Abstract
Anterior-posterior (AP) limb patterning is directed by sonic hedgehog (SHH) signaling from the posteriorly located Zone of Polarizing Activity (ZPA). GLI3 and GLI2 are the transcriptional mediators generally utilized in SHH signaling, and each can function as an activator (A) and repressor (R). Although GLI3R has been suggested to be the primary effector of SHH signaling during limb AP patterning, a role for GLI3A or GLI2 has not been fully ruled out, nor has it been determined whether Gli3 plays distinct roles in limb development at different stages. By conditionally removing Gli3 in the limb at multiple different time points, we uncovered four Gli3-mediated functions in limb development that occur at distinct but partially over-lapping time windows; AP patterning of the proximal limb, AP patterning of the distal limb, regulation of digit number and bone differentiation. Furthermore, by removing Gli2 in Gli3 temporal conditional knock-outs, we uncovered an essential role for Gli2 in providing the remaining posterior limb patterning seen in Gli3 single mutants. To test whether GLIAs or GLIRs regulate different aspects of AP limb patterning and/or digit number, we utilized a knock-in allele in which GLI1, which functions solely as an activator, is expressed in place of the bifunctional GLI2 protein. Interestingly, we found that GLIAs contribute to AP patterning specifically in the posterior limb, whereas GLIRs predominantly regulate anterior patterning and digit number. Since GLI3 is a more effective repressor, our results explain why GLI3 is required only for anterior limb patterning and why GLI2 can compensate for GLI3A in posterior limb patterning. Taken together, our data suggest that establishment of a complete range of AP positional identities in the limb requires integration of the spatial distribution, timing, and dosage of GLI2 and GLI3 activators and repressors.
Keywords: digit patterning, sonic hedgehog, polydactyly, Gli1
Introduction
A long-standing question in developmental biology is how the vertebrate limb acquires a stereotyped polarity and number of skeletal elements along the anterior-posterior (AP) axis within each segment of the proximal-distal axis (stylopod, zeugopod, autopod). The secreted factor sonic hedgehog (SHH) is responsible for mediating the AP organizing function of the zone of polarizing activity (ZPA), a posterior cell-population that promotes digit formation and AP polarity (Bastida and Ros, 2008). The mechanism by which SHH signaling determines digit number in the autopod (distal to the wrist) appears to involve regulating proliferation and survival of digit precursor cells (Towers et al., 2008; Zhu et al., 2008). However, studies focused on Shh have not fully delineated the mechanism by which SHH signaling specifies AP pattern. Whereas gain-of-function studies have provided evidence for a classical morphogen gradient model in which AP positional identities are defined by the concentration of SHH protein (Riddle et al., 1993; Tickle, 1981; Tickle et al., 1975), fate-mapping studies and experimental manipulations have suggested that integration of temporal SHH exposure and responsiveness influence AP identity (Ahn and Joyner, 2004; Harfe et al., 2004; Scherz et al., 2007). In addition, experiments addressing whether regulation of digit number and specification of digit identity by SHH signaling are temporally separable can be interpreted to mean either that proliferation of limb mesenchymal cells is coincident with AP patterning (Towers et al., 2008), or that Shh is required only transiently for AP specification, after which it is required only for mesenchymal expansion (Zhu et al., 2008). We reasoned that additional insight into the mechanisms of AP patterning could be gained through better understanding how the key intracellular mediators of SHH signaling temporally regulate this process.
The downstream effectors of SHH signaling include the GLI family of transcription factors, each with a distinct biochemical activity. Whereas Gli2 and Gli3 are required for patterning most organs and survival past birth, Gli1 is dispensable in the mouse (Jiang and Hui, 2008). GLI2 and GLI3 are bifunctional transcription factors that have activator function (GLI2A/GLI3A) only in the presence of SHH, and otherwise undergo proteolytic cleavage to produce repressors (GLI2/GLI3R) (Pan et al., 2006; Wang et al., 2000). In contrast, Gli1 is a direct transcriptional target of GLI2A/3A (Bai et al., 2004) that functions only as an activator (Dai et al., 1999; Park et al., 2000; Sasaki et al., 1997). Although both GLI2 and GLI3 can form activators and repressors, genetic experiments in mice have demonstrated that GLI3 predominates in its repressor form, whereas GLI2 is the main activator induced by SHH (Fuccillo et al., 2006). Since Gli1 can rescue all the Gli2−/− phenotypes when expressed from the Gli2 locus (Bai and Joyner, 2001; Bai et al., 2004), it is possible that any repressor function of GLI2 is masked by GLI3R. Conversely, as Gli3 cannot rescue most Gli2−/− phenotypes (Bai et al., 2004), any GLI3A function would appear to be weak. Given the distinct activities of GLI2 and GLI3, and their overlapping expression throughout most of limb development (Buscher and Ruther, 1998; Zulch et al., 2001), it was surprising to find that only Gli3 is required for limb AP patterning (Hui and Joyner, 1993; Mo et al., 1997; Park et al., 2000; Bai et al, 2002). Since the posteriorly restricted expression of Shh in the limb results in a gradient of GLI3 transcriptional activity along the AP limb axis (Wang et al., 2000), it is possible that AP positional identities are specified by different levels of GLI3R. Consistent with this, the autopods of Shh−/− limbs, in which the only form of GLI3 (and GLI2) present is the repressor, develop only a single digit thought to retain digit 1 identity (anterior-most digit) (Kraus et al., 2001; Litingtung et al., 2002), whereas Gli3−/− limbs develop polydactyly and lack digit 1 identity (Johnson, 1967). Furthermore, removal of Gli3 rescues the digit loss observed in Shh−/− autopods and results in polydactyly (Litingtung et al., 2002; te Welscher et al., 2002b). However, since some pattern remains in the posterior autopod of Gli3 mutants (Hill et al., 2009; Panman et al., 2005; Zuniga and Zeller, 1999), and Gli2−/−; Gli3−/− embryos die before the onset of skeletogenesis (Mo et al., 1997), it remains an open question whether indeed GLI3R alone is sufficient to pattern the entire AP axis of the limb, or whether instead Gli2 contributes to limb patterning.
Efforts at determining the mechanism by which GLI3 patterns the AP limb axis downstream of SHH have been confounded by the expression of Gli3 during multiple stages of limb development. Prior to expression of Shh in the ZPA at embryonic day (E) ~10.5 (or ~32 somites) in the hindlimbs, GLI3R (expressed in the anterior limb) posteriorly restricts the expression of several AP patterning genes, setting up a “pre-pattern” that could be required for normal AP limb morphology (te Welscher et al., 2002a; Zuniga and Zeller, 1999). Subsequently, during the time Shh is expressed from the ZPA (E10.5-E12.5 in the hindlimbs), the extracellular SHH gradient present along the AP limb axis (highest posterior) generates an opposing intracellular gradient of GLI3R (highest anterior) that could provide a mechanism for specifying AP positional values (Wang et al., 2000) (Fig. 1). Furthermore, Gli3 persists after Shh is downregulated (after ~E12.5) and acts downstream of indian hedgehog (IHH) in chondrocyte differentiation of the digital rays (Hilton et al., 2005; Koziel et al., 2005). However, since Gli3 is also expressed in the interdigital mesenchyme (IDM) (Buscher and Ruther, 1998) and digit identity can be manipulated by signals emanating from the IDM late in chick limb development (Dahn and Fallon, 2000), it is possible that GLI3 plays a role in regulating digit identity during this late developmental stage. Given the multiple roles Gli3 could play during AP limb patterning and its apparent status within the GLI family as the principle mediator of SHH signaling in the limb, resolving the temporal specific roles of Gli3 during limb development would provide new understanding of the mechanisms by which SHH signaling patterns the limb.
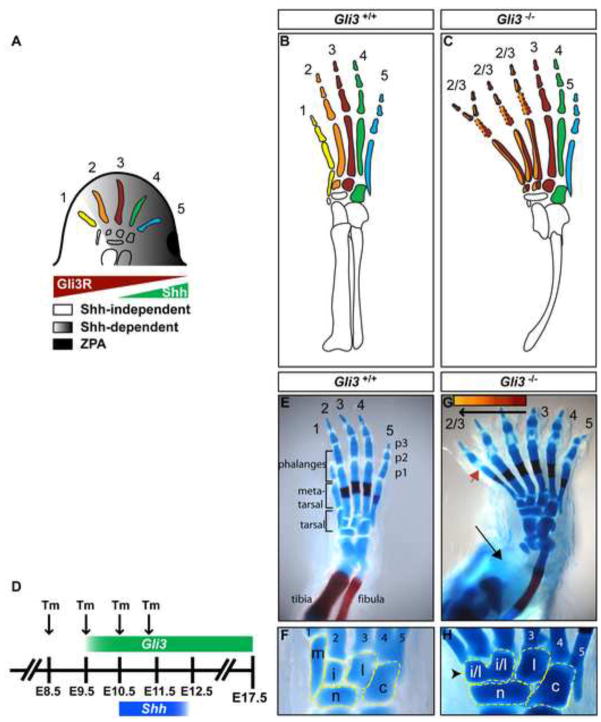
Fig. 1.
Gli3−/− hindlimbs develop polydactyly and severe AP patterning defects only in the anterior zeugopod and autopod. (A) Schematic of AP patterning mediated by Shh signaling. Grey shading represents a gradient of Shh signaling. (B,C) Schematic representations of wild type and Gli3−/− hindlimbs with each digit identity represented by a different color. Dotted lines denote bones that are variably lost. (D) Experimental design for CKO study showing when Tm was administered relative to Gli3 expression (green) and Shh expression (blue) in the hindlimb. (E,F) Wild type E17.5 hindlimb. (G,H) Gli3−/− hindlimbs develop digit polydactyly, with variable phalangeal bone duplications, variable abnormal p1 differentiation (red arrow-head), tibial agenesis (black arrow) and posteriorization of structures anterior to digit 3, including loss of digit 1 metatarsal morphology and ossification center, loss of digit 1 tarsal bone (black arrowhead) and anterior elongation of the navicular bone. Colored bar at the top of panel (G) indicates that digit and tarsal bones anterior to digit 3 adopt progressively more anterior pattern, but exclude digit 1 pattern. For all panels anterior is left, posterior right. m: medial cuneiform, i: intermediate cuneiform, l: lateral cuneiform, c: cuboid, n: navicular.
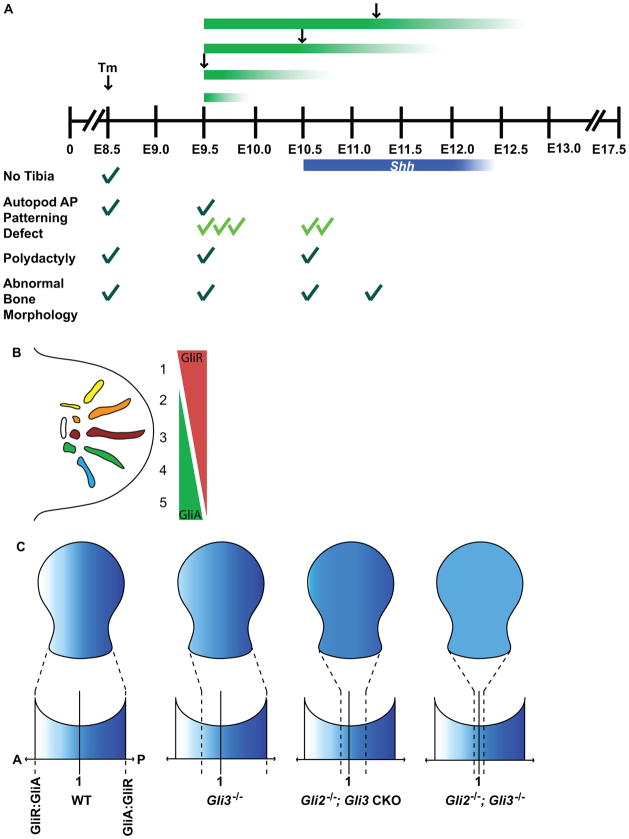
Using a Hoxb6-CreER transgenic line and Tamoxifen (Tm) to conditionally inactivate Gli3 in the hindlimb mesenchyme before, during, and after Shh expression, we have uncovered a temporal sequence of Gli3 requirements, first in specifying AP pattern sequentially along the proximal-distal axis, then in regulating digit number, and finally in sustaining normal bone morphogenesis. Importantly, by removing Gli2 in Gli3 conditional limb mutants, we have uncovered a role for Gli2 in providing AP pattern to Gli3 mutant limbs throughout the AP axis. Furthermore, we show that although regulation of AP patterning and digit number are temporally uncoupled after ~E12.0 in Gli3 conditional mutants (our results and (Lopez-Rios et al.)), the two processes remain coupled in Gli2/3 double mutants. Finally, by replacing the bifunctional GLI2 protein with the GLI1 activator, we show that GLI repressors pattern the anterior autopod, whereas posterior autopod patterning involves a combination of activators and repressors or a weak activator. Thus, the mechanism of GLI-mediated limb patterning is complex, and depends on the integration of timing and dosage of total Gli expression, as well as the distribution of GLI2R/3R and GLI2A/3A functions within spatially distinct domains.
