Summary
Propagation of gene expression patterns through the cell cycle requires the existence of an epigenetic mark that re-establishes the chromatin architecture of the parental cell in the daughter cells. We devised assays to determine which potential epigenetic marks associate with epigenetic maintenance elements during DNA replication in Drosophila embryos. Histone H3 trimethylated at lysine 4 or 27 are present during transcription, but surprisingly are replaced by non-methylated H3 following DNA replication. Methylated H3 is detected on DNA only in nuclei not in S phase. In contrast, the TrxG and PcG proteins Trithorax and Enhancer-of-Zeste that are H3K4 and H3K27 methylases, and Polycomb continuously associate with their response elements on the newly replicated DNA. We suggest that histone modification enzymes may re-establish the histone code on newly assembled unmethylated histones, and thus may act as epigenetic marks.
Introduction
Epigenetic inheritance requires the existence of a molecule that identifies the transcriptional status of a gene, is stable to DNA replication, and that re-establishes the expression pattern, termed an epigenetic mark (Brown, 1984). Epigenetic inheritance requires the existence of a molecule that identifies the transcriptional status of a gene, and that reestablishes the expression pattern, termed an epigenetic mark (Brown, 1984). Since transcription occurs in both G1 and G2, this suggests that epigenetic marks are necessary in both G1 and G2, and that they have to be stably transmitted through DNA replication and mitosis, respectively. Here, we examine only marks established in G2, as early Drosophila embryos lack G1. During DNA replication, chromatin structure throughout the genome is reorganized as a consequence of the passage of the replication fork (RF) and/or progression of the DNA polymerase complex (Corpet and Almouzni, 2009). Any epigenetic mark must be stable to replication or rapidly recruited to nascent DNA, and parental levels of the mark must be re-established in each daughter cell. DNA methylation provides the best example of an epigenetic mark that meets these criteria (Sharif et al., 2007).
The PCNA protein acts as a platform to recruit DNA Polymerase and many other proteins to the single-stranded DNA (ssDNA) located downstream of the RF and acts as a processivity factor for the DNA polymerase (reviewed in (Moldovan et al., 2007). PCNA is stably associated with DNA on both leading and lagging strands (See Figure 3A). Despite our increasing knowledge of DNA-replication coupled events it has been difficult to establish the order and timing of recruitment of proteins and modified histones to nascent DNA in vivo at specific sequences because methods with sufficient sensitivity have not been available.
Figure 3. The size and composition of the replicating DNA fragments.
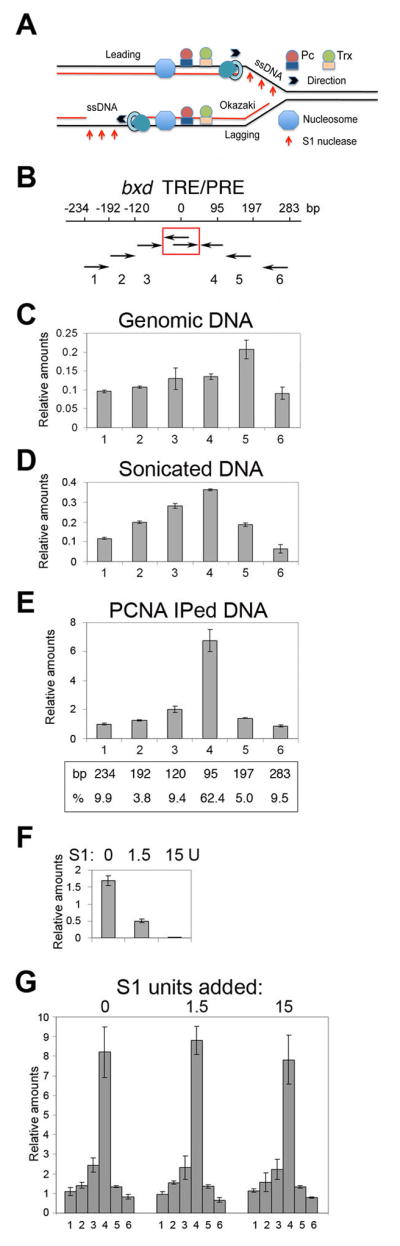
(A) Schematic diagram showing location of parental dsDNA (two black lines), parental ssDNA (one black line), and replicated nascent DNA (one red and one black line) and the replication fork (RF). The direction of travel of the RF, and of the DNA polymerase complexes on the leading and lagging strands (PCNA is indicated as a torus, and the polymerase as a tear drop) are indicated.
(B) Map of the bxd ME region tested. The primer sets are indicated below the map. Fixed primers in the center are boxed in red.
(C) The relative efficiency of primer sets on genomic DNA. The raw data in D,E,G were normalized by multiplying the reciprocal of the efficiencies of amplification for each primer pair relative to primer pair 6. Data are represented as mean +/− SEM.
(D) The relative abundance of sonicated DNA amplified in PCR using the primer sets in shown in B. Data are represented as mean +/− SEM.
(E) The relative abundance of DNA fragments immunoprecipitated with PCNA following sonication that amplifies in PCR reactions. Below the graph is shown the percentage of the population of amplified fragments, determined after correcting for overlap. Data are represented as mean +/− SEM.
(F) Carrier ssDNA (0.5 μg) was digested with 0, 1.5 and 15 units of S1 nuclease, to determine the titer necessary for complete digestion. Data are represented as mean +/− SEM.
(G) The relative abundance of DNA immunoprecipitated with PCNA and digested with 0, 1.5, and 15 units of S1 nuclease. 0.5 μg of carrier ssDNA was added to each reaction. The experiments were performed at least three times. Data are represented as mean +/− SEM..
The best-studied proteins required for maintenance of gene expression patterns are chromatin proteins of the trithorax and Polycomb groups (TrxG and PcG). TrxG and PcG proteins are associated with complex DNA elements termed Trithorax and Polycomb response elements (TREs and PREs) (reviewed in (Muller and Kassis, 2006; Ringrose and Paro, 2007). In this study we examine elements that combine both TREs and PREs, and we will call them maintenance elements (ME). The TrxG protein Trithorax (Trx) is a histone methyltransferase (HMT) that trimethylates H3K4 (Smith et al., 2004). The PcG protein Enhancer of zeste, E(z), is also an HMT that trimethylates H3K27 (Cao et al., 2002; Czermin et al., 2002; Muller et al., 2002).
TrxG and PcG proteins are attractive candidates to be the epigenetic marks required for maintenance, consistent with observations that PcG proteins of the PRC1 complex are stable to DNA replication in vitro (Francis et al., 2009). If the HMT E(z) and Trx were stable to passage of the RF and DNA polymerase, they could methylate newly synthesized histones to re-establish the parental pattern of histone modifications on nascent DNA. Consistent with this model Set8, an H4K20 monomethylase, is targeted to replication forks through a direct interaction with PCNA (proliferating cell nuclear antigen) (Huen et al., 2008), and a complex containing SetDB1-MBD1-CAF-1 that trimethylates H3K9 is recruited by hemimethylated DNA (Sarraf and Stancheva, 2004).
An attractive alternative model is based on observations that parental histones are transferred randomly to nascent DNA (Jackson and Chalkley, 1985). To ensure that the daughter DNA has the correct nucleosome content, newly synthesized unmodified histones must also be deposited on nascent DNA. It is proposed that post-translationally modified parental histones are transferred immediately to nascent DNA during DNA replication (Corpet and Almouzni, 2009) and recruit the appropriate HMTs or other histone-modifying TrxG and PcG proteins (Chang et al., 2010; Hansen et al., 2008), which will then restore the pattern of parental modifications to the newly synthesized histones. Thus, modified histones, especially those carrying H3K27me3 and H3K4me3 established by E(z) and Trx respectively, could be epigenetic marks (Ng and Gurdon, 2008). Hansen et al. (2008) showed in mammalian cells that H3K27me3 recruits the PRC2 complex to repress transcription, and proposed that H3K27me3 was an epigenetic mark.
If trimethylation of histone H3 is an epigenetic mark, then one would predict that these modifications would be detected shortly after DNA replication. To our knowledge, no one has determined how soon after DNA replication H3K4me3 or H3K27me3 modifications can be detected on nascent DNA in vivo. A recent study demonstrates that PcG and methylated H3 are enriched at PREs in early S phase prior to their replication. In late S phase, when PREs are replicated, the amount of methylated H3K7 drops (Lanzuolo et al., 2011).
In this work we developed assays to assess which proteins or modified histones are candidates to be the epigenetic mark in early Drosophila embryos. In all assays, parental trimethylated H3K4 and H3K27 are replaced by the unmethylated histone H3 downstream of the DNA polymerase. In contrast, Trx, Pc and E(z) remain associated with DNA following the passage of the DNA polymerase. These data suggest that HMTs and other TrxG or PcG proteins may be epigenetic marks, but that H3K4me3 and H3K27me3 are unlikely to be epigenetic marks at this stage of Drosophila development.
