Abstract
Quantitative high throughput assays of eosinophil-mediated activities in fluid samples from patients in a clinical setting have been limited to ELISA assessments for the presence of the prominent granule ribonucleases, ECP and EDN. However, the demonstration that these ribonucleases are expressed by leukocytes other than eosinophils, as well as cells of non-hematopoietic origin, limits the usefulness of these assays. Two novel monoclonal antibodies recognizing eosinophil peroxidase (EPX) were used to develop an eosinophil-specific and sensitive sandwich ELISA. The sensitivity of this EPX-based ELISA was shown to be similar to that of the commercially available ELISA kits for ECP and EDN. More importantly, evidence is also presented confirming that among these granule protein detection options, EPX-based ELISA is the only eosinophil-specific assay. The utility of this high throughput assay to detect released EPX was shown in ex vivo degranulation studies with isolated human eosinophils. In addition, EPX-based ELISA was used to detect and quantify eosinophil degranulation in several in vivo patient settings, including bronchoalveolar lavage fluid obtained following segmental allergen challenge of subjects with allergic asthma, induced sputum derived from respiratory subjects following hypotonic saline inhalation, and nasal lavage of chronic rhinosinusitis patients. This unique EPX-based ELISA thus provides an eosinophil-specific assay that is sensitive, reproducible, and quantitative. In addition, this assay is adaptable to high throughput formats (e.g., automated assays utilizing microtiter plates) using the diverse patient fluid samples typically available in research and clinical settings.
Keywords: EPX, eosinophilia, granule proteins, allergic inflammation
INTRODUCTION
Diagnosis and monitoring of allergic diseases often uses biomarkers thought to reflect levels of tissue infiltrating eosinophils. ELISA-based detection assays specific for the abundant eosinophil secondary granule proteins ECP and EDN are generally perceived as easy to perform as high throughput assessments reflective of eosinophils (reviewed in (Metso et al., 2002)). However, the literature is also replete with studies that both support and in other cases, provide evidence against the use of these biomarkers as either eosinophil-specific metrics (see for example (Peters et al., 1986) vs. (Sur et al., 1998)) or whether they are reflective of eosinophil-mediated disease pathology (reviewed in (Koh et al., 2007)). In addition, the more generalized dangers of a surrogate marker approach to assess disease are highlighted by the use of exhaled nitrous oxide (FENO) to diagnose and monitor symptomatic severity in asthma patients (Smith et al., 2005). That is, despite its increasingly common use, discrepancies between pulmonary inflammation, eosinophils, and levels of FENO have been observed, questioning the usefulness of this surrogate marker (Berlyne et al., 2000). Thus, the direct assessment of eosinophil numbers and/or eosinophil degranulation in patients remains the only reproducibly unambiguous metrics to quantify eosinophil mediated events in disease. Currently, assessments of eosinophil tissue infiltration are typical histological evaluations performed in the context of patient diagnostic strategies (see for example (Katzenstein, 2000)). Alternative evaluations such as assessments of eosinophil degranulation also exist but are restricted primarily to the use of commercially available ELISA kits detecting the presence of the two prominent granule ribonucleases, ECP (Reimert et al., 1991) or EDN (Morioka et al., 2000). These assays are ubiquitously used in research oriented clinical studies of patients (see for example (Tang and Chen, 2001)) but technical and logistical issues have curtailed their widespread use as diagnostic biomarkers in patient care. Specifically, the assay ranges of these ELISA kits are small and need to be carefully verified each time the assays are performed (Reimert et al., 1991; Morioka et al., 2000). In addition, studies since the identification and characterization of these eosinophil granule components have suggested that neither ECP nor EDN is eosinophil-specific (reviewed in (Metso et al., 2002)). That is, expression of both ribonucleases have been demonstrated in other leukocytes such as neutrophils (Sur et al., 1998) and, in the case of EDN, expression also extends to non-hematopoietic cell types such as the liver (Sorrentino et al., 1992). Collectively, these limitations have significantly narrowed the use of high throughput assays of ECP and EDN as diagnostic metrics of disease in many clinical settings.
This report describes the adaptation and validation of our mouse eosinophil peroxidase detection strategy (Ochkur et al., 2012) as an easy-to-use high throughput ELISA to detect and quantify the levels of human eosinophil peroxidase in patient samples/biopsies. The EPX-based ELISA we developed was shown to be a sensitive and, more importantly, a uniquely eosinophil specific assay. Comparative studies are presented showing that the commercially available assays detecting ECP and EDN detect these proteins produced by cells other than the eosinophil. In contrast, the signal derived from the eosinophil peroxidase (EPX)-based ELISA is restricted to the presence/absence of eosinophils. The EPX-based ELISA also displayed a similar level of sensitivity relative to the assays for ECP and EDN, doing so with a large response range. Moreover, we demonstrated the utility of this EPX-based assay as a means of assessing patients in a variety of clinical settings, including bronchoalveolar lavage (BAL) samples following segmental bronchiole-allergen challenge, induced sputum following inhalation of hypotonic saline, and nasal lavage derived from chronic rhinosinusitis patients. Thus, this direct biomarker of eosinophils and eosinophil-mediated activities represents a novel metric for ex vivo studies of mixed cell populations as well as providing a diagnostic assessment tool to evaluate patients. Moreover, we demonstrated in a companion manuscript the specificity and use of this EPX-based ELISA as a reliable diagnostic metric with which to manage the care of respiratory patients (Nair et al., 2012). In summary, these reports suggest that measures of EPX provide a needed assay that is eosinophil-specific, sensitive, and useful as a high throughput format in a variety of clinical settings.
MATERIALS AND METHODS
2.1 Antibodies
EPX specific mouse monoclonal antibodies were generated by immunizing eosinophil peroxidase knockout mice (EPX−/− (Denzler et al., 2001)) as previously described (Protheroe et al., 2009). The resulting hybridomas (~2000) were screened for the IgG isotype and for immune reactivity to EPX using a single dimensional format. The hybridomas surviving these initial screens underwent further selection on the basis of their secreted monoclonal antibody being a human EPX specific reagent as determined by immunohistochemistry with formalin-fixed paraffin embedded biopsies (Protheroe et al., 2009). These final monoclonal antibodies (~10) were assessed for their functionality in both western blots of cell/tissue extracts and in a soluble sandwich ELISA format (unpublished observations and (Protheroe et al., 2009), respectively). From these, two monoclonal antibodies were selected (clone MM25-429.1.1 as the capture antibody and clone MM25-82.2.1 as the detection antibody) for the development of a soluble format ELISA (i.e., sandwich ELISA) to detect EPX. The detection antibody was biotinylated using an EZ-Link NHS-LC-Biotin kit (Pierce, Rockford, IL (USA)) that had a reproducible addition efficiency of 8–12 molecules of biotin per molecule of immunoglobulin. The overall strategy of EPX purification, the generation of specific mouse monoclonal antibodies, and the subsequent identification of an antibody pair for use in an EPX-specific ELISA for human clinical fluid samples is schematically summarized in Figure 1.
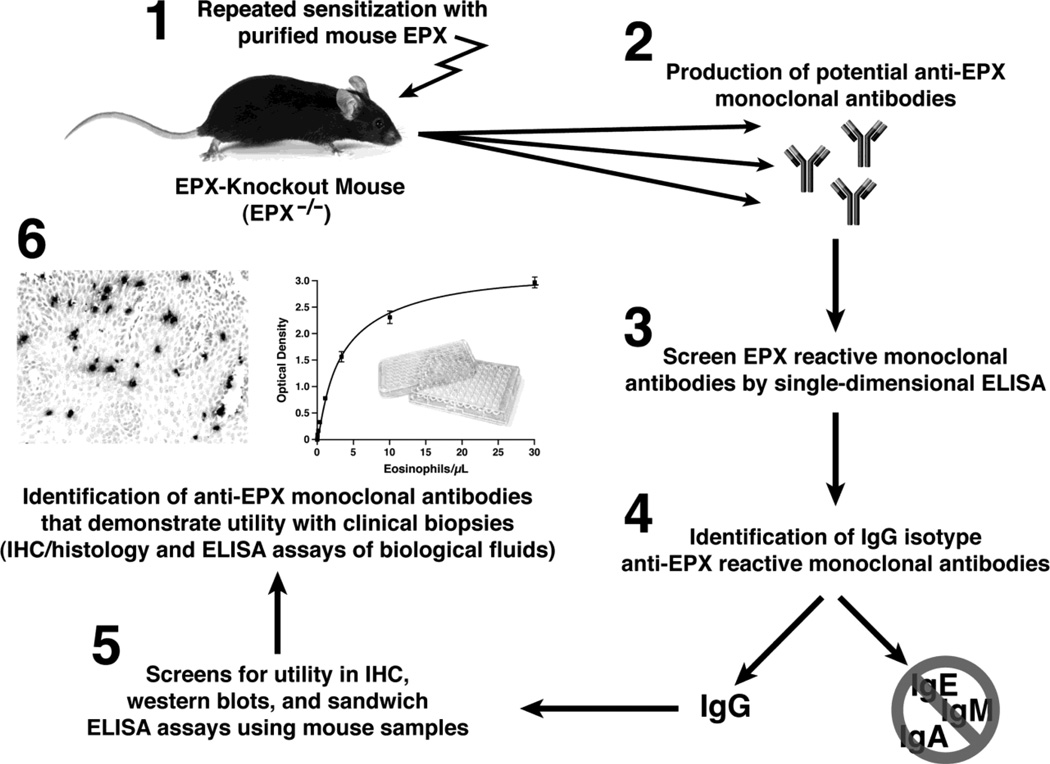
Figure 1. The generation of mouse anti-EPX monoclonal antibodies and the development of an EPX-specific sandwich ELISA.
