Abstract
Combining high mass accuracy mass spectrometry, isobaric tagging, and novel software for multiplexed, large-scale protein quantification, we report deep proteomic coverage across multiple biological replicates and cell lines. We applied this method to study four human embryonic stem cell and four induced pluripotent stem cell lines in biological triplicate, a 24-sample comparison resulting in the largest set of identified proteins and phosphorylation sites in pluripotent cells to date. The statistical analysis afforded by this approach revealed, for the first time, subtle but reproducible differences in protein and protein phosphorylation between embryonic stem cells and induced pluripotent cells. Merging these results with RNA-seq analyses, we found functionally related differences across each tier of regulation. Finally, we introduce the Stem Cell–Omics Repository (SCOR), a resource that collates and displays quantitative information across multiple planes of measurement, including mRNA, protein, and post-translational modifications.
For practical and ethical reasons, induced pluripotent stem (iPS) cells hold great potential for therapeutic and research purposes. Based on morphology, capacity to self-renew, and developmental potential, iPS cells are nearly indistinguishable from their embryonic stem (ES) cell counterparts1–3; however, their degree of similarity on the molecular level remains controversial4–6. While various studies have stressed the overall similarity of gene expression programs between ES and iPS cells1, 2, 5, 7, a handful of studies have reported subtle differences in RNA levels, DNA methylation, and the efficiency of many iPS lines to differentiate into neural lineages6, 8–10. Meanwhile, similarity on the protein level remains completely unexplored. These analyses are critical, as many forms of regulation are enforced post-transcriptionally or through post-translational modifications (PTMs).
To address the proteomic and phosphoproteomic similarity between ES and iPS cells, we employed a method that combines isobaric tagging, high mass accuracy mass spectrometry, and novel software. Applying this method to the comparison of two ES, one iPS, and one fibroblast cell line we identified 7,952 proteins and 10,499 phosphorylation sites. Leveraging the multiplexing nature of our approach, we then examined protein and their phosphorylation sites in four ES and four iPS cell lines in biological triplicate (24 samples total) and identified 6,761 proteins and 19,122 phosphorylation sites. Rigorous statistical analysis revealed statistically significant and functionally related differences between proteins and phosphorylation sites in human ES and iPS cells, which may reflect residual regulation characteristic of iPS cells’ somatic origin. Finally, we introduce a queryable online resource for large-scale data related to pluripotency.
RESULTS
Peptide identification and quantitation
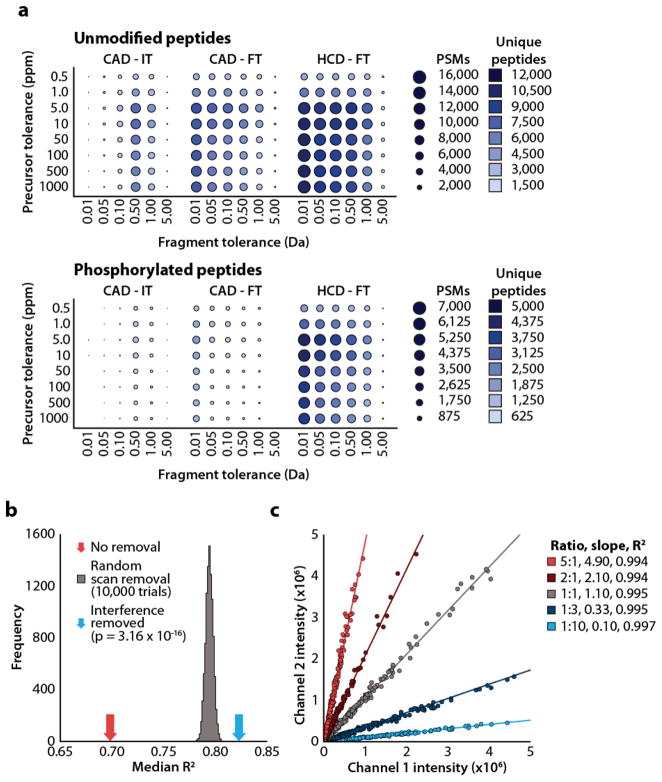
To remove the limitation of low mass cutoff, imposed by resonant excitation CAD, we employed beam-type collision-activated dissociation (HCD) with high mass accuracy detection of fragment ions11,12–14. As shown in Figure 1a these methods increase peptide identification over 60% and phosphopeptide identifications over 260% compared to CAD with low mass accuracy fragment ion detection. We attribute these increases to greater specificity in database searches and fewer sequence-directed cleavage events. Importantly, HCD is compatible with isobaric tagging strategies for multiplexed peptide quantitation. Isobaric tags can compare up to eight samples in a single experiment and facilitate analysis of biological replicates and multiple cell lines15–17. However, this form of quantitation is subject to a unique and widespread source of quantitative error arising from the co-isolation of multiple peptide precursors prior to fragmentation18. We therefore developed novel software, TagQuant, which identifies mass spectra compromised by interference and excludes these data points from peptide and protein quantitation19. This filtering method resulted in a statistically significant increase in quantitative precision (permutation testing, P < 3×10−16; Fig. 1b). TagQuant also incorporates mathematic correction of tag impurities, summing of reporter ion intensities, and exclusion of low intensity reporter ions (see supplemental materials and methods)20, 21. We tested our complete workflow using whole-cell lysate from S. cerivisae. Separate pools of protein were labeled with isobaric tags, combined in known ratios, and analyzed via mass spectrometry. The observed results match closely to the expected ratios for the range of mixtures tested (R2 > 0.99; Figure 1c).
Figure 1. Figures of merit for peptide identification and quantitation.
(a) Peptide identifications as a function of precursor and product mass tolerance. We performed liquid chromatography tandem mass spectrometry for each combination of dissociation method and mass analyzer. We searched data using a range of fragment ion tolerances ranging from 0.01 to 5.0 Daltons, filtered results by precursor mass tolerances ranging from 0.5 to 1,000 ppm, and filtered identifications to a achieve 1%FDR. We performed experiments in triplicate and averaged the results. The number of peptide spectrum matches (PSM) is proportional to circle size while unique peptides are represented by circle color. (b) We used permutation testing and the data from the 4-plex experiment to test the benefit of interference filtering. R2 values for all peptides in each protein were calculated as a metric for quality of quantitation. The median R2 increases from 0.70 (red arrow) to 0.82 (blue arrow) with filtering. Since random removal of spectra also increases R2 values, we used permutation testing to test the statistical significance of the increase in R2 value resulting from interference filtering. By fitting a Gaussian curve to the distribution we estimated the statistical significance of the increase in R2 due to interference filtering (P = 3.16 × 10−16). (c) Characterization of iTRAQ quantitation. Each circle represents reporter ion intensities for a single protein mixed in the indicated ratios.