Results
Differential localization of modified histones in replicating cells in vivo
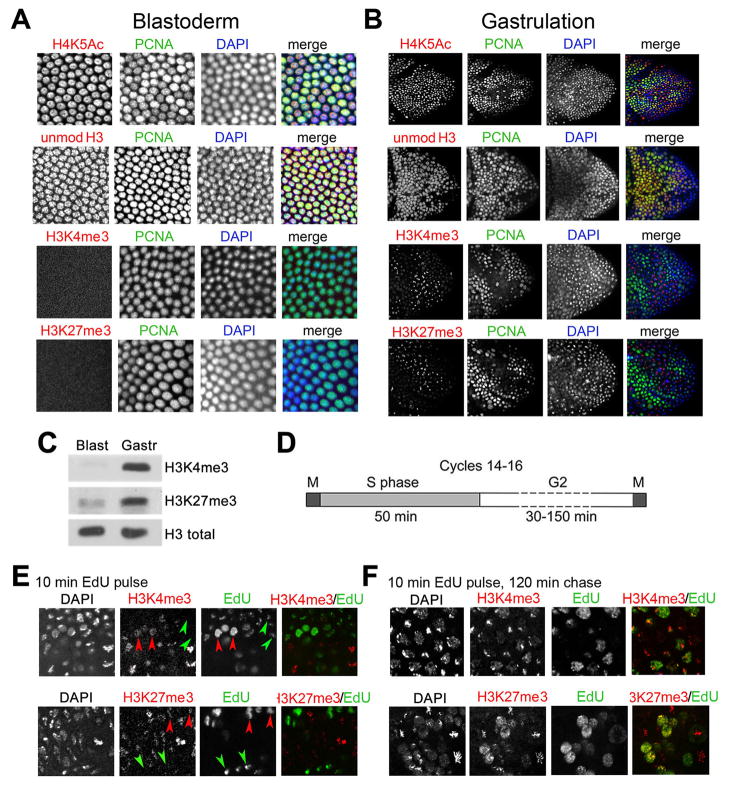
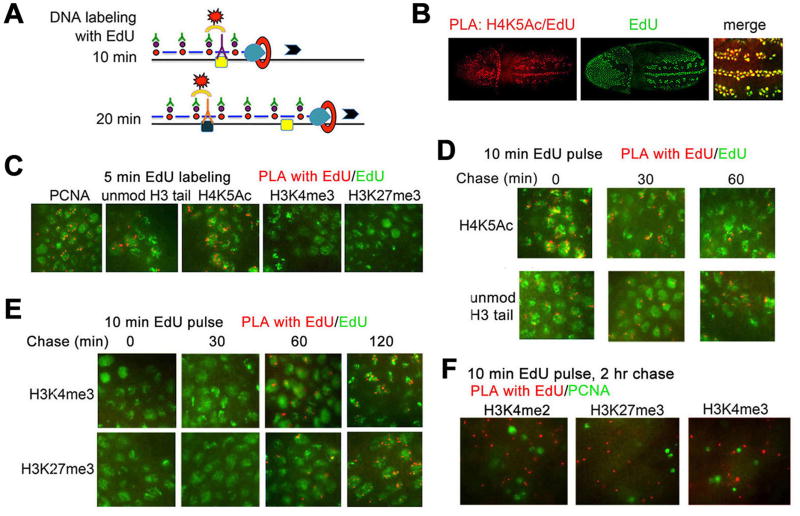
We examined whether the distribution of modified histones is changed during the cell cycle in Drosophila embryos. Acetylation of H4 at K5 (H4K5Ac) correlates with DNA replication (Corpet and Almouzni, 2009). As expected H4K5Ac colocalizes with DAPI and PCNA staining at all examined embryonic stages starting from syncytial blastoderm (Figure 1A,B and not shown), and to both the S-phase and mitotic nuclei in gastrulating embryos (Figure 1B). We also examined the H3K4me3 and H3K27me3 modifications required for gene activation and silencing. Antibodies to H3K27me3 and H3K4me3 do not recognize the corresponding mono- and di-methylated forms of H3 (Figure S1A). None of these antibodies show any significant cross-reactivity (Figure S1B). We also used antibody that was generated against the first 20 N-terminal amino acids of the unmodified H3 (Figure 1A,B). This antibody binds the N-terminal peptide of H3, the recombinant H3 and H3 from acid-acid purified histones (Figure S1B,C).
Figure 1. Methylated histones are weakly associated with replicating nuclei.
(A, B) Double-immunostaining of Drosophila embryos was performed with indicated antibodies against PCNA (green) and H4K5Ac, H3K4me3, H3K27me3 and antibody to unmodified N-terminus (aa 1–20) of histone H3. Nuclei were stained with DAPI. (A) cellular blastoderm, (B) early gastrulating Drosophila embryos. See also Figure S1.
(C) Quantification of H3K4me3 and H3K27me3 in the cellular blastoderm and gastrulating embryos by Western blotting.
(D) Schematic diagram showing the length of the cell cycle phases in mid-staged Drosophila embryos (adopted from (Shermoen et al., 2010).
(E) Gastrulating embryos were labeled with EdU for 10 min and immunostained for EdU (green) and for H3K4me3 or H3K27me3 (red). Red arrows indicate early replicating nuclei, and green arrows indicate late replicating nuclei as described in (Shermoen et al., 2010).
(F) Gastrulating embryos were pulse labeled with EdU for 10 min and kept for 120 min prior to fixation. Embryos were immunostained for EdU (green) and for H3K4me3 or H3K27me3 (red).
Unmodified H3 was detected in the blastoderm embryos (Figure 1A), and mostly in the replicating cells of the gastrulating embryo (Figure 1B). Interestingly, there is very little H3K4me3 and H3K27me3 at cellular blastoderm (Figure 1A,C), suggesting that these post-translational modifications (PTMs) of histone H3 may not be essential for DNA replication in very early embryos. During gastrulation, both methylated forms of H3 are found predominantly in nuclei that do not contain PCNA (Figure 1B). To confirm this observation, we analyzed distribution of histones in embryos that were labeled for 10 min with EdU. Widespread foci and restricted patterns of EdU incorporation in nuclei correspond to early and late S phase, respectively (Shermoen et al., 2010). Figure 1E shows that while small amounts of methylated H3 forms are detected during gastrulation in the early S phase nuclei (red arrows), these proteins are not detected in the late S phase nuclei (green arrows), in agreement with (Lanzuolo et al., 2011) who detected a decrease of H3K27me3 in the late S phase of cell lines. Pulse-labeling with EdU for 10 min followed by chase for 120 min ensures that all labeled nuclei transit to the G2 phase (see Figure 1D for a scheme of the cell cycle in early embryos). In these embryos we observed very significant co-localization of trimethylated H3K4 and H3K27 with labeled DNA (Figure 1F).
These results show that at these embryonic stages, different histone PTMs are present at different stages of the cell cycle, and that H3K4me3 and H3K27me3 forms are present in low amounts on replicating DNA, especially at late stages of the S phase. These results support the model that H4K5Ac is required for histone deposition on nascent DNA, and suggest that unmodified H3, but not methylated H3 forms are associated with the newly replicated bulk DNA. Instead, methylated H3 mostly accumulates in the transcriptional G2 phase and in mitosis. This result suggests that at these embryonic stages, methylated H3 forms may not have a role in epigenetic inheritance in Drosophila.
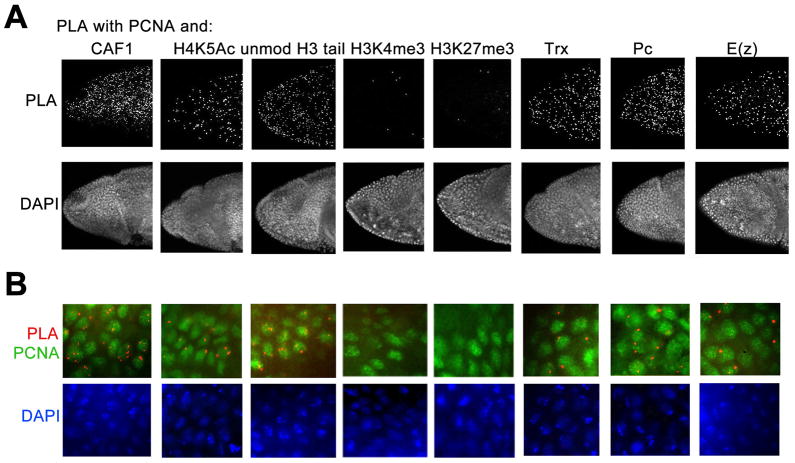
Identification of proteins in close proximity to the DNA replication complex
Any protein that is near the RF will be close to PCNA in vivo. We used the Proximity Ligation Assay (PLA, Olink Bioscience) to identify which proteins or histone PTM associate with PCNA in vivo. In the PLA assay, antibodies to the proteins or PTM of interest made from different species are detected with secondary antibodies linked to oligonucleotides that form a closed circle after addition and ligation of complementary linkers. Rolling circle amplification in the presence of fluorescently-labeled oligonucleotides generates a fluorescent signal if they are in close proximity. The sensitivity of the PLA makes it possible to identify single molecule interactions in vivo. Embryos were counterstained with antibodies to PCNA to detect nuclei in S phase. The control experiments show very low background with all antibodies used in PLA assays (Figure S2).
