Abstract
The human CCND1 gene, encoding a cell cycle protein cyclin D1, is one of the most frequently amplified genes in human cancers. Cyclin D1 activates cyclin dependent kinases CDK4 and CDK6 and drives cell proliferation. Beyond the cell cycle role, the full repertoire of cyclin D1 functions in cancer cells is still unclear. Emerging evidence indicates that cyclin D1 may play a role in DNA damage response. In this review we discuss observations linking cyclin D1 to DNA damage repair, and we summarize our recent findings, which demonstrate cyclin D1 function in homologous recombination-mediated DNA repair.
Cyclin D1 in cell cycle progression
D-type cyclins are components of the core cell cycle machinery. The D-cyclin family is composed of three proteins, cyclin D1, D2 and D3, which are expressed in proliferating cells (1). The gene encoding cyclin D1 represents the second most frequently amplified locus in the human cancer genome (2). The protein product of this locus, cyclin D1, binds and activates cyclin-dependent kinases CDK4 and CDK6 (1). During cell cycle progression, cyclin D1-CDK4 and D1-CDK6 complexes phosphorylate the retinoblastoma protein, pRB, pRB-related p107 and p130 proteins, as well as Smad3 and FOXM1 transcription factors (1, 3, 4). By far the best-documented function of cyclin D1 is its ability to drive cell cycle progression through phosphorylation of pRB, p107 and p130. In their hypophosphorylated forms pRB, p107 and p130 inhibit the transcriptional activity of E2F transcription factors. Phosphorylation of these three proteins by cyclin D1-CDK4/6 kinase releases and de-represses E2Fs, thereby allowing G1→S phase progression (1) (Figure 1A). In addition to this kinase-dependent function, cyclin D1-CDK4/6 complexes sequester cell cycle inhibitors p27Kip1 and p21Cip1 away from cyclin E-CDK2, thereby contributing to activation of cyclin E-CDK2 kinase (1). Lastly, there is growing evidence that cyclin D1 plays cell cycle-independent roles which are also independent of CDK4 and CDK6 (5).
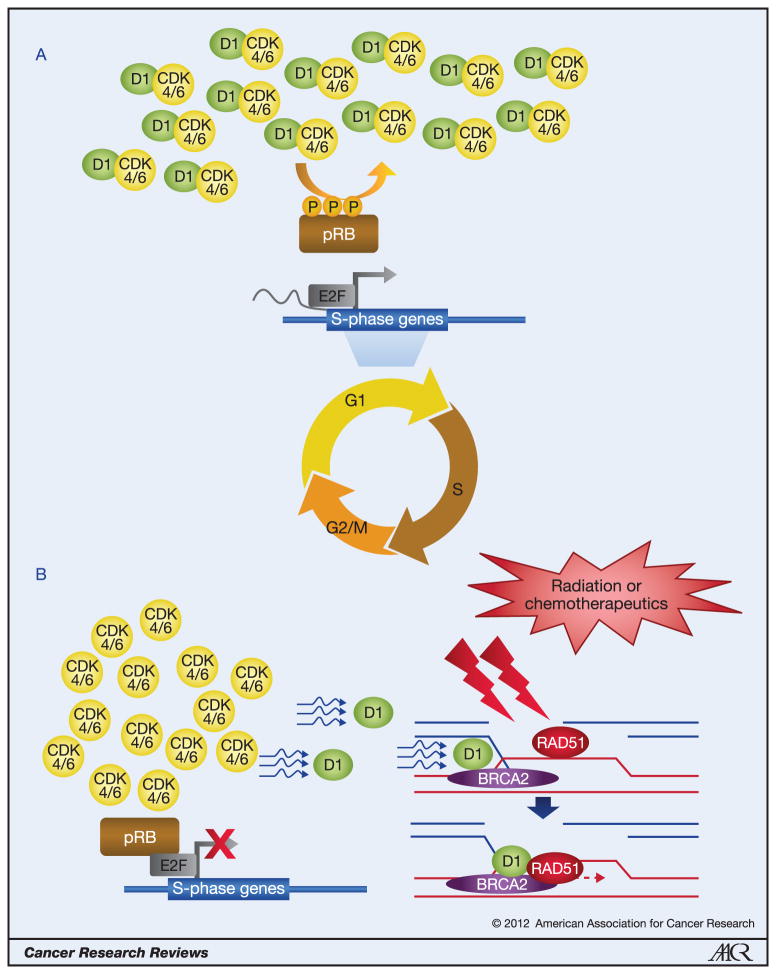
Figure 1. A model summarizing cyclin D1 function in cell proliferation and in DNA repair.
