Abstract
Purpose
The significance of mTOR activation in uterine leiomyosarcoma (ULMS) and its potential as a therapeutic target were investigated. Furthermore, given that effective therapies likely require combination mTOR blockade with inhibition of other targets, coupled with recent observations suggesting that Aurora-A kinase (Aurk-A) deregulations commonly occur in ULMS, the preclinical impact of dually targeting both pathways was evaluated.
Experimental design
Immunohistochemical staining was used to evaluate expression of activated mTOR componentry in a large (>200 samples) ULMS tissue microarray. Effects of mTOR blockade (using rapamycin) and Aurk-A inhibition (using MLN8237) alone and in combination on human ULMS cell growth, cell-cycle progression, and apoptosis were assessed in cellular assays. Drug interactions were determined via combination index (CI) analyses. The anti-tumor effects of inhibitors alone or in combination were evaluated in vivo.
Results
Enhanced mTOR activation was seen in human ULMS samples. Increased pS6RP and p4EBP1 expression correlated with disease progression; p4EBP1 was found to be an independent prognosticator of patient outcome. Rapamycin inhibited growth and cell cycle progression of ULMS cell strains/lines in culture. However, only a cytostatic effect on tumor growth was found in vivo. Combining rapamycin with MLN8237 profoundly (and synergistically) abrogated ULMS cells’ growth in culture; interestingly, these effects were seen only when MLN8237 was pre-administered. This novel therapeutic combination and scheduling regimen resulted in marked tumor growth inhibition in vivo.
Conclusions
mTOR and Aurk-A pathways are commonly deregulated in ULMS. Preclinical data support further exploration of dual mTOR and Aurk-A therapeutic blockade for human ULMS.
Keywords: uterine leiomyosarcoma, TMA, mTOR, Aurora kinase A, MLN8237
Introduction
Leiomyosarcoma (LMS) is an uncommon but highly aggressive malignancy with smooth muscle cell differentiation (1); LMSs arising from the uterine myometrium are designated uterine LMS (ULMS). While long-term survival approaches 90% for the much more common endometrial adenocarcinoma (2), ULMS 5-year survival rates are less favorable, ranging between 8–60% for the past several decades (3, 4). While initially confined to the uterus, their location within the blood-vessel-rich myometrium facilitates early vascular invasion and metastasis (5). More than 50% of early-stage disease patients recur after “curative” therapy (6); more ominously, 81% of stage III patients will recur, and only 8% of advanced disease patients survive five years (4, 7). Early stage disease therapy is primarily surgical (8). While adjuvant chemotherapy and/or radiotherapy are used in cases deemed high risk for recurrence, the impact of these strategies is currently not well defined (8). Limited therapeutic options are available for patients with locally advanced or metastatic ULMS where the treatment approach is that of other genetically complex soft tissue sarcoma; i.e., radiation to local recurrences and systemic chemotherapy for advanced disease (9). Unfortunately, these approaches are confounded by relative chemo- and radio-resistance (10). Several ULMS Phase II clinical trials failed to reach response rates of 10% using conventional chemotherapies; e.g., doxorubicin (11) or paclitaxel (12). Recently, the Gynecologic Oncology Group (GOG) reported that gemcitabine combined with docetaxel induced an objective response in 35.8% of metastatic chemo-naive ULMS patients (9); a slightly lower response rate (27%) was observed when an identical regimen was used for recurrent disease (13). While encouraging, it is uncertain whether or not this therapeutic regimen will improve patient survival. Clearly, there is an ongoing and pressing need for more effective ULMS therapies.
Mammalian target of rapamycin (mTOR) is a serine/threonine kinase playing a pivotal role in cell growth control, proliferation, survival, and metabolism (14). mTOR is a component of two distinct protein complexes: mTORC1, which directly phosphorylates and activates S6 kinase (S6K) and 4E-binding protein 1 (4EBP1), and mTORC2, which phosphorylates AKT (14). Aberrant mTOR signaling plays a critical role in many malignancies, making this axis an important anticancer therapeutic target (15). Rapamycin and rapamycin derivatives (“rapalogs”) that block the mTORC1 complex demonstrate variable antitumor effects in preclinical models and are currently under clinical investigation; such therapies have been established as safe and demonstrate promising, albeit cytostatic, anti-tumor effects against several different types of cancers (16). While not specifically evaluated in ULMS, aberrant mTOR signaling has recently been demonstrated in human LMS (17); enhanced mTOR activation was shown to correlate with worse clinical outcome (17). A role for mTOR deregulation in leiomyosarcomagenesis and progression was suggested using mice with smooth muscle lineage-specific knockout of PTEN, a negative regulator of the mTOR pathway, whose loss results in enhanced mTOR signaling (18). These mice developed widespread smooth muscle cell hyperplasia and abdominal leiomyosarcomas; tumor growth was abrogated using the rapamycin derivative everolimus. These data supported a recent study evaluating the rapalog temsirolimus in a small cohort of advanced LMS patients (19); tumor control was observed in three of six patients. Similarly, several additional trials testing rapamycin and rapalogs for LMS (as part of a larger soft tissue sarcoma population) demonstrated beneficial responses in ~1/3 of patients (20).
The current study focused specifically on ULMS, examining the expression and prognostic value of activated mTOR componentry in a relatively large panel of human ULMS samples. Furthermore, the anti-ULMS mTORC1 blockade effects were evaluated. Seeking potential novel anti-ULMS therapeutic strategies and based on recent insights supporting a role for Aurora-A kinase (Aurk-A) in ULMS (21), we sought to evaluate Aurk-A as a potential co-target for combination therapy with mTORC1 inhibitors. Aurk-A is a the most studied member of the aurora kinase family of serine/threonine kinases, consisting of Aurk-A, -B, and -C, sharing ~70% sequence homology (22). These kinases function primarily by regulating cell cycle progression and are critical for mitosis (22). Aurora A kinase localizes to centrosomes and has been implicated in diverse mitotic-associated processes, including mitotic entry, centrosome duplication, maturation and separation, microtubule–kinetochore attachment, bipolar spindle assembly, chromosome alignment, and cytokinesis (23). Several lines of evidence support a role for Aurk-A in cancer: 1) it was found to be amplified and/or over-expressed in multiple cancers including breast, ovarian, and hepatic carcinomas(24), 2) forced Aurk-A expression resulted in NIH3T3 fibroblast oncogenic transformation (25) and induced aneuploidy in a nearly diploid breast cancer cell line (25), and 3) Aurk-A interacts with and modifies the function of several key cancer-associated molecules such as BRCA1 and p53 (23). These insights have resulted in much interest in developing small molecule therapeutic Aurk-A inhibitors including MLN8237, which has shown significant anti-cancer effects in pediatric cancer models.(26) Several other inhibitors have already shown preclinical efficacy and are undergoing clinical investigation (27). The current study evaluates the impact of mTORC1 inhibition alone and combined mTORC1 and Aurk-A dual blockade on ULMS growth in vitro and in vivo.
Materials and Methods
Tissue microarray (TMA) construction and immunohistochemistry
With Institutional Review Board (IRB) approval, the University of Texas MD Anderson Cancer Center (UTMDACC) Pathology Archive was searched for formalin-fixed, paraffin embedded ULMS specimens. Two hundred and forty three tumor blocks (representing 208 lesions retrieved from 109 patients) containing sufficient viable tumor tissue adequate for analytic purposes were selected for TMA construction. These included 18 primary lesions, 66 recurrent lesions, and 124 metastatic lesions. In addition, FFPE blocks of 10 healthy gastrointestinal smooth muscle specimens, 15 healthy myometrium samples, and 10 benign leiomyomas were identified as controls. ULMS patient clinical information including demographic, therapeutic, tumor, and clinical outcome variables were retrieved from institutional medical records and tabulated for correlative analyses. A tissue microarray (TMA) was constructed as previously described (28). Information regarding antibodies, IHC procedure, and scoring is provided in Supplemental data.
Cell culture and reagents
Information regarding cellular models and reagents is provided in supplemental data file.
Cellular assays
A panel of in vitro cell culture-based assays was utilized. Information is provided in Supplementary data file.