Comparison of ES and iPS cell proteomes
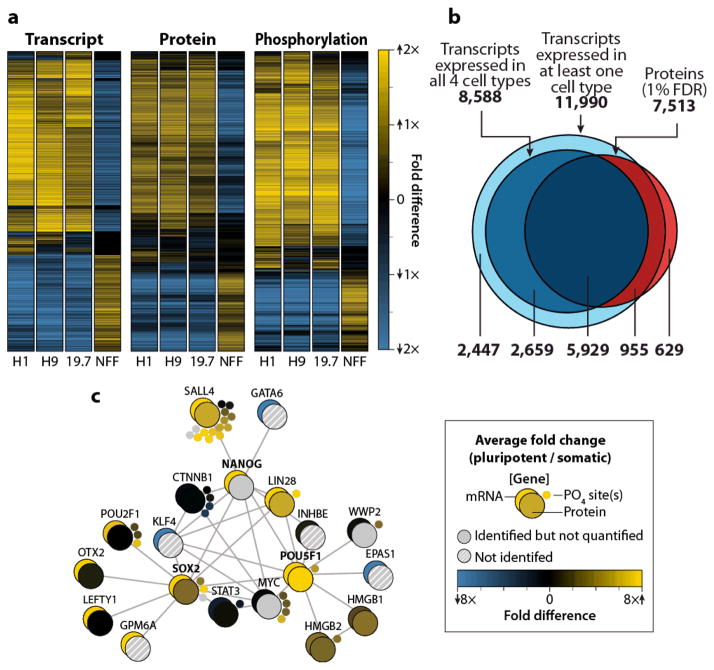
We first compared transcripts, proteins, and phosphorylation sites across two human ES (H1 and H9), one iPS (DF19.7), and one fibroblast (newborn foreskin fibroblasts, NFF) cell line (Supplementary Fig. 1) using isobaric tags. With less than two weeks of instrument analysis, we identified 7,952 proteins (1% false discovery rate (FDR); Supplementary Table 1) and 10,499 sites of phosphorylation (localized with 95% confidence; Supplementary Table 2). We validated measurements for selected, representative proteins by Western blots (Supplementary Fig. 2). Identified proteins include key regulators of pluripotency, such as OCT4/POU5F1, NANOG, and SOX2 (Fig. 2e), and nearly every major component of the developmentally related epigenetic regulators, polycomb group and trithorax proteins (Supplementary Fig. 3).
Figure 2. A transcriptomic, proteomic and phosphoproteomic comparison of two ES (H1 and H9), one iPS (19.7), and one fibroblast (NFF) line.
(a) Heatmaps depicting all quantified transcripts, proteins, and phosphorylation sites. Values were median normalized. (b) The overlap between transcripts and proteins detected in the 4-plex experiment. We considered transcripts present if the reads per kilobase of exon per million mapped reads (RPKM) value was greater than one for all four cell types while we determined protein identification via P-value filtering (1% FDR). (c) Cytoscape schematic of mRNA, protein, and phosphorylation quantitation from the 4-plex experiment for genes known to interact with NANOG, SOX2, or POU5F1 (STRING database, confidence score > 0.90).
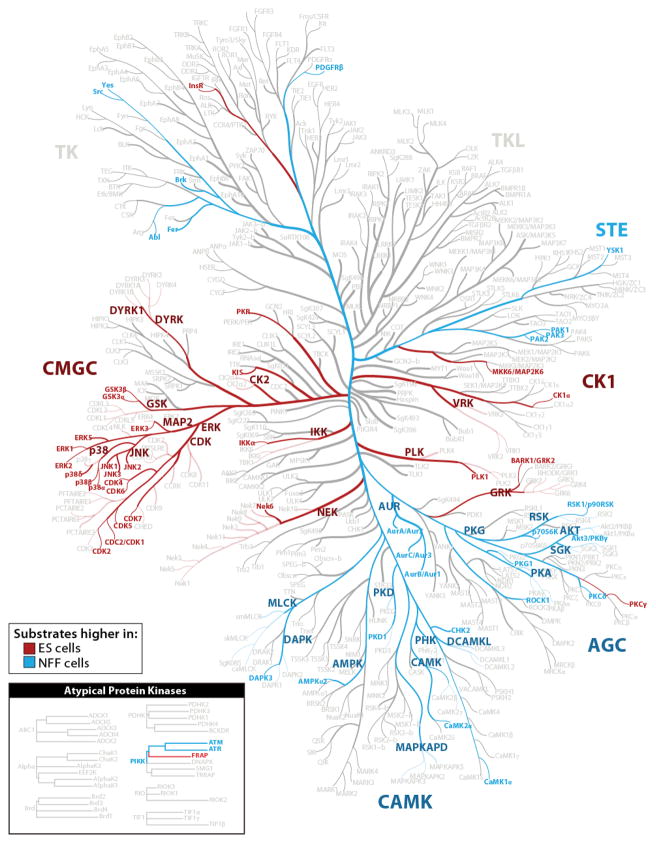
Comparing ES and NFF cell lines revealed that 35% of proteins and 59% of phosphorylation sites differed by at least two-fold in abundance. The genes corresponding to these differentially regulated proteins and phosphorylation sites were functionally related and representative of the two cell states. For example, proteins found at higher levels (two-fold) in ES cells were enriched for cell cycle-related processes (e.g., DNA replication, cell division, etc.), reflecting the rapid proliferation and shorter doubling times characteristic of pluripotent cells (Supplementary Table 3)22. Conversely, proteins observed at higher levels in the NFFs were enriched for processes pertinent to differentiated cell types. Differential regulation of phosphorylation sites was likewise apparent. Phosphorylation sites that were at least twofold higher in either ES or NFF cells were enriched for a number of different amino acid motifs (Supplemental Table 4). To test whether this reflected differences in kinase activity between the two cell types, we mapped potential kinases to each phosphorylated site using the Group-based Prediction System software23. We then used Fisher’s exact test to determine if substrates for particular kinases were enriched in sets of phosphorylation sites that were at least two-fold different between ES and NFF cells and mapped them to the human kinome tree (Figure 3, adapted from Manning et al.24). Entire kinase families appear highly active in distinct cell types. For example, targets of CMGC kinases were more highly phosphorylated in the ES cells relative to NFFs, while substrates of CAMK and AGC kinases were more heavily occupied in the NFFs (P < 0.05, Fisher’s Exact Test with Benjamini-Hochberg correction)25. The high number of differences and their functional enrichment confirm that two sample comparisons, without replicate analysis, are sufficient to characterize major differences between highly dissimilar cell types.
