grant MCB-0114653 to Drs. Albrecht von Arnim and Carl Johnson as part of the Arabidopsis 2010 project and by the National Institutes of General Medical Science (R01 GM065467) to Carl Johnson.
Footnotes
Coelenterazine is sensitive to light, so store the powder and stock solutions in the dark at −70°C with desiccant. For preparation of stock solutions (e.g., 250 µM), dissolve powder with 95% EtOH, distribute 40–120 µl aliquots to a set of microcentrifuge tubes, and dry down in a Speed-Vac. After the samples are dried into the bottom of the microcentrifuge tubes, replace the air in the tubes with gaseous N2 or Ar (Gently! Don't blow off the coelenterazine! We place all the tubes in a box and flow N2 gas into the box for 20 min to replace all the air. Argon gas is helpful because it is heavier than air and will settle in the tubes, but it is more expensive). The tubes are then capped and returned to −70°C for long-term storage. To make a working solution, dissolve the dried coelenterazine in one or more tubes with a minimal amount of 95% EtOH, and then dilute to 10 µM with distilled water or medium. Be sure to prepare a fresh working solution before each experiment - keep the working solution on ice and in the dark (we wrap the tubes with aluminum foil).
ViviRen™ (Promega) is a modified version of the coelenterazine analog, coelenterazine-h, to which ester groups have been attached. The design intention of ViviRen™ was to develop an inactive RLUC substrate that would not release oxidation-induced autoluminescence in a serum-containing extracellular medium, but that would permeate into cells where intracellular esterases cleave the ester groups to generate active coelenterazine-h intracellularly. EnduRen™ (Promega) is a related substrate that is very stable and can be used for longer term measurement of luminescence and therefore BRET (17). In our experience, the luminescence intensity with EnduRen™ is much lower than with ViviRen™, but this limitation can potentially be counteracted by a very sensitive detection system.
We used a modified electron bombardment-charge coupled device (EB-CCD) camera (Hamamatsu Photonic Systems, Bridgewater NJ, USA); the modifications were a GaAsP photocathode with low ion feedback and increased pho-tocathode cooling to −25°C. The low ion feedback was achieved by a special modification to the EB-CCD camera by Hamamatsu to remove the aluminum mask from the sensor that is normally included to avoid the “double focus phenomenon.” In the case of low light imaging, this problem is negligible. In addition, the camera is using full frame transfer CCD, so it is possible to remove the mask. As a result, the sensor gets the same gain at a lower acceleration voltage. This low acceleration voltage reduces the ion feedback phenomenon drastically, improving performance for very low light level imaging. Finally, the cooling of the photocathode to −25°C reduces the dark current of the photo-cathode. The acquisition software was Photonics-WASABI (Hamamatsu).
The Dual-View™ micro-imager (Optical Insights, LLC) is an image splitter that is coupled to the EB-CCD camera to allow the simultaneous acquisition of luminescence images at two wavelengths. It consists of a dichroic mirror (in our case, to split the emission at 505 nm using Q505LPxr) and interference filters to refine wavelengths (i) below 505 nm (HQ505SP, short-pass filter; “Blue”) and (ii) above 505 nm (HQ505LP, long-pass filter; “Yellow”). For BRET imaging, the ratio of emission in the two wavelength ranges can be calculated without the complications due to possible changes in the total luminescence signal over the time course of the exposure (this kind of problem could occur with long exposure time if we exposed the camera to one wavelength and subsequently to a second wavelength - if the total luminescence).
An epifluorescence attachment (EX 500/20 nm, EM 520LP) is connected to the IX-71 inverted microscope to allow the measurement of fluorescence images. For low-power imaging (e.g., Arabidopsis cotyledons, roots etc.), we used an Olympus Macro XLFLuor 2× objective, NA 0.14 (working distance of 16.3 mm). For higher magnification imaging, an Olympus UPlanFl 40× objective, NA 1.30 (oil immersion) was used for Arabidopsis suspension culture cells, or an Olympus Plan Apo 60× objective, NA 1.45 (oil immersion) was used for mammalian cells.
