Abstract
Enucleated oocytes have the remarkable ability to reprogram somatic nuclei back to totipotency. Here we investigate genome-scale DNA methylation patterns after nuclear transfer and compare them to the dynamics at fertilization. We identify specific targets for DNA demethylation after nuclear transfer such as germ-line associated promoters, as well as unique limitations that include certain repetitive element classes.
Mammalian DNA methylation generally shows limited global dynamics except during pre-implantation and primordial germ cell development. The observed global demethylation of the paternal genome upon fertilization is mediated by the oocyte and is critical for establishment of totipotency and developmental competence1. An enucleated oocyte can reprogram the epigenetic and transcriptional identity of somatic cells through a procedure known as somatic cell nuclear transfer (SCNT), though successful reprogramming occurs at low efficiency and is likely affected in part by the retention of somatically-conferred epigenetic lesions2,3. To date, the oocyte’s intrinsic capacity to erase somatic DNA methylation patterns to a true zygotic signature remains incompletely characterized. Here, we generated genome-scale single basepair resolution maps of DNA methylation from donor fibroblasts and SCNT reconstructed mouse embryos using reduced representation bisulfite sequencing (RRBS) 4 (Supplementary Fig. 1a). We conducted two independent rounds of nuclear transfer into enucleated BDF1 (C57/DBA F1) oocytes, each consisting of two biological replicates. We used both BDF1 x Cast hybrid and 129X1 inbred tail tip fibroblasts (referred to as Cast and X1, respectively) to serve as controls for low input RRBS by capturing over 10,000 hybrid SNPs and to ensure that dynamics observed were consistent across strain identity (Supplementary Fig. 1b–e). Using our stringently collected samples and the first genome-scale measurement in any nuclear transfer experiment, we detected a low level (~15%) of the host oocyte genome (see Supplementary Methods). This affected ~35% of loci in the X1 samples and likely ~13% of loci in the embryos using the hybrid donors. Given the complexity inherent to the protocol and the number of cells required, the presence of residual host DNA may be unavoidable; however, the low frequency with which it is sampled is unlikely to consistently capture the same locus across experiments. Accordingly, we present the SCNT embryo data without compensation for residual host DNA methylation.
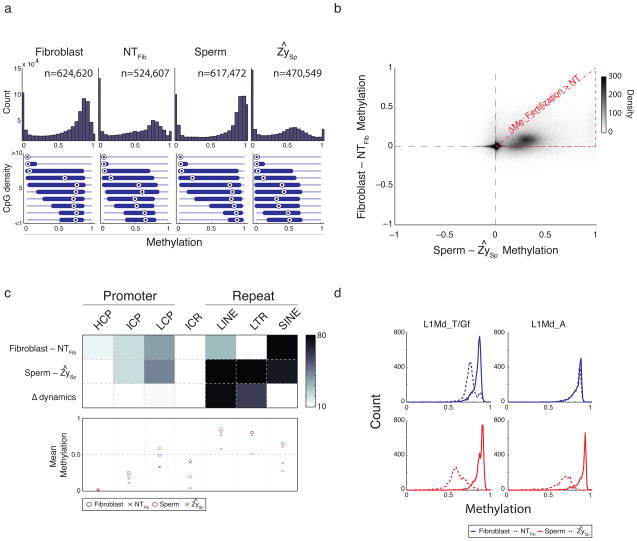
We compared methylation profiles between the donor cells and reconstructed embryos to the methylation dynamics observed during fertilization 5. DNA methylation patterns of donor fibroblasts and sperm exhibit a conventional somatic bimodality that depends upon relative CpG density. After nuclear transfer, a shift in the fibroblast methylation landscape resembles the demethylation that occurs within the paternal genome upon fertilization (Fig. 1a). Though many regions are affected in both processes, demethylation occurs at a smaller magnitude after SCNT (Fig. 1b). Globally, reconstructed embryos more closely resemble donor fibroblasts than the paternal genome after fertilization or the pre-implantation embryo (Supplementary Fig. 1f). Importantly, SCNT embryos are more similar to each other regardless of experimental round or donor strain than they are to either fibroblasts or the early embryo, suggesting the majority of methylation changes conferred by SCNT are consistent across experiments (Supplementary Fig. 1g). The difference in reprogramming response appears to be partly due to genomic context. For instance, we find that methylation levels of different repetitive element classes change by different magnitudes (Fig. 1c, top). While SINE elements appear similarly demethylated in both processes, methylation levels at LINEs and LTRs only slightly decrease or do not change after nuclear transfer (Fig. 1c, bottom, Supplementary Fig. 2a). Upon closer inspection of LINE families, we found that methylation at L1Md_A elements remains almost completely static, whereas the evolutionarily younger L1Md_T/Gf families appear slightly more dynamic6,7 (Fig.1d). The predominance of LINE and LTR element classes in the genome and their recalcitrance to demethylation could explain the striking retention of somatic methylation patterns after SCNT. Taken together, we conclude that repetitive elements, which account for a large proportion of demethylation events during fertilization, appear more resistant to change when the ooplasm is confronted with a somatic nucleus. By the nature of the experiment we cannot rule out the possibility that some of the global dynamics observed in SCNT embryos may be due to the presence of residual host oocyte DNA. However, we observed similar demethylation at CpGs associated with Cast alleles compared to C57 in our hybrid fibroblast experiments, which can only result from reprogramming of the donor fibroblast genome (Supplementary Fig. 2b). It is technically not possible to extend this SNP analysis to repetitive elements, but the reasonable association between Cast exclusive demethylation dynamics and the C57 haplotype argue that demethylation is in fact an observable and measurable event during nuclear transfer in our data.