Figure 3. Kinase substrate analysis.
Adapted from Manning et al.24. We predicted potential kinases for every phosphorylation site using the Group-based Prediction system. We applied Fisher’s exact test (followed by Benjamini-Hochberg adjustment) to test for enrichment of kinase substrates in sets of phosphorylation sites that were changing by more than two-fold between ES and NFF cells kinase substrates enriched in ES cells are highlighted in red (P < 0.05). Kinase substrates enriched in ES cells are highlighted in blue (P < 0.05).
Of course it is often necessary to perform large-scale comparison of more similar proteomes. ES and iPS cells offer one such example, and a complete map of their similarities and differences will be key for both fundamental science and clinical applications. Single replicate comparison of one ES and one iPS cell line, however, revealed two-fold or greater differences in less than 1% of proteins and phosphorylation sites. This small set of proteins and phosphorylation sites showed no functional commonality (i.e., gene ontology terms26, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways27, or phosphorylation motifs). Moreover, comparing ES and iPS cells yielded roughly the same number of differences as a comparison between two ES cell lines, regardless of fold-difference (Supplementary Fig. 4). Together, these data suggested an overall inability to differentiate between ES and iPS cells at the protein level.
Next we examined RNA-seq data, which was acquired concomitantly with protein-level experiments (Supplementary Table 5). Consistent with proteomic results, large differences were detected between ES and NFF cells: 48% of transcripts differed by two-fold or more. While 9% of transcripts differed by greater than two-fold when comparing ES (H9) and iPS transcripts for a single replicate, the two ES cell lines showed even greater variation (12% of transcripts). This suggested that it was not possible to distinguish differences between cell types from line-to-line variability. Unlike the initial proteomic experiments, however, the RNA measurements were carried out in biological triplicate. Statistical analysis afforded by replicates enabled us to move beyond arbitrary fold-cutoffs and establish statistical significance. Using Student’s t-test with Benjamini-Hochberg correction (P < 0.05), we observed 623 differentially regulated transcripts between ES (H9) and iPS cells. From these data we reasoned that proteomic differences likely existed between these similar cell types but were subtle and therefore masked by our inability to perform statistical analyses.
To test this hypothesis we leveraged the multiplexing capabilities of 8-plex isobaric tags to compare proteins and phosphorylation sites across four ES (H1, H7, H9, H14) and four iPS (DF4.7, DF6.9, DF19.11, DF19.7) cell lines in biological triplicate (Supplementary Fig. 1). To facilitate comparison between all 24 samples, reporter ion intensities were median normalized. Proteomic and phosphoproteomic analyses took less than six weeks to acquire and resulted in the identification of 6,761 proteins (<1% FDR; Supplementary Table 1) and 19,122 sites of phosphorylation (localized with at least 95% confidence; Supplementary Table 2). 4,742, 3,396, and 2,234 proteins were quantified in at least one, two, or three replicates respectively while 14,162, 8,217, and 4,564 localized phosphorylation sites were quantified in at least one, two, or three replicates respectively. Accompanying mRNA analysis was again performed for each of the samples using an Illumina Genome Analyzer IIx.
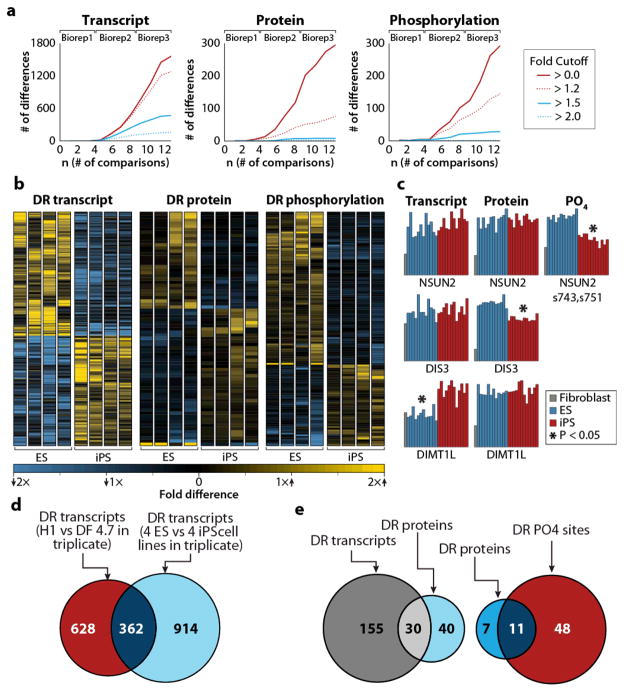
Analysis within a single biological replicate (8 cell lines) revealed only 1 transcript, 5 proteins, and 4 phosphorylation sites that were statistically different (P < 0.05, Student’s t-test with Benjamini-Hochberg correction; Fig. 4a). However, inclusion of two more biological replicates permitted detection of numerous differentially regulated elements – 1560 transcripts, 293 proteins, and 292 phosphoisoforms differed significantly between ES and iPS cells (P < 0.05, Student’s t-test with Benjamini-Hochberg correction; Fig. 4a, Supplementary Table 6). Greater than 90% of the differentially regulated transcripts, proteins, and phosphorylation sites differed by less than two fold. These minor deviations were only detectable through biological replicate analysis, which increased sample size, and with it, statistical power.
Figure 4. Comparison of four ES and four iPS cell lines.
(a) Differentially regulated transcripts, proteins, and phosphorylation sites are shown as a function of the number of comparisons. We performed differential expression analysis using subsets of the data. For example, the n = 2 value reflects the number of differences detected from comparing just two ES lines and two iPS lines without biological replicate whereas n = 12 represents the differences detected from comparing all four ES lines and all four iPS lines in biological triplicate. The number of differentially regulated elements for a given fold-difference is indicated by different colors. (b) Heatmaps depicting differentially regulated transcripts, proteins, and phosphorylation sites (P < 0.05, Student’s t-test, with Benjamini-Hochberg correction). Only transcripts exhibiting at least a 1.5-fold difference and protein and phosphorylation sites exhibiting at least a 1.2-fold difference are shown. (c) Randomly selected examples of differentially regulated transcripts, proteins, and phosphorylation sites. Asterisks indicate statistically significant differences between ES and iPS cells. (d) Differentially regulated transcripts detected based on either a comparison between biological triplicates of H1 and DF4.7 cell lines (blue) or a comparison of biological triplicates of all four ES and all four iPS cell lines (red). (e) The overlap between differentially regulated proteins and transcripts (left) and differentially regulated proteins and phosphorylation sites (right). Only genes with both a quantified protein and transcript were included. Only genes with both a quantified protein and phosphorylation site were included.