Light-tight boxes for imaging: #1. For imaging larger samples, e.g., plant seedlings that are 1- to 3-cm long, we used a light-tight “box setup.” For plant tissues, like tobacco or Arabidopsis seedlings, non-infinity-corrected objectives (Plan 4×, NA=0.13 DL, 160/-; or Plan 10×, NA=0.30 DL, 160/0.17; both manufactured by Nikon) were attached directly to the Dual-View™ image splitter, which was placed through a hole in the box (the hole was sealed with black felt to prevent light leaks). The EB-CCD was coupled to the Dual-View™ outside the box. The box was in a room whose temperature was set at 22°C. Under these conditions, spontaneous YFP fluorescence or auto-fluorescence in the absence of coelenterazine (as might occur if there was a light leak into the box) could not be detected for 10- to 30-min exposures - therefore, the box setup was confirmed to be light-tight. After treating the transgenic seedlings with coelenterazine, images could be detected by the EB-CCD camera for exposures of 5 min or less. See images in Fig. 1.4.
#2. For microscopic imaging of mammalian cells (36°C) or plant cells (22°C), the IX-71 microscope was encased in a temperature-controlled (22–37°C) light-tight box with the Dual-View™ and EB-CCD attached to the bottom port of the microscope (Dual-View™ and EB-CCD camera were outside of the box). See images in Fig. 1.1.
A spectrofluorimeter (QuantaMaster QM-7/SE) was used for spectral measurements of BRET emission. For luminescence spectrum acquisition, the excitation beam was blocked and the slit width was set to 16 nm.
BRET experiments should always include a positive control - in our experiments, we used either RLUC-EYFP (for the plant studies) or hRLUC-Venus (for the mammalian cell studies). See Figs. 1. 1f–j and 1.4f–j. The production of the hRLUC-Venus construct is described in the next note (see Note 9).
CCAAT/Enhancer Binding Protein α (C/EBPα) is a tran-scriptional factor that localizes to heterochromatin in the nucleus and dimerizes. To test BRET signal detection in the nucleus, we made two fusion proteins with the nuclear transcriptional factor C/EBPα, one with hRluc and another with Venus. C/EBP was amplified from the EYFPC1r-C/EBPalpha244 plasmid (obtained from Dr. R.N. Day) using XhoI and HindIII linkers. The PCR product was fused to the C-terminus of hRluc by insertion into the XhoI/HindIII site of PCMV::hRluc to give PCMV::hRluc-C/EBP. For the other fusion protein (Venus-C/EBP), Venus (YFP) was amplified from its original plasmid Venus/pCS2 (obtained from Dr. Roger Tsien). The amplicon with NheI and HindIII linkers was inserted into the XhoI/ HindIII site of the EYFPC1r-C/EBPalpha244 plasmid by replacing the EYFP with Venus at the N-terminus of C/EBP forming a Venus-C/EBP fusion construct. For an image of GHFT1 cells expressing hRLUC-C/EBPα + Venus-C/EBPα, see Fig. 1.1p–t.
After germination in calli induction medium, the young seedlings grow with two normal cotyledons (= embryonic “leaves”), a short hypocotyl (= embryonic stem), and tiny roots. Calli and single cells could be easily detected with the microscope. For subculturing, material that floats in the flask (enriched single cells and small calli) was transferred to fresh liquid medium. The green color of newly induced suspension lines turns to light brown after several subculture passages. To isolate single Arabidopsis suspension cells, the cell culture solution can be passed through a sterilized Nylon mesh filter.
Cells were grown in two different types of 35-mm dishes. We used normal 35 mm dishes (NunclonR, InterMed) for cell viability and spectral assays. We used 35-mm dishes with cover-glass bottoms (MatTek Corporation) for BRET imaging.
Longer incubation times may decrease the activity of lipofectamine.
The 20 min incubation is to allow DNA-lipofectamine 2000 complexes to form; this complex is stable for 6 h at room temperature.
After adding coelenterazine to the assay buffer, start measurements immediately. Our measurements were manually controlled for exposure duration and frequency.