EPX-specific monoclonal antibodies with utilities in immunohistochemical and an ELISA format were generated by the sensitization of EPX knockout mice (EPX−/−) with purified mouse EPX (panel 1). The generation and screening of EPX-specific monoclonal antibodies (panels 2 – 4) were described earlier (Protheroe et al., 2009). The monoclonal antibodies surviving these screens were evaluated for their usefulness in immunohistochemistry (IHC), western blot, and ELISA using samples derived from mouse cells/tissues (panel 5). Monoclonal antibodies of defined utilities were further evaluated for similar applicability with human biopsies and fluid samples to define reagents for use in clinical settings (panel 6).
2.2 EPX ELISA Required Reagents and Disposables
The development of the EPX-based sandwich ELISA was similar to methods we described earlier (Ochkur et al., 2012). In order to eliminate any potential interference from the activity associated with EPX it was necessary to avoid peroxidase-based detection systems. For example, the commonly used substrate in these systems (i.e., TMB (3,3', 5,5"-tetramethylbenzidine)) is readily converted by the peroxidase activity of EPX into the same colored product that is measured by the detection system itself (our unpublished observations). The consequences are obvious as an ELISA based on this detection method would appear more sensitive and would not accurately quantify the level of EPX actually present in a given sample. These logistical issues were resolved here by focusing our efforts on an alkaline phosphatase-based detection strategy. The EPX-based ELISA was created with KPL (Gaithersburg, MD (USA)) reagents optimized for alkaline phosphatase based sandwich ELISA in combination with BluePhos™ substrate. The following reagents were also utilized as part of the final ELISA protocol: 10X Coating Solution Concentrate, 10% BSA Diluent/Blocking Solution Kit, 20X Wash Solution Concentrate, BluePhos™ Microwell Phosphatase Substrate System, Streptavidin-Alkaline Phosphatase (Strep-AP) from RD (R&D Systems, Minneapolis, MN (USA)), and Trizma hydrochloride buffer solution (Sigma-Aldrich, St. Louis MO (USA)) - used to prepare Streptavidin-AP Diluent. Solid phase 96 well Nunc-Immuno Plates with MaxiSorp surface (Thermo Scientific San Diego, CA (USA)) were used as the support structure to perform this ELISA in a high throughput format. BioTek µQuant Microplate Spectrophotometer with KC4™ Data Analysis Software from Bio-Tek (Winooski, VT (USA)), was used for OD, max OD, and spectrum reading of the ELISA plates.
2.3 Isolation of Human Eosinophils
Whole blood was collected into EDTA containing Venous Blood Collection Tubes (Becton Dickerson (BD), Franklin Lakes, NJ (USA)). Lymphocytes and granulocytes were separated from red blood cells using Histopaque 1077 and 1119 (Sigma-Aldrich, St. Louis, MO (USA)) following the manufacturer’s instructions. The layer containing the granulocytes was washed with PBS and subjected to hypotonic lysis to remove any remaining red blood cells. The granulocytes are resuspended in 0.5% BSA, 2mM EDTA in PBS. Eosinophils were purified (>98%) by selective depletion through positive selection and removal of other leukocytes using a Human Eosinophil Purification Kit (Miltenyi Biotec GmbH, Bergisch Gladbach, Germany), again following the manufacturer’s instructions.
2.4 EPX and Eosinophil Standards
Human eosinophil peroxidase (>98% purity) was purchased from Lee Biosciences (St. Louis, MO (USA)) and serially diluted in Diluent/Blocking Solution that was itself previously diluted 1:15. This series of purified EPX samples was used to create assay standard curves. In some cases, whole cell extracts were prepared from purified human eosinophils. Briefly, at room temperature purified eosinophils (>98%) were re-suspended in PBS, subsequently centrifuged at 400g for 5 minutes at 4 °C, and this supernatant (80% of the total volume) was replaced with solution containing 0.22% hexadecyltrimethylammonium bromide (Sigma-Aldrich, St. Louis, MO (USA)) and 0.3M sucrose. This suspension was vortexed for 1 minute at room temperature and one time use only aliquots were prepared and flash-frozen in liquid nitrogen (i.e., LN2). Aliquots of eosinophil extracts were stored −80°C until used. Serial dilutions of these extracts were used for assay optimization, generation of standard curves, and quality control in further experiments.
2.5 Human EPX ELISA protocol
The basic Sandwich/Capture ELISA protocol was created as a modification of our previously published protocol for the detection and quantification of EPX in fluid samples from mouse models of human disease (Ochkur et al., 2012). This original protocol was used as a reference point for the development of the final optimized assay of EPX in human fluid samples from patients in a health care setting(s):
A micro-titer plate is pre-treated with 4µg/ml anti-EPX monoclonal antibody MM25-429.1.1 (capture antibody) in 100µl of Coating Solution at 4 °C overnight.
Coated wells of the micro-titer plate are cleared of unbound antibody with 4–5 cycles of rinsing using Wash Solution.
Potential areas of non-specific binding in each well of the plate are blocked by a 30 minute room temperature pre-incubation with 300µl of Diluent/Blocking Solution that was itself previously diluted 1:10.
Following the incubation to block the wells, the Diluent/Blocking Solution is removed and 100µl of sample (and/or standard) are added and the plate is incubated at room temperature without shaking for 1.5–2 hours.
The wells of the micro-titer plate are cleared of unbound target antigen with 4–5 cycles of rinsing using Wash Solution. 100µl of biotinylated anti-EPX monoclonal antibody MM25-82.2.1 (detection antibody) are added to each well at a final concentration of 0.8µg/ml and the plate is incubated at room temperature for 1.5–2 hours.
The wells of the micro-titer plate are cleared of unbound detection antibody with 4–5 cycles of rinsing using Wash Solution. 100µl of Strep-AP (diluted 1/500 in 1% BSA, 0.05% Tween 20, 0.025M Tris, 0.5M NaCl (pH 7.4)) are added to each well and the plate is incubated at room temperature for 30 minutes.
The wells of the micro-titer plate are cleared of unbound Strep-AP with 4–5 cycles of rinsing using Wash Solution. 100µl of BluePhos™ substrate are added to each well and the plate is incubated at 37°C for 1 hour with gentle rotation.
The colorimetric reaction is terminated with the addition of 100µl of Stop Solution. Absorbance of individual wells of the plate is determined at a wavelength of 630nm.
2.6 Eosinophil Stimulation Ex Vivo
Human peripheral blood eosinophils were collected and purified (as described earlier) prior to a manual count using a hemacytometer and final resuspension in Phenol Red-free RPMI (final cell concentration = 0.5 × 106 cells/mL. 200µl aliquots (1 × 105 eosinophils) were prepared in 96-well microtiter plates and incubated for 30 minutes in an atmosphere of 5% CO2 and 95% humidity following the addition of either 50ng/ml PAF-C18 (Alexis Biochemicals, San Diego, CA (USA)) or 50ng/ml PAF-C18 plus 1µM Ionomycin (Sigma-Aldrich, St. Louis MO (USA)); eosinophil viability following incubation was ~95% as determined by Trypan Blue (Sigma-Aldrich, St. Louis MO (USA)) exclusion. DMSO (Sigma-Aldrich, St. Louis MO (USA)) alone was used as a vehicle control. The cells in each well were removed by centrifugation (1300 × g, 5 minutes) and the recovered supernatants were further cleared of any cell or organelle (i.e., secondary granules) contamination by an additional high speed centrifugation (13,000 × g, 5 minutes). These final cell and organelle-free supernatants were stored at −80°C until use.