Though biological replicates provide the statistical power to detect differences, they may not always distinguish pervasive differences between cell types from variance between cell lines. This is best illustrated by considering just H1-ES and DF4.7-iPS cell lines. Biological triplicate analysis of transcripts from these lines indicates 990 differentially regulated transcripts (P < 0.05, Student’s t-test with Benjamini-Hochberg correction; Fig. 4d). However, most (63%) of these differences did not overlap with differentially regulated transcripts as determined by the full 24-sample comparison. Moreover, 72% of the differences detected by analysis of all eight cell lines in biological triplicate were not detected by comparison between H1 and DF4.7 cells alone (Fig. 4d). We conclude that analyzing multiple cell lines is an essential addition to biological replicates.
Despite the subtlety of the differences observed here, their functional enrichment suggests a consistent distinction in regulation between ES and iPS cells. Transcripts, proteins, and phosphorylation sites found at higher levels in iPS cells were enriched for many biological processes required for somatic cell function, including system process, organ development, blood circulation, and muscle system process (Supplementary Table 7). However, motif analysis of differentially regulated phosphorylation sites did not implicate any specific kinases or phosphatases in these differences. Despite the functional relationship of the differentially regulated elements, the differences at each level of regulation (transcript, protein, and phosphorylation) often did not correspond to the same genes (Fig. 4e).
To determine if differences between ES and iPS cells represented incomplete reprogramming, we contrasted ES and iPS cell comparisons with ES and NFF cell comparisons. Based on gene enrichment analysis, three biological processes showed enrichment at every level in iPS cells compared to ES cells (transcript, protein, and phosphorylation): muscle system process, muscle contraction, and wound healing. These terms reflect cellular function characteristic of mesodermal lineages and may represent the NFF origin of the iPS cells. Further supporting this hypothesis, all three terms were enriched in the transcripts, proteins, and phosphorylation sites that were at least two-fold higher in NFF cells than ES cells (Supplementary Table 8). In fact, more than half of the gene ontology terms enriched among transcripts, proteins, and phosphorylation sites that were significantly higher in iPS compared to ES cells were also enriched in NFF compared to ES cells. Among this dataset were multiple phosphorylation events on NSUN2, a proto-oncogene implicated in cellular proliferation28(Fig. 4c). Transcript and total protein levels for NSUN2 were not different between ES and iPS cells, suggesting the changes are not simply a matter of protein abundance. Further, phosphorylation of these sites in iPS cells was similar to the levels observed in fibroblast cells, which may reflect residual regulation from kinases and phosphatases more characteristic of the differentiated NFF cells. NSUN2 acts downstream of c-MYC28, one of a handful of factors commonly used to improve reprogramming efficiency. At the transcript level, the set of mRNAs more abundant in iPS cells, which included TBX15 and PITX2, were enriched for developmental function and exposed a connection to mesoderm differentiation29, 30. All of these results suggest that somatic cell programs are not completely silenced during reprogramming. While this has been observed before in gene expression studies31, this is the first evidence that incomplete silencing is also reflected in regulation of proteins and post-translational modifications10, 32.
Data resource and sharing
To facilitate integration of these results with other data sets we created the Stem Cell-Omics Repository (SCOR, http://coongroup.chem.wisc.edu/scor/ username: POU5F1; password: txn_factor), a web-based resource that collates quantitative biological analyses of ES and iPS. A key feature of SCOR is the ability to visualize quantitative information for transcripts, proteins, and PTMs from numerous sources (Supplementary Figure 5). Included in the database are several large-scale analyses from other labs, all of which are queried during standard searches. To ensure that SCOR remains relevant, we’ve added an option to submit published data for inclusion on the website. Our intention is that the resource will expand as the field grows. A separate tab in the tools section provides open-access, downloadable programs used for post-acquisition data processing, including the interference filtering program, TagQuant. All datasets are downloadable at the SCOR database and have also been deposited in Tranche (http://www.proteomecommons.org/dev/dfs/users/index.html).
To demonstrate the value of this resource, we applied the SCOR database to evaluate results from this and several other microarray and RNA seq experiments1,4. This analysis, encompassing iPS cells derived using integrating, viral vectors and non-integrating, episomal vectors, identified a number of transcripts that were consistently different in ES cells versus iPS cells (Supplementary Table 9). To include data from outside laboratories we intersected our results with a similar data set4 (Supplementary Table 9). Contained in this data set were two transcripts (TCERG1L and FAM19A5) that were consistently higher in ES cells relative to iPS cells. Interestingly, recent work reported that both of these genes exhibit promoter hypermethylation and ultimately lower expression in a number of iPS cell lines10. These and other genes that show consistent differential regulation are of great interest for further studies. As more proteomic studies of ES and iPS cells become available, SCOR will facilitate similar inter-laboratory comparisons to determine the most pervasive transcriptomic, proteomic, and phosphoproteomic discrepancies.
DISCUSSION
This transcriptomic, proteomics, and phosphoproteomic comparison of ES and iPS cells offers important insights into the nature of reprogrammed cells. One subtle but critical conclusion is the remarkable similarity between ES and iPS cells, which is highlighted by the technical rigor required to detect even minor differences. While the exact biological relevance of these differences remains unknown, functional similarity of the genes that contribute to them suggest that iPS cells retain residual regulation characteristic of the cells from which they were derived. These differences do not appear to appreciably alter cellular function in the pluripotent state, but instead may surface during differentiation as cells invoke gene expression programs needed for development. Although iPS cells are capable of producing mesoderm, endoderm, and ectoderm, the process of reprogramming selects for cells predisposed to the pluripotent state, not necessarily for cells that differentiate with equal efficiency to all lineages. For example, recent studies have reported that ES lines differentiate into neural lineages with higher efficiency than most iPS lines33. From our data, many transcripts lower in iPS cells relative to ES cells, like NEURONATIN and SOX11, were also functionally related through their role in neural development34, 35. On the PTM level, phosphorylation states were consistently lower in iPS cells for a number of microtubule-related proteins that are directly (DPYSL236) or indirectly (FAM29A37) implicated in neural differentiation and development. Understanding how these genes contribute to neural differentiation in both ES and iPS cells will be the subject of further study.