Materials and methods
Mouse lines
Hoxb6-CreERT1 transgenic mice (Nguyen et al., 2009), R26lox-STOP-lacZ (R26R) (Soriano, 1999) and Gli3XtJ (Maynard et al., 2002), Gli2zfd (Mo et al., 1997), and Gli21ki (Bai and Joyner, 2001) alleles were genotyped as previously described. Gli3rec and Gli3fl (Blaess et al., 2008) alleles were detected using the following primers: 5′-GGAAAGTCCTCTACAGTCTG-3′ and 5′-CAGTAGTAGCCTGGTTACAG-3′. Gli3 CKO embryos were generated by mating Hoxb6-CreERT1; Gli3XtJ/+ compound heterozygous males to Gli3fl/fl homozygous females. Gli3 CKO; Gli2zfd/zxfd embryos were generated by mating Hoxb6-CreERT1; Gli3XtJ/+; Gli2zfd/+ compound heterozygous males to Gli2zfd/+; Gli3fl/fl females. Gli3 CKO; Gli21ki/zfd embryos were generated by mating Hoxb6-CreERT1; Gli3XtJ/+; Gli2zfd/+ compound heterozygous males to Gli21ki/+; Gli3fl/fl females.
Embryo and limb staging
Embryonic day 0.5 (E0.5) was defined as noon the day a vaginal plug was detected. For the Shh and dHand expression analysis, limbs were stage-matched by somite number and limb morphology, as described in (Wanek et al., 1989).
Tamoxifen administration and dosage
Pregnant females were administered 200μg/g Tm admixed with 50μg/g progesterone dissolved in corn oil via oral gavage.
Skeletal staining
E17.5 embryos were processed for alizarin red/alcian blue skeletal staining as described (Lufkin et al., 1992).
Western blot analysis
Embryos were dissected and lysates prepared from individual hindlimb buds, electrophoresed and probed with anti-GLI3 antibody as previously described (Chen et al., 2004; Wang et al., 2000). Anti-vinculin was used as a control for total protein loading.
RNA in situ
Standard whole mount RNA in situ hybridization analysis was performed as described on the Joyner Lab website: http://www.mskcc.org/mskcc/html/75282.cfm. Shh (Echelard et al., 1993), Hoxd11/12 (Herault et al., 1999), Pax9 (Neubuser et al., 1995), Tbx2 (Gibson-Brown et al., 1996), dHand (Fernandez-Teran et al., 2000).
Detection of β-gal activity
Embryos were fixed in 0.2% gluteraldehyde/4% formaldehyde, processed for cryosectioning at 14 μm, stained for beta-galactosidase (β-gal) activity at 37 C in X-gal solution for ~12 hr and counterstained with Fast Red. Detailed protocols can be found on the Joyner Lab website.
Autopod phenotype analysis
Polydactyly
Digit number of E17.5 embryos was determined as the number of complete metatarsal bones that developed. If phalanges were duplicated but associated with the same metatarsal bone, it was counted as one digit, as in the first digit of Fig 3E. Cartilagenous structures that developed in the position of a metatarsal bone, but did not develop into complete metatarsal bones, were not counted as digits, as in the structures that formed anterior to the first digit in Fig 5E. In the text, “digit” therefore refers to the entire digital ray, a proximal-distal unit that includes a metatarsal bone and its associated phalanges.
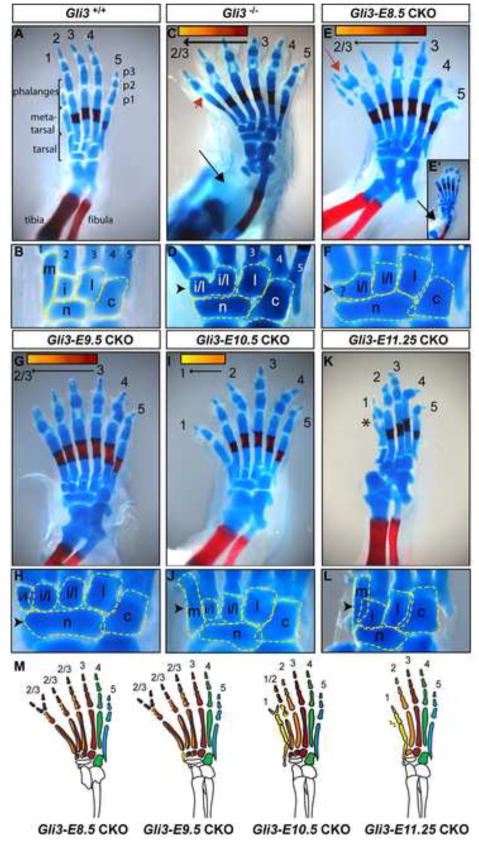
Fig. 3.
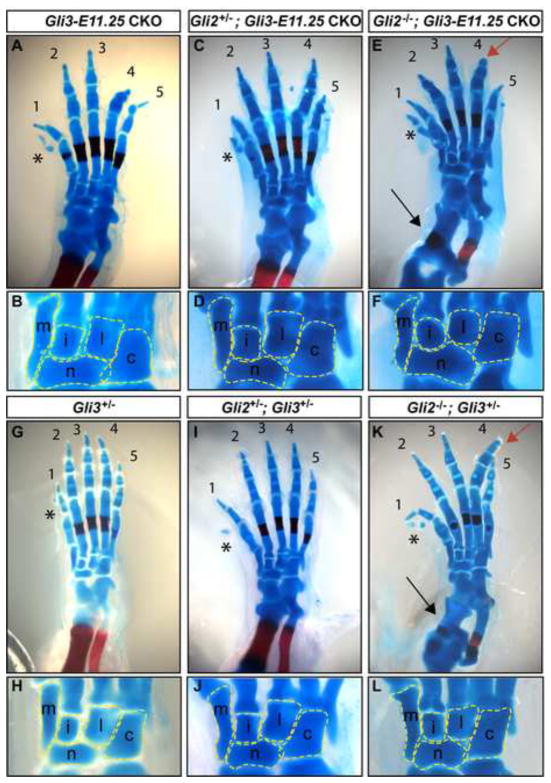
Hoxb6-CreER mediated Gli3 conditional knockout in the hindlimb reveals temporal-specific roles for Gli3 during limb patterning. The autopod shown in inset (E′) is an example of a Gli3-E8.5 CKO hindlimb with tibial bone loss (black arrow) similar to Gli3−/−. Black arrowhead draws attention to the anterior tarsal bone phenotype, which suggests digit 1 identity in J and L (compare to B), and correlates with the absence of digit 1 identity in D, F, and H. Note the retention of wild-type morphology in the cuboid and lateral cuneiform bones in all Gli3 mutants. (M) Schematic representation of E17.5 Gli3 CKO hindlimbs in which each digit identity is represented by a different color. The appearance of dislocation of the distal fibula and tibia in (E) and (K) is a consequence of tissue processing rather than a mutant phenotype. The appearance of digit 4 and 5 truncations in (E) and (K) is due to the angle of the photo, and is not a mutant phenotype. Red arrow: example of phalangeal duplication. Asterix: preaxial partial digit 1 duplication typical of Gli3+/− hindlimbs.See Fig. 1 for additional symbols.
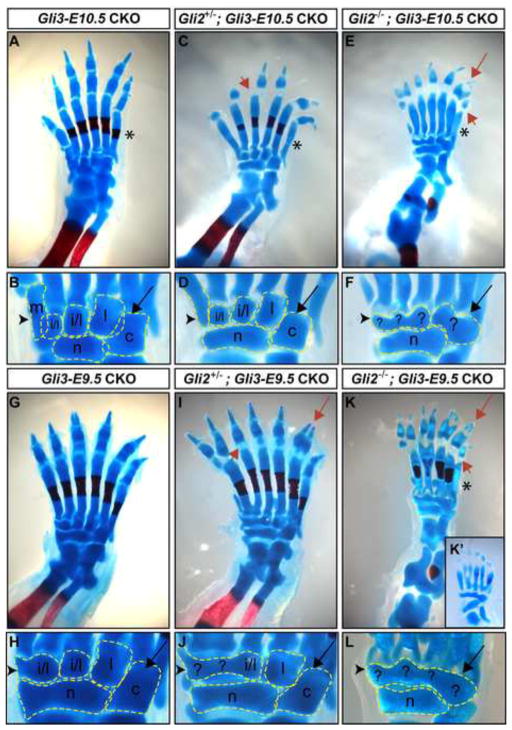
Fig. 5.
GLI2 provides AP patterning in autopods lacking Gli3 after E11.0. (A,B) Gli3-E10.5 CKO autopods develop polydactyly but retain a complete complement of normal digit identity. (C, D) Posteriorization of the anterior autopod of Gli2+/−; Gli3-E10.5 CKO mutants is suggested by lengthening of the first metatarsal bone and failure of the medial cuneiform bone to completely form. (E,F) Complete posteriorization of the anterior autopod and loss of posterior autopod patterning occurs in Gli2−/−; Gli3-E10.5 CKOs. (G, H) Gli3-E9.5 CKOs and (I, J) Gli2+/−; Gli3-E9.5 CKOs have similar anterior autopod patterning phenotypes. (K, L) Loss of anterior and posterior autopod patterning is evident in Gli2−/−; Gli3-E9.5 CKO tarsal bones. (K′) A Gli2−/−; Gli3-E9.5 CKO exhibiting nearly complete loss of AP polarity. Black arrowhead draws attention to the anterior tarsal bone of each mutant. Black arrow points to the two posterior tarsal bones. Red arrowhead: examples of abnormal p1 differentiation. Red arrow: examples of phalangeal duplications. Asterix: lack of complete metatarsal ossification center staining. For all panels anterior is left, posterior right. m: medial cuneiform, i: intermediate cuneiform, l: lateral cuneiform, c: cuboid, n: navicular.