2.7 Preparation of Clinical Fluid Samples for Assessment of EPX Levels
Segmental bronchiole-allergen challenge and BAL of subjects with mild allergic asthma
The bronchoscopy studies were approved by the University of Wisconsin Institutional Review Board of the Human Subjects Committee. Written informed consent was obtained from all subjects prior to their participation. At least one month prior to bronchoscopy, subjects underwent a graded whole-lung allergen inhalation challenge to determine the provocative dose of allergen required to achieve a 20% reduction (AgPD20) in forced expiratory volume in one second (FEV1) (Chai et al., 1975). The total dose for segmental allergen change was 30% of the AgPD20; a dose of 10% was instilled into one bronchopulmonary segment and 20% was instilled into a second segment as previously described (Liu et al., 2002). Allergens included short ragweed (Ambrosia artemisiifolia), house dust mite (Dermatophagoides farinae) and standardized cat hair (Felis catus domesticus) extracts, obtained from Greer Laboratories (Lenoir, NC (USA)). Forty-eight hours after segmental allergen challenge, bronchoscopy and BAL were performed in the two antigen-challenged segments. BAL fluid kept on ice, pooled, mixed thoroughly, and stored in 1 mL aliquots at −80°C until used. Total BAL cell numbers were determined as a manual cell count with a hemocytometer using Turk’s counting solution containing acetic acid and methylene blue. Two slides from each BAL cell preparation were prepared by cytospin and stained with the Wright Giemsa-based Hema 3 system (Fisher Diagnostics, Middletown, VA (USA)). In total, 500 cells/slide (1000 cells/sample) were counted.
Induction and Collection of Sputum from Subjects with Allergic Asthma
The respiratory patients with asthma were defined based on a PC20 following methacholine challenge of <8 mg/ml or a >15% (and 200ml) improvement in FEV1 after inhaling a short-acting bronchodilator. Otherwise-healthy volunteers served as control subjects. Sputum from each individual was induced by inhalation of hypertonic saline, selected from the expectorate, and processed as described earlier (Pizzichini et al., 1996). Briefly, the recovered sputum was treated with dithiothreitol (DTT) to disperse the mucus for processing by cytocentrifugation. A manual total cell count (expressed as cells/gram of sputum × 105) was performed using a hemacytometer (trypan blue was used to determine cell viability). Cell differential assessments were determined from Wright stained cytospin preparations, counting >200 cells. The sputum supernatants were storing at −70°C until use. All subjects provided consent to use their sputum samples for biomarker discovery and the studies were approved by the Firestone Institute for Respiratory Health, St. Joseph’s Healthcare & McMaster University Hospital Research Ethics Board.
Lavage and Recovery of Fluid Samples from Sinus Patients
Subjects with chronic rhinosinusitis (CRS) were recruited from the allergy and otolaryngology clinics at Northwestern University Feinberg School of Medicine. Nasal lavage fluids were obtained in the clinic or in the operating room as previously described (Kato et al., 2008). Specimens from patients without CRS were obtained in the allergy clinic or in the OR during the performance of skull base tumor excisions, facial fracture repair, lacrimal duct surgery and orbital decompression surgery. All samples were collected with informed consent and the protocol used was approved by the Northwestern Investigational Review Board.
Regardless of the origin of the biological fluid to be assayed, all samples were thawed on ice, subjected to vortexing for 10–20 seconds, and pulse-spun for 30 seconds at 10,000 × g, prior to dilution in Diluent/Blocking Solution that was itself previously diluted 1:15. The extent of dilution was established by trial and error and reflected either sample availability and/or the EPX levels typically found in a given sample type: BAL fluid = 1:2, Sputum Supernatant = 1:3, Nasal lavage fluid = 1:5
2.8 Statistical Analysis
GraphPad Prism 5 (GraphPad Software, Inc. La Jolla, CA (USA)) was used for basic analysis, including the generation of graphs and plots. JMP (SAS Institute, Cary, NC (USA)) was used as a design of experiment statistical platform. Statistical analysis for comparisons between groups was performed using ANOVA and a pair-wise Student's T test. Data are expressed as the mean ± SEM. Differences between mean values were considered significant when P<0.05.
3. RESULTS
3.1 The development and optimization of a human EPX-based ELISA
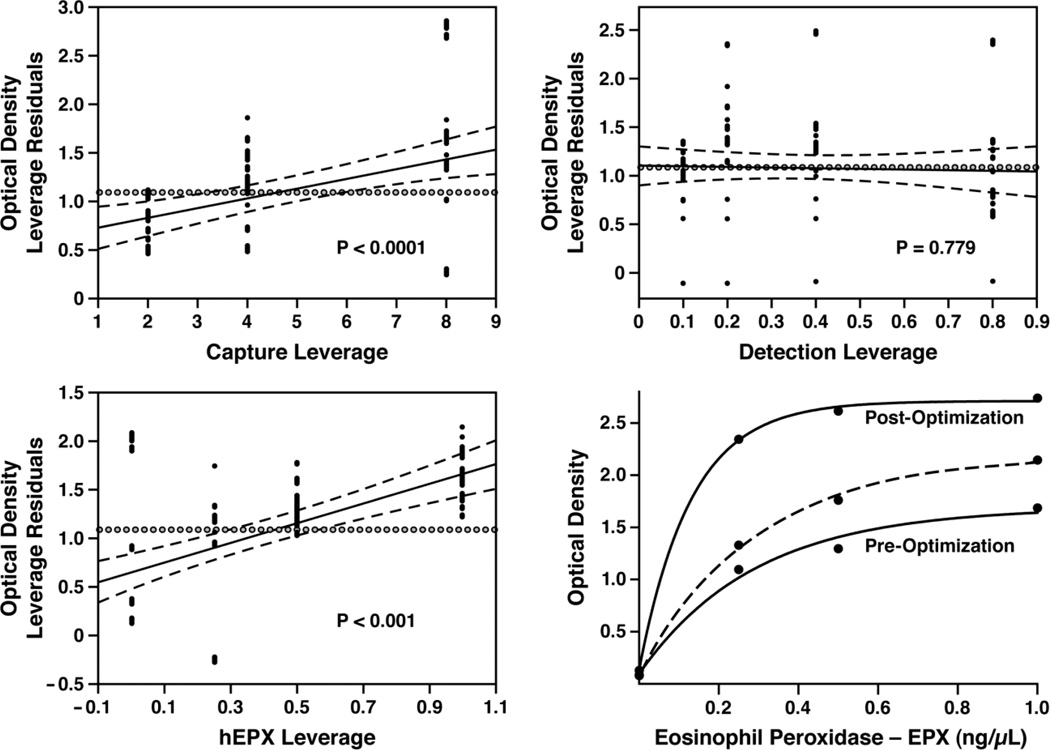
A full-factorial approach (see Materials and Methods) to optimize an EPX-based ELISA (Figure 2) generated the initial dose response curve in the lower right-hand panel (Pre-Optimization), an assay maximizing signal strength (dose response curve in the lower right-hand panel labeled Post-Optimization), and finally a logistical compromise of maximizing signal to noise response using antibody concentrations that were practical for routine use as a high throughput assay (dose response curve drawn with a dashed line in the lower right-hand panel). This final assay has a limit of detection (i.e., measureable signal three standard deviations above background) of 1.4 ± 0.2ng/mL.
Figure 2. Full factorial analysis of the EPX-specific antibody pair optimizing the responsiveness of the EPX-specific sandwich ELISA.
The significance of each factor, (capture antibody, detection antibody and eosinophil concentrations) as well as the effect of each data point are presented as leverage plots obtained in the final experiment. The horizontal lines ( ) in each plot represent the overall average. Predicted values derived from the fitted curves are shown as solid lines with 95% CI (confidence interval) as flanking dashed lines. The distance from each data point (●) to the line of fit is the error or residual for that point. The distance from each data point (●) to the horizontal line (
) in each plot represent the overall average. Predicted values derived from the fitted curves are shown as solid lines with 95% CI (confidence interval) as flanking dashed lines. The distance from each data point (●) to the line of fit is the error or residual for that point. The distance from each data point (●) to the horizontal line ( ) is what the error would be if you removed the effects this point has in the fitted curve model. The data points farther from the middle of the plot in the horizontal direction have more effect/leverage on the fit. As a result, the strength of the effect of varying each parameter is shown by how the line of fit is suspended away from the horizontal by the data points. If the 95% confidence curves cross the horizontal reference line (
) is what the error would be if you removed the effects this point has in the fitted curve model. The data points farther from the middle of the plot in the horizontal direction have more effect/leverage on the fit. As a result, the strength of the effect of varying each parameter is shown by how the line of fit is suspended away from the horizontal by the data points. If the 95% confidence curves cross the horizontal reference line ( ), then the effect is significant; if the curves do not cross, variations of this parameter failed to achieve statistical significance (at the 5% level). The standard curves derived from the ELISA under the initial assay conditions (2µg/ml of capture and 0.4µg/ml of detection antibody, Pre-Optimization) and the final assay conditions (16µg/ml of capture and 1.6µg/ml of detection antibody, Post-Optimization) are shown by optical density as a function of input purified eosinophil peroxidase (ng/µl). Subsequent optimization experiments maximizing the assay's signal to noise response with antibody concentrations that were practical in a high throughput format (4µg/ml of capture and 0.8µg/ml of detection antibody) resulted in the final EPX-based ELISA assay shown (dashed line in the lower right-hand panel).