A major advantage of combining multiple planes of measurement is the ability to dissect regulatory mechanisms not apparent in a single dimension. For instance, many of the protein kinases whose substrates exhibited significant differences in phosphorylation levels exhibited little to no change at the transcript or protein level. For example, while mRNA and protein levels of CDK2 are largely unchanged (< two-fold) in pluripotent cells relative to NFF cells, CDK2 substrates, were more highly phosphorylated in pluripotent cells. A possible explanation for this observation was apparent in our global PTM data. Phosphorylation of CDK2 at threonine 160, a mark required for kinase activity38, is up-regulated by nearly 6-fold in all three pluripotent cell lines. Likewise, CDK4, CDK5, and CDK6 are all found at similar levels in the pluripotent cells, but the motifs they target show a significant increase in phosphorylation. In contrast, the higher transcript and protein expression levels of PKA and PKC in NFF cells may explain the correspondingly high levels of substrate phosphorylation. Taken together these data suggest multiple mechanisms for the regulation of kinases. For instance, proteins involved in transitory functions, like the aforementioned cell cycle-related kinases, may be regulated via rapid and dynamic signals (i.e., phosphorylation and dephosphorylation) rather than by slower and longer lasting transcriptional and translational changes.
The results presented here highlight the importance of including multiple biological replicates to overcome biological and technical variability and to establish statistical significance. Moreover, evaluating multiple cell lines or subjects ensures that observed differences are persistent and not merely single sample aberrations. This study incorporated 24 different samples, though we recognize the importance of expanding the comparison of ES and iPS cells to cover as many lines, reprogramming methods, and growth conditions as possible. To date, 75 ES cell lines are listed on the NIH-approved registry and innumerable iPS lines are available from diverse sources. Comparing all of these cell lines is a daunting task for a single research group. We therefore created SCOR, an open-access resource to collate, visualize, and analyze large-scale datasets related to pluripotency. As research expands, the SCOR website will bring datasets together and facilitate cross-laboratory comparisons at every tier of regulation.
METHODS
Cell Growth and Lysis
We maintained human embryonic stem cells (lines H1, H7, H9, and H14) and induced pluripotent cells (lines DF4.7, DF6.9, DF19.7, and DF19.11) in a feeder independent system, as previously described39. We karyotyped all ES and iPS cell lines prior to experiments using standard G-banding chromosome analysis (WiCell Research Institute). Upon reaching 70% confluency, we passaged cells enzymatically using dispase (Invitrogen) at a 1:4 splitting ratio. We cultured human newborn foreskin fibroblasts (Cat# CRL-2097™, ATCC) essentially according to ATCC recommendations. We maintained cells in 10% fetal bovine serum (Hyclone Laboratories Incorporated), 1 mM L-glutamine (Invitrogen), 0.1 mM beta-mercaptoethanol (Sigma-Aldrich), and 0.1 mM non-essential amino acids in DMEM (both from Invitrogen). We passaged cells ar roughly 70% confluency at a 1:3 splitting ratio, using Tryp-LE (Invitrogen).
For proteomics experiments, we harvested all cells by individualizing for 10 minutes with an adequate volume of pre-warmed (37 °C), 0.05% Tryp-LE to cover the culture surface. Following cell detachment, we added an equivalent volume of either ice-cold growth media, in the case of NFF cells, or ice-cold DPBS (Invitrogen), in the case of ES cells, before collecting the cells. We subsequently washed cell pellets were twice in ice-cold DPBS and stored at −80 °C. We collected approximately 108 cells for each analysis. We lysed samples via sonication in lysis buffer containing 8M Urea, 40 mM NaCl, 50 mM Tris (pH 8), 2 mM MgCl2, 50 mM NaF, 50 mM b-glyceradelhyde phosphate, 1 mM sodium orthovanadate, 10 mM sodium pyrophosphate, 1X mini EDTA-free protease inhibitor (Roche Diagnostics), and 1X phosSTOP phosphatase inhibitor (Roche Diagnostics).
For RNA-seq analysis, we washed celled twice in pre-warmed (37°C) DPBS and lysed them on the culture dish using Trizol reagent (Invitrogen). We added Chloroform (Sigma) to a final concentration of 16.7% (v/v) and the sample was centrifuged for 15 minutes at 12,000×g at 4°C. We combined the resulting supernatant with an equal volume of 70% ethanol and processed it using the Qiagen RNeasy kit with on column DNAse digestion. We linearly amplified poly A+ RNAs using a modified T7 amplification method40 that retains directionality of the transcripts. This protocol generates Illumina RNA-Seq libraries with uniform coverage of the entire length of the mRNAs. Samples were run on an Illumina Genome Analyzer IIx. We then aligned each lane to the genome and the exon splice sites database using bowtie41, allowing up to ten multiple matches and three mismatches. For data processing, we filtered 42bp reads to remove adapters in each lane. We used ERANGE42 to obtain expression values in RPKM (reads per kilobase of exon model per million mapped reads).
mRNA analysis
We performed microarray raw data processing and normalization as previously described1, 3. We performed the assessment of the ES and iPS specificity of transcripts as follows. First, we fit a linear model to estimate all the fold changes across the iPS and ES lines, and then applied Bayesian smoothing to the standard errors among the same type of cell lines. Finally, we calculated a p-value based on the moderated t-statistics for the differentially expressed genes and then adjusted them based on Benjamini and Hochberg’s method to control the false discovery rate25. Second, we required the fold change to be at least 3-fold different between the two cell types, with an adjusted p-value of less than or equal to 0.05. The data in Supplementary Table 7 was generated from 15 microarrays for ES cells, 25 microarrays for iPS cells, and three microarrays for differentiated cell types (NFF and IMR90) pooled from the work of Junying Yu et al. 2009 and Junying Yu et al. 20071, 3.
Western Blot analysis
To confirm quantitation determined by mass spectrometry, we analyzed several proteins for western blot analysis (Supplementary Fig. 3). Following cell lysis, we loaded equal amounts of total protein from H1, H9, iPS and NFF cells onto a 4–15% acrylamide gel (Biorad). We used the following primary antibodies to detect the indicated protein: mouse, anti-human OCT4 monoclonal antibody (1:2000, sc-5279, Santa Cruz Biotechnology), goat, anti-human DNMT3B (1:1000, sc-10235, Santa Cruz Biotechnology), mouse, anti-human GAPDH (1:2,000, MAB374, Chemicon), mouse, anti-human CD44 (1:10, 550989, Pharmingen-BD). We used the following horseradish peroxidase-linked secondary antibodies: goat, anti-mouse IgG (1:2,000, sc-2005, Santa Cruz Biotechnology), donkey, anti-goat IgG (1:2,000, sc-2056, Santa Cruz Biotechnology). We loaded a biotin labeled ladder according to the manufacturer’s specification (Cell Signaling). We used a Super Signal West Pico Chemiluminescent Substrate (Thermo Scientific Pierce) according to protocol to image blots on a LAS-3000 Imaging System (Fujifilm Life Science). We determined quantitation according to manufacturer’s instructions with MultiGauge software, ver 2.0 (Fujifilm Life Science). Between detections, we stripped the membrane using Restore Western Blot Stripping Buffer (Thermo Scientific Pierce).