A, During normal cell cycle progression, cyclin D1 (D1) forms complexes with CDK4 or CDK6 to regulate G1 phase progression by phosphorylating pRB, p107 and p130 proteins. pRB phosphorylation releases E2F transcription factors, and allows E2F-dependent transcription of S-phase genes.
B, After DNA damage caused by radiation or by chemotherapeutic agents, cyclin D1 protein levels are significantly reduced, and CDK4 kinase activity is diminished. Hypophosphorylated pRB inhibits E2F’s ability to transactivate genes, thereby contributing to cell cycle arrest. Part of the remaining pool of cyclin D1 protein is recruited to DNA damage sites, in a BRCA2-dependent manner, to facilitate RAD51 localization or to stabilize RAD51 at the DNA damage sites, thereby assisting the homologous recombination-mediated DNA repair. Note that it is unclear whether CDK4 or CDK6 are recruited to DNA damage sites along with cyclin D1.
Degradation of cyclin D1 upon DNA damage
Proliferating cells usually respond to DNA damage by arresting their cell cycle progression. Several independent reports pointed to downregulation of cyclin D1 as one of the mechanisms that underlie this cell cycle arrest (6–8). DNA damage was shown to activate GSK3β, which phosphorylates cyclin D1 on Thr-286. Phosphorylated cyclin D1 is then exported from the nucleus, polyubiquitinated by the SCFFbx4-aBcrystallin E3 ubiquitin ligase, and degraded by the proteasome (7, 9). Strikingly, a related cyclin D2 does not undergo phosphorylation on the corresponding residue following DNA damage, suggesting that cyclin D1 may play a non-redundant role in transmitting post-radiation growth-arresting signals to the core cell cycle machinery (7, 9). The activity of ATM was shown to be required for cyclin D1 phosphorylation and degradation triggered by double stranded DNA breaks, while the ATR kinase mediates the effect on cyclin D1 following UV irradiation (7, 10, 11). In contrast to these findings implicating F-box protein Fbx4 and cofactor αB crystallin in degradation of cyclin D1, another group postulated that Thr-286-phosphorylated cyclin D1 interacts with and is targeted for degradation by an F-box protein FBXO31 (8). Moreover, DNA damage was shown to cause proteolysis of cyclin D1 by the anaphase promoting complex/cyclosome (APC/C). This effect is mediated by the destruction box in cyclin D1 and it was shown to be independent of cyclin D1 phosphorylation on Thr-286 (6). It is possible that these different scenarios reflect distinct modes of cyclin D1 degradation in particular cell types. Overall, these reports point to cyclin D1 degradation as an important molecular mechanism which arrests cell proliferation following DNA damage. Persistent high expression of cyclin D1 in cells which accumulated double-stranded DNA breaks leads to radio-resistant DNA synthesis (7). Moreover, downregulation of cyclin D1 following UV damage was shown to be required for efficient DNA repair, and forced overexpression of cyclin D1 prevented DNA repair (12).
To complicate this picture, some reports documented an increase of cyclin D1 levels following DNA damage (13–16). These findings hint that cyclin D1 may play an active role in DNA damage repair.
Interaction of cyclin D1 with DNA damage proteins
The first indications that cyclin D1 may play a direct role in DNA damage repair came from the observations functionally linking cyclin D1 with proteins involved in DNA repair. Richard Pestell’s group showed that cyclin D1 tethered to chromatin can recruit RAD51, a protein that plays an essential role in homologous recombination process (17). Intriguingly, recruitment of RAD51 by cyclin D1 took place after DNA damage, but not in naive cells, suggesting a functional relevance of this interaction for DNA repair (17). The same group demonstrated a link between cyclin D1 and another DNA repair protein, BRCA1, by showing that cyclin D1 antagonizes BRCA1-mediated repression of estrogen receptor α transcriptional activity (18). The effect was mediated through an ability of cyclin D1 to compete with BRCA1 for ERα binding (18). A direct link between cyclin D1 and BRCA1 was provided by the observation that cyclin D1-CDK4 kinase phosphorylates BRCA1 on Ser-632; this event was shown to inhibit recruitment of BRCA1 to target promoters (19). BRCA1, as well as BRCA1 splice variant BRCA1-IRIS were shown to transcriptionally upregulate cyclin D1 (20, 21), while cyclin D1 was demonstrated to induce BRCA1 expression, through the pRB→E2F pathway (22).