Trx and Pc proteins are detected early in embryogenesis. If PcG proteins are stable to DNA replication (Francis et al., 2009) then Pc, E(z), and perhaps Trx proteins could co-localize with PCNA in vivo. As shown in Figure 2A,B, Trx, Pc and E(z) are clearly detected in PLA assays with PCNA in gastrulating embryos in replicating nuclei. These results suggest that Trx, Pc and E(z) are either in close proximity to, or directly interact with PCNA at the replicating DNA polymerase complex. The unmodified H3 tail, and H4K5Ac also associate with PCNA, consistent with deposition of de novo synthesized histones (Figure 2A,B). Surprisingly, tri-methylated H3K27 and H3K4 are not detected in PLA experiments with PCNA (Figure 2A,B). These results strongly suggest that methylated histones are not in close contact with replication machinery, and are not transferred to nascent DNA soon after DNA replication. Interestingly, we easily detect association of PCNA and CAF-1 in these assays (Figure 2A,B), suggesting that while CAF-1 is likely to load H3 with an unmodified amino tail, it is unlikely to load histones H3K4me3 and H3K27me3 on nascent DNA as proposed (Corpet and Almouzni, 2009). Together, these results support the results in Figure 1.
Figure 2. Trx, Pc and E(z) transiently interact with PCNA.
(A) Top panel, PLA signals between PCNA antibody and antibodies indicated on the top. Bottom, DAPI staining on the same embryos. Photos are taken at 20X magnification. See also Figure S1.
(B) 100X magnification of the embryos from the same experiments. PLA signals are in red, EdU is in green. DAPI is shown at the bottom.
PCNA immunoprecipitates short DNA fragments behind the replication fork
To determine which proteins or PTM are retained on DNA during DNA replication, we developed a sequential chromatin immunoprecipitation (re-ChIP) in Drosophila embryos. For most experiments, we used immunoprecipitation with PCNA as a marker for replicating DNA. To interpret the re-ChIP experiments, it is essential to know the size of DNA that is immunoprecipitated with PCNA, and to show that parental chromatin is not present (see model in Figure 3A). First, we developed a method to allow us to estimate the relative abundance of amplicons obtained from immunoprecipitated DNA fragments when sample was limiting. We used q-PCR with fixed central and flanking primers within the core of the well established ME in the bxd region of the homeotic gene Ubx (Figure 3B). To account for differences in amplification frequency between different primer pairs, we determined the relative amplification frequency for each primer pair on genomic DNA (Figure 3C), and used this information to normalize the data reported in Figures 3D, E, G (details are provided in Extended Experimental Procedures). As shown, after sonication DNA fragments of about 200 bp or less are represented more frequently than fragments of greater than 200 bp (Figure 3D), as expected from the results of (Schwartz et al., 2005) who showed the this region is sensitive to sonication. We next determined the relative abundance of amplified DNA following ChIP with antibodies to PCNA, and found that the ratio between DNA fragments that are larger and smaller that 200 bp is approximately 30% and 70%, respectively (Figure 3E).
Any PCNA-immunoprecipitated DNA fragment that contains parental and nascent DNA will also contain ssDNA between the parental and nascent regions of dsDNA (Figure 3A). Therefore treatment of such fragments with S1 nuclease will cleave these fragments into two smaller pieces and thus increase the relative frequency of short fragments in our assay. We digested PCNA-immunoprecipitated DNA with an amount of S1 nuclease that is sufficient to completely digest a large quantity of the carrier ssDNA (Figure 3F). Digestion with S1 nuclease does not significantly affect the lengths of the PCNA-immunoprecipitated DNA fragments from the ME (Figure 3G). Therefore, ssDNA is not present in PCNA-precipitated DNA, showing that there is no parental DNA present. Similar results were obtained with other ME regions (not shown). These results imply that DNA breaks at or near the ss/dsDNA border during sonication. Therefore, any DNA fragments recovered after immunoprecipitation with PCNA must come from dsDNA from immediately after progression of the RF (see model in Figure 3A). The re-ChIP experiments do not distinguish between proteins that are stably associated with DNA during replication, or rapidly re-recruited to nascent DNA, so we will use “stably associated” to include either possibility.
Histones are associated with DNA behind the RF
Nucleosomes associate with nascent DNA shortly after passage of the RF (Kriegstein and Hogness, 1974). Sequential ChIP of newly synthesized DNA provided the opportunity to determine the modification status of histones on nascent DNA near the DNA polymerase complex. In these and subsequent experiments, we used fixed chromatin prepared from 1 ml of embryos 2–15 h after egg lay (AEL), which is 200 times more than usually used for ChIP experiments. The second immunoprecipitation uses the same amount of starting material as regular ChIP (Figure S3), consistent with the yield from the PCNA IP being 0.5% of the starting material (see Figure 5A). All ChIP and re-ChIP experiments were performed with the same batches of antibodies using the same sonicated chromatin. Thus, the ratios between the values for different proteins in these linked ChIP/re-ChIP assays provide a true comparison of the relative amounts of these proteins in the bulk and replicating DNA.
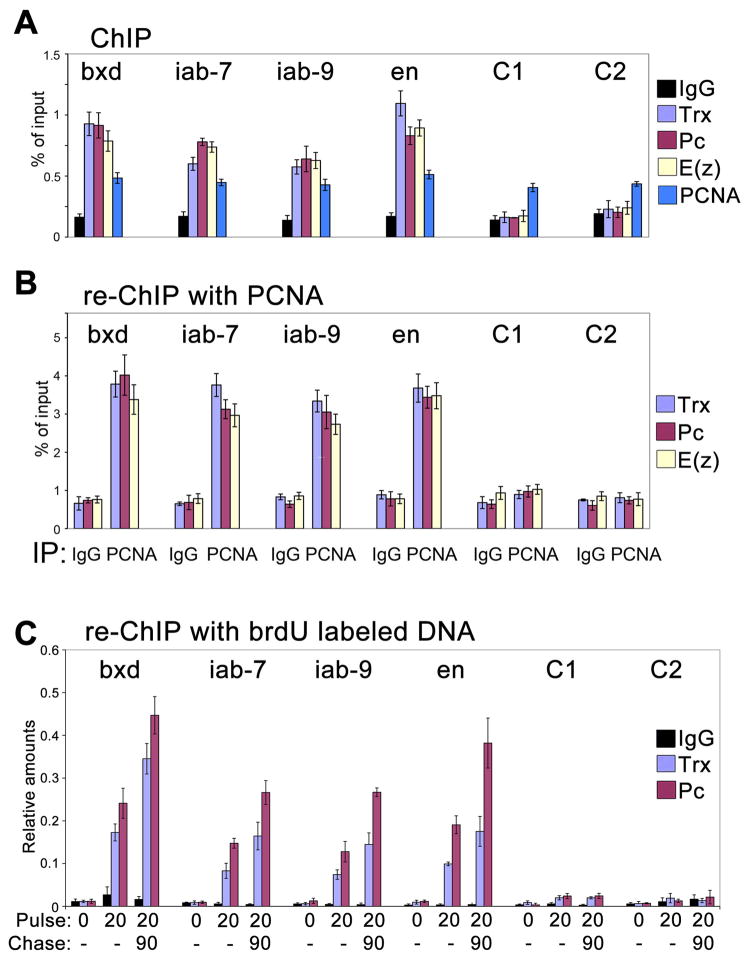
Figure 5. Trx, Pc and E(z) are specifically associated with their MEs on nascent DNA from embryos.
(A) ChIP assays with IgG (black), Trx (purple), Pc (maroon), E(z) (light yellow) and PCNA (blue) antibodies from chromatin prepared from 2–15 h AEL embryos. The central primer sets shown in Figure 4A were used. Data are represented as mean +/− SEM.
(B) Sequential re-ChIP assays using either IgG or PCNA antibodies (indicated at the bottom) in the first immunoprecipitation step, and Trx, Pc and E(z) antibodies in the second immunoprecipitation step. Percent of input for re-ChIP experiments was calculated using the amount of material eluted from the beads following immunoprecipitation with PCNA as 100% input. Data are represented as mean +/− SEM.