), then the effect is significant; if the curves do not cross, variations of this parameter failed to achieve statistical significance (at the 5% level). The standard curves derived from the ELISA under the initial assay conditions (2µg/ml of capture and 0.4µg/ml of detection antibody, Pre-Optimization) and the final assay conditions (16µg/ml of capture and 1.6µg/ml of detection antibody, Post-Optimization) are shown by optical density as a function of input purified eosinophil peroxidase (ng/µl). Subsequent optimization experiments maximizing the assay's signal to noise response with antibody concentrations that were practical in a high throughput format (4µg/ml of capture and 0.8µg/ml of detection antibody) resulted in the final EPX-based ELISA assay shown (dashed line in the lower right-hand panel).
3.2 The sensitivity of the EPX-based ELISA is comparable to commercially available assays detecting ECP and EDN but unlike these other assays, EPX-based ELISA is eosinophil-specific
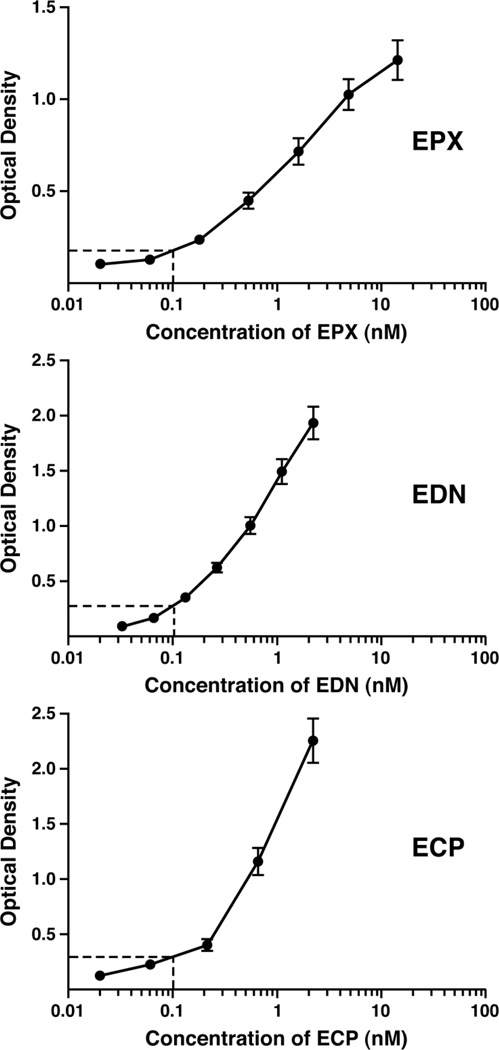
Side-by-side comparisons of our EPX-based ELISA and the commercially available ELISA kits (www.mblintl.com) detecting ECP and EDN were performed using purified protein in each case to determine the relative sensitivities of the assays (Figure 3). The dose response data derived from these comparisons demonstrated that the EPX-based ELISA displayed comparable assay responsiveness in the lower range of detection relative to the ECP and EDN ELISA kits with limits of 18.2pM, 6.7pM, and 33.3pM, respectively.
Figure 3. EPX-based ELISA is an assay whose sensitivity is comparable to the commercially available ELISA kits assessing ECP and EDN.
The assay response of EPX-based ELISA was evaluated in a side by side comparison with the commercially available ECP and EDN ELISA kits. In each panel, ELISA responses (optical density) were plotted as a function of input purified eosinophil granule protein (nM). The identified point within each curve (dashed lines) was chosen arbitrarily to provide a visible comparative reference of the relative sensitivities of each assay at an input of 0.1nM of each eosinophil granule protein. All assays were run as either duplicate or triplicate samples in two independent experiments. The data are presented as means ± SEM.
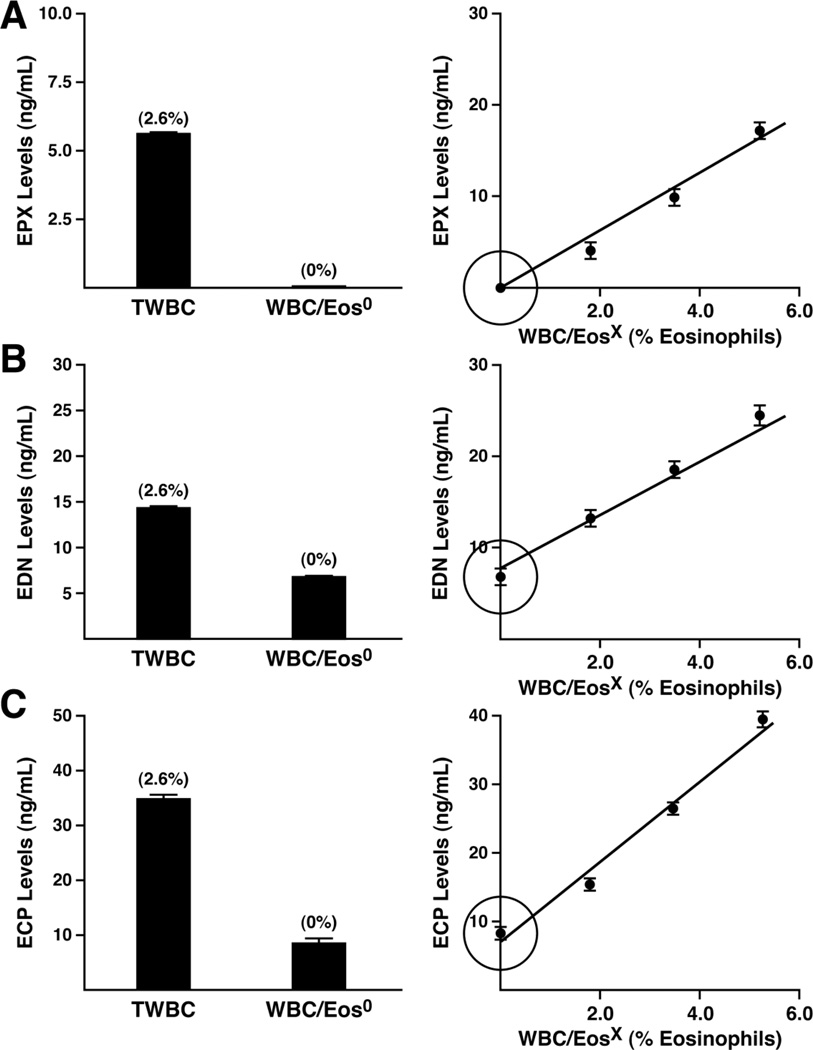
The eosinophil specificities of EPX-based ELISA relative to the commercial assays detecting ECP and EDN were determined based on the responsiveness of these assays using cell extracts of mixed peripheral white blood cells derived from healthy control donors (Figure 4). In particular, we compared ELISA responses using extracts derived from total white blood cells (TWBC) relative to white blood cells following three complete rounds of magnetic bead selection/column chromatography generating a white blood cell population devoid of eosinophils (WBC/Eos0). Moreover, the ELISA signals from each of these assays were also determined in eosinophil-depleted white blood cell populations subsequently enriched with defined numbers of eosinophils (WBC/EosX). In all cases, manual cell counts and differentials (>500 cells/slide) were used to determine the presence vs. absence of eosinophils as well as the absolute number of eosinophils present in each population. EPX-based ELISA assessments of peripheral white blood cells clearly demonstrated the eosinophil specificity of this assay with virtually no response above the assay's detection limit using cell extracts devoid of eosinophils (Figure 4(A), left panel). In addition, ELISA-based EPX levels increased linearly as a function of the number of eosinophils present in the enriched white blood cell populations examined (WBC/EosX), independently demonstrating the eosinophil-specificity of this assay (Figure 4(A), right panel). It is also noteworthy that this lack of signal in white blood cell populations devoid of eosinophils demonstrated that the EPX-based ELISA is EPX specific, showing no cross-reactivity with the other prominent leukocyte-derived peroxidase, MPO (Winterbourn et al., 2000; Klebanoff, 2005).
Figure 4. EPX-based ELISA uniquely represents an eosinophil-specific assay among the available ELISA assessments of eosinophil granule proteins.