A possible functional link between cyclin D1 and another DNA repair protein, BRCA2 was suggested by a study demonstrating that cyclin D1, BRCA2 as well as RAD51 physically interact with Sp1 transcription factor (23, 24).
Cyclin D1 was also shown to physically interact with two other proteins involved in DNA repair, PCNA (25, 26) and replication factor C (RFC, ref. 27). Both PCNA and RFC functionally interact with BRCA1 during repair of UV-induced DNA damage (28). Collectively, all these observations point to multiple functional links between cyclin D1 and DNA repair proteins suggesting that cyclin D1 may play a role in DNA damage repair.
Consistent with this view, several studies noted that elevated levels of cyclin D1 confer relative radiation-resistance to cancer cells (29–33). A correlation was noted between high levels of cyclin D1 and unfavorable response to radiotherapy (34, 35). Conversely, knock-down of cyclin D1 sensitized cancer cells to radiation or to DNA damaging agent cisplatin (33, 36). Moreover, antisense-mediated downregulation of cyclin D1 expression increased sensitivity of zebrafish embryos to radiation (37).
It should be noted, however, that many studies reached the opposite conclusion. For example, Coco-Martin et al. (38) demonstrated that ectopic overexpression of cyclin D1 sensitized cancer cells to irradiation, by rendering cells more susceptible to radiation-induced apoptosis. Similar conclusion was reached by Trent et al. (39) who proposed that the cyclin D1-driven enhancement of radiation sensitivity is CDK-independent, and it is mediated, at least in part, through the ability of cyclin D1 to transcriptionally induce expression of a heat shock protein HSPB8. According to some reports, cyclin D1 overexpression was shown to correlate with increased radiosensitivity and with favorable response to radio- or chemotherapy in squamous cell carcinoma of head and neck (40), oral squamous cell carcinoma (41) and in early-stage larynx cancer (42). It is possible that these conflicting observations reflect different types of tumors studied. Alternatively, these findings may illustrate a dual role for cyclin D1 in DNA damage response. Thus, following an acute DNA damage cyclin D1 levels need to be lowered to trigger cell cycle arrest, while the remaining pool of cyclin D1 may play an important, positive function in promoting DNA damage repair. Hence, strong overexpression of cyclin D1 to levels which cannot be efficiently reduced following DNA damage would compromise cell cycle checkpoints, and would impair cell survival, while elevated levels of cyclin D1 after DNA damage-induced cell cycle arrest might augment DNA repair. Likewise, a complete knock-down of cyclin D1 might compromise DNA repair, by depleting a protein that is needed for DNA repair.
A direct role of cyclin D1 in homologous recombination-based DNA repair
In a recent study (43) we utilized immunoaffinity purification of cyclin D1-containing complexes, followed by high-throughput shotgun mass spectrometry, to decipher the identity of cyclin D1-interacting proteins “cyclin D1-interactome” in several human cancer cell lines. Among cyclin D1-interactors we observed a strong enrichment for proteins belonging to “DNA damage repair” category.
One of these proteins was RAD51, an essential DNA recombinase which mediates homologous recombination-based DNA repair (44). An interaction between endogenous RAD51 and cyclin D1 was verified by immunoprecipitation-western blotting in a wide panel of human cancer cell lines, indicating that the interaction is not cell type-specific. Importantly, the interaction is strongly enhanced following irradiation of cells, suggesting that DNA damage might induce posttranslational modification of cyclin D1 and/or RAD51, which then stabilizes cyclin D1-RAD51 binding (43).