(C) Sequential re-ChIP assays using chromatin prepared from 2–7 h AEL embryos that were labeled with brdU for 20 min and for 20 min followed by a pulse of 1.5 h. Chromatin was first immunoprecipitated with Trx Pc or IgG antibodies. In the second step, DNA purified from the resulting material was immunoprecipitated with the brdU antibody. Chromatin from unlabeled embryos was used as a control. Since multiple samples from the first step are shown on the same graph, relative amounts are shown instead of the percent of input which is specific for each sample. The percentages of input for brdU immunoprecipitation from the Pc and Trx samples in different regions range from 1 to 4%, respectively. Calculations used DNA purified from the material following immunoprecipitation with Trx and Pc antibodies as 100% input. Data are represented as mean +/− SEM.
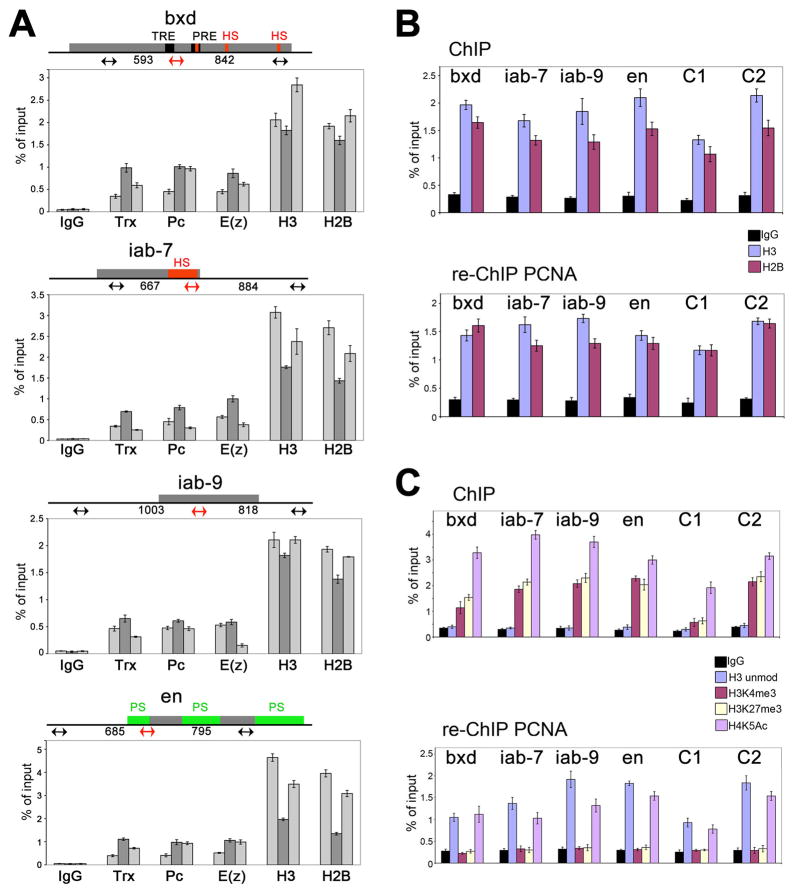
For these experiments, we chose four DNA regions that contain MEs: three regions of the BX-C, bxd, iab-7 and iab-9, and a known ME region of the Engrailed (En) gene. Figure 4A indicates known functional elements in these MEs (see legend). Based on the results of our ChIP assays (Figure 4A) we chose one primer set which shows a highest levels of Trx, Pc and E(z) in these ME sequences for further analysis. These regions contain somewhat lower amounts of histones compared to flanking regions (Figure 4A) consistent with rapid histone turnover observed for epigenetic elements (Deal et al., 2010), but we readily detect enough histone for reliable analysis. We also included two regions, C1 (located in +39 kb to the Dpr12 gene) and C2 (located 7 kb upstream of the bxd PRE in the BX-C), which do not bind TrxG and PcG proteins (see Figure 5A) below) as controls for cross-reaction of the antibodies.
Figure 4. Methylated histones H3 are replaced by unmodified histone H3 on nascent DNA.
(A) ChIP assays from chromatin prepared from 2–15 h AEL embryos. Antibodies are indicated at the bottom. Locations of the primer sets within four tested TRE/PRE sequences are shown in the schemes on the top of each graph. Mapped TRE and PRE sequences in the bxd region are shown in black (Tillib et al., 1999). Known DNase I hypersensitive sites in the bxd (Dellino et al., 2002) and iab-7 (Mishra et al., 2001) elements of the BX-C are shown in red. Mapped pairing sensitive regions in the en PRE are shown in green (Kassis, 1994). Primer sets in the iab-9 PRE are chosen based on data from (Beisel et al., 2007). The central primer sets (shown in red on the maps and as dark grey bars in the graphs) were used in all experiments in this and subsequent figures. Data are represented as mean +/− SEM.
(B) Top, ChIP assays with IgG (black), H3 (purple) and H2B (maroon) antibodies from chromatin prepared from 2–15 h AEL embryos. Bottom, sequential re-ChIP assays using chromatin prepared from 2–15 h AEL embryos. Chromatin was immunoprecipitated with PCNA antibody, eluted and precipitated with IgG, H3 and H2B antibodies. Percent of input for re-ChIP experiments was calculated using as a standard the material eluted from the beads following immunoprecipitation with PCNA. See also Figure S3. Coordinates of the primer sets to four MEs, bxd, iab-7, iab-9, en and two non-ME regions, C1 and C2, are given in Extended Experimental Procedures. All ChIP and re-ChIP experiments in this paper were done at least in triplicate, and standard error bars are shown in these and subsequent figures. Data are represented as mean +/− SEM.
(C) Top, ChIP assays with IgG (black), and antibodies to unmodified N-terminus of H3 (purple), H3K4me3 (maroon), H3K27me3 (light yellow) and H4K5Ac (magenta) were performed on bulk chromatin prepared from 2–15 h AEL embryos at 6 loci. Bottom, sequential re-ChIP assays using PCNA in the first immunoprecipitation step, using the same antibodies as in ChIP in the second immunoprecipitation step. The same regions as in Figure 2 were tested in A and B. See also Figure S4. Data are represented as mean +/− SEM.
We first used polyclonal antibodies generated against the large unmodified portions of histones H3 and H2B that will recognize major forms of these histones using the IgG antibody as a negative control in conventional ChIP assays. We found that as expected, H3 and H2B are associated with all tested regions of DNA (Figure 4B, top panel). We also detected both histones H3 and H2B in all tested regions in the re-ChIP assays using PCNA in the first immunoprecipitation step (Figure 4B, bottom panel). Thus, histone components of both H2A-H2B and H3-H4 dimers are detected immediately after the passage of the RF.
Association of modified histones with replicating chromatin
Developing Drosophila embryos contain a mixture of cells in which different BX-C genes and en are either activated by TrxG proteins or repressed by PcG proteins. Therefore, both silencing and activating PTMs, as well as TrxG and PcG proteins can be detected in the same ChIP experiments. Consistent with this, both H3K4me3 and H3K27me3 were detected in ChIP experiments in bulk chromatin at all four tested MEs and in the C2 region (Figure 4C, top panel). The C1 control region contains lower amounts of these H3 forms, and may be transcriptionally inert. In bulk chromatin, antibody to the N-terminus of unmodified H3 failed to detect this histone form at any tested DNA region (Figure 4C, top panel). In ChIP experiments, H4K5Ac was detected in all tested DNA regions (Figure 4C, top panel).
H4K5Ac, as well as other acetylated histones, is required for deposition of histones by chaperones on nascent DNA, and therefore should be detected in our re-ChIP experiments with PCNA. As expected, H4K5Ac was detected in all regions where it was initially detected by conventional ChIP (Figure 4C, bottom panel). Unexpectedly, in re-ChIP experiments we did not detect H3K4me3 and H3K27me3 in any tested DNA region (Figure 4C, bottom panel). This is not caused by the low affinity of these antibodies when small amounts of chromatin are used, as we detect total H3, H3K4me3 and H3K27me3 equally well using comparable amounts of chromatin in ChIP assays (Figure S4). These results suggest that these parental modified histones are not recruited quickly to nascent DNA after the passage of the approaching DNA polymerase complex.
However, we detected H3 with an unmodified N-terminus in all DNA regions that were immunoprecipitated with PCNA. Since H3 antibody was generated against the first 20 N-terminal amino acids of histone H3, these results suggest that during DNA synthesis displaced H3K4me3 is replaced by H3 that is not methylated at Lys-4. No antibody to unmethylated K27 region of H3 is available, so we cannot determine directly if H3 that is unmethylated on H3K4 also lacks K27 methylation. The observation that H3K27 methylation is not seen in re-ChIP suggests that the newly deposited H3 is not modified at either K4 or K27, consistent with the PLA results above. We suggest that progression of the RF is accompanied by the dissociation of the methylated histone H3 forms, and that unmethylated H3 histone is rapidly recruited to nascent DNA. These results confirm that contamination with parental chromatin does not occur in our re-ChIP experiments, because otherwise we would detect trimethylated forms of H3. In reciprocal re-ChIP experiments, we detected PCNA only in the material that was immunpoprecipitated with antibody against the non-methylated H3 tail. No signals for PCNA were detected in the material that was pulled down with antibodies against H3K4me3 and H3K27me3 (Figure S5). This key result shows that the re-ChIP experiments are specific, and that the replicating chromatin is not contaminated with parental chromatin.