Digestion and iTRAQ labeling
We reduced cysteine residues with 5mM dithiothreitol, alkylated them using 10mM iodoacetamide, and digested proteins in a two-step process. We added proteinase Lys-C (Wako Chemicals) (enzyme:protein ratio = 1:100) and incubated for approximately 2 hours at 37°C in lysis buffer. We then diluted samples with 50 mM Tris pH 8 until the urea concentration was 1.5 M and digested them with trypsin (Promega) (enzyme:protein ratio = 1:50) at 37°C overnight. We quenched reactions using trifluoroacetic acid (TFA). We dried samples to completion after purification using C18 solid phase extraction (SPE) columns (SepPak, Waters). We performed iTRAQ labeling according to manufacturer supplied protocols (Applied Biosystems)16, 17. To ensure that each of the samples contained the same amount of protein we prepared a small 1:1:1:1 (1:1:1:1:1:1:1:1 for 8-plex experiment) aliquot and analyzed it by mass spectrometry. We used summed reporter ion ratios from this experiment to inform mixing ratios of the remaining labeled digests. Once mixed, we dried samples to completion and purified by them by sold phase extraction (SPE).
Fractionation
We resuspended the labeled peptides in strong cation exchange (SCX) buffer A [5 mM KH2PO4, 30% acetonitrile (pH 2.65)] and injected them onto a polysulfoethylaspartamide column (9.4 × 200 mm; PolyLC). We performed separations using a Surveyor liquid chromatography quaternary pump (Thermo Scientific) at a flow rate of 3.0 mL/min. We used the following gradient for separation: 0–2 min, 100% buffer A, 2–5 min, 0–15% buffer B, 5–35 min, 15–100% buffer B. Buffer B was held at 100% for 10 minutes. Finally, the column was washed extensively with buffer C and water prior to recalibration. We used the following buffers: buffer A [5 mM KH2PO4, 30% acetonitrile (pH 2.65)], buffer B [5 mM KH2PO4, 30% acetonitrile, 350 mM KCl (pH 2.65)], buffer C [50 mM KH2PO4, 500 mM KCl (pH 7.5)]. We collected the samples by hand and desalted them by SPE.
Phosphopeptide enrichment
Following SCX fractionation, we enriched phosphopeptides using magnetic beads (Qiagen). We washed the beads 3× with water, 3× with 40 mM EDTA (pH 8.0) for 30 minutes with shaking, and 3× with water again. We then incubated beads with 100 mM FeCl3 for 30 minutes with shaking. Finally, we resuspended beads in 1 mL 1:1:1 (acetonitrile/methanol/0.01% acetic acid) and washed them 3 times with 80% acetonitrile/0.1% TFA. We resuspended samples in 80% acetonitrile/0.1% TFA and incubated them with beads for 30 minutes with shaking. We washed the beads 6 times with 200 μL 80% acetonitrile/0.1% TFA, and eluted the peptides using 1:1 acetonitrile:5% NH4OH in water. We acidified eluted phosphopeptides immediately with 4% formic acid, lyophilized them to ~10 μL, and diluted them with 50 mM phosphate buffer prior to analysis.
Mass spectrometry
We performed tandem mass spectrometry using a NanoAcquity ultra high-pressure liquid chromatography system (Waters) coupled to a dcQLT-orbitrap (Thermo Fisher Scientific). Samples were loaded onto a precolumn (75 μm inner diameter, packed with 5 cm C18 particles, Alltech) for 10 min at a flow rate of 1 μm/min. Samples were then eluted over an analytical column (50 μm ID, packed with 15 cm C18 particles, Alltech) using a 120 min linear gradient from 1% to 35% acetonitrile with 0.2% formic acid and a flow rate of 300 nL/min. An additional 30 min were used for column washing and equilibration. We constructed columns as previously described12.
All mass spectrometer instrument methods consisted of one MS1 (resolving power = 30,000 – 60,000) scan followed by data dependent MS2 scans (resolving power = 7,500) of the ten most intense precursors. Protein identification experiments used exclusively beam-type CAD (HCD) with orbitrap mass analysis. Some phosphopeptide identification experiments included alternating HCD and electron transfer dissociation (ETD) MS2 scans. We quantified any peptides identified by ETD using the corresponding HCD scan. We used an exclusion list for 60 s using a window of −0.55 Th to 2.55 Thompson. We excluded precursors with unassigned charges states or charge states of one (and two for ETD scans). We used automatic gain control target values of 1,000,000 for MS1 analysis and 50,000 for orbitrap MS2 analysis. To maximize quantified identifications we employed QuantMode for some analyses.
Database search and FDR filtering
We used DTA generator to extract peak information from .RAW files and print it into a searchable text file43. This software removed fragment ions related to the iTRAQ reagents and as well as charged reduced precursors. We searched spectra against the International Protein Index (IPI) human database version 3.75 with full enzyme specificity using The Open Mass Spectrometry Search Algorithm (OMSSA; version 2.1.4) 44, 45. We used a mass tolerance of ±4.5 Dalton precursors and a monoisotopic mass tolerance of ±0.01 Dalton for fragments ions. We set carbamidomethylation of cysteines, iTRAQ 4-plex on the N-terminus, and iTRAQ (4-plex or 8-plex) on lysines as fixed modifications, and oxidation of methionines and iTRAQ (4-plex or 8-plex) on tyrosines as variable modifications. For phosphopeptide searches we included variable phosphorylation of Serine, Threonine, and Tyrosine as variable modifications. We used the COMPASS software suite to filter peptides to a 1% FDR. COMPASS groups peptides into proteins following the rules previously established46. COMPASS multiplies peptide level P-scores for unique peptides corresponding to each protein to obtain protein P-Scores and then filters proteins by this score to achieve a 1% FDR at the protein level.
Peptide and Protein Quantitation
We used custom software, TagQuant, to perform iTRAQ quantification. TagQuant is written in C# programming language and istributed along with COMPASS software suite. TagQuant extracts reporter ion intensities and multiplies them by injection times to determine counts. TagQuant performs purity correction as previously described20. TagQuant normalizes intensities such that the total signal from each channel is equal. We summed reporter ion intensities for each channel for all peptides in a given protein with three exceptions; (1) scans corresponding to peptides found in multiple protein groups were not used for quantification (2) peptides found to be phosphorylated were not used for protein quantification and (3) if peaks not related to the precursor were present in the MS1 scan within +/− 1.8 Thompson of the selected precursor at an intensity greater than 25% of the selected precursor the resulting MS2 scan was not used for quantitation. We median normalized protein and phosphorylation site quantitation in order to compare across all three replicate experiments.