In vivo therapeutic experiments
All animal procedures/care were approved by UTMDACC Institutional Animal Care and Usage Committee. Animals received humane care as per the Animal Welfare Act and the NIH "Guide for the Care and Use of Laboratory Animals." Animal models were utilized as previously described (29). Viable SKLMS1 cells were confirmed using trypan blue staining, and 2×106cells/0.1mL RPMI/mouse were used. Cell suspensions were injected subcutaneously into the flank of 6–8 week old female hairless SCID mice (n = 7–8/group) and growth was measured twice weekly; after establishment of palpable lesions (average diameter ~4–7mm depending on the study) mice were assigned to one of the following treatment groups: in the first set of experiments: 1) vehicle control and 2) rapamycin (3.75 mg/kg/d, five days a week, per gavage) and in the second: 1) vehicle control; 2) rapamycin (3.75 mg/kg/d, five days a week, per gavage); 3) MLN8237 (15mg/kg/bid, every day, per gavage); or 4) combination of both agents. Treatment was repeated as per the dose/schedule above until study termination. Rapamycin dose followed previously published studies (30); MLN8237 dose was selected based on the company’s recommendation and previously published data demonstrating that the maximal tolerated dose of the compound in most mouse strains (continuous dosing for ~21 days) is approximately 20mg/kg/bid (i.e. a total of 40mg/kg/d) and anti-tumor efficacy is observed with a total dose of 30mg/kg/d (31). Of note, MLN8237 was administered alone on day one of treatment while rapamycin treatment was initiated on day two. Mice were followed for tumor size, well being, and body weight, and sacrificed when control group tumors reached an average of 1.5 cm in their largest dimension (21 days of treatment). Tumors were resected, weighed, and frozen or fixed in formalin and paraffin-embedded for immunohistochemical studies. Additional information is included in Supplemental Data.
Statistical analyses
To score each gene expression profile of ULMS or normal myometrium for similarity to a predefined gene transcription “signature” of the PI3K/Akt/mTOR pathway, we derived a "t score" for the sample profile in relation to the signature patterns as previously described (32–34). In brief, the PI3K mRNA t score was defined as the two-sided t statistic comparing the average of the PI3K-induced genes with that of the repressed genes within each tumor (after normalizing the log-transformed values to standard deviations from the median across samples). The mapping of transcripts or genes between the two array datasets was made on the Entrez Gene identifier; where multiple human array probe sets referenced the same gene, one probe set with the highest variation represented the gene. Fisher exact test was used to determine the correlation between biomarkers’ expression and tissue-associated variables such as histology and disease-status. Correlation between the different biomarkers was evaluated using Spearman's correlation coefficient analyses. To evaluate the correlation of TMA biomarker expression and patient disease specific survival (DSS) each independent variable was examined separately in a univariable Cox proportional hazards model. Independent variables that had p-values of 0.10 or less in the univariable Cox model analysis were further examined in multivariable Cox models; p≤0.05 was set as the cutoff. All computations were performed using SAS for Windows (release 9.2; SAS Institute, Cary, NC).
Cell culture-based assays were repeated at least twice; mean ± SD was calculated. Cell lines were examined separately. For outcomes that were measured at a single time point, two-sample t-tests were used to assess differences. To determine whether the cytotoxic interactions of rapamycin and MLN8237 in SKLMS1 cells were synergistic, additive, or antagonistic, drug effects were examined using the combination index (CI) method of Chou and Talalay (35, 36). Briefly, the fraction affected (Fa) was calculated from cell viability assays, and CIs were generated using CalcuSyn software (Biosoft, Cambridge, UK). CI values <0.9 are considered synergistic, 0.9–1.1 additive, and >1.1 antagonistic. Additional information regarding this methodology, the isobologram, and fraction affected graphs can be found in reference(36). Differences in xenograft growth in vivo were assessed using a Two-way ANOVA (using log-transformed values; p<0.01) and a two-tailed Student's t-test was used to determine differences in tumor volume and weight at the termination of the studies (p≤0.05).
Results
The AKT/mTOR pathway is highly activated in human ULMS
A recent study suggested a role for mTOR pathway targeting as an anti-ULMS therapeutic strategy (37). To determine if this axis is activated in human ULMS, recently obtained gene expression profiles of 12 ULMS specimens (FIGO stage 1) and 10 healthy myometrium samples (21) were compared bioinformatically to three previously reported different PI3K/AKT/mTOR-related gene expression signatures: 1) the CMap signature, which identified commonly deregulated genes using expression profiles generated from several different cell lines treated with PI3K inhibitors (32); 2) the Saal profile, based on gene expression in breast cancers exhibiting PTEN loss (and therefore, highly active mTOR pathway (33); and 3) the Majumder gene set which examines gene regulation in mice over expressing AKT1 (34). Each profiled ULMS or myometrium sample was scored for PI3K pathway activation based on pathway-specific transcriptional targets. For each signature evaluated, ULMS tumors had higher PI3K/AKT/mTOR pathway activation scores versus myometrium (p<0.01, each signature, two-sided t-test) (Fig 1A), providing initial evidence of AKT/mTOR deregulation in ULMS.
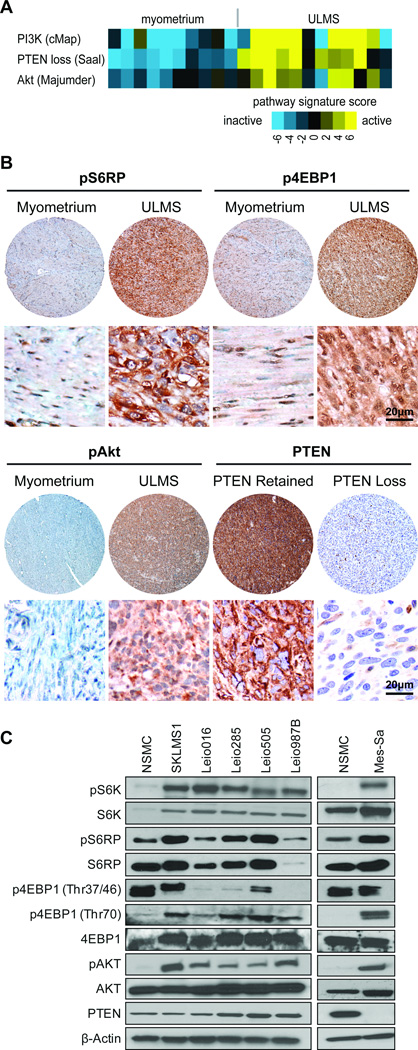
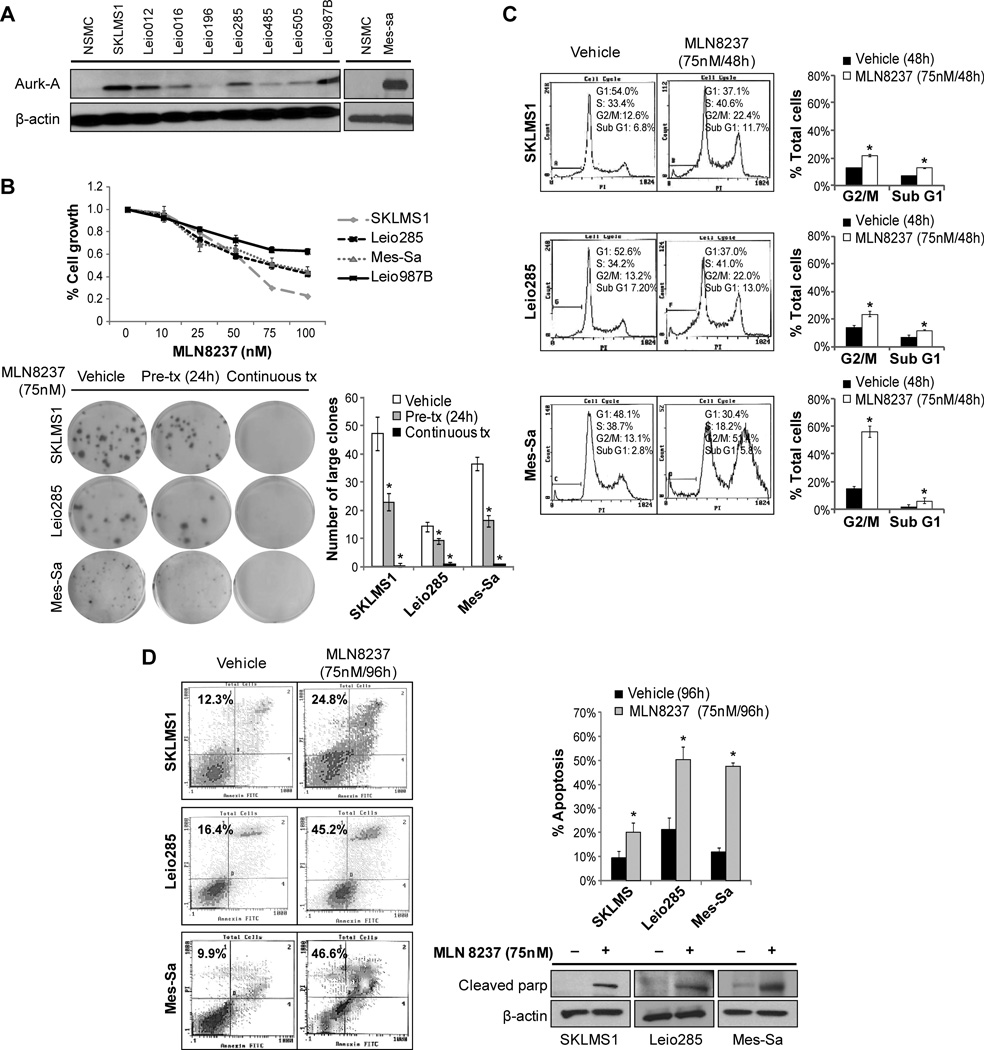
Figure 1. mTOR pathway is commonly activated in ULMS.