Figure 1. Classifying common and distinct DNA methylation dynamics during fertilization and nuclear transfer.
a. Histogram of methylation (top) and boxplots of methylation by CpG density (bottom) for 100 bp tiles in fibroblast, nuclear transfer reconstructed embryos (NTFib, mean of 4 replicates), sperm, and the inferred sperm value in zygote (ZySp; see Supplementary Methods). Fibroblasts and sperm show a global methylation pattern typical of somatic tissues while SCNT embryos (NTFib) exhibit a similar global shift as ZySp. Bulls-eye indicates the median, edges the 25th/75th percentile and whiskers the 2.5th/97.5th percentile.
b. Scatterplot comparing global methylation dynamics between nuclear transfer (Fibroblast – NTFib) and fertilization (Sperm – ZySp). While demethylated regions appear to occur in common sites (upper right quadrant), the magnitude of demethylation is larger during fertilization as indicated by the dense cloud below the diagonal. The red triangle outlines the region where demethylation after SCNT is smaller than during fertilization.
c. Heatmap (top) depicting genomic features that significantly change (dark) in Fibroblast-NTFib or Sperm-ZySp transitions, and the comparison of changes between the two transitions (Δ dynamics). Promoters are partitioned to high (H), intermediate (I) or low (L) CpG density and known imprint control regions (ICRs) are included as a control set. Color is the –log p-value. The mean methylation value for each feature set is shown in the bottom panel. Most features appear to change similarly in both processes with the exception of LINE and LTR features, which are comparably resistant to change after nuclear transfer.
d. Histogram of methylation for elements in the L1Md_T and L1Md_Gf (left) and L1Md_A (right) families of the LINE-1 class. Both families are dynamic during fertilization, but only the L1Md_T/Gf families, which show larger demethylation during fertilization, show any detectable change after nuclear transfer.
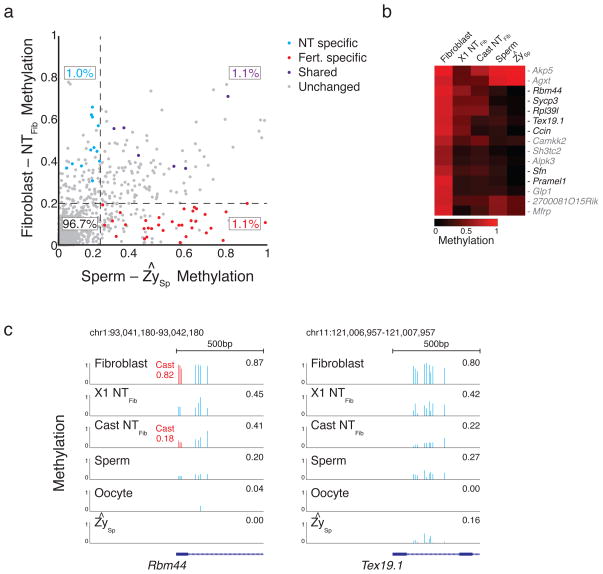
We next compared methylation dynamics in promoters during both processes. Given the likely stochastic models for demethylation during SCNT and the potential effect of residual host DNA, we applied stringent criteria for identifying changing promoters such that hypermethylated promoters (>0.5) were required to (1) change by > 0.2 in at least three replicates, (2) change by >0.2 in donor-normalized X1 replicates, and (3) could not have contradictory dynamics in Cast-tracked CpGs (see Supplementary Methods). This strategy identifies the most consistently changing promoters during the nuclear transfer procedure but excludes promoters that change either less efficiently or are targeted less frequently for DNA demethylation. We then classified promoter dynamics as being either SCNT specific, shared with fertilization, or unique to fertilization (Fig. 2a, Supplementary Table 1). We identified a stringent set of 15 SCNT specific promoters, which include several genes that function during meiosis and are typically hypomethylated in both gametes and the early embryo (Fig. 2b). The specific demethylation observed in the SCNT embryos suggests the presence of defined, targeting factors within the ooplasm that ensure the unmethylated status of these sites (Fig. 2c) 8. We observed that approximately two thirds of fertilization specific demethylated targets retained considerable DNA methylation after nuclear transfer, indicating that only regions in certain contexts are equivalently demethylated in both processes (Supplementary Fig. 2c,d). These likely represent somatically retained epigenetic information that could affect embryonic development. We also investigated 102 previously identified differentially methylated promoters that are hypermethylated in the oocyte and transiently remain methylated on the maternal allele during pre-implantation (Supplementary Fig. 2e) 5. The set is unmethylated in fibroblasts and remains so after nuclear transfer, representing another example of the inequivalence between SCNT reconstructed embryos and true embryos in which maternally encoded regulatory information is not conferred in SCNT.