Phosphorylation Analysis
We filtered phosphopeptides to a 1% FDR based on unique peptides as described above. To avoid over-reporting of phosphorylation sites, we combined phosphorylated peptides and non-phosphorylated peptides and grouped them into proteins together, following previously established rules46.
We used the program phosphinator software to localize phosphorylation sites47. The algorithm calculates theoretical fragment ion m/z ratios for all possible permutations of phosphopeptide isoforms given the sequence and number of phosphorylations. The algorithm then compares the experimental spectrum against the theoretical product ions for each candidate phosphopeptide isoform, using a product mass tolerance of ±0.02 Th. Two criteria are required for localization. First, the candidate with the highest number of matching product ions must have at least one more matching product ion than the second highest. Second, the algorithm performs a statistical test to determine the significance of the observed product ions supporting phosphorylation at a specific residue. We take the null hypothesis to be that there is no evidence that a given phosphorylation is localized, and that any site-determining fragments observed are merely spurious matches. We calculate a probability value (p-value) that represents the likelihood of obtaining the observed number of site-determining fragments or more based on random chance, using the following equation, the cumulative distribution function for a binomial distribution:
where P is the p-value, N is the number of possible site-determining fragment ions, n is the number of observed site-determining fragment ions, and p is the probability of a single spurious fragment ion match. The algorithm calculates p as the product of the number of observed MS/MS peaks and the twice the product mass tolerance (±), divided by the MS/MS m/z range.
The algorithm performs this significance test twice for every phosphorylation site in the top isoform — once on each side of the phosphorylated residue. The site-determining fragment ions are those between the phosphorylation site and the closest amino acid residue that could be phosphorylated but are not in the top isoform. The algorithm considers doubly charged products for +3 and higher precursors when the product is comprised of a sufficient portion of the peptide. Phosphinator converts the p-value to a human-readable score by taking −10 log10 (P). We only consider sites where this score is above 13 (i.e., p < 0.05) on both the left and right side of the residue to be localized, and we only use peptides with all phosphorylations localized for quantitative analysis.
Next, we counted phosphorylation sites. We summed quantitative information from all phosphopeptides that contained the same sites in order to get the most accurate quantitation for each site or combination of sites. We grouped peptides containing multiple sites with other peptides containing the exact same combination of sites. Therefore, we presented a list of phosphorylation isoforms rather than a list of phosphorylated sites. Phosphorylation isoforms can have information regarding one site or a combination of multiple sites. We only counted redundant sites that were found in more than one isoform once in the final count of phosphorylation sites.
Enrichment Analysis
We performed two-tailed Student’s t-test assuming equal variance in Microsoft Excel. To correct for multiple-hypothesis testing, we applied Benjamini-Hochberg adjustment using the R statistics package. We used a local gene ontology MySQL database installation for analysis of function and cellular location and another local MySQL database populated with information from the KEGG API web services for pathway analysis. We determined putative kinase targets using the Group-based prediction system software. To perform Fisher’s exact test and subsequent Benjamini-Hochberg correction, we wrote custom software in the C# programming language and interfaced to the R statistics package through the R COM library.
Supplementary Material
Acknowledgments
We gratefully acknowledge AJ Bureta for illustrations, Alicia Williams and Krista Eastman for editing, and Graeme McAlister for assistance with instrumentation. We thank Junying Yu (Department of Cell & Regenerative Biology, University of Wisconsin, Madison, Wisconsin, USA) for providing transcriptomic data prior to publication and Thomson lab members for critical reading and discussion of this manuscript. This work was supported by the University of Wisconsin, the Beckman Foundation, and National Institutes of Health Grants R01GM080148 (to J.J.C.) and P01GM081629 (to J.A.T. and J.J.C.). D.H.P. acknowledges support from a National Institutes of Health predoctoral traineeship—the Genomic Sciences Training Program, National Institutes of Health Grant 5T32HG002760.
Footnotes
D.H.P. designed research, prepared samples, performed mass spectrometry, wrote software, analyzed data and wrote the manuscript. J.B. designed research, grew cells, prepared samples, analyzed data and wrote the manuscript. C.D.W. wrote software. S.T. analyzed data. M.D.P. grew cells. D.J.B. designed websites. D.L.S. helped with phosphorylation analysis. M.A.T. optimized labeling procedure. J.M.B. performed RNA-sequencing. V.R. analyzed data. R.S. designed research and analyzed data. J.A.T. designed research and wrote the manuscript. J.J.C. designed research and wrote the manuscript.
Declaration of interests
J.A.T. is a founder, stockowner, consultant and board member of Cellular Dynamics International (CDI), and serves as scientific advisor to and has financial interests in Tactics II Stem Cell Ventures.