Autopod AP patterning
In the wild-type mouse autopod, the most anterior tarsal bone (medial cuneiform) underlying digit 1 is distinguishable from the tarsal bones underlying digits 2 and 3 due to its elongated morphology. The two rectangular tarsal bones underlying digits 2 and 3 (intermediate and lateral cuneiform, respectively) have similar shapes, but the tarsal bone underling digit 2 is smaller. The most posterior tarsal bone (cuboid) is shared by digits 4 and 5, and has a trapezoidal shape. The more proximal tarsal bone (navicular) is positioned directly beneath the tarsal bones underlying digits 2 and 3. The lengths of the metatarsal bones were used to distinguish digit 1 and digit 5 from the remaining digits, as digit 1 has the smallest metatarsal, and digit 5 the next smallest, whereas digits 2, 3, and 4 are larger and similar in size. The smaller metatarsal ossification sizes of digits 1 and 5 were similarly used to distinguish these digits from 2, 3 and 4. Within each autopod, there is a trend for the size of the metatarsal and ossification center to decrease from posterior to anterior within these three digits (4>3>2). Since Gli3−/− autopods exhibit severe abnormalities in phalanx and joint development, individual phalanges could not be unequivocally discerned, and phalangeal bone number was not used as part of the criteria for analyzing AP patterning in the Gli3 mutants generated in this study. AP patterning of Gli3−/− and Gli3 CKO autopods was therefore evaluated using (1) the morphology and size of the distal tarsal bones (at the position of the cuneiform and cuboid bones in wild-type hindlimbs) (2) the anterior extent of the position of the navicular bone (3) the length of the metatarsal bone associated with each digit and (4) the relative size of the metatarsal ossification center of each digit. Digit identity was assigned only when all four criteria were coincident and the digit in question was located in the correct AP position for the assigned identity. For example, digit 1 identity was assigned when (1) the tarsal bone developed a medial cuneiform morphology, (2) the navicular bone was not located directly underneath the tarsal bone, (3) the metatarsal length and (4) metatarsal ossification center size was similar to wild-type digit 1 and (5) if the digit was located in the most anterior position, as in the first digit of Fig. 1E. The same criteria inferred digit 3, 4, and 5 identity to the last three digits of all Gli3 mutants analyzed in this study. The variable lack of, or delay in, ossification in the metatarsals of Gli3 CKO; Gli2−/− autopods precluded the use of metatarsal ossification center size to evaluate AP patterning in these mutants. Analysis of AP autopod patterning in Gli3 CKO; Gli2−/− embryos was therefore restricted to tarsal bone morphologies.
Results
Removal of Gli3 from hindlimb mesenchyme by E10.0 can reproduce the Gli3−/− phenotype
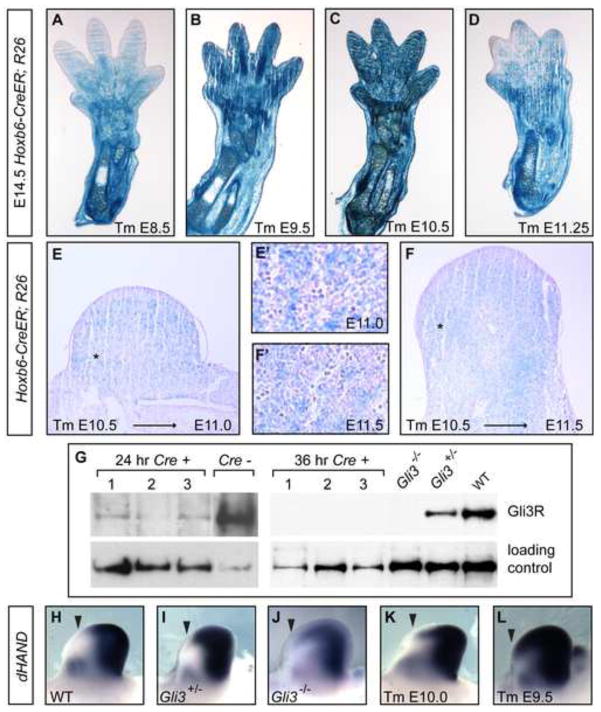
To determine whether there are sequential requirements for Gli3 in regulating digit number and AP limb patterning, we took a conditional mutagenesis approach using a Hoxb6-CreER transgene that induces recombination throughout the hindlimb mesenchyme (Zhu et al., 2008), and a Gli3fl conditional allele (Blaess et al., 2008). Analysis of lacZ expression from the R26R locus (based on β-gal activity) in E14.5 hindlimbs of Hoxb6-CreER; R26R embryos exposed to Tm on E8.5, E9.5, E10.5, and E11.25 revealed that the pattern and efficiency of recombination induced by Hoxb6-CreER is similar for each time-specific Tm administration and most mesenchymal cells express lacZ between 12 and 24 hours. (Fig. 2A–F). Western blot detection of GLI3 protein in Gli3 CKO limbs showed that GLI3 protein was reduced 24 hours after Tm administration, and undetectable 36 hours after Tm administration (Fig. 2G). Consistent with the timing of loss of GLI3 protein, anterior expansion of the GLI3R target gene dHand (Vokes et al., 2008) was similar to that seen in Gli3−/− embryos by ~36 to 48 hr post Tm administration (Fig. 2H–L). Thus, Gli3 protein can be considered ablated by 36 hr after Tm administration, although the effects of GLI3 removal on target gene expression likely begin between 24 and 36 hr, since protein levels are already substantially reduced by 24 hrs.
Fig. 2.
Hoxb6-CreER mediated recombination efficiency in the hindlimb mesenchyme. (A–F) Xgal staining in cross-sections of Hoxb6-CreER: R26R hindlimbs in embryos exposed to Tm at indicated stages and harvested at E14.5 (A–D), 36 hours post-gavage (E) and 48 hours post-gavage (F). Insets E′ and F′ are taken from regions of E and F indicated by *, respectively. (G) Western blots show GLI3R and control protein from hindlimb bud lysates of different single Gli3-E9.5 CKO mutants (Cre+, numbered) and sibling (Cre−) control embryos with genotypes as indicated, collected at 24 hours or at 36 hours after Tm treatment. Vinculin detection was used as a loading control. In the 24 hour blot, the Gli3fl/−; Cre negative sibling control sample lane was spliced to remove irrelevant lanes. The 24 hour and 36 hour examples shown are representative of results of 3 independent experiments (total of 7 mutant embryos analyzed at 24 hours post Tm treatment and 5 mutants analyzed at 36 hours post Tm treatment). (H–L) dHand in situs of E11.5 limb buds with indicated genotypes. The dHand domain is expanded anteriorly in Gli3−/− (n=10/10) hindlimbs compared to Gli3+/− (n=8/8). Anterior expansion is partial in Gli3 CKO limbs 36 hours post-gavage (6/8) and complete within 48 hours (7/8).
We first asked whether removing Gli3 before hindlimb outgrowth results in a hindlimb phenotype similar to Gli3 null mutants. In our Swiss Webster outbred background, the Gli3−/− hindlimb phenotype consisted of invariable agenesis of the tibia bone, posteriorization of the anterior autopod, polydactyly, distal phalangeal bone (p3) duplications and disrupted proximal (p1) and middle (p2) phalangeal bone development (n=10/10) (Panman et al., 2005; Zuniga and Zeller, 1999). Morphological characteristics of digit 1 pattern were missing (defined in Experimental Procedures). Instead, digits anterior to digit 3 developed digit 2 and/or 3-like pattern (Fig. 1, Table S1). Significantly, we found that some Hoxb6-CreER; Gli3fl/− CKO embryos administered Tm at E8.5 (Gli3-E8.5 CKOs) and analyzed at E17.5 completely recapitulated the Gli3−/− phenotype (n=2/6 limbs) (Figs. 2E,F). The remaining Gli3-E8.5 CKO embryos developed all aspects of the Gli3−/− phenotype except agenesis of the tibia bone (n=4/6). Our ability to recapitulate the Gli3−/− hindlimb phenotype in Gli3-E8.5 CKO embryos demonstrates that the Tm dosage was optimized to induce recombination at the Gli3 locus to a degree that any remaining GLI3 protein is not sufficient to sustain Gli3 function. Furthermore, in the Gli3-E8.5 CKO embryos that developed a tibia bone it is possible that GLI3 protein was ablated slightly later, suggesting that Gli3 is only required until shortly after hindlimb outgrowth is initiated (E10.0) for tibia bone development.
Tibia development is restored when Gli3 is expressed until ~E11
Since Gli3 expression in the limb both precedes and overlaps with Shh, we reasoned that if there were differential requirements for Gli3 function before versus during Shh expression, they would be revealed through comparing the phenotypes produced by removing Gli3 prior to Shh expression with those produced by removing Gli3 after initiation of Shh. RNA in situ hybridization analysis of Shh in our genetic background established that Shh mRNA is detectable in the hindlimb ZPA starting at ~E10.5 (~32 somites) (Fig. S1). To achieve depletion of GLI3 protein before ~E11.0, Gli3-E9.5 CKO mutants were produced. Unlike Gli3-E8.5 CKOs, which did not consistently develop a tibia, all E17.5 Gli3-E9.5 CKO hindlimbs had a complete tibia (n=20/20 limbs). Nevertheless, the autopod AP patterning phenotype of the majority of Gli3-E9.5 CKOs was similar to Gli3-E8.5 CKOs (n=13/20) (Fig. 3,Table S1). The remaining Gli3-E9.5 CKOs developed either a partial (n=5/20) or complete (n=2/20) digit 1 medial cuneiform bone, however the navicular bone still extended anteriorly to underlie the first digit in all (7/20) of these mutants, suggesting a partially posteriorized pattern. All Gli3-E9.5 CKOs developed polydactyly, p3 duplications and abnormal p1 and p2 development (20/20). In summary, our analysis of Gli3-E9.5 CKO embryos compared to Gli3-E8.5 CKOs demonstrates that Gli3 is required before E11.0 for development of the tibial bone and after E11.0 for normal AP patterning of the autopod, regulation of digit number, and regulation of phalangeal bone development
Digit patterning is restored when Gli3 is expressed until ~E12
To further address the requirement for Gli3 during the time Shh is expressed in the ZPA, Gli3 was removed just before downregulation of Shh, which occurs by E12.5 (Buscher et al., 1997). In contrast to Gli3-E9.5 CKOs, most Gli3-E10.5 CKOs developed all characteristics of a normal digit 1 pattern (n= 17/20 limbs) and clear AP polarity of the entire autopod. Moreover, despite persistent polydactyly, all morphological traits of digit 2 pattern were observed in the digit and underlying tarsal bones anterior to digit 3 (n= 20/20) (Figs. 2I, J, Table S1). Thus, the morphologies of all structures formed in Gli3-E10.5 CKO autopods suggest a full complement of wild type digit identities. However, all Gli3-E10.5 CKO hindlimbs developed polydactyly (n=20/20), p3 duplications (n=7/20), and abnormal p1 and p2 development, although less severe than in Gli3-E9.5 CKOs (n=19/20). These results show that Gli3 is not required after ~E12.0 to regulate digit identity in the hindlimb, but continues to be required after this stage to regulate digit number and phalangeal bone development. This result is consistent with a recent study that found that when Gli3 is removed by E11.75 in the distal mesenchyme of the forelimb, which develops earlier than the hindlimb, digit identity is maintained but mild polydactyly is observed (HoxD13Cre/+; Gli3 flox/flox; mice; (Lopez-Rios et al.)). Thus, the requirement for Gli3 in regulating digit number can be temporally uncoupled from its requirement in regulating AP autopod patterning.
After Shh is down-regulated in the ZPA Gli3 is no longer required to regulate digit number
To determine whether Gli3 is required to regulate digit number and/or phalangeal bone development after Shh is down-regulated in the limb, Gli3-E11.25 CKO embryos were generated (Tm administered at 9:00 am on E11.5). In contrast to Gli3-E10.5 CKO autopods, the digit number phenotype of Gli3-E11.25 CKOs was similar to Gli3+/− mice (Figs. 2K,L). Interestingly, the one observable difference between Gli3-E11.25 CKO and Gli3+/− hindlimbs was the persistence of dysmorphology in the phalanges of Gli3-E11.25 CKO autopods (n=6/8 limbs), although it was less severe than in Gli3-E10.5 CKOs (Table S1). Our analysis of Gli3-E11.25 CKO hindlimbs compared to earlier temporal mutants shows that Gli3 is not required beyond ~E12.75 to regulate hindlimb digit number, but continues to be required after this stage for normal phalangeal bone development.