The levels (ng/mL) of EPX (A), EDN (B), and ECP (C) in white blood cell extracts demonstrate that unlike ELISA assessments of EDN and ECP, the EPX-based ELISA is eosinophil-specific. The histograms shown in the left panels directly compare the assay responses of each granule protein ELISA using cell extracts (167 cells/µl) from total white blood cells (TWBC) vs. cell extracts from white blood cell populations in which eosinophils had been selectively removed (WBC/Eos0). The percentage of eosinophils identified in each population (derived from a manual cell count/differential of 500 cells/slide) is shown in parentheses above each histogram. The linear plots on the right panels show the responses from these assays as a function of the percent eosinophils in a series of white blood cell extracts generated from cell populations with equal cell concentrations (167 cells/µl) created by adding purified eosinophils back to the white blood cell population previously devoid of eosinophils (WBC/EosX). All assays were run as either duplicate or triplicate samples in two independent experiments. The data are presented as means ± SEM. *P < 0.05
In contrast to EPX-based ELISA, neither of the two commercially available assays detecting EDN and ECP displayed eosinophil-specific responses (Figure 4(B) and (C), respectively). These assays (left panels) detected the presence of significant amounts of EDN and ECP even in white blood cell populations devoid of eosinophils (WBC/Eos0), 46.5% of TWBC and 23.6% of TWBC, respectively. Nonetheless, these assays displayed linear increases in signal as a function of eosinophil number in the enriched white blood cell populations examined (right panels). Thus, significant levels of EDN and ECP are clearly found in eosinophils; however, the EDN and ECP assays also detect the presence of these proteins in one or more other white blood cell lineages (i.e., they are quantitatively responsive to the presence of eosinophils but they are not eosinophil-specific, particularly in cases where eosinophils are only a minority population).
3.3 EPX-based ELISA allows for the detection and quantification of eosinophil degranulation using ex vivo cell culture assays
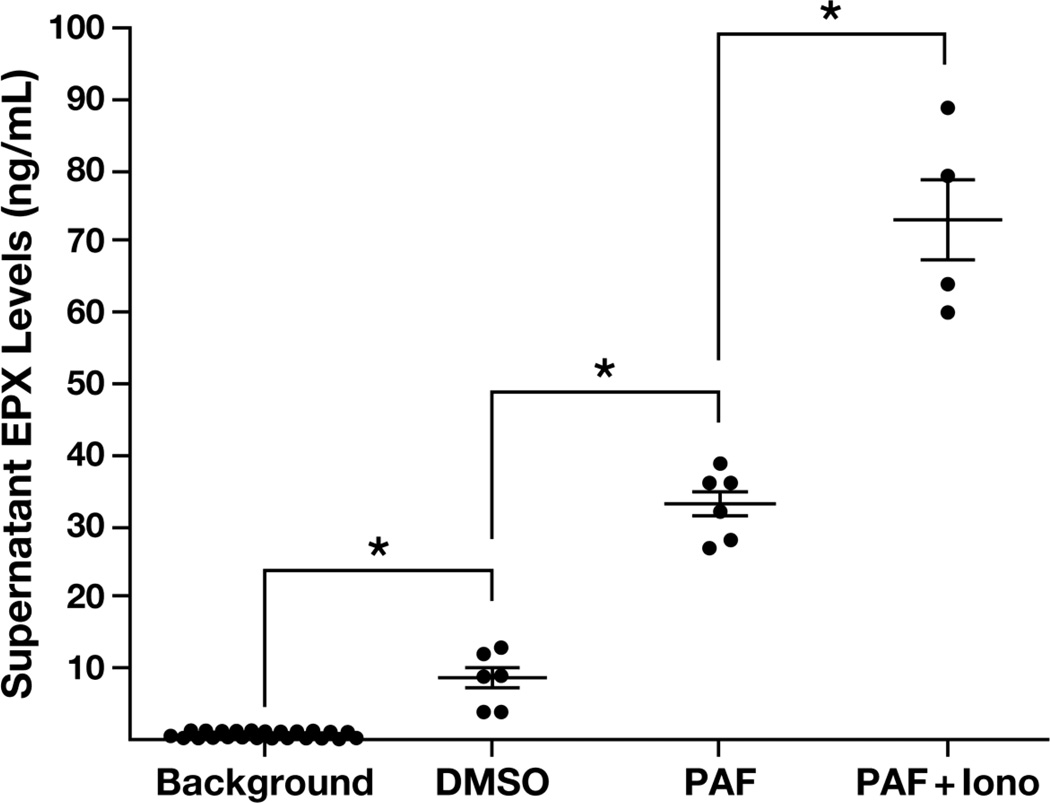
Eosinophils were isolated from the blood of healthy control donors and stimulated in culture to elicit degranulation (Figure 5). The EPX-based ELISA allowed for the detection (i.e., relative to background control (BKG)) of induced EPX release from eosinophils following stimulation with PAF alone. In addition, this ELISA displayed a >2-fold increase above this PAF signal following stimulation with PAF+Iono (33ng/mL vs. 73ng/mL, respectively), suggesting that this assay is capable of quantifying the degranulation events occurring in response to various experimental stimuli. Interestingly, the EPX-based ELISA was also capable of detecting and quantifying degranulation from eosinophils cultured without stimulation (i.e, media with vehicle (DMSO)). It is also noteworthy that the concentrations of PAF and/or Ionomycin used to stimulate eosinophils were at the very low end of the range (<10%) typically used (see for example (Dyer et al., 2010)) and yet the assay’s sensitivity was sufficient to quantify the induced degranulation. An additional consequence of the absolute cell specificity of EPX-based ELISA is an ability to use this assay to estimate the concentration of EPX per eosinophil. That is, it is possible to use the standard curves derived from the EPX-based ELISA representing assay responsiveness with eosinophil cell extracts vs. purified protein as a means of approximating the EPX content of individual eosinophils. Compilation of data derived from the blood leukocytes of five subjects (2 healthy controls, 1 allergic asthma patient, 1 subject with scleroderma and 1 atopic dermatitis patient) showed that the estimated concentration of EPX in peripheral blood eosinophils is 33 ± 8.6pg/eosinophil.
Figure 5. EPX-based ELISA provides an easy-to-use high throughput assay for ex vivo assessments of eosinophil degranulation.
Scatter plots of cell-free EPX released by stimulating eosinophils with the secretagogues platelet activating factor (PAF) or PAF and Ionomycin (PAF+Iono) are presented relative the assay's background response with media alone (BKG). Culture supernatants from eosinophils incubated in DMSO containing media served as a vehicle control for secretagogue-induced EPX release (DMSO). The resulting absolute values of EPX (ng/mL) corresponding to each assessment were derived from assay-specific standard curves using purified EPX. All assays were run as either duplicate or triplicate samples in three independent experiments. The data are presented as means ± SEM. *P < 0.05
3.4 EPX-based ELISA provides a sensitive and quantitative assay for the detection of eosinophil degranulation using clinically-derived fluid samples from patients
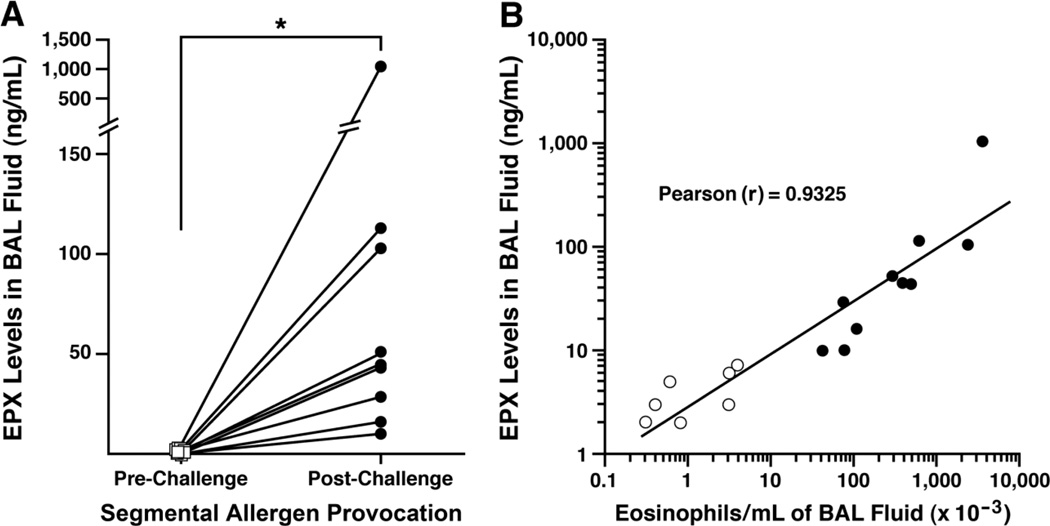
Provocation of asthma patients revealed that EPX levels in BAL fluid following segmental allergen challenge are a surrogate biomarker for the induced airways eosinophilia: The responsiveness of EPX-based ELISA was demonstrated (Figure 6) in a post hoc assessment of BAL fluid obtained from asthma subjects 48 hours after segmental allergen challenge (Liu et al., 2002). This assessment showed that in contrast to the nearly undetectable pre-challenge EPX levels, the EPX-based ELISA demonstrated significant increases in the same airway segments following allergen challenge (Figure 6(A)). BAL levels of EPX following segmental allergen challenge also increased as a function of the induced BAL eosinophilia (r = 0.93), suggesting that EPX-based ELISA is a reproducibly accurate metric representative of the number of airway eosinophils (Figure 6(B)).
Figure 6. EPX levels correlate with the observed airways eosinophilia induced by allergen challenge of asthmatic patients.