Trx, Pc and E(z) are associated with their MEs during and following the passage of the RF
Epigenetic proteins that create and recognize histone PTMs are likely candidates for epigenetic marks. Therefore, we focused on the TrxG and PcG proteins Trx and E(z), that are H3K4me3 and H3K27me3 HMTases, respectively, and the PcG protein Pc which is the component of the PcG PRC1 complex that recognizes the silencing H3K27me3 mark. Trx, Pc and E(z) were detected in ChIP assays on bulk DNA at all four tested MEs, and were not detected in the C1 and C2 regions (Figures 4A and 5A). Importantly, in re-ChIP experiments with PCNA, all these proteins were detected in all four MEs (Figure 5B). These results were confirmed using a second PCNA antibody (not shown). We also used IgG instead of PCNA for the first immunoprecipitation step, followed by immunoprecipitation with Trx, Pc or E(z) antibodies. As shown in Figure 5B, immunoprecipitation with IgG does not result in detection of Trx, Pc and E(z) at significant levels. In bulk chromatin, we recover more than 2% of input chromatin after immunoprecipitation with antibodies to H3K4me3 and H3K27me3, which is approximately twice the recovery with antibodies to Trx and Pc. However, in re-ChIP assays we recover 3–4% of immunoprecipitated chromatin in the second immunoprecipitation step with antibodies to Trx, Pc and E(z), whereas H3K4me3 and H3K27me3 are recovered at background levels (Figures 4, 5). These results suggest that Trx, Pc and E(z) remain associated with their response elements during or immediately after the passage of the DNA replication complex.
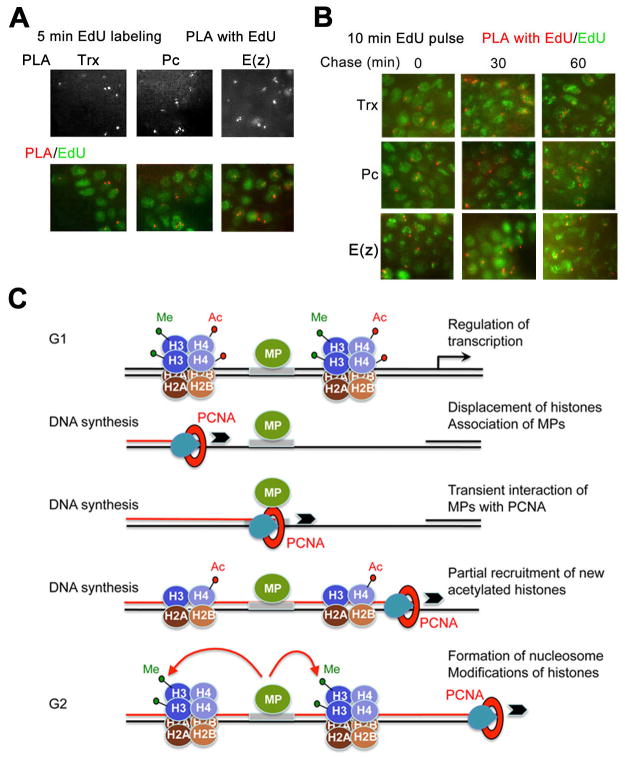
The PLA assays (Figure 2) detected Trx, Pc and E(z) in close proximity to PCNA and re-ChIP assays confirm that these proteins are found at their MEs on short fragments of nascent DNA (Figure 5B). Both assays use PCNA as a marker of the vicinity of the RF. To determine if these proteins associate with nascent DNA independent of PCNA, we employed the re-ChIP assay using brdU labeling of nascent DNA that was introduced previously for cultured Drosophila cells (Francis et al., 2009). To enhance the efficiency of labeling, 2–7 h AEL embryos were mechanically disrupted prior to incubation with brdU. Chromatin from these embryos was immunoprecipitated with antibodies against Trx and Pc. DNA purified from this material was further immunoprecipitated with brdU antibody and analyzed by PCR. Figure 5C shows that Trx and Pc are specifically associated with their MEs on long stretches of nascent DNA labeled with brdU for 20 min. The amounts of these proteins increase approximately two fold following the additional chase for 1.5 h (Figure 5C), suggesting that the full complement of these proteins is restored at the late stages of the S phase or in the interphase. These results agree with similar experiments in Drosophila S2 cells (Francis et al., 2009), and show that Trx and Pc are associated with nascent DNA long after the passage of the RF, and that this association is independent of PCNA.
Tri-methylated histones H3 are not associated with nascent DNA during S phase
The re-ChIP experiments detect proteins associating with DNA about 200 bp from the RF. To extend the re-ChIP results to assay DNA further from the RF, we developed a technique that we call the “Chromatin Assembly Assay” (CAA). Its design is diagrammed in Figure 6A. Nascent DNA is labeled with EdU, and chemically conjugated with biotin. The biotin and protein of interest are detected with appropriate antibodies, and the PLA assay (Olink Bioscience) described above is used to examine whether the protein of interest is associated with nascent DNA labeled with EdU in vivo in a time and tissue specific manner. Given a DNA replication rate of 30–50 bp sec−1, 5 min of EdU incorporation will detect at most 9–15 kb of DNA. This method provides an overview of the events that occur in many genomic locations and thus generalizes the results seen by re-ChIP in specific DNA sequences. Figure 6B shows that the signals from the CAA reaction between H4K5Ac required for deposition of histones on nascent DNA and the EdU labeled DNA occur only in nuclei that are labeled with EdU, demonstrating the specificity and low background of this reaction.
Figure 6. Chromatin assembly on nascent DNA.
(A) Schematic representation of the CAA. Incorporated EdU and conjugated biotin are shown as red and purple circles on nascent DNA, respectively. The assembled proteins are shown as squares. Amplified PLA signal is shown in red.
(B) PLA signals between EdU and H4K5Ac (red). The same embryo is immunostained for EdU (green) after the PLA reaction to identify replicating nuclei. The magnified merged image of the part of this embryo is on the right.
(C) PLA signals in red resulting from CAA between EdU and the histone proteins indicated on the top. Gastrulating embryos were labeled with EdU for 5 min. In C–E, following the PLA reaction embryos were immunostained for EdU (green).
(D) Pulse-chase CAA for H4K5Ac and unmodified H3 tail. Gastrulating embryos were pulse labeled with EdU for 10 min, washed extensively to remove EdU and kept for 0, 30 and 60 min prior to fixation. PLA signals in red between EdU and the proteins indicated to the left.
(E) Pulse-chase CAA for methylated H3 forms (shown on the left). Experiments were performed as in D.
(F) 2 hr pulse-chase CAA for methylated H3 forms. Following PLA reaction embryos were immunostained for PCNA (green) to distinguish replicating nuclei.
Using CAA, we addressed the timing of protein assembly on nascent DNA in gastrulating Drosophila embryos. During gastrulation, the S phase is about 50 min and is followed by the G2 phases of variable lengths in different nuclei during cell cycles 14–16 (Shermoen et al., 2010) (Figure 1D). Association of H4K5Ac, and unmodified H3 tail with nascent DNA were detected after 5 min of EdU incorporation. Interestingly, the number of H4K5Ac CAA signals is significantly decreased after 30 min of chase (Figure 6D), consistent with the previous data suggesting that the amount of this histone form is decreased 20–60 min after DNA replication (Jackson et al., 1976). In contrast, H3K4me3 and H3K27me3 were not detected after 5 min of EdU labeling (Figure 6C), consistent with the re-ChIP assays. These results suggest that methylated H3 is not present in the fully assembled nucleosomes on nascent DNA that is several kb from the RF.
To understand when H3K4me3 and K3K27me3 modifications appear on DNA following DNA replication, we performed pulse-chase experiments after 10 min of EdU incorporation. H3K4me3 and H3K27me3 were not detected in CAA assays at 0 and 30 min of chase (Figure 6E). However, they become detectable after 1 hr of chase. The same results were obtained in later, post cell cycle 16 embryos (not shown). Interestingly, the number of signals of all methylated H3 forms significantly increases after 2 hr of chase (Figure 6E), and these signals were detected only in cells that are not undergoing DNA replication as evidenced by counter-staining for PCNA (Figure 6F). We conclude that accumulation of methylated H3 forms occurs after S phase.