References
- 1.Yu JY, et al. Human Induced Pluripotent Stem Cells Free of Vector and Transgene Sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 3.Yu JY, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 4.Chin MH, et al. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell. 2009;5:111–123. doi: 10.1016/j.stem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guenther MG, et al. Chromatin structure and gene expression programs of human embryonic and induced pluripotent stem cells. Cell Stem Cell. 2010;7:249–257. doi: 10.1016/j.stem.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chin MH, Pellegrini M, Plath K, Lowry WE. Molecular analyses of human induced pluripotent stem cells and embryonic stem cells. Cell Stem Cell. 2010;7:263–269. doi: 10.1016/j.stem.2010.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bock C, et al. Reference Maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 2011;144:439–452. doi: 10.1016/j.cell.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stadtfeld M, et al. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse inducedpluripotent stem cells. Nature. 2010;465:175–181. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doi A, et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet. 2009;41:1350–1353. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lister R, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011 doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olsen JV, et al. Higher-energy C-trap dissociation for peptide modification analysis. Nature Methods. 2007;4:709–712. doi: 10.1038/nmeth1060. [DOI] [PubMed] [Google Scholar]
- 12.McAlister GC, Phanstiel D, Wenger CD, Lee MV, Coon JJ. Analysis of Tandem Mass Spectra by FTMS for Improved Large-Scale Proteomics with Superior Protein Quantification. Analytical Chemistry. 2010;82:316–322. doi: 10.1021/ac902005s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olsen JV, et al. A Dual Pressure Linear Ion Trap Orbitrap Instrument with Very High Sequencing Speed. Molecular & Cellular Proteomics. 2009;8:2759–2769. doi: 10.1074/mcp.M900375-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagaraj N, D’Souza RCJ, Cox J, Olsen JV, Mann M. Feasibility of Large-Scale Phosphoproteomics with Higher Energy Collisional Dissociation Fragmentation. Journal of Proteome Research. 2010;9:6786–6794. doi: 10.1021/pr100637q. [DOI] [PubMed] [Google Scholar]
- 15.Thompson A, et al. Tandem mass tags: A novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Analytical Chemistry. 2003;75:1895–1904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- 16.Ross PL, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Molecular & Cellular Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 17.Choe L, et al. 8-plex quantitation of changes in cerebrospinal fluid protein expression in subjects undergoing intravenous immunoglobulin treatment for Alzheimer’s disease. Proteomics. 2007;7:3651–3660. doi: 10.1002/pmic.200700316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ow SY, et al. iTRAQ Underestimation in Simple and Complex Mixtures: “The Good, the Badand the Ugly”. Journal of Proteome Research. 2009;8:5347–5355. doi: 10.1021/pr900634c. [DOI] [PubMed] [Google Scholar]
- 19.Wenger CD, Phanstiel DH, Lee MV, Bailey DJ, Coon JJ. COMPASS: A suite of pre- and post-search proteomics software tools for OMSSA. Proteomics. 2011 doi: 10.1002/pmic.201000616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shadforth IP, Dunkley TPJ, Lilley KS, Bessant C. i-Tracker: For quantitative proteomics using iTRAQ (TM) Bmc Genomics. 2005:6. doi: 10.1186/1471-2164-6-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Griffin TJ, et al. iTRAQ reagent-based quantitative proteomic analysis on a linear ion trap mass spectrometer. Journal of Proteome Research. 2007;6:4200–4209. doi: 10.1021/pr070291b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Becker KA, Stein JL, Lian JB, van Wijnen AJ, Stein GS. Establishment of histone gene regulation and cell cycle checkpoint control in human embryonic stem cells. J Cell Physiol. 2007;210:517–526. doi: 10.1002/jcp.20903. [DOI] [PubMed] [Google Scholar]
- 23.Xue Y, et al. GPS 2.0, a tool to predict kinase-specific phosphorylation sites in hierarchy. Mol Cell Proteomics. 2008;7:1598–1608. doi: 10.1074/mcp.M700574-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 25.Benjamini Y, Hochberg Y. Controlling the false discovery rate - A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B-Methodological. 1995;57:289–300. [Google Scholar]
- 26.Ashburner M, et al. Gene Ontology: tool for the unification of biology. Nature Genetics. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Research. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frye M, Watt FM. The RNA methyltransferase Misu (NSun2) mediates Myc-induced proliferation and is upregulated in tumors. Current Biology. 2006;16:971–981. doi: 10.1016/j.cub.2006.04.027. [DOI] [PubMed] [Google Scholar]
- 29.Singh MK, et al. The T-box transcription factor Tbx15 is required for skeletal development. Mech Dev. 2005;122:131–144. doi: 10.1016/j.mod.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 30.Dong F, et al. Pitx2 promotes development of splanchnic mesoderm-derived branchiomeric muscle. Development. 2006;133:4891–4899. doi: 10.1242/dev.02693. [DOI] [PubMed] [Google Scholar]
- 31.Kim K, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Polo JM, et al. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat Biotechnol. 2010;28:848–855. doi: 10.1038/nbt.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu BY, et al. Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc Natl Acad Sci U S A. 2010;107:4335–4340. doi: 10.1073/pnas.0910012107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Siu IM, et al. Coexpression of neuronatin splice forms promotes medulloblastoma growth. Neuro Oncol. 2008;10:716–724. doi: 10.1215/15228517-2008-038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hargrave M, et al. Expression of the Sox11 gene in mouse embryos suggests roles in neuronal maturation and epithelio-mesenchymal induction. Dev Dyn. 1997;210:79–86. doi: 10.1002/(SICI)1097-0177(199710)210:2<79::AID-AJA1>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 36.Kawano Y, et al. CRMP-2 is involved in kinesin-1-dependent transport of the Sra-1/WAVE1 complex and axon formation. Mol Cell Biol. 2005;25:9920–9935. doi: 10.1128/MCB.25.22.9920-9935.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu H, Coppinger JA, Jang CY, Yates JR, 3rd, Fang G. FAM29A promotes microtubule amplification via recruitment of the NEDD1-gamma-tubulin complex to the mitotic spindle. J Cell Biol. 2008;183:835–848. doi: 10.1083/jcb.200807046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bourke E, Brown JAL, Takeda S, Hochegger H, Morrison CG. DNA damage induces Chk1-dependent threonine-160 phosphorylation and activation of Cdk2. Oncogene. 2010;29:616–624. doi: 10.1038/onc.2009.340. [DOI] [PubMed] [Google Scholar]
- 39.Ludwig TE, et al. Derivation of human embryonic stem cells in defined conditions. Nat Biotechnol. 2006;24:185–187. doi: 10.1038/nbt1177. [DOI] [PubMed] [Google Scholar]
- 40.Sengupta S, et al. Highly consistent, fully representative mRNA-Seq libraries from ten nanograms of total RNA. Biotechniques. 2010;49:898–904. doi: 10.2144/000113556. [DOI] [PubMed] [Google Scholar]
- 41.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biology. 2009:10. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 43.Good DM, et al. Post-acquisition ETD spectral processing for increased peptide identifications. J Am Soc Mass Spectrom. 2009;20:1435–1440. doi: 10.1016/j.jasms.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Geer LY, et al. Open mass spectrometry search algorithm. Journal of Proteome Research. 2004;3:958–964. doi: 10.1021/pr0499491. [DOI] [PubMed] [Google Scholar]
- 45.Kersey PJ, et al. The International Protein Index: An integrated database for proteomics experiments. Proteomics. 2004;4:1985–1988. doi: 10.1002/pmic.200300721. [DOI] [PubMed] [Google Scholar]
- 46.Nesvizhskii AI, Aebersold R. Interpretation of shotgun proteomic data - The protein inference problem. Molecular & Cellular Proteomics. 2005;4:1419–1440. doi: 10.1074/mcp.R500012-MCP200. [DOI] [PubMed] [Google Scholar]
- 47.Swaney DL, Wenger CD, Thomson JA, Coon JJ. Human embryonic stem cell phosphoproteome revealed by electron transfer dissociation tandem mass spectrometry. Proc Natl Acad Sci U S A. 2009;106:995–1000. doi: 10.1073/pnas.0811964106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.