For autoluminescence in phenol-red free DMEM only: we added substrate to 100 µl medium ± serum to a final concentration of 10 µM. The total luminescence was measured using the FB12 luminometer. In our tests, we found that ViviRen™ has less autoluminescence than native coelenterazine in serum-containing medium for the first 15–20 min, but that its autoluminescence level steadily increases so that after 20 min, it has more autoluminescence than native coelenterazine (see Fig. 1.3a). In serum-free medium, native coelenterazine has a brief burst of autoluminescence upon addition but thereafter both native and ViviRen™ coelenterazine have low autoluminescence. The autoluminescence of native and ViviRen™ coelen-terazines in the simple salt medium (1/2 MS) was comparable to that of serum-free DMEM (see Fig. 1.3a). For brightness tests, we used HEK293 cells transfected with PCMV::hRluc-Venus and tobacco seedlings transfected with P35 S::RLUC-EYFP. HEK293 cells transfected with BRET constructs have brighter luminescence when using ViviRen™ than when using native coelenterazine in serum-containing medium during the interval 5–50 min after addition (see Fig. 1.3b). Therefore, for mammalian cells in serum-containing medium, ViviRen™ has a better tradeoff of brightness vs. autoluminescene than native coelenter-azine (compare Fig. 1.3a with 3b). On the other hand, ViviRen™ does not appear to be useful for BRET in plant seedlings - native coelenterazine has very low auto-luminescence in simple salt medium (1/2 MS), and the brightness signal for native coelenterazine is higher than for ViviRen™ in plant seedlings, in addition to being well sustained for over 100 min (see Fig. 1.3b). For improving the stability of ViviRen™ in serum-containing medium, we used PBI 1419 (obtained from Erika Hawkins at Promega) along with ViviRen™. PBI 1419 stabilizes the signal of ViviRen™-dependent luminescence (see Fig. 1.3c; the structure of PBI 1419 is shown in the inset). Note that PBI 1419 is not commercially available as of this printing, but Dr. Erika Hawkins at Promega says that Promega will provide PBI 1419 to interested users (Dr. Erika Hawkins, personal communication).
The cells were incubated for one or 6 h with different substrates: native coelenterazine, coelenterazine-h, or ViviRen™. Coelenterazine and coelenterazine-h were dissolved in ethanol and ViviRen™ was dissolved in DMSO. Final concentrations were 5 µM for coelenterazine and coelenterazine-h and 10 µM for ViviRen™. After the incubation, the cells were harvested and 0.1 ml 0.4% (w/v) Try-pan Blue was added to 0.1 ml of the cell suspension from each sample. The stained and unstained cells were counted using a hemacytometer. Viable cells exclude the Trypan Blue dye. Therefore, blue-stained cells were scored as nonviable and unstained cells were scored as viable. Therefore “Percent Viability” = number of viable cells ÷ total number of cells. The viability of cells was not significantly affected by treatment for 1–6 h with either native coelenterazine or ViviRen™ as compared with the solvent controls (see Fig. 1.3d.)
For focusing the samples in the “box setup,” a scissors-jack was used to support the sample under the non-infinity-corrected objective (e.g., Plan 4×, NA=0.13 DL, 160/-; or Plan 10×, NA=0.30 DL, 160/0.17; Nikon) that was attached to the Dual-View™ and EB-CCD. Therefore this scissors-jack provided a moveable stage which was moved up and down to focus the sample. The sample was placed on the jack on top of a white paper background.
The Dual-View™ micro-imager needs to be precisely aligned before use. To set up “Full View” (Bypass Mode), pull the filter holder half-way out of the Dual-View™ tube to image normal brightfield.
Float the plant seedling on a drop of assay buffer and do a preliminary focusing. Then, aspirate the extra buffer while keeping the seedling surrounded by a thin film of buffer. Fine-focus and acquire the BRET image in darkness. Bright-field (and fluorescence) images can be acquired after the BRET image is taken. Plant seedling images can be acquired in the “box setup” to capture the entire seedling’s image (as in Fig. 1.4) or by using the inverted microscope setup with the Macro XLFLuor 2× objective (NA 0.14, working distance of 16.3 mm).
For mammalian cells, 20 sequential 100-ms exposures were acquired and then placed into a “stack” in ImageJ (version WCIF). These stacks were integrated to improve signal:noise by choosing the median value for each pixel over the sequence of 20 exposures and generating an integrated image that was used for subsequent analyses.
When RLUC-EYFP is expressed in E. coli and mammalian cells, the emission peaks at 480 nm vs. 530 nm are approximately equal (as in Fig. 1.2c), while the BRET ratio of RLUC-EYFP expressed in green plant tissue was usually greater than 1.0 (as in Fig. 1.5b). When we measured the emission spectrum of RLUC-EYFP in etiolated tobacco seedlings, we indeed found the emission spectrum was closer to 1:1 for 480 nm: 530 nm (see Fig. 1.5b, c). This is probably due to the greater absorption of plant pigments at 480 nm than that at 530 nm in green tissue, which can be visualized by the normalization of luminescence spectra to 530 nm and by absorption spectra of extracted plant tissue (see Fig. 1.5a). The correction factor can be used to correct the image for the differential absorption (compare Fig. 1.4d with 4e and 1.4i with 4j).