TrxG and PcG proteins are associated with nascent DNA in vivo following the passage of the RF
We also examined association of Trx, Pc and E(z) with nascent DNA labeled with EdU. As seen in Figure 7A, Trx, Pc and E(z) are detected on nascent DNA of cell cycle 14–15 nuclei after EdU incorporation for 5 min. These results provide independent in vivo confirmation of the results of the PLA and PCNA re-ChIP assays, using a method that depends on their association with nascent DNA, similar to the brdU re-ChIP assay. CAA signals for association of Trx, Pc and E(z) with nascent DNA remained the same for all chase times from 0–60 min (Figure 7B), suggesting that there is no sensitivity problem at early labeling time points. Thus, Trx, Pc and E(z) are detected in vivo by CAA long after the passage of the RF. Taken together with the results of the re-ChIP assays and PLA assays, this suggests that Trx, Pc and E(z) are retained at their response elements during the passage of the DNA replication complex and remain stably associated with nascent DNA throughout the S phase. The existence of replication factories may be why we observe relatively few signals per nucleus in the CAA assay. This observation has been made by others using standard immunohistochemical approaches or PLA to detect replication proteins (Hervouet et al., 2010).
Figure 7. Trx, Pc and E(z) are associated with nascent DNA throughout S phase.
(A) PLA signals in red resulting from CAA between EdU and Trx, Pc and E(z). Gastrulating embryos were labeled with EdU for 5 min. Following the PLA reaction embryos were immunostained for EdU (green). Only red channel is shown on the top.
(B) Pulse-chase CAA for Trx, Pc and E(z). Gastrulating embryos were pulse labeled with EdU for 10 min, washed extensively to remove EdU and kept for 0, 30 and 60 min prior to fixation. PLA signals in red between EdU and the proteins indicated to the left. Following the PLA reaction, embryos were immunostained for EdU (green).
(E) A model for the re-constitution of chromatin structure during DNA replication. MP, maintenance proteins (TrxG and PcG); RF, replication fork. Note that full nucleosomes may not be assembled immediately after passage of DNA polymerase.
Discussion
The key logical problem that has prevented identification of epigenetic marks is the difficulty of distinguishing effects on transcriptional regulation from heritable effects. Any change to transcriptional regulation will affect inheritance, and vice versa. Reasoning that any protein or PTM that is not stable to replication is unlikely to be the epigenetic mark, we have focused on identification of proteins or PTM that are stable to DNA replication. To do so, we asked what proteins or PTM are closely associated with the RF based on association with proteins found near the replication fork, using PLA to assess in vivo protein-protein interactions and re-ChIP to examine protein associations on specific nascent DNA sequences. To examine events at longer distances from the RF, we have investigated protein and PTM association with nascent DNA labeled with EdU or BrdU using CAA or reverse re-ChIP to ensure that we are looking at protein-DNA interactions rather than transient interactions with the replication machinery. Together, these approaches give consistent, repeatable results, allowing us to assay events at different distances from the RF.
The results of PLA assays show that unmodified H3 and H4K5Ac are in close proximity to PCNA and CAF-1, but H3K4me3 and H3K27me3 are not (Figure 2), agreeing with our results from re-ChIP with PCNA. Electron micrographs of replication bubbles from cleavage stage Drosophila embryos show less than 200 bp of nucleosome-free DNA adjacent to the RF on one strand, and on the other strand show partial nucleosome assembly very close to the RF (Kriegstein and Hogness, 1974). These micrographs, together with our observation that 70% of DNA fragments after sonication are less than 200 bp suggest that the majority of the histones we detect in re-ChIP assays with PCNA are associated with DNA. However, we cannot rule out the possibility that we are examining protein-protein and protein-DNA interactions in the PLA assays, as we may detect small amounts of histones that are associated with CAF-1 prior to deposition rather than histones bound to nascent DNA.
It is possible that parental modified H3 forms are recruited to nascent DNA with some delay. This would not be detected in our re-ChIP experiments because the fragment sizes are short. Therefore, we developed the CAA assay to detect direct association of modified histones on much larger fragments of nascent DNA in vivo. Surprisingly, we do not detect H3K4me3 and H4K27me3 PTM for at least 30 m after passage of the RF, and detect the first clear signal at 1 hr (Figure 6). This agrees with our observation that H3K4me3 and H4K27me3 are present at low amounts at cellular blastoderm (Figure 1A,C) or are undetectable in S phase during gastrulation (Figure 6E, F). Taken together, the results of CAA, PLA and re-ChIP suggest that methylated H3 forms are not associated with nascent DNA from the time of the passage of the replication fork to the end of the S phase. This is in contrast to unmodified H3, which is easily detected by re-ChIP and CAA on nascent DNA of any size (Figure 6E, D).
Our data suggest that in embryos, CAF-1 may deposit acetylated histones and unmodified H3, but does not transfer parental H3K4me3 and H3K4me27 to nascent DNA soon after replication as proposed (Corpet and Almouzni, 2009). In yeast, experiments in which parental histones can be distinguished from de novo synthesized histones show that parental histones are deposited within 400 bp of their original locations (Radman-Livaja et al., 2011). These experiments did not consider PTM, or timing of deposition, but make the important point that if parental histones retaining PTM are deposited at a different location on nascent DNA, then these would be “epi-mutations” because the same mark would be at a different location. Considering the yeast and our data, we propose that parental histones are dissociated from parental DNA, lose their trimethyl PTM, and are then transferred to nascent DNA along with the newly synthesized histones. This suggestion is consistent with earlier results showing that methylation of lysines 9 and 27 of histone H3 is lost during DNA synthesis (Gullerova and Proudfoot, 2008; Hernandez-Munoz et al., 2005), and that most histone methylation occurs after deposition (Loyola et al., 2006).
Together our results suggest that in embryos, trimethylated parental histones are not transferred to nascent DNA, that trimethylation of H3K4 and H3K27 occur after deposition, and support the previous suggestion that trimethylation of bound, unmodified H3 is regulated (Scharf et al., 2009). These data suggest that H3K4me3 and H3K27me3 are unlikely to be epigenetic marks in Drosophila embryos.
In contrast to methylated histones, we detect Trx, Pc and E(z) associated with PCNA in PLA and re-ChIP assays, and stably bound to labeled nascent DNA in CAA assays and re-ChIP assays with brdU. Our results show that Trx, Pc and E(z) are stable to DNA replication, are constitutively associated with nascent DNA through the S phase, consistent with previous observations that Psc and Pc are stable to replication in an SV40 in vitro replication system (Francis et al., 2009). Stability to DNA replication and the ability to restore the structure of chromatin required for transcriptional regulation are prerequisites for any putative epigenetic mark. Thus Trx and E(z) fulfill these criteria for epigenetic marks, although functional analysis will be needed to confirm this suggestion. ME have been proposed to be “cellular memory modules” because they are sufficient to retain gene expression pattern (Cavalli and Paro, 1998). The observation that Trx, E(z) and Pc are retained at the ME after DNA replication supports the model that ME are cellular memory modules, and is consistent with the possibility that these proteins could be epigenetic marks. However, our observations do not rule out the possibility that other proteins not tested in these experiments are epigenetic marks.
Our data imply that Trx, Pc and E(z) remain bound or rapidly rebind to nascent DNA in the absence of trimethylated histones. There are many reports of trimethylated histone-independent binding of PcG proteins and of MLL1 (Francis et al., 2009; Puschendorf et al., 2008; Tavares et al., 2012) and see (Henikoff and Shilatifard, 2011) for a recent review. Our data do not preclude a role for modified histones in binding of PcG and TrxG proteins during transcriptional regulation, but suggest that their retention on nascent DNA does not require trimethylation in Drosophila embryos. A recent report shows that in mammalian cell lines, EZH2 associates with PCNA and DNA labeled with BrdU for 5 min, that H3K27me3 is needed to propagate transcriptional repression at a reporter locus, and that H3K27me3 is required for recruitment of PRC2 in interphase in mammalian cells (Hansen et al., 2008). However this paper did not directly assay the role of H3K27me3 for recruitment of EZH2 in S phase. It is possible that chromatin assembly in Drosophila embryos differs from that in mammalian cells.
The mechanism of retention of Trx and Pc during DNA replication requires association with the ssDNA following unwinding by DNA helicase, and transfer from the ssDNA to the nascent dsDNA following passage of the DNA polymerase. The preSET domains of Trx and E(z) bind very tightly to ssDNA (Krajewski et al., 2005), thus providing a plausible explanation for retention of these proteins on the ssDNA found immediately downstream of helicase (Figure 7C). It is not known how TrxG and PcG proteins are transferred to nascent DNA. Stable association of the core components of the PRC1 PcG complex, including Pc, with replicating SV40 DNA in vitro may occur by mass-action (Francis et al., 2009). Alternatively, Trx, Pc and E(z) may be retained on replicating DNA by transient interaction with components of the DNA replication complex.