A) PI3K/AKT/mTOR pathway is more active in ULMS, based on gene expression profiling data. Three previously described gene transcription signatures for the PI3K/AKT/mTOR pathway were applied to expression profiles of ULMS samples; each sample was scored for relative signature activity (heatmap is depicted: yellow = more active, blue = less active); B) Representative photographs of ULMS tissue microarray pS6RP, p4EBP1, and pAKT immunostaining (original images were captured at 400× magnification) depicting differences in expression between tumor and normal smooth muscle. Staining for PTEN is also shown and includes an example of PTEN expression loss as compared to positive staining – the later was observed in most evaluable samples; C) Western blot (WB) analyses demonstrate increased phosphorylation of the mTOR downstream effectors, S6K, S6RP, and 4EBP1, and the mTOR upstream regulator, AKT, in protein extracts of human ULMS cell strains/lines as compared to expression noted in normal smooth muscle cells (NSMC). Only Mes-Sa cells exhibited loss of PTEN expression.
Next, we evaluated expression levels of the activated (phosphorylated) mTOR downstream effectors S6RP and 4EBP1 in a large human ULMS sample panel assembled into a TMA; normal smooth muscle, myometrium, and leiomyoma samples served as controls (Fig 1B, FigS1, and TableS1A). Only samples representing different lesions were included in the final analysis. All ULMS specimens expressed pS6RP: low levels were observed in 28% (n=55), moderate expression was noted in 49% (n=97), and high expression was found in 24% (n=48). An average of 60.7% (±20) of tumor cells per sample exhibited positive pS6RP staining. While pS6RP expression was observed in all controls, staining intensity was found to be significantly lower (p<0.0001). No differences in the expression of this marker were identified between normal myometrium and leiomyoma. p4EBP1 was scored for both cytoplasmic and nuclear expression intensity and distribution. All ULMS samples expressed cytoplasmic p4EBP1: low levels were observed in 22% (n=43), moderate expression was noted in 59% (n=114), and high expression was found in 19% (n=36). An average of 72.5% (±14.4) of tumor cells per sample exhibited positive cytoplasmic p4EBP1 staining. Similarly, nuclear p4EBP1 expression was found in all ULMS samples: low levels were observed in 39% (n=75), moderate expression was noted in 45% (n=87), and high expression was found in 16% (n=30). An average of 70.8% (±13.7) of tumor cells per sample exhibited positive nuclear p4EBP1 staining. All controls exhibited cytoplasmic and nuclear p4EBP1 staining, albeit at markedly lower levels compared to ULMS (p<0.0001 and p=0.0005, respectively). Interestingly, significantly increased pS6RP and cytoplasmic and nuclear p4EBP1 expression levels were identified in recurrent and metastatic ULMS samples compared to primary lesions (p=0.0005, 0.05, and 0.0088, respectively; Table S1B).
Expression of pAKT, the major upstream regulator of mTOR, was next evaluated (Fig 1B, FigS1, and Table S1A). All ULMS exhibited pAKT expression at varying degrees: low levels were observed in 41% (n=76), moderate in 47% (n=87), and high in 12% (n=23). An average of 66.4% (±20.4) of tumor cells per sample exhibited positive pAKT staining. In contrast, all control samples exhibited only low levels of pAKT expression (p<0.0001). No difference between pAKT expression levels was noted between primary, recurrent, and/or metastatic ULMS specimens (Table S1B). Loss of the PTEN tumor suppressor was suggested as a potential common molecular LMS deregulation, contributing to mTOR activation (18). Interestingly, immunohistochemical analysis identified loss of PTEN expression in only 7% of ULMS samples (n=15); 9% of controls (two normal smooth muscle and one leiomyoma) exhibited no PTEN expression. Further, we found that pS6RP, cytoplasmic and nuclear p4EBP1, and pAKT expression all statistically correlated with one another but none significantly correlated with PTEN expression status (Table S2).
A univariable Cox model was utilized to determine whether the expression level and distribution (i.e., % positive cells) of any of the mTOR-related biomarkers was associated with clinical outcome; only localized ULMS specimens were included (n=57). A univarable analysis for DSS (Table S3) identified only increased percentage of cells expressing nuclear p4EBP1 as significantly correlating with decreased DSS (p<0.05 set as the cutoff point). Multivariable models fitted to include variables with a p<0.1 (including the clinical variables: age, tumor size, disease status, margin status) identified cytoplasmic and nuclear p4EBP1 expression and distribution as prognosticators of unfavorable DSS (Table S3). Taken together, these data demonstrate mTOR deregulation as commonly occurring in human ULMS, thereby supporting further consideration of mTOR as a potential ULMS therapeutic target.
mTOR blockade inhibits ULMS growth in vitro and in vivo
Next, we evaluated whether cultured human ULMS cells recapitulated the clinical scenario by over-expressing activated mTOR downstream effectors (pS6K, pS6RP, and p4EBP1). Cells tested included SKLMS1, a well characterized LMS cell line of gynecological origin (ATCC), and four different ULMS primary cultures (cell strains) recently isolated in our laboratory; normal smooth muscle cell primary cultures (NSMC) were used as controls (Fig 1C). Mes-Sa, a commercially available (ATCC), poorly differentiated uterine sarcoma cell line was also included; whether this cell line represents a ULMS or another uterine sarcoma histological subtype is difficult to ascertain, but it was included as a relevant positive control (we have previously demonstrated Mes-Sa loss of PTEN expression; (38). Western blots (WBs) demonstrated increased pS6K, pS6RP, and p4EBP1 expression in tumor cells vs. control (Fig 1C); increased levels of pAKT were observed in tumor cells (Fig 1C). Excluding Mes-Sa, none of the ULMS cell strains exhibited loss of PTEN expression (Fig 1C). Together, these data confirm that (similar to the observations in human specimens) activation of the mTOR axis is observed in ULMS cells growing in culture, rendering this a relevant model to test the effects of mTOR blockade.