Surprisingly, we detected cyclin D1 at the sites of double stranded DNA breaks where it co-localized with RAD51, suggesting that cyclin D1 may directly participate in the repair of damaged DNA. We also observed that RAD51 recruitment to DNA damage sites was significantly reduced in cells depleted of cyclin D1. Moreover, knock-down of cyclin D1 reduced the rate of homologous recombination-mediated DNA repair (43). These findings are consistent with and fully support the observations of Li et al. (17) who demonstrated that cyclin D1, when targeted to chromatin, can recruit RAD51 following DNA damage. Collectively these findings suggest a model in which cyclin D1 localizes to double stranded DNA breaks following DNA damage, and helps to recruit RAD51 (Figure 1B).
How is then cyclin D1 recruited to broken DNA? Among cyclin D1-interacting proteins, detected in our screen, we observed BRCA2. Like RAD51, BRCA2 is also recruited to DNA damage sites. BRCA2 localization at the DNA damage foci represents a crucial step for homologous recombination, and it occurs prior to the recruitment of RAD51 (45). BRCA2 displaces the single stranded DNA (ssDNA) binding protein RPA from single stranded regions generated by end-resection, and facilitates loading of RAD51 onto ssDNA (45). We observed that depletion of cyclin D1 had no effect on BRCA2 recruitment to DNA damage foci. However, knock-down of BRCA2 decreased loading of cyclin D1, suggesting that BRCA2 is responsible for recruiting cyclin D1 to DNA damage sites. Consistent with the observed association between BRCA2 and cyclin D1 in cancer cells, we found that purified recombinant cyclin D1 binds to purified BRCA2 fragments, in an in vitro binding assay.
We propose that cyclin D1 is recruited to double stranded DNA breaks via BRCA2. Cyclin D1 then helps either to recruit RAD51, or to stabilize RAD51 on the repair foci, thereby contributing to the homologous recombination process (Figure 1B).
Of note, we showed that the function of cyclin D1 in DNA repair is independent of its role in cell cycle progression. All our DNA repair analyses were performed in pRB-negative cancer cells, which do not require cyclin D1 for proliferation (46, 47). Moreover, the DNA repair role of cyclin D1 is independent of cyclin D1’s ability to activate CDK4 and CDK6.
Possible implications for cancer treatment
Inhibition of cyclin D-associated kinase activity is currently being entertained as an attractive strategy for treatment of several cancer types (48), and inhibitors of cyclin D-CDK4 and D-CDK6 kinases are currently in clinical trials. Demonstration that cyclin D1 plays a CDK-independent function in DNA damage repair suggests that cyclin D1 protein (rather than cyclin D1-associated kinase) might represent a more effective anti-cancer target, as inhibition of cyclin D1 is expected to impair both cell proliferation, as well as DNA repair.
Another implication from our study is the function of cyclin D1 in pRB-negative cancer cells. It is very well documented that cancer cells that lost pRB no longer require D-cyclins for proliferation (46, 47). Consequently, depletion of cyclin D1, or inhibition of CDK4/6 kinase activity has no impact on proliferation of pRB-negative cancer cells. However, our study raises a possibility that targeting cyclin D1 might have a therapeutic value also in pRB-negative tumors, where it is expected to decrease the efficiency of DNA damage repair.
Unresolved issues
A direct role for cyclin D1 in DNA damage repair was detected in a screen which utilized human cancer cells. It remains to be seen whether cyclin D1 plays role in DNA repair also in normal cells. Consistent with this possibility, we observed that immortalized mouse embryonic fibroblasts lacking all three D-cyclins are more susceptible to radiation-induced DNA damage as compared to their wild-type counterparts.
Another unresolved issue is whether other D-type cyclins (D2 and D3) play a similar role in DNA repair. It will be interesting to determine whether these proteins also interact with RAD51 and with BRCA2, and are recruited to DNA damage sites.
A more fundamental question is whether D-cyclins, or other components of mammalian core cell cycle machinery affect the choice of DNA repair pathway. Mammalian cells can repair double stranded DNA breaks through error-prone non-homologous end joining (NHEJ), or through relatively faithful homologous recombination. The identity of molecules/signaling pathways that determine the choice of DNA repair pathway is not fully understood. Several reports showed that bona fide cell cycle CDKs can phosphorylate DNA repair proteins, and may determine the choice of DNA repair pathway. In budding yeast, it was shown that homologous recombination is controlled at an early step called DNA-end resection, by an activity of Cdc28 (a yeast homolog of mammalian CDKs) (49). Cdc28 phosphorylates Sae2 and a nuclease Dna2, key proteins in DNA-resection, thereby allowing progression of the DNA end-resection process, which represents a prerequisite for homologous recombination (50, 51). Conversely, inhibition of yeast CDK activity during DNA damage causes yeast to employ NHEJ to repair DNA (51). A similar regulation was observed in human cells (52). However, the processes governing DNA repair pathways is mammalian cells seem to be more complex. For instance, cyclin A-CDK2 (and possibly cyclin B-CDK1) kinase was reported to block interaction between the C-terminus of BRCA2 and RAD51, and likely to inhibit homologous recombination, by phosphorylating Ser 3291 on BRCA2 (53). It is clear that the cell cycle and DNA repair machinery intersect at several points, and more work is needed to fully understand the functional interplay between these pathways.