We determined the average ratio over the entire sample along with standard deviation calculation between samples to demonstrate the reproducibility of replicate samples. In the case of RLUC being directly fused to EYFP, the expected BRET ratio is quantifiable (e.g., for RLUC-EYFP, the 530:480 ratio should be ~1:1). In situations where RLUC and EYFP are fused to interacting proteins (e.g., RLUC-COP1, YFP-COP1 or C/EBP fusions) and are expressed from independent promoters, it is better to show the averaged BRET ratio (or ratio in a given region). When using the same promoter to drive the expression of both constructs, the estimated relative amount of RLUC and EYFP fusion proteins varied by less than 10% in our plant and mammalian cell studies.
Several programs or software can be used to analyze BRET images. We prefer to use the open architecture “ImageJ” program. There are also other commercially available programs that work well, such as “Image Pro Plus” or “Auto-quant.”
References
- 1.Mendelsohn AR, Brent G. Protein biochemistry: protein interaction methods-toward an endgame. Science. 1999;284:1948–1950. doi: 10.1126/science.284.5422.1948. [DOI] [PubMed] [Google Scholar]
- 2.Periasamy A, Day RN. Molecular imaging: FRET microscopy and spectroscopy. New York, NY: Oxford University Press; 2005. p. 321. [Google Scholar]
- 3.Hoshino H, Nakajima Y, Ohmiya Y. Luciferase-YFP fusion tag with enhanced emission for single-cell luminescence imaging. Nat. Methods. 2007;4:637–639. doi: 10.1038/nmeth1069. [DOI] [PubMed] [Google Scholar]
- 4.Xu Y, Piston DW, Johnson CH. A bioluminescence resonance energy transfer (BRET) system: application to interacting circadian clock proteins. Proc. Natl. Acad. Sci. USA. 1999;96:151–156. doi: 10.1073/pnas.96.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pfleger KDG, Eidne KA. Illuminating insights into protein–protein interactions using bioluminescence resonance energy transfer (BRET) Nat. Methods. 2006;3:165–174. doi: 10.1038/nmeth841. [DOI] [PubMed] [Google Scholar]
- 6.Subramanian C, Kim BH, Lyssenko NN, Xu XD, Johnson CH, von Arnim AG. The Arabidopsis repressor of light signaling, COP1, is regulated by nuclear exclusion: mutational analysis by bioluminescence resonance energy transfer. Proc. Natl. Acad. Sci. USA. 2004;101:6798–6802. doi: 10.1073/pnas.0307964101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gales C, Rebois RV, Hogue M, Trieu P, Breit A, Hebert TE, Bouvier M. Real-time monitoring of receptor and G-protein interactions in living cells. Nat. Methods. 2005;2:177–184. doi: 10.1038/nmeth743. [DOI] [PubMed] [Google Scholar]
- 8.James JR, Oliveira MI, Carmo AM, Iaboni A, Davis SJ. A rigorous experimental framework for detecting protein oligomerization using bioluminescence resonance energy transfer. Nat. Methods. 2006;3:1001–1006. doi: 10.1038/nmeth978. [DOI] [PubMed] [Google Scholar]
- 9.Subramanian C, Woo J, Cai X, Xu XD, Servick S, Johnson CH, Nebenfuhr A, von Arnim AG. A suite of tools and application notes for in vivo protein interaction assays using bioluminescence resonance energy transfer (BRET) Plant J. 2006;48:138–152. doi: 10.1111/j.1365-313X.2006.02851.x. [DOI] [PubMed] [Google Scholar]
- 10.De A, Gambhir SS. Noninvasive imaging of protein–protein interactions from live cells and living subjects using bioluminescence resonance energy transfer. FASEB J. 2005;19:2017–2019. doi: 10.1096/fj.05-4628fje. [DOI] [PubMed] [Google Scholar]
- 11.Pfleger KD, Seeber RM, Eidne KA. bioluminescence resonance energy transfer (BRET) for the real-time detection of protein–protein interactions. Nat. Protoc. 2006;1:337–345. doi: 10.1038/nprot.2006.52. [DOI] [PubMed] [Google Scholar]