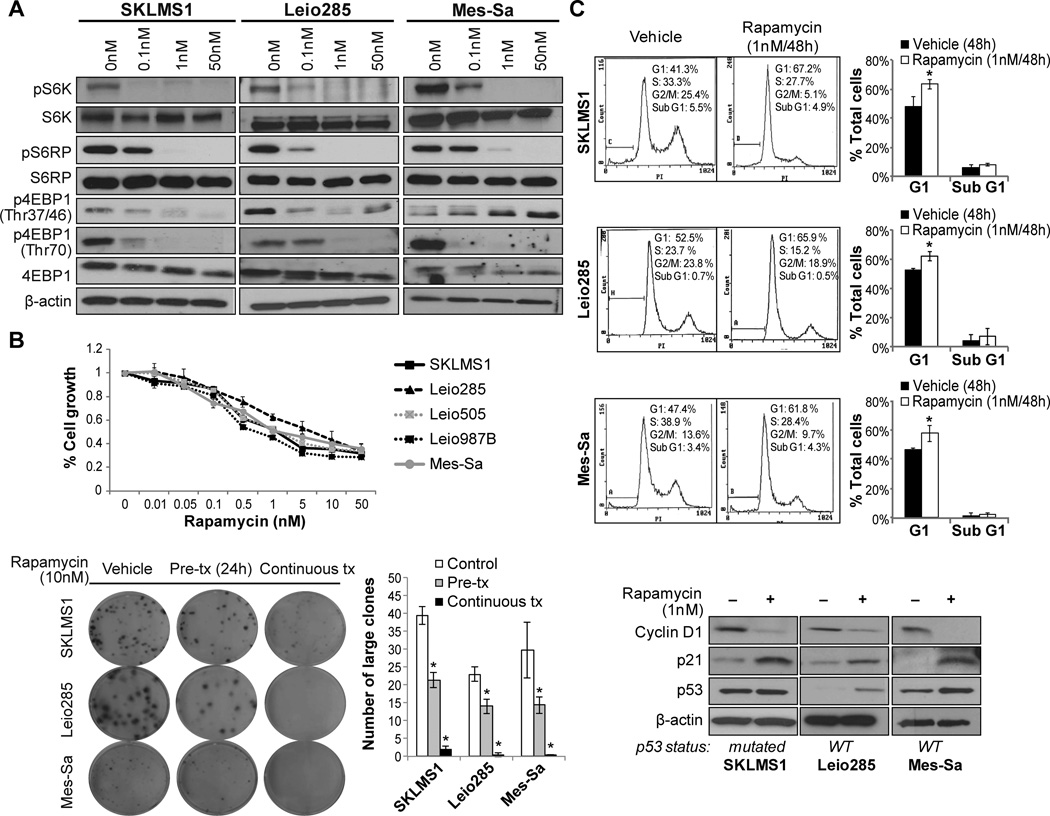
Rapamycin was utilized to evaluate mTORC1 inhibition of ULMS cell growth. Tumor cells were treated with incremental drug doses for 4h; decreased phosphorylation of S6K, S6RP and 4EBP1 were observed even at the lowest dose (0.1nM) tested (Fig 2A). Functionally, a dose-dependent decrease in tumor cell growth in response to rapamycin (0.01–50nM/96h) was observed in all tested cell lines/strains with GI50 levels of ~ 1nM (Fig 2B). Similarly, mTORC1 blockade inhibited colony formation capacities of ULMS cells; rapamycin pre-treatment (24h) significantly reduced the number of large colonies by 40–50%; when applied continuously, rapamycin nearly abrogated clonogenicity (Fig 2B). Next, the effects of rapamycin on cell cycle progression and apoptosis were evaluated. Rapamycin treatment (1nM/48h) resulted in a G1 cell cycle arrest (Fig 2C).This effect could partly be secondary to the observed decrease in cyclin D1 expression and increased p21 expression in treated cells (Fig 2C). Interestingly, G1 cell cycle arrest was independent of p53 mutational status and occurred in both wild-type p53 cell strains/lines (Leio285 and Mes-Sa) where an increase in p53 protein expression was noted, as well as in p53 mutated cells (SKLMS1) where increased p21 levels were observed independent of enhanced p53 expression (Fig 2C). Notably, no increased sub-G1 fractions were noted in response to rapamycin per PI staining FACS analysis (Fig 2C). Furthermore, Annexin-V/PI staining FACS analysis, conducted after rapamycin treatment for 96h, failed to demonstrate significant apoptosis (data not shown).
Figure 2. mTOR blockade using rapamycin inhibit ULMS cell growth and induces G1 cell cycle arrest.
A) Rapamycin (0.1–50nM/4h) blocks the activation of the mTOR downstream targets S6K, pS6RP, and 4EBP1 (WB analyses); B) MTS assays demonstrating a rapamycin (96h) dose-dependent decrease in ULMS cell growth (upper graph). In addition, rapamycin (both as pre- and continuous treatments) inhibits the colony formation capacity of ULMS cells (lower panel); C) Rapamycin treatment (1nM/48h) results in a G1 cell cycle arrest in ULMS cells. WB analyses demonstrate decrease in cyclin D1 and increased p21 expression in treated cells, independent of p53 mutational status. Increased p53 protein expression was found in cells harboring wild-type p53. [Graphs represent the average of at least two repeated experiments ±SD; * denotes statistically significant effects (p<0.05)]
Based on the above findings, we next sought to evaluate whether rapamycin effects could also be observed in vivo. While ULMS primary cultures can be utilized for experimental studies in vitro, none of the cell strains available to us reproducibly grow in vivo. In contrast, SKLMS1 cells reproducibly grow as xenografts when injected into immunocompromised mice; therefore, this experimental model was selected for therapeutic testing. Rapamycin (or vehicle control) treatment was initiated after tumor establishment (~4–5mm in larger diameter; average tumor volumes in each group at the initiation of treatment were control: 57.5mm3±25 and rapamycin: 55.7mm3±25.6). Mice in both groups were followed for tumor size and toxicity; treatment was terminated when tumors in control group reached an average of 1.5cm in largest dimension. Treatment with rapamycin resulted in a statistically significant tumor growth delay compared to vehicle-treated tumors (Fig 3A; two-factor ANOVA by time<0.01). Average tumor volumes recorded at termination of the study were control group: 1370mm3±1311 vs. rapamycin group: 510mm3±353 (Fig 3A). While a statistical significance in growth over time was found, differences in tumor volumes at the end of the study were not statistically significant (ttest p>0.5). Together, these data demonstrate a potential cytostatic effect of rapamycin on ULMS growth. Similarly a decrease in tumor weight was noted (control = 1g±0.8 vs 0.3g±0.2) although it did not reach statistical significance (p=0.52). To confirm these results the experiment was repeated when tumors were larger on average: control: 95mm3±64 and rapamycin: 97mm3±62. Same as above, treatment with rapamycin resulted in a statistically significant tumor growth delay compared to vehicle-treated tumors (Fig 3A; two-factor ANOVA by time<0.05). Average tumor volumes recorded at termination of the study were control group: 1361mm3±354 vs. rapamycin group: 895mm3±449 (p=0.0519; Fig 3A). Similarly a decrease in tumor weight was noted (control = 1.97g±1.11 vs 1.1g±0.72) although it did not reach statistical significance (p=0.11).
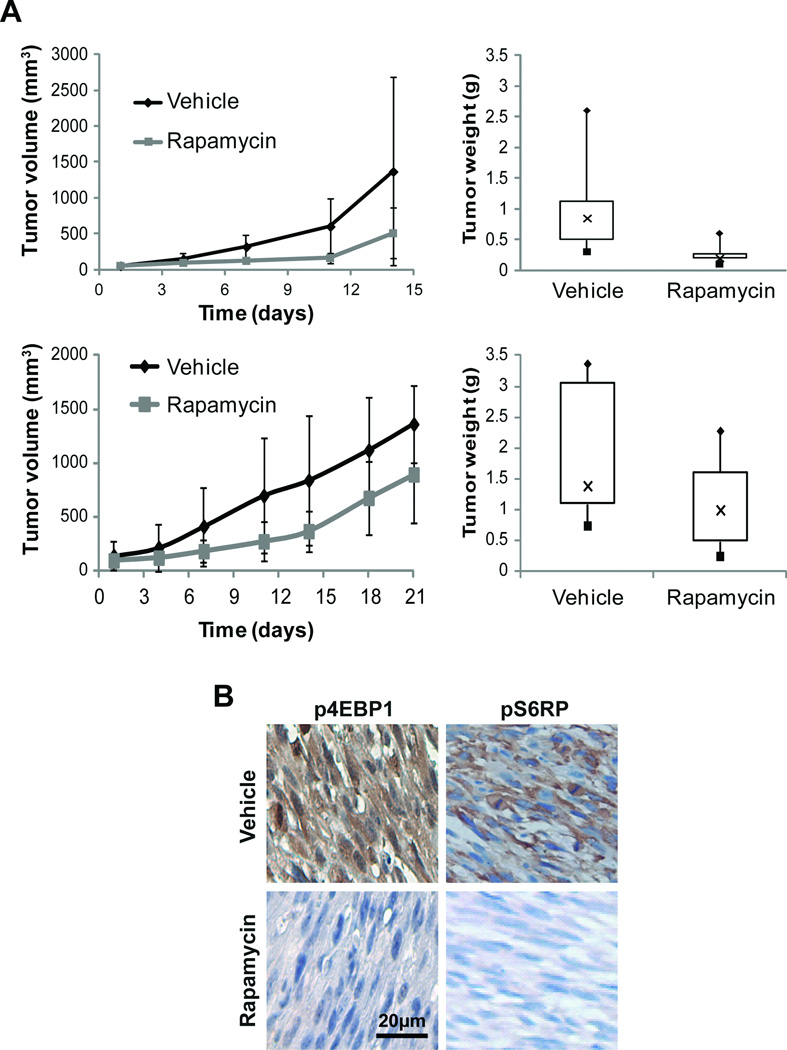
Figure 3. Rapamycin treatment delays the growth of ULMS xenografts.
A) Treatment with rapamycin (3.75mg/kg/d, 5 days per week) resulted in SKLMS1 xenograft tumor growth delay compared to control vehicle-treated tumors. Two separate experiments are shown depicting effect of treatment on growth and tumor weight at study termination: B) Immunohistochemical (IHC) staining confirmed decreased p4EBP1 and pS6RP expression in rapamycin treated tumors (original images were captured at 400× magnification).