Gli2 and Gli3 act together to regulate digit identity in the anterior autopod
To explore the possible contribution of Gli2 to anterior AP autopod patterning in Gli3 CKO mutants, Hoxb-6CreER; Gli3fl/−; Gli2+/− and Hoxb6-CreER; Gli3fl/−; Gli2−/− CKO embryos were generated. We first administered Tm at the late time points when Gli3 is not required for digit patterning, and found that AP patterning and digit number in Gli2+/−; Gli3-E11.25 CKO (n=2/2 limbs) and Gli2−/−; Gli3-E11.25 CKO (n=2/2 limbs) hindlimbs were similar to Gli3-E11.25 CKOs (Fig. 4). Thus, Gli2 and Gli3 do not regulate digit identity or number after ~E12.75. In contrast, removal of only one allele of Gli2 in Gli3-E10.5 CKO mutants resulted in posteriorization of the anterior autopod. Whereas nearly all Gli3-E10.5 CKO hindlimbs developed all characteristics of a normal digit 1 pattern (n=17/20 limbs), only 6/16 Gli2+/−; Gli3-E10.5 CKO hindlimbs did. The remainder developed either a partial medial cuneiform bone (n=7/16), or none at all (n=3/16) (Figs. 4C,D, S3, Table S1). The relative sizes of metatarsal ossification centers suggested a 1, 1/2, 2, 3, 4, 5 digit identity pattern similar to Gli3-E10.5 CKO hindlimbs (n=13/16). However, the anterior-most metatarsal appeared longer than normal (n=10/16) and the navicular bone was extended anteriorly to underlie the anterior-most tarsal bone (n=11/16). These morphological changes suggest that removing one copy of Gli2 in Gli3-E10.5 CKOs results in partial posteriorization of the anterior autopod.
Fig. 4.
Gli2 is not required for AP autopod patterning when Gli3 is removed by ~E13. (A–F) Both digit number and AP autopod patterning are normal in all Gli3-E11.25 CKO limbs, with and without Gli2. (G–L) Controls demonstrating pre-axial partial digit 1 duplication (asterix), digit 4 p3 duplication (red arrow) and shortened zeugopods bones (black arrow) typical of Gli3+/− (G,H), Gli2+/−; Gli3+/− (I,J) and Gli2−/−; Gli3+/− (K,L) hindlimbs. See Figs. 1 and 2 for additional symbols.
Remarkably, there was no obvious polarity in the length of digits in Gli2−/−; Gli3-E10.5 CKOs; instead, there was mirror image symmetry around the middle digit, which tended to be longest. However, unlike Gli2+/−; Gli3-E10.5 CKOs or Gli3-E10.5 CKOs, alizarin red failed to consistently stain the metatarsal ossification centers of Gli2−/−; Gli3-E10.5 CKO hindlimbs, suggesting either a defect in chondrocyte differentiation, or a general delay in mineralization of the bone, as has been described in Gli2−/− autopods (Mo et al., 1997). Since this phenotype precluded the use of metatarsal ossification center size to determine digit identity, we focused on the morphology of the tarsal bones to assess autopod AP patterning. The digit 1 medial cuneiform bone was only partially formed (n=7/14 limbs) or completely absent (n=3/14) in the majority of Gli2−/−; Gli3-E10.5 CKO hindlimbs (Figs. 4E,F, S3, Table S1). The anterior-most metatarsal bone was also longer than normal (n=6/14) and the navicular bone was extended anteriorly to underlie the anterior-most tarsal bone (n=10/14). The more extreme anterior tarsal and metatarsal bone posteriorization observed in Gli2+/−; Gli3-E10.5 CKO and Gli2−/−; Gli3-E10.5 CKO autopods compared to Gli3-E10.5 CKOs reveal a requirement for Gli2 in providing anterior autopod pattern to Gli3 CKO hindlimbs after ~E12. Since both digit number and AP pattern are abnormal in Gli2−/−; Gli3-E10.5 CKOs, GLI-mediated regulation of digit number and AP pattern remains coupled until after E12.0 when both Gli2 and Gli3 are absent.
A further requirement for Gli2 in anterior autopod patterning was demonstrated by removing Gli3 at an early stage in Gli2 null mutants. Gli2+/−; Gli3-E9.5 CKO and Gli2−/−; Gli3-E9.5 CKO hindlimbs also consistently had a less polarized phenotype than Gli3-9.5 CKOs. In most Gli2+/−; Gli3-E9.5 CKO autopods the digit 1 medial cuneiform bone was either completely absent (n=18/20 limbs) or only partially formed (n=2/20), the navicular bone extended to underlie the anterior-most tarsal (n=20/20), and the anterior-most metatarsal was longer than normal (n=20/20) (Fig. 5I,J, Table S1). Likewise, in all Gli2−/−; Gli3-E9.5 CKOs the medial cuneiform bone was completely absent (n=6/7 limbs) or partially formed (n=1/7), the anterior-most metatarsal was longer than normal (n=7/7), and the navicular bone extended anteriorly to underlie the anterior-most tarsal bone (n=7/7) (Figs. 4K,L, Table S1). Similar to Gli2−/−; Gli3-E10.5 CKOs, alizarin red failed to consistently stain the metatarsal ossification centers of Gli2−/−; Gli3-E9.5 CKO limbs (Fig. 5K). These results further emphasize a requirement for Gli2 in anterior autopod patterning and underscore the sensitivity of AP polarity to the duration of Gli3 expression, especially when Gli2 is reduced or absent.
Gli2 does not play a major role in regulating digit number
Similar to Gli3-E10.5 CKOs, Gli2+/−; Gli3-E10.5 CKO and Gli2−/−; Gli3-E10.5 CKO hindlimbs developed polydactyly and p3 duplications (Fig. 5, Table S1). While Gli2+/−; Gli3-E10.5 CKO and Gli2−/−; Gli3-E10.5 CKO mutants appeared to be more likely than Gli3-E10.5 CKOs to develop 7 rather than 6 complete digits, this trend was not statistically significant (Fig. S2). Curiously, Gli2−/−; Gli3-E9.5 CKO hindlimbs were significantly more likely than Gli3-E9.5 CKOs to develop 5 rather than 6 complete digits, however the phalangeal bone duplications were more severe, tending to involve p1 as well as p3 (Fig. 5K, Table S1). In addition, Gli2−/−; Gli3-E11.25 mutants did not have polydactyly. Consistent with a previous report of redundancy between Gli2 and Gli3 in regulation of long-bone length (Mo et al., 1997), the zeugopod bones of Gli2−/−; Gli3 CKOs were shorter than in Gli3 CKOs. In addition, loss of p1 differentiation and dysmorphology of p2 was more severe in both Gli2+/−; Gli3 CKOs and Gli2−/−; Gli3 CKOs than in Gli3 CKOs (Table S1).
Gli2 and Gli3 act together to pattern the posterior autopod
To determine whether redundancy between Gli2 and Gli3 accounts for the normal posterior autopod pattern in the Gli3 null mutants, we next analyzed patterning of the posterior autopod of double mutants. Strikingly, in contrast to Gli3−/− mutants and Gli3 temporal CKOs both the Gli2−/−; Gli3-E10.5 CKO and Gli2−/−; Gli3-E9.5 CKO hindlimbs had AP patterning defects in the posterior autopod. Instead of developing as two distinctly identifiable bones, the two most posterior tarsals in Gli2−/−; Gli3-E10.5 CKOs were fused and morphologically similar to one another and to medial tarsal bones (n= 12/14 limbs) (Figs. 4E,F, Table 1). Moreover, a more complete loss of polarity of tarsals was observed in all Gli2−/−; Gli3-E9.5 CKO autopods (n=7/7 limbs) (Figs. 4K,L). In one case the morphology of the tarsal bones was almost completely homogeneous and fused along the entire AP axis of the autopod (Fig. K′). Thus, Gli2 appears to be required to provide AP pattern to both the anterior and posterior regions of the autopod in Gli3 mutant hindlimbs. These results suggest that Gli2 and Gli3 cooperate to pattern the entire AP axis of the autopod. In addition, removal of one copy of Gli2 from Gli3 CKOs results in partial posteriorization of the anterior autopod, but has no effect on patterning of the posterior tarsal bones. This result provides evidence that AP patterning of the anterior autopod is more sensitive than the posterior autopod to total Gli2/3 gene dosage.
Table 1.
Summary of Limb Phenotypes in Gli3 CKOs and Gli2−/−; Gli3 CKOs
| Phenotype | Tm E8.5 | Tm E9.5 | Tm E10.5 | Tm E11.25 | ||||
|---|---|---|---|---|---|---|---|---|
| No Tibia | √ | ND | ||||||
| Loss of anterior* autopod polarity | √√ | ND | √ | XXX | XX | |||
| Loss of posterior* autopod polarity | ND | XX | X | |||||
| Polydactyly | √ | ND | √ | X | √ | X | ||
| Abnormal Bone Morphology | √ | ND | √ | X | √ | X | √ | X |
√: Gli3 CKO embryos have the indicated phenotype
X: Gli2−/−; Gli3 CKO have the indicated phenotype
: The number of √s or Xs indicates the relative severity of the noted phenotypes when Gli3 is removed at sequentially later embryonic stages, and when Gli2 is removed in Gli3 CKO limbs. No √ or X indicates wild type phenotype.
ND: not determined
Attempts at generating Gli2−/−; Gli3-E8.5 CKOs were unsuccessful. From 32 matings, only 6 litters survived to E17.5 after Tm administration at E8.5. The one Gli2+/−; Gli3-E8.5 CKO that survived had a similar phenotype to Gli2+/−; Gli3-E9.5 CKOs (data not shown). A constitutive Hoxb6-Cre was then utilized to generate Hoxb6-Cre; Gli2−/−; Gli3fl/− limbs, however Hoxb6-Cre; Gli3fl/− limbs did not consistently have the full Gli3−/− phenotype, therefore this genetic approach could not be used to test whether removal of Gli3 prior to E11.0 results in complete loss of AP polarity at E17.5 (data not shown).
GLI activators contribute to AP patterning of the posterior autopod
Given that full-length GLI2 possesses strong activator function, our finding that the posterior autopod, which receives the highest levels of SHH, develops partially anteriorized morphology when both copies of Gli2 are removed in Gli3 CKOs raises the question of whether GLI activator function contributes to limb AP patterning. To address this question, we took advantage of a Gli2 knock-in (ki) allele in which mouse GLI1 is expressed in place of GLI2 (Gli21ki) (Bai and Joyner, 2001). Since GLI1 functions only as a transcriptional activator, the Gli21ki allele effectively replaces the potentially bifunctional GLI2 protein with a GLI protein that only has constitutive activator function. Therefore, if only loss of GLI repressors is responsible for the AP limb patterning and/or digit number defects observed in Gli2−/−; Gli3 CKOs, then introducing a Gli21ki allele should not rescue them. If, however, any of the phenotypes are rescued by the Gli21ki allele, then GLI activator function must contribute to limb development.