Acknowledgments
This work was supported by P01 CA080111 (to D.M.L and P.S).
References
- 1.Sherr CJ, Roberts JM. Living with or without cyclins and cyclin-dependent kinases. Genes Dev. 2004;18:2699–2711. doi: 10.1101/gad.1256504. [DOI] [PubMed] [Google Scholar]
- 2.Beroukhim R, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anders L, Ke N, Hydbring P, Choi YJ, Widlund HR, Chick JM, Zhai H, Vidal M, Gygi SP, Braun P, Sicinski P. A Systematic Screen for CDK4/6 Substrates Links FOXM1 Phosphorylation to Senescence Suppression in Cancer Cells. Cancer Cell. 2011;20:620–634. doi: 10.1016/j.ccr.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matsuura I, Denissova NG, Wang G, He D, Long J, Liu F. Cyclin-dependent kinases regulate the antiproliferative function of Smads. Nature. 2004;430:226–231. doi: 10.1038/nature02650. [DOI] [PubMed] [Google Scholar]
- 5.Fu M, Wang C, Li Z, Sakamaki T, Pestell RG. Minireview: Cyclin D1: normal and abnormal functions. Endocrinology. 2004;145:5439–5447. doi: 10.1210/en.2004-0959. [DOI] [PubMed] [Google Scholar]
- 6.Agami R, Bernards R. Distinct initiation and maintenance mechanisms cooperate to induce G1 cell cycle arrest in response to DNA damage. Cell. 2000;102:55–66. doi: 10.1016/s0092-8674(00)00010-6. [DOI] [PubMed] [Google Scholar]
- 7.Pontano LL, Aggarwal P, Barbash O, Brown EJ, Bassing CH, Diehl JA. Genotoxic stress-induced cyclin D1 phosphorylation and proteolysis are required for genomic stability. Mol Cell Biol. 2008;28:7245–7258. doi: 10.1128/MCB.01085-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Santra MK, Wajapeyee N, Green MR. F-box protein FBXO31 mediates cyclin D1 degradation to induce G1 arrest after DNA damage. Nature. 2009;459:722–725. doi: 10.1038/nature08011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pontano LL, Diehl JA. DNA damage-dependent cyclin D1 proteolysis: GSK3beta holds the smoking gun. Cell Cycle. 2009;8:824–827. doi: 10.4161/cc.8.6.7889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choo DW, Baek HJ, Motoyama N, Cho KH, Kim HS, Kim SS. ATM is required for rapid degradation of cyclin D1 in response to gamma-irradiation. Biochem Biophys Res Commun. 2009;378:847–850. doi: 10.1016/j.bbrc.2008.11.132. [DOI] [PubMed] [Google Scholar]
- 11.Hitomi M, Yang K, Stacey AW, Stacey DW. Phosphorylation of cyclin D1 regulated by ATM or ATR controls cell cycle progression. Mol Cell Biol. 2008;28:5478–5493. doi: 10.1128/MCB.02047-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pagano M, Theodoras AM, Tam SW, Draetta GF. Cyclin D1-mediated inhibition of repair and replicative DNA synthesis in human fibroblasts. Genes Dev. 1994;8:1627–1639. doi: 10.1101/gad.8.14.1627. [DOI] [PubMed] [Google Scholar]
- 13.Casafont I, Palanca A, Lafarga V, Berciano MT, Lafarga M. Effect of ionizing radiation in sensory ganglion neurons: organization and dynamics of nuclear compartments of DNA damage/repair and their relationship with transcription and cell cycle. Acta Neuropathol. 2011;122:481–493. doi: 10.1007/s00401-011-0869-0. [DOI] [PubMed] [Google Scholar]
- 14.Choi HS, Cho MC, Lee HG, Yoon DY. Indole-3-carbinol induces apoptosis through p53 and activation of caspase-8 pathway in lung cancer A549 cells. Food Chem Toxicol. 2010;48:883–890. doi: 10.1016/j.fct.2009.12.028. [DOI] [PubMed] [Google Scholar]