To confirm that rapamycin inhibits mTORC1 activity in vivo, formalin-fixed, paraffin-embedded tumor sections from mice of both study arms were immunohistochemically evaluated. Decreased p4EBP1 and pS6RP expression was observed in rapamycin treated tumor samples (Fig 3B). Of note, a significant (p=0.001) decrease in the number of Ki67 (a nuclear marker for proliferation) expressing tumor cells was observed in rapamycin related samples. A small but significant increase in TUNEL expression was noted with rapamycin treatment (p=0.005). While no apoptosis can be found after rapamycin treatment in vitro, the small increase noted in vivo might be the result of the anti-vascular effects of rapamycin. IHC for CD31 identified a decrease in large blood vessels in rapamycin treated tumors (9.5±0.99) compared to controls (12.4±4.5), although this difference did not not reach statistical significance. For IHC images see Figure 6. Taken together, observations made in this preclinical model recapitulate effects noted in human clinical studies, supporting identification of additional ULMS molecular aberrations therapeutically targetable in combination with mTOR.
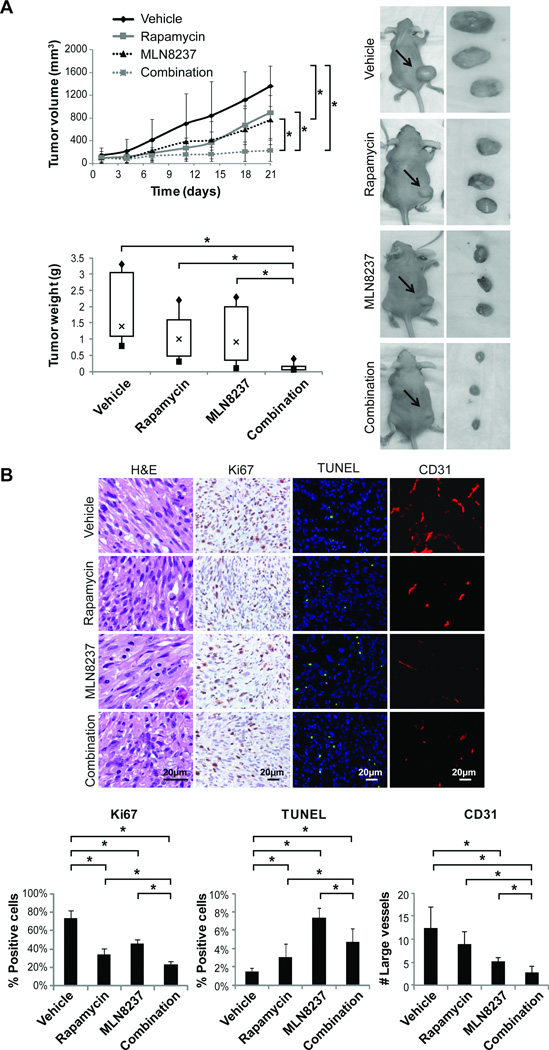
Figure 6. Combined mTOR and Aurk-A targeting results in superior anti-ULMS effects in vivo.
A) The impact of combined therapy (Rapamycin: 3.75mg/kg/d, five days a week and MLN8237: 15mg/kg/bid, every day. Of note MLN8237 was administered alone on day 1) was assessed in vivo using SKLMS1 xenografts growing in hairless SCID mice. MLN8237 as a single agent significantly inhibited tumor growth as compared to control. Most importantly, combination therapy resulted in significant growth abrogation as compared to rapamycin, MLN8237, or vehicle. Combination treated mice exhibited the most significant decrease in tumor weight as compared to all other therapeutic groups. B) IHC analyses demonstrated decreased Ki67 positive staining cells in all treatment groups, the most pronounced was in combination treatment tumors. An increase in TUNEL positive cells was noted in all treated tumors. Moreover, combination treated tumors exhibited the greatest decrease in CD31 positivity. [* denotes statistically significant effects (p<0.05)].
The Aurora kinase A inhibitor, MLN 8237, inhibits ULMS cell growth and induces G2 cell cycle arrest and apoptosis
We recently showed that dysregulated centrosome function and spindle assembly is a dominant feature of ULMS, highlighting a potential therapeutic targeting role for Aurora-A kinase (Aurk-A), a protein involved in these events (21). As demonstrated in Fig 4A, ULMS cell strains/lines expressed increased levels of Aurk-A compared to normal smooth muscle cells. Therefore, we aimed to evaluate whether combining mTOR blockade with an Aurk-A inhibitor could enhance the anti-ULMS effects observed using rapamycin alone. We acquired a novel investigational orally bioavailable selective Aurk-A inhibitor, MLN8237 (Millennium Pharmaceuticals), and first examined its anti-tumor effects in our in vitro ULMS model. MTS assays demonstrated marked MLN8237 dose-dependent (0–100nM/96h) ULMS cell growth inhibition; estimated GI50 levels of ~ 75nM were observed in all cell lines/strains tested except for Leio987B (Fig 4B). For this later cell strain which exhibits a longer doubling time a higher MLN8237 dose was needed to achieve 50% growth inhibition at 96h. Furthermore, MLN8237 abrogated ULMS colony formation capacity: 24h pre-treatment with MLN8237 resulted in markedly reduced numbers of colonies; under continuous treatment almost no colonies were observed (Fig 4B). Next, the effects of MLN8237 on cell cycle progression were evaluated. MLN8237 treatment (75nM/48h) resulted in a G2/M cell cycle arrest (Fig 4C); increased sub-G1 fraction was noted. To determine the impact of MLN8237 on ULMS apoptotic cell death, Annexin-V/PI staining FACS analyses were conducted (Fig 4D) after 96h of treatment. An increase (~2–4 fold) in apoptosis was observed in MLN8237 treated cells compared to vehicle treated controls. Increased cleaved PARP was noticed after 48h of treatment, providing evidence of MLN8237-induced apoptosis. Together, these data confirm Aurk-A as a candidate anti-ULMS therapeutic target and demonstrate potential efficacy of MLN8237.
Figure 4. The Aurk-A inhibitor, MLN8237, inhibits ULMS cell growth inducing G2/M cell cycle arrest and apoptosis.
A) WB analysis demonstrating increased Aurk-A protein expression in a panel of ULMS cell strains/lines as compared to normal smooth muscle cells (NSMC); B) MTS assays demonstrating marked MLN8237 dose-dependent (0–100nM/96h) ULMS cell growth inhibition (upper graph). In addition, MLN8237 (both as pre- and continuous-treatment) abrogates the colony formation capacity of ULMS cells (lower panel); C) MLN8237 treatment (75nM/48h) results in a G2/M cell cycle arrest in ULMS cells. Furthermore, increased sub-G1 fraction is observed; D) An increase (~2–4 fold) in apoptosis was observed in MLN8237 treated cells compared to vehicle treated controls (Annexin-V/PI staining FACS analysis). WB analyses further demonstrate increased cleaved PARP in response to treatment. [Graphs represent the average of at least two repeated experiments ±SD; * denotes statistically significant effects (p<0.05)]
Combining rapamycin and MLN8237 results in superior (synergistic) anti-ULMS effects
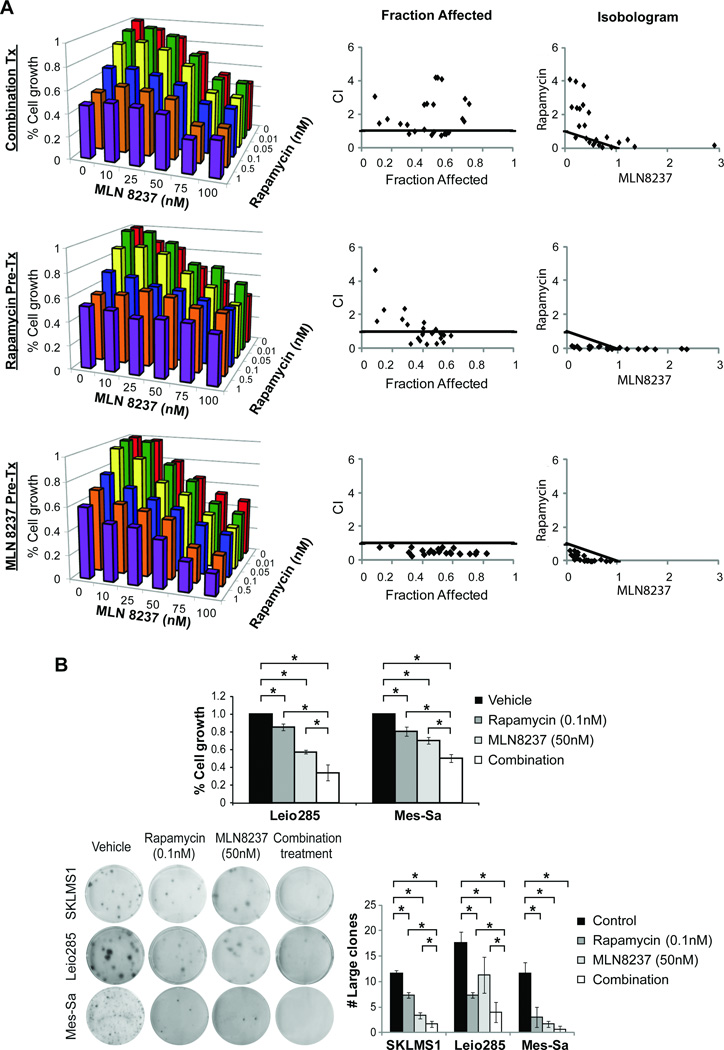
Based on these results, we sought to determine ULMS growth effects of combined rapamycin and MLN8237. MTS assays of treated SKLMS1 cells were conducted to determine the combination index (CI), enabling assessment of potential interactions between these drugs (Figs 5A and S2, Table S4). Several scheduling regimens were tested: 1) simultaneous co-administration of rapamycin (increasing doses; 0–1nM) and MLN8237 (increasing doses; 0–100nM) for 96h; 2) 24h pre-treatment with rapamycin followed by co-treatment with and MLN 8237 for 72 hours; 3) 24h MLN8237 pretreatment followed by 72hr co-treatment with rapamycin. Interestingly, while the first two regimens failed to demonstrate synergism between the drugs (for some doses antagonistic responses were even observed; CI>1.1), isobologram analyses revealed that growth-inhibitory effects of the drug combination were synergistic when administered per the third schedule (CI<0.9; Fig 5A and Table S4). Similarly, superior anti-growth effects were observed when combining low dose rapamycin and MLN8237 (as per the above schedule) in Leio285 and Mes-Sa compared to either agent alone (p<0.05; Fig 5B). Similarly, combination therapy induced a superior inhibitory effect on colony formation compared to either agent alone (Fig 5B).