(A) Post hoc assessments of bronchoalveolar lavage (BAL) fluid from asthma patients (n = 10) undergoing segmental allergen provocation as part of an investigatory study in a pulmonary clinic setting (Liu et al., 2002). EPX levels (ng/mL) in BAL fluid increased significantly 48 hours following segmental allergen challenge (Post-Challenge (●)) relative to the levels observed in these same patients prior to allergen challenge (Pre-Challenge (□)). *P<0.01 (Pre-Challenge Mean ± SEM/Median = 0.3 ± 0.3/0.0; Post-Challenge Mean ± SEM/Median = 146.8 ± 100.3/44.5). (B) EPX levels (ng/mL) observed in BAL fluid recovered from asthma patients (pre- and post- aeroallergen challenge) were plotted relative to the number of eosinophils in these same BAL fluids (identified by manual cell counts and differentials of >300 cells/slide). The data points represent individual patients ((○) = normal and non-allergen challenged control allergic asthmatics and (●) = allergic asthmatics 48 hours following segmental allergen challenge). All assays were run as duplicate samples performed as a single experiment. The line derived from a linear regression of the collective data set comparing BAL EPX levels vs. BAL eosinophilia (r = 0.93, p = 0.001).
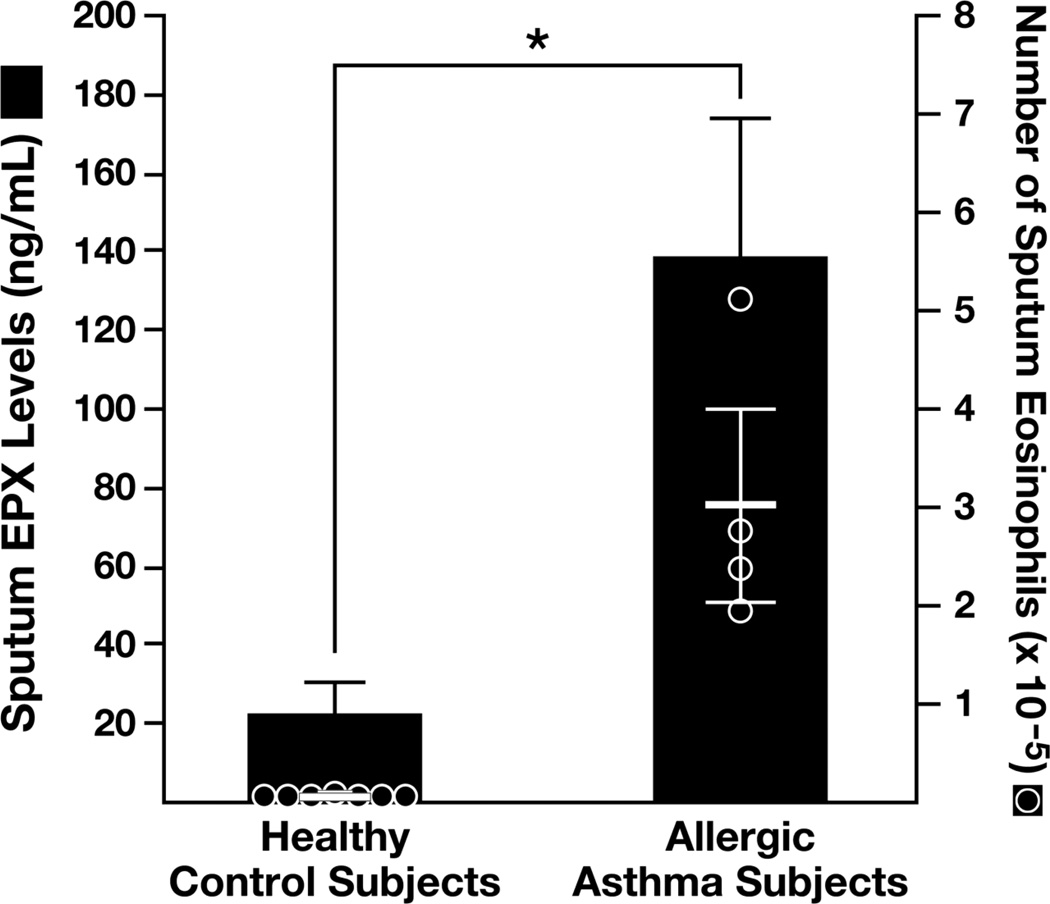
EPX levels in sputum derived from subjects with allergic asthma correlate with airways eosinophilia
EPX-based ELISA was performed as a post hoc assessment of sputum from respiratory subjects (Pizzichini et al., 1996). These assessments showed that EPX levels in cell-free supernatants were significantly higher in allergic asthma patients relative to otherwise healthy volunteer subjects (Figure 7). These data further showed that the increased sputum EPX levels observed in asthma patients were a valid surrogate biomarker of the significant airways eosinophilia that occurred in these subjects.
Figure 7. ELISA-based assessments of sputum EPX levels represent a surrogate biomarker representative of the airways eosinophilia occurring in asthmatic patients.
Post hoc assessments of induced sputum from asthma patients (n = 4) and otherwise healthy control subjects (n = 7) recovered as part of therapeutic clinical studies in a hospital/medical center setting (Pizzichini et al., 1996). The histograms presented showed that EPX levels in cell-free sputum supernatants (ng/mL-gram of sputum) were significantly higher than the levels observed in healthy control subjects. The overlaid scatter plots (●) of sputum eosinophil numbers observed in these patients (identified by manual cell counts and differentials of >200 cells/slide) demonstrated that the increased sputum EPX levels correlated with the induced airways eosinophilia occurring in asthma patients. All assays were run as duplicate samples performed as a single experiment. *P<0.05
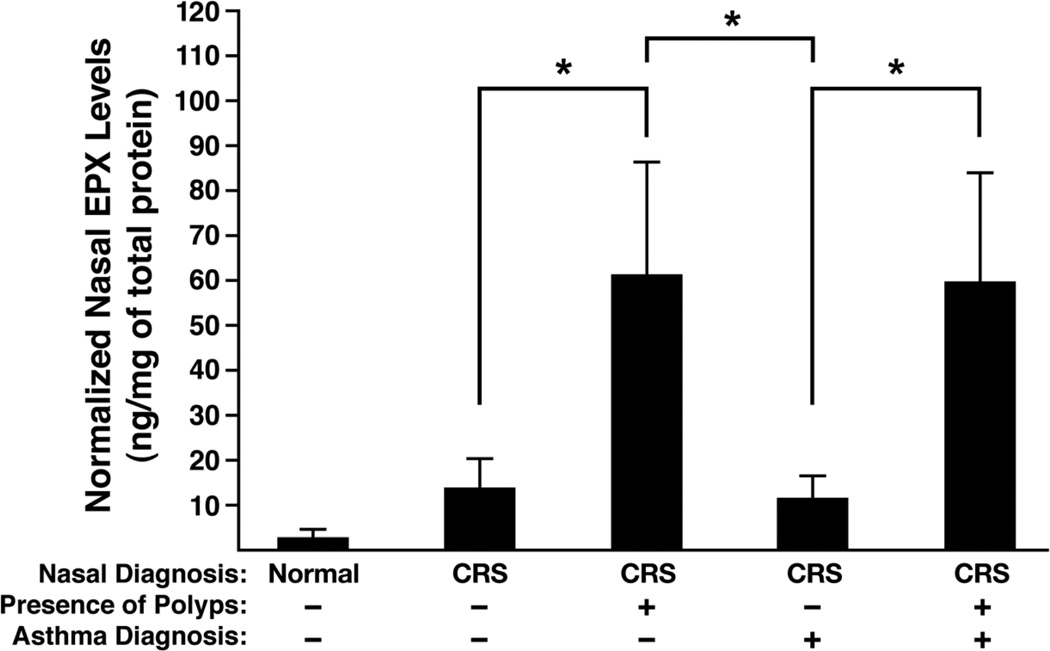
EPX levels in nasal lavage of chronic rhinosinusitis patients are a potentially useful surrogate biomarker for the presence of eosinophilia in nasal polyps
The utility of EPX-based ELISA as a discriminatory marker stratifying rhinitis patients was demonstrated (Figure 8) in a post hoc assessment of nasal lavage fluid derived from cohorts of subjects in a study of chronic rhinosinusitis (CRS) patients (Kato et al., 2008; Schleimer et al.). These data showed that EPX levels in CRS patients who lacked nasal polyps were increased five-fold relative to the nominal levels observed in nasal lavage from healthy control subjects. Moreover, the presence vs. absence of an asthma diagnosis in patients with non-polypoid disease did not affect the observed levels of EPX in these nasal lavage samples. These assessments, however, showed that the presence of nasal polyps in CRS patients (irrespective of an asthma diagnosis) was linked with significant increases in nasal EPX levels, and in turn eosinophils, confirming observations first identified in the original study of CRS patients (Schleimer et al.).