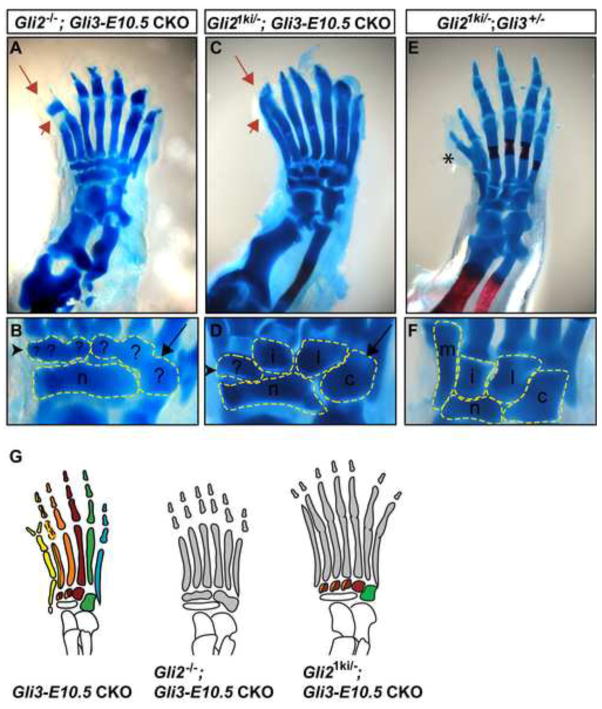
Indeed, one copy of Gli1 expressed in place of Gli2 was not able to support anterior autopod patterning when Gli3 was removed at ~E12 and instead resulted in a more complete posterior transformation of the anterior autopod. The digit 1 medial cuneiform bone was completely absent (n=5/8 limbs) or only partially formed (n=3/8) in all Gli21ki/−; Gli3-E10.5 CKOs (Figs. 5C,D, Table S1) compared to either Gli2−/−; Gli3-E10.5 or Gli2+/−; Gli3-E10.5 CKOs, in which the medial cuneiform was completely preserved in a small proportion of hindlimbs (4/14 and6/16, respectively). In contrast, patterning of the posterior autopod of Gli21ki/−; Gli3-E10.5 CKOs was partially rescued, as the lateral cuneiform and cuboid bones developed more distinct morphologies in all mutants (n=8/8). In addition, the tarsal bones were not fused. Thus, consistent with GLIAs normally being present in the posterior limb bud, Gli1 can partially restore pattern to the posterior autopod in limbs lacking both Gli2 and Gli3. These genetic complementation experiments also indicate that loss of GLI2A is primarily responsible for the tarsal bone fusion observed in Gli2−/−; Gli3-E10.5 CKO autopods (Fig. 6). However, since posterior autopod patterning was not completely restored in Gli21ki/−; Gli3-E10.5 CKOs, it is possible that both GLIAs and GLIRs are required to pattern the posterior autopod, or that the level of activator function of GLI1 is not optimal. Since patterning of the anterior autopod was not rescued, but was instead more consistently posteriorized in Gli21ki/−; Gli3-E10.5 CKO hindlimbs than in Gli2−/−; Gli3-E10.5 CKOs, the anterior autopod not only requires GLIR for normal patterning, but GLIA can reverse GliR effects.
Fig. 6.
GLIR patterns the anterior autopod and the posterior autopod is patterned by both GLIR and GLIA. (A,B) Gli2−/−; Gli3-E10.5 CKO autopods develop severely posteriorized anterior tarsal bones (black arrowhead) and anteriorized posterior tarsal bones (black arrow). (C,D) Gli21ki/−; Gli3-E10.5 CKO autopods develop severely posteriorized anterior tarsal bones, nearly normal posterior tarsal bone morphology. (E,F) Gli21ki/−; Gli3+/− are similar to Gli3+/− but have a more complete digit 1 duplication (asterix). Red arrowhead refers to loss of p1 in A and a gain-of-function phenotype associated with the Gli21ki allele, which causes the metatarsal bones to appear fused to the phalanges in C. Red arrow indicates examples of phalangeal duplications. (G) Schematic of autopod tarsal bone patterning in Gli CKOs.
GLI1 has gain-of-function activities in regulation of digit number and phalangeal bone development
Interestingly, polydactyly appeared more severe in Gli21ki/−; Gli3-E10.5 CKOs than in Gli2−/−; Gli3-E10.5 and Gli2+/−; Gli3-E10.5 CKOs, with a much greater tendency to develop 7 complete digits (n=6/8 limbs) rather than 6 (n=2/8) and phalanx duplications (n=6/8), although this tendency was not statistically significant (Fig. S2). This result is in line with recent studies in which constitutive activator forms of GLI3 (Wang et al., 2007) or GLI2 (Pan et al., 2009) were expressed in the mouse and resulted in preaxial polydactyly.
Some of the bone differentiation phenotypes that developed in Gli21ki/−; Gli3 CKOs suggested additional gain-of-function effects of the Gli21ki allele. Instead of the loss of p1 differentiation seen in Gli2−/−; Gli3-E10.5 CKO hindlimbs, Gli21ki/−; Gli3-E10.5 CKO digits developed as single, non-segmented bones (n=8/8). This phenotype was also observed in Gli21ki/+; Gli3-E10.5 CKOs, although it was less severe (n=6/6) (Table S1, data not shown), and was not observed in Gli21ki/−; Gli3+/− autopods (Figs. 5E,F). Tibia shortening was less severe in Gli21ki/−; Gli3-E10.5 CKO zeugopods than in Gli2−/−; Gli3-E10.5 CKOs, but Gli21ki/−; Gli3-E10.5 CKO tibias were thicker and dysmorphic (n=8/8). These results show that expression of GLI1A in the absence of both Gli3 and Gli2 causes abnormalities in cartilage/bone differentiation in the phalanges and long bones not observed in the double Gli2/3 loss-of-function mutants.
Changes in gene expression confirm altered AP polarity in Gli3 CKO limbs
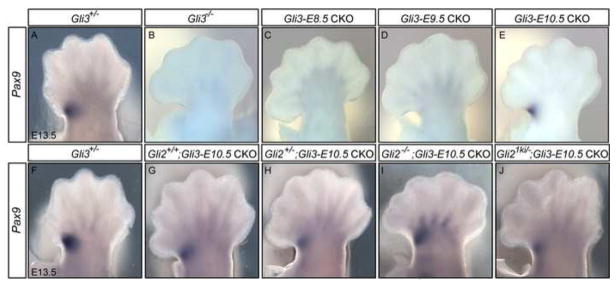
As a means to analyze the molecular patterning of Gli3 CKO limbs, we evaluated the expression of genes restricted to the anterior or posterior limb after E12.5. Both the anteriorly expressed Alx4 (Qu et al., 1997) and Pax9 genes have been reported to be dependent on Gli3 for their activation. However, we found that expression of Alx4 was not anteriorly restricted as late as E13.5 (data not shown), rendering it inappropriate for following AP patterning changes in Gli3 temporal CKOs. Expression of Pax9 in the limb, however, is restricted to the anterior mesenchyme until at least E13.5 (McGlinn et al., 2005; Neubuser et al., 1995). Analyses of Pax9 expression in Shh−/−; Gli3−/− and Gli3−/− mouse limbs and in response to SHH protein-soaked beads in chick limb buds suggest that Pax9 transcription is positively regulated by the action of GLI3R and might also be indirectly repressed by GLIA (McGlinn et al., 2005). Consistent with this hypothesis, we observed dramatic down-regulation of Pax9 expression in Gli3-E8.5 CKO (n=2/2) and Gli3-E9.5 CKO (n=2/2) hindlimbs, similar to Gli3−/− mutants (Fig. 7A–D). Interestingly, in Gli3-E10.5 CKO hindlimbs Pax9 expression was either similar to Gli3+/− embryos or only mildly reduced (n=4 limbs) (Fig. 7E). In addition, Pax9 was further reduced in the majority of Gli21ki/−; Gli3-E10.5 CKO hindlimbs (3/5), consistent with GliA resulting in repression of Pax9 expression (Fig. 7F–J). Although Pax9 expression appeared normal in Gli2−/− hindlimbs (n=3) and in Gli2+/−; Gli3-E10.5 CKO hindlimbs, in half of the Gli2−/−; Gli3-E10.5 CKO hindlimbs Pax9 expression was expanded posteriorly (4/8 limbs), while appearing similar to Gli3-E10.5 CKOs in the remainder. These findings suggest that Gli3R is required at least up to ~E11 for anterior hindlimb expression of Pax9 and that GLIA is normally involved in repressing Pax9 expression in much of the remainder of the limb.
Fig. 7.
Pax9 expression is restored in Gli3-E10.5 CKO limbs and correlates with the lack of AP polarity in Gli2; Gli3 CKO double mutant limbs. Dorsal view of Pax9 RNA in situs in E13.5 hindlimbs of the genotypes indicated. (A) Pax9 is expressed in the anterior proximal autopod in Gli3+/− limbs. Anterior Pax9 is lacking in Gli3−/− (B), Gli3-E8.5 CKO (C) and Gli3-E9.5 CKOs (D). Pax9 is expressed in Gli3-E10.5 CKOs, although reduced in some mutants (E, G). Pax9 appears reduced in some Gli2+/−; Gli3-E10.5 CKO hindlimbs (H) and in Gli21ki/−; Gli3-E10.5 CKOs (J), but is posteriorly expanded in some Gli2−/−; Gli3-E10.5 CKOs (I). Anterior is to the left in all images.
We next analyzed HoxD11 and HoxD12, since they are normally expressed in the posterior limb mesenchyme and mis-expressed anteriorly in Gli3 null mutants. It was recently shown that in E11.75 Hoxa13Cre/+; Gli3 flox/flox forelimbs, in which Gli3 is removed before E11.75, HoxD12 is expressed normally (Lopez-Rios et al.). Curiously, in our Gli3-E10.5 CKO hindlimbs (n=3/3 limbs), as well as Gli3-E8.5 CKO (n=2/2) and Gli3-E9.5 CKO (n=3/3) hindlimbs, at E13.5 HoxD12 expression was similar to Gli3−/− mutants (Fig. S3). Normal expression of HoxD12 was restored in Gli3-E11.25 CKOs (n=2/2). The lack of rescue of HoxD12 expression in our Gli3-E10.5 CKO hindlimbs compared to Hoxa13Cre/+; Gli3fl/fl forelimbs could be due to a slight difference in the timing of the knockouts relative to limb developmental stage, with our ablation occurring earlier, or because we analyzed our mutants 1.5–3 days after loss of GLI3 protein rather than immediately afterwards. Indeed, Hoxa13Cre/+; Gli3fl/fl forelimbs had a normal morphology at E11.75 whereas our Gli3-E10.5 CKO hindlimbs had an expanded anterior handplate at E13.5. Alternatively, HoxD12 expression might be normal in Hoxa13Cre/+; Gli3fl/fl forelimbs because unlike in our mutants Gli3 is only deleted in the most distal limb mesenchyme. A similar anterior expansion of HoxD11 expression was observed in Gli3−/− (n=2/2), Gli3-E8.5 CKO (n=3/3), Gli3-E9.5 CKO (n=3/3), and Gli3-E10.5 CKO (n=3/3) hindlimbs at E13.5, with restoration of posteriorly restricted expression in Gli3-E11.25 CKOs (n=3/3).
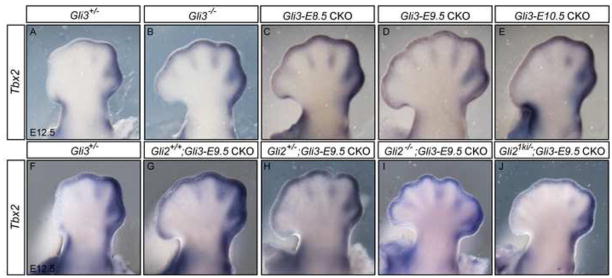
We next analyzed expression of the gene encoding T box transcription factor 2 (Tbx2) since it is also expressed in a posterior domain in the hindlimb mesenchyme (Gibson-Brown et al., 1996; Suzuki et al., 2004). By E12.5 the highest levels of Tbx2 are found in the interdigital mesenchyme (IDM) posterior to digits 4 and 5, as well as in the mesenchyme just proximal to the apical ectodermal ridge (AER) posterior to digit 1 (Fig. 8) and (Gibson-Brown et al., 1996). Tbx2 can be induced in the anterior limb by ectopic SHH expression, and Tbx2 expression has been considered in AP patterning studies as a marker of posterior mesenchyme in mutant limbs (Gibson-Brown et al., 1998; Zhu et al., 2008). However, we observed an anterior expansion of Tbx2 expression in Gli3 null mutants and all Gli3 CKO hindlimbs at E12.5, both in the IDM and in the sub-AER mesenchyme. Anterior expansion was also observed in Gli2+/−; Gli3-E9.5 CKOs and was most extreme in the IDM of Gli2−/−; Gli3-E9.5 CKO hindlimbs (Fig. 8). One possible explanation of the results is that GLI3R is required to repress Tbx2 in the anterior limb. Curiously, IDM expression appeared more posteriorly restricted in Gli21ki/−; Gli3-E9.5 CKOs than in Gli3-E9.5 CKOs. Thus, just as with Pax9 expression, polarized expression of Tbx2 in the autopod appears the most disrupted in Gli2−/−; Gli3-E9.5 CKOs correlating with the mutant with the least morphological polarity.