Figure 5. Combined mTOR and Aurk-A targeting results in superior (synergistic) anti-ULMS effects in vitro.
A) MTS assays were conducted using increasing doses of both rapamycin and MLN8237, three different scheduling regimens were used: 1. simultaneous co-administration of rapamycin (increasing doses; 0–1nM) and MLN8237 (increasing doses; 0–100nM) for 96h (upper graphs), 2. 24h pre-treatment with rapamycin followed by co-treatment with and MLN 8237 for 72 hours (middle graphs), 3. 24h MLN8237 pre-treatment followed by 72hr co-treatment with rapamycin (bottom graphs). Isobologram analyses revealed that growth-inhibitory effects of the drug combination were synergistic when administered per the third schedule (CI<0.9; graphs represent three separate experiments; individual assay results can be found in Fig S2); B) Similarly, superior anti-growth effects are observed in Leio285 and Mes-Sa cells in response to dual mTOR/Aurk-A inhibition (administered in low doses as per the aforementioned schedule) compared to either agent alone (upper panel). Furthermore, combination therapy induces a superior inhibitory effect on colony formation compared to either agent alone (lower panel). [All graphs represent the average of three repeated experiments ±SD; * denotes statistically significant effects (p<0.05)]
Lastly, to determine whether the effects noted in vitro could be recapitulated in vivo, a four-armed therapeutic study was conducted comparing the effect of combination treatment to each drug alone or vehicle control. Of note, in accord with in vitro findings, mice were treated with MLN8237 and rapamycin sequentially. Average tumor volumes at the initiation of the study were control group: 96mm3±66 rapamycin group: 97mm3±63, MLN8237: 101mm3±76, and combination: 101mm3±96. No major side effects or discomforts were noted; average mouse weights at the termination of the study were: control − 24.9g±0.7, rapamycin − 24.3g±2, MLN8237 − 23.6g±2.2, and combination − 22.9g±2.3. Both compounds and their combination delayed tumor growth over time in a statistically significant manner (two-way ANOVA<0.01). Tumor volume at study termination was significantly reduced in MLN8237 treated mice as compared to control (Average volumes: MLN8237 − 765mm3±429 and control - 1361mm3±354, p=0.012; Fig 6A). Most importantly, combination therapy resulted in significant decrease in tumor volume compared to rapamycin, MLN8237, or control alone (average of combination treatment tumor volumes at study termination was 227mm3±185; p=0.0016, =0.0047, and <0.0001, respectively). Average tumor weights recorded at termination of the study were control group: 2.0g±1.1; rapamycin group: 1.1g±0.72; MLN8237 group: 1.2g±0.99; and combination group: 0.15g±0.13 (Fig 6A). To confirm these results the experiment was repeated demonstrating reproducible results (Fig S3). Of note, in this second experiment control, rapamycin, and MLN8237 mice were euthanized when control mice tumors reached 15mm on average while combination therapy continued for an additional two weeks demonstrating slight increase in growth (Fig S3). IHC analysis demonstrated decreased Ki67 positive staining cells in all treatment groups, most pronounced in combination treatment tumors (Fig 6B). An increase in TUNEL positive cells was noted in all treated tumors. Interestingly, less apoptotic cells were observed in combination- as compared to MLN8237 alone-treated tumors. As combination treated tumors are significantly smaller it is possible that cells affected by treatment have undergone apoptosis and death and thus not observed in the remaining specimen while a proportion of viable cells are arrested in G1 secondary to rapamycin treatment and are thus less affected by the pro-apoptotic effects of MLN8237. Finally, combination treated tumors exhibited the greatest decrease in CD31 positivity. Taken together, these data suggest that mTOR blockade in combination with Aurora-A kinase inhibition results in significant anti-ULMS effects in vitro and in vivo, a finding of potential clinical utility.
Discussion
Enhanced mTOR signaling, serving as a convergence point for multiple upstream molecular deregulations, is commonly observed in many malignancies, making this axis an attractive anti-cancer therapeutic target (15). Recent data suggest a role for this pathway in LMS (17, 18); studies here extend these observations to ULMS, a particularly devastating LMS subset, in which we have demonstrated increased mTOR activation. Enhanced mTOR signaling occurred with disease progression, and activation of the mTOR downstream effector 4EBP1 correlated with poor prognosis in localized ULMS. Notably, in accord with data demonstrating a low PTEN mutation rate (~5%) in ULMS (39), loss of PTEN expression was not demonstrable as the prominent mechanism underling mTOR activation in our tumors. mTOR activation can be due to multiple upstream molecular derangements acquired throughout the tumorigenic process; e.g., among possible mechanisms, over-expression and activation of tyrosine kinase receptors (TKRs) can contribute to mTOR activation (40). While knowledge is limited, several studies suggest that TKRs such as PDGFR-α (41) and IGF-1R (42) are over-expressed in ULMS. Regardless of the underlying mechanisms, our study demonstrates that mTOR activation contributes to ULMS cell growth and cell cycle progression. Importantly, and of therapeutic relevance, these effects are independent of p53 mutational status. The latter finding is of clinical relevance given that p53 mutations occur in human ULMS (43) and that p53 mutation can contribute to conventional chemotherapeutic resistance (44). While encouraging, these data also demonstrate the limitation of single agent mTOR inhibitors as treatment for ULMS, resulting in cytostatic effects that might not be sufficient to improve patient outcomes, reminiscent of mTOR blockade-based trials in other human solid tumors and in LMS (19). Identifying additional targets for inhibition in combination with mTOR is thus urgently needed, a strategy currently being evaluated in conjunction with conventional chemotherapy-, radiotherapy-, and molecular-based therapies in the context of several malignancies (45).
Recently we found that ULMS over-express gene products regulating centrosome structure and function, including Aurk-A (21). In our recent study we identified Aurk-A to be over-expressed in ULMS and demonstrated that the Aurk-A specific inhibitor MK-5108 exhibits anti-ULMS effects in vitro and in vivo (21). Together, these data suggest a role for Aurk-A as a novel anti-ULMS therapeutic target. Aware that a critical next step in utilizing mTOR inhibition for ULMS treatment mandates developing effective drug combinations and taking into account our finding that Aurk-A might be a novel ULMS therapeutic target, we evaluated dual mTOR and Aurk-A blockade in our preclinical models. To the best of our knowledge, this therapeutic combination has not been previously reported. We selected a novel Aurk-A inhibitor, MLN8237, an orally bioavailable, small molecule inhibitor with 200-fold selectivity for Aurk-A compared to Aurk-B or -C; reports indicate no significant cross-reactivity with a diverse panel of receptors and ion channels (31, 46). Encouraging anti-cancer effects have been noted in several solid and hematological malignancy preclinical models in which MLN8237 promoted mitotic spindle abnormality, induced G2/M cell cycle arrest and polyploidy, ultimately enhancing tumor cell death (47). These insights formed the basis for ongoing early phase clinical studies(48); initial results have shown promising antitumor activity and prolonged disease stabilization (49). We found that ULMS cells are sensitive to MLN8237 at doses similar to those previously reported (31), observing significant treatment-induced growth inhibition and apoptosis. Most importantly, our data demonstrate that MLN8237 synergizes with rapamycin to induce superior in vitro and in vivo anti-ULMS effects compared to either agent alone (although MLN8237 was given at a dose lower than the maximal tolerated dose (MTD; (46)). While the exact mechanism underlying these synergistic effects are yet to be determined, it has previously been shown that AurkA might act as an mTOR pathway activator (50) and our findings might reflect the interaction between these pathways. Notably, synergism was found only when MLN8237 was administered prior to rapamycin treatment, highlighting the importance of scheduling sequences in development of therapeutic combination regimens. These results may reflect the differential impact of these compounds on cell cycle progression; AurkA inhibition results in G2 cell cycle arrest while mTOR blockade in G1 arrest. Future studies to evaluate if other G2 cell cycle regulators enhance mTOR blockade effects are currently undergoing. In summary, ULMS is an aggressive malignancy with a poor prognosis. Our findings suggest that enhanced mTOR signaling and Aurk-A deregulation play important roles in ULMS tumorigenicity and provide supporting justification for clinical trials to evaluate combined mTOR and Aurk-A inhibitors to enhance the anti-ULMS effects observed with either inhibitor alone.