Figure 2. Promoter dynamics during SCNT include demethylation of gamete-specific genes.
a. Scatterplot of promoter dynamics between donor fibroblasts and SCNT embryos compared to those observed at fertilization. The majority of promoters are unchanged in either process, with 1.0%, 1.1% and 1.1% of promoters dynamic in either NT specific, shared, or fertilization specific contexts. Colored dots refer to promoters that were observed to change consistently across SCNT experiments and/or during demethylation of the paternal genome after fertilization. “Other” includes promoters that either did not change or did not pass the stringent criteria for being called as dynamic.
b. Dynamics specific to the Fibroblast-NT transition include several promoters that function specifically in gametes and are already hypomethylated in sperm and oocyte.
c. The germ-line associated genes RNA binding motif protein 44 (Rbm44) and Testes expressed gene 19.1 (Tex19.1) are hypomethylated and expressed during gametogenesis, the early embryo, and pluripotent cell lines but de novo methylated upon gastrulation/differentiation 4. In fibroblasts, both gene promoters are hypermethylated and show a strong demethylation after either NT round. The level of demethylation suggests erasure in a large proportion of transplanted cells. Blue bars highlight single CpGs that are captured in all stages, red bars highlight 3 CpGs that can be associated with the Cast allele, with the mean allele-specific methylation value highlighted in red.
The demethylation events observed after nuclear transfer are similar to those at fertilization, though often at lower magnitudes, which may indicate less efficient or stochastic targeting. In turn, this may affect the number of SCNT embryos that successfully reactivate sufficient developmental loci upon zygotic activation9. Certain repeats, such as LTRs and L1Md_As, appear completely refractory to demethylation in nuclear transfer and may be protected within the epigenetic context of somatic cells. Demethylation of the paternal genome at fertilization is accompanied by global rechromatinization1, which may provide a unique window of opportunity for parasitic genomic elements to either initiate demethylation or escape methylation machinery. It has been observed, for instance, that cytoplasmic injection of chromatinized round spermatids generates live embryos at equivalent rates to injection of terminally differentiated, protamine-compacted spermatozoa but does not co-occur with an apparently equivalent global erasure of DNA methylation10. Our genome-scale profiling strategy confirms previous locus specific bisulfite sequencing and global immunostaining data which have shown that DNA demethylation after nuclear transfer is not as dramatic as that observed in the paternal genome7. Presumably, the interplay between histone exchange, transcription, and active demethylation may limit the magnitude and targets for DNA demethylation observed after nuclear transfer or lead to aberrant signatures11. Tet3 mediated hydroxymethylation may also be more robustly targeted upon fertilization than after nuclear transfer, where it is not specifically recruited to a single pronucleus but rather distributed at restricted levels across the entire diploid genome12,13. It is interesting that demethylated loci after nuclear transfer resemble the genomic features and promoter classes enriched for hydroxymethylation within mouse embryonic stem cells, namely repetitive LINE families and germ-line gene promoters, which suggests that at least a portion of the demethylation events involved with induced pluripotency may be specifically targeted14,15. Technical improvements to other genomic profiling strategies should soon identify the targets and dynamics of histone deposition, as well as the distribution of hydroxymethylcytosine15, and should comprehensively define the full reprogramming capacity of the mammalian ooplasm.