- 15.He YY, Council SE, Feng L, Chignell CF. UVA-induced cell cycle progression is mediated by a disintegrin and metalloprotease/epidermal growth factor receptor/AKT/Cyclin D1 pathways in keratinocytes. Cancer Res. 2008;68:3752–3758. doi: 10.1158/0008-5472.CAN-07-6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu S, Gonzalez J, Hwang BJ, Steinberg ML. Induction of cyclin D1 by arsenite and UVB-irradiation in human keratinocytes. J Health Care Poor Underserved. 2011;22:110–121. doi: 10.1353/hpu.2011.0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Z, Jiao X, Wang C, Shirley LA, Elsaleh H, Dahl O, Wang M, Soutoglou E, Knudsen ES, Pestell RG. Alternative cyclin D1 splice forms differentially regulate the DNA damage response. Cancer Res. 2010;70:8802–8811. doi: 10.1158/0008-5472.CAN-10-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang C, Fan S, Li Z, Fu M, Rao M, Ma Y, Lisanti MP, Albanese C, Katzenellenbogen BS, Kushner PJ, Weber B, Rosen EM, Pestell RG. Cyclin D1 antagonizes BRCA1 repression of estrogen receptor alpha activity. Cancer Res. 2005;65:6557–6567. doi: 10.1158/0008-5472.CAN-05-0486. [DOI] [PubMed] [Google Scholar]
- 19.Kehn K, Berro R, Alhaj A, Bottazzi ME, Yeh WI, Klase Z, Van Duyne R, Fu S, Kashanchi F. Functional consequences of cyclin D1/BRCA1 interaction in breast cancer cells. Oncogene. 2007;26:5060–5069. doi: 10.1038/sj.onc.1210319. [DOI] [PubMed] [Google Scholar]
- 20.Nakuci E, Mahner S, Direnzo J, ElShamy WM. BRCA1-IRIS regulates cyclin D1 expression in breast cancer cells. Exp Cell Res. 2006;312:3120–3131. doi: 10.1016/j.yexcr.2006.06.021. [DOI] [PubMed] [Google Scholar]
- 21.Welcsh PL, Lee MK, Gonzalez-Hernandez RM, Black DJ, Mahadevappa M, Swisher EM, Warrington JA, King MC. BRCA1 transcriptionally regulates genes involved in breast tumorigenesis. Proc Natl Acad Sci U S A. 2002;99:7560–7565. doi: 10.1073/pnas.062181799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang A, Schneider-Broussard R, Kumar AP, MacLeod MC, Johnson DG. Regulation of BRCA1 expression by the Rb-E2F pathway. J Biol Chem. 2000;275:4532–4536. doi: 10.1074/jbc.275.6.4532. [DOI] [PubMed] [Google Scholar]
- 23.Opitz OG, Rustgi AK. Interaction between Sp1 and cell cycle regulatory proteins is important in transactivation of a differentiation-related gene. Cancer Res. 2000;60:2825–2830. [PubMed] [Google Scholar]
- 24.Tapias A, Ciudad CJ, Roninson IB, Noe V. Regulation of Sp1 by cell cycle related proteins. Cell Cycle. 2008;7:2856–2867. doi: 10.4161/cc.7.18.6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsuoka S, Yamaguchi M, Matsukage A. D-type cyclin-binding regions of proliferating cell nuclear antigen. J Biol Chem. 1994;269:11030–11036. [PubMed] [Google Scholar]
- 26.Xiong Y, Zhang H, Beach D. D type cyclins associate with multiple protein kinases and the DNA replication and repair factor PCNA. Cell. 1992;71:505–514. doi: 10.1016/0092-8674(92)90518-h. [DOI] [PubMed] [Google Scholar]
- 27.van der Kuip H, Carius B, Haque SJ, Williams BR, Huber C, Fischer T. The DNA-binding subunit p140 of replication factor C is upregulated in cycling cells and associates with G1 phase cell cycle regulatory proteins. J Mol Med (Berl) 1999;77:386–392. doi: 10.1007/s001090050365. [DOI] [PubMed] [Google Scholar]