Supplementary Material
Statement of translational relevance.
Uterine leiomyosarcoma (ULMS) are characterized by marked chemoresistance, frequent relapses, and poor outcome, hence, the urgent need for more effective therapeutic strategies. Studies reported here identifed mTOR pathway activation to commonly occur in ULMS and unraveled a role for pS6RP and p4EBP1 as molecular disease prognosticators. However, while rapamycin abrogates ULMS cell growth and cell cycle progression in culture, it induces only growth delay in vivo. Given that effective therapies will most likely combine mTOR blockade with inhibitors of other molecular targets, coupled with recent observations suggesting that Aurora-A kinase (Aurk-A) deregulations commonly occur in ULMS, the impact of combining rapamycin and MLN8237 (an investigational selective Aurk-A inhibitor) on ULMS growth was assessed. Combined therapy resulted in pronounced (synergistic) growth inhibition; interestingly, these superior effects were noted only when MLN8237 was administered first. Together, these data support further exploration of dual mTOR and Aurk-A blockade for human ULMS.
Acknowledgments
We wish to thank Dr. Jeffrey A. Ecsedy (Millennium Pharmaceuticals, Cambridge, MA) for kindly providing MLN8237. Ms. Kim Vu is thanked for her aid in figure preparation and Mr. Juan Cuevas for his aid with manuscript submission.
Grant/Funding Support: The research described in this manuscript was supported in part by a NIH/NCI RO1CA138345 (to DL), an Amschwand Foundation Seed Grant (to DL), a NIH/NCI 5T32CA009599-21 training grant (supporting KL), and a Deutsche Forschungsgemeinschaft training grant (supporting MPHG). MDACC cell-line characterization and immuohistochemistry core facilities were further supported by an NCI Cancer Center Support Grant (CA#16672).
Footnotes
Conflicts of Interest: None to declare. It is noted that Millennium Pharmaceuticals (Cambridge, MA) has kindly provided the investigational Aurora-A kinase inhibitor MLN8237.
Supplemental Data: This word document includes supplementary Figures and Tables.
References
- 1.Schwartz LB, Diamond MP, Schwartz PE. Leiomyosarcomas: clinical presentation. Am J Obstet Gynecol. 1993;168:180–183. doi: 10.1016/s0002-9378(12)90910-2. [DOI] [PubMed] [Google Scholar]
- 2.Smith DC, Macdonald OK, Lee CM, Gaffney DK. Survival impact of lymph node dissection in endometrial adenocarcinoma: a surveillance, epidemiology, and end results analysis. Int J Gynecol Cancer. 2008;18:255–261. doi: 10.1111/j.1525-1438.2007.01020.x. [DOI] [PubMed] [Google Scholar]
- 3.Zivanovic O, Leitao MM, Iasonos A, Jacks LM, Zhou Q, Abu-Rustum NR, et al. Stage-specific outcomes of patients with uterine leiomyosarcoma: a comparison of the international Federation of gynecology and obstetrics and american joint committee on cancer staging systems. J Clin Oncol. 2009;27:2066–2072. doi: 10.1200/JCO.2008.19.8366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salazar OM, Bonfiglio TA, Patten SF, Keller BE, Feldstein M, Dunne ME, et al. Uterine sarcomas: natural history, treatment and prognosis. Cancer. 1978;42:1152–1160. doi: 10.1002/1097-0142(197809)42:3<1152::aid-cncr2820420319>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 5.Bodner K, Bodner-Adler B, Kimberger O, Czerwenka K, Leodolter S, Mayerhofer K. Evaluating prognostic parameters in women with uterine leiomyosarcoma. A clinicopathologic study. J Reprod Med. 2003;48:95–100. [PubMed] [Google Scholar]
- 6.Zagouri F, Dimopoulos AM, Fotiou S, Kouloulias V, Papadimitriou CA. Treatment of early uterine sarcomas: disentangling adjuvant modalities. World J Surg Oncol. 2009;7:38. doi: 10.1186/1477-7819-7-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gadducci A, Landoni F, Sartori E, Zola P, Maggino T, Lissoni A, et al. Uterine leiomyosarcoma: analysis of treatment failures and survival. Gynecol Oncol. 1996;62:25–32. doi: 10.1006/gyno.1996.0185. [DOI] [PubMed] [Google Scholar]
- 8.Giuntoli RL, 2nd, Metzinger DS, DiMarco CS, Cha SS, Sloan JA, Keeney GL, et al. Retrospective review of 208 patients with leiomyosarcoma of the uterus: prognostic indicators, surgical management, and adjuvant therapy. Gynecol Oncol. 2003;89:460–469. doi: 10.1016/s0090-8258(03)00137-9. [DOI] [PubMed] [Google Scholar]
- 9.Hensley ML, Blessing JA, Mannel R, Rose PG. Fixed-dose rate gemcitabine plus docetaxel as first-line therapy for metastatic uterine leiomyosarcoma: a Gynecologic Oncology Group phase II trial. Gynecol Oncol. 2008;109:329–334. doi: 10.1016/j.ygyno.2008.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D'Angelo E, Prat J. Uterine sarcomas: a review. Gynecol Oncol. 2010;116:131–139. doi: 10.1016/j.ygyno.2009.09.023. [DOI] [PubMed] [Google Scholar]
- 11.Omura GA, Blessing JA, Major F, Lifshitz S, Ehrlich CE, Mangan C, et al. A randomized clinical trial of adjuvant adriamycin in uterine sarcomas: a Gynecologic Oncology Group Study. J Clin Oncol. 1985;3:1240–1245. doi: 10.1200/JCO.1985.3.9.1240. [DOI] [PubMed] [Google Scholar]
- 12.Sutton G, Blessing JA, Ball H. Phase II trial of paclitaxel in leiomyosarcoma of the uterus: a gynecologic oncology group study. Gynecol Oncol. 1999;74:346–349. doi: 10.1006/gyno.1999.5463. [DOI] [PubMed] [Google Scholar]
- 13.Hensley ML, Blessing JA, Degeest K, Abulafia O, Rose PG, Homesley HD. Fixed-dose rate gemcitabine plus docetaxel as second-line therapy for metastatic uterine leiomyosarcoma: a Gynecologic Oncology Group phase II study. Gynecol Oncol. 2008;109:323–328. doi: 10.1016/j.ygyno.2008.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 15.Wan X, Helman LJ. The biology behind mTOR inhibition in sarcoma. Oncologist. 2007;12:1007–1018. doi: 10.1634/theoncologist.12-8-1007. [DOI] [PubMed] [Google Scholar]
- 16.Fasolo A, Sessa C. mTOR inhibitors in the treatment of cancer. Expert Opin Investig Drugs. 2008;17:1717–1734. doi: 10.1517/13543784.17.11.1717. [DOI] [PubMed] [Google Scholar]
- 17.Setsu N, Yamamoto H, Kohashi K, Endo M, Matsuda S, Yokoyama R, et al. The Akt/mammalian target of rapamycin pathway is activated and associated with adverse prognosis in soft tissue leiomyosarcomas. Cancer. 2011 doi: 10.1002/cncr.26448. [DOI] [PubMed] [Google Scholar]
- 18.Hernando E, Charytonowicz E, Dudas ME, Menendez S, Matushansky I, Mills J, et al. The AKT-mTOR pathway plays a critical role in the development of leiomyosarcomas. Nat Med. 2007;13:748–753. doi: 10.1038/nm1560. [DOI] [PubMed] [Google Scholar]
- 19.Italiano A, Kind M, Stoeckle E, Jones N, Coindre JM, Bui B. Temsirolimus in advanced leiomyosarcomas: patterns of response and correlation with the activation of the mammalian target of rapamycin pathway. Anticancer Drugs. 2011;22:463–467. doi: 10.1097/CAD.0b013e3283442074. [DOI] [PubMed] [Google Scholar]
- 20.Buckner JC, Forouzesh B, Erlichman C, Hidalgo M, Boni JP, Dukart G, et al. Phase I, pharmacokinetic study of temsirolimus administered orally to patients with advanced cancer. Invest New Drugs. 2010;28:334–342. doi: 10.1007/s10637-009-9257-1. [DOI] [PubMed] [Google Scholar]