Fig. 8.
Tbx2 expression is expanded anteriorly in Gli3 conditional mutant limbs. (A) In E12.5 Gli3+/− hindlimbs, Tbx2 is detected in the anterior mesenchyme posterior to digit 3 and in the mesenchyme proximal to the AER posterior to digit 1. (B) Tbx2 expression is expanded anteriorly in Gli3−/− hindlimbs, posterior expression and distal mesenchymal expression is maintained. Similar anterior expansion of Tbx2 is seen in Gli3-E8.5 CKO (C), Gli3-E9.5 CKO (D) and Gli3-E10.5 CKO hindlimbs (E). In Gli2+/−; Gli3-E9.5 CKO (H) and Gli21ki/−; Gli3-E9.5 (J) CKO hindlimbs Tbx2 expression is expanded similar to in Gli3-E9.5 CKOs (G), but more extensively in Gli2−/−; Gli3-E9.5 CKOs (I). Anterior is to the left in all images.
Discussion
Using limb specific temporal conditional gene inactivation, we have provided new insights into the mechanism by which SHH functions through GLI2 and GLI3 to pattern the limb by elucidating the time when particular developmental processes require GLI3 in the hindlimb and by uncovering distinct roles for the GLI2 activator and repressor in limb patterning. Interestingly, Gli3 is required for tibia development only before E11.0, and is required for AP patterning of the autopod only up until E12.0, whereas Gli3 continues to be required during the time SHH is expressed to regulate digit number (before E12.75), and is involved in regulating bone morphogenesis after E12.75 (Table 1). Since the majority of Gli3-E8.5 CKOs analyzed developed a tibia, the requirement for Gli3 in tibia development is likely to occur before Shh is first detected in the hindlimb ZPA (~E10.5), implying that tibia development is a SHH-independent process mediated by GLI3R. There is genetic evidence implicating the transcription factors dHAND and PLZF as partners of GLI3 during this process (Barna et al., 2005; Galli et al.). The ongoing requirement for GLI3 after E11.0 observed in Gli3-E9.5 CKOs suggests that the limb prepattern set up by GLI3 prior to ZPA formation, which was proposed to establish the initial competence of the mesenchyme to respond to SHH signaling (Chiang et al., 2001; te Welscher et al., 2002a), is not sufficient for normal autopod AP patterning to occur. Rather, it is critical that GLI3 continues to be expressed during a portion of the time SHH is present for a full complement of wild type digit identities to develop, consistent with the idea that a gradient of GLI3R activity generated by SHH signaling is important for setting up AP limb polarity. The later requirement for GLI3 in regulating phalangeal bone morphology observed in Gli3-E11.25 CKOs could reflect a role for inter-digitally expressed GLI3 in phalangeal bone morphogenesis, or may be a consequence of GLI3 acting downstream of IHH during chondrogenesis (Hilton et al., 2005; Koziel et al., 2005). Our temporal series of Gli3 conditional mutants has therefore established that the times when GLI3 is involved in specifying AP limb pattern, regulating digit number, and sustaining normal bone morphogenesis are separable, although occurring in overlapping temporal windows.
A previous study provided evidence that the stylopod-zeugopod-autopod segments are progressively determined in a proximal-to-distal sequence, since disaggregated limb mesenchyme cells become restricted to progressively more distal fates the later they are harvested, re-aggregated and grafted to a host embryo (Dudley et al., 2002; Sato et al., 2007). If limb mesenchymal cell fates are indeed determined in a proximal-to-distal temporal sequence, then proximal limb segments should become refractory to changes in gene expression (i.e. the consequences of removing Gli3) before correspondingly more distal limb segments. Indeed, comparison of the hindlimb phenotypes resulting from removal of Gli3 at sequentially later embryonic stages revealed a proximal to distal sequence of sensitivity to loss of Gli3 function. Formation of a normal tibial bone requires Gli3 for the least amount of time, followed by the anterior-most tarsal bone (medial cuneiform), then the anterior-most metatarsal, and finally, the phalanges. Thus, we have provided in vivo genetic evidence in support of a proximal-to-distal wave of determination guiding normal limb development.
In addition to dissecting Gli3 function in limb development, we have identified an essential role for the related gene Gli2 in providing the remaining AP pattern observed in Gli3 mutant limbs. We have therefore demonstrated that when GLI2 is absent GLI3 is critical for AP patterning until approximately the end of expression of SHH in the ZPA. Intriguingly, both the anterior autopod, which is posteriorized in Gli3−/− mutants, and posterior autopod, which retains normal polarity in Gli3−/− hindlimbs, depend on Gli2 for proper AP patterning in Gli3 CKOs. This was demonstrated in double mutants by loss of distinct anterior and posterior bone morphologies at E17.5, lack of limb AP morphological polarity at E12.5/E13.5 (see Fig. 7,8) and loss of polarized expression of the anterior marker Pax9 and the posterior marker Tbx2 at these stages. Furthermore, loss of AP polarity progressively worsens in Gli2 mutants the earlier Gli3 is removed, indicating that both timing and dosage of GLI function influence AP polarity. Therefore, our results reveal that the threshold of GLI-function required to set up AP polarity is calculated by integrating the duration with the total level of cellular exposure to GLI2 and GLI3 function.
By replacing Gli2 with Gli1 in Gli3-E10.5 CKO hindlimbs, we provide genetic evidence for an active role for GLI activators in AP limb patterning and show that GLIAs are specifically required in the posterior autopod. This finding is in accordance with the expected spatial distribution of GLIA function, since fate mapping studies have shown that cells expressing the HH target gene Gli1 contribute most extensively to digits 3–5, only minimally to digit 2, and are excluded from digit 1 (Ahn and Joyner, 2004). One explanation for the incomplete rescue of posterior tarsal bone patterning observed in Gli21k/−; Gli3-E10.5 CKOs is that both GLIRs and GLIAs are required for normal AP polarity in the posterior autopod. Another possibility is that GLI1A does not have correct level of activator function. In contrast, GLI1A exaggerates the anterior tarsal bone patterning defects observed in Gli2−/−; Gli3-E10.5 CKOs. Therefore, it is likely that anterior autopod AP patterning is mediated only by GLIRs, coinciding with the spatial predominance of GLIR anteriorly (Wang et al., 2000). Interestingly, one copy of Gli2 was sufficient to pattern the posterior autopod in Gli2+/−; Gli3 CKOs, but was not sufficient to pattern the anterior autopod. The lack of compensation for GLI3 in the anterior limb might be explained by significantly lower levels of GLI2R compared to GLI3R being present in mouse embryos (Pan et al., 2006) and the highest levels of GLIA being in the posterior limb (Ahn and Joyner, 2004). The fact that one allele of Gli2 is sufficient (or required) to pattern the posterior autopod might indicate that only a low level of activator is necessary for posterior limb pattern.
An AP patterning role for GLI3A was previously suggested based on the finding that AP polarity in limbs co-expressing two mutant forms of GLI3, a SHH-independent GLI3A (Gli3P1-4) and a constitutive repressor (Gli3D699) had a partially rescued phenotype compared to Gli3P1-4/P1-4 or Gli3D699/D699 limbs (Wang et al., 2007). However, it was recently reported that Gli3P1-4/D699 limbs are indistinguishable from Gli3D699/− (Hill et al., 2009), which raised the possibility that GLI3R alone is responsible for the nearly normal AP polarity observed in Gli3P1-4/D699 limbs. The interpretation of both studies is potentially confounded by the presence of Gli2, which likely contributes to aspects of the normal AP polarity observed in the Gli3 deletion mutants analyzed. In our study, GLI1A was added to a Gli2−/−; Gli3 CKO background, in which neither GLI2/3As nor GLI2/3Rs were present (after E12.0), allowing us to observe the extent to which a GLIA alone can pattern a limb devoid of functionally redundant paralogues. Therefore, our results provide definitive genetic evidence that a simple GLI activator is sufficient to provide some AP pattern, and therefore that both GLIAs and GLIRs participate in regulating AP limb patterning during normal development.
One prediction of our findings is that loss of autopod AP polarity should be more severe in Shh; Gli3 double mutant limbs compared to Gli3 single mutants, since Shh is required for induction of GLIA function. Two recent studies have addressed this question. In one study, conditional removal of dHand from the forelimb of Gli3−/− mutants resulted in complete loss of AP polarity (Galli et al.). Since dHAND is required for induction of Shh in the ZPA, dHand−/−; Gli3−/− limbs lack GLIA function in addition to GLI3. Reduced AP polarity in dHand−/−; Gli3−/− double mutants compared to Gli3−/− supports a role for GLIAs in posterior autopod polarity. Alternatively, dHAND could be affecting autopod AP polarity independent of SHH. Second, a comparison of Gli3D699/− limbs (expressing GLI3R only), to Gli3D699/−; Shh−/− limbs concluded that GLI3A is unlikely to play a role in AP patterning, since removal of Shh was interpreted as having no affect on AP asymmetry in Gli3D699/− mutants (Hill et al., 2009). However, the morphology of digits and carpal bones in the posterior Gli3D699/−; Shh−/− forelimb autopods compared to Gli3D699/− suggests that the distinctions between digits 3, 4, and 5 are retained in Gli3D699/− mutants, but are partially lost in Gli3D699/−; Shh−/− double mutant limbs. As we have observed in Gli2−/−; Gli3 CKOs, the posterior carpal bones in Gli3D699/−; Shh−/− forelimbs (the forelimb-equivalent to tarsal bones in the hindlimb autopod) appeared fused and homogenous in morphology, whereas their patterning was normal in Gli3D699/− limbs (Fig. 6 of (Hill et al., 2009). The overall digit length also appeared similar in the three posterior-most digits of Gli3D699/−; Shh−/− double mutants, further suggesting loss of posterior polarity. Our interpretation of the Gli3D699/−; Shh−/− morphological data presented in the Hill et al. (2009) study is therefore consistent with a model of GLI-mediated AP patterning in which GLI repressors alone are not sufficient to pattern the posterior autopod.
An on-going debate about the role of SHH signaling during vertebrate limb development concerns whether SHH-mediated AP limb patterning and regulation of digit number are temporally uncoupled, as indicated in the mouse (Zhu et al., 2008), or if these two processes take place at the same time, as suggested in the chick (Towers et al., 2008). In our study, depending on the type of conditional mutant, we observed either uncoupled or coupled patterning and digit number regulation. In Gli3-E10.5 CKOs digit number and AP pattern was temporally uncoupled, consistent with a biphasic model in which AP pattern is specified by SHH signaling early in limb development, but regulation of digit number occurs over an extended time (Zhu et al., 2008). However, temporal uncoupling was not observed in Gli2−/− mutants lacking Gli3 after ~E12. Given that only 6–12 hours of Shh expression is required to specify AP pattern (Zhu et al., 2008), yet Gli2 and Gli3 function is required to regulate AP polarity until between ~E12.0 and ~E12.75, it could be interpreted that a mechanism for differentially regulating GLI2 and GLI3 activity across the limb independent of SHH must take over after ~E11. One possibility is that GLI3-binding partners (HOXD12, SMAD, beta-catenin) can modify GLI3 transcriptional activity independent of late SHH signaling (Chen et al., 2004; Liu et al., 1998; Ulloa et al., 2007), as a high percentage of regulatory regions bound by GLI3R in the limb do not contain GLI-recognition sites, and are therefore likely to require GLI3 binding partners to regulate gene expression in the limb or are inert (Vokes et al., 2008). Our results suggest that whether or not temporal uncoupling of regulation of digit number and identity by SHH signaling is observed depends, fundamentally, on the assay employed.