- 28.Pathania S, Nguyen J, Hill SJ, Scully R, Adelmant GO, Marto JA, Feunteun J, Livingston DM. BRCA1 is required for postreplication repair after UV-induced DNA damage. Mol Cell. 2011;44:235–251. doi: 10.1016/j.molcel.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boothman DA, Meyers M, Odegaard E, Wang M. Altered G1 checkpoint control determines adaptive survival responses to ionizing radiation. Mutat Res. 1996;358:143–153. doi: 10.1016/s0027-5107(96)00115-7. [DOI] [PubMed] [Google Scholar]
- 30.Milas L, Akimoto T, Hunter NR, Mason KA, Buchmiller L, Yamakawa M, Muramatsu H, Ang KK. Relationship between cyclin D1 expression and poor radioresponse of murine carcinomas. Int J Radiat Oncol Biol Phys. 2002;52:514–521. doi: 10.1016/s0360-3016(01)02693-1. [DOI] [PubMed] [Google Scholar]
- 31.Shimura T. Acquired radioresistance of cancer and the AKT/GSK3beta/cyclin D1 overexpression cycle. J Radiat Res (Tokyo) 2011;52:539–544. doi: 10.1269/jrr.11098. [DOI] [PubMed] [Google Scholar]
- 32.Shimura T, Kakuda S, Ochiai Y, Kuwahara Y, Takai Y, Fukumoto M. Targeting the AKT/GSK3beta/cyclin D1/Cdk4 survival signaling pathway for eradication of tumor radioresistance acquired by fractionated radiotherapy. Int J Radiat Oncol Biol Phys. 2011;80:540–548. doi: 10.1016/j.ijrobp.2010.12.065. [DOI] [PubMed] [Google Scholar]
- 33.Shimura T, Kakuda S, Ochiai Y, Nakagawa H, Kuwahara Y, Takai Y, Kobayashi J, Komatsu K, Fukumoto M. Acquired radioresistance of human tumor cells by DNA-PK/AKT/GSK3beta-mediated cyclin D1 overexpression. Oncogene. 2010;29:4826–4837. doi: 10.1038/onc.2010.238. [DOI] [PubMed] [Google Scholar]
- 34.Chang AR, Wu HG, Park CI, Jun YK, Kim CW. Expression of epidermal growth factor receptor and cyclin D1 in pretreatment biopsies as a predictive factor of radiotherapy efficacy in early glottic cancer. Head Neck. 2008;30:852–857. doi: 10.1002/hed.20788. [DOI] [PubMed] [Google Scholar]
- 35.Lai JP, Tong CL, Hong C, Xiao JY, Tao ZD, Zhang Z, Tong WM, Betz CS. Association between high initial tissue levels of cyclin d1 and recurrence of nasopharyngeal carcinoma. Laryngoscope. 2002;112:402–408. doi: 10.1097/00005537-200202000-00036. [DOI] [PubMed] [Google Scholar]
- 36.Wang MB, Yip HT, Srivatsan ES. Antisense cyclin D1 enhances sensitivity of head and neck cancer cells to cisplatin. Laryngoscope. 2001;111:982–988. doi: 10.1097/00005537-200106000-00010. [DOI] [PubMed] [Google Scholar]
- 37.McAleer MF, Duffy KT, Davidson WR, Kari G, Dicker AP, Rodeck U, Wickstrom E. Antisense inhibition of cyclin D1 expression is equivalent to flavopiridol for radiosensitization of zebrafish embryos. Int J Radiat Oncol Biol Phys. 2006;66:546–551. doi: 10.1016/j.ijrobp.2006.05.040. [DOI] [PubMed] [Google Scholar]
- 38.Coco Martin JM, Balkenende A, Verschoor T, Lallemand F, Michalides R. Cyclin D1 overexpression enhances radiation-induced apoptosis and radiosensitivity in a breast tumor cell line. Cancer Res. 1999;59:1134–1140. [PubMed] [Google Scholar]
- 39.Trent S, Yang C, Li C, Lynch M, Schmidt EV. Heat shock protein B8, a cyclin-dependent kinase-independent cyclin D1 target gene, contributes to its effects on radiation sensitivity. Cancer Res. 2007;67:10774–10781. doi: 10.1158/0008-5472.CAN-07-1475. [DOI] [PubMed] [Google Scholar]