- 12.James JR, Oliveira MI, Carmo AM, Iaboni A, Davis SJ. A rigorous experimental framework for detecting protein oligomerization using bioluminescence resonance energy transfer. Nat. Methods. 2006;3:1001–1006. doi: 10.1038/nmeth978. [DOI] [PubMed] [Google Scholar]
- 13.Carriba P, Navarro G, Ciruela F, Ferre S, Casado V, Agnati L, Cortes A, Mallol J, Fuxe K, Canela EI, Lluis C, Franco R. Detection of heteromerization of more than two proteins by sequential BRET-FRET. Nat. Methods. 2008;5:727–733. doi: 10.1038/nmeth.1229. [DOI] [PubMed] [Google Scholar]
- 14.De A, Loening AM, Gambhir SS. An improved bioluminescence resonance energy transfer strategy for imaging intracellular events in single cells and living subjects. Cancer Res. 2007;67:7175–7183. doi: 10.1158/0008-5472.CAN-06-4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dixit R, Cyr R, Gilroy S. Using intrinsically fluorescent proteins for plant cell imaging. Plant J. 2006;45:599–615. doi: 10.1111/j.1365-313X.2006.02658.x. [DOI] [PubMed] [Google Scholar]
- 16.Xu XD, Soutto M, Xie QG, Servick S, Subramanian C, von Arnim A, Johnson CH. Imaging protein interactions with BRET in plant and mammalian cells and tissues. Proc. Natl. Acad. Sci. USA. 2007;104:10264–10269. doi: 10.1073/pnas.0701987104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pfleger KD, Dromey JR, Dalrymple MB, Lim EM, Thomas WG, Eidne KA. Extended bioluminescence resonance energy transfer (eBRET) for monitoring prolonged protein–protein interactions in live cells. Cell Signal. 2006;18:1664–1670. doi: 10.1016/j.cellsig.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 18.Loening AM, Fenn TD, Wu AM, Gambhir SS. Consensus guided mutagenesis of Renilla luciferase yields enhanced stability and light output. Protein Eng. Des. Sel. 2006;19:391–400. doi: 10.1093/protein/gzl023. [DOI] [PubMed] [Google Scholar]
- 19.Loening AM, Wu AM, Gambhir SS. Red-shifted Renilla reniformis luciferase variants for imaging in living subjects. Nat. Methods. 2007;4:641–643. doi: 10.1038/nmeth1070. [DOI] [PubMed] [Google Scholar]
- 20.Loening AM, Fenn TD, Gambhir SS. Crystal structures of the luciferase and green fluorescent protein from Renilla reniformis. J. Mol. Biol. 2007;374:1017–1028. doi: 10.1016/j.jmb.2007.09.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu Y, Piston D, Johnson CH. Green fluorescent protein: applications and protocols (methods in molecular biology series) Totowa, NJ: Humana Press; 2002. BRET assays for protein–protein interactions in living cells; pp. 121–133. [DOI] [PubMed] [Google Scholar]
- 22.Angers S, Salahpour A, Joly E, Hilairet S, Chelsky D, Dennis M, Bouvier M. Detection of beta 2-adrenergic receptor dimerization in living cells using bioluminescence resonance energy transfer (BRET) Proc. Natl. Acad. Sci. USA. 2000;97:3684–3689. doi: 10.1073/pnas.060590697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soutto M, Xu Y, Johnson CH. Molecular imaging: FRET microscopy and spectroscopy. New York, NY: Oxford University Press; 2005. Bioluminescence RET (BRET): techniques and potential; pp. 260–271. [Google Scholar]
- 24.Mathur J, Koncz C. Arabidopsis protocols. Totowa, NJ: Humana Press; 1998. Establishment and maintenance of cell suspension cultures; pp. 27–30. [DOI] [PubMed] [Google Scholar]
- 25.Siegel RM, Chan FK-M, Zacharias DA, Swofford R, Holmes KL, Tsien RY, Lenardo MJ. Measurement of molecular interactions in living cells by fluorescence resonance energy transfer between variants of the green fluorescent protein. Sci. STKE. 2000;2000:PL1. doi: 10.1126/stke.2000.38.pl1. [DOI] [PubMed] [Google Scholar]