Conclusion
Using a combination of temporal conditional knock-out and knock-in mutant analyses we have demonstrated that the task of translating SHH signaling into normal digit number and AP pattern is distributed between GLI3 and GLI2, and requires both GLIR and GLIA function. Spatially, the dependence of the anterior autopod on GLIR, and the posterior autopod on both GLIA and GLIR corresponds to the regional distributions of the proteins in the limb (Fig. 9B). Interestingly, as the tarsal bone morphologies become increasingly homogenous along the AP axis upon progressively decreasing the time and dose of GLI function, expression of AP polarity markers become progressively homogenous as well. The tarsal bone morphologies appear to converge upon a morphology resembling the bone normally located in the middle of the autopod. Since the middle of the autopod is the region most likely to experience a similar dose of GLIR and GLIA function, the ‘medial’ morphology of the tarsal bones in Gli2−/−; Gli3 CKO hindlimbs might correspond to the AP limb polarity produced when the ratio of GLI3R and GLI3A function is close to 1 (Fig. 9C). The results of our genetic studies demonstrate that the mechanism by which GLI-transcription factors provide a full range of polarized positional identities along the autopod AP axis involves integrating spatial, temporal, and dosage requirements for both GLIR and GLIA functions.
Fig. 9.
Summary and model for GLI function in limb AP patterning. (A) Temporal conditional inactivation of Gli3 reveals the sequence of Gli3-mediated events during limb patterning. Removal of Gli2 in Gli3 CKOs reveals the requirement for Gli2 during AP patterning. Removal of Gli3 at sequentially earlier stages in Gli2 mutants emphasizes the combined role of duration and dosage of total GLI function to AP patterning. Black arrows demarcate the embryonic stage when Tm was administered. Green and blue bars indicate the expected duration of Gli3 and Shh expression, respectively. Dark green checks indicate phenotypes observed in Gli3 mutants, light green checks indicate phenotypes observed in Gli2; Gli3 CKOs. The number of checks represents the severity of each phenotype listed. (B) AP patterning of the anterior autopod is dependent on GliRs (red), corresponding to the spatial domain containing predominantly GliRs. AP patterning of the posterior autopod requires both GliA (green), and GliR function, which corresponds to the spatial domain of strong GliA function (Ahn and Joyner, 2004). (C) Polarity of the limb is altered in different Gli-mutant backgrounds. The range of polarity is decreased in Gli3−/− limbs, excluding the anterior-most positional identities. Removal of Gli2 in Gli3 CKO mutants further restricts the range of polarity so that distinct posterior positional identities are lost, and the morphology of tarsal bones converge upon a state that could represent the consequence of GliR:GliA=1. This predicts that a hypothetical Gli2−/−; Gli3−/− limb would completely lack AP polarity. Blue shading represents the range of positional identities present in a wild type limb. Dotted lines indicate the range of positional identities available in each genetic background.
Supplementary Material
Shh is induced in the ZPA at ~E10.5. (A–D) Shh RNA in situ showing the range of limb morphology in litters collected on E10.5. Shh is first detectable in stage 2 limbs (~32 somites).
GliR regulates digit number and anterior tarsal bone patterning in Gli3 mutant hindlimbs. (A) Quantification of medial cuneiform bone phenotype in Gli3-E10.5 CKO hindlimbs in different Gli2 mutant backgrounds, comparing the percentage of hindlimbs with a complete bone (yellow), partial bone (red) or no bone (yellow). †: Statistically significant differences according to Fisher’s exact test. (B) Quantification of digit number phenotype in Gli3-E10.5 CKO hindlimbs in different Gli2 mutant backgrounds, comparing the percentage of hindlimbs with 6 digits (blue) versus 7 digits (red). *: Statistically significant differences according to Fisher’s exact test.
Posteriorly restricted HoxD11 and Hox12 expression is restored only in Gli3-E11.25 CKOs. HoxD11 and HoxD12 expression is excluded from the digit 1 region in wild type and Gli3+/− hindlimbs at E13.5 (A, G). In Gli3−/− hindlimbs HoxD11/12 expression is expanded anteriorly to include the region of the anterior-most digit (B, H). HoxD11/12 expression is similar to Gli3−/− in Gli3-E8.5 CKOs (C, I), Gli3-E9.5 CKOs (D, J) and Gli3-E10.5 CKOs (E, K), and similar to Gli3+/− in Gli3-E11.25 CKOs (F, L).
Highlights.
GLI3 sequentially regulates AP limb patterning, digit number and bone differentiation
Gli2 is essential for regulating autopod AP patterning in limbs lacking Gli3
GLI repressors provide anterior pattern, GLI activators contribute posterior pattern
regulation of AP patterning and digit number remains coupled in Gli2/3 double mutants
AP polarity integrates the dose, duration and domain of GLI activators and repressors
Acknowledgments
We would like to thank Dr. Cindy Loomis for advice and insightful comments throughout the studies and Dr. Sandra Wilson for critical reading of the manuscript. We would like to thank Dr. Virginia E Papaioannou and Dr. Cindy Loomis for generously providing the Tbx2 and Pax9 in situ probes, respectively. This work was funded by NIH grants R01 CA128158 and R01 DE016779 to ALJ and by the Center for Cancer Research, National Cancer Institute to SM.
Abreviations
- AP
anterior-posterior
- ZPA
zone of polarizing activity
- A
activator
- R
repressor
- p
phalanx
- AER
apical ectodermal ridge
- IDM
interdigital mesenchyme
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahn S, Joyner AL. Dynamic changes in the response of cells to positive hedgehog signaling during mouse limb patterning. Cell. 2004;118:505–16. doi: 10.1016/j.cell.2004.07.023. [DOI] [PubMed] [Google Scholar]
- Bai CB, Joyner AL. Gli1 can rescue the in vivo function of Gli2. Development. 2001;128:5161–72. doi: 10.1242/dev.128.24.5161. [DOI] [PubMed] [Google Scholar]
- Bai CB, Stephen D, Joyner AL. All mouse ventral spinal cord patterning by hedgehog is Gli dependent and involves an activator function of Gli3. Dev Cell. 2004;6:103–15. doi: 10.1016/s1534-5807(03)00394-0. [DOI] [PubMed] [Google Scholar]
- Barna M, Pandolfi PP, Niswander L. Gli3 and Plzf cooperate in proximal limb patterning at early stages of limb development. Nature. 2005;436:277–81. doi: 10.1038/nature03801. [DOI] [PubMed] [Google Scholar]
- Bastida MF, Ros MA. How do we get a perfect complement of digits? Curr Opin Genet Dev. 2008;18:374–80. doi: 10.1016/j.gde.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Blaess S, Stephen D, Joyner AL. Gli3 coordinates three-dimensional patterning and growth of the tectum and cerebellum by integrating Shh and Fgf8 signaling. Development. 2008;135:2093–103. doi: 10.1242/dev.015990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buscher D, Bosse B, Heymer J, Ruther U. Evidence for genetic control of Sonic hedgehog by Gli3 in mouse limb development. Mech Dev. 1997;62:175–82. doi: 10.1016/s0925-4773(97)00656-4. [DOI] [PubMed] [Google Scholar]
- Buscher D, Ruther U. Expression profile of Gli family members and Shh in normal and mutant mouse limb development. Dev Dyn. 1998;211:88–96. doi: 10.1002/(SICI)1097-0177(199801)211:1<88::AID-AJA8>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Chen Y, Knezevic V, Ervin V, Hutson R, Ward Y, Mackem S. Direct interaction with Hoxd proteins reverses Gli3-repressor function to promote digit formation downstream of Shh. Development. 2004;131:2339–47. doi: 10.1242/dev.01115. [DOI] [PubMed] [Google Scholar]
- Chiang C, Litingtung Y, Harris MP, Simandl BK, Li Y, Beachy PA, Fallon JF. Manifestation of the limb prepattern: limb development in the absence of sonic hedgehog function. Dev Biol. 2001;236:421–35. doi: 10.1006/dbio.2001.0346. [DOI] [PubMed] [Google Scholar]
- Dahn RD, Fallon JF. Interdigital regulation of digit identity and homeotic transformation by modulated BMP signaling. Science. 2000;289:438–41. doi: 10.1126/science.289.5478.438. [DOI] [PubMed] [Google Scholar]
- Dai P, Akimaru H, Tanaka Y, Maekawa T, Nakafuku M, Ishii S. Sonic Hedgehog-induced activation of the Gli1 promoter is mediated by GLI3. J Biol Chem. 1999;274:8143–52. doi: 10.1074/jbc.274.12.8143. [DOI] [PubMed] [Google Scholar]
- Dudley AT, Ros MA, Tabin CJ. A re-examination of proximodistal patterning during vertebrate limb development. Nature. 2002;418:539–44. doi: 10.1038/nature00945. [DOI] [PubMed] [Google Scholar]
- Echelard Y, Epstein DJ, St-Jacques B, Shen L, Mohler J, McMahon JA, McMahon AP. Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of CNS polarity. Cell. 1993;75:1417–30. doi: 10.1016/0092-8674(93)90627-3. [DOI] [PubMed] [Google Scholar]
- Fernandez-Teran M, Piedra ME, Kathiriya IS, Srivastava D, Rodriguez-Rey JC, Ros MA. Role of dHAND in the anterior-posterior polarization of the limb bud: implications for the Sonic hedgehog pathway. Development. 2000;127:2133–42. doi: 10.1242/dev.127.10.2133. [DOI] [PubMed] [Google Scholar]
- Galli A, Robay D, Osterwalder M, Bao X, Benazet JD, Tariq M, Paro R, Mackem S, Zeller R. Distinct roles of Hand2 in initiating polarity and posterior Shh expression during the onset of mouse limb bud development. PLoS Genet. 6:e1000901. doi: 10.1371/journal.pgen.1000901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson-Brown JJ, Agulnik SI, Chapman DL, Alexiou M, Garvey N, Silver LM, Papaioannou VE. Evidence of a role for T-box genes in the evolution of limb morphogenesis and the specification of forelimb/hindlimb identity. Mech Dev. 1996;56:93–101. doi: 10.1016/0925-4773(96)00514-x. [DOI] [PubMed] [Google Scholar]
- Gibson-Brown JJ, Agulnik SI, Silver LM, Niswander L, Papaioannou VE. Involvement of T-box genes Tbx2-Tbx5 in vertebrate limb specification and development. Development. 1998;125:2499–509. doi: 10.1242/dev.125.13.2499. [DOI] [PubMed] [Google Scholar]
- Harfe BD, Scherz PJ, Nissim S, Tian H, McMahon AP, Tabin CJ. Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell. 2004;118:517–28. doi: 10.1016/j.cell.2004.07.024. [DOI] [PubMed] [Google Scholar]
- Herault Y, Beckers J, Gerard M, Duboule D. Hox gene expression in limbs: colinearity by opposite regulatory controls. Dev Biol. 1999;208:157–65. doi: 10.1006/dbio.1998.9179. [DOI] [PubMed] [Google Scholar]