Our results suggest that the appropriate amounts of Trx and Pc are replenished late in the S phase or in the interphase (Figure 5C), but the mechanism remains unknown. One possibility is that this may occurs through interactions between these proteins themselves, for example through dimerization of the SET domain of Trx that we have demonstrated previously (Rozovskaia et al., 2000). Interestingly, Trx binding can withstand assembly of nucleosomes and interferes with the formation of regular nucleosomal arrays (Krajewski et al., 2005). Thus, Trx may specify a particular nucleosome structure in the MEs. This is a likely possibility given recent discovery of a specific nucleosome which associates with Gal4 at its binding sites. Importantly, this complex works as a barrier that is essential for establishing specific chromatin architecture of the region surrounding Gal4 binding sites (Floer et al., 2010).
PRC2 activity is cell cycle regulated (Hansen et al., 2008; Kaneko et al., 2010). Several authors have examined retention of PcG and TrxG proteins in mitosis with varying results (Buchenau et al., 1998; Dietzel et al., 1999; Fanti et al., 2008; Fonseca et al., 2012). Our results suggest that in future it will be interesting to monitor DNA binding and enzymatic activity of PcG and TrxG protein to determine how epigenetic marks are propagated in G2 and M phases of the cell cycle.
We propose a model for the re-constitution of chromatin structure during DNA replication in Drosophila embryos (Figure 7C). We suggest that Trx, Pc and E(z), and likely other TrxG and PcG proteins are stably associated with their response elements during the progression of the RF, potentially through direct interactions with components of the DNA polymerase complex. Importantly, the stability of the TrxG and PcG proteins during replication ensures sequence specificity in association of these proteins with their response elements after replication. During replication, methylated histones are rapidly replaced by unmethylated histones. The continuous presence of histone-modifying TrxG and PcG proteins may result in histone modification, leading to restoration of the specific chromatin structure that allows either activation or repression of the target gene in the corresponding cells.
Experimental Procedures
Chromatin immunoprecipitations (ChIP) and sequential ChIP (re-ChIP)
Approximately 1 ml of 2–12 hr staged wild-type strain Oregon R embryos were cross-linked in fixing solution containing 2% formaldehyde. Nuclei were isolated and sonicated to shear DNA to an average size of approximately 500 bp. Fragmented chromatin extract was diluted to the final volume of 1 ml, and was divided for ChIP and re-ChIP experiments: 5–10 μl of material were used for ChIP with each antibody, and all remaining material was used for re-ChIP experiments. In re-ChIP experiments, precipitated complexes were eluted by incubation for 30 min at 37°C in TE buffer containing 10 mM DTT and 0.05%SDS. 5% of material was removed, crosslinks were reversed, and DNA purified to serve as a second input or as material for S1 nuclease treatment (see Extended Experimental Procedures). The remaining material was subjected to the second ChIP procedure with different antibodies. Immunoprecipitated chromatin was assayed by real time PCR. Antibodies, a list of primers used and other details of these procedures are described in the Extended Experimental Procedures.
EdU labeling of Drosophila embryos
For EdU (5-ethynyl-2′-deoxyuridine) incorporation staged embryos were dechorionated and permeabilized by incubation in octane. Embryos were transferred to Schneider’s Drosophila medium with 250 μM EdU (Invitrogen) for 5–10 min, washed and fixed in 10% formaldehyde using standard procedures. EdU-labeled embryos were subjected to a “click-iT reaction” using Click-iT Cell Reaction Buffer Kit (Invitrogen) to attach biotin to EdU. Embryos were then incubated with mouse anti-biotin antibody for PLA assays and/or immunostaining.
brdU labeling of Drosophila embryos
1ml of 2–5 h AEL embryos were dechorionated and homogenized gently with a Teflon pestle in 10 ml of Schneider’s Drosophila medium. Cells were incubated with 50 μM BrdU (Sigma) for 20 min, washed and fixed in 1% formaldehyde. Cells were sonicated to shear DNA to an average size of approximately 500 bp and used for first immunoprecipitation with antibodies to TrxG or PcG proteins. Following immunoprecipitation, DNA was purified and used for sequential immunoprecipitation with brdU antibody followed by PCR (for details, see Extended Experimental Procedures).
Immunostaining of Drosophila embryos
Whole-mount embryos were double-immunostained using standard protocols. The primary antibodies and their dilutions are described in the Extended Experimental Procedures.
Proximity Ligation Reaction (PLA)
Fixed embryos were incubated with primary antibodies as for immunostaining of embryos. The PLA assay was performed according to manufacturer’s instruction (Olink Bioscience): after incubation with primary mouse and rabbit antibodies, secondary antibodies with conjugated oligonucleotides (PLA probes MINUS and PLUS) were incubated for 1 h. This was followed by addition of ligase and two oligonucleotides which hybridize to the two PLA probes and join them into a closed circle if they are in close proximity. Subsequently PCR amplification solution containing fluorescent labeled oligonucleotides which hybridize to the rolling circle amplification product was added. After PLA reactions, PCNA or EdU were detected in embryos by incubation with FITC-conjugated anti-mouse antibody.
Chromatin Assembly Assay
Edu-labeled embryos were subjected to Click-iT to add biotin to the EdU as described above, washed and incubated with mouse monoclonal anti-biotin antibody and the rabbit antibody of interest. This was followed by the PLA reaction as described above. After PLA reactions, EdU was detected in embryos by incubation with FITC-conjugated anti-mouse antibody.
Dot Blot and Western blot
Details of antibodies and procedures are given in the Extended Experimental Procedures.
Supplementary Material
Research Highlights.
Parental H3K4me3 and H3K27me3 are not transferred to original sites on nascent DNA
De novo methylation of H3 occurs only in the next G phase
Trx, Pc and E(z) are transiently associated with PCNA
Trx, Pc and E(z) are transferred to their response elements during DNA replication
Acknowledgments
We thank Valerio Orlando, Nicole Francis, Richard Jones and Yi Zhang for antibodies. This work was supported by the following grants: NIH R01GM075141, NIH P01CA129242 to A.M, a grant from the Canadian Institutes of Health Research to H.W.B.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Beisel C, Buness A, Roustan-Espinosa IM, Koch B, Schmitt S, Haas SA, Hild M, Katsuyama T, Paro R. Comparing active and repressed expression states of genes controlled by the Polycomb/Trithorax group proteins. Proc Natl Acad Sci U S A. 2007;104:16615–16620. doi: 10.1073/pnas.0701538104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DD. The role of stable complexes that repress and activate eukaryotic genes. Philos Trans R Soc Lond B Biol Sci. 1984;307:297–299. doi: 10.1098/rstb.1984.0130. [DOI] [PubMed] [Google Scholar]
- Buchenau P, Hodgson J, Strutt H, Arndt-Jovin DJ. The distribution of polycomb-group proteins during cell division and development in Drosophila embryos: impact on models for silencing. J Cell Biol. 1998;141:469–481. doi: 10.1083/jcb.141.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Cavalli G, Paro R. The Drosophila Fab-7 chromosomal element conveys epigenetic inheritance during mitosis and meiosis. Cell. 1998;93:505–518. doi: 10.1016/s0092-8674(00)81181-2. [DOI] [PubMed] [Google Scholar]
- Chang PY, Hom RA, Musselman CA, Zhu L, Kuo A, Gozani O, Kutateladze TG, Cleary ML. Binding of the MLL PHD3 finger to histone H3K4me3 is required for MLL-dependent gene transcription. J Mol Biol. 2010;400:137–144. doi: 10.1016/j.jmb.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet A, Almouzni G. Making copies of chromatin: the challenge of nucleosomal organization and epigenetic information. Trends Cell Biol. 2009;19:29–41. doi: 10.1016/j.tcb.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- Deal RB, Henikoff JG, Henikoff S. Genome-wide kinetics of nucleosome turnover determined by metabolic labeling of histones. Science. 2010;328:1161–1164. doi: 10.1126/science.1186777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellino GI, Tatout C, Pirrotta V. Extensive conservation of sequences and chromatin structures in the bxd polycomb response element among Drosophilid species. Int J Dev Biol. 2002;46:133–141. [PubMed] [Google Scholar]