Supplementary Material
Editorial Summary.
Alex Meissner and colleagues report base-pair resolution methylation maps from donor fibroblasts and nuclear transfer reconstructed mouse embryos. They compare methylation profiles to normal fertilization and find that specific promoters and repeat elements exhibit differential dynamics.
Acknowledgments
We would like to thank all the members of the Meissner and Regev labs. Manuel Garber, Nir Yosef, Jimmie Ye, Richard Koche, Hongcang Gu, Andreas Gnirke, and Tarjei Mikkelsen for technical advice and discussion. All members of the Broad Sequencing Platform in particular Fontina Kelley and James Meldrim, Tim Fennel, Kathleen Tibbetts and Jennifer Fostel. We also thank Stuart Levine, Michael Gravina, and Kevin Thai from the MIT BioMicro Center. D.E. is supported by the NYSCF. A.R. is an investigator of the Merkin Foundation for Stem Cell Research at the Broad Institute and supported by an NIH Pioneer Award (5DP1OD003958) and a NHGRI CEGS grant (1P50HG006193), the Burroughs Wellcome Career Award at the Scientific Interface and HHMI. A.M. is supported by the Pew Charitable Trusts, Human Frontiers Science Program and the NIH (U01ES017155, P01GM099117 and 1P50HG006193).
Footnotes
Author contributions
MMC, ZDS, DE, and AM conceived and designed the study. DE performed SCNT, ZDS performed methylation profiling, and MMC performed all analysis. MMC, ZDS, AR and AM interpreted the data. MMC, ZDS and AM wrote the paper with input from the other authors.
The authors declare no competing financial interests.
Data Access: RRBS data is deposited at the Gene Expression Omnibus under accession number GSE38711.
References
- 1.Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 2.Rideout WM, 3rd, Eggan K, Jaenisch R. Nuclear cloning and epigenetic reprogramming of the genome. Science. 2001;293:1093–1098. doi: 10.1126/science.1063206. [DOI] [PubMed] [Google Scholar]
- 3.Wakayama T, Perry AC, Zuccotti M, Johnson KR, Yanagimachi R. Full-term development of mice from enucleated oocytes injected with cumulus cell nuclei. Nature. 1998;394:369–374. doi: 10.1038/28615. [DOI] [PubMed] [Google Scholar]
- 4.Meissner A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith ZD, et al. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 2012;484:339–44. doi: 10.1038/nature10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goodier JL, Ostertag EM, Du K, Kazazian HH., Jr A novel active L1 retrotransposon subfamily in the mouse. Genome Res. 2001;11:1677–1685. doi: 10.1101/gr.198301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wossidlo M, et al. Dynamic link of DNA demethylation, DNA strand breaks and repair in mouse zygotes. Embo J. 2010;29:1877–1888. doi: 10.1038/emboj.2010.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Egli D, et al. Reprogramming within hours following nuclear transfer into mouse but not human zygotes. Nat Commun. 2011;2:488. doi: 10.1038/ncomms1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dean W, et al. Conservation of methylation reprogramming in mammalian development: aberrant reprogramming in cloned embryos. Proc Natl Acad Sci U S A. 2001;98:13734–13738. doi: 10.1073/pnas.241522698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Polanski Z, Motosugi N, Tsurumi C, Hiiragi T, Hoffmann S. Hypomethylation of paternal DNA in the late mouse zygote is not essential for development. Int J Dev Biol. 2008;52:295–298. doi: 10.1387/ijdb.072347zp. [DOI] [PubMed] [Google Scholar]
- 11.Ng RK, Gurdon JB. Epigenetic memory of an active gene state depends on histone H3.3 incorporation into chromatin in the absence of transcription. Nat Cell Biol. 2008;10:102–109. doi: 10.1038/ncb1674. [DOI] [PubMed] [Google Scholar]
- 12.Gu TP, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- 13.Wossidlo M, et al. 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat Commun. 2011;2:241. doi: 10.1038/ncomms1240. [DOI] [PubMed] [Google Scholar]
- 14.Branco MR, Ficz G, Reik W. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat Rev Genet. 2011 doi: 10.1038/nrg3080. [DOI] [PubMed] [Google Scholar]
- 15.Booth MJ, et al. Quantitative Sequencing of 5-Methylcytosine and 5-Hydroxymethylcytosine at Single-Base Resolution. Science. 2012 doi: 10.1126/science.1220671. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.