- Hill P, Gotz K, Ruther U. A SHH-independent regulation of Gli3 is a significant determinant of anteroposterior patterning of the limb bud. Dev Biol. 2009;328:506–16. doi: 10.1016/j.ydbio.2009.02.017. [DOI] [PubMed] [Google Scholar]
- Hilton MJ, Tu X, Cook J, Hu H, Long F. Ihh controls cartilage development by antagonizing Gli3, but requires additional effectors to regulate osteoblast and vascular development. Development. 2005;132:4339–51. doi: 10.1242/dev.02025. [DOI] [PubMed] [Google Scholar]
- Jiang J, Hui CC. Hedgehog signaling in development and cancer. Dev Cell. 2008;15:801–12. doi: 10.1016/j.devcel.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DR. Extra-toes: anew mutant gene causing multiple abnormalities in the mouse. J Embryol Exp Morphol. 1967;17:543–81. [PubMed] [Google Scholar]
- Koziel L, Wuelling M, Schneider S, Vortkamp A. Gli3 acts as a repressor downstream of Ihh in regulating two distinct steps of chondrocyte differentiation. Development. 2005;132:5249–60. doi: 10.1242/dev.02097. [DOI] [PubMed] [Google Scholar]
- Kraus P, Fraidenraich D, Loomis CA. Some distal limb structures develop in mice lacking Sonic hedgehog signaling. Mech Dev. 2001;100:45–58. doi: 10.1016/s0925-4773(00)00492-5. [DOI] [PubMed] [Google Scholar]
- Litingtung Y, Dahn RD, Li Y, Fallon JF, Chiang C. Shh and Gli3 are dispensable for limb skeleton formation but regulate digit number and identity. Nature. 2002;418:979–83. doi: 10.1038/nature01033. [DOI] [PubMed] [Google Scholar]
- Liu F, Massague J, Ruiz i Altaba A. Carboxy-terminally truncated Gli3 proteins associate with Smads. Nat Genet. 1998;20:325–6. doi: 10.1038/3793. [DOI] [PubMed] [Google Scholar]
- Lopez-Rios J, Speziale D, Robay D, Scotti M, Osterwalder M, Nusspaumer G, Galli A, Hollander GA, Kmita M, Zeller R. GLI3 Constrains Digit Number by Controlling Both Progenitor Proliferation and BMP-Dependent Exit to Chondrogenesis. Dev Cell. 22:837–48. doi: 10.1016/j.devcel.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lufkin T, Mark M, Hart CP, Dolle P, LeMeur M, Chambon P. Homeotic transformation of the occipital bones of the skull by ectopic expression of a homeobox gene. Nature. 1992;359:835–41. doi: 10.1038/359835a0. [DOI] [PubMed] [Google Scholar]
- Maynard TM, Jain MD, Balmer CW, LaMantia AS. High-resolution mapping of the Gli3 mutation extra-toes reveals a 51.5-kb deletion. Mamm Genome. 2002;13:58–61. doi: 10.1007/s00335-001-2115-x. [DOI] [PubMed] [Google Scholar]
- McGlinn E, van Bueren KL, Fiorenza S, Mo R, Poh AM, Forrest A, Soares MB, de Bonaldo MF, Grimmond S, Hui CC, Wainwright B, Wicking C. Pax9 and Jagged1 act downstream of Gli3 in vertebrate limb development. Mech Dev. 2005;122:1218–33. doi: 10.1016/j.mod.2005.06.012. [DOI] [PubMed] [Google Scholar]
- Mo R, Freer AM, Zinyk DL, Crackower MA, Michaud J, Heng HH, Chik KW, Shi XM, Tsui LC, Cheng SH, Joyner AL, Hui C. Specific and redundant functions of Gli2 and Gli3 zinc finger genes in skeletal patterning and development. Development. 1997;124:113–23. doi: 10.1242/dev.124.1.113. [DOI] [PubMed] [Google Scholar]
- Neubuser A, Koseki H, Balling R. Characterization and developmental expression of Pax9, a paired-box-containing gene related to Pax1. Dev Biol. 1995;170:701–16. doi: 10.1006/dbio.1995.1248. [DOI] [PubMed] [Google Scholar]
- Nguyen MT, Zhu J, Nakamura E, Bao X, Mackem S. Tamoxifen-dependent, inducible Hoxb6CreERT recombinase function in lateral plate and limb mesoderm, CNS isthmic organizer, posterior trunk neural crest, hindgut, and tailbud. Dev Dyn. 2009;238:467–74. doi: 10.1002/dvdy.21846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Bai CB, Joyner AL, Wang B. Sonic hedgehog signaling regulates Gli2 transcriptional activity by suppressing its processing and degradation. Mol Cell Biol. 2006;26:3365–77. doi: 10.1128/MCB.26.9.3365-3377.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Wang C, Wang B. Phosphorylation of Gli2 by protein kinase A is required for Gli2 processing and degradation and the Sonic Hedgehog-regulated mouse development. Dev Biol. 2009;326:177–89. doi: 10.1016/j.ydbio.2008.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panman L, Drenth T, Tewelscher P, Zuniga A, Zeller R. Genetic interaction of Gli3 and Alx4 during limb development. Int J Dev Biol. 2005;49:443–8. doi: 10.1387/ijdb.051984lp. [DOI] [PubMed] [Google Scholar]
- Park HL, Bai C, Platt KA, Matise MP, Beeghly A, Hui CC, Nakashima M, Joyner AL. Mouse Gli1 mutants are viable but have defects in SHH signaling in combination with a Gli2 mutation. Development. 2000;127:1593–605. doi: 10.1242/dev.127.8.1593. [DOI] [PubMed] [Google Scholar]
- Qu S, Niswender KD, Ji Q, van der Meer R, Keeney D, Magnuson MA, Wisdom R. Polydactyly and ectopic ZPA formation in Alx-4 mutant mice. Development. 1997;124:3999–4008. doi: 10.1242/dev.124.20.3999. [DOI] [PubMed] [Google Scholar]
- Riddle RD, Johnson RL, Laufer E, Tabin C. Sonic hedgehog mediates the polarizing activity of the ZPA. Cell. 1993;75:1401–16. doi: 10.1016/0092-8674(93)90626-2. [DOI] [PubMed] [Google Scholar]
- Sasaki H, Hui C, Nakafuku M, Kondoh H. A binding site for Gli proteins is essential for HNF-3beta floor plate enhancer activity in transgenics and can respond to Shh in vitro. Development. 1997;124:1313–22. doi: 10.1242/dev.124.7.1313. [DOI] [PubMed] [Google Scholar]
- Sato K, Koizumi Y, Takahashi M, Kuroiwa A, Tamura K. Specification of cell fate along the proximal-distal axis in the developing chick limb bud. Development. 2007;134:1397–406. doi: 10.1242/dev.02822. [DOI] [PubMed] [Google Scholar]
- Scherz PJ, McGlinn E, Nissim S, Tabin CJ. Extended exposure to Sonic hedgehog is required for patterning the posterior digits of the vertebrate limb. Dev Biol. 2007;308:343–54. doi: 10.1016/j.ydbio.2007.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–1. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Takeuchi J, Koshiba-Takeuchi K, Ogura T. Tbx Genes Specify Posterior Digit Identity through Shh and BMP Signaling. Dev Cell. 2004;6:43–53. doi: 10.1016/s1534-5807(03)00401-5. [DOI] [PubMed] [Google Scholar]
- te Welscher P, Fernandez-Teran M, Ros MA, Zeller R. Mutual genetic antagonism involving GLI3 and dHAND prepatterns the vertebrate limb bud mesenchyme prior to SHH signaling. Genes Dev. 2002a;16:421–6. doi: 10.1101/gad.219202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Welscher P, Zuniga A, Kuijper S, Drenth T, Goedemans HJ, Meijlink F, Zeller R. Progression of vertebrate limb development through SHH-mediated counteraction of GLI3. Science. 2002b;298:827–30. doi: 10.1126/science.1075620. [DOI] [PubMed] [Google Scholar]
- Tickle C. The number of polarizing region cells required to specify additional digits in the developing chick wing. Nature. 1981;289:295–8. doi: 10.1038/289295a0. [DOI] [PubMed] [Google Scholar]
- Tickle C, Summerbell D, Wolpert L. Positional signalling and specification of digits in chick limb morphogenesis. Nature. 1975;254:199–202. doi: 10.1038/254199a0. [DOI] [PubMed] [Google Scholar]
- Towers M, Mahood R, Yin Y, Tickle C. Integration of growth and specification in chick wing digit-patterning. Nature. 2008;452:882–6. doi: 10.1038/nature06718. [DOI] [PubMed] [Google Scholar]
- Ulloa F, Itasaki N, Briscoe J. Inhibitory Gli3 activity negatively regulates Wnt/beta-catenin signaling. Curr Biol. 2007;17:545–50. doi: 10.1016/j.cub.2007.01.062. [DOI] [PubMed] [Google Scholar]
- Vokes SA, Ji H, Wong WH, McMahon AP. A genome-scale analysis of the cis-regulatory circuitry underlying sonic hedgehog-mediated patterning of the mammalian limb. Genes Dev. 2008;22:2651–63. doi: 10.1101/gad.1693008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanek N, Muneoka K, Holler-Dinsmore G, Burton R, Bryant SV. A staging system for mouse limb development. J Exp Zool. 1989;249:41–9. doi: 10.1002/jez.1402490109. [DOI] [PubMed] [Google Scholar]
- Wang B, Fallon JF, Beachy PA. Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell. 2000;100:423–34. doi: 10.1016/s0092-8674(00)80678-9. [DOI] [PubMed] [Google Scholar]
- Wang C, Ruther U, Wang B. The Shh-independent activator function of the full-length Gli3 protein and its role in vertebrate limb digit patterning. Dev Biol. 2007;305:460–9. doi: 10.1016/j.ydbio.2007.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Nakamura E, Nguyen MT, Bao X, Akiyama H, Mackem S. Uncoupling Sonic hedgehog control of pattern and expansion of the developing limb bud. Dev Cell. 2008;14:624–32. doi: 10.1016/j.devcel.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zulch A, Becker MB, Gruss P. Expression pattern of Irx1 and Irx2 during mouse digit development. Mech Dev. 2001;106:159–62. doi: 10.1016/s0925-4773(01)00411-7. [DOI] [PubMed] [Google Scholar]
- Zuniga A, Zeller R. Gli3 (Xt) and formin (ld) participate in the positioning of the polarising region and control of posterior limb-bud identity. Development. 1999;126:13–21. doi: 10.1242/dev.126.1.13. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Shh is induced in the ZPA at ~E10.5. (A–D) Shh RNA in situ showing the range of limb morphology in litters collected on E10.5. Shh is first detectable in stage 2 limbs (~32 somites).
GliR regulates digit number and anterior tarsal bone patterning in Gli3 mutant hindlimbs. (A) Quantification of medial cuneiform bone phenotype in Gli3-E10.5 CKO hindlimbs in different Gli2 mutant backgrounds, comparing the percentage of hindlimbs with a complete bone (yellow), partial bone (red) or no bone (yellow). †: Statistically significant differences according to Fisher’s exact test. (B) Quantification of digit number phenotype in Gli3-E10.5 CKO hindlimbs in different Gli2 mutant backgrounds, comparing the percentage of hindlimbs with 6 digits (blue) versus 7 digits (red). *: Statistically significant differences according to Fisher’s exact test.
Posteriorly restricted HoxD11 and Hox12 expression is restored only in Gli3-E11.25 CKOs. HoxD11 and HoxD12 expression is excluded from the digit 1 region in wild type and Gli3+/− hindlimbs at E13.5 (A, G). In Gli3−/− hindlimbs HoxD11/12 expression is expanded anteriorly to include the region of the anterior-most digit (B, H). HoxD11/12 expression is similar to Gli3−/− in Gli3-E8.5 CKOs (C, I), Gli3-E9.5 CKOs (D, J) and Gli3-E10.5 CKOs (E, K), and similar to Gli3+/− in Gli3-E11.25 CKOs (F, L).