- Dietzel S, Niemann H, Bruckner B, Maurange C, Paro R. The nuclear distribution of Polycomb during Drosophila melanogaster development shown with a GFP fusion protein. Chromosoma. 1999;108:83–94. doi: 10.1007/s004120050355. [DOI] [PubMed] [Google Scholar]
- Fanti L, Perrini B, Piacentini L, Berloco M, Marchetti E, Palumbo G, Pimpinelli S. The trithorax group and Pc group proteins are differentially involved in heterochromatin formation in Drosophila. Chromosoma. 2008;117:25–39. doi: 10.1007/s00412-007-0123-7. [DOI] [PubMed] [Google Scholar]
- Floer M, Wang X, Prabhu V, Berrozpe G, Narayan S, Spagna D, Alvarez D, Kendall J, Krasnitz A, Stepansky A, et al. A RSC/nucleosome complex determines chromatin architecture and facilitates activator binding. Cell. 2010;141:407–418. doi: 10.1016/j.cell.2010.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca JP, Steffen PA, Muller S, Lu J, Sawicka A, Seiser C, Ringrose L. In vivo Polycomb kinetics and mitotic chromatin binding distinguish stem cells from differentiated cells. Genes Dev. 2012;26:857–871. doi: 10.1101/gad.184648.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis NJ, Follmer NE, Simon MD, Aghia G, Butler JD. Polycomb proteins remain bound to chromatin and DNA during DNA replication in vitro. Cell. 2009;137:110–122. doi: 10.1016/j.cell.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gullerova M, Proudfoot NJ. Cohesin complex promotes transcriptional termination between convergent genes in S. pombe. Cell. 2008;132:983–995. doi: 10.1016/j.cell.2008.02.040. [DOI] [PubMed] [Google Scholar]
- Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A, Rappsilber J, Lerdrup M, Helin K. A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol. 2008;10:1291–1300. doi: 10.1038/ncb1787. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Shilatifard A. Histone modification: cause or cog? Trends Genet. 2011;27:389–396. doi: 10.1016/j.tig.2011.06.006. [DOI] [PubMed] [Google Scholar]
- Hernandez-Munoz I, Taghavi P, Kuijl C, Neefjes J, van Lohuizen M. Association of BMI1 with polycomb bodies is dynamic and requires PRC2/EZH2 and the maintenance DNA methyltransferase DNMT1. Mol Cell Biol. 2005;25:11047–11058. doi: 10.1128/MCB.25.24.11047-11058.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervouet E, Lalier L, Debien E, Cheray M, Geairon A, Rogniaux H, Loussouarn D, Martin SA, Vallette FM, Cartron PF. Disruption of Dnmt1/PCNA/UHRF1 interactions promotes tumorigenesis from human and mice glial cells. PLoS One. 2010;5:e11333. doi: 10.1371/journal.pone.0011333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen MS, Sy SM, van Deursen JM, Chen J. Direct interaction between SET8 and proliferating cell nuclear antigen couples H4-K20 methylation with DNA replication. J Biol Chem. 2008;283:11073–11077. doi: 10.1074/jbc.C700242200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson V, Chalkley R. Histone segregation on replicating chromatin. Biochemistry. 1985;24:6930–6938. doi: 10.1021/bi00345a027. [DOI] [PubMed] [Google Scholar]
- Jackson V, Shires A, Tanphaichitr N, Chalkley R. Modifications to histones immediately after synthesis. J Mol Biol. 1976;104:471–483. doi: 10.1016/0022-2836(76)90282-5. [DOI] [PubMed] [Google Scholar]
- Kaneko S, Li G, Son J, Xu CF, Margueron R, Neubert TA, Reinberg D. Phosphorylation of the PRC2 component Ezh2 is cell cycle-regulated and up-regulates its binding to ncRNA. Genes Dev. 2010;24:2615–2620. doi: 10.1101/gad.1983810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassis JA. Unusual properties of regulatory DNA from the Drosophila engrailed gene: three “pairing-sensitive” sites within a 1.6-kb region. Genetics. 1994;136:1025–1038. doi: 10.1093/genetics/136.3.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krajewski WA, Nakamura T, Mazo A, Canaani E. A motif within SET-domain proteins binds single-stranded nucleic acids and transcribed and supercoiled DNAs and can interfere with assembly of nucleosomes. Mol Cell Biol. 2005;25:1891–1899. doi: 10.1128/MCB.25.5.1891-1899.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegstein HJ, Hogness DS. Mechanism of DNA replication in Drosophila chromosomes: structure of replication forks and evidence for bidirectionality. Proc Natl Acad Sci U S A. 1974;71:135–139. doi: 10.1073/pnas.71.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzuolo C, Lo Sardo F, Diamantini A, Orlando V. PcG complexes set the stage for epigenetic inheritance of gene silencing in early S phase before replication. PLoS Genet. 2011;7:e1002370. doi: 10.1371/journal.pgen.1002370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loyola A, Bonaldi T, Roche D, Imhof A, Almouzni G. PTMs on H3 variants before chromatin assembly potentiate their final epigenetic state. Mol Cell. 2006;24:309–316. doi: 10.1016/j.molcel.2006.08.019. [DOI] [PubMed] [Google Scholar]
- Mishra RK, Mihaly J, Barges S, Spierer A, Karch F, Hagstrom K, Schweinsberg SE, Schedl P. The iab-7 polycomb response element maps to a nucleosome-free region of chromatin and requires both GAGA and pleiohomeotic for silencing activity. Mol Cell Biol. 2001;21:1311–1318. doi: 10.1128/MCB.21.4.1311-1318.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O’Connor MB, Kingston RE, Simon JA. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- Muller J, Kassis JA. Polycomb response elements and targeting of Polycomb group proteins in Drosophila. Curr Opin Genet Dev. 2006;16:476–484. doi: 10.1016/j.gde.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Ng RK, Gurdon JB. Epigenetic inheritance of cell differentiation status. Cell Cycle. 2008;7:1173–1177. doi: 10.4161/cc.7.9.5791. [DOI] [PubMed] [Google Scholar]
- Petruk S, Sedkov Y, Riley KM, Hodgson J, Schweisguth F, Hirose S, Jaynes JB, Brock HW, Mazo A. Transcription of bxd noncoding RNAs promoted by trithorax represses Ubx in cis by transcriptional interference. Cell. 2006;127:1209–1221. doi: 10.1016/j.cell.2006.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puschendorf M, Terranova R, Boutsma E, Mao X, Isono K, Brykczynska U, Kolb C, Otte AP, Koseki H, Orkin SH, et al. PRC1 and Suv39h specify parental asymmetry at constitutive heterochromatin in early mouse embryos. Nat Genet. 2008;40:411–420. doi: 10.1038/ng.99. [DOI] [PubMed] [Google Scholar]
- Radman-Livaja M, Verzijlbergen KF, Weiner A, van Welsem T, Friedman N, Rando OJ, van Leeuwen F. Patterns and mechanisms of ancestral histone protein inheritance in budding yeast. PLoS Biol. 2011;9:e1001075. doi: 10.1371/journal.pbio.1001075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringrose L, Paro R. Polycomb/Trithorax response elements and epigenetic memory of cell identity. Development. 2007;134:223–232. doi: 10.1242/dev.02723. [DOI] [PubMed] [Google Scholar]
- Rozovskaia T, Rozenblatt-Rosen O, Sedkov Y, Burakov D, Yano T, Nakamura T, Petruck S, Ben-Simchon L, Croce CM, Mazo A, et al. Self-association of the SET domains of human ALL-1 and of Drosophila TRITHORAX and ASH1 proteins. Oncogene. 2000;19:351–357. doi: 10.1038/sj.onc.1203307. [DOI] [PubMed] [Google Scholar]
- Sarraf SA, Stancheva I. Methyl-CpG binding protein MBD1 couples histone H3 methylation at lysine 9 by SETDB1 to DNA replication and chromatin assembly. Mol Cell. 2004;15:595–605. doi: 10.1016/j.molcel.2004.06.043. [DOI] [PubMed] [Google Scholar]
- Scharf AN, Barth TK, Imhof A. Establishment of histone modifications after chromatin assembly. Nucleic Acids Res. 2009;37:5032–5040. doi: 10.1093/nar/gkp518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz YB, Kahn TG, Pirrotta V. Characteristic low density and shear sensitivity of cross-linked chromatin containing polycomb complexes. Mol Cell Biol. 2005;25:432–439. doi: 10.1128/MCB.25.1.432-439.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- Shermoen AW, McCleland ML, O’Farrell PH. Developmental control of late replication and S phase length. Curr Biol. 2010;20:2067–2077. doi: 10.1016/j.cub.2010.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ST, Petruk S, Sedkov Y, Cho E, Tillib S, Canaani E, Mazo A. Modulation of heat shock gene expression by the TAC1 chromatin-modifying complex. Nat Cell Biol. 2004;6:162–167. doi: 10.1038/ncb1088. [DOI] [PubMed] [Google Scholar]
- Tavares L, Dimitrova E, Oxley D, Webster J, Poot R, Demmers J, Bezstarosti K, Taylor S, Ura H, Koide H, et al. RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell. 2012;148:664–678. doi: 10.1016/j.cell.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillib S, Petruk S, Sedkov Y, Kuzin A, Fujioka M, Goto T, Mazo A. Trithorax- and Polycomb-group response elements within an Ultrabithorax transcription maintenance unit consist of closely situated but separable sequences. Mol Cell Biol. 1999;19:5189–5202. doi: 10.1128/mcb.19.7.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.