- 21.Shan W, Akinfenwe P, Brewer Savannah K, Kolomeyevskaya N, Laucirica R, Thomas D, et al. A small-molecular inhibitor targeting the mitotic spindle checkpoint impairs the growth of uterine leiomyosarcoma. Clin Cancer Res. 2012;18:3352–3365. doi: 10.1158/1078-0432.CCR-11-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vader G, Lens SM. The Aurora kinase family in cell division and cancer. Biochim Biophys Acta. 2008;1786:60–72. doi: 10.1016/j.bbcan.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 23.Karthigeyan D, Prasad SB, Shandilya J, Agrawal S, Kundu TK. Biology of Aurora A kinase: Implications in cancer manifestation and therapy. Med Res Rev. 2010 doi: 10.1002/med.20203. [DOI] [PubMed] [Google Scholar]
- 24.Katayama H, Brinkley WR, Sen S. The Aurora kinases: role in cell transformation and tumorigenesis. Cancer Metastasis Rev. 2003;22:451–464. doi: 10.1023/a:1023789416385. [DOI] [PubMed] [Google Scholar]
- 25.Zhou H, Kuang J, Zhong L, Kuo WL, Gray JW, Sahin A, et al. Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat Genet. 1998;20:189–193. doi: 10.1038/2496. [DOI] [PubMed] [Google Scholar]
- 26.Carol H, Houghton PJ, Morton CL, Kolb EA, Gorlick R, Reynolds CP, et al. Initial testing of topotecan by the pediatric preclinical testing program. Pediatr Blood Cancer. 2010;54:707–715. doi: 10.1002/pbc.22352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheung CH, Coumar MS, Hsieh HP, Chang JY. Aurora kinase inhibitors in preclinical and clinical testing. Expert Opin Investig Drugs. 2009;18:379–398. doi: 10.1517/13543780902806392. [DOI] [PubMed] [Google Scholar]
- 28.Lazar AJ, Tuvin D, Hajibashi S, Habeeb S, Bolshakov S, Mayordomo-Aranda E, et al. Specific mutations in the beta-catenin gene (CTNNB1) correlate with local recurrence in sporadic desmoid tumors. Am J Pathol. 2008;173:1518–1527. doi: 10.2353/ajpath.2008.080475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang L, Hannay JA, Liu J, Das P, Zhan M, Nguyen T, et al. Vascular endothelial growth factor overexpression by soft tissue sarcoma cells: implications for tumor growth, metastasis, and chemoresistance. Cancer Res. 2006;66:8770–8778. doi: 10.1158/0008-5472.CAN-06-1217. [DOI] [PubMed] [Google Scholar]
- 30.Verheul HM, Salumbides B, Van Erp K, Hammers H, Qian DZ, Sanni T, et al. Combination strategy targeting the hypoxia inducible factor-1 alpha with mammalian target of rapamycin and histone deacetylase inhibitors. Clin Cancer Res. 2008;14:3589–3597. doi: 10.1158/1078-0432.CCR-07-4306. [DOI] [PubMed] [Google Scholar]
- 31.Sells TB, Ecsedy J, Stroud SG, Janowick DA, Hoar KM, LeRoy PJ, et al. MLN8237:an Orally Active Small Molecule Inhibitor of Aurora A Kinase in Phase I Clinical Trials. AACR. 2008 [Google Scholar]
- 32.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 33.Saal LH, Johansson P, Holm K, Gruvberger-Saal SK, She QB, Maurer M, et al. Poor prognosis in carcinoma is associated with a gene expression signature of aberrant PTEN tumor suppressor pathway activity. Proc Natl Acad Sci U S A. 2007;104:7564–7569. doi: 10.1073/pnas.0702507104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Majumder PK, Febbo PG, Bikoff R, Berger R, Xue Q, McMahon LM, et al. mTOR inhibition reverses Akt-dependent prostate intraepithelial neoplasia through regulation of apoptotic and HIF-1-dependent pathways. Nat Med. 2004;10:594–601. doi: 10.1038/nm1052. [DOI] [PubMed] [Google Scholar]
- 35.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 36.Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010;70:440–446. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 37.Wong TF, Takeda T, Li B, Tsuiji K, Kitamura M, Kondo A, et al. Curcumin disrupts uterine leiomyosarcoma cells through AKT-mTOR pathway inhibition. Gynecol Oncol. 2011;122:141–148. doi: 10.1016/j.ygyno.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 38.Zhu QS, Ren W, Korchin B, Lahat G, Dicker A, Lu Y, et al. Soft tissue sarcoma cells are highly sensitive to AKT blockade: a role for p53-independent up-regulation of GADD45 alpha. Cancer Res. 2008;68:2895–2903. doi: 10.1158/0008-5472.CAN-07-6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Amant F, de la Rey M, Dorfling CM, van der Walt L, Dreyer G, Dreyer L, et al. PTEN mutations in uterine sarcomas. Gynecol Oncol. 2002;85:165–169. doi: 10.1006/gyno.2002.6601. [DOI] [PubMed] [Google Scholar]
- 40.Hay N. The Akt-mTOR tango and its relevance to cancer. Cancer Cell. 2005;8:179–183. doi: 10.1016/j.ccr.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 41.Adams SF, Hickson JA, Hutto JY, Montag AG, Lengyel E, Yamada SD. PDGFR-alpha as a potential therapeutic target in uterine sarcomas. Gynecol Oncol. 2007;104:524–528. doi: 10.1016/j.ygyno.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 42.Gloudemans T, Pospiech I, Van Der Ven LT, Lips CJ, Schneid H, Den Otter W, et al. Expression and CpG methylation of the insulin-like growth factor II gene in human smooth muscle tumors. Cancer Res. 1992;52:6516–6521. [PubMed] [Google Scholar]
- 43.de Vos S, Wilczynski SP, Fleischhacker M, Koeffler P. p53 alterations in uterine leiomyosarcomas versus leiomyomas. Gynecol Oncol. 1994;54:205–208. doi: 10.1006/gyno.1994.1194. [DOI] [PubMed] [Google Scholar]
- 44.Bush JA, Li G. Cancer chemoresistance: the relationship between p53 and multidrug transporters. Int J Cancer. 2002;98:323–330. doi: 10.1002/ijc.10226. [DOI] [PubMed] [Google Scholar]
- 45.Meric-Bernstam F, Gonzalez-Angulo AM. Targeting the mTOR signaling network for cancer therapy. J Clin Oncol. 2009;27:2278–2287. doi: 10.1200/JCO.2008.20.0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gorgun G, Calabrese E, Hideshima T, Ecsedy J, Perrone G, Mani M, et al. A novel Aurora-A kinase inhibitor MLN8237 induces cytotoxicity and cell-cycle arrest in multiple myeloma. Blood. 2010;115:5202–5213. doi: 10.1182/blood-2009-12-259523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Manfredi MG, Ecsedy JA, Chakravarty A, Silverman L, Zhang M, Hoar KM, et al. Characterization of Alisertib (MLN8237), an investigational small-molecule inhibitor of aurora A kinase using novel in vivo pharmacodynamic assays. Clin Cancer Res. 2011;17:7614–7624. doi: 10.1158/1078-0432.CCR-11-1536. [DOI] [PubMed] [Google Scholar]
- 48.Bethesda (MD): National Cancer Institute (US); ClinicalTrials.gov [homepage on the Internet] c2010 - updated 2012 June 25; cited 2012 June 30]. Available from http://clinicaltrials.gov/ct2/show/NCT01154816. [Google Scholar]
- 49.Maris JM, Morton CL, Gorlick R, Kolb EA, Lock R, Carol H, et al. Initial testing of the aurora kinase A inhibitor MLN8237 by the Pediatric Preclinical Testing Program (PPTP) Pediatr Blood Cancer. 2010;55:26–34. doi: 10.1002/pbc.22430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang X, Zhou YX, Qiao W, Tominaga Y, Ouchi M, Ouchi T, et al. Overexpression of aurora kinase A in mouse mammary epithelium induces genetic instability preceding mammary tumor formation. Oncogene. 2006;25:7148–7158. doi: 10.1038/sj.onc.1209707. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.