Figure 8. ELISA-based assessments of EPX levels represent an accurate biomarker for the presence of nasal polyps in subjects with chronic sinusitis (CRS).
EPX levels in nasal lavage fluid (ng/mg of total protein) recovered from CRS patients were assessed (relative to healthy control patients (Normal) in a post hoc assessment of a previous study of these patients (Kato et al., 2008; Schleimer et al.). The assessment was performed using a factorial strategy examining subjects with (+) and without (−) polyps as well as with (+) and without (−) an asthma diagnosis. Mean ± SEM/Median EPX levels for each group: Normal = 2.8 ± 2.3/0.0; CRS/Polyps(−)/Asthma(−) = 14.0 ± 6.4/0.0; CRS/Polyps(+)/Asthma(−) = 60.7 ± 25.5/21.5; CRS/Polyps(−)/Asthma(+) = 11.0 ± 4.7/7.7; CRS/Polyps(+)/Asthma(+) = 59.4 ± 24.7/22.1; All assays were run as duplicate samples performed as a single experiment. *P<0.02
4. DISCUSSION
Eosinophil-associated diseases appear to be ever-increasingly common, expanding in some cases at almost epidemic proportions (Bloom and Cohen, 2007; Pleis and Lethbridge-Cejku, 2007). However, our understanding of eosinophils and their associated effector functions have also increased exponentially. Thus, this more complete understanding of the varied contributions of eosinophils in health and disease, together with the higher prevalence of eosinophil-associated diseases, has dramatically increased demand for assays detecting the presence vs. absence of these cells and/or their activities. Two prominent examples highlighting this increase are the direct assessments of airway eosinophils as a diagnostic metric in the management of asthma patients (reviewed in (Hargreave and Nair, 2011)) and a near total dependence on identifying eosinophils in biopsies for the diagnosis of eosinophilic esophagitis (Furuta et al., 2007). It is noteworthy that this greater appreciation of the importance of eosinophils has also led to the adoption and, in some cases, wide use of surrogate biomarkers for eosinophil mediated inflammation such as assessments of FENO in asthma patients (reviewed in (Smith et al., 2005)).
Unfortunately, logistical/technical uncertainties have been linked with all of the available assays assessing eosinophils and their activities, including questions regarding assay specificity (Metso et al., 2002) as well as the correlative significance of particular surrogate biomarkers (Berlyne et al., 2000). These uncertainties have renewed interest among investigators for alternative diagnostic strategies based on the detection and quantification of eosinophils. Our continued investigations of eosinophil-specific expression of EPX in mouse models of human diseases (Denzler et al., 2001; Lee et al., 2004; Ochkur et al., 2012) and the subsequent characterization of EPX-based diagnostics in the characterization of patients in clinical settings (Protheroe et al., 2009; Willetts et al., 2011) have been driven, in part, because of this renewed interest. These evolving studies provide an alternative strategy for the unambiguous detection and quantification of eosinophils. Specifically, our development of a sensitive high throughput EPX-based ELISA solves many of the logistical/technical issues that have plagued the currently available assays for the assessment of eosinophils and their activities from patient-derived fluid samples:
EPX-based ELISA has a large response range whose assay sensitivity was equivalent to that of the only available commercial kits for other prominent eosinophil granule proteins, ECP and EDN.
EPX-based ELISA is the only assay among these detection options to display an unambiguous specificity for eosinophils. In particular, the data presented showed that ECP and EDN were also present in one or more other leukocyte populations. Significantly, the wider pattern of cell expression exhibited by ECP and EDN did not affect the utility of these assays to quantify eosinophils/activities when these granulocytes were present in large numbers in the populations examined. However, clearly when the number of eosinophils is limited, the contribution from other sources dominated the response derived from these assays.
A direct consequence of the unique eosinophil-specificity displayed by EPX-based ELISA is that this assay does not recognize the myeloid cell specific myeloperoxidase prominent in neutrophils and monocytes/macrophages (reviewed in (Winterbourn et al., 2000; Klebanoff, 2005)). Thus, the EPX-based ELISA alone represents a unique opportunity to assess even nominal eosinophil-mediated activities from mixed cell populations that were previously not amenable because of the lack of cell specificity (i.e., cross-reactivity) of the other available ELISA (i.e., ECP and EDN) or enzymatic (i.e., ribonucleases and peroxidase activities) assay systems.
Human fluid samples with diverse biochemical compositions were sampled as part of this report, demonstrating the unique ability and, in turn, the utility of EPX-based ELISA in a variety of clinical settings.
A demonstrably significant value of EPX-based ELISA was its utility as a surrogate biomarker representative of eosinophil infiltration/activation in clinical settings. Three distinct clinical applications linked with the care of upper (nasal) and lower (lung) respiratory patients were presented that demonstrated usefulness of this assay to overcome several confounding issues, including assessments of samples derived from mixed cell populations, responsiveness in samples where the range of eosinophil presence varied by order of magnitudes, and an ability not only to detect but to quantify the level of eosinophil activity in different patient samples. The extent of this assay’s utility to stratify these patients as part-of-point of care strategies is yet to be fully realized thus warranting validation assessments in controlled studies as a means of potentially identifying unique subpopulations of patients to direct better available therapeutic treatment options. There is also no reason a priori to assume that the utility of this assay is limited to respiratory patients. Indeed, we expect that this assay will fulfill a critical need for the assessment of eosinophils and/or eosinophil activities in a variety of subjects suffering with eosinophil-associated diseases.
HIGHLIGHTS.
The development of a clinically relevant high-throughput eosinophil peroxidase (EPX)-based ELISA to detect and quantify eosinophil activities in patient-derived samples as part of either ex vivo studies of degranulation or in vivo assessments of biological fluids.
The EPX-based ELISA was eosinophil-specific. In particular, this assay did not display cross-reactivity with other human peroxidases such as the myeloperoxidase (i.e., MPO) found in neutrophils/monocytes.
Assessments using EPX-based ELISA demonstrated the utility of this assay as uniquely capable (i.e., relative to other available assay systems) of identifying eosinophils and/or assessing the level of eosinophil activities in patient samples.
The sensitivity and reproducibility of EPX-based ELISA highlights this strategy as an eosinophil-specific high throughput assay that was also shown to have utility with numerous and diverse patient-derived fluid samples.
ACKNOWLEDGMENTS
The authors wish to thank the members of all the participating laboratories as well as colleagues within the greater eosinophil community for insightful discussions and critical comments that directly led to the development of an EPX-based ELISA with clinically utility. We also wish to acknowledge the invaluable assistance of the Mayo Clinic Arizona Statistical support group (Amylou Dueck, PhD, Yu-Hui J. Chang, and Joseph Hentz), our staff medical graphic artist (Marv Ruona), the Mayo Clinic Arizona Phlebotomy System Staff (Marian Gannon and Graciela Snyder), the members of the Northwestern Sinus Center, and the excellent administrative support provided to Lee Laboratories by Linda Mardel and Shirley (“Charlie”) Kern. The Mayo Foundation for Medical Education and Research and grants from the United States National Institutes of Health [NAL (HL058723), JJL (HL065228, RR0109709), RPS (HL068546, HL078860), NNJ (HL088594, RR025011, HL56396, RR03186), GTF (RR025780)], the American Heart Association [NAL (05556392) and JJL (0855703)], The Bazley Trust (RPS), the Canadian Institutes of Health Research [RM and PL (MOP89748)], the Lung Association of Alberta [JDK], and a Canada Research Chair in Airway Inflammometry (PN) were the sources of funding used in the performance of studies including data analysis and manuscript preparation. These funding sources had no involvement in study design, data collection (including analysis and interpretation), the writing of the manuscript, or the decision to submit for publication.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Berlyne GS, Parameswaran K, Kamada D, Efthimiadis A, Hargreave FE. A comparison of exhaled nitric oxide and induced sputum as markers of airway inflammation. J. Allergy Clin. Immunol. 2000;106:638–644. doi: 10.1067/mai.2000.109622. [DOI] [PubMed] [Google Scholar]