- 40.Akervall J, Brun E, Dictor M, Wennerberg J. Cyclin D1 overexpression versus response to induction chemotherapy in squamous cell carcinoma of the head and neck--preliminary report. Acta Oncol. 2001;40:505–511. doi: 10.1080/028418601750288244. [DOI] [PubMed] [Google Scholar]
- 41.Shintani S, Mihara M, Ueyama Y, Matsumura T, Wong DT. Cyclin D1 overexpression associates with radiosensitivity in oral squamous cell carcinoma. Int J Cancer. 2001;96:159–165. doi: 10.1002/ijc.1014. [DOI] [PubMed] [Google Scholar]
- 42.Yoo SS, Carter D, Turner BC, Sasaki CT, Son YH, Wilson LD, Glazer PM, Haffty BG. Prognostic significance of cyclin D1 protein levels in early-stage larynx cancer treated with primary radiation. Int J Cancer. 2000;90:22–28. doi: 10.1002/(sici)1097-0215(20000220)90:1<22::aid-ijc3>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 43.Jirawatnotai S, Hu Y, Michowski W, Elias JE, Becks L, Bienvenu F, Zagozdzon A, Goswami T, Wang YE, Clark AB, Kunkel TA, van Harn T, Xia B, Correll M, Quackenbush J, Livingston DM, Gygi SP, Sicinski P. A function for cyclin D1 in DNA repair uncovered by protein interactome analyses in human cancers. Nature. 2011;474:230–234. doi: 10.1038/nature10155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baumann P, West SC. Role of the human RAD51 protein in homologous recombination and double-stranded-break repair. Trends Biochem Sci. 1998;23:247–251. doi: 10.1016/s0968-0004(98)01232-8. [DOI] [PubMed] [Google Scholar]
- 45.Yuan SS, Lee SY, Chen G, Song M, Tomlinson GE, Lee EY. BRCA2 is required for ionizing radiation-induced assembly of Rad51 complex in vivo. Cancer Res. 1999;59:3547–3551. [PubMed] [Google Scholar]
- 46.Bates S, Parry D, Bonetta L, Vousden K, Dickson C, Peters G. Absence of cyclin D/cdk complexes in cells lacking functional retinoblastoma protein. Oncogene. 1994;9:1633–1640. [PubMed] [Google Scholar]
- 47.Lukas J, Parry D, Aagaard L, Mann DJ, Bartkova J, Strauss M, Peters G, Bartek J. Retinoblastoma-protein-dependent cell-cycle inhibition by the tumour suppressor p16. Nature. 1995;375:503–506. doi: 10.1038/375503a0. [DOI] [PubMed] [Google Scholar]
- 48.Shapiro GI. Cyclin-dependent kinase pathways as targets for cancer treatment. J Clin Oncol. 2006;24:1770–1783. doi: 10.1200/JCO.2005.03.7689. [DOI] [PubMed] [Google Scholar]
- 49.Aylon Y, Liefshitz B, Kupiec M. The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. Embo J. 2004;23:4868–4875. doi: 10.1038/sj.emboj.7600469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen X, Niu H, Chung WH, Zhu Z, Papusha A, Shim EY, Lee SE, Sung P, Ira G. Cell cycle regulation of DNA double-strand break end resection by Cdk1-dependent Dna2 phosphorylation. Nat Struct Mol Biol. 2011;18:1015–1019. doi: 10.1038/nsmb.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huertas P, Cortes-Ledesma F, Sartori AA, Aguilera A, Jackson SP. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature. 2008;455:689–692. doi: 10.1038/nature07215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huertas P, Jackson SP. Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J Biol Chem. 2009;284:9558–9565. doi: 10.1074/jbc.M808906200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Esashi F, Christ N, Gannon J, Liu Y, Hunt T, Jasin M, West SC. CDK-dependent phosphorylation of BRCA2 as a regulatory mechanism for recombinational repair. Nature. 2005;434:598–604. doi: 10.1038/nature03404. [DOI] [PubMed] [Google Scholar]