- Bloom B, Cohen RA. Summary health statistics for U.S. children: National Health Interview Survey 2006. Vital Health Stat. 2007;10:1–79. [PubMed] [Google Scholar]
- Chai H, Farr RS, Froehlich LA, Mathison DA, McLean JA, Rosenthal RR, Sheffer AL, Spector SL, Townley RG. Standardization of bronchial inhalation challenge procedures. J. Allergy Clin. Immunol. 1975;56:323–327. doi: 10.1016/0091-6749(75)90107-4. [DOI] [PubMed] [Google Scholar]
- Denzler KL, Borchers MT, Crosby JR, Cieslewicz G, Hines EM, Justice JP, Cormier SA, Lindenberger KA, Song W, Wu W, Hazen SL, Gleich GJ, Lee JJ, Lee NA. Extensive eosinophil degranulation and peroxidase-mediated oxidation of airway proteins do not occur in a mouse ovalbumin-challenge model of pulmonary inflammation. J. Immunol. 2001;167:1672–1682. doi: 10.4049/jimmunol.167.3.1672. [DOI] [PubMed] [Google Scholar]
- Dyer KD, Percopo CM, Xie Z, Yang Z, Kim JD, Davoine F, Lacy P, Druey KM, Moqbel R, Rosenberg HF. Mouse and human eosinophils degranulate in response to platelet-activating factor (PAF) and lysoPAF via a PAF-receptor-independent mechanism: evidence for a novel receptor. J. Immunol. 2010;184:6327–6334. doi: 10.4049/jimmunol.0904043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta GT, Liacouras CA, Collins MH, Gupta SK, Justinich C, Putnam PE, Bonis P, Hassall E, Straumann A, Rothenberg ME. Eosinophilic esophagitis in children and adults: a systematic review and consensus recommendations for diagnosis and treatment. Gastroenterology. 2007;133:1342–1363. doi: 10.1053/j.gastro.2007.08.017. [DOI] [PubMed] [Google Scholar]
- Hargreave FE, Nair P. Point: Is measuring sputum eosinophils useful in the management of severe asthma? Yes. Chest. 2011;139:1270–1273. doi: 10.1378/chest.11-0618. [DOI] [PubMed] [Google Scholar]
- Kato A, Peters A, Suh L, Carter R, Harris KE, Chandra R, Conley D, Grammer LC, Kern R, Schleimer RP. Evidence of a role for B cell-activating factor of the TNF family in the pathogenesis of chronic rhinosinusitis with nasal polyps. J. Allergy Clin. Immunol. 2008;121:1385–1392. doi: 10.1016/j.jaci.2008.03.002. 1392 e1-2 2802261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzenstein AL. Diagnostic features and differential diagnosis of Churg-Strauss syndrome in the lung. A review. Am. J. Clin. Pathol. 2000;114:767–772. doi: 10.1309/F3FW-J8EB-X913-G1RJ. [DOI] [PubMed] [Google Scholar]
- Klebanoff SJ. Myeloperoxidase: friend and foe. J. Leukoc. Biol. 2005;77:598–625. doi: 10.1189/jlb.1204697. [DOI] [PubMed] [Google Scholar]
- Koh GC, Shek LP, Goh DY, Van Bever H, Koh DS. Eosinophil cationic protein: is it useful in asthma? A systematic review. Respir. Med. 2007;101:696–705. doi: 10.1016/j.rmed.2006.08.012. [DOI] [PubMed] [Google Scholar]
- Lee JJ, Dimina D, Macias MP, Ochkur SI, McGarry MP, O'Neill KR, Protheroe C, Pero R, Nguyen T, Cormier SA, Lenkiewicz E, Colbert D, Rinaldi L, Ackerman SJ, Irvin CG, Lee NA. Defining a link with asthma in mice congenitally deficient in eosinophils. Science. 2004;305:1773–1776. doi: 10.1126/science.1099472. [DOI] [PubMed] [Google Scholar]
- Liu LY, Sedgwick JB, Bates ME, Vrtis RF, Gern JE, Kita H, Jarjour NN, Busse WW, Kelly EAB. Decreased Expression of Membrane IL-5 Receptor {alpha} on Human Eosinophils: I. Loss of Membrane IL-5 Receptor {alpha} on Airway Eosinophils and Increased Soluble IL-5 Receptor {alpha} in the Airway After Allergen Challenge. J. Immunol. 2002;169:6452–6458. doi: 10.4049/jimmunol.169.11.6452. [DOI] [PubMed] [Google Scholar]
- Metso T, Venge P, Haahtela T, Peterson CG, Seveus L. Cell specific markers for eosinophils and neutrophils in sputum and bronchoalveolar lavage fluid of patients with respiratory conditions and healthy subjects. Thorax. 2002;57:449–451. doi: 10.1136/thorax.57.5.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morioka J, Kurosawa M, Inamura H, Nakagami R, Mizushima Y, Chihara J, Yokoseki T, Kitamura S, Omura Y, Shibata M. Development of a novel enzyme-linked immunosorbent assay for blood and urinary eosinophil-derived neurotoxin: a preliminary study in patients with bronchial asthma. Int. Arch. Allergy Immunol. 2000;122:49–57. doi: 10.1159/000024358. [DOI] [PubMed] [Google Scholar]
- Nair P, Ochkur SI, Protheroe CA, Radford K, Efthimiadis A, Lee NA, Lee JJ. Assessments of Eosinophil Peroxidase in Sputum Represents a Unique Biomarker for the Diagnosis of Respiratory Patients. J. Allergy Clin. Immunol. 2012 doi: 10.1111/all.12206. Submitted as companion to Ochkur, et al, 2012 JACI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochkur SI, Kim JD, Protheroe CA, Colbert D, Moqbel R, Lacy P, Lee JJ, Lee NA. The Development of a Sensitive and Specific ELISA for Mouse Eosinophil Peroxidase: Assessment of Eosinophil Degranulation Ex Vivo and in Models of Human Disease. Journal of Immunological Methods. 2012;375:138–147. doi: 10.1016/j.jim.2011.10.002. NIHMSID 331312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters MS, Rodriguez M, Gleich GJ. Localization of human eosinophil granule major basic protein, eosinophil cationic protein, and eosinophil-derived neurotoxin by immunoelectron microscopy. Laboratory Investigation. 1986;54:656–662. [PubMed] [Google Scholar]
- Pizzichini E, Pizzichini M, Efthimiadis A, Hargreave F, Dolovich J. Measurement of inflammatory indices in induced sputum: effects of selection of sputum to minimize salivary contamination. Eur. Respir. J. 1996;9:1174–1180. doi: 10.1183/09031936.96.09061174. [DOI] [PubMed] [Google Scholar]
- Pleis JR, Lethbridge-Cejku M. Summary health statistics for U.S. adults: National Health Interview Survey, 2006. Vital Health Stat. 2007;10:1–153. [PubMed] [Google Scholar]
- Protheroe CA, Woodruff SA, DePetris G, Mukkada V, Ochkur SI, Janarthanan S, Lewis JC, Pasha S, Lunsford T, Harris L, Sharma VK, McGarry MP, Lee NA, Furuta GT, Lee JJ. A novel histological scoring system to evaluate mucosal biopsies from patients with eosinophilic esophagitis. Clinical Gastroenterology and Hepatology. 2009;7:749–755. doi: 10.1016/j.cgh.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimert CM, Venge P, Kharazmi A, Bendtzen K. Detection of eosinophil cationic protein (ECP) by an enzyme-linked immunosorbent assay. J. Immunol. Methods. 1991;138:285–290. doi: 10.1016/0022-1759(91)90177-h. [DOI] [PubMed] [Google Scholar]
- Schleimer RP, Kato A, Kern R. Eosinophils and Chronic Rhinosinusitis. In: Lee JJ, Rosenberg H, editors. Eosinophils in Health and Disease. Waltham, MA: Elsevier; 2012. In press. [Google Scholar]
- Smith AD, Cowan JO, Brassett KP, Herbison GP, Taylor DR. Use of exhaled nitric oxide measurements to guide treatment in chronic asthma. N. Engl. J. Med. 2005;352:2163–2173. doi: 10.1056/NEJMoa043596. [DOI] [PubMed] [Google Scholar]
- Sorrentino S, Glitz DG, Hamann KJ, Loegering DA, Checkel JL, Gleich GJ. Eosinophil-derived neurotoxin and human liver ribonuclease. Identity of structure and linkage of neurotoxicity to nuclease activity. J. Biol. Chem. 1992;267:14859–14865. [PubMed] [Google Scholar]
- Sur S, Glitz DG, Kita H, Kujawa SM, Peterson EA, Weiler DA, Kephart GM, Wagner JM, George TJ, Gleich GJ, Leiferman KM. Localization of eosinophil-derived neurotoxin and eosinophil cationic protein in neutrophilic leukocytes. J. Leukoc. Biol. 1998;63:715–722. doi: 10.1002/jlb.63.6.715. [DOI] [PubMed] [Google Scholar]
- Tang RB, Chen SJ. Serum levels of eosinophil cationic protein and eosinophils in asthmatic children during a course of prednisolone therapy. Pediatr. Pulmonol. 2001;31:121–125. doi: 10.1002/1099-0496(200102)31:2<121::aid-ppul1019>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- Willetts LW, Parker K, Wesselius LJ, protheroe CA, Jaben E, Graziano P, Moqbel R, Lacy P, Leslie KO, lee NA, Lee JJ. Immunodettection of occult eosinophils in lung tissue biopsies may help predict survival of acute lung injury. Respiratory Research. 2011;12:116. doi: 10.1186/1465-9921-12-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winterbourn CC, Vissers MC, Kettle AJ. Myeloperoxidase. Curr. Opin. Hematol. 2000;7:53–58. doi: 10.1097/00062752-200001000-00010. [DOI] [PubMed] [